-
Články
- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
C-di-GMP Regulates Motile to Sessile Transition by Modulating MshA Pili Biogenesis and Near-Surface Motility Behavior in
The human pathogen Vibrio cholerae causes the debilitating disease cholera through ingestion of contaminated food and water. V. cholerae is a natural inhabitant of aquatic environments. Transmission of V. cholerae to the human host is dependent on survival of the pathogen in aquatic reservoirs where it is challenged with many stressors, including changes in the physiochemical parameters of environments and predation by protozoa and phages. One method utilized to endure these assaults is to form a multicellular community called a biofilm. The signaling molecule cyclic dimeric guanosine monophosphate (c-di-GMP) is utilized to induce biofilm formation in many bacteria, including V. cholerae. We demonstrate that c-di-GMP promotes the production of a cell surface structure called MshA pili by binding the molecular motor responsible for polymerizing pilus subunits. These pili are adhesive appendages that are essential for attachment to surfaces. This study identifies a novel mechanism for c-di-GMP regulation of pilus production through interactions with the molecular motor responsible for pilus assembly. Since many bacteria utilize pili for attachment to surfaces and c-di-GMP as a pro-biofilm signaling molecule, the mechanism for pilus regulation and biofilm formation described here may be widespread among many pathogens.
Published in the journal: . PLoS Pathog 11(10): e32767. doi:10.1371/journal.ppat.1005068
Category: Research Article
doi: https://doi.org/10.1371/journal.ppat.1005068Summary
The human pathogen Vibrio cholerae causes the debilitating disease cholera through ingestion of contaminated food and water. V. cholerae is a natural inhabitant of aquatic environments. Transmission of V. cholerae to the human host is dependent on survival of the pathogen in aquatic reservoirs where it is challenged with many stressors, including changes in the physiochemical parameters of environments and predation by protozoa and phages. One method utilized to endure these assaults is to form a multicellular community called a biofilm. The signaling molecule cyclic dimeric guanosine monophosphate (c-di-GMP) is utilized to induce biofilm formation in many bacteria, including V. cholerae. We demonstrate that c-di-GMP promotes the production of a cell surface structure called MshA pili by binding the molecular motor responsible for polymerizing pilus subunits. These pili are adhesive appendages that are essential for attachment to surfaces. This study identifies a novel mechanism for c-di-GMP regulation of pilus production through interactions with the molecular motor responsible for pilus assembly. Since many bacteria utilize pili for attachment to surfaces and c-di-GMP as a pro-biofilm signaling molecule, the mechanism for pilus regulation and biofilm formation described here may be widespread among many pathogens.
Introduction
Vibrio cholerae, the causative agent of the human intestinal disease cholera, is a natural inhabitant of aquatic ecosystems [1]. Cholera infection results from consumption of food and water contaminated with V. cholerae. Subsequently the bacteria turn on regulatory networks that facilitate bacterial growth and survival during the infection process [2]. They also activate production of virulence factors including the toxin-coregulated pilus (TCP), essential for intestinal colonization, and the cholera toxin (CT), responsible for production of massive watery diarrhea that results in dissemination of V. cholerae back to aquatic ecosystems [3]. V. cholerae’s ability to cause epidemics is tied to its dissemination and survival in aquatic habitats and its transmission to the human host. One critical factor for dissemination, environmental survival and transmission of the pathogen is its ability to form matrix enclosed surface-associated communities termed biofilms [4–6]. V. cholerae forms biofilms on the surfaces of phytoplankton and zooplankton [7], and exists in the surface waters of cholera endemic areas in matrix-enclosed aggregates thought to arise from biofilm-like populations of V. cholerae present in human stools [5]. Removal of particles >20 μm in diameter from water can reduce cholera incidence by 48% [8]. Additionally, growth in a biofilm induces a hyper-infectious phenotype [9]. Collectively, these studies highlight the importance of the biofilm growth mode in both the intestinal and aquatic phases of the V. cholerae life cycle.
Biofilm formation by V. cholerae begins with surface attachment, and subsequent development of microcolonies and mature biofilm structures [10–16]. The biofilm matrix is primarily composed of Vibrio exopolysaccharide (VPS) [17], extracellular DNA [18], and biofilm matrix proteins (RbmA, RbmC, and Bap1) [13,14,16,19], which are required for cell-cell and cell-surface interactions and development of mature V. cholerae biofilms [15].
Two cell-surface structures, a single polar flagellum and type IVa mannose-sensitive hemagglutanin pili (MshA), are critical for initial attachment and biofilm formation [11,20]. Type IVa pili have the ability to rapidly extend and retract, contributing to twitching and swarming motility in many bacteria [21]. Though V. cholerae produces the type IVa MshA pilus, twitching motility has not been reported. Genes required for biogenesis of MshA pilus are clustered into 16.7 kb region in V. cholerae chromosome-I and organized into two operons: the first operon harbors 9 genes from mshI-mshF predicted to encode proteins required for assembly and secretion and the second harboring 7 genes from mshB—mshQ encoding pilus structural components mshB-Q [22]. The pilus is comprised of repeats of the major pilin, MshA. Predicted function of the proteins encoded by MSHA gene locus are included in Table 1.
Tab. 1. Extragenic suppressors of Δ<i>cdgJ</i> motility phenotype. 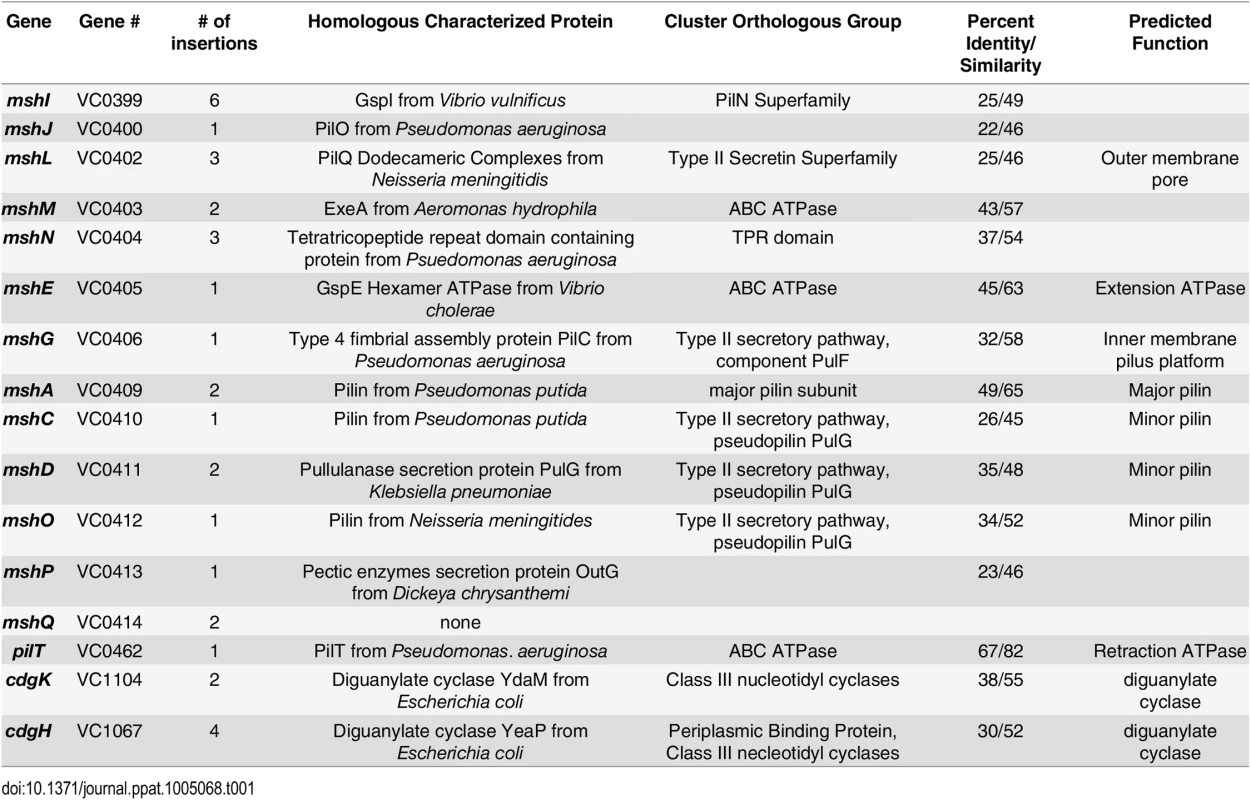
MshA pili and flagellum are also crucial for two distinct near-surface motility trajectories of V. cholerae: ‘roaming’ and ‘orbiting’, [10]. Low curvature ‘roaming’ trajectories meander over large distances and result from weak MshA-surface interactions. In contrast, orbiting trajectories repeatedly trace out tight circular tracks over the same region, and are the result of strong MshA-surface interactions. Cells that attach to the surface come from the orbiting subpopulation, while the roaming subpopulation pass over the surface without attaching. That orbiting motility is ablated when a mannose derivative is added to the medium to saturate MshA pilus binding further indicates that interactions between MshA pili with the surface are important [10].
The second messenger cyclic dimeric guanosine monophosphate (c-di-GMP) is an important promoter of the switch from motile planktonic growth mode to biofilm growth mode [23–25]. c-di-GMP synthesis is catalyzed by diguanylate cyclases (DGC) harboring a GGDEF domain while degradation is catalyzed by phosphodiesterases (PDE) harboring an EAL or HD-GYP domain. Subsequently, c-di-GMP is sensed by different classes of receptor proteins or RNAs and thereby converted to specific phenotypic outputs affecting motility, biofilm formation, and virulence. Elevated intracellular levels of c-di-GMP inhibit motility both by post-transcriptional and transcriptional mechanisms. In Salmonella enterica and Escherichia coli, the PilZ class of c-di-GMP receptor protein YcgR affects flagellar motor functions through interaction with FliG and FliM subunits of the flagellar rotor or the stator subunit MotA [26,27]. In Pseudomonas aeruginosa and V. cholerae, c-di-GMP inhibits motility by repressing transcription of flagellar genes through the AAA+ ATPase enhancer binding class of c-di-GMP receptor and transcriptional regulators FleQ [28] and FlrA [29], respectively. In addition to the regulation of flagellum production and activity by c-di-GMP, there are reports of c-di-GMP regulating the assembly and activity of Type IV pili. In Klebsiella pneumoniae, c-di-GMP is bound by PilZ class of c-di-GMP receptor protein MrkH, which upregulates the transcription of the fimbrial subunit mrkA [30–32]. In P. aeruginosa, the degenerate GGDEF-EAL domain class of c-di-GMP receptor FimX modulates Type IV pili production in an intracellular c-di-GMP concentration-dependent manner [33,34].
The V. cholerae genome encodes 31 GGDEF domain, 12 EAL domain and 10 dual GGDEF/EAL domain proteins [35]. Systematic analysis of in-frame deletion mutants of all V. cholerae genes encoding proteins with GGDEF and/or EAL domains for motility phenotypes revealed that four DGCs (CdgH, CdgK, CdgL, and CdgD) and two PDEs (CdgJ and RocS) affect motility in an LB soft agar motility assay [36]. Though deletion of the PDE cdgJ affected motility, no difference in intracellular c-di-GMP concentration was observed between the ΔcdgJ mutant and WT [36]. This measurement was conducted from a population, so there may be subcellular localized differences or population differences that affect motility. The molecular mechanism of c-di-GMP mediated motility repression and contribution of these DGCs and PDEs to switch from motile to surface-associated lifestyle remains elusive. In this study, we demonstrate that c-di-GMP inversely regulates motility and biofilm formation through direct regulation of the assembly and activity of the MshA pilus. Swimming motility is impaired in strains lacking the phosphodiesterase cdgJ, and disruption of the assembly or disassembly of the MshA pilus restores motility to WT levels by reducing the interactions with surfaces. Quantitative measurements indicate that c-di-GMP leads to increased production of MshA pili, which in turn bind surfaces and reduce motility. We demonstrate that the ATPase responsible for pilus polymerization, MshE, functions as a c-di-GMP receptor thereby providing an input for the c-di-GMP signal into the assembly of the MshA pilus. Collectively, this study elucidates how type IV pili and swimming motility are regulated by c-di-GMP in V. cholerae by presenting the first characterization of the complex involved in the assembly and disassembly of the MshA pilus and how c-di-GMP regulates the production and function of this complex.
Results
MshA pilus impacts CdgJ mediated motility
CdgJ is a PDE and a cdgJ mutant displays a decrease in motility, enhanced VPS production, and increased biofilm formation compared to WT [36]. To begin investigating the mechanism by which CdgJ impacts motility, we performed transposon mutagenesis in a cdgJ deletion mutant (ΔcdgJ) and screened the resulting mutants for enhanced motility phenotype using LB soft agar motility assay. We screened 7054 transposon mutants and identified 42 extragenic suppressor mutants with increased motility and mapped the transposon insertions site to 22 different genes (Table 1, Fig 1). As previously reported, we found that mutations in the DGC encoding genes cdgH and cdgK in a ΔcdgJ strain enhance motility [36]. We also found that insertion in the gene encoding a pilus retraction motor, PilT, and insertions into different genes predicted to be required in mannose-sensitive hemagglutinin type IV pilus (MshA) biogenesis restored swimming motility.
Fig. 1. Schematic representation of the transposon insertions in the Msh operons. 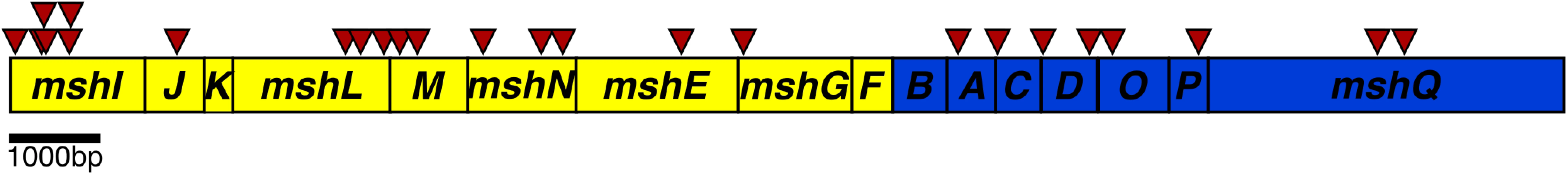
The operon encoding genes required for assembly and secretion (mshI-F) is displayed in yellow and the operon encoding pilus structural components mshB-Q is displayed in blue. Transposon insertions are indicated by red arrowheads above the operon. There were no identical insertions observed; arrows are shifted up for clarity indicating two insertions near each other. To further investigate the suppression of a ΔcdgJ motility phenotype, we generated in-frame deletions of several of the genes identified in the transposon screen in wild-type and ΔcdgJ strains and analyzed the mutants for motility phenotype using LB soft agar motility assay. We focused on mshA (encoding major pilin subunit), mshE (encoding putative polymerizing ATPase), and pilT (encoding putative depolymerizing ATPase) as they are crucial for production and function of MshA pili. As previously reported, ΔcdgJ has a significant motility defect compared to the parental WT strain [36] (Fig 2A). Deletion of mshA, mshE, and pilT in a ΔcdgJ background restored motility similar to the WT strain, confirming that those mutations mediate suppression of the flagellar motility defect in the ΔcdgJ mutant. Deletion of pilU (predicted to encode a second copy of putative depolymerizing ATPase) in ΔcdgJ had no effect on the motility compared to the parental ΔcdgJ mutant. Mutants of mshA, mshE, and pilT in a WT background were assayed for motility to determine if their enhanced motility in the ΔcdgJ background is dependent on this mutation, or if this could be a case of bypass suppression. The ΔmshA, ΔmshE, and ΔpilT mutants exhibited enhanced motility compared to the WT strain (Fig 2A). The ΔpilU strain had a similar motility phenotype to the WT strain, suggesting that the functions or expression profiles of pilT and pilU are different. These data demonstrate that deletion of mshA, mshE, and pilT enhances motility regardless of the presence of a wild-type copy of cdgJ. However, we could not rule out the possibility that cdgJ could directly or indirectly control the production or function of the MshA pilus.
Fig. 2. Mutations in mshA, mshE, and pilT increase motility. 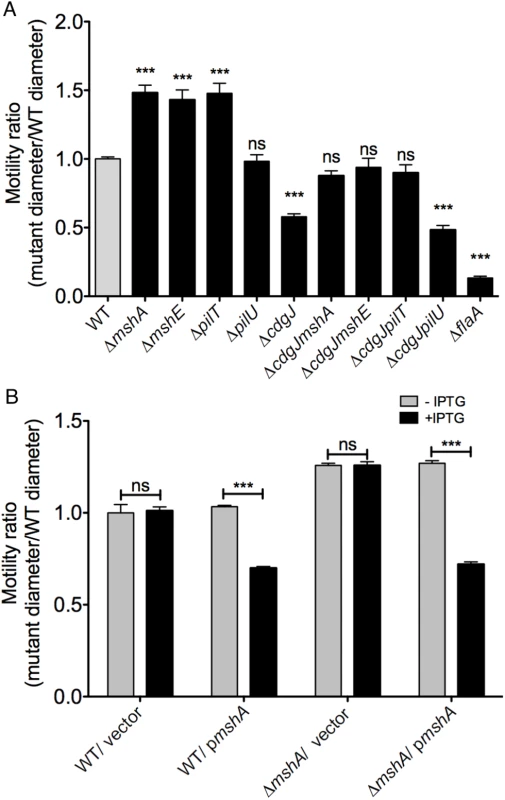
A. The diameters of migration zones of the WT and mutants were measured after 16 h of incubation at 30°C on LB soft agar motility plates and normalized to the motility of the WT strain. Four biological replicates were performed in quadruplicate. Statistical significance determined with Oneway ANOVA followed by Dunnett’s multiple comparison test comparing to the WT strain. (ns—not significant, *** p≤0.001) B. Expression of mshA reduces motility in soft agar. The diameters of migration zones were measured after 16 hours of growth at 30°C with or without IPTG induction and normalized to the motility of the WT strain. Two biological replicates were performed in triplicate. Induced strains compared to uninduced strains with Student’s t-test. (ns—not significant, *** p≤0.001). To determine if mshA-mediated suppression of the flagellar motility phenotype is specific to the cdgJ mutation or if it occurs in other PDE deletion backgrounds, we mutated mshA in a ΔrocS strain. RocS has both GGDEF and EAL domains and is predicted to function mainly as a PDE as ΔrocS mutants have reduced motility along with enhanced VPS production and biofilm formation [36–38]. We generated a ΔrocSmshA double mutant and determined that this mutant exhibited a wild-type motility phenotype (S1 Fig). These findings suggest that MshA negatively impacts V. cholerae flagellar motility and is involved in general c-di-GMP mediated repression of motility.
To evaluate further the ability of MshA to repress motility, we analyzed the effect of the expression of a WT copy of mshA provided in trans in an expression plasmid with an IPTG-inducible promoter. Motility assays confirmed that expression of mshA, upon induction with IPTG, significantly reduced motility in a ΔmshA strain (Fig 2B). Additionally, expression of mshA in the WT strain reduced motility, suggesting that overproduction of MshA impairs motility. IPTG had no effect on motility in strains harboring an empty vector control.
mshA, mshE, and pilT mutants are deficient in biofilm formation
Since the MshA pilus is critical for initial stages of surface attachment and subsequent biofilm formation [10,11,20], we hypothesized that the ΔmshE and ΔpilT mutants would phenocopy the reduced biofilm phenotype of a ΔmshA mutant. Biofilms of these mutants were grown using a flow cell system, imaged using confocal microscopy, and analyzed using the COMSTAT image analysis software package to evaluate biofilm structural properties. As expected, the ΔmshA mutant attached poorly to the substrate and formed biofilms with low biomass (Fig 3A and 3B). The ΔmshE and ΔpilT mutants grew biofilms with significantly less biomass, thickness, and substrate coverage than WT. COMSTAT analysis revealed that while biofilm biomass of ΔpilT and ΔmshA was similar, the biomass of ΔmshE was significantly less than the ΔpilT strain (Fig 3B). Additionally, surface coverage of ΔpilT mutant was greater than that of ΔmshA and ΔmshE. These differences suggest that though the ΔmshA, ΔmshE, and ΔpilT mutants are deficient at forming biofilms, there are subtle differences in the phenotypes of the strains. The ΔpilU mutant produced biofilms that were indistinguishable from WT.
Fig. 3. mshA, mshE, and pilT mutants are impaired in biofilm formation. 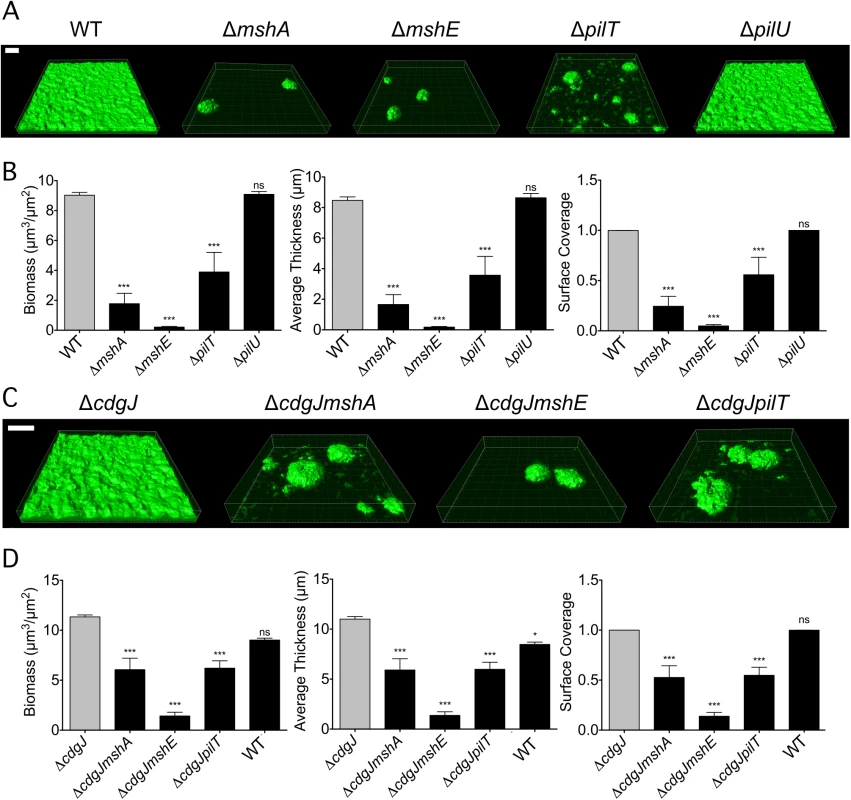
A. Three-dimensional biofilm structures of WT, ΔmshA, ΔmshE, ΔpilT, and ΔpilU strains formed 24 h post inoculation in a flow cell system. Scale bar represents 20μm. B. COMSTAT quantitative analysis of biomass, average thickness, and surface coverage of biofilms from A. Three images from each of two independent experiments were analyzed. Significance was determined with a Oneway ANOVA followed by Dunnett’s Multiple Comparison test comparing to WT. (ns = not significant, *** p≤0.001) C. Three-dimensional biofilm structures of WT, ΔcdgJ, ΔcdgJmshA, ΔcdgJmshE, and ΔcdgJpilT, strains formed after 24 h post inoculation in a flow cell system. Scale bar represents 20μm. D. COMSTAT quantitative analysis of biomass, average thickness, and surface coverage of biofilms from C. Three images from each of two independent experiments were analyzed. Significance was determined with a Oneway ANOVA followed by Dunnett’s Multiple Comparison test comparing to ΔcdgJ. (ns = not significant, * p≤0.05, *** p≤0.001). Since our initial interest in the MshA pilus was sparked by the discovery that mutations in pilus genes can suppress the motility defect in mutants of the PDE cdgJ, we determined whether the MshA pilus mutations were epistatic to the cdgJ mutation. As previously reported, the ΔcdgJ strain produced thicker biofilms than the WT strain (Fig 3C and 3D) [36]. The ΔcdgJmshA, ΔcdgJmshE, and ΔcdgJpilT strains formed biofilms with significantly less biomass, thickness, and surface coverage than the ΔcdgJ strain. These findings are consistent with the hypothesis that these proteins are involved in formation of the MshA pilus and that a functional MshA pilus is required for biofilm formation, surface attachment, and the inhibition of flagellar motility. Biofilms formed by the ΔcdgJmshE mutant had a significant reduction in biomass, thickness, and surface coverage compared to the ΔcdgJmshA or ΔcdgJpilT strains.
mshA, mshE, and pilT mutants are defective in surface attachment and near surface motility
In addition to using bulk differences in biofilm formation as an index of the transition between motile and sessile behavior, we also directly monitored surface attachment of V. cholerae with single cell resolution using high-speed microscopy and cell tracking (at 5 ms frame rate) to elucidate how genes involved in MshA pilus production impact microscopic outcomes such as initial surface attachment. ΔmshA mutants do not attach to the surface in significant numbers, with no attached cells observed in the first 15min after inoculation. In contrast, well over 100 cells of the WT strain attach to the surface over the same time interval (Fig 4A). Using the same metrics, the ΔmshE mutant was also unable to attach to the surface, exhibiting binding profiles that were similar to the ΔmshA strain. The ΔpilT strain was able to attach to the surface more than the ΔmshA; however, it is unable to attach with the same efficiency as the WT strain. This suggests the following hierarchy of behavioral categories for comparison with non-WT backgrounds: the strong binding strain (WT), the intermediate binding strain (ΔpilT), and the weak binding strains (ΔmshA and ΔmshE). To dissect the origin of this aggregate statistical behavior of surface attachment, we needed more single-cell metrics for bacterial behavior near a surface. We examined the temporal aspects of single bacterium interactions with the surface in the form of single cell residence time (number of seconds that each stationary cell remained associated with the surface, Fig 4B). The WT strain formed prolonged associations with the surface, with a mean residence time of 2.6 seconds and a maximum of 78.9 seconds. The ΔmshA, ΔmshE, and ΔpilT mutant strains demonstrated more transient interactions with the surface, with reduced mean residence times compared to WT (0.69, 0.58, and 0.58 seconds, respectively). This is most evident in the inset of Fig 4B, where the entire adherent populations of these mutants have residence times of less than 6 sec, while the WT residence times extend out to nearly 80 seconds.
Fig. 4. Early surface interactions are reduced in mshA, mshE, and pilT mutants and increased in cdgJ mutant. 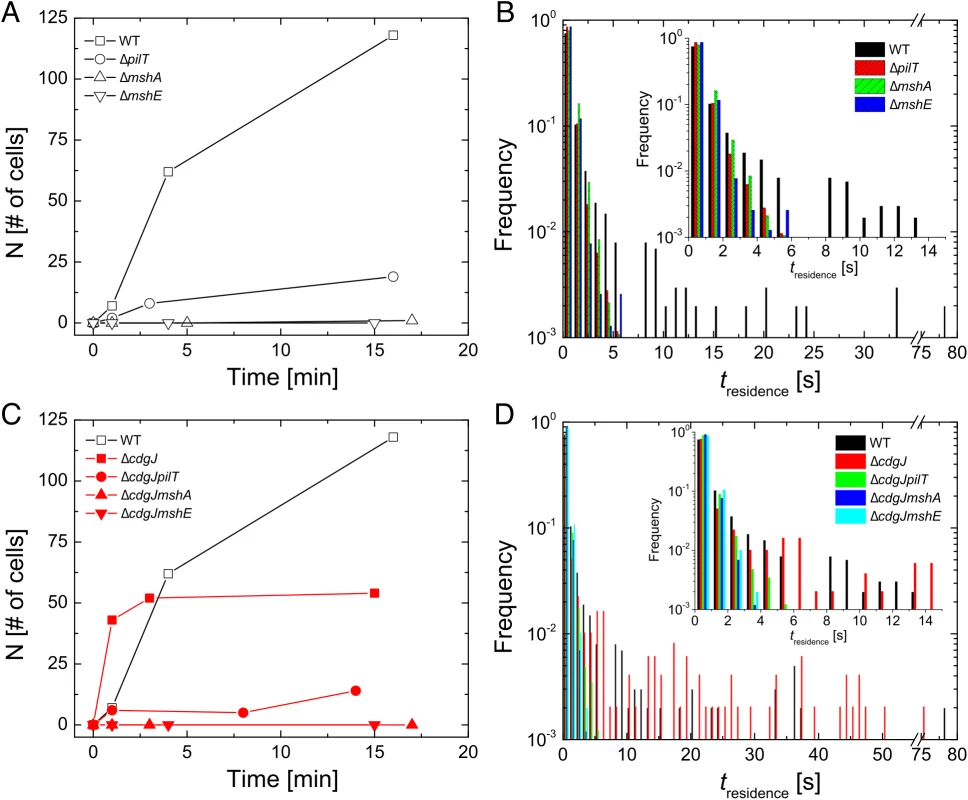
A, C. The number of surface-adhered, stationary cells were counted at the indicated times after inoculation in a flow cell. B, D. Residence times were measured for each cell that interacted with the surface during three high-speed movies of 82s in length within the first 15 minutes post inoculation. mshA, mshE, and pilT mutants are defective in near surface motility
We used high-speed microscopy and near-surface cell tracking to record the trajectories of cells within one micrometer of the coverslip surface in a microscopy chamber (Fig 5). All strains with flagella are capable of swimming motility in 3D, and can exhibit trajectories that come in and out of focus. Consistent with previous reports, the WT strain exhibits orbiting and roaming behavior with regards to near surface motility [10]. All of the surface attached cells come from the orbiting subpopulation. The ΔmshA mutant does not show orbiting or roaming behavior, consistent with the model that MshA pili-surface interactions are responsible for these near-surface motility phenotypes. Moreover, also consistent with the model, the mutant shows greatly reduced surface attachment (Fig 4A) [10]. The ΔmshE strain tracks phenocopy ΔmshA, exhibiting predominantly a swimming phenotype with little observable attachment. The ΔpilT mutant has an intermediate phenotype; a few cells appear to exhibit behavior similar to WT orbiting, but has greatly reduced attachment compared to WT. These data are consistent with the biofilm and attachment data described in Figs 2 and 3.
Fig. 5. Mutations in mshA, mshE, pilT, and cdgJ affect near surface motility. 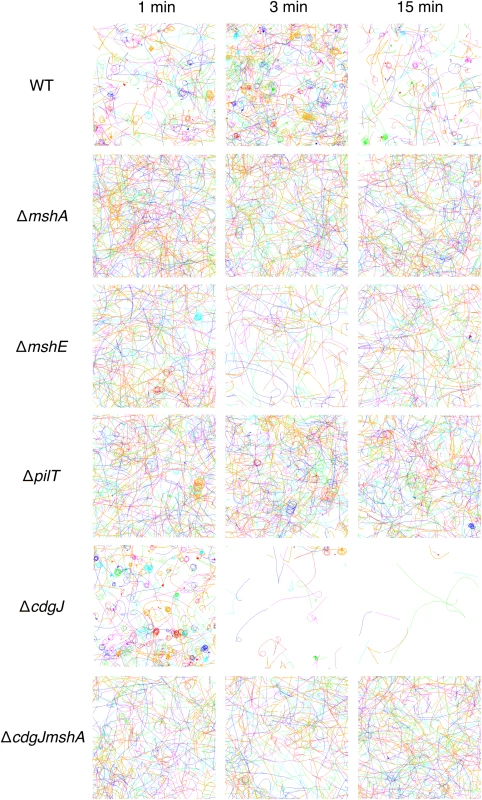
Measured cell tracks at the indicated times post-inoculation. Bacterial populations were imaged for 82s with a 5 ms resolution, and a representative subset of individual cell tracks are presented for clarity. The field of view is 160μm x 160μm. Each line represents the motion of one cell and different colors are used to denote different cells. Non-motile cells have been filtered out of the data set at each time point for clarity. mshA, mshE and pilT mutations are epistatic to a cdgJ mutation with regards to surface attachment
To investigate the role of the PDE CdgJ on initial surface attachment, we observed mshA, mshE, pilT mutants in a ΔcdgJ background with high-speed microscopy using similar experiments. We determined that the ΔcdgJ mutant exhibits strong attachment to the surface, with a sharp increase in the number of attached cells as a function of time during the initial few minutes compared to WT. (Fig 4C, time = 1). This strain rapidly associated with the surface, with nearly all cells binding within the first two minutes of observation. As predicted from the biofilm and motility data (Figs 1 and 2), the ΔcdgJmshA, ΔcdgJmshE, and ΔcdgJpilT strains exhibited reduced surface attachment compared to the parental ΔcdgJ strain and WT (Fig 4C). As in the WT background, the ΔcdgJpilT strain exhibited an intermediate level of attachment and the ΔcdgJmshA and ΔcdgJmshE strains were poor at attachment. The ΔcdgJmshA, ΔcdgJmshE, and ΔcdgJpilT strains exhibited short residence times with mean values of 0.43, 0.53, and 0.57 s, respectively (Fig 4D). By contrast, the ΔcdgJ strain exhibits much longer mean surface residence times (3.59 s with and a maximum of 75.8 seconds), which surpass even WT (Fig 4D, red vs black bars). These longer residence times indicate a strong tendency of the ΔcdgJ strain to associate with surfaces, which is also evident in the rapid decrease in density of tracks of the ΔcdgJ over time (Fig 5): since only motile cells are displayed in each image, the density of tracks diminishes over time in the ΔcdgJ strain due to the increasing proportion of adherent, nonmotile cells. These results demonstrate the ΔcdgJ strain adheres to surfaces more rapidly than WT, and that mshA is epistatic to cdgJ.
Production of an extracellular MshA pilus is altered in ΔmshE, ΔpilT, and ΔcdgJ
The motility and biofilm phenotypes of the ΔmshE and ΔpilT mutants, combined with homology to known Type IV pilus motor proteins suggests that these genes are involved in the production of a functional MshA pilus. MshE shares 75% amino acid similarity with the Type IV extension ATPase PilB of P. aeruginosa and 77% similarity to the Type II extension ATPase EpsE of V. cholerae, suggesting that MshE is the extension ATPase of the MshA pilus (S2 and S3 Figs). We investigated the role of these genes in the production of MshA pili using a surface MshA pilin ELISA (Fig 6A), which detects only assembled pili. ΔmshA, ΔmshE, and ΔpilT mutants produced significantly less surface MshA pili than the WT strain. This supports the hypothesis that MshE and PilT are involved in the production of a functional MshA pilus. The ΔpilT strain produced significantly more surface pili than ΔmshA or ΔmshE, correlating with the intermediate phenotype of this strain observed in biofilm and near surface motility assays (Fig 3A and 3A). Surface MshA pilus production was determined in deletion mutants of other genes in the secretory operon with multiple transposon insertions (S4 Fig). Deletion of mshL, which encodes the putative outer membrane pore protein, resulted in no surface pili. While deletion of mshM (predicted to encode an ATPase) or mshN (predicted to encode a tetratricopeptide repeat domain) resulted in similar pilus production to the WT strain, pilus production was increased in a ΔmshI mutant, The specific mechanisms by which lack of these genes results in suppression of a motility defect in a ΔcdgJ strain is yet to be determined. The ΔcdgJ mutant produced significantly more surface MshA pili than WT. Surface MshA pili were reduced in ΔcdgJmshA, ΔcdgJmshE, and ΔcdgJpilT mutants to the level of the ΔmshA strain, indicating that none of these strains could produce a functional pilus. Whole cell western analysis indicated that all of the strains except ΔmshA and ΔcdgJmshA produced similar amounts of MshA protein, suggesting that the altered surface MshA is not a result of altered MshA production (Fig 6B and 6C). These data support the hypothesis that a functional MshA pilus inhibits motility and enhances surface attachment and biofilm formation. This effect is likely due to the interactions between the pili and surfaces, however, there may be additional mechanisms affecting motility and surface attachment, as discussed below. Additionally, the elevated production of MshA pili by the ΔcdgJ mutant could explain the motility and biofilm phenotypes observed in this mutant.
Fig. 6. Production of surface MshA pili is MshE and PilT dependent and repressed by CdgJ. 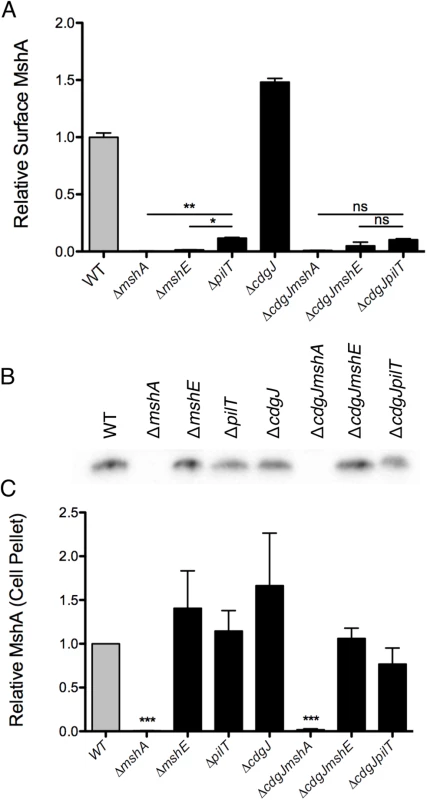
A. Surface MshA pilin production determined via ELISA with MshA-specific antibody. Two biological replicates were assayed in quadruplicate and normalized to the average of the WT strain. Oneway ANOVA followed by Bonferroni’s Multiple Comparison test. (ns = not significant, * p≤0.05, ** p≤0.01) B. Western blots detect MshA production from cell pellets using α-MshA antibody. Blots were performed in triplicate, with one representative image included. C. Densitometry of Western blots quantified MshA production in various strains. Densitometry analysis of blots was performed in triplicate and detection was normalized to the WT sample on each blot. Statistical significance determined using a one sample t-test to compare to WT. (*** p≤0.001). To confirm further that mshE was responsible for the lack of pili in a ΔmshE mutant, we generated chromosomal replacements at the native locus with either the WT mshE or a sequence encoding a mutation in the Walker A ATPase active site (K329A) (S5 Fig). Pilus production was restored to WT levels when the WT sequence was inserted, however the K329A produced no detectable surface pili. These data confirm that MshE, and specifically an intact ATPase domain, are required for MshA pilus production.
We also utilized transmission electron microscopy (TEM) to assess presence of MshA on the cell surface (Fig 7). We determined that WT produces several pili along the cell body (range 2–5) that were about one half to one cell body length. These pili were not present in the mshA mutant, suggesting that the pili observed are in fact MshA pili. Similarly, the ΔmshE strain produced no visible pili, which is consistent with the prediction that MshE is the motor protein responsible for extension of the MshA pilus. These images reveal that there were no differences observed between pili produced by WT and the ΔpilT, ΔpilU and ΔcdgJ mutants. Due to the fragile nature of MshA pili, quantitative measurements were not possible with TEM though these images observing the presence or absence of MshA pili support the quantitative measurements observed by ELISA.
Fig. 7. MshA pilus production is altered in an mshE mutant. 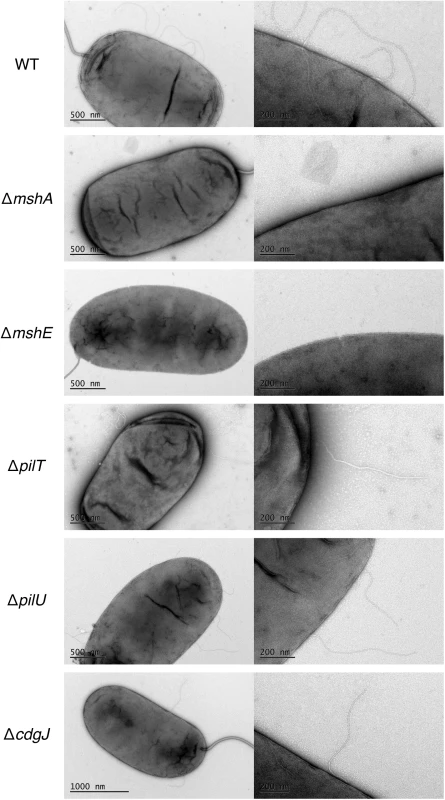
Transmission Electron Microscopy of negative stained cells in exponential growth reveals MshA pili. Scale bars included for reference. Representative image presented of at least 8 images analyzed for each strain. Right panel is increased magnification to clearly visualize pili. MshE binds c-di-GMP
The pilus motor proteins MshE and PilT contain Walker A ATPase domains, which are utilized to energize the assembly and disassembly of pili. Baraquet et al. demonstrated that the activity of the P. aeruginosa regulator FleQ is modulated by binding c-di-GMP at its Walker A site [39]. We hypothesized that one or both of the Msh pilus motor proteins could function as a c-di-GMP receptor. Isothermal calorimetry was utilized to investigate the interaction of these proteins with c-di-GMP. We determined that MshE binds c-di-GMP, while PilT and PilU were unable to bind c-di-GMP (Fig 8A). ATPase activity of purified MshE, PilT, and PilU confirmed that these preparations contain functional protein (S6 Fig). VpsT was purified and included as a positive control, as it has been demonstrated to bind c-di-GMP [40]. Fitting the data to a single binding site model indicates that MshE has a slightly lower affinity for c-di-GMP (K = 1.14x105 ± 3.24x104 M-1) than VpsT (K = 9.9x104 ± 2.06x104 M-1). These data suggest that c-di-GMP does affect MshA pilus production through interactions with MshE.
Fig. 8. MshE binds c-di-GMP. 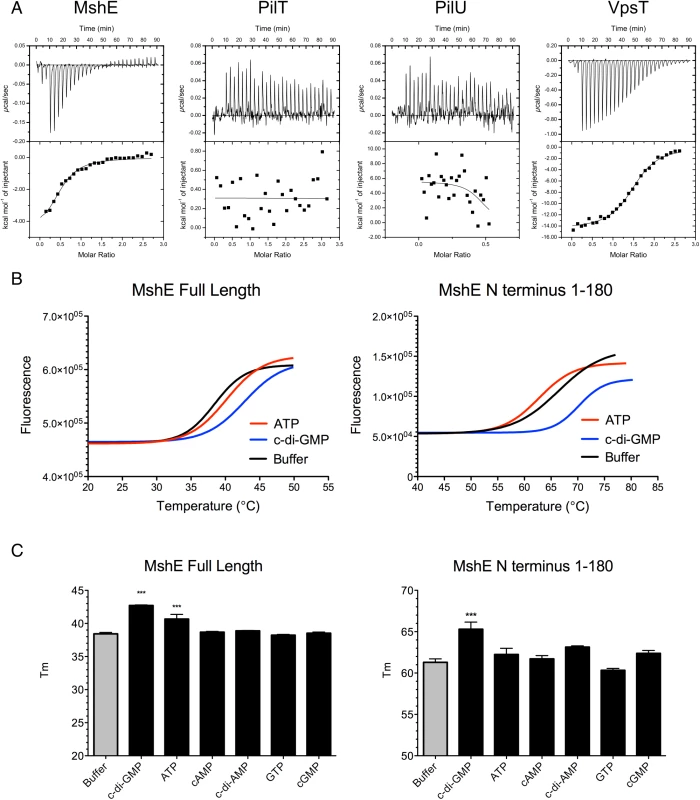
A. Isothermal Calorimetry was performed on purified proteins to determine the binding affinity for c-di-GMP. Three independent runs were included in the analysis, one representative run is depicted. VpsT is included as a positive control for c-di-GMP binding. B. Fluorescence thermal shift was utilized to investigate domains of MshE responsible for binding c-di-GMP. Fluorescence melt curves are displayed in the presence of 2mM ATP or c-di-GMP. Three independent experiments were performed in triplicate, with one representative experiment shown. C. Midpoints of melt curves are displayed in the presence of 2mM nucleotides or a buffer control. Three independent experiments were performed in triplicate, Significance was determined with a Oneway ANOVA followed by Dunnett’s Multiple Comparison test comparing to Buffer control. (*** p≤0.001, all others not significant). Since MshE, PilT, and PilU contain conserved Walker A ATPase domains, but only MshE bound c-di-GMP, we purified the N terminal domain of MshE (amino acids 1–180) for analysis of the interactions with c-di-GMP. This domain lacks homology with either PilT or PilU and therefore is a likely candidate for binding c-di-GMP. Fluorescence thermal shift assays were utilized to determine the interaction between MshE or MshE-N terminal domain and nucleotide. Purified proteins are stabilized by a bound ligand; therefore the temperature midpoint of unfolding (Tm) of the protein in the presence of ligand is higher than the Tm of the protein in buffer [41,42]. We observed that the Tm of full-length MshE in buffer was 38.43 ± 0.52°C (Fig 8B). When full-length MshE was incubated with ATP or c-di-GMP, the Tm increased to 40.15 ±1.02°C and 42.77±0.70°C, respectively. This indicates that full-length MshE binds both ATP and c-di-GMP. The N-terminal domain of MshE had a Tm of 66.36 ± 0.79°C in buffer. There was no increase in Tm in the presence of ATP (Tm = 62.72 ±0.46°C). In contrast, incubation of the MshE N-terminal domain with c-di-GMP increased the Tm to 70.18 ±0.55°C, indicating that this domain binds c-di-GMP (Fig 8C). Additionally, these interactions are specific, as neither the full-length nor the N-terminus of MshE bind to cAMP, c-di-AMP, GTP, or cGMP in thermal shift assays (Fig 8B and 8C).
c-di-GMP promotes MshA pilus production
To further evaluate the link between c-di-GMP production and pilus assembly, we performed a surface pilin ELISA to determine the production of MshA pili over a range of c-di-GMP concentrations (Fig 9). To generate a range of c-di-GMP concentrations, we introduced a construct harboring an IPTG-inducible copy of the DGC VCA0956 (Ptac0956) into the chromosome of the V. cholerae O1 El Tor strain A1552. This strain was grown in the presence of varying concentrations of IPTG, followed by detection of extracellular MshA with the surface pilin ELISA. We observed that upon induction of the expression of VCA0956 with IPTG, there was an increase in intracellular c-di-GMP (Fig 9, Grey bars). Additionally, the surface pilin ELISA detected increased amounts of extracellular MshA that coincided with the increased c-di-GMP (Fig 9, Black bars), though the total MshA production was not increased (S7B Fig) suggesting that the increased surface pilin is due to enhanced assembly. There was a significant correlation between elevated c-di-GMP and increased surface MshA (Pearson correlation, p = 0.0025, R2 = 0.9199). Increased c-di-GMP resulted in reduced motility and increased biofilm maximum thickness compared to WT (S7 Fig). Collectively, these data indicate that c-di-GMP promotes assembly of the MshA pilus. Induction of VCA0956 in a ΔmshE strain does not increase the production of surface MshA pili, further supporting that MshE is required for the c-di-GMP-dependent increase of MshA pili (S8 Fig).
Fig. 9. MshA pilus production correlates with c-di-GMP. 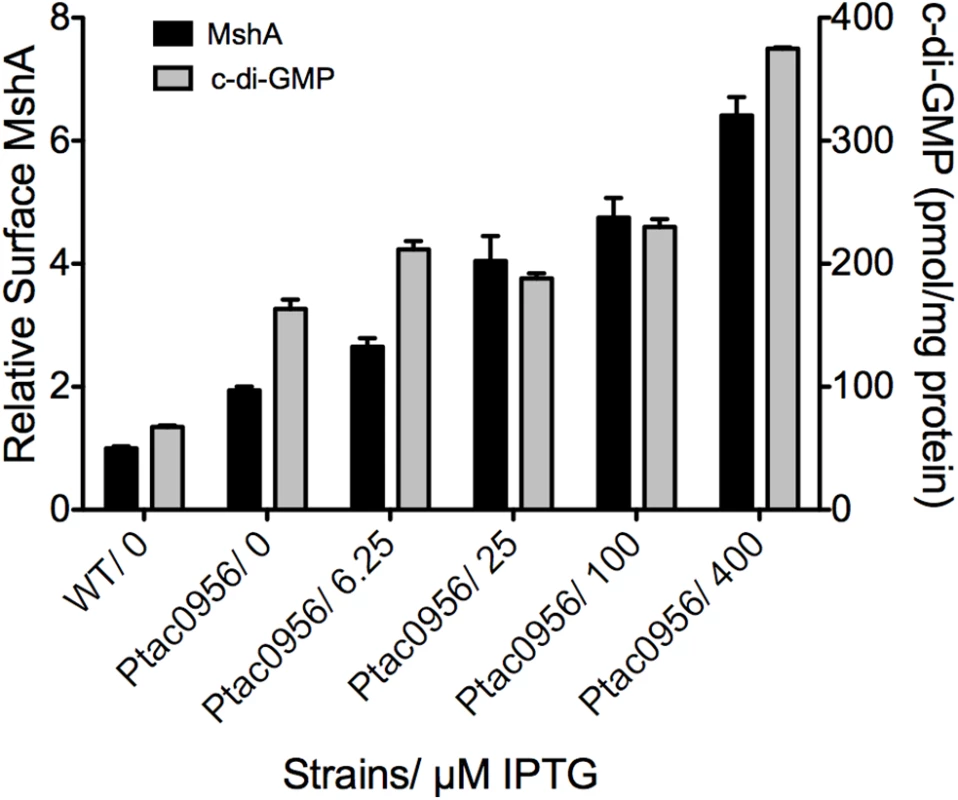
Expression of the DGC VCA0956 was induced with varying amounts of IPTG. Intracellular c-di-GMP level was determined with LC-MS/MS and normalized to total protein in the extract (grey bars, right Y axis). Four independent samples were used for each induction condition. Oneway ANOVA followed by Dunnett’s Multiple Comparison Test determined significance. Surface MshA pilin was determined by ELISA (black bars, left Y axis). Three biological replicates were tested in triplicate. Results were normalized to MshA production in the WT strain. Surface MshA pili were significantly increased in all conditions compared to WT (Oneway ANOVA, Dunnett’s Multiple Comparison Test.) Also, there was a significant correlation between increased c-di-GMP and increased surface MshA pili (Pearson correlation, p = 0.0025, R2 = 0.9199). Discussion
High c-di-GMP levels inhibit motility of bacteria and studies have highlighted some of the mechanisms involved. c-di-GMP can repress transcription of flagellar genes, or can act post-transcriptionally to regulate flagellar reversals by interactions with particular flagellar motor proteins or by altering the chemotaxis signal transduction system [26–29,43–45]. Our work revealed a role for c-di-GMP in the regulation of MshA pilus production with effects on near surface motility, motile to sessile transition, and biofilm formation via a post-translational mechanism. c-di-GMP also promotes the assembly and activity of Type IV pili in P. aeruginosa [34,46]. In this example, ectopic expression of a DGC results in elevated amounts of c-di-GMP and increased surface pili. We demonstrate that production of the MshA pilus in V. cholerae is increased in response to high concentrations of c-di-GMP. These similar results in two organisms suggest that c-di-GMP may promote type IV pili assembly and activity as a more general mechanism of pilus regulation than previously identified. Many bacteria rely on type IV pili for motility and attachment, so integration of c-di-GMP in post-translational control of these structures could be a conserved mechanism across species.
The V. cholerae genome encodes 31 GGDEF domain proteins but we have found that only a subset of these impact motility, biofilm formation, or both. We previously reported that four DGCs CdgH, CdgK, CdgL, and CdgD and two PDEs CdgJ and RocS affect motility. Furthermore, a strain lacking all four DGC-encoding genes (ΔcdgDΔcdgHΔcdgKΔcdgL) has a markedly high motility phenotype, suggesting the effect of these proteins on motility is additive [36]. Screening for suppressor mutants of ΔcdgJ that restored swimming motility identified CdgH and CdgK, suggesting that c-di-GMP produced by these DGCs could be a substrate for CdgJ. To test if CdgJ and DGCs that control motility physically interact, we analyzed interactions of CdgJ with CdgH, CdgK, CdgL, and CdgD using the commercially available Bacterial Adenylate Cyclase Two-Hybrid System (BACTH) (Euromedex Strasbourg, France). We did not observe any interaction between DGCs and CdgJ or between CdgJ and MshE or PilT, suggesting that these proteins do not need to be in physical contact, or the interaction is too weak or brief to be detected with this method.
Pili are dynamic structures that are generated by the assembly and disassembly of pilin subunits. The motor proteins responsible for this process have been characterized in many other systems. The two putative motor proteins of interest in this report, MshE and PilT, were identified based on homology to the PilT motor protein in P. aeruginosa. These proteins belong to AAA+ ATPase family proteins and are responsible for energizing the addition and disassembly of pilin subunits. Previous studies revealed presence of a bacterial AAA+ ATPase enhancer binding class of c-di-GMP receptors [28]. Here, we demonstrate that MshE binds to c-di-GMP. This is an important finding, as it establishes that ATPases beyond enhancer binding proteins are also capable of binding c-di-GMP. We note that PilT and PilU, which both harbor AAA+ ATPase domain, are unable to bind to c-di-GMP under the conditions tested. We also showed that N-terminal domain of MshE, which is not present in PilT and PilU is capable of binding to c-di-GMP. Future studies will elucidate the effect of c-di-GMP binding by MshE, as well as the specific mechanisms of this interaction.
We have also characterized several proteins necessary for the production of a functional MshA pilus. Although the function of members of the MshA operons were predicted based on homology to proteins of known function in other bacteria, function of these genes in MshA biogenesis were not characterized [22]. We have demonstrated that MshE is responsible for the assembly of MshA pilin subunits into a functional pilus. These data indicate that the decreased attachment and biofilm phenotypes, as well as the enhanced motility of the mshA, mshE and pilT mutants relies on the dynamic nature of the MshA pilus. If the presence of a pilus could enhance attachment and biofilm formation, a ΔpilT strain would in principle produce biofilms that had similar, or even greater biomass than the WT strain. The observation that the ΔpilT strain phenocopies ΔmshA and ΔmshE, which lack extracellular pili, suggests that both extension and retraction of the pili are critical for normal substrate attachment. The ELISA indicates that the ΔpilT strain produces fewer pili than WT, though the TEM images confirm that there are pili on the surface. Future studies will further investigate the role of PilT in the retraction of the MshA pilus. It is important to note that PilT has already been characterized as the retraction pilus of the ChiRP pilus (chitin-induced competence), suggesting that PilT can function in more than one type IV pilus system [47]. This could be a mechanism for genomic conservation, where one promiscuous retraction ATPase is encoded, while several extension ATPases allow for specificity of the system in regulation. Future studies will address the mechanism of co-regulation of these systems to determine whether pilT is expressed constitutively while the specific extension ATPases are regulated, or if there is overlap in the regulation between the extension and retraction ATPases.
Several studies have investigated how bacteria sense and respond to surfaces, often times by rapidly upregulating production of adhesins and polysaccharides [48–50]. This regulation is typically mediated by c-di-GMP [23,28,40,50,51]. Many bacteria also regulate motility in response to surfaces. E. coli uses the resistance to flagellar rotation as a mechanosensor and adapt by adding force-generating motor subunits to the stator complex [43]. This allows the bacterium to adjust the force of flagellar rotation to match the viscosity of the environment. An additional example of “stator swapping” to modulate flagellar force was recently published for P. aeruginosa [44]. In this example, c-di-GMP represses motility by excluding the swarming proficient MotC/D proteins from the stator complex in favor of the swarming deficient MotA/B proteins. P. aeruginosa also utilizes the altered chemotaxis protein WspA as a surface sensor, which results in production of c-di-GMP by the DGC WspR upon interaction with a surface [52,53]. In B. subtilis, production of flagella, and therefore swarming motility, is regulated by Lon-dependent proteolysis of the master regulator of flagellar biosynthesis SwrA upon surface contact [54]; and flagellar function is modulated by EpsE which synergizes exopolysaccharide biosynthesis with flagellar motility by acting as a clutch through interaction with the flagellar protein FliG to limit rotation and therefore motility [55].
This study presents a possible mechanism for how c-di-GMP production affects motility and biofilm formation through modulating MshA pilus production. Both pili and flagella contribute to near surface motility and initial attachment [10]. Besides the generation of near-surface motility modes conducive to surface attachment, it is known that van der Waals forces depend crucially on the extent of surface contact [56]. That V. cholerae has a comma-like helicoid shape with smaller surface contact areas implies that adhesive forces between the surface and the cell body will be decreased relative to more cylindrically symmetrical species such as P. aeruginosa for most cell orientations, so adhesive contributions from appendages like MSHA to ‘anchor’ the cell on surface will be comparatively more important. In fact, recent measurements of TFP adhesive forces show that they can be quite strong, in the hundreds of pico-Newton (pN) range, and are surface chemistry dependent, in agreement with our results [57]. That V. cholera select for surfaces that interact with MSHA strongly (and thereby generate ‘orbiting’ motility) implies that the existence of more functional MSHA induced by c-di-GMP can better anchor a cell mechanically and mitigate against flagellum driven motion. This work further strengthens the notion that there is a mechanistic link between c-di-GMP and initial attachment through modulation of flagellar motility and pilus activity.
Materials and Methods
Bacterial strains, plasmids, and culture conditions
The bacterial strains and plasmids used in this study are listed in S1 Table. All V. cholerae and Escherichia coli strains were grown aerobically, at 30°C and 37°C, respectively, unless otherwise noted. All cultures were grown in Luria-Bertani (LB) broth (1% Tryptone, 0.5% Yeast Extract, 1% NaCl), pH 7.5. LB agar medium contains 1.5% (wt/vol) granulated agar (BD, Sparks, MD). Concentrations of antibiotics and inducers used, where appropriate, were as follows: ampicillin, 100 μg/ml; rifampicin, 100 μg/ml; gentamicin, 50 μg/ml, kanamycin, 50 μg/ml, and arabinose, 0.2% (w/v), 6.25–400μM IPTG. In-frame deletion and GFP-tagged strains were generated according to protocols previously published [13,14].
Recombinant DNA techniques
DNA manipulations were carried out by standard molecular techniques according to manufacturer’s instructions. Restriction and DNA modification enzymes were purchased from New England Biolabs (Ipswitch, MA). Polymerase chain reactions (PCR) were carried out using primers purchased from Bioneer Corporation (Alameda, CA) and the Phusion High-Fidelity PCR kit (New England Biolabs, Ipswitch, MA), unless otherwise noted. Sequences of the primers used in the present study are available upon request. Sequences of constructs were verified by DNA sequencing (UC Berkeley DNA Sequencing Facility, Berkeley, CA).
Generation of a c-di-GMP overproducing strain
A region encompassing the PlacIq-lacI and the Ptac promoter elements was amplified from the pMAL-c5x plasmid (New England Biolabs, Ipswitch, MA) by PCR. The amplified product was joined by overlapping PCR to amplicons of ~500 bp that correspond to sequences upstream and downstream of the VCA0956 translational start site. The resulting amplicon was cloned into the suicide plasmid pGP704sacB28 and mobilized into Vibrio cholerae A1552 by biparental mating. The selection of double recombinants with the desired insertion of the PlacIq-lacI and Ptac promoter elements was performed as described in [14]. Sequences of constructs were verified by DNA sequencing (UC Berkeley DNA Sequencing Facility, Berkeley, CA).
Transposon mutagenesis
To generate a library of transposon mutants, V. cholerae O1 El Tor strain A1552 ΔcdgJ was conjugated with the donor E. coli S-17-l λpir containing the Mariner transposon on the pSC189 backbone [58]. Transconjugants were selected on LB agar containing kanamycin 50μg/ml and rifampicin 100μg/ml. A total of 7054 mutants were isolated and screened for motility phenotypes on LB soft agar (0.3%) motility plates.
Plate motility assay
Motility plates consist of LB containing 0.3% agar supplemented with 100μM IPTG where appropriate. Plates were poured and allowed to dry at room temperature for 4 h prior to inoculation. Colonies from overnight LB agar plates grown at 30°C were transferred to motility plates and incubated for 16 h at 30°C. Motility diameter was measured and normalized to the average of WT on each plate. Experiments were performed with three biological replicates in triplicate and data were analyzed with a Oneway ANOVA followed by Dunnett’s multiple comparison test.
Confocal laser scanning microscopy (CLSM) and flow cell biofilm studies
Inoculation of flow cells was done by diluting overnight-grown cultures to an OD600 of 0.04 and injecting into a μ-Slide VI0.4 (Ibidi, Martinsried, Germany). To inoculate the flow cell surface, bacteria were allowed to adhere at room temperature for 1 h. Flow of 2% v/v LB (0.02% tryptone, 0.01% yeast extract, 1% NaCl; pH 7.5) was initiated at a rate of 7.5 ml/h and continued for 24 h. Confocal images were obtained on a Zeiss LSM 5 PASCAL Laser Scanning Confocal microscope. Images were obtained with a 40X dry objective and were processed using Imaris (Bitplane, Zurich, Switzerland). Quantitative analyses were performed using the COMSTAT software package [59]. Statistical significance was determined using Oneway ANOVA with Dunnett’s Multiple Comparison test. Three biological replicates were performed in triplicate. Images presented are from one representative experiment.
High-speed imaging and cell tracking
Bacteria were cultured in full strength Luria–Bertani (LB) broth overnight under shaking at 30°C. Immediately prior to inoculation, cultures were diluted into 2% LB (containing 171 mM NaCl) to an OD600 0.01–0.03. The V. cholerae cells were then injected into a sterile flow-cell containing the same media and imaged immediately.
Imaging was performed with a Phantom V12.1 high-speed camera (Vision Research) collecting ~20,000 bright-field images at 5 ms resolution with a 100x oil objective on an IX71 Olympus microscope. All movies were recorded at the same frame rate, for the same duration, after the first, third and 15th minute post inoculation.
For cell-tracking algorithms and analysis protocol, every frame of a movie was preprocessed in Matlab (Mathworks) by subtracting the background, scaling, smoothing and thresholding. Image processing this way causes the bacteria appear as bright regions. Tracking is done by locating all bright objects that overlap objects in the next frame by combining the two frames into a three-dimensional (3D) matrix and then by locating 3D connected components.
Results are stored in a tree-like data structure with multiple roots; every newly detected bacterium that appears is recorded as a ‘root’ of the tree. When bacteria interact, they are recorded as a ‘node’ of the tree; when they depart, they are recorded as a ‘leaf’. Each root or node stores the sequence of pixel lists that comprise the bacterium in all frames until the next interaction or detachment event. We measure the instantaneous shape properties of the bacteria using the Matlab regionprops function [10].
Surface Pilin ELISA
Surface pili composed of MshA were quantified using an ELISA based on a previously published protocol [46]. Briefly, overnight culture was diluted 1 : 100 in fresh LB medium and grown to OD600 0.5 at 30°C. Cells (125μL) were added to a 96-well plate (Greiner Bio-One, Monroe, NC) and incubated at 30°C for one hour. Cells were fixed with 100μL of methanol for 10 minutes at room temperature, then washed twice with PBS. Samples were blocked in 5% nonfat dry milk and immunoblotted with polyclonal rabbit anti-MshA (1 : 1000 dilution, gift of J. Zhu) and horseradish peroxidase (HRP)-conjugated secondary antibody (Santa Cruz Biotechnology, Santa Cruz, CA). After three washes in PBS, 100μL of TMB (eBioscience, San Diego, CA) was added and incubated for 30 minutes at room temperature followed by the addition of 100μL of 2N H2SO4. Absorbance was recorded at 490nm and the samples were normalized to the change in WT. Three biological replicates were assayed in triplicate and statistical significance was determined with a Oneway ANOVA followed by a Dunnett’s Multiple Comparison test.
Western blot
Samples were grown to mid-exponential phase (OD6000.5) in LB or LB with IPTG. Cells were collected via centrifugation and cell pellets were resuspended in 2% SDS and boiled for 5 minutes. Lysates were cleared via centrifugation and total protein was quantified via BCA assay (Pierce, Rockford, IL). Two hundred μg of protein was separated on a 12% SDS PAGE gel and transferred to a PVDF membrane using a semi-dry transfer apparatus (Bio-Rad, Hercules, CA). Blots were blocked in 5% nonfat dry milk and immunoblotted with polyclonal rabbit anti-MshA (1 : 2000 dilution, gift of J. Zhu) and horseradish peroxidase (HRP)-conjugated secondary antibody (Santa Cruz Biotechnology, Santa Cruz, CA). Chemiluminescence was detected with the SuperSignal West Pico reagents (Pierce, Rockford, IL) on the ChemiDoc MP Imager (Bio-Rad Hercules, CA). Densitometry was performed using the Image Lab software v4.0.1 (Bio-Rad, Hercules, CA) using the band in the WT lane as a reference. Blots were performed in triplicate for densitometry analysis and a representative image is shown.
Transmission Electron Microscopy
Bacteria were prepared for electron microscopy by inoculating a single colony of each V. cholerae strain in LB broth grown overnight at 30°C with shaking at 200 rpm after which each culture was diluted 1 : 100 in LB broth and allowed to grow similarly to OD 0.4. An aliquot of each culture was diluted to yield an optical density of 0.1–0.2 and then applied to a 300 mesh carbon-coated Formvar grid (Electron Microscopy Sciences, Hatfield, PA). After 2 minutes, each grid was washed five times with deionized water, and negatively stained with 2% (w/v) aqueous uranyl acetate solution for 90 seconds. Imaging was performed with a JEOL JEM-1400 transmission microscope.
Protein purification
E. coli BL21 harboring plasmids for gene expression were grown to an OD600 of 0.4 at 30°C in LB containing 100μg/mL ampicillin. Cultures were shifted to 18°C and IPTG was added to a final concentration of 100μM. 16h post induction, cells were harvested by centrifugation at 10,000 x g for 15 minutes and stored at -80°C.
Cell pellets were resuspended in GST Lysis Buffer (50mM Tris (pH 8.0), 1M NaCl, 0.5% Tween-20 containing PI cocktail tablets (Roche Life Science, Indianapolis, IN). Cells were lysed by sonication and cell lysate was cleared via centrifugation. Cleared lysate was loaded onto GST FPLC column as follows.
GSTPrep FF16/10 column (GE Healthcare, Piscataway, NJ) was equilibrated in lysis buffer at 1mL/minute using a BioLogic DuoFlow FPLC system (Bio-Rad, Hurcules, CA). Sample was loaded and washed with 1 column volume of GST Lysis Buffer (20mL). Subsequent washes were performed with 20mL of Wash Buffer 2 (50mM Tris (pH 8.0), 0.25M NaCl, 0.5% Tween-20, 0.5mM DTT) and 3 (50mM Tris (pH 8.0), 0.25M NaCl, 0.5mM DTT). Bound protein was eluted with 80mL of GST Elution Buffer 4 (50mM Tris (pH 8.0), 0.25M NaCl, Glutathione 1.5g/L) and collected in 15mL fractions.
These fractions were pooled and concentrated to approximately 10mL using an Amicon 10KDa cutoff spin fliter (EMD Millipore, Darmstadt, Germany). Samples were dialyzed against ITC buffer (25mM Tris-HCl, 150mM NaCl, 250μM DTT, pH 7.5) overnight using 12 kDa cutoff dialysis tubing (Fisherbrand, Pittsburgh, PA). An aliquot of dialyzed protein was diluted in 6M guanidinium HCl and concentration determined via A280.
Isothermal calorimetry
MshE (18.9μM), PilT (20.2μM), PilU (21.1μM), VpsT (19.5μM) and c-di-GMP (250μM) were prepared in 25mM TrisHCl pH 7.5, 150mM NaCl, and 200μM DTT and degassed prior to analysis. ITC was performed in with a VP-ITC (MicroCal, Northampton, MA) with the following parameters: 3 initial injections of 2μL followed by 40 injections of 10μL spaced at 180 seconds. The data were normalized to a run injecting c-di-GMP into buffer to account for the heat of dilution. Data were processed in Origin v7.0 software (OriginLab, Northampton, MA) and fit to a single site model.
Fluorescence thermal shift
Thermal shift assays were performed as previously described with modifications [41,42]. Briefly, purified protein was added to the reaction to a final concentration of 3μM in the presence or absence of 2mM concentration of the indicated nucleotide in buffer (25Mm TrisHCl pH 7.5, 100mM NaCl, 1 : 1000 dilution of SYPRO Orange Dye (Invitrogen), and 0.2mM MgCl2. A melt curve protocol was run on an Applied Biosystems ViiA7 qPCR instrument. The fluorescence was measured using the ROX reporter with a temperature gradient of 20–95°C in 0.5°C increments at 30 second intervals. Melt curve data were trimmed to three data points after maximum and the data were plotted with Boltzmann model to obtain the temperature midpoint of unfolding (Tm) of the protein in each condition using Prism 5.0 software (GraphPad). The fluorescence baseline of each sample was normalized to the buffer control for visualization purposes. Three biological replicates were assayed in triplicate and statistical significance was determined with a Oneway ANOVA followed by a Dunnett’s Multiple Comparison test.
ATPase assay
ATPase activity of purified proteins was determined by measuring the production of inorganic phosphate from ATP using the Enzchek Phosphate Assay Kit (Invitrogen). The standard reaction mixture was prepared with the addition of 2mM MgCl2, 10mM KCl, and 1mM DTT. Purified protein in buffer (25mM TrisHCl pH 7.5, 100mM NaCl) was added to the standard reaction mixture to a final concentration of 5μM. After a 10 minute incubation at room temperature, ATP was added to a final concentration of 10mM and reactions were incubated at 22°C for 30 minutes. Production of inorganic phosphate was monitored by reading OD360 and compared to a standard curve of solutions of KH2PO4. The data are reported as specific activity (nmol Pi/min/mg of protein). BSA was included as a negative control. Three independent experiments were run in triplicate.
c-di-GMP measurement
c-di-GMP extraction was performed as described previously [36]. Briefly, 40 ml of culture grown to OD600 ~0.4 was centrifuged at 3220 x g for 30 min. Cell pellets were allowed to dry briefly then re-suspended in 1 ml extraction solution (40% acetonitrile, 40% methanol, 0.1% formic acid, 19.9% water), and incubated on ice for 5 min. Samples were then centrifuged at 16,100 g for 5 min and 800 μl of supernatant was dried under vacuum and lyophilized. Samples were re-suspended in 50 μl of 184 mM NaCl and analyzed by liquid chromatography-tandem mass spectrometry (LC-MS/MS) on a Thermo-Electron Finnigan LTQ mass spectrometer coupled to a surveyor HPLC (Thermo, Waltham, MA). The Synergin Hydro 4u Fusion-RP 80A column (150 mm x 2.00 mm diameter; 4-μm particle size) (Phenomenex, Torrance, CA) was used for reverse-phase liquid chromatography. Solvent A was 0.1% acetic acid in 10 mM ammonium acetate, solvent B was 0.1% formic acid in methanol. The gradient used was as follows: time (t) = 0–4 min, 98% solvent A, 2% solvent B; t = 10–15 minutes, 5% solvent A, 95% solvent B. The injection volume was 20 μl and the flow rate for chromatography was 200 μl/minutes.
The amount of c-di-GMP in samples was calculated with a standard curve generated from pure c-di-GMP suspended in 184 mM NaCl (Biolog Life Science Institute, Bremen, Germany). Concentrations used for standard curve generation were 50 nM, 100 nM, 500 nM, 2 μM, 3.5 μM, 5 μM, 7.5 μM, and 10 μM. The assay is linear from 50 nM to 10 μM with an R2 of 0.999. c-di-GMP levels were normalized to total protein per ml of culture.
To determine protein concentration, 4 ml from each culture was pelleted, the supernatant was removed, and cells were lysed in 1 ml of 2% sodium dodecyl sulfate. Total protein in the samples was estimated with BCA assay (Pierce, Rockford, IL) using bovine serum albumin (BSA) as standards. Each c-di-GMP quantification experiment was performed with four biological replicates. Levels of c-di-GMP were compared to WT with Oneway ANOVA followed by a Dunnett’s Multiple Comparison test.
Supporting Information
Zdroje
1. Kaper JB, Morris JG, Levine MM. Cholera. Clin Microbiol Rev. 1995 Jan;8(1):48–86. 7704895
2. Matson JS, Withey JH, DiRita VJ. Regulatory networks controlling Vibrio cholerae virulence gene expression. Infect Immun. 2007 Dec;75(12):5542–9. 17875629
3. Taylor RK, Miller VL, Furlong DB, Mekalanos JJ. Use of phoA gene fusions to identify a pilus colonization factor coordinately regulated with cholera toxin. Proc Natl Acad Sci USA. 1987 May;84(9):2833–7. 2883655
4. Islam MS, Jahid MIK, Rahman MM, Rahman MZ, Islam MS, Kabir MS, et al. Biofilm acts as a microenvironment for plankton-associated Vibrio cholerae in the aquatic environment of Bangladesh. Microbiol Immunol. 2013 Nov 14;51(4):369–79.
5. Faruque SM, Biswas K, Udden SMN, Ahmad QS, Sack DA, Nair GB, et al. Transmissibility of cholera: in vivo-formed biofilms and their relationship to infectivity and persistence in the environment. Proc Natl Acad Sci USA. 2006 Apr 18;103(16):6350–5. 16601099
6. Alam M, Sultana M, Nair GB, Siddique AK, Hasan NA, Sack RB, et al. Viable but nonculturable Vibrio cholerae O1 in biofilms in the aquatic environment and their role in cholera transmission. Proc Natl Acad Sci USA. 2007 Nov 6;104(45):17801–6. 17968017
7. Tamplin ML, Gauzens AL, Huq A, Sack DA, Colwell RR. Attachment of Vibrio cholerae serogroup O1 to zooplankton and phytoplankton of Bangladesh waters. Appl Environ Microbiol. 1990 Jun;56(6):1977–80. 2383016
8. Colwell RR, Huq A, Islam MS, Aziz KMA, Yunus M, Khan NH, et al. Reduction of cholera in Bangladeshi villages by simple filtration. Proc Natl Acad Sci USA. 2003 Feb 4;100(3):1051–5. 12529505
9. Tamayo R, Patimalla B, Camilli A. Growth in a biofilm induces a hyperinfectious phenotype in Vibrio cholerae. Infect Immun. 2010 Aug;78(8):3560–9. doi: 10.1128/IAI.00048-10 20515927
10. Utada AS, Bennett RR, Fong JCN, Gibiansky ML, Yildiz FH, Golestanian R, et al. Vibrio cholerae use pili and flagella synergistically to effect motility switching and conditional surface attachment. Nat Commun. 2014;5 : 4913. doi: 10.1038/ncomms5913 25234699
11. Watnick PI, Kolter R. Steps in the development of a Vibrio cholerae El Tor biofilm. Mol Microbiol. 1999 Nov;34(3):586–95. 10564499
12. Moorthy S, Watnick PI. Genetic evidence that the Vibrio cholerae monolayer is a distinct stage in biofilm development. Mol Microbiol. 2004 Mar 4;52(2):573–87. 15066042
13. Fong JCN, Karplus K, Schoolnik GK, Yildiz FH. Identification and characterization of RbmA, a novel protein required for the development of rugose colony morphology and biofilm structure in Vibrio cholerae. J Bacteriol. 2006 Feb;188(3):1049–59. 16428409
14. Fong JCN, Yildiz FH. The rbmBCDEF gene cluster modulates development of rugose colony morphology and biofilm formation in Vibrio cholerae. J Bacteriol. 2007 Mar;189(6):2319–30. 17220218
15. Berk V, Fong JCN, Dempsey GT, Develioglu ON, Zhuang X, Liphardt J, et al. Molecular architecture and assembly principles of Vibrio cholerae biofilms. Science. 2012 Jul 13;337(6091):236–9. doi: 10.1126/science.1222981 22798614
16. Absalon C, Van Dellen K, Watnick PI. A communal bacterial adhesin anchors biofilm and bystander cells to surfaces. PLoS Pathog. 2011 Aug 25;7(8):e1002210. doi: 10.1371/journal.ppat.1002210 21901100
17. Yildiz FH, Schoolnik GK. Vibrio cholerae O1 El Tor: Identification of a gene cluster required for the rugose colony type, exopolysaccharide production, chlorine resistance, and biofilm formation. Proc Natl Acad Sci USA. 1999 Mar 30;96(7):4028–33. 10097157
18. Seper A, Fengler VHI, Roier S, Wolinski H, Kohlwein SD, Bishop AL, et al. Extracellular nucleases and extracellular DNA play important roles in Vibrio cholerae biofilm formation. Mol Microbiol. 2011 Oct 27;82(4):1015–37. doi: 10.1111/j.1365-2958.2011.07867.x 22032623
19. Teschler JK, Zamorano-Sánchez D, Utada AS, Warner CJA, Wong GCL, Linington RG, et al. Living in the matrix: assembly and control of Vibrio cholerae biofilms. Nat Rev Micro. 2015 Apr 16;13(5):255–68.
20. Watnick PI, Fullner KJ, Kolter R. A role for the mannose-sensitive hemagglutinin in biofilm formation by Vibrio cholerae El Tor. J Bacteriol. 1999 Jun;181(11):3606–9. 10348878
21. Merz AJ, So M, Sheetz MP. Pilus retraction powers bacterial twitching motility. Nature. 2000 Sep 7;407(6800):98–102. 10993081
22. Marsh JW, Taylor RK. Genetic and transcriptional analyses of the Vibrio cholerae mannose-sensitive hemagglutinin type 4 pilus gene locus. J Bacteriol. 1999 Feb;181(4):1110–7. 9973335
23. Hengge R. Principles of c-di-GMP signalling in bacteria. Nat Rev Micro. 2009 Apr;7(4):263–73.
24. Sondermann H, Shikuma NJ, Yildiz FH. You’ve come a long way: c-di-GMP signaling. Curr Opin Microbiol. 2012 Apr;15(2):140–6. doi: 10.1016/j.mib.2011.12.008 22226607
25. Römling U, Galperin MY, Gomelsky M. Cyclic di-GMP: the first 25 years of a universal bacterial second messenger. Microbiol Mol Biol Rev. 2013 Mar;77(1):1–52. doi: 10.1128/MMBR.00043-12 23471616
26. Paul K, Nieto V, Carlquist WC, Blair DF, Harshey RM. The c-di-GMP binding protein YcgR controls flagellar motor direction and speed to affect chemotaxis by a “backstop brake” mechanism. Mol Cell. 2010 Apr 9;38(1):128–39. doi: 10.1016/j.molcel.2010.03.001 20346719
27. Boehm A, Kaiser M, Li H, Spangler C, Kasper CA, Ackermann M, et al. Second messenger-mediated adjustment of bacterial swimming velocity. Cell. 2010 Apr 2;141(1):107–16. doi: 10.1016/j.cell.2010.01.018 20303158
28. Hickman JW, Harwood CS. Identification of FleQ from Pseudomonas aeruginosa as a c-di-GMP-responsive transcription factor. Mol Microbiol. 2008 Jul;69(2):376–89. doi: 10.1111/j.1365-2958.2008.06281.x 18485075
29. Srivastava D, Hsieh M - L, Khataokar A, Neiditch MB, Waters CM. Cyclic di-GMP inhibits Vibrio cholerae motility by repressing induction of transcription and inducing extracellular polysaccharide production. Mol Microbiol. 2013 Nov 8;90(6):1262–76. doi: 10.1111/mmi.12432 24134710
30. Wilksch JJ, Yang J, Clements A, Gabbe JL, Short KR, Cao H, et al. MrkH, a novel c-di-GMP-dependent transcriptional activator, controls Klebsiella pneumoniae biofilm formation by regulating type 3 fimbriae expression. PLoS Pathog. 2011 Aug;7(8):e1002204. doi: 10.1371/journal.ppat.1002204 21901098
31. Johnson JG, Murphy CN, Sippy J, Johnson TJ, Clegg S. Type 3 fimbriae and biofilm formation are regulated by the transcriptional regulators MrkHI in Klebsiella pneumoniae. J Bacteriol. 2011 Jul;193(14):3453–60. doi: 10.1128/JB.00286-11 21571997
32. Yang J, Wilksch JJ, Tan JWH, Hocking DM, Webb CT, Lithgow T, et al. Transcriptional activation of the mrkA promoter of the Klebsiella pneumoniae type 3 fimbrial operon by the c-di-GMP-dependent MrkH protein. PLoS ONE. 2013;8(11):e79038. doi: 10.1371/journal.pone.0079038 24244411
33. Navarro MVAS, De N, Bae N, Wang Q, Sondermann H. Structural analysis of the GGDEF-EAL domain-containing c-di-GMP receptor FimX. Structure. 2009 Aug 12;17(8):1104–16. doi: 10.1016/j.str.2009.06.010 19679088
34. Jain R, Behrens AJ, Kaever V, Kazmierczak BI. Type IV pilus assembly in Pseudomonas aeruginosa over a broad range of cyclic di-GMP concentrations. J Bacteriol. 2012 Jul 27;194(16):4285–94. doi: 10.1128/JB.00803-12 22685276
35. Beyhan S, Odell LS, Yildiz FH. Identification and characterization of cyclic diguanylate signaling systems controlling rugosity in Vibrio cholerae. J Bacteriol. 2008 Nov;190(22):7392–405. doi: 10.1128/JB.00564-08 18790873
36. Liu X, Beyhan S, Lim B, Linington RG, Yildiz FH. Identification and characterization of a phosphodiesterase that inversely regulates motility and biofilm formation in Vibrio cholerae. J Bacteriol. 2010 Sep;192(18):4541–52. doi: 10.1128/JB.00209-10 20622061
37. Rashid MH, Rajanna C, Ali A, Karaolis DKR. Identification of genes involved in the switch between the smooth and rugose phenotypes of Vibrio cholerae. FEMS Microbiol Lett. 2003 Oct 10;227(1):113–9. 14568156
38. Lim B, Beyhan S, Meir J, Yildiz FH. Cyclic-diGMP signal transduction systems in Vibrio cholerae: modulation of rugosity and biofilm formation. Mol Microbiol. 2006 Apr;60(2):331–48. 16573684
39. Baraquet C, Harwood CS. Cyclic diguanosine monophosphate represses bacterial flagella synthesis by interacting with the Walker A motif of the enhancer-binding protein FleQ. Proc Natl Acad Sci USA. 2013 Nov 12;110(46):18478–83. doi: 10.1073/pnas.1318972110 24167275
40. Krasteva PV, Fong JCN, Shikuma NJ, Beyhan S, Navarro MVAS, Yildiz FH, et al. Vibrio cholerae VpsT regulates matrix production and motility by directly sensing cyclic di-GMP. Science. 2010 Feb 12;327(5967):866–8. doi: 10.1126/science.1181185 20150502
41. Ouyang S, Song X, Wang Y, Ru H, Shaw N, Jiang Y, et al. Structural analysis of the STING adaptor protein reveals a hydrophobic dimer interface and mode of cyclic di-GMP binding. Immunity. 2012 Jun;36(6):1073–86. doi: 10.1016/j.immuni.2012.03.019 22579474
42. Huynh K, Partch CL. Analysis of protein stability and ligand interactions by thermal shift assay. Curr Protoc Protein Sci. 2015;79 : 28.9.1–28.9.14.
43. Lele PP, Hosu BG, Berg HC. Dynamics of mechanosensing in the bacterial flagellar motor. Proc Natl Acad Sci USA. 2013 Jul 16;110(29):11839–44. doi: 10.1073/pnas.1305885110 23818629
44. Kuchma SL, Delalez NJ, Filkins LM, Snavely EA, Armitage JP, O'Toole GA. Cyclic di-GMP-mediated repression of swarming motility by Pseudomonas aeruginosa PA14 requires the MotAB stator. J Bacteriol. 2015 Feb;197(3):420–30. 25349157
45. Beyhan S, Tischler AD, Camilli A, Yildiz FH. Transcriptome and phenotypic responses of Vibrio cholerae to increased cyclic di-GMP level. J Bacteriol. 2006 May;188(10):3600–13. 16672614
46. Kazmierczak BI, Lebron MB, Murray TS. Analysis of FimX, a phosphodiesterase that governs twitching motility in Pseudomonas aeruginosa. Mol Microbiol. 2006 May;60(4):1026–43. 16677312
47. Seitz P, Blokesch M. DNA-uptake machinery of naturally competent Vibrio cholerae. Proc Natl Acad Sci USA. 2013 Oct 14;110(44):17987–92. doi: 10.1073/pnas.1315647110 24127573
48. Hinsa SM, Espinosa-Urgel M, Ramos JL, O'Toole GA. Transition from reversible to irreversible attachment during biofilm formation by Pseudomonas fluorescens WCS365 requires an ABC transporter and a large secreted protein. Mol Microbiol. 2003 Aug;49(4):905–18. 12890017
49. Hickman JW, Tifrea DF, Harwood CS. A chemosensory system that regulates biofilm formation through modulation of cyclic diguanylate levels. Proc Natl Acad Sci USA. 2005 Oct 4;102(40):14422–7. 16186483
50. Li G, Brown PJB, Tang JX, Xu J, Quardokus EM, Fuqua C, et al. Surface contact stimulates the just-in-time deployment of bacterial adhesins. Mol Microbiol. 2011 Nov 22;83(1):41–51. doi: 10.1111/j.1365-2958.2011.07909.x 22053824
51. Newell PD, Boyd CD, Sondermann H, O'Toole GA. A c-di-GMP effector system controls cell adhesion by inside-out signaling and surface protein cleavage. PLoS Biol. 2011;9(2):e1000587. doi: 10.1371/journal.pbio.1000587 21304920
52. Guvener ZT, Harwood CS. Subcellular location characteristics of the Pseudomonas aeruginosa GGDEF protein, WspR, indicate that it produces cyclic-di-GMP in response to growth on surfaces. Mol Microbiol. 2007 Dec;66(6):1459–73. 18028314
53. O'Connor JR, Kuwada NJ, Huangyutitham V, Wiggins PA, Harwood CS. Surface sensing and lateral subcellular localization of WspA, the receptor in a chemosensory-like system leading to c-di-GMP production. Mol Microbiol. 2012 Nov;86(3):720–9. doi: 10.1111/mmi.12013 22957788
54. Mukherjee S, Bree AC, Liu J, Patrick JE, Chien P, Kearns DB. Adaptor-mediated Lon proteolysis restricts Bacillus subtilis hyperflagellation. Proc Natl Acad Sci USA. 2015 Jan 6;112(1):250–5. doi: 10.1073/pnas.1417419112 25538299
55. Guttenplan SB, Blair KM, Kearns DB. The EpsE flagellar clutch is bifunctional and synergizes with EPS biosynthesis to promote Bacillus subtilis biofilm formation. PLoS Genet. 2010 Dec 9;6(12):e1001243. doi: 10.1371/journal.pgen.1001243 21170308
56. Parsegian VA. Van der Waals Forces. Cambridge University Press; 2005.
57. Beaussart A, Baker AE, Kuchma SL, El-Kirat-Chatel S, O'Toole GA, Dufrêne YF. Nanoscale adhesion forces of Pseudomonas aeruginosa type IV pili. ACS Nano. 2014 Oct 28;8(10):10723–33. doi: 10.1021/nn5044383 25286300
58. Chiang SL, Rubin EJ. Construction of a mariner-based transposon for epitope-tagging and genomic targeting. Gene. 2002 Aug 21;296(1–2):179–85. 12383515
59. Heydorn A, Nielsen AT, Hentzer M, Sternberg C, Givskov M, Ersbøll BK, et al. Quantification of biofilm structures by the novel computer program COMSTAT. Microbiology. 2000 Oct;146 (Pt 10):2395–407. 11021916
Štítky
Hygiena a epidemiologie Infekční lékařství Laboratoř
Článek TRIM21 Promotes cGAS and RIG-I Sensing of Viral Genomes during Infection by Antibody-Opsonized VirusČlánek Effector OspB Activates mTORC1 in a Manner That Depends on IQGAP1 and Promotes Cell ProliferationČlánek Fundamental Roles of the Golgi-Associated Aspartyl Protease, ASP5, at the Host-Parasite InterfaceČlánek Modulation of the Surface Proteome through Multiple Ubiquitylation Pathways in African Trypanosomes
Článek vyšel v časopisePLOS Pathogens
Nejčtenější tento týden
2015 Číslo 10- Stillova choroba: vzácné a závažné systémové onemocnění
- Perorální antivirotika jako vysoce efektivní nástroj prevence hospitalizací kvůli COVID-19 − otázky a odpovědi pro praxi
- Jak souvisí postcovidový syndrom s poškozením mozku?
- Diagnostika virových hepatitid v kostce – zorientujte se (nejen) v sérologii
- Infekční komplikace virových respiračních infekcí – sekundární bakteriální a aspergilové pneumonie
-
Všechny články tohoto čísla
- Expression of Concern: Misregulation of Underlies the Developmental Abnormalities Caused by Three Distinct Viral Silencing Suppressors in Arabidopsis
- Preparing for the Next Epidemic with Basic Virology
- Effectively Communicating the Uncertainties Surrounding Ebola Virus Transmission
- Translating Basic Research into Clinical Applications: Malaria Research at an NIH Lab
- A Gut Odyssey: The Impact of the Microbiota on Spore Formation and Germination
- Papillomavirus E6 Oncoproteins Take Common Structural Approaches to Solve Different Biological Problems
- Chronobiomics: The Biological Clock as a New Principle in Host–Microbial Interactions
- Dimensions of Horizontal Gene Transfer in Eukaryotic Microbial Pathogens
- Addressing the Complications of Ebola and Other Viral Hemorrhagic Fever Infections: Using Insights from Bacterial and Fungal Sepsis
- Time for Chocolate: Current Understanding and New Perspectives on Cacao Witches’ Broom Disease Research
- Ganglioside and Non-ganglioside Mediated Host Responses to the Mouse Polyomavirus
- Crosslinking of a Peritrophic Matrix Protein Protects Gut Epithelia from Bacterial Exotoxins
- Structure Elucidation of Coxsackievirus A16 in Complex with GPP3 Informs a Systematic Review of Highly Potent Capsid Binders to Enteroviruses
- CD39 Expression Identifies Terminally Exhausted CD8 T Cells
- Abiotic Stresses Antagonize the Rice Defence Pathway through the Tyrosine-Dephosphorylation of OsMPK6
- Dissociation of Tissue Destruction and Bacterial Expansion during Bubonic Plague
- Interferon-γ: The Jekyll and Hyde of Malaria
- CCR2 Inflammatory Dendritic Cells and Translocation of Antigen by Type III Secretion Are Required for the Exceptionally Large CD8 T Cell Response to the Protective YopE Epitope during Infection
- A New Glycan-Dependent CD4-Binding Site Neutralizing Antibody Exerts Pressure on HIV-1
- The Suramin Derivative NF449 Interacts with the 5-fold Vertex of the Enterovirus A71 Capsid to Prevent Virus Attachment to PSGL-1 and Heparan Sulfate
- Trans-generational Immune Priming Protects the Eggs Only against Gram-Positive Bacteria in the Mealworm Beetle
- Peripheral Vγ9Vδ2 T Cells Are a Novel Reservoir of Latent HIV Infection
- Respiratory Syncytial Virus Disease Is Mediated by Age-Variable IL-33
- TRIM21 Promotes cGAS and RIG-I Sensing of Viral Genomes during Infection by Antibody-Opsonized Virus
- Modeling the Effects of Vorinostat Reveals both Transient and Delayed HIV Transcriptional Activation and Minimal Killing of Latently Infected Cells
- Identification of a Novel Lipoprotein Regulator of Spore Germination
- Calcium Regulation of Hemorrhagic Fever Virus Budding: Mechanistic Implications for Host-Oriented Therapeutic Intervention
- Antigen-Specific Th17 Cells Are Primed by Distinct and Complementary Dendritic Cell Subsets in Oropharyngeal Candidiasis
- Comparative Life Cycle Transcriptomics Revises Genome Annotation and Links a Chromosome Duplication with Parasitism of Vertebrates
- The Autophagy Receptor TAX1BP1 and the Molecular Motor Myosin VI Are Required for Clearance of Salmonella Typhimurium by Autophagy
- Carcinogenic Parasite Secretes Growth Factor That Accelerates Wound Healing and Potentially Promotes Neoplasia
- Effector OspB Activates mTORC1 in a Manner That Depends on IQGAP1 and Promotes Cell Proliferation
- Dengue Virus Infection of Requires a Putative Cysteine Rich Venom Protein
- Distinct Viral and Mutational Spectrum of Endemic Burkitt Lymphoma
- Fundamental Roles of the Golgi-Associated Aspartyl Protease, ASP5, at the Host-Parasite Interface
- Phenotypic and Functional Alterations in Circulating Memory CD8 T Cells with Time after Primary Infection
- Systematic Identification of Cyclic-di-GMP Binding Proteins in Reveals a Novel Class of Cyclic-di-GMP-Binding ATPases Associated with Type II Secretion Systems
- Influenza Transmission in the Mother-Infant Dyad Leads to Severe Disease, Mammary Gland Infection, and Pathogenesis by Regulating Host Responses
- Myeloid Cell Arg1 Inhibits Control of Arthritogenic Alphavirus Infection by Suppressing Antiviral T Cells
- The White-Nose Syndrome Transcriptome: Activation of Anti-fungal Host Responses in Wing Tissue of Hibernating Little Brown Myotis
- Influenza Virus Reassortment Is Enhanced by Semi-infectious Particles but Can Be Suppressed by Defective Interfering Particles
- Identification of the Mechanisms Causing Reversion to Virulence in an Attenuated SARS-CoV for the Design of a Genetically Stable Vaccine
- Differentiation-Dependent KLF4 Expression Promotes Lytic Epstein-Barr Virus Infection in Epithelial Cells
- The Histone Acetyltransferase Hat1 Regulates Stress Resistance and Virulence via Distinct Chromatin Assembly Pathways
- C-di-GMP Regulates Motile to Sessile Transition by Modulating MshA Pili Biogenesis and Near-Surface Motility Behavior in
- Modulation of the Surface Proteome through Multiple Ubiquitylation Pathways in African Trypanosomes
- Crystal Structure of the Human Cytomegalovirus Glycoprotein B
- Depletion of . GlmU from Infected Murine Lungs Effects the Clearance of the Pathogen
- Immunologic Control of Papillomavirus Type 1
- Requires Host Rab1b for Survival in Macrophages
- Structure Analysis Uncovers a Highly Diverse but Structurally Conserved Effector Family in Phytopathogenic Fungi
- PD-L1 Expression on Retrovirus-Infected Cells Mediates Immune Escape from CD8 T Cell Killing
- Phospho-dependent Regulation of SAMHD1 Oligomerisation Couples Catalysis and Restriction
- IL-4 Induced Innate CD8 T Cells Control Persistent Viral Infection
- Crystal Structures of a Piscine Betanodavirus: Mechanisms of Capsid Assembly and Viral Infection
- BCG Skin Infection Triggers IL-1R-MyD88-Dependent Migration of EpCAM CD11b Skin Dendritic cells to Draining Lymph Node During CD4+ T-Cell Priming
- Antigenic Characterization of the HCMV gH/gL/gO and Pentamer Cell Entry Complexes Reveals Binding Sites for Potently Neutralizing Human Antibodies
- Rescue of a Plant Negative-Strand RNA Virus from Cloned cDNA: Insights into Enveloped Plant Virus Movement and Morphogenesis
- Geminivirus Activates to Accelerate Cytoplasmic DCP2-Mediated mRNA Turnover and Weakens RNA Silencing in
- Disruption of Sphingolipid Biosynthesis Blocks Phagocytosis of
- The Fungal Exopolysaccharide Galactosaminogalactan Mediates Virulence by Enhancing Resistance to Neutrophil Extracellular Traps
- The Timing of Stimulation and IL-2 Signaling Regulate Secondary CD8 T Cell Responses
- Structural and Functional Analysis of Murine Polyomavirus Capsid Proteins Establish the Determinants of Ligand Recognition and Pathogenicity
- The Dual Role of an ESCRT-0 Component HGS in HBV Transcription and Naked Capsid Secretion
- PLOS Pathogens
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Chronobiomics: The Biological Clock as a New Principle in Host–Microbial Interactions
- Interferon-γ: The Jekyll and Hyde of Malaria
- Crosslinking of a Peritrophic Matrix Protein Protects Gut Epithelia from Bacterial Exotoxins
- Antigen-Specific Th17 Cells Are Primed by Distinct and Complementary Dendritic Cell Subsets in Oropharyngeal Candidiasis
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



