-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaCTCF Prevents the Epigenetic Drift of EBV Latency Promoter Qp
The establishment and maintenance of Epstein-Barr Virus (EBV) latent infection requires distinct viral gene expression programs. These gene expression programs, termed latency types, are determined largely by promoter selection, and controlled through the interplay between cell-type specific transcription factors, chromatin structure, and epigenetic modifications. We used a genome-wide chromatin-immunoprecipitation (ChIP) assay to identify epigenetic modifications that correlate with different latency types. We found that the chromatin insulator protein CTCF binds at several key regulatory nodes in the EBV genome and may compartmentalize epigenetic modifications across the viral genome. Highly enriched CTCF binding sites were identified at the promoter regions upstream of Cp, Wp, EBERs, and Qp. Since Qp is essential for long-term maintenance of viral genomes in type I latency and epithelial cell infections, we focused on the role of CTCF in regulating Qp. Purified CTCF bound ∼40 bp upstream of the EBNA1 binding sites located at +10 bp relative to the transcriptional initiation site at Qp. Mutagenesis of the CTCF binding site in EBV bacmids resulted in a decrease in the recovery of stable hygromycin-resistant episomes in 293 cells. EBV lacking the Qp CTCF site showed a decrease in Qp transcription initiation and a corresponding increase in Cp and Fp promoter utilization at 8 weeks post-transfection. However, by 16 weeks post-transfection, bacmids lacking CTCF sites had no detectable Qp transcription and showed high levels of histone H3 K9 methylation and CpG DNA methylation at the Qp initiation site. These findings provide direct genetic evidence that CTCF functions as a chromatin insulator that prevents the promiscuous transcription of surrounding genes and blocks the epigenetic silencing of an essential promoter, Qp, during EBV latent infection.
Published in the journal: . PLoS Pathog 6(8): e32767. doi:10.1371/journal.ppat.1001048
Category: Research Article
doi: https://doi.org/10.1371/journal.ppat.1001048Summary
The establishment and maintenance of Epstein-Barr Virus (EBV) latent infection requires distinct viral gene expression programs. These gene expression programs, termed latency types, are determined largely by promoter selection, and controlled through the interplay between cell-type specific transcription factors, chromatin structure, and epigenetic modifications. We used a genome-wide chromatin-immunoprecipitation (ChIP) assay to identify epigenetic modifications that correlate with different latency types. We found that the chromatin insulator protein CTCF binds at several key regulatory nodes in the EBV genome and may compartmentalize epigenetic modifications across the viral genome. Highly enriched CTCF binding sites were identified at the promoter regions upstream of Cp, Wp, EBERs, and Qp. Since Qp is essential for long-term maintenance of viral genomes in type I latency and epithelial cell infections, we focused on the role of CTCF in regulating Qp. Purified CTCF bound ∼40 bp upstream of the EBNA1 binding sites located at +10 bp relative to the transcriptional initiation site at Qp. Mutagenesis of the CTCF binding site in EBV bacmids resulted in a decrease in the recovery of stable hygromycin-resistant episomes in 293 cells. EBV lacking the Qp CTCF site showed a decrease in Qp transcription initiation and a corresponding increase in Cp and Fp promoter utilization at 8 weeks post-transfection. However, by 16 weeks post-transfection, bacmids lacking CTCF sites had no detectable Qp transcription and showed high levels of histone H3 K9 methylation and CpG DNA methylation at the Qp initiation site. These findings provide direct genetic evidence that CTCF functions as a chromatin insulator that prevents the promiscuous transcription of surrounding genes and blocks the epigenetic silencing of an essential promoter, Qp, during EBV latent infection.
Introduction
Epstein-Barr Virus (EBV) is a human gamma herpesvirus that establishes latent infection in more than 90% of the adult population world-wide [1], [2]. The ∼170 kb genome encodes ∼90 viral genes but only a few of these are expressed during latent infection. The latent infection is a cofactor in several human malignancies and may play an essential causative role in the endemic forms of Burkitt's lymphoma (BL) and nasopharyngeal carcinoma (NPC), as well as diffuse B-cell lymphomas in HIV-AIDS and iatrogenic immunosuppressed individuals [3]. Remarkably, the viral gene expression patterns vary in each tumor type suggesting that EBV can establish multiple forms of latency [4]. These different gene expression programs have been referred to as latency types and may also correlate with the changes in host-cell differentiation state and tissue origin [4], [5]. Changes in EBV latency type may also be important for evasion of host-immune recognition [6].
EBV latency gene expression programs have been categorized into four different types based primarily on the differential expression of the EBNA and LMP gene transcripts [4]. Type 0 latency is defined as the absence of expression of any viral genes, and is thought to exist in quiescent memory B-cells [5], [7]. Type I latency is characterized by the expression of the EBNA1 gene only, and is observed in proliferating memory B-cells in normal hosts, and found predominantly in Burkitt lymphoma tissue and derived cell lines [8], [9], [10]. Type II latency is characterized by the expression of EBNA1 and LMP2 expression, with some variable expression of LMP1. This pattern of gene expression is observed in epithelial cell derived tumors including NPC and gastric carcinomas [11], [12], [13]. Type III latency is characterized by the expression of EBNA-1, -2, -3A, -3B, -3C,-LP, LMP1, LMP2. This more permissive gene expression program is observed upon primary infection of naïve B-cells and is associated with B-cell proliferation and immortalization [14]. Type III latency is observed in immortalized B-cells in culture and diffuse B-cell lymphomas in immunosuppressed individuals. The natural history of EBV infection suggests that type III latency progresses to type I latency during B-cell maturation, and that viral lytic occurs in terminally differentiating plasma B-cells [5], [15].
Latency type gene expression is regulated largely through differential promoter utilization [16], [17], [18], [19], [20]. The promoters for type III infection are activated upon primary infection by B-cell specific factors [21]. Initial transcription from Wp promoter allows the expression of EBNA2, which functions as a transcriptional activator or the Cp promoter which drives expression of EBNA-LP, EBNA-2, EBNA-3A, -3B, -3C and EBNA1 [22], [23], [24], [25]. EBNA2 also activates LMP1 and LMP2 transcription to maintain type III gene expression [25]. Type II and Type I gene expression arise through mechanisms that are not completely understood, but involve the epigenetic silencing of the Cp and LMP1 promoter by DNA methylation and histone deacetylation [26], [27], [28], [29], [30], [31], [32], [33]. EBNA1 expression is required for the replication and maintenance of the viral latent genome [34], [35], [36], [37]. EBNA1 mRNA expression is maintained in type III latency by Cp promoter utilization and mRNA processing, while in type I latency EBNA1 expression is driven largely through the Q promoter (Qp) [17], [31], [38], [39], [40]. EBNA1 protein binds to two sites located at the +10–+57 position relative to the Qp transcription initiation site and restrict its usage in type III cells where EBNA1 proteins levels are elevated [41], [42]. Thus, EBNA1 can autoregulate its own expression levels through promoter selection, and help to coordinate the switch between latency types.
The epigenetic control of EBV promoter utilization and latency type is evident in the differential pattern of DNA methylation between latency types, and by the ability of DNA methylation inhibitors to stimulate type III gene transcription from type I latently infected cells [26], [43]. It is also apparent that histone acetylation occurs at the Cp promoter during type III latency where they are transcribed, but not in type I latency where Cp promoter is silenced [44]. However, little else is known about the epigenetic controls the determine promoter utilization and gene expression during the different latency types. LMP1 and Cp activation depend on the enhancer functions of EBNA2 and EBNA1. EBNA1 binds to the EBV origin of plasmid (OriP) replication and is essential for both viral replication and plasmid maintenance, as well as for transcriptional enhancement of EBNA2 and LMP1 [45], [46], [47]. The mechanism through which EBNA1 activates Cp and LMP1 from OriP, which is located over 2 kb from each promoter is not clear. It is also not known whether EBNA1 binding at Qp may also regulate transcription of type III promoters.
Cellular factors that regulate communication between promoters and enhancers, have also been implicated in the organization of chromatin structure [48], [49], [50], [51]. The chromatin insulator protein CTCF has been implicated in segregating active from inactive chromatin domains, as well as in mediating long-distance interactions between transcriptional regulatory regions [52], [53]. At least one CTCF site has been mapped to a region between OriP and Cp, and its binding was found to correlate with the inhibition of Cp transcription in type I latency [54], [55]. Other CTCF sites in EBV chromosome are known to exist, but their function has not been explored in detail [54]. In this work, we use a genome-wide ChIP assays to explore the epigenetic landscape of the EBV genome in type I and type III infected cells. We found that CTCF binding sites are positioned in key regulatory locations throughout the viral genome. We investigate in detail the function of a high affinity CTCF site positioned immediately upstream of the EBNA1 binding sites in Qp. We find that CTCF binding is required to maintain the transcriptional activity and prevent the epigenetic silencing of Qp in proliferating cells.
Results
Epigenetic differences between type I and type III genomes
An EBV genome-wide real-time PCR array was used to compare the patterns of several epigenetic marks between type I and type III latent virus genomes using the chromatin immunoprecipitation (ChIP) assay (Fig. 1). For these experiments we used a 384-well array that covers the entire EBV genome at a density of ∼400 bp between primer sets. In a previous study, we used a similar approach to analyze the first 60 kb of the EBV genome for various histone modifications and protein factor binding sites [54]. In the present study, we compared a type I latently infected Burkitt lymphoma cell line, Mutu I, with a lymphoblastoid cell line derived from Mutu I viral DNA (Mutu-LCL), ensuring that these two cell types were isogenic with respect to EBV genomes. We compared the pattern of CTCF binding sites to those of histone H3me3K9 (H3mK9) and H3me2K4 (H3mK4) modifications and also that of cytosine methylation (mCpG) using methyl DNA immunoprecipitation (MeDIP). Several patterns were noteable. Peaks of histone H3mK9 and H3mK4 were complementary and non-overlapping. Major peaks of H3mK4 were detected at the regions surrounding the RNA polymerase III transcribed EBERS and the Bam A microRNA cluster in both cell types. H3mK4 peaks regions surrounding the LMP1 promoter and Cp promoter were elevated in type III relative to type I cells. H3mK9 methylation was elevated over the Cp and W repeats in type I latency, but not type III, correlating with transcription repression in type I. CTCF binding sites were located at multiple positions, with only a few differences in type I and type III. CTCF sites tended to exist between clusters of H3mK4 and H3mK9, as can be seen at a newly discovered peak at the region 3′ of the W repeats. All the CTCF binding sites found in our assay are listed in Table S1. mCpG patterns were also different between type I and III latency. High levels of mCpG were detected across the Cp and LMP2 regions in type I, but not in type III, correlating with transcription silencing of Cp/Wp in type I cells. Interestingly, CTCF sites tended to demarcate boundaries of mCpG, which also correlated well with H3mK9 in both cell types.
Fig. 1. EBV genome-wide analysis of histone and DNA methylation patterns in different latency types. 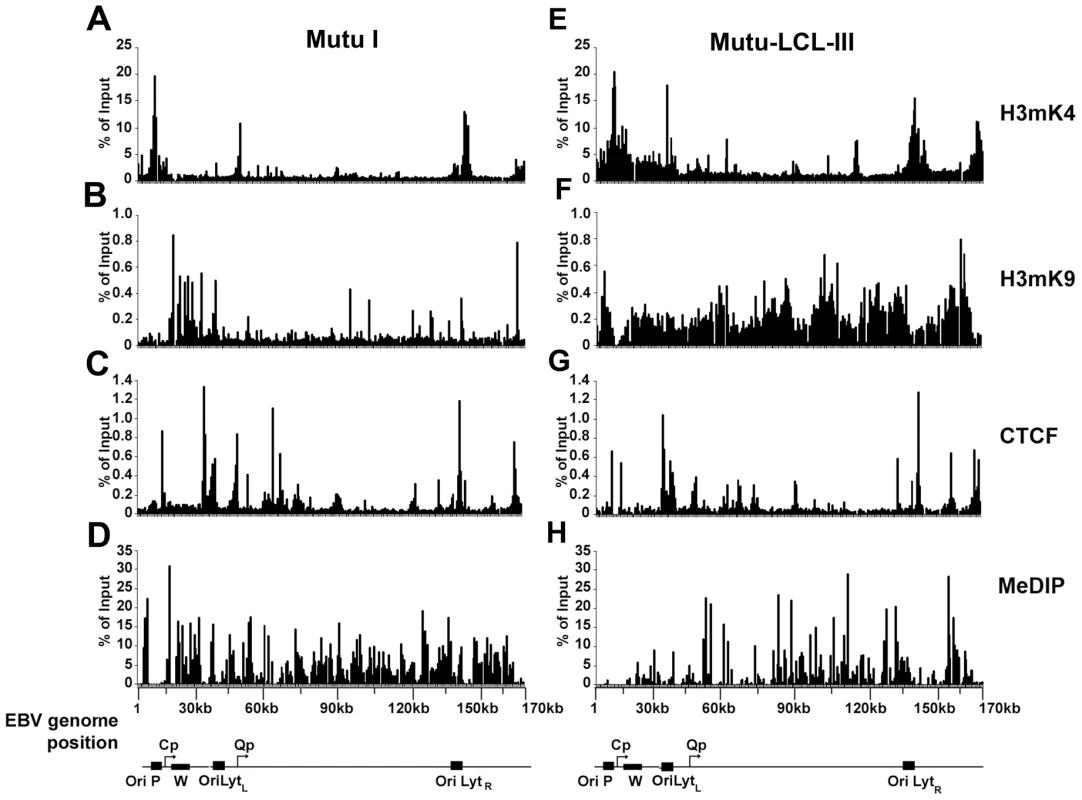
ChIP assays were performed with Mutu I (A–D), or Mutu-LCL (E–H) using antibodies for histone H3me2K4 (A and E), H3me3K9 (B and F), CTCF (C and G), or methyl cytosine (MeDIP) (D–H). ChIP DNA was assayed by real-time PCR using a genome wide array of 384 primers spaced ∼400 bp across the EBV genome. Approximate EBV genome positions are indicated in the schematic below each column of graphs. Graphs represent an average of three independent experiments. Standard deviation was less than 10 percent of the mean for all data points. A more detailed examination of the regions surrounding the major latency promoter elements reveals other features relevant to epigenetic regulation (Fig. 2). While most CTCF sites are bound similarly in each cell type, the CTCF site upstream of the EBERS was significantly reduced in Mutu I cells (Fig. 2A). Interestingly, this region is enriched in mCpG in MutuI, relative to type III cells (Fig. 2B). One possible explanation for the loss of CTCF binding at this region in type I cells is that enriched DNA methylation replaces and blocks CTCF binding. In contrast, this same region is elevated in H3mK9 in type III latency, where CTCF occupies a 3′ boundary upstream of the EBERS (Fig. 2D and 2J). Another striking feature is the elevated H3mK4 across the EBNA2 transcript and BHRF1 miRNA cluster in type III latency, but not in type I (Fig. 2C). This correlates well with the difference in RNA polymerase II transcription across this region in these two cell types. The CTCF site at this position just 3′ of W repeats provides a 5′ border for the high H3mK4 in type III latency (Fig. 2I), and a 3′ border for the high H3mK9 (Fig. 2E) and mCpG (Fig. 2G) in type I latency.
Fig. 2. Analysis of epigenetic patterns in the EBV latency control region. 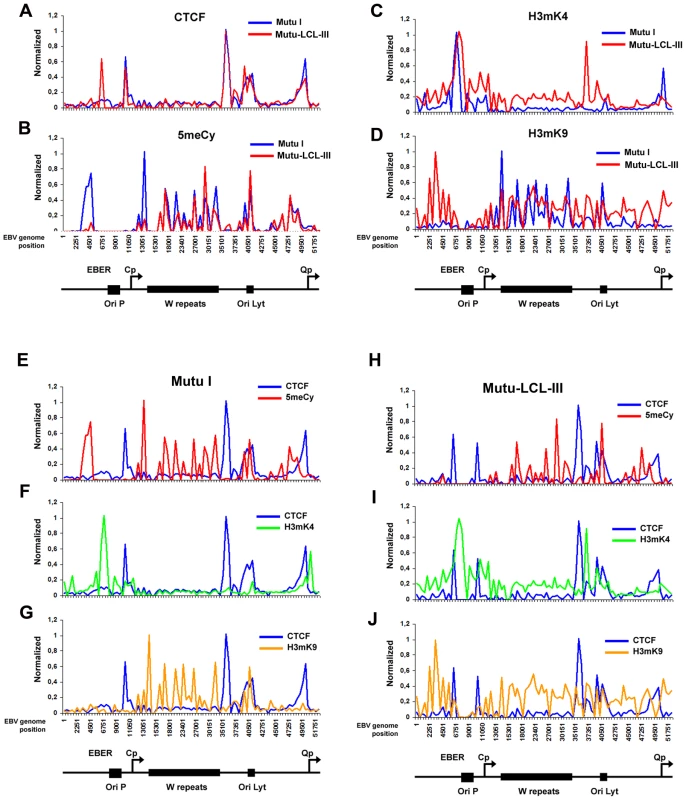
A–D) EBV genome-wide ChIP data from figure 1 was reanalyzed at EBV positions 1–60 kb as direct comparison between Mutu I (blue) and Mutu-LCL (red) for CTCF (panel A), methyl cytosine (panel B), H3me2K4 (panel C), H3me3K9 (panel D). E–G) Mutu I cells compared for CTCF (blue) vs methyl cytosine (red); CTCF (blue) vs H3me2K4 (green); CTCF (blue) vs H3me3K9 (yellow). H–J) Mutu-LCL-III cells compared for CTCF (blue) vs methyl cytosine (red); CTCF (blue) vs H3me2K4 (green); CTCF (blue) vs H3me3K9 (yellow). CTCF binds consistently at Qp in both cell types (Fig. 2A). In type I cells, the Qp CTCF site separates a 3′ H3mK4 peak from a 5′ region enriched in H3mK9 and mCpG (Fig. 2E–G). In type III cells, the CTCF site appears to spare Qp from surrounding regions of elevated mCpG and H3 mK9 (Fig. 2H and J). The region is also reduced in H3mK4, corresponding to a reduction in EBNA1 binding and transcription from Qp (Fig. 2I). These marks are largely consistent with known transcription properties of Qp in which it is active in type I, and repressed in type III (Figure S1).
Mapping the CTCF site at Qp
To identify the specific sequence element bound by CTCF near Qp, we first examined the region for candidate CTCF binding sites using a prediction algorithm (http://insulatordb.uthsc.edu). At least two candidate CTCF sites were identified at positions 43739 and 50082 (Fig. 3A). These sites were synthesized as DNA oligonucleotide probes and tested by EMSA for binding to purified recombinant CTCF protein (Fig. 3B and C). We found that CTCF bound efficiently to the 50082 binding site, but not to the 49739 sequence or to a control oligonucleotide (from EBV 49901) that lacked any candidate CTCF binding site (Fig. 3C). The precise nucleotide binding site of CTCF was mapped by DNase I footprinting using the entire Qp control region as a probe and purified recombinant CTCF (Fig. 3D). We found that CTCF protected a ∼20 bp regions between 50082–50102. In the same DNase footprinting reaction we included recombinant EBNA1 protein. EBNA1 bound to two sites in Qp covering EBV nucleotides 50142 to 50189.
Fig. 3. Identification of a CTCF binding site upstream of Qp. 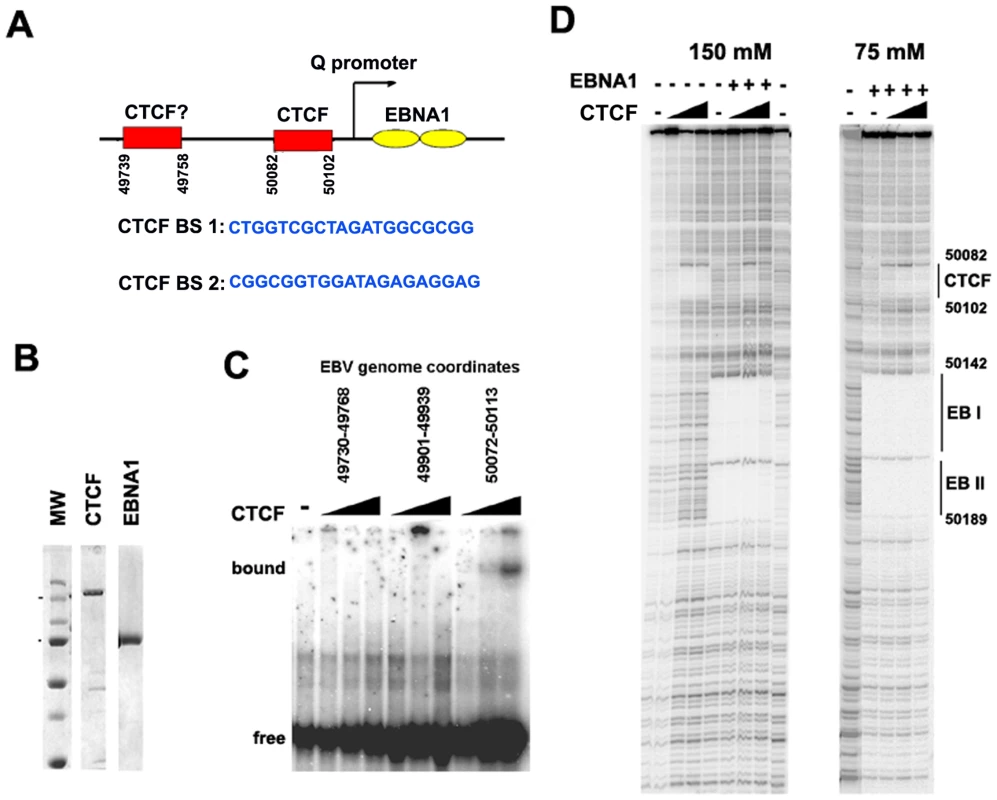
A) Schematic of CTCF and EBNA1 binding site organization at Qp, and the sequence of candidate CTCF binding sties (BS) 1 and 2. B) Coomassie stain of purified recombinant CTCF and EBNA1 proteins derived from baculovirus expression system. C) EMSA analysis of purified CTCF (10–100 ng) binding to DNA probes for EBV regions 49730–49768 (BS1), 49901–49939, or 50072–50113 (BS2). D) DNase I footprinting assay of purified CTCF protein (30–300 ng) in the absence (-) or addition (+) of 30 ng purified EBNA1, in buffer containing 150 mM (left panel) or 75 mM (right panel) NaCl. The DNase I footprinting assays demonstrate that CTCF and EBNA1 can bind simultaneously to Qp, and that potential interactions between these proteins may regulate Qp. However, detailed biochemical analysis of these interactions were limited by the different salt sensitivities of the purified proteins in the DNA binding assays (Fig. 3D). The physiological significance of this differential salt sensitivity is not clear.
Generation of an EBV Bacmid with CTCF mutations at Qp
To investigate the functional significance of CTCF binding at Qp in cell-based assays, we engineered a substitution mutation in the CTCF binding site at Qp in EBV bacmids using recombineering with GALK gene insertion and gene replacement [56] (http://recombineering.ncifcrf.gov) (Fig. 4A). GALK insertion, CTCF substitution mutation (ΔCTCF), and Wild-type (Wt) rescue mutants in EBV bacmids were validated by restriction enzyme digestion (Fig. 4B), PCR across the junctions (Fig. 4C) and sequencing of the insertions (data not shown). Bacmid DNA for ΔCTCF and Wt rescue control was introduced into 293 cells and stable transformants were selected for hygromycin and GFP expression. After 8 weeks of selection, stable cell pools were assayed by ChIP assay to validate that the substitution mutation disrupted CTCF binding in living cells (Fig. 4D). As expected, CTCF failed to bind to Qp in the ΔCTCF mutant (Fig. 4D, top panel). We also found that EBNA1 binding to Qp was reduced in the ΔCTCF mutant (Fig. 4D, lower panel), suggesting that CTCF facilitates EBNA1 binding at Qp in living cells. Stable cell pools were also assayed at 8 weeks for their relative expression GFP, EBNA1, CTCF and PCNA (Fig. 4E). We found no apparent differences in the expression of these proteins after 8 weeks of selection in 293 cell pools.
Fig. 4. Mutagenesis of CTCF binding site in Qp. 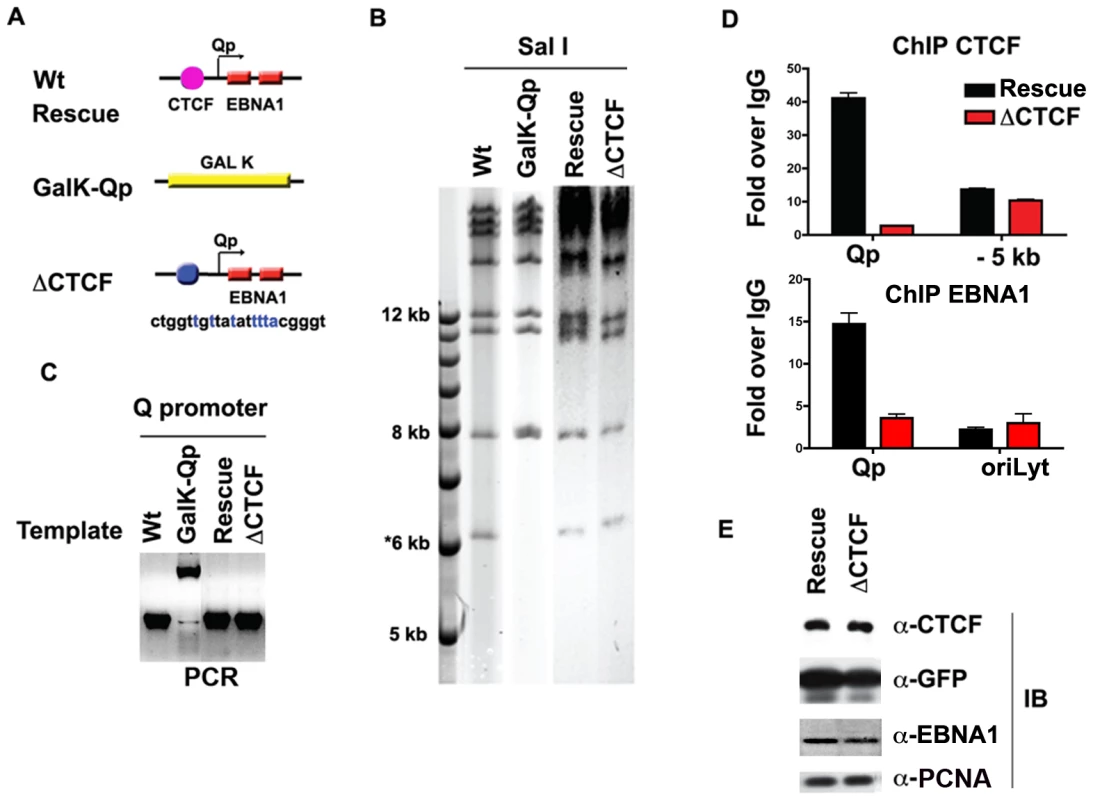
A) Schematic of mutations introduced into the Qp region of EBV bacmid. B) Purified bacmid DNA for EBV Wt, GAL K, Wt rescue, and ΔCTCF was analyzed by Sal I restriction digest and 0.7% agarose gel electrophoresis. DNA was visualized by ethidium bromide staining. C) PCR amplification of the region encompassing Qp for EBV Wt, GAL K, Wt rescue and ΔCTCF. D and E) ChIP assay of Wt rescue, or ΔCTCF bacmids in stable 293 cell pools after 8 weeks of hygromycin selection with antibody for CTCF (top panel), or EBNA1 (lower panel). CTCF ChIP was analyzed at Qp or a region −5 kb to Qp. EBNA1 ChIP was analyzed at Qp, or at OriLyt control region. E) Western blot analysis of CTCF, GFP, EBNA1, and PCNA protein levels for Wt rescue or ΔCTCF 293 cell pools. CTCF binding site at Qp is required for stable maintenance of EBV episomes in 293 cells
While early passage cell pools showed little difference in GFP and EBNA1 expression between ΔCTCF and Wt rescue genomes, we observed a marked loss of GFP expression in the ΔCTCF relative to Wt rescue genomes after longer passages in culture (Fig. 5A and B). At 4 and 8 weeks after transfection, ΔCTCF and Wt rescue had nearly identical percentage of GFP positive cells. In contrast, at 16 weeks ΔCTCF pools were ∼9% positive, while Wt rescue was ∼72% GFP positive, as measured by FACS (Fig. 5B). EBV DNA copy number per cell was determined by real time PCR for 293 cell pools at 8 and 16 weeks (Fig. 5C). We found that ΔCTCF cell pools had ∼50% less EBV DNA per cell than Wt rescue containing pools as measured at 8 and 16 weeks. Isolation of EBV episomes by Hirt lysis revealed that ΔCTCF cell pools had ∼3 fold lower DNA than WT rescue at 8 weeks, and both pools were reduced at 16 weeks, indicating that ΔCTCF episomes are lost at a greater rate than Wt episomes (Fig. 5D). These data suggest that the Qp CTCF binding site is important for the stable maintenance of EBV episomes in selected 293 cell pools.
Fig. 5. CTCF binding site at Qp is required for stable maintenance of EBV episome in 293 cells. 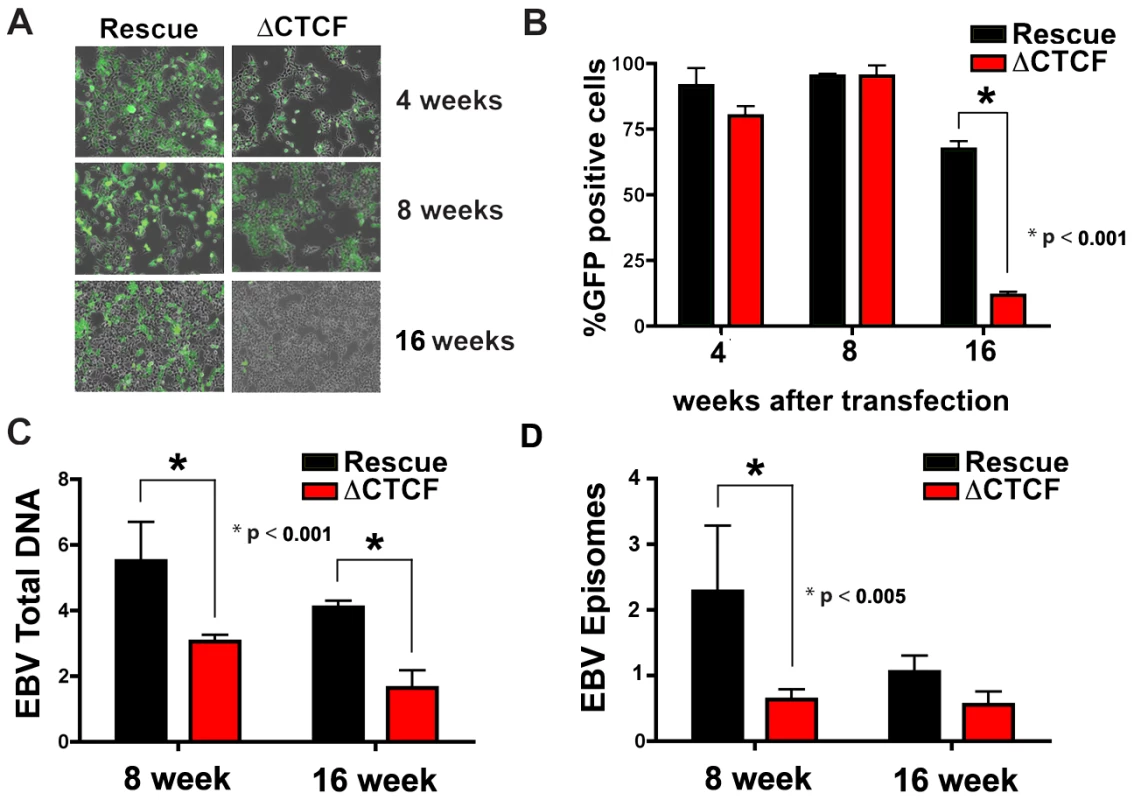
A) Photomicrographs of GFP fluorescence of EBV bacmid Wt rescue or ΔCTCF in 293 cell pools after 4, 8 and 16 weeks post-transfection. B) EBV episome maintenance in Wt rescue or ΔCTCF 293 cell pools was assayed by FACS analysis as the percentage of GFP positive cells at the indicated weeks. Bars represent the average of three independent experiments. C) EBV genome copy number was assayed by real time PCR analysis in Wt rescue or ΔCTCF 293 cell pools at the indicated weeks. Bars represent the average of three independent experiments. D) Episomal viral DNA from Wt rescue or ΔCTCF 293 cell pools was isolated at the indicated weeks by Hirt extraction and assayed by quantitative real time PCR using the viral DNA from Raji cells as reference (ΔΔCt method). Bars represent the rate of episome lost of three independent experiments. All error bars indicate the standard deviation from the mean. ΔCTCF genomes have altered transcription and promoter usage
The loss of GFP expression and episome stability in 293 cell pools could be due to a deregulation of viral gene expression. Others have shown that EBV establishes a restricted pattern of latency gene expression in 293 cells, resembling a type I program with stable Qp utilization for EBNA1 expression [57]. To assess viral gene expression patterns in transfected 293 cells, we first assay mRNA expression of EBNA1, EBNA2, EBNA3A, and EBNA3C in 293 cell pools after 4, 8, and 16 weeks of selection (Fig. 6). RNA expression was measured by quantitative RT-PCR and normalized to bacmid expression of GFP mRNA. At 4 weeks, EBNA1 and EBNA2 mRNA levels were expressed at lower levels in ΔCTCF compared to Wt rescue cell pools (Fig. 6C, top panel). At 8 weeks, EBNA1 levels were similar, while EBNA2, EBNA3A, and EBNA3C levels were higher in ΔCTCF relative to Wt rescue cell pools (Fig. 6C, middle panel). By 16 weeks, EBNA1 mRNA levels were maintained in the Wt rescue, but almost undetectable in ΔCTCF cell pools (Fig. 6C, lower panel). EBNA2, EBNA3A, and EBNA3C were expressed at very low levels in both Wt rescue and ΔCTCF cell pools at these later passages in culture. These observations are consistent with a previous study showing that EBV initially expresses EBNA2, but eventually adopts an EBNA1 only, type I latency in 293 cells [57].
Fig. 6. RNA expression and promoter utilization in Qp mutated bacmids. 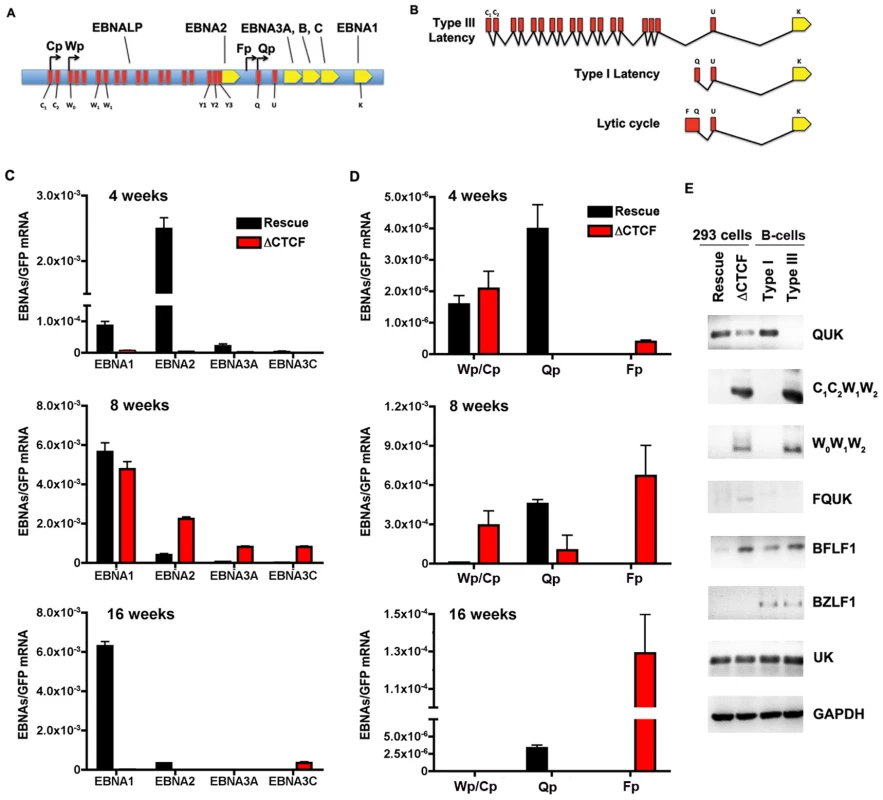
A) Schematic representation of the EBV latency genes and promoters. Promoters are indicated by arrows. The position of the six EBNAs ORFs are indicated. B) Schematic representation of different EBNA1 transcripts. Exons present at 5′ end of EBNA1 mRNA are indicated in red. C) Quantitative RT-PCR was used to measure the abundance of EBNA2, EBNA3A and EBNA3C mRNA relative to bacmid GFP for Wt rescue or ΔCTCF bacmids in 293 cell pools at 4, 8, and 16 weeks after transfection, as indicated. D) Same as in C, except EBNA1-transcripts initiating from either Cp/Wp, Qp, or Fp were measured relative to GFP in Wt rescue or ΔCTCF bacmids in 293 cell pools at 4, 8, and 16 weeks after transfection. E) RT-PCR was measured for Wt rescue or ΔCTCF bacmids in 293 cell pools at 8 weeks post-transfection, as well as for type I (Mutu I) or type III (Mutu-LCL) controls. RNA was analyzed for the junction specific transcripts QUK (Qp initiation), C1C2W1W2 (Cp initiation), W0W1W2 (Wp initiation), BFLF1 (lytic gene adjacent to Qp), UK (EBNA1 mRNA in both type I and type III), and control cellular GAPDH. To better understand the failure of ΔCTCF bacmids to sustain EBNA1 mRNA expression, we investigated the promoter utilization at 4, 8, and 16 weeks after transfection (Fig. 6D). EBNA1 mRNA has been shown to initiate from Wp/Cp in most type III latency, from Qp in most type I latency, and from Fp during lytic cycle gene expression (Fig. 6A and B). We assayed the utilization of Wp/Cp, Qp, and Fp using quantitative RT-PCR with primers specific for each promoter (Table S2). At 4 weeks, we found that Wp/Cp was utilized at similar levels in Wt rescue and ΔCTCF containing cell pools (Fig. 6D, top panel). Remarkably, Qp was utilized at relatively high levels in Wt rescue, but nearly undetectable in ΔCTCF. Interestingly, Fp utilization was detected in ΔCTCF, but undetectable in Wt rescue. At 8 weeks, Wt rescue containing cells utilized Qp predominantly, while ΔCTCF cells utilized all three promoters, with Fp dominating (Fig. 6D, middle panel). By 16 weeks, Wt rescue genomes utilized Qp exclusively, while ΔCTCF genomes utilized Fp exclusively, although ∼ 10 fold less than Fp utilization at 8 weeks (Fig. 6D, lower panel). Since Fp is typically associated with lytic gene activity, we tested whether ΔCTCF containing cells were expressing the lytic immediate early gene BZLF1 (Fig. S3 and Fig. 6E). BZLF1 expression was undetectable at all time points tested for Wt or ΔCTCF 293 cell pools, as measured by quantitative RT-PCR (Fig.S3) or by conventional PCR (Fig. 6E). These findings suggest that latency promoter utilization is deregulated in ΔCTCF genomes.
To better understand the ΔCTCF defects in viral gene expression, we assayed viral mRNA using exon specific PCR for transcripts (Table S3), initiating at Qp (QUK), Cp (C1C2W1W2), Wp (W0W1W2), or within the lytic transcripts of BFLF1 [58] (Fig. 6E). In addition, we measured the UK intron junction for EBNA1, which is expressed in type I and type III cells. We also measure BZLF1 expression as an indicator of lytic cycle gene expression. As a control we assayed the expression levels of cellular GAPDH. RNA was isolated from pools after 8 weeks in culture. We found that QUK was expressed at slightly higher levels in Wt rescue relative to ΔCTCF cell pools (Fig. 6E, top panel). C1C2W1W2, W0W1W2 and FQUK transcripts were elevated in ΔCTCF relative to Wt rescue, consistent with RT-PCR data showing elevated levels of Wp/Cp and Fp utilization in bacmid lacking the CTCF binding site. UK transcripts were similar in ΔCTCF and Wt rescue, consistent with observations from quantitative RT-PCR (Fig. 6C) showing that EBNA1 mRNA levels were similar at 8 weeks after transfection. BZFL1 was not detected in either bacmid 293 cell pool, indicating that lytic gene activation or DNA replication was not indirectly responsible for these differences in gene expression. These exon-specific PCR studies further substantiate the real-time PCR data, and support the conclusion that CTCF mutations in Qp deregulate the latency type transcription pattern and promoter utilization.
CTCF binding site is required for the maintenance of epigenetic patterns at Qp
CTCF has been implicated in several functions, including chromatin insulation and boundary functions. To determine if the disruption of CTCF site at Qp altered the normal pattern of epigenetic marks surrounding Qp, we performed MeDIP and ChIP assays with a set of primers that probe the regions −1000, −500, +1, and +800 relative to the Qp initiation site (Fig. 7A–C). At 8 weeks post-transfection, MeDIP revealed that mCpG was enriched at −1000 and −500 position in Wt rescue, but undetectable in ΔCTCF genomes (Fig. 7A, top panel). At 16 weeks post-transfection, mCpG was elevated at −1000 in WT rescue, but not at +1 or +800, consistent with high levels of transcription initiating at Qp in these cell pools (Fig. 6). In contrast, mCpG was elevated at +1 position in ΔCTCF genomes, consistent with the lack of Qp transcription initiation at 16 weeks in ΔCTCF cells (Fig. 6). To examine potential changes in euchromatic or heterochromatic histone modifications surrounding Qp, we focused on H3mK4 or H3mK9, respectively. At 8 weeks post-transfection, we found high levels of H3mK4 at the +1 and +800 positions in both Wt rescue and ΔCTCF genomes (Fig. 7B, top panel). At 16 weeks, H3mK4 was elevated primarily in Wt rescue (Fig. 7B, lower panel), consistent with persistent transcription from Qp in these cells (Fig. 6). The heterochromatic mark for histone H3mK9 was more revealing since it showed elevated levels upstream of Qp (−1000 and −500) in Wt rescue genomes at 8 weeks (Fig. 7C, top panel), and low levels in ΔCTCF where Fp is active. At 16 weeks, H3mK9 was highly enriched at +1 site of Qp in ΔCTCF genomes (Fig. 7C, lower panel), consistent with the transcription inactivity of Qp in these cells at this time after transfection. The epigenetic pattern surrounding Cp and Wp was also examined (Fig. S5). We found that Cp and Wp were hypermethylated and enriched on H3mK9 both at 8 and 16 weeks in Wt rescue, consistent with a previous study demonstrating that Cp and Wp undergo transcription silencing in 293 cells [57]. In ΔCTCF genomes Cp and Wp are hypomethylated and enriched on H3mK4 at 8 weeks, consistent with transcription initiation. However, by 16 weeks, Cp and Wp show elevated mCpG and H3mK9, consistent with the extinction of transcription initiation from these promoters at later passages. Thus, Cp and Wp undergo similar epigenetic silencing with Wt and ΔCTCF genomes, but Qp is protected from epigenetic silencing only in genomes were the CTCF site is intact (Fig. 7D).
Fig. 7. Changes in epigenetic patterns in Qp mutated bacmids. 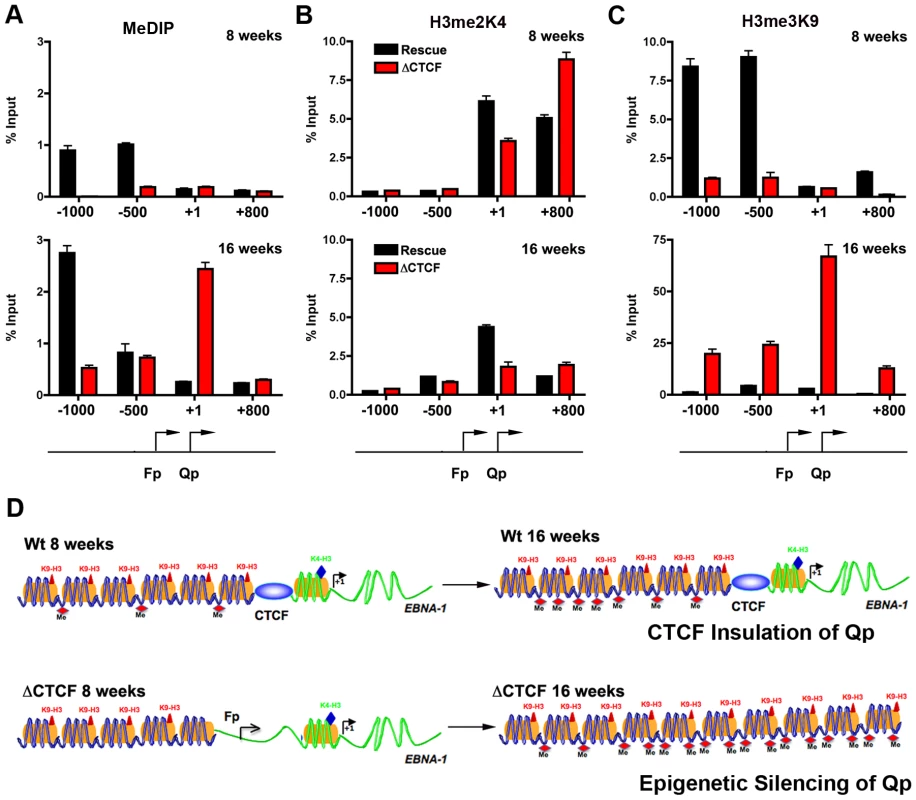
A) The epigenetic pattern of Qp region in Wt rescue and ΔCTCF bacmids in 293 cell pools was analyzed by MeDIP assay (A), H3me2K4 (B) and H3me3K9 (C) ChIp assay at 8 weeks (top panel) or 16 weeks (lower panel) after transfection. A schematic representation of Qp region is shown. D) Model of CTCF function in the chromatin organization at EBV Q promoter. CTCF is positioned as a barrier to the 5′ encroachment of H3me3K9 and mCpG in Wt rescue 293 cells (top panel). In ΔCTCF 293 cells, H3me2K4 is enriched throughout Qp and Fp at 8 weeks (lower panel, left), but is converted to H3me3K9 and mCpG at later times (16 weeks) (lower panel, right), indicating that CTCF is required to prevent this epigenetic drift at Qp. Discussion
Stable gene expression programs, like those associated with cell-type differentiation, correlate with heritable epigenetic changes to the cellular chromosome. The meta-stable gene expression programs associated with Epstein-Barr virus latency types have also been shown to correlate with epigenetic changes in the viral genome. The most consistently observed epigenetic difference between type I and type III latency is the DNA methylation of the Cp and LMP1 promoter regions [26], [43], [59]. We and others have also explored histone modification patterns at the major latency control regions for EBV, and observed distinct patterns between cell lines carrying EBV genomes with either type I and type III latency [44], [54], [60]. In this work, we extended this approach to examine the pattern of histone H3 K4 and K9 methylation, DNA methylation, and CTCF binding across the complete EBV genome in cells carrying stable type I or type III EBV latent infections (Figs 1 and 2, Figs. S1 and S2). We found that CTCF binding sites were located at or near regulatory regions, and commonly marked boundaries between euchromatic and heterochromatic marks (Fig. 2). The heterochromatic mark was typically H3mK9 in type III latency (e.g. EBV regions 2–6 kb in Fig. 2D), and mCpG, especially in type I latency (e.g. EBV regions 2–6 kb in Fig. 2B). This may reflect the natural history of epigenetic silencing where a partially repressive histone modification, like H3mK9 in type III, may eventually evolve into the more stable silencing modification associated with DNA methylation in type I. In some cases, the emergence of extensive CpG methylation correlates with the loss of CTCF binding (e.g. EBV region ∼6 kb in Fig. 2E and 2H). CTCF binding is known to be sensitive to CpG methylation [61]. These observations suggest that CTCF plays a key role in organizing epigenetic marks along the EBV genome, and that CTCF binding and epigenetic patterns change in different latency types.
To directly test the function of CTCF and other sequence specific binding factors in maintaining EBV latency type gene expression programs, we focused on the EBV Q promoter. We found that CTCF bound to Qp, and we used DNase I footprinting to map this binding site to a ∼20 bp region located ∼40 bp upstream of the EBNA1 binding sites (Fig. 3). In type I cells, CTCF appeared to separate a 3′ enrichment of H3mK4 that overlaps the Q transcription initiation site, from a 5′ enrichment of CpG methylation, that covers the neighboring lytic BFLF1 gene (Fig. 2E–G, Fig. S1). In type III cells, CTCF appears to spare the Qp transcription initiation site from surrounding H3 K9 methylation (Fig. 2I). To directly test the function of CTCF binding at Qp, we used recombineering methods to engineer a mutation that disrupts CTCF binding in EBV bacmids (Fig. 4). Disruption of CTCF binding site at Qp caused a loss of stable GFP expression and loss of bacmid episomes after multiple cell divisions (Fig. 5). CTCF site disruption also caused an increase in Fp promoter utilization, with no other evidence of lytic gene activation (Fig. 6C–E). Consistent with changes in gene expression, CTCF site disruption allowed for the formation of mCpG and H3mK9 methylation at the Qp initiation site (Fig. 7). These finding strongly suggest that CTCF contributes to the establishment and maintenance of an epigenetic pattern at Qp which is required for consistent expression of EBNA1 and episomal persistence in 293 cells pools. These findings also suggest that CTCF provides a barrier function that normally prevents Fp activation (upstream) and Qp silencing (downstream) during latent infection (Fig. 7D).
CTCF has been implicated in several gene regulatory and chromatin organizing activities [52], [53]. At the H19/Igf2 imprinted loci, CTCF functions as an enhancer blocker [62]. At the paternal allele, DNA methylation prevents CTCF binding, and allows enhancer activation of the Igf2 promoter. In contrast, at the unmethylated maternal allele, CTCF binds to a cluster of sites and prevents enhancer activation of the Igf2 promoter. Our findings here and previously [55] suggest that CTCF may have enhancer blocking activity at the sites surrounding OriP (e.g. the 5′ site at ∼6 kb and the 3′ site at 10 kb on the EBV genome). These sites are positioned to physically block OriP interactions with LMP1/2 and Cp, respectively. Interestingly, we found that CTCF binding at the 5′ site (∼6 kb on EBV) is reduced in type I latency where the sites have been partially subject to CpG DNA methylation. Thus, latency type-specific DNA methylation patterns may regulate CTCF binding at some regulatory regions. Conversely, CTCF binding may prevent DNA methylation at other regulatory elements. The region surrounding the Qp initiation site has been shown to lack CpG methylation in all latency types [43]. The CTCF site we identified may function to prevent the spread of CpG methylation which is normally elevated in the regions upstream of Qp (BFLF1 ORF) in both type I and type III latency. Disruption of the CTCF site caused a significant increase in CpG methylation immediately over the Qp initiation site (Fig. 7A). Thus, an essential function of CTCF may be to prevent CpG methylation at the Qp initiation site. This is consistent with the observation that CpG methylation is never detected at Qp in any EBV latency type [43].
Chromatin boundary factors, like CTCF, are thought to prevent the spread of processive histone modifications [48]. Our data suggests that CTCF has chromatin boundary activity at Qp that prevents inactivating heterochromatin, like H3mK9 from invading Qp. Elevation in H3mK9 at Qp was evident at 4 and 8 weeks post-transfection in ΔCTCF, suggesting that this mark is the first to cross into the Qp promoter region. At 16 weeks, mCpG is highly elevated at Qp, thus following H3mK9 and more completely silencing Qp transcription. Remarkably, the CTCF boundary functions in the reciprocal direction since the loss of CTCF leads to elevated transcription of Fp and BFLF1, along with a corresponding increase in histone H3mK4, and decrease in the normal H3mK9 and CpG methylation. It remains unclear what drives each modification at the specific sites. Our data strongly suggests that H3mK9 precedes the formation of mCpG, and that CTCF helps keep each modification in its proper place. Precisely what sets the pattern on each side of CTCF is not clear. CTCF may function in the recruitment of RNA polymerase II to Qp, and this may help to establish the correct orientation of the chromatin boundary at Qp. Loss of this boundary function leads to the loss of Qp transcription and the inappropriate regulation of other viral genes. CTCF also affected EBNA1 binding to Qp in vivo (Fig. 4D), raising the possibility that EBNA1 may also contribute to some of these changes in chromatin and transcription at Qp. Furthermore, it is not known if CTCF or EBNA1 provide additional structural features that directly affect these other promoters, or if these are indirect effects of altering EBNA1 levels in the 293 cell model of latency
DNA methylation, like histone modifications, can also spread across chromosomal regions to alter gene expression programs [63]. Epigenetic silencing due to promoter CpG methylation commonly arises at sights that have been actively repressed by histone deacetylation and K9 trimethylation. The mechanisms that restrict the drift of CpG methylation have not been completely elucidated. Our findings provide clear genetic evidence that CTCF can prevent the encroachment of CpG methylation at the Qp promoter of EBV. Our study shows that CpG methylation arises only after multiple generations at a region that is initially euchromatic and transcriptionally active. In the absence of CTCF, transcription initiation favors an alternative upstream promoter, Fp, which may prevent Qp utilization. Thus, CTCF may also facilitate promoter selection perhaps through its reported ability to interact with RNA polymerase II [64]. While the precise mechanism through which CTCF directs promoter selection and maintains chromatin boundaries remains to be discovered, our findings clearly indicate that CTCF provides an essential function in maintaining the epigenetic patterns at Qp. Our findings also indicate that protection of Qp by CTCF is essential for EBV genome stability during long-term latent infection. These findings also provide a framework for understanding the role of CTCF at other viral and cellular genes where protection from epigenetic drift and transcription silencing is critical for stable gene expression programs.
Materials and Methods
Cells
D98/HR1, HeLa and 293 cells were cultured in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum and antibiotics in a 5% CO2 incubator at 37°C. EBV positive Mutu I, Mutu-lymphoblastoid cell lines (Mutu-LCL), Sav I and Sav III cells were cultured in suspension in RPMI 1640 medium supplemented with 10% fetal bovine serum and antibiotics in a 5% CO2 incubator at 37°C. Mutu-LCL were established by primary infection of peripheral blood mononuclear cells (PBMCs) with EBV virions derived from stimulated Mutu I cells.
Bacmids, plasmids and recombinant proteins
EBV bacmid was a generous gift of Dr. H. –J. Delecluse [65]. Mutations in EBV bacmid were generated by recombineering using the GalK marker gene insertion and negative selection method for its substitution as described previously (http://recombineering.ncifcrf.gov/) [56]. The GalK gene was recombined into the Qp region at EBV coordinates 49927–50185. The CTCF site at 50082 ggtcgctagatggcgcgggtgagg was mutated by single substitutions to ggtTgTtaTatTTTAcgggtgagg. The ΔEBNA1 binding site at 50142 gaaaag[gcgggatagcgtgcgctaccggatggcgggtaatacatgct]atccttaca was mutated by deletion and substitution with gaaag[tgcttgaaaaggcgcgg]atccttaca. Plasmid containing the Qp region (49712–50250) was subcloned by PCR into pBKSII using Asp718 and HinDIII sites. Recombinant human CTCF protein was expressed as an N-terninal hexa-histidine tagged fusion protein from a baculovirus expression virus in sf9 cells, as described previously[55].
Chromatin Immunoprecipitation (ChIP) assay
ChIP assay followed the protocol provided by Upstate Biotechnology, Inc., with minor modifications as previously described [54]. Additional modifications are as follows. DNAs were sonicated to between 200 - and 350-bp DNA fragments on a Diagenode Bioruptor according to manufacturer's protocol, and real-time PCR was performed with SYBER green probe in an ABI Prism 7900 using 1/100 to 1/2,500 of the ChIP DNA according to manufacturer's specified parameters. Primer sequences for the EBV genome array are available upon request. Antibodies for H3 me2 K4, H3 me3 K9, CTCF, were purchased from Upstate Biotechnology. Primary antibodies to EBNA1 (Advanced Biotechnologies, Inc.), CTCF (Millipore), GFP (Santa Cruz Biotecnology) and PCNA (Santa Cruz Biotecnology) were used according to manufacturer's specifications
Electrophoretic mobility shift assay
EMSA assays with CTCF were described previously [55]. In a 20 µl reaction purified CTCF (∼100 ng) was added to a reaction mixture containing 0.5 µg poly(dI–dC), 5% glycerol, 0.1 mM ZnSO4, and 10,000 cpm of 32P-labeled DNA probe (∼0.1 ng). Reaction mixtures were incubated for 30 min at 25°C, electrophoresed in a 5% nondenaturing, polyacrylamide gel at 110 V, and visualized by PhosphorImager.
DNase I footprinting analysis
DNase I footprinting was performed as described previously [66]. 5′-end labeled DS probe was generated using 30 µCi of [−32P]dATP (6,000 Ci/mmol; Perkin-Elmer) and 2 U of Klenow fragment (Roche) for 30 min at 25°C. Purified proteins were incubated in a reaction mixture containing 1XPBS, 5 mM MgCl2, 0.1 mM ZnSO4, 1 mM dithiothreitol, 0.1% NP-40, 10% glycerol, 1 µg bovine serum albumin, 0.4 µg poly(dI–dC), and 10,000 cpm of 32P-labeled probe. The protected probe was digested with different dilutions of DNase I (Sigma) and purified by phenol-chloroform extraction following proteinase K digestion. The DNA samples were then electrophoresed on a 7% denaturing, polyacrylamide sequencing gel at 33 mA and visualized by PhosphorImager.
RNA extraction and RT-PCR
RNA was extract from 5×106 cells using Qiagen RNA extraction Kit according to manufacturer's protocol (Qiagen). After the extraction the RNA was incubated with 2 U DNAse I at 37°C for 30 minutes, following by the inactivation of the enzyme at 65°C for 10 minutes. The RNA was quantified and 2 µg of RNA was reverse transcribed using Super Script II Reverse Transcriptase from Invitrogen. 50 ng of cDNA was then analyzed by real time or conventional PCR. Primer sequences used for real time and conventional PCR are listened in tables 2 and 3 (Tables S2 and S3).
EBV genome copy number quantification
For EBV genome quantification Namalwa titration was used, assuming that Namalwa cell lines contain 2 copies of EBV genome and each cell contain 6.6×10−12 g of DNA. Namalwa cells and EBV positive cells were lysed in SDS lysis buffer (20 mM Tris pH 8, 4 mM EDTA, 20 mM NaCl, 1% SDS) following by incubation with Proteinase K for 2 h at 50°C. The DNA was extracted by phenol-chloroform extraction and precipitated by ethanol. Titration of 6.6×10−7–6.6×10−12 g of Namalwa DNA were used to obtain a calibration curve using primers for EBV genome (48779 – 48834) and β-Actin. 6.6×10−9 g of DNA from EBV positive cells were then analyzed by real time PCR for EBV genome copy quantification. Primer sequences used for real time PCR were listed in Tables S2 and S3.
Methylated-DNA immunoprecipitation
10×106 cells were resuspended in Lysis Buffer (20 mM Tris pH 8, 4 mM EDTA, 20 mM NaCl, 1% SDS) plus 0.7 µg/µl Proteinase K and incubated at 50°C overnight. The DNA was extracted by twice phenol-chloroform extraction followed by ethanol precipitation. The DNA was resuspended in 300 µl of TE buffer containing 20 µg/ml RNAse A and incubated at 37°C for 1 hr followed by DNA sonication to between 750 – 500 bp and DNA purification by phenol-chloroform extraction and ethanol precipitation. 8 µg of DNA were resuspended in Immunoprecipitation Buffer, IP buffer, [10 mM Na-Phosphate pH 7, 140 mM NaCl, 0.05% Triton X-100, and Proteinase inhibitor cocktail (Sigma)], denaturated at 95°C for 10 min and incubated with 5 µg of 5-methylCytodine antibody (Abcam) or 5 µg mouse IgG (Upstate), overnight at 4°C. The immunocomplexes were precipited by adding 50 µl of Protein G Dynabeads (Invitrogen) for 2 hr at 4°C. The beads were collected by a magnetic rack and washed twice for 10 min with 1 ml of IP buffer. The DNA was eluted by incubating the beads with 250 µl of Proteinase digestion buffer (50 mM Tris pH 8, 10 mM EDTA, 0.5% SDS, 0.3 µg/µl Proteinase K) at 50°C for 3 hr with shacking. The DNA was then purified and then analyzed by real time PCR. Primer sequences are listed in Table S4. 1 µg of genomic DNA was used as Input material.
Quantitation of episomal DNA
Viral episomal DNA was extracted using the Hirt lysis method [67] Briefly, Wt rescue and ΔCTCF 293 cells were pelleted by centrifugation and then resuspended in 800 µl Hirt's lysis buffer (0.6% SDS, 10 mM EDTA, 10 mM Tris-HCl, pH 7.4) containing 500 µg Proteinase K. The samples were incubated at 37°C for 1 h, followed by addition of 200 µl of 5 M NaCl and incubated at 37°C overnight. The samples were centrifuged at 14000 g for 20 minute and the supernatant was transferred in a new tube. DNA was then purified by phenol/chloroform/isoamyl alcohol extraction and ethanol precipitation. The DNA was resuspended in 50 µl of TE buffer and analyzed by real Time PCR by ΔΔCt method, using mitochondrial DNA as endogenous control and viral DNA extracted from Raji cell as a standard.
Supporting Information
Zdroje
1. KieffE
2007 Epstein-Barr Virus and its replication.;
FieldsBN
KnipeDM
HowleyPM
Philadelphia Wolters Kluwer Health/Lippincott Williams & Wilkins 2 v. (xix, 3091, 3086 p.) p
2. RickinsonAB
KieffE
2007 Epstein-Barr Virus.;
FieldsBN
KnipeDM
HowleyPM
Philadelphia Wolters Kluwer Health/Lippincott Williams & Wilkins 2 v. (xix, 3091, 3086 p.) p
3. YoungLS
RickinsonAB
2004 Epstein-Barr virus: 40 years on. Nat Rev Cancer 4 757 768
4. RoweM
RoweDT
GregoryCD
RickinsonAB
1987 Differences in B-cell growth phenotype reflect novel patterns of Epstein-Barr virus latent gene expression in Burkitt's lymphoma cells. EMBO J 6 2743 2751
5. BabcockGJ
HochbergD
Thorley-LawsonAD
2000 The expression pattern of Epstein-Barr virus latent genes in vivo is dependent upon the differentiation stage of the infected B cell. Immunity 13 497 506
6. Thorley-LawsonDA
2001 Epstein-Barr virus: exploiting the immune system. Nat Rev Immunol 1 75 82
7. MiyashitaEM
YangB
LamKM
CrawfordDH
Thorley-LawsonDA
1995 A novel form of Epstein-Barr virus latency in normal B cells in vivo. Cell 80 593 601
8. ChenF
ZouJZ
di RenzoL
WinbergG
HuLF
1995 A subpopulation of normal B cells latently infected with Epstein-Barr virus resembles Burkitt lymphoma cells in expressing EBNA-1 but not EBNA-2 or LMP1. J Virol 69 3752 3758
9. QuL
RoweDT
1992 Epstein-Barr virus latent gene expression in uncultured peripheral blood lymphocytes. J Virol 66 3715 3724
10. TierneyRJ
StevenN
YoungLS
RickinsonAB
1994 Epstein-Barr virus latency in blood mononuclear cells: analysis of viral gene transcription during primary infection and in the carrier state. J Virol 68 7374 7385
11. FahraeusR
FuHL
ErnbergI
FinkeJ
RoweM
1988 Expression of Epstein-Barr virus-encoded proteins in nasopharyngeal carcinoma. Int J Cancer 42 329 338
12. ShibataD
WeissLM
1992 Epstein-Barr virus-associated gastric adenocarcinoma. Am J Pathol 140 769 774
13. YoungLS
DawsonCW
ClarkD
RupaniH
BussonP
1988 Epstein-Barr virus gene expression in nasopharyngeal carcinoma. J Gen Virol 69 (Pt 5) 1051 1065
14. FarrellPJ
1995 Epstein-Barr virus immortalizing genes. Trends Microbiol 3 105 109
15. Thorley-LawsonDA
GrossA
2004 Persistence of the Epstein-Barr virus and the origins of associated lymphomas. N Engl J Med 350 1328 1337
16. WoisetschlaegerM
StromingerJL
SpeckSH
1989 Mutually exclusive use of viral promoters in Epstein-Barr virus latently infected lymphocytes. Proc Natl Acad Sci U S A 86 6498 6502
17. WoisetschlaegerM
JinXW
YandavaCN
FurmanskiLA
StromingerJL
1991 Role for the Epstein-Barr virus nuclear antigen 2 in viral promoter switching during initial stages of infection. Proc Natl Acad Sci U S A 88 3942 3946
18. LinaIY
SpeckSH
2000 Regulation of EBNA gene expression. Epstein-Barr Virus Report 7 175 185
19. RoweM
LearAL
Croom-CarterD
DaviesAH
RickinsonAB
1992 Three pathways of Epstein-Barr virus gene activation from EBNA1-positive latency in B lymphocytes. J Virol 66 122 131
20. NonkweloC
SkinnerJ
BellA
RickinsonA
SampleJ
1996 Transcription start sites downstream of the Epstein-Barr virus (EBV) Fp promoter in early-passage Burkitt lymphoma cells define a fourth promoter for expression of the EBV EBNA-1 protein. J Virol 70 623 627
21. TierneyR
KirbyH
NagraJ
RickinsonA
BellA
2000 The Epstein-Barr virus promoter initiating B-cell transformation is activated by RFX proteins and the B-cell-specific activator protein BSAP/Pax5. J Virol 74 10458 10467
22. YooL
SpeckSH
2000 Determining the role of the Epstein-Barr virus Cp EBNA2-dependent enhancer during the establishment of latency by using mutant and wild-type viruses recovered from cottontop marmoset lymphoblastoid cell lines. J Virol 74 11115 11120
23. LingPD
RawlinsDR
HaywardSD
1993 The Epstein-Barr virus immortalizing protein EBNA2 is targeted to DNA by a cellular enhancer binding protein. Proc Natl Acad Sci, USA 90 9237 9241
24. HenkelT
LingPD
HaywardSD
PetersonMG
1994 Mediation of Epstein-Barr virus EBNA2 transactivation by recombination signal-binding protein J kappa. Science 265 92 95
25. AbbotSD
RoweM
CadwalladerK
RickstenA
GordonJ
1990 Epstein-Barr virus nuclear antigen 2 induces expression of the virus-encoded latent membrane protein. J Virol 64 2126 2134
26. AmbinderRF
RobertsonKD
TaoQ
1999 DNA methylation and the Epstein-Barr virus. Semin Cancer Biol 9 369 375
27. FalkKI
SzekelyL
AlemanA
ErnbergI
1998 Specific methylation patterns in two control regions of Epstein-Barr virus latency: the LMP-1-coding upstream regulatory region and an origin of DNA replication (oriP). J Virol 72 2969 2974
28. RobertsonKD
MannsA
SwinnenLJ
ZongJC
GulleyML
1996 CpG methylation of the major Epstein-Barr virus latency promoter in Burkitt's lymphoma and Hodgkin's disease. Blood 88 3129 3136
29. RobertsonKD
HaywardSD
LingPD
SamidD
AmbinderRF
1995 Transcriptional activation of the Epstein-Barr virus latency C promoter after 5-azacytidine treatment: evidence that demethylation at a single CpG site is crucial. Mol Cell Biol 15 6150 6159
30. JinXW
SpeckSH
1992 Identification of critical cis elements involved in mediating Epstein-Barr virus nuclear antigen 2-dependent activity of an enhancer located upstream of the viral BamHI C promoter. J Virol 66 2846 2852
31. WoisetschlaegerM
YandavaCN
FurmanskiLA
StromingerJL
SpeckSH
1990 Promoter switching in Epstein-Barr virus during the initial stages of infection of B lymphocytes. Proc Natl Acad Sci U S A 87 1725 1729
32. AlldayMJ
KunduD
FinertyS
GriffinBE
1990 CpG methylation of viral DNA in EBV-associated tumours. Int J Cancer 45 1125 1130
33. ErnbergI
FalkK
MinarovitsJ
BussonP
TurszT
1989 The role of methylation in the phenotype-dependent modulation of Epstein-Barr nuclear antigen 2 and latent membrane protein genes in cells latently infected with Epstein-Barr virus. J Gen Virol 70 (Pt 11) 2989 3002
34. HummeS
ReisbachG
FeederleR
DelecluseHJ
BoussetK
2003 The EBV nuclear antigen 1 (EBNA1) enhances B cell immortalization several thousandfold. Proc Natl Acad Sci U S A 100 10989 10994
35. KennedyG
SugdenB
2003 EBNA-1, a Bifunctional Transcriptional Activator. Mol Cell Biol 23 6901 6908
36. WangJ
LindnerSE
LeightER
SugdenB
2006 Essential elements of a licensed, mammalian plasmid origin of DNA synthesis. Mol Cell Biol 26 1124 1134
37. LindnerSE
SugdenB
2007 The plasmid replicon of Epstein-Barr virus: mechanistic insights into efficient, licensed, extrachromosomal replication in human cells. Plasmid 58 1 12
38. SchaeferBC
StromingerJL
SpeckSH
1995 Redefining the Epstein-Barr virus-encoded nuclear antigen EBNA-1 gene promoter and transcription initiation site in group I Burkitt lymphoma cell lines. Proc Natl Acad Sci U S A 92 10565 10569
39. TsaiCN
LiuST
ChangYS
1995 Identification of a novel promoter located within the Bam HI Q region of the Epstein-Barr virus genome for the EBNA 1 gene. DNA Cell Biol 14 767 776
40. NonkweloC
RufIK
SampleJ
1997 The Epstein-Barr virus EBNA-1 promoter Qp requires an initiator-like element. J Virol 71 354 361
41. YoshiokaM
CrumMM
SampleJT
2008 Autorepression of Epstein-Barr virus nuclear antigen 1 expression by inhibition of pre-mRNA processing. J Virol 82 1679 1687
42. RufIK
MoghaddamA
WangF
SampleJ
1999 Mechanisms that regulate Epstein-Barr virus EBNA-1 gene transcription during restricted latency are conserved among lymphocryptoviruses of Old World primates. J Virol 73 1980 1989
43. MinarovitsJ
2006 Epigenotypes of latent herpesvirus genomes. Curr Top Microbiol Immunol 310 61 80
44. AlazardN
GruffatH
HiriartE
SergeantA
ManetE
2003 Differential hyperacetylation of histones H3 and H4 upon promoter-specific recruitment of EBNA2 in Epstein-Barr virus chromatin. J Virol 77 8166 8172
45. AltmannM
PichD
RuissR
WangJ
SugdenB
2006 Transcriptional activation by EBV nuclear antigen 1 is essential for the expression of EBV's transforming genes. Proc Natl Acad Sci U S A 103 14188 14193
46. ReismanD
SugdenB
1986 trans activation of an Epstein-Barr viral transcriptional enhancer by the Epstein-Barr viral nuclear antigen 1. Mol Cell Biol 5 3838 3846
47. PuglielliMT
WoisetschlaegerM
SpeckSH
1996 oriP is essential for EBNA gene promoter activity in Epstein-Barr virus-immortalized lymphoblastoid cell lines. J Virol 70 5758 5768
48. BusheyAM
DormanER
CorcesVG
2008 Chromatin insulators: regulatory mechanisms and epigenetic inheritance. Mol Cell 32 1 9
49. WestAG
GasznerM
FelsenfeldG
2002 Insulators: many functions, many mechanisms. Genes Dev 16 271 288
50. WallaceJA
FelsenfeldG
2007 We gather together: insulators and genome organization. Curr Opin Genet Dev 17 400 407
51. CapelsonM
CorcesVG
2004 Boundary elements and nuclear organization. Biol Cell 96 617 629
52. PhillipsJE
CorcesVG
2009 CTCF: master weaver of the genome. Cell 137 1194 1211
53. OhlssonR
RenkawitzR
LobanenkovV
2001 CTCF is a uniquely versatile transcription regulator linked to epigenetics and disease. Trends Genet 17 520 527
54. DayL
ChauCM
NebozhynM
RennenkampAJ
ShoweM
2007 Chromatin Profiling Of Epstein-Barr Virus Latency Control Region. J Virol
55. ChauCM
ZhangXY
McMahonSB
LiebermanPM
2006 Regulation of Epstein-Barr virus latency type by the chromatin boundary factor CTCF. J Virol 80 5723 5732
56. CopelandNG
JenkinsNA
CourtDL
2001 Recombineering: a powerful new tool for mouse functional genomics. Nat Rev Genet 2 769 779
57. PaulsonEJ
FingerothJD
YatesJL
SpeckSH
2002 Methylation of the EBV genome and establishment of restricted latency in low-passage EBV-infected 293 epithelial cells. Virology 299 109 121
58. TrivediP
SpinsantiP
CuomoL
VolpeM
TakadaK
2001 Differential regulation of Epstein-Barr virus (EBV) latent gene expression in Burkitt lymphoma cells infected with a recombinant EBV strain. J Virol 75 4929 4935
59. LiH
MinarovitsJ
2003 Host cell-dependent expression of latent Epstein-Barr virus genomes: regulation by DNA methylation. Adv Cancer Res 89 133 156
60. ChauCM
LiebermanPM
2004 Dynamic chromatin boundaries delineate a latency control region of Epstein-Barr virus. J Virol 78 12308 12319
61. HarkAT
SchoenherrCJ
KatzDJ
IngramRS
LevorseJM
2000 CTCF mediates methylation-sensitive enhancer-blocking activity at the H19/Igf2 locus. Nature 405 486 489
62. KanduriC
PantV
LoukinovD
PugachevaE
QiCF
2000 Functional association of CTCF with the insulator upstream of the H19 gene is parent of origin-specific and methylation-sensitive. Curr Biol 10 853 856
63. BergerSL
2007 The complex language of chromatin regulation during transcription. Nature 447 407 412
64. ChernukhinI
ShamsuddinS
KangSY
BergstromR
KwonYW
2007 CTCF interacts with and recruits the largest subunit of RNA polymerase II to CTCF target sites genome-wide. Mol Cell Biol 27 1631 1648
65. DelecluseHJ
HilsendegenT
PichD
ZeidlerR
HammerschmidtW
1998 Propagation and recovery of intact, infectious Epstein-Barr virus from prokaryotic to human cells. Proc Natl Acad Sci U S A 95 8245 8250
66. DengZ
LezinaL
ChenCJ
ShtivelbandS
SoW
2002 Telomeric proteins regulate episomal maintenance of Epstein-Barr virus origin of plasmid replication. Mol Cell 9 493 503
67. HirtB
1967 Selective extraction of polyoma DNA from infected mouse cell cultures. J Mol Biol 26 365 369
Štítky
Hygiena a epidemiologie Infekční lékařství Laboratoř
Článek vyšel v časopisePLOS Pathogens
Nejčtenější tento týden
2010 Číslo 8- Stillova choroba: vzácné a závažné systémové onemocnění
- Diagnostický algoritmus při podezření na syndrom periodické horečky
- Diagnostika virových hepatitid v kostce – zorientujte se (nejen) v sérologii
- Perorální antivirotika jako vysoce efektivní nástroj prevence hospitalizací kvůli COVID-19 − otázky a odpovědi pro praxi
- Infekční komplikace virových respiračních infekcí – sekundární bakteriální a aspergilové pneumonie
-
Všechny články tohoto čísla
- Dissecting the Genetic Architecture of Host–Pathogen Specificity
- The Battle for Iron between Bacterial Pathogens and Their Vertebrate Hosts
- Global Genotype-Phenotype Correlations in
- Burkholderia Type VI Secretion Systems Have Distinct Roles in Eukaryotic and Bacterial Cell Interactions
- Chitin Synthases from Are Involved in Tip Growth and Represent a Potential Target for Anti-Oomycete Drugs
- Distinct Merkel Cell Polyomavirus Molecular Features in Tumour and Non Tumour Specimens from Patients with Merkel Cell Carcinoma
- Biological and Structural Characterization of a Host-Adapting Amino Acid in Influenza Virus
- Functional Characterisation and Drug Target Validation of a Mitotic Kinesin-13 in
- CTCF Prevents the Epigenetic Drift of EBV Latency Promoter Qp
- The Human Fungal Pathogen Escapes Macrophages by a Phagosome Emptying Mechanism That Is Inhibited by Arp2/3 Complex-Mediated Actin Polymerisation
- Bim Nuclear Translocation and Inactivation by Viral Interferon Regulatory Factor
- Cyst Wall Protein 1 Is a Lectin That Binds to Curled Fibrils of the GalNAc Homopolymer
- Reciprocal Analysis of Infections of a Model Reveal Host-Pathogen Conflicts Mediated by Reactive Oxygen and imd-Regulated Innate Immune Response
- A Subset of Replication Proteins Enhances Origin Recognition and Lytic Replication by the Epstein-Barr Virus ZEBRA Protein
- Damaged Intestinal Epithelial Integrity Linked to Microbial Translocation in Pathogenic Simian Immunodeficiency Virus Infections
- Kaposin-B Enhances the PROX1 mRNA Stability during Lymphatic Reprogramming of Vascular Endothelial Cells by Kaposi's Sarcoma Herpes Virus
- Direct Interaction between Two Viral Proteins, the Nonstructural Protein 2C and the Capsid Protein VP3, Is Required for Enterovirus Morphogenesis
- A Novel CCR5 Mutation Common in Sooty Mangabeys Reveals SIVsmm Infection of CCR5-Null Natural Hosts and Efficient Alternative Coreceptor Use
- Micro RNAs of Epstein-Barr Virus Promote Cell Cycle Progression and Prevent Apoptosis of Primary Human B Cells
- Enterohemorrhagic Requires N-WASP for Efficient Type III Translocation but Not for EspF-Mediated Actin Pedestal Formation
- Host Imprints on Bacterial Genomes—Rapid, Divergent Evolution in Individual Patients
- UNC93B1 Mediates Host Resistance to Infection with
- The Transcription Factor Rbf1 Is the Master Regulator for -Mating Type Controlled Pathogenic Development in
- Protective Efficacy of Cross-Reactive CD8 T Cells Recognising Mutant Viral Epitopes Depends on Peptide-MHC-I Structural Interactions and T Cell Activation Threshold
- Bacteriophage Lysin Mediates the Binding of to Human Platelets through Interaction with Fibrinogen
- Insecticide Control of Vector-Borne Diseases: When Is Insecticide Resistance a Problem?
- Immune Modulation with Sulfasalazine Attenuates Immunopathogenesis but Enhances Macrophage-Mediated Fungal Clearance during Pneumonia
- PKC Signaling Regulates Drug Resistance of the Fungal Pathogen via Circuitry Comprised of Mkc1, Calcineurin, and Hsp90
- A Multi-Step Process of Viral Adaptation to a Mutagenic Nucleoside Analogue by Modulation of Transition Types Leads to Extinction-Escape
- “Everything You Always Wanted to Know about Sex (but Were Afraid to Ask)” in after Two Decades of Laboratory and Field Analyses
- Norovirus Gastroenteritis, Carbohydrate Receptors, and Animal Models
- Variations in TcdB Activity and the Hypervirulence of Emerging Strains of
- SWAN-1 Binds to EGL-9 and Regulates HIF-1-Mediated Resistance to the Bacterial Pathogen PAO1
- Conformational Adaptation of Asian Macaque TRIMCyp Directs Lineage Specific Antiviral Activity
- The Proteasome Active Site Threonine Is Essential for Persistence Yet Dispensable for Replication and Resistance to Nitric Oxide
- Characterization of Oseltamivir-Resistant 2009 H1N1 Pandemic Influenza A Viruses
- The Pneumococcal Serine-Rich Repeat Protein Is an Intra-Species Bacterial Adhesin That Promotes Bacterial Aggregation and in Biofilms
- Regulatory T Cell Suppressive Potency Dictates the Balance between Bacterial Proliferation and Clearance during Persistent Infection
- Structural Alterations in a Component of Cytochrome Oxidase and Molecular Evolution of Pathogenic in Humans
- A Limited Number of Antibody Specificities Mediate Broad and Potent Serum Neutralization in Selected HIV-1 Infected Individuals
- Spliced Leader Trapping Reveals Widespread Alternative Splicing Patterns in the Highly Dynamic Transcriptome of
- Contribution of Coagulases towards Disease and Protective Immunity
- Early Severe Inflammatory Responses to Uropathogenic Predispose to Chronic and Recurrent Urinary Tract Infection
- PLOS Pathogens
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- The Transcription Factor Rbf1 Is the Master Regulator for -Mating Type Controlled Pathogenic Development in
- PKC Signaling Regulates Drug Resistance of the Fungal Pathogen via Circuitry Comprised of Mkc1, Calcineurin, and Hsp90
- Contribution of Coagulases towards Disease and Protective Immunity
- Early Severe Inflammatory Responses to Uropathogenic Predispose to Chronic and Recurrent Urinary Tract Infection
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



