-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaPhylodynamics and Human-Mediated Dispersal of a Zoonotic Virus
Understanding the role of humans in the dispersal of predominately animal pathogens is essential for their control. We used newly developed Bayesian phylogeographic methods to unravel the dynamics and determinants of the spread of dog rabies virus (RABV) in North Africa. Each of the countries studied exhibited largely disconnected spatial dynamics with major geo-political boundaries acting as barriers to gene flow. Road distances proved to be better predictors of the movement of dog RABV than accessibility or raw geographical distance, with occasional long distance and rapid spread within each of these countries. Using simulations that bridge phylodynamics and spatial epidemiology, we demonstrate that the contemporary viral distribution extends beyond that expected for RABV transmission in African dog populations. These results are strongly supportive of human-mediated dispersal, and demonstrate how an integrated phylogeographic approach will turn viral genetic data into a powerful asset for characterizing, predicting, and potentially controlling the spatial spread of pathogens.
Published in the journal: . PLoS Pathog 6(10): e32767. doi:10.1371/journal.ppat.1001166
Category: Research Article
doi: https://doi.org/10.1371/journal.ppat.1001166Summary
Understanding the role of humans in the dispersal of predominately animal pathogens is essential for their control. We used newly developed Bayesian phylogeographic methods to unravel the dynamics and determinants of the spread of dog rabies virus (RABV) in North Africa. Each of the countries studied exhibited largely disconnected spatial dynamics with major geo-political boundaries acting as barriers to gene flow. Road distances proved to be better predictors of the movement of dog RABV than accessibility or raw geographical distance, with occasional long distance and rapid spread within each of these countries. Using simulations that bridge phylodynamics and spatial epidemiology, we demonstrate that the contemporary viral distribution extends beyond that expected for RABV transmission in African dog populations. These results are strongly supportive of human-mediated dispersal, and demonstrate how an integrated phylogeographic approach will turn viral genetic data into a powerful asset for characterizing, predicting, and potentially controlling the spatial spread of pathogens.
Introduction
Every year approximately 55,000 people die from rabies [1]. Over 99% of these deaths occur in developing countries where rabies virus (RABV; negative-sense RNA virus, family Rhabdoviridae) is endemic in the domestic dog [2]. Rabies has been neglected across much of Asia and Africa, despite becoming an increasing problem in the recent decades [1], [3], [4]. Although the history of rabies in Africa prior to the 20th century is uncertain [5], the currently circulating dog rabies virus is thought to have emerged during the 19th and 20th centuries [6], [7]. Despite the importance of dogs as vectors for human rabies, little is known about the spatial and temporal dynamics of rabies in this major reservoir species, or the processes responsible for its maintenance in specific geographic localities. In particular, the role of human activities in mediating the spread of dog RABV is unclear, nor is it known how landscape characteristics, including human infrastructures such as roads, affect RABV dispersal within dog populations. However, such information is critical to revealing the determinants of RABV transmission and hence for its control in the domestic dog.
We used a recently developed probabilistic approach [8] to determine the spatial and temporal dynamics of dog RABV transmission from a large-scale gene sequence study. We encode different phylogeographic scenarios of viral spread, as well as different landscape features, in a model-based approach, and choose among these models in a quantitatively rigorous fashion. Our focus was on dog populations in North Africa where RABV has been endemic for more than a century, and our key aim was to determine how ecological, anthropogenic and evolutionary dynamics shape the spatial distribution and spread of this important zoonotic pathogen [9], [10], [11], [12].
Results/Discussion
We first inferred the evolutionary history of 287 RABV sequences (3080 nt; encompassing the whole N, P and intergenic G-L region) sampled from Algeria, Morocco, Tunisia and the Spanish territories from North Africa (Ceuta and Melilla) between 1986 and 2008. All these viruses are assigned to the Africa 1 genotype (relevant epidemiological information for all RABV isolates analysed in this study is presented in Table S1 in Supporting Information S1). We estimated the timescale of this evolutionary history using a Bayesian Markov chain Monte Carlo (MCMC) approach [13]. The most recent common ancestor of all the North African RABV sampled here was estimated to have existed between 1878–1945, supporting previous suggestions that dog RABV was periodically responsible for local sporadic epidemics in the middle of the 19th century [14], and that rabies became enzootic in this entire region during the 20th century. More generally, this timescale is consistent with the expanding European colonial influence in North Africa [7]. This analysis also revealed distinct phylogenetic lineages in Algeria, Morocco and Tunisia, indicating that viruses generally grouped according to their country of origin (Figure 1). This result is unexpected if the virus is only dispersed through the local movement of animals as observed in wildlife rabies [9], [10] as these would not respect geo-political boundaries. Indeed, we found only a few exceptions to the country-specific clustering, such as two Algerian sequences within the Moroccan clade and four Moroccan sequences in the Algerian clade. The Africa 1 clade is therefore consistent with the general phylogeographic pattern observed for dog RABV at reasonably large geographic scales; a series of spatially distinct clusters that experience relatively little contact among them [6], [7].
Fig. 1. MCC tree of 287 sequences of the Africa 1 clade, estimated from the N, P and G-L genes and intergenic regions of dog RABV, and showing the spatial structure of the viral lineages. 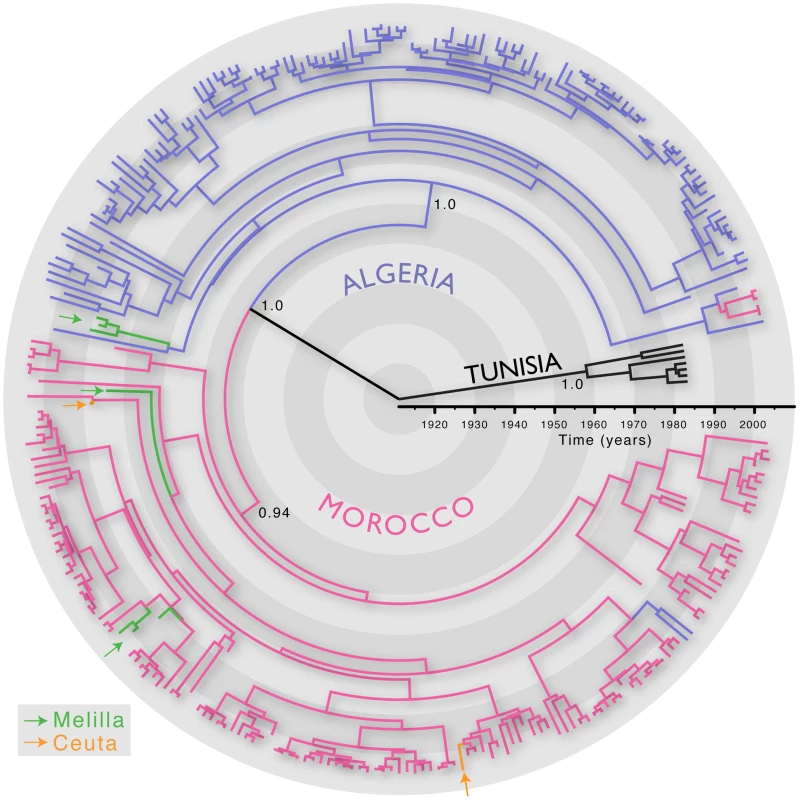
Branches are colored according to the locations of viral sampling. Branch lengths are estimated in time units (years) as indicated by the time bar. Posterior clade probability values (>0.9) are shown for key nodes. Although Melilla and Ceuta are considered Spanish locations they are in fact Moroccan enclaves. As such they do not represent virus dispersal across the strait of Gibraltar. To analyze intra-country patterns of viral transmission in more detail, we considered a stochastic diffusion process among the 20 (Algeria) and 28 (Morocco) sampling localities for which most data were available (Figure 2 and Figure S1 in Supporting Information S1). We quantified the degree of spatial admixture using a modified Association Index (AI, [8], [15]), and by summarizing the number of inferred transitions to each location within Algeria and Morocco (Table 1) based on an analysis in which rates of diffusion between each pair of locations were estimated. Although these analyses reveal that there is still significant spatial structure within each country (p<0.001), the AIs are considerably higher (0.67 [0.62–0.73] and 0.55 [0.51–0.63] for Algeria and Morocco, respectively) than those found for rabies at a larger spatial scale (e.g., 0.087 [0.043–0.132] for the Africa 2 lineage in Central and West Africa) [8], indicating weaker spatial structure at the within-country level. The summaries of transitions to each location generally identify multiple independent introductions of viruses in each location from which several samples were obtained (Table 1). Overall, the number of independent transitions to densely sampled locations is lower in Morocco than Algeria, in agreement with the lower AI for Morocco. Taken together with the strong spatial structure across countries, these results suggest that a relatively fluid RABV diffusion process within countries is restricted by geopolitical boundaries at larger scales.
Fig. 2. Road network, accessibility and sampling locations of RABV samples. 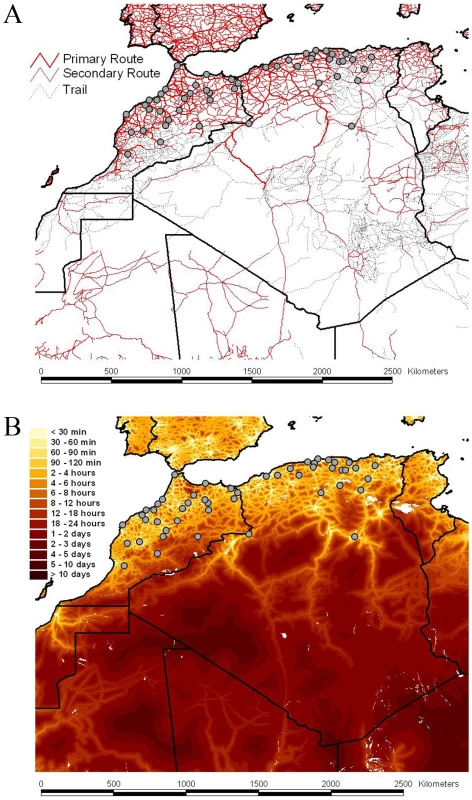
The road network and the accessibility are shown in panels A and B, respectively The border between the countries is indicated by dark lines. Specific locations were unavailable for Tunisian samples. The color gradient indicates the estimated travel time to the nearest city of population greater than 50,000 people, with yellow at one extreme indicating low travel times (<30 min) and red at the other extreme indicating long travel times (>10 days). The data are available for download from http://gem.jrc.ec.europa.eu/. Tab. 1. Transition summaries to each location in Algeria and Morocco. 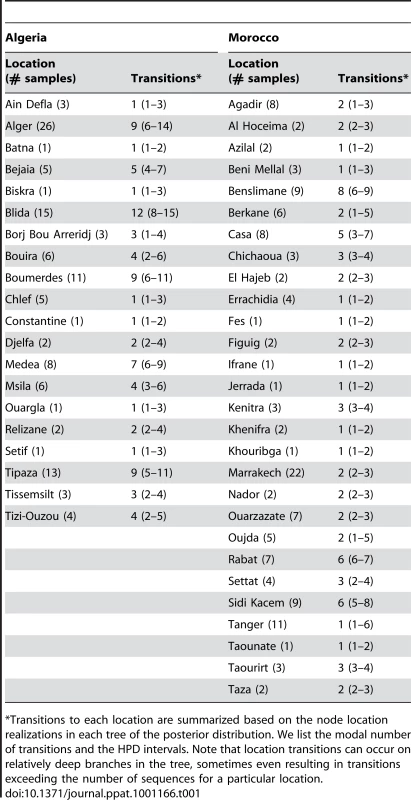
*Transitions to each location are summarized based on the node location realizations in each tree of the posterior distribution. We list the modal number of transitions and the HPD intervals. Note that location transitions can occur on relatively deep branches in the tree, sometimes even resulting in transitions exceeding the number of sequences for a particular location. To identify the factors that may explain RABV spread, we incorporated several potential predictors as relative diffusion rates among each pair of locations, and tested these against equal rates of diffusion. Specifically, we considered geographical distances (great-circle distances), human population size, road distances and spatial accessibility measures. Road distances were derived from transport network data (Figure 2) and demonstrated a strong correlation with great-circle distances (r = 0.96). More detailed landscape features, which may imply multiple, direct and indirect pathways connecting the different localities sampled in this study, were represented by accessibility data. These data reflect the travel time to the nearest major city using road/track-based travel [16] and were less correlated with geographical distances (r = 0.61). We employed circuit theory to translate the accessibility landscape into an origin-destination distance matrix (the so-called ‘isolation by resistance model’) [17]. We also tested a simple gravity model of viral spread that, in the absence of real dog population sizes for the locations involved, was based on human population sizes for the discrete as a proxy. Finally, we also used population sizes in a landscape approach, similar to accessibility measures, to construct a population surface matrix [18].
Marginal likelihood estimates of the model fit of these different predictors suggested that RABV spatial dynamics are best described by road distances (Table 2). This was consistent across both countries and again supports human-assisted dispersal of rabies-infected dogs. As expected by their high correlation, geographical distances provided only a marginally lower fit compared to road distances. Only the population surface provided inconsistent results between both countries; whereas this model competes with road distances in Algeria, the population surface did not provide a good fit to the Moroccan data (Figure S2 in Supporting Information S1). Although accessibility did not seem to explain RABV diffusion as well, we note that all samples were obtained from relatively accessible parts of Morocco and Algeria.
Tab. 2. Marginal (log) likelihood estimates for the fit of different phylogeographic diffusion predictors in Algeria and Morocco. 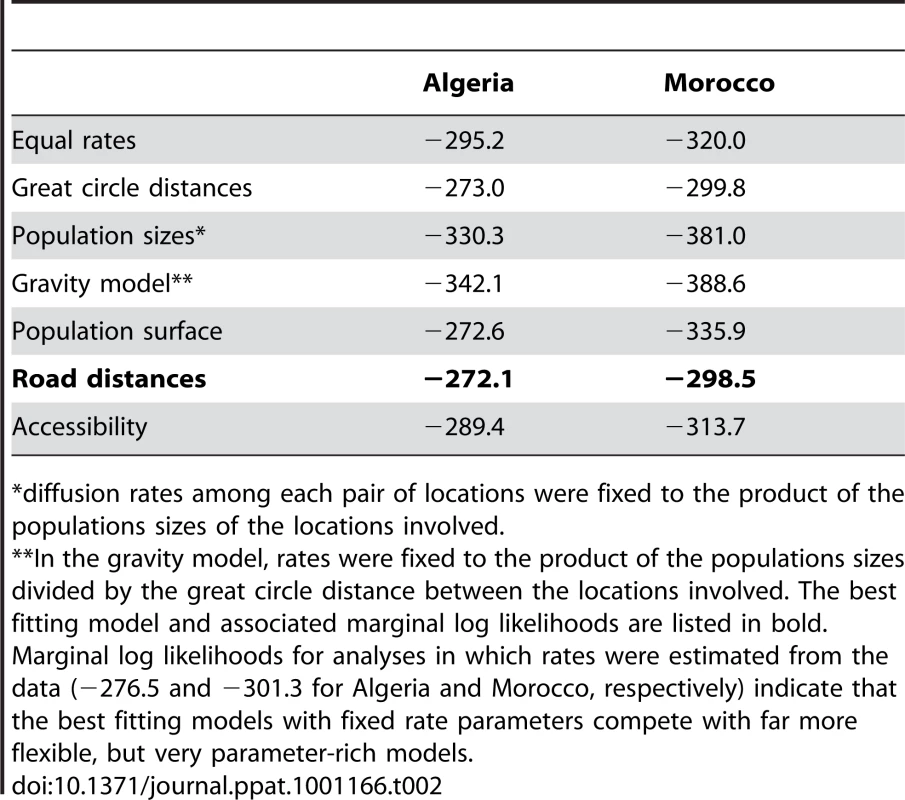
*diffusion rates among each pair of locations were fixed to the product of the populations sizes of the locations involved. To quantify and compare the dissemination process with previous results, we estimated the rate of RABV gene flow among the sampled isolates using ‘Markov jump’ counts [19] of location state transitions and their reward-associated distances between locations across each branch. The posterior average rate of viral gene flow among localities estimated for Algeria was 26 km/yr (95% highest probability density interval: 18–34) and 33 (23–43) km/yr based on great circle distances and road distances, respectively. Somewhat higher viral gene flow rate estimates were obtained for Morocco with 42 (26–58) km/yr and 51 (34–72) km/yr for great circle distances and road distances, respectively. We note that the rates of viral gene flow estimated here are highly dependent of the scale of sampling such that comparison may only prove useful at the same geographic scale. However, these estimates were 2.7 to 4.4 times higher than those recorded in established enzootic situations in wildlife animals [9], again suggestive of human-mediated transmission. Although it is theoretically possible that these relatively high rates reflect epidemic waves periodically moving through this geographical region [20], particularly since similar rates have been observed in wild carnivores during epidemic spread [9], [10], such waves were not observed in the geographical areas studied here and where the virus appears to be largely enzootic.
The occasional mixing of sequences from different locations at the tips of the inferred tree (Figure 1) is suggestive of long distance spread in relatively little time (6 months to one year). To quantify such rapid and long distance spread, we summarized the posterior distribution of distances covered along individual branches (Figure 3). We focused on branches along which inferred location state changes occurred in a time period of less than 1 year, and between 1 and 2 years. As a control, we analysed the branches without inferred state changes; as expected these all had negligible Markov jump count distances (not shown). Across the posterior distribution of trees, we observed between 5 and 13 branches per tree that have a time length less than 2 years and cover a distance of more than 200 km, and 12 to 13 branches that cover a distance of about 100 km (Figure 3). Importantly, our ability to clearly detect long-distance movement is limited to branches representing short evolutionary times; longer branches could also harbour such events, providing an explanation for the relatively high average rates of viral gene flow.
Fig. 3. Short-time virus gene flow events in Algeria and Morocco. 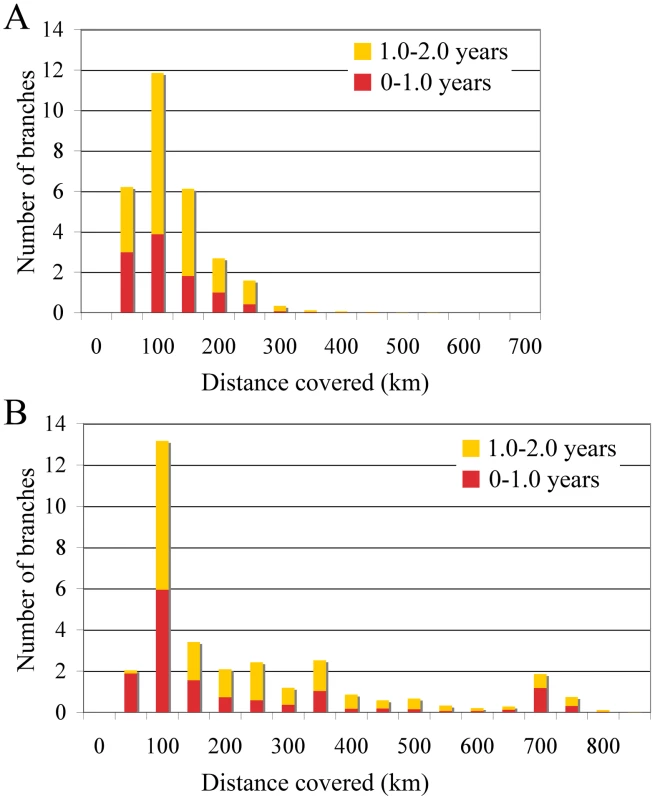
Branches that harbor an expected state change (different location at parent and daughter node) were binned according to the distance covered along that branch in Algeria (A) and Morocco (B). This was carried out separately for branches along which state changes occurred in less than 1 year, and between 1 and 2 years. The rates of viral gene flow we estimate among the sampled isolates from Algeria and Morocco contrast with those of spatial RABV movement in an African dog population that should experience very limited human-mediated dissemination of rabid dogs [21]. In this case, the spatial dispersal of single RABV infections was estimated to be predominantly less than 2 km (and always smaller than 20 km). Considering that the average incubation period of RABV is between 22 to 29 days [20], [21], [22], [23], [24], it is clear that such long distances as those recorded in our study could only be achieved with at least some human intervention. To investigate more formally how the RABV distribution we observe in Algeria and Morocco contrasts with the patterns of spread we would expect from transmission dynamics in African dogs alone (i.e. without human intervention), we simulated a phylogeodynamic process based on epidemiological parameters obtained from detailed analyses of rabies transmission biology [20], [21]. In particular, we considered epidemiologically informed virus movement over all evolutionary histories in the posterior distribution resulting from our phylodynamic inference (see Supplementary Information). In our spatial simulation we analyze cases in which (i) each new infection takes a random direction in continuous space, or (ii) subsequent infections consistently take the same direction (Figure 4). Although the latter may not be very realistic, it should resemble virus movement along roads. For both Algeria and Morocco, spatial diffusion is initiated at the centre of the sampling locations, such that the process has the largest probability to cover these locations a priori. When assuming up to one year of movement these simulations clearly show that RABV could not have spread to the same extent as shown by the current sampling in Algeria and Morocco if the virus was simply being transmitted by dog dispersal alone. Even if we enforce a year of successive RABV transmissions in the same direction, which is highly implausible given the observed dynamics of dog RABV in a local setting [21], the simulations still do not attain the observed spatial RABV spread. In addition, the distances realized by dispersal in random directions along branches less than 2 years were all less than 60 km, which is far more restricted than estimated for the real data (Figure 3).
Fig. 4. Spatial simulation based on the inferred evolutionary histories and epidemiological parameters of rabies spread in Algeria (A) and Morocco (B). 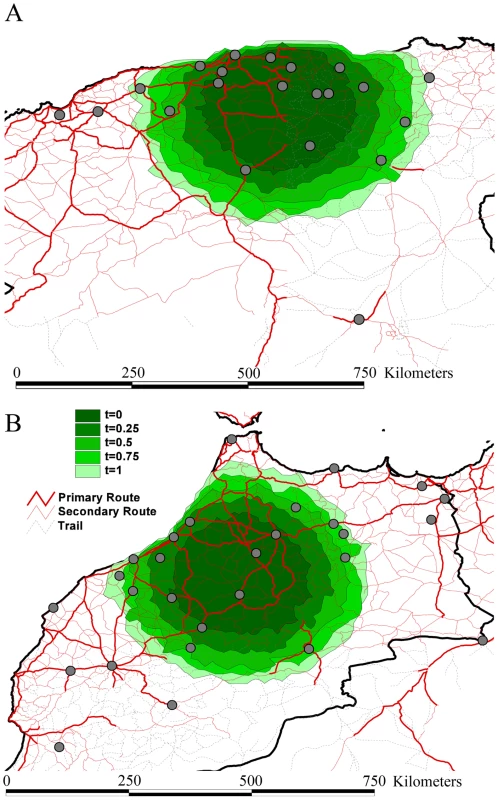
We report the simulated spread using contours represented under the form of polygons. Different contours represent different simulation scenarios: for time = 0.25 to time = 1.0, we force the virus to move consistently in the same direction for successive transmissions during a time period of three months to one year. The dark green contour in the centre represents simulated spread with each infection taking a new and random direction, while the increasingly lighter ones represent time = 0.25 to time = 1.0. Rabies is a prime example of an infectious disease in which dispersal can be exacerbated by animal movement mediated by humans. This is illustrated by raccoon rabies in Virginia, USA [25], dog rabies in Indonesia in Flores Island [26], in Bali (F.X. Meslin, Personal communication) and in parts of Europe [27]. Each epidemic resulted in enormous expenditure on rabies post exposure prophylaxis in humans and animal vaccination programs [28], [29], [30], [31]. Importantly, our study allows us to quantify rates of viral gene flow among sampled dog isolates (between 18 and 72 km/yr) in a mixed geographic and socio-economic landscape, such as those characterized by Algeria and Morocco where there is currently little dog vaccination. In addition, our analysis suggests that the human-mediated dispersal of infected dogs is likely to continue to play a major role in the transmission of RABV in geographical areas where it has been present for many years. Indeed, our observations of administrative borders that restrict a relatively fluid pattern of spread, the occasional long-distance movement of viruses to particular countries, and the fit between spatial dynamics and road distances, all point to the displacement of rabies-infected dogs by humans.
Understanding the frequency and distance of movements of potentially infected animals is of paramount importance in predicting the spread of viral infections [32], [33]. In addition, such information has important implications for disease control; understanding the conditions under which the containment of wildlife [34] and dog rabies can reliably be achieved will assist in the long term goal of eliminating animal RABV. In particular, that humans mediate the transmission of RABV among dogs in North Africa requires that intervention procedures are implemented more rapidly than in situations in which humans play little or no role in viral transmission. The high cost associated with surveillance underscores the importance of sampling design and the development of cost-effective monitoring and testing approaches [9], [12], [35]. In addition, this study illustrates the power of phylogeographic approaches [8] to identify the factors responsible for the spread of major animal and zoonotic pathogens. By integrating spatial dynamics with temporal inferences, the Bayesian analysis utilized here constitutes a powerful new tool that may complement traditional epidemiological methods in studying the effects of human behaviour on the evolution of zoonotic viruses.
Materials and Methods
Data selection
The Office of Veterinary Public Health Services of the different countries coordinates follow-up per animal bites. The respective state health departments (in conjunction with qualified laboratories that they designate) conduct all collection, observations, and euthanization (if necessary) of animals suspected of rabies, according to established national standardized protocols. A total of 287 isolates sampled from Morocco, Algeria, Tunisia and Spain (Ceuta and Melilla) were collected by the authors within the framework of these qualified laboratories, and sequenced. These samples were collected from dead animals suspected of rabies so that submission for laboratory rabies diagnosis is mandatory. Spatial co-ordinates and time of sampling, covering a period of 22 years (1986–2007), were available for the majority of these isolates. Relevant epidemiological information and GenBank accession numbers for all RABV isolates analysed in this study are presented in Table S1 in Supporting Information S1. All the locations sampled in this study experienced cases every year. As such, our sampling does not focus on areas that have been free of rabies in the recent past.
To perform a reliable evolutionary analysis of dog RABV circulating in North Africa, we aimed at selecting a genetic region with sufficient phylogenetic information. To this end, we sequenced a total of 3080 nt encompassing the N, P and intergenic G-L region. Total RNA from the original brain samples was extracted using Trizol reagent (Invitrogen) according to the manufacturer's instructions. RT-PCRs and sequencing reactions were performed as described previously [6], [36]. All sequences obtained have been deposited in GenBank (accession numbers GU798102–GU798962).
Additional primers used in this study were N1280 (5′ - AGTCAGTTCTAATCATCAAGC-3′), M138(5′-AAGTTCCTYATGTTYTTCTTGC-3′), G(5′GACTTGGGTCTCCCGAACTGGGG-3′) and L (5′-CAA AGG AGA GTT GAG ATT GTA GTC-3′), at positions 1348–1368, 2632–2653, 4666–4688 and 5512–5535, respectively, of the lyssavirus genome [37].
Multiple sequence alignment was performed using MUSCLE available through the Muscle web interface (http://www.ebi.ac.uk/Tools/muscle/index.html) [38]. All alignments are available from the authors on request.
Bayesian evolutionary analyses of spatiotemporal RABV dynamics
To investigate the evolutionary relationships and time to common ancestry among RABV lineages circulating in north Africa, we reconstructed the phylogenetic history for the entire data set using Bayesian Markov chain Monte Carlo (MCMC) analysis implemented in the BEAST package [13]. BEAST incorporates sampling time information to estimate evolutionary rates and a posterior distribution of time-scaled trees. We employed a GTR model of nucleotide substitution with gamma-distributed rate variation among sites and a relaxed (uncorrelated log-normal) molecular clock model [39]. We specified a Bayesian skyline plot model as flexible tree prior [40]. All chains were run for a sufficient length and convergence was diagnosed using Tracer (http://tree.bio.ed.ac.uk/software/tracer/) ignoring 10% of the chain as burn-in. Evolutionary history was summarized using an annotated Maximum Clade Credibility (MCC) phylogenetic tree. Posterior probability values provide an assessment of the degree of support for each node on the tree.
To reconstruct the spatial dynamics of dog-associated RABV spread and investigate the role of different diffusion predictors in shaping the epidemic in both Morocco and Algeria, we extracted two data sets with their specific spatial and temporal co-ordinates: (i) A total of 117 Algerian sequences (3080 nt) collected from dogs in 20 cities over 7 years (from 2001 to 2008), and (ii) a total of 133 Moroccan sequences (3080 nt) sampled from dogs in 28 cities between 2004 and 2008. For all these isolates precise dates (month) of sampling and geographical localities (city) are available (Table S2 in Supporting Information S1).
As an additional component in the fully probabilitistic Bayesian inference framework, we consider a discretized diffusion process among the sampling locations in both countries, formalized as a continuous time Markov chain (CTMC) model [8]. A CTMC is fully characterized using a matrix that describes the rate of movement from location state i to j for every pair of locations. To efficiently estimate the diffusion process from a single observation (a single location realization for each sequence), we restrict the parameterization to a sparse set of rates that adequately explains the phylogeographic dispersal process using Bayesian Stochastic search variable selection (BSSVS). This BSSVS procedure also allows us to employ Bayes factor testing in the identification of the most parsimonious description of the diffusion process [8]. We used a modified Association Index (AI) to assess the degree of spatial structure in the phylogeographic data [8], [15]. This reports the posterior distribution of association values relative to those obtained by randomizing the tip locations. In addition, we summarize the number of transitions to each sampling location in the posterior tree distribution based on the location realizations at the nodes. The latter provide a conservative estimate of the number of independent introductions in each location.
To quantify the dissemination process, we estimated the rate of rabies spread among the sampled isolates using ‘Markov jump’ counts [19] of location state transitions for all possible states along the phylogeny. Markov jump counts measure the expected number of transitions along each branch conditional on the observed data. By multiplying the expected number of transitions between each pair of locations by the geographical distance between these two locations, we arrive at the expected distance travelled within the time elapsed on each branch. This approach, implemented in BEAGLE [41] a library that can be used in conjunction with BEAST), integrates over all uncertainty in the evolutionary tree and offers a degree of robustness to model misspecification [42].
To test different scenarios of phylogeographic diffusion, we fix the CTMC relative rate parameters to the normalized pairwise location measures that represent different diffusion predictors and perform Bayesian model selection using marginal likelihood approximations [43]. We consider; (i) geographical distances, specifically great-circle distances that represent the shortest path on the surface of the Earth between two points, (ii) human population size, obtained from http://en.wikipedia.org/ and http://www.mongabay.com (Table S3 in Supporting Information S1, rates between each pair of locations were fixed to the normalized products of the population sizes), (iii) road distances, (iv) a gravity model, (v) spatial accessibility, and (vi) ‘population conductivity’ measures. The accessibility estimates are derived from a range of spatial data sets, road type and network data [44], satellite derived and cover information, settlement database locations and sizes, and satellite derived topography. They are combined to create a ‘friction surface’ where each 1×1 km square represents the difficulty (or travel time) in crossing it. These estimates provide a representation of the difficulty in travel between all the locations. Using a circuit theory approach, an origin-destination distance matrix was estimated from this accessibility landscape [17]. A simple gravity model was constructed by fixing the rates to the normalized product of the population sizes divided by the great circle distance between the locations involved. As an alternative, population sizes were also mapped in a landscape (Figure S2 in Supporting Information S1) [18] and again translated to an origin-destination distance matrix using circuit theory. To assess model fit, marginal likelihood approximations are obtained using an importance sampling estimator [43], [45], which employs a mixture of model prior and posterior samples [46].
Spatial simulation
To contrast the spatial distribution of our rabies samples in Algeria and Morocco with the patterns of spread we would expect from local transmission dynamics in African dogs as the sole maintenance population, we performed a simulation analysis that integrates phylodynamic and epidemiological parameters. Specifically, we consider a spatial process based on epidemiological parameters obtained from a detailed analysis of rabies transmission biology in African dogs [20], and simulate virus movement accordingly over all evolutionary histories in the posterior distribution resulting from our phylodynamic inference. The latter characterizes the successful ancestral transmission history of the viruses we sampled and provides a time-scale for the spatial process we would like to simulate.
For each tree in the posterior distribution, we consider the ancestral virus at the root to start spreading from the mid-point of our available samples (average of longitudes and latitudes). We recursively visit all branches from root to tip, each time simulating a number of successive infections, which jointly encompass the entire time length for each branch. Each time interval t (in days) between successive infections follows:(1)where a is a random incubation time, b is the random period of infectiousness, and f is a random fraction drawn from a uniform[0,1] distribution. Following the results of the comprehensive study by Hampson et al. [21], we consider(2)and(3)Each new infection is moved a random distance d (in m) away from its source case; the distribution for d follows a previously described spatial infection kernel [21]:(4)In the spatial simulation process, we assume that each new distance takes a random direction in continuous space, but we also explore subsequent infections consistently taking the same direction. To achieve a realistic distribution in the relevant geographic area, we prohibit new infections to invade water areas. The tree heights used for simulation for Algeria and Morocco were 33 (23–46) years and 28 (18–39) years respectively (as estimated from the country-specific data), whereas the tree lengths encompassed 623 (490–789) years and 532 (367–706) years respectively.
This simulation procedure yields location realizations in continuous space for all tips and all trees in the posterior distribution. We summarize this spatial distribution using two-dimensional contours. Because the spatial simulation for each tree in the posterior distribution may cover a different area, it is important to note that the contour representing the process over all trees depicts the maximum area that can be covered for the set of epidemiological parameters we consider.
Supporting Information
Zdroje
1. KnobelDL
CleavelandS
ColemanPG
FevreEM
MeltzerMI
2005 Re-evaluating the burden of rabies in Africa and Asia. Bull World Health Organ 83 360 368
2. CleavelandS
MeslinFX
BreimanR
2006 Dogs can play useful role as sentinel hosts for disease. Nature 440 605
3. FevreEM
KaboyoRW
PerssonV
EdelstenM
ColemanPG
2005 The epidemiology of animal bite injuries in Uganda and projections of the burden of rabies. Trop Med Int Health 10 790 798
4. LyS
BuchyP
HengNY
OngS
ChhorN
2009 Rabies situation in cambodia. PLoS Negl Trop Dis 3 e511
5. NelLH
RupprechtCE
2007 Emergence of lyssaviruses in the Old World: the case of Africa. Curr Top Microbiol Immunol 315 161 193
6. BourhyH
ReynesJM
DunhamEJ
DacheuxL
LarrousF
2008 The origin and phylogeography of dog rabies virus. J Gen Virol 89 2673 2681
7. TalbiC
HolmesEC
de BenedictisP
FayeO
NakouneE
2009 Evolutionary history and dynamics of dog rabies virus in western and central Africa. J Gen Virol 90 783 791
8. LemeyP
RambautA
DrummondAJ
SuchardMA
2009 Bayesian phylogeography finds its roots. PLoS Comput Biol 5 e1000520
9. BiekR
HendersonJC
WallerLA
RupprechtCE
RealLA
2007 A high-resolution genetic signature of demographic and spatial expansion in epizootic rabies virus. Proc Natl Acad Sci U S A 104 7993 7998
10. BourhyH
KissiB
AudryL
SmreczakM
Sadkowska-TodysM
1999 Ecology and evolution of rabies virus in Europe. J Gen Virol 80 ( Pt 10) 2545 2557
11. RealLA
BiekR
2007 Spatial dynamics and genetics of infectious diseases on heterogeneous landscapes. J R Soc Interface 4 935 948
12. RealLA
HendersonJC
BiekR
SnamanJ
JackTL
2005 Unifying the spatial population dynamics and molecular evolution of epidemic rabies virus. Proc Natl Acad Sci U S A 102 12107 12111
13. DrummondAJ
RambautA
2007 BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol Biol 7 214
14. Ben NéfissaK
MoulinAM
DellagiK
2007 La rage en Tunisie au XIX siècle: recrudescence ou émergence? Gesnerus 64 173 192
15. WangTH
DonaldsonYK
BrettleRP
BellJE
SP
2001 Identification of shared populations of human immunodeficiency virus type 1 infecting microglia and tissue macrophages outside the central nervous system. J Virol 75 11686 11699
16. GrayRR
TatemAJ
LamersS
HouW
LaeyendeckerO
2009 Spatial phylodynamics of HIV-1 epidemic emergence in east Africa. Aids 23 F9 F17
17. McRaeBH
BeierP
2007 Circuit theory predicts gene flow in plant and animal populations. Proc Natl Acad Sci U S A 104 19885 19890
18. BalkDL
DeichmannU
YetmanG
PozziF
HaySI
2006 Determining global population distribution: methods, applications and data. Adv Parasitol 62 119 156
19. MininVN
SuchardM
2008 Counting labeled transitions in continuous-time Markov models of evolution. Journal of Mathematical Biology 56 391 412
20. HampsonK
DushoffJ
BinghamJ
BrucknerG
AliYH
2007 Synchronous cycles of domestic dog rabies in sub-Saharan Africa and the impact of control efforts. Proc Natl Acad Sci U S A 104 7717 7722
21. HampsonK
DushoffJ
CleavelandS
HaydonDT
KaareM
2009 Transmission dynamics and prospects for the elimination of canine rabies. PLoS Biol 7 e53
22. KitalaPM
McDermottJJ
ColemanPG
DyeC
2002 Comparison of vaccination strategies for the control of dog rabies in Machakos District, Kenya. Epidemiol Infect 129 215 222
23. WandelerAI
MatterHC
KappelerA
BuddeA
1993 The ecology of dogs and canine rabies: a selective review. Rev Sci Tech 12 51 71
24. ZinsstagJ
DurrS
PennyMA
MindekemR
RothF
2009 Transmission dynamics and economics of rabies control in dogs and humans in an African city. Proc Natl Acad Sci U S A 106 14996 15001
25. JenkinsSR
WinklerWG
1987 Descriptive epidemiology from an epizootic of raccoon rabies in the Middle Atlantic States, 1982–1983. Am J Epidemiol 126 429 437
26. WindiyaningsihC
WildeH
MeslinFX
SurosoT
WidarsoHS
2004 The rabies epidemic on Flores Island, Indonesia (1998–2003). J Med Assoc Thai 87 1389 1393
27. BourhyH
DacheuxL
StradyC
MaillesA
2005 Rabies in Europe in 2005. Euro Surveill 10 213 216
28. CleavelandS
KaareM
TiringaP
MlengeyaT
BarratJ
2003 A dog rabies vaccination campaign in rural Africa: impact on the incidence of dog rabies and human dog-bite injuries. Vaccine 21 1965 1973
29. ColemanPG
FevreEM
CleavelandS
2004 Estimating the public health impact of rabies. Emerg Infect Dis 10 140 142
30. KayaliU
MindekemR
HuttonG
NdoutamiaAG
ZinsstagJ
2006 Cost-description of a pilot parenteral vaccination campaign against rabies in dogs in N'Djamena, Chad. Trop Med Int Health 11 1058 1065
31. LardonZ
WatierF
BrunetA
BernèdeC
GoudalM
2010 Imported Episodic Rabies Increases Patient Demand for and Physician Delivery of Antirabies Prophylaxis. Plos Neglected Tropical Diseases in press
32. HufnagelL
BrockmannD
GeiselT
2004 Forecast and control of epidemics in a globalized world. Proc Natl Acad Sci U S A 101 15124 15129
33. RussellCA
SmithDL
WallerLA
ChildsJE
RealLA
2004 A priori prediction of disease invasion dynamics in a novel environment. Proc Biol Sci 271 21 25
34. HendersonJC
BiekR
HanlonCA
O'DeeS
RealLA
2008 Rabies virus in raccoons, Ohio, 2004. Emerg Infect Dis 14 650 652
35. TatemAJ
RogersDJ
HaySI
2006 Global transport networks and infectious disease spread. Adv Parasitol 62 293 343
36. KissiB
TordoN
BourhyH
1995 Genetic polymorphism in the rabies virus nucleoprotein gene. Virology 209 526 537
37. DelmasO
HolmesEC
TalbiC
LarrousF
DacheuxL
2008 Genomic diversity and evolution of the lyssaviruses. PLoS ONE 3 e2057
38. EdgarRC
2004 MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32 1792 1797
39. DrummondAJ
HoSY
PhillipsMJ
RambautA
2006 Relaxed phylogenetics and dating with confidence. PLoS Biol 4 e88
40. DrummondAJ
RambautA
ShapiroB
PybusOG
2005 Bayesian coalescent inference of past population dynamics from molecular sequences. Mol Biol Evol 22 1185 1192
41. SuchardMA
RambautA
2009 Many-core algorithms for statistical phylogenetics. Bioinformatics 25 1370 1376
42. O'BrienJD
MininVN
SuchardM
2009 Learning to count: robust estimates for labeled distances between molecular sequences. Molecular Biology and Evolution 26 801 814
43. SuchardM
KitchenC
SinsheimerJ
WeissR
2003 Hierarchical phylogenetic models for analyzing multipartite sequence data. Syst Biol 52 649 664
44. 2000 Road network data. VMAP0 (Vector Map Level 0) Highlands Ranch, Colorado LandInfo Worldwide Mapping
45. RedelingsBD
SuchardMA
2005 Joint Bayesian estimation of alignment and phylogeny. Syst Biol 54 401 418
46. NewtonM
RafteryA
1994 Approximate Bayesian inference with the weighted likelihood bootstrap. Journal of the Royal Statistical Society Series B 56 3 48
Štítky
Hygiena a epidemiologie Infekční lékařství Laboratoř
Článek The Combined Effect of Environmental and Host Factors on the Emergence of Viral RNA RecombinantsČlánek Functional Interchangeability of Late Domains, Late Domain Cofactors and Ubiquitin in Viral BuddingČlánek Parvovirus Minute Virus of Mice Induces a DNA Damage Response That Facilitates Viral ReplicationČlánek Phenolglycolipid-1 Expressed by Engineered BCG Modulates Early Interaction with Human Phagocytes
Článek vyšel v časopisePLOS Pathogens
Nejčtenější tento týden
2010 Číslo 10- Stillova choroba: vzácné a závažné systémové onemocnění
- Diagnostika virových hepatitid v kostce – zorientujte se (nejen) v sérologii
- Autoinflamatorní onemocnění: prognózu zlepšuje včasná diagnostika a protizánětlivá terapie
- Infekční komplikace virových respiračních infekcí – sekundární bakteriální a aspergilové pneumonie
- Kompletní remise ALK-pozitivního karcinomu plic u pacienta po mnoha liniích cílené léčby – kazuistika
-
Všechny články tohoto čísla
- Social Media and Microbiology Education
- Antimicrobial Peptides: Primeval Molecules or Future Drugs?
- Phylodynamics and Human-Mediated Dispersal of a Zoonotic Virus
- Retroviral RNA Dimerization and Packaging: The What, How, When, Where, and Why
- Distinct Clones of Caused the Black Death
- Strain-Specific Differences in the Genetic Control of Two Closely Related Mycobacteria
- The Combined Effect of Environmental and Host Factors on the Emergence of Viral RNA Recombinants
- MHC Class I Bound to an Immunodominant Epitope Demonstrates Unconventional Presentation to T Cell Receptors
- Stimulates Immune Gene Expression and Inhibits Development in
- Crystal Structure of DotD: Insights into the Relationship between Type IVB and Type II/III Secretion Systems
- Cytomegalovirus microRNAs Facilitate Persistent Virus Infection in Salivary Glands
- Strategies to Avoid Killing by Human Neutrophils
- Transforming Growth Factor-β: Activation by Neuraminidase and Role in Highly Pathogenic H5N1 Influenza Pathogenesis
- Autoimmunity in Arabidopsis Is Mediated by Epigenetic Regulation of an Immune Receptor
- Functional Interchangeability of Late Domains, Late Domain Cofactors and Ubiquitin in Viral Budding
- Dengue Virus Ensures Its Fusion in Late Endosomes Using Compartment-Specific Lipids
- Nucleocapsid Promotes Localization of HIV-1 Gag to Uropods That Participate in Virological Synapses between T Cells
- Host Genetics and HIV-1: The Final Phase?
- Variations in the Hemagglutinin of the 2009 H1N1 Pandemic Virus: Potential for Strains with Altered Virulence Phenotype?
- High-Resolution Functional Mapping of the Venezuelan Equine Encephalitis Virus Genome by Insertional Mutagenesis and Massively Parallel Sequencing
- Viral Replication Rate Regulates Clinical Outcome and CD8 T Cell Responses during Highly Pathogenic H5N1 Influenza Virus Infection in Mice
- Calcineurin Inhibition at the Clinical Phase of Prion Disease Reduces Neurodegeneration, Improves Behavioral Alterations and Increases Animal Survival
- Parvovirus Minute Virus of Mice Induces a DNA Damage Response That Facilitates Viral Replication
- -Induced Inactivation of the Macrophage Transcription Factor AP-1 Is Mediated by the Parasite Metalloprotease GP63
- Epstein Barr Virus-Encoded EBNA1 Interference with MHC Class I Antigen Presentation Reveals a Close Correlation between mRNA Translation Initiation and Antigen Presentation
- Fidelity Variants of RNA Dependent RNA Polymerases Uncover an Indirect, Mutagenic Activity of Amiloride Compounds
- The Spread of Tomato Yellow Leaf Curl Virus from the Middle East to the World
- Concerted Action of Two Formins in Gliding Motility and Host Cell Invasion by
- Requirements for Receptor Engagement during Infection by Adenovirus Complexed with Blood Coagulation Factor X
- Release of Intracellular Calcium Stores Facilitates Coxsackievirus Entry into Polarized Endothelial Cells
- Gene Annotation and Drug Target Discovery in with a Tagged Transposon Mutant Collection
- Nuclear Export and Import of Human Hepatitis B Virus Capsid Protein and Particles
- Retention and Loss of RNA Interference Pathways in Trypanosomatid Protozoans
- Identification and Genome-Wide Prediction of DNA Binding Specificities for the ApiAP2 Family of Regulators from the Malaria Parasite
- Controlling Cellular P-TEFb Activity by the HIV-1 Transcriptional Transactivator Tat
- Direct Visualization of Peptide/MHC Complexes at the Surface and in the Intracellular Compartments of Cells Infected by
- Phenolglycolipid-1 Expressed by Engineered BCG Modulates Early Interaction with Human Phagocytes
- In Vitro and In Vivo Studies Identify Important Features of Dengue Virus pr-E Protein Interactions
- Inhibition of Nipah Virus Infection In Vivo: Targeting an Early Stage of Paramyxovirus Fusion Activation during Viral Entry
- PLOS Pathogens
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Retroviral RNA Dimerization and Packaging: The What, How, When, Where, and Why
- Viral Replication Rate Regulates Clinical Outcome and CD8 T Cell Responses during Highly Pathogenic H5N1 Influenza Virus Infection in Mice
- Antimicrobial Peptides: Primeval Molecules or Future Drugs?
- Crystal Structure of DotD: Insights into the Relationship between Type IVB and Type II/III Secretion Systems
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Revma Focus: Spondyloartritidy
nový kurz
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



