-
Články
- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Shade Avoidance Components and Pathways in Adult Plants Revealed by Phenotypic Profiling
Because plants depend on light for photosynthesis, neighboring plant shade can be detrimental to survival. Many plants sense and respond to neighbor shade to compete for light. Although shade causes responses throughout the plant (collectively known as the shade avoidance syndrome or SAS), most SAS studies have been limited to single-gene analyses in seedlings. Here we move beyond these analyses by taking a multi-gene, multi-trait study of SAS across developmental stages. Recently, whole-genome studies examining large mutant collections have been exploited to determine the pathways and their interactions that combine to determine complex phenotypes. This type of analysis (phenotypic profiling) typically uses thousands of mutants and robotic phenotyping for assaying many characters in the multitude of mutant lines. In this paper, we develop a directed alternative that allows us to take a similar approach to understanding SAS. To reduce the number of mutants required for such an approach, we used a logical selection procedure to define mutants of interest by over-representation analysis of shade-responsive genes. We found at least three different subgroups of shade responses, and that each subgroup had both shared and separate pathways. Also, we found eighteen novel genes involved in SAS. Therefore, our method is useful for multi-dimensional phenotypic profiling without expensive robots.
Published in the journal: . PLoS Genet 11(4): e32767. doi:10.1371/journal.pgen.1004953
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1004953Summary
Because plants depend on light for photosynthesis, neighboring plant shade can be detrimental to survival. Many plants sense and respond to neighbor shade to compete for light. Although shade causes responses throughout the plant (collectively known as the shade avoidance syndrome or SAS), most SAS studies have been limited to single-gene analyses in seedlings. Here we move beyond these analyses by taking a multi-gene, multi-trait study of SAS across developmental stages. Recently, whole-genome studies examining large mutant collections have been exploited to determine the pathways and their interactions that combine to determine complex phenotypes. This type of analysis (phenotypic profiling) typically uses thousands of mutants and robotic phenotyping for assaying many characters in the multitude of mutant lines. In this paper, we develop a directed alternative that allows us to take a similar approach to understanding SAS. To reduce the number of mutants required for such an approach, we used a logical selection procedure to define mutants of interest by over-representation analysis of shade-responsive genes. We found at least three different subgroups of shade responses, and that each subgroup had both shared and separate pathways. Also, we found eighteen novel genes involved in SAS. Therefore, our method is useful for multi-dimensional phenotypic profiling without expensive robots.
Introduction
Plant canopy shade limits available light for photosynthesis. Because plants are sessile, this presents a particular challenge. Perhaps as a consequence plants developed a light-quality sensory system for canopy shade; perception of foiliar shade and/or reflection from neighbor plants (“neighbor detection”) can induce the shade avoidance syndrome (SAS) is collection of responses to canopy shade in plants. These SAS responses can be seen in all developmental stages from seeds to adult plants [1]. Various plant organs elongate under shade, including the hypocotyl (stem) of young seedlings, and the internodes, and leaf petioles of older plants. Furthermore shade induces upward leaf movement, accelerates flowering time (the developmental transition form vegetative phase to reproductive phase), suppresses shoot branching, and alters resource allocation [1]. All of these responses can be helpful for promoting survival when there is competition for light from neighboring plants.
Foliar shade, which has reduced photosynthetically active radiation (PAR) can be detected both by cryptochrome photoreceptors due to its reduced intensity of blue light and by phytochrome photoreceptors due to its reduced ratio of red to far-red light [1]. Remarkably, plants can perceive nearby neighbors even before true shading and the concomitant reduction in PAR. This type of neighbor detections is possible because, even though PAR is not reduced, light reflected from neighbors has a reduced ratio of red to far-red light detectable by phytochromes [1,2]. Here we use a neighbor-detection protocol to focus on phytochrome-mediated responses. Detailed analysis of shade induced hypocotyl elongation has revealed that light perception activates transcription factors (TF) that, in turn, modulate plant hormone pathways to promote organ growth. For example, the PHYTOCHROME-INTERACTING FACTOR (PIF) 5 TF protein is stabilized under shade and induces transcription of genes important for synthesis of the growth-promoting hormone auxin [3,4]. Another example is that upon shade treatment PIF7 is dephosphorylated and activated to induce YUCCA (YUC) 2, YUC5, YUC8, and YUC9 auxin biosynthetic genes [5].
Plant hormones regulate many aspects of development and growth. At least five plant hormone pathways are related to SAS; auxin, brassinosteroid (BR), gibberellic acid (GA), ethylene, and cytokinin (CK). Many auxin or BR responsive genes are induced by end-of-day far-red treatment (EODFR, a proxy for shade treatment) and both auxin (big and shade avoidance 3 (sav3) / tryptophan aminotransferase of Arabidopsis 1 (taa1)) and BR (rotundifolia3 (rot3)) mutant showed reduced shade-induced or EODFR-induced petiole elongation as well as shade-induced gene expression [6,7]. There is some evidence that GA and CK are involved in leaf SAS [8,9,10]. Shade also influences jasmonic-acid (JA) mediated plant immune system [11] and reduced volatile JA levels [12]. However, the entire network of light signaling and hormone pathways in regulation of shade avoidance are unclear. Also, the extent of shared and separate pathways for each shade avoidance response is not currently known. In part this is because each SAS mutant has been tested under different experimental conditions making it difficult to compare the phenotypic consequences of each mutant.
Phenotypic profiling of genetic mutants with high-throughput phenotyping is a powerful method to tease out complex gene networks [13]. Systematic phenotypic profiling of multiple traits originated with bacterial studies, followed by studies on single eukaryotic cells such as yeast and cultured animal cells [14]. Multi-dimensional phenotypic profiling of gene-perturbed multi-cellular organisms had been done both in invertebrates and vertebrates [15,16,17]. In plants phenotypic profiling of recombinant inbred lines or natural population has been conducted for QTL analysis or genome-wide association studies [18], but it has not yet been applied to induced mutants in plants.
Advances of automatic and robotic technologies made it possible to conduct high-throughput phenotyping. In plant, high-throughput robotic phenotyping systems had been reported (reviewed in [19,20]), but its use in research has only recently been published [21]. Profiling of multiple phenotypes in selected gene-perturbed plants has been reported in root epidermal cell patterning study [22] and red-light signaling [23,24], which showed effectiveness of reverse-genetics approaches to recover mutants of interest from transcriptome data.
Here we extend this approach to multiple phenotypes to develop a systems-level understanding of shade avoidance. We took advantage of a newly developed semi-automated leaf shape measurement system for throughput measurement of shade avoidance in leaves [25]. Furthermore, we aimed to narrow down candidate genes involved in SAS to be screened instead of screening of entire knockout mutant collection or mutagenized population. For this purpose, we selected candidate mutants based on over-representation analysis of shade-responsive genes in leaves. Our broad phenotypic profiling of hypocotyl, leaf, and flowering time in selected mutants has allowed us to dissect the complex shade avoidance syndrome network.
Results and Discussion
Shared and separate gene sets are enriched in shade-responsive genes in Arabidopsis seedling and leaf/apical region
To begin to define the genes required for SAS in leaf/apical region, after the seedling stage we performed an expression profiling experiment to find gene induces or repressed by simulated shade. Previous SAS expression profiling experiments have used microarrays and focused on seedlings or specifically on the petiole or leaf blade after EODFR treatment [6,7,26]. To obtain a broader view of expression changes in older plants, we harvested leaf and shoot apex tissue and used RNAseq (statistics are presented in S1 Table) since the most common Arabidopsis microarray (Affymetrix ATH1) only covers about 70% of defined genes in the transcriptome. We compared gene expression in samples treated with simulated shade (white light supplemented with far-red light to achieve R/FR of 0.5 and 80–100 μE PAR) for 1 hour or 4 hours to untreated control samples (R/FR = 1.9 and 80–100 μE PAR) and found a total of 164 and 97 genes to be differentially expressed (FDR <0.001; S2 Table see Materials and Methods). Most known shade induced genes in leaves were found in our list (bold text in S2 Table, 1 hour after onset of shade treatment) [3,4,7,27,28,29]. There is a high correlation of expression fold changes by shade treatment between our data and published microarray data in leaf [6] (S1 Fig), confirming that our RNA-seq based transcriptional analysis is reliable. Significant correlation of shade-responsive genes between seedlings and leaves indicates common mechanisms exist between these two organs (S1 Fig). In addition we identified 38 (1 hour treatment) and 19 (4 hour treatment) genes not present on the Arabidopsis ATH1 microarray, including the known shade induced gene PHYTOCHROME-INTERACTING-FACTOR-3 LIKE I (PIL1) [3,30], while 50 (1 hour) and 68 (4 hour) genes were on ATH1 but not previously found as shade-responsive genes in EODFR treated petiole or leaf (S2 Table).
GO enrichment analysis showed that plants respond to 1 hour and 4 hour shade treatment differently (Tables 1 and 2). Two GO terms common to both time points were GO:0009733 (response to auxin stimulus) and GO:0009753 (response to JA stimulus). Known shade avoidance related genes were also enriched in the 1 hour treatment, but not in the 4 hour treatment. Plant immune related pathways (GO:0009611 (response to wounding) and GO:0009617 (response to bacterium), which are related to the JA pathway, are enriched in the 4 hour treatment.
Tab. 1. GO category analysis of 1 hour shade-responsive genes in leaf/apical region. 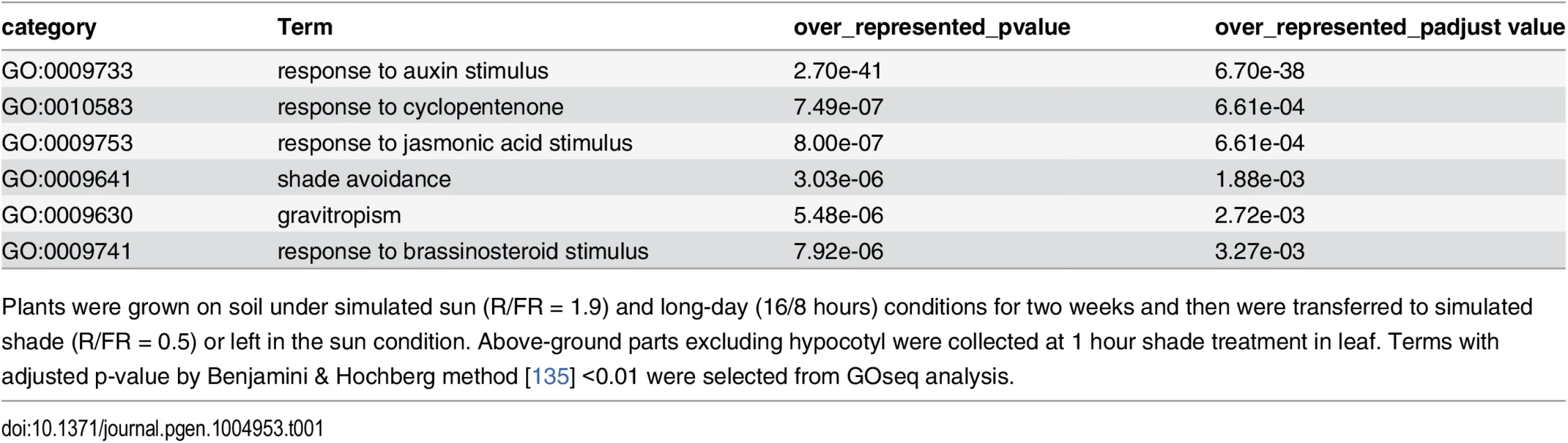
Plants were grown on soil under simulated sun (R/FR = 1.9) and long-day (16/8 hours) conditions for two weeks and then were transferred to simulated shade (R/FR = 0.5) or left in the sun condition. Above-ground parts excluding hypocotyl were collected at 1 hour shade treatment in leaf. Terms with adjusted p-value by Benjamini & Hochberg method [135] <0.01 were selected from GOseq analysis. Tab. 2. GO category analysis of 4 hour shade-responsive genes in leaf/apical region. 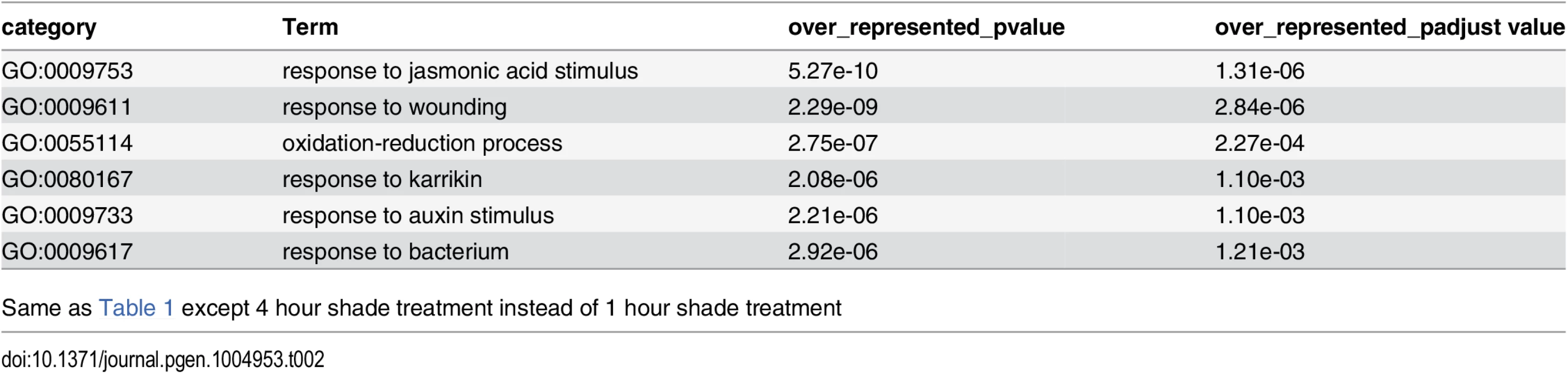
Same as Table 1 except 4 hour shade treatment instead of 1 hour shade treatment It has been described that some plant hormone pathways, such as auxin, BR, and GA, are involved in SAS [1]. To gain a better understanding of involvement of hormone pathways in SAS, over-representation analysis (ORA) was done to test if any hormone-responsive genes were enriched among the shade-regulated genes (Table 3). Consistent with ORA with previous microarray data of leaf upon EODFR treatment, auxin, BR, and JA pathways were enriched [6]. In addition, we found that ethylene and abscisic acid (ABA) pathways were enriched in our data sets. Involvement of ethylene in SAS was suggested because shade increases ambient ethylene levels in Sorghum [31] and tobacco [32]. However ethylene production is required only for early response to shade in petiole and stem, but not response in leaf angle [32], although ethylene induces leaf hyponasty [33], hypocotyl elongation [34], and stem elongation [35]. Involvement of ABA in SAS is known for shade-suppressed branching [36], but has not been reported to be involved in the shade-avoidance responses examined in this paper. Interestingly leaves of four-day shade treated tomato plants have increase level of ethylene precursor and ABA [37]. In summary, the significant differences in the shade-responsive transcriptome at these two time-points reflected dynamic temporal changes of early SAS signaling cascade.
Tab. 3. Over-representation analysis of shade-responsive genes and hormone responsive genes. 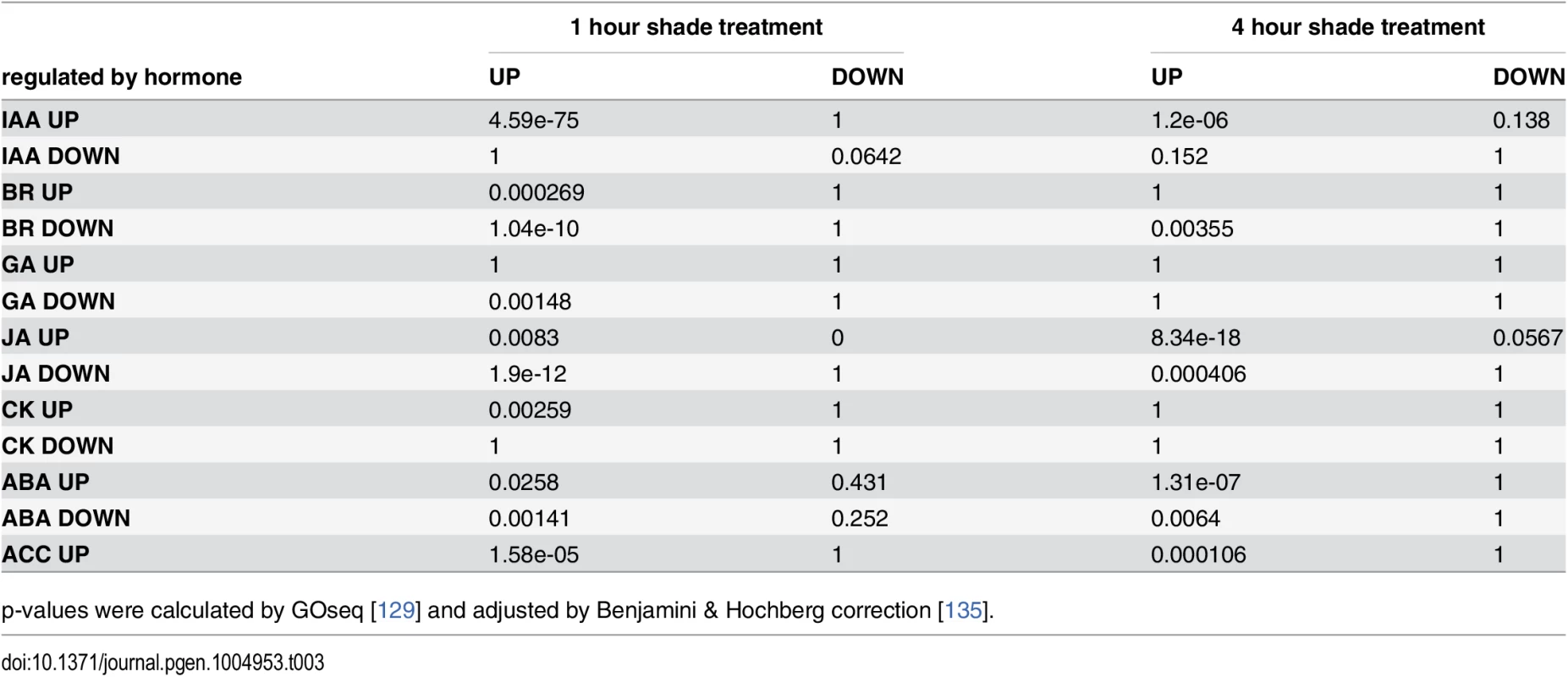
p-values were calculated by GOseq [129] and adjusted by Benjamini & Hochberg correction [135]. Next we asked if the previous seedling shade transcriptome data [7] shows the same trends as our data. We found that there were both common and specific GO terms between the hypocotyl and our leaf/apical region data sets (Table 1, 2, and 4). Of the common terms, we focused on GO:0009733 (response to auxin stimulus), GO:0009741 (response to BR stimulus), and GO:0009641 (shade avoidance). Among the leaf/apical region-specific terms GO:0009753 (response to JA stimulus) is of particular interest because the role of JA pathways in morphological aspects of SAS is not fully understood. Based on our ORA we chose to include mutants of genes in these categories in our phenotypic profiling (see below).
Tab. 4. GO category analysis of 1 hour shade-responsive genes in hypocotyl. 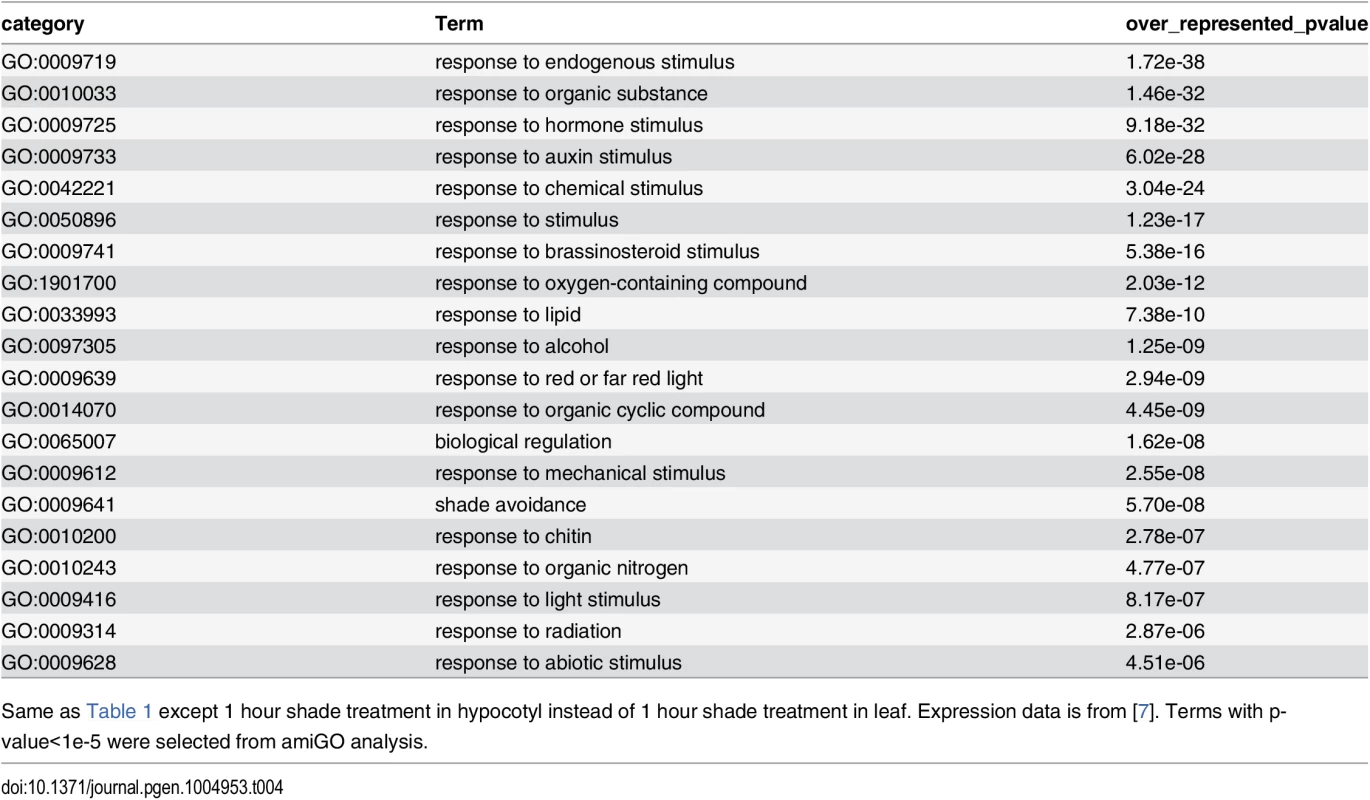
Same as Table 1 except 1 hour shade treatment in hypocotyl instead of 1 hour shade treatment in leaf. Expression data is from [7]. Terms with p-value<1e-5 were selected from amiGO analysis. Leaf phenotype profiling of fifty-nine mutant lines
We hypothesized that shade-responsive genes and/or pathways are required for proper SAS because among the 34 causal genes in known hypocotyl SAS mutants 11 (TAA1, PIN3, YUC2, YUC5, YUC8, YUC9, BIM1, GAI, PHYB, PAR1, HAT3) of them have shade-responsive transcripts (S3 Table). Based on our differentially expressed gene list and previous knowledge, we chose 59 mutant lines encompassing 59 mutant genes (although some lines have more than one mutant gene) from nine categories (auxin, GA, JA, BR, light signaling, shade avoidance, flowering time, leaf size, and unknown shade responsive genes) (Fig 1). We prioritized auxin and JA pathways because both pathways were enriched in both time points of our transcriptome analysis (see above).
Fig. 1. Mutants used in SAS phenotypic profiling. 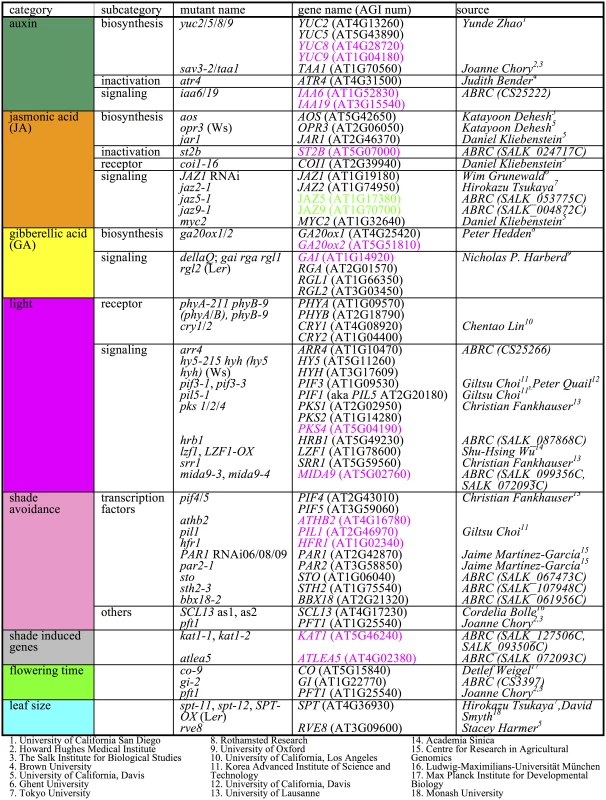
Shade-induced genes are shown in magenta bold and shade-repressed are shown in green. Background genotype for almost all mutants is Col (other genotypes are show in parentheses). Petiole elongation is an important component of SAS, but most leaf phenotype measurement software does not report petiole and blade length. Phenotyping of mutants/overexpressors with a pathway of interest is a direct method to test if the given gene in the pathway is involved in phenotype of your interest. At the first step towards high throughput petiole and blade phenotyping, we developed LeafJ, an ImageJ plug-in, which is more accurate and faster measurement system than manual method [25].
To normalize petiole elongation between genotypes with different leaf size, we calculated ratio of petiole length to blade length. Statistical significance between Col under sun treatment and shade treatment effects was examined by a mixed effects model (Fig 2). Mutants that showed a statistically different response to shade when compared to the corresponding wild-type (P<0.05; see Methods) were considered to have a significant SAS phenotype. In Col, we could detect significant shade-induced petiole elongation and an increase of the petiole length to leaf blade length ratio (S2F Fig and S3 Table), but not in other leaf blade parameters (length, width, and area) (S2C–S2E Fig). Some other studies have reported that leaf area does increase or decrease upon shade treatment; the differences between these studies and ours may be due to differences in plant growth conditions or because the developmental stage of our leaves might be too young to show these responses [7,38,39,40]. Comparing the kinetics of leaf development under both light condition could tell us when shade-responsive organ elongation happens and should be examined in future studies.
Fig. 2. Phenotypic profiling of 59 mutants/overexpressors. 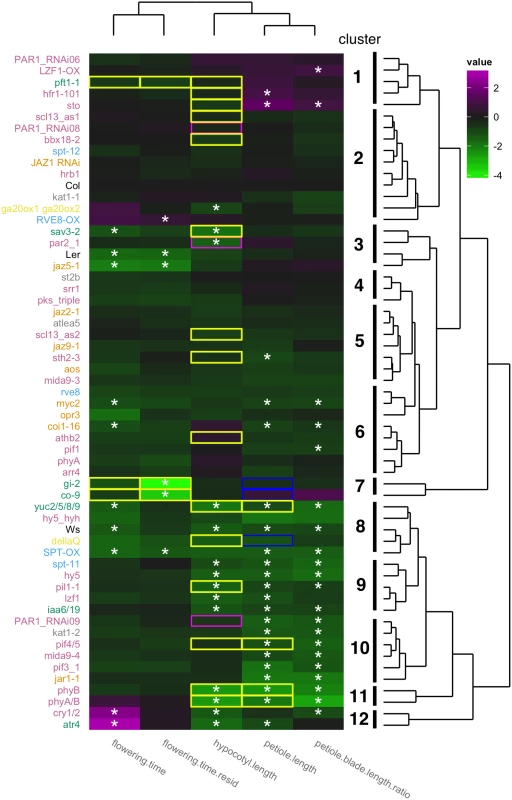
For hypocotyl phenotype, plants were grown under continuous simulated sun conditions (R/FR = 1.3) for four days and further grown for three days under either simulated sun or simulated shade (R/FR = 0.5). For leaf phenotypes, plants were grown under long day conditions (16 hour light/8 hour dark) with approximately 90 μE PAR (R/FR = 1.9). Two week old plants were further grown for 12 days under either simulated sun (R/FR = 1.9) or simulated shade (R/FR = 0.5). For flowering time phenotype plants were grown in the same condition with leaf phenotyping and days after stratification at bolted was used for flowering time index. For phenotype clustering heatmap, differences between shade and sun values (except flowering time) were normalized and centered on Col (i.e., Col value = 0) and visualized with color coding (magenta indicates larger response than Col while green indicates reduced response relative to Col). For flowering time, responses to shade were normalized against flowering time under sun condition to eliminate strong dependencies of response on flowering time under sun condition (see S2H Fig for normalized data and S2G Fig for non normalized data). Asterisks (*) from hypocotyl.length to flowering.time indicate a significant difference from corresponding wild type (p-value<0.05). Although SPT_ox appeared to be a SAS mutant in flowering time (residual method, S3H Fig), it was eliminated from SAS mutants in flowering time (residual method) because its background genotype (Ler) also showed a similar shift from the regression line. Colors of mutant names correspond to groups found in Fig 1. Clustering of mutants according to its phenotype is shown in the dendgrogram on the right. Clustering of traits is show on the top dendrogram. Known phenotypes for hypocotyl, petiole, or flowering time are shown in colored boxes (yellow-green for less response, blue for normal response, and magenta for exaggerated response). Screening these 59 mutant lines, we found 33 mutants that showed differences in at least one trait from the background ecotype (Fig 2, and S3 Table). These 33 mutants include genes in the auxin, jasmonic acid, and light signaling pathways. Involvement of auxin in petiole elongation upon far-red light treatment has been previously reported [5], whereas there have not been any reports of JA involvement in petiole shade avoidance. Details of these mutants will be discussed below. It is important to note that we assay a leaf series from each plant where some leaves are still expanding and others are mature. As a consequence, the mutations that we have identified could be affecting leaf development itself or developmental timing (the proportion of expanding to expanded leaves).
Shade-induced acceleration of flowering time of fifty-nine mutant lines
Compared to other SAS phenotypes, only seven mutant lines for acceleration of flowering time have been described; phytochrome mutants (phytochromeB (phyB) phyD, phyB/D/E) [41,42], known flowering time mutants (constans (co) and gigantea (gi)) [43], a circadian clock component (early flowering 3 (elf3)) [44], an auxin-related mutant (big1) [45], and a mediator complex mutant (phytochrome and flowering time 1 (pft1)) [46]. Our SAS phenotypic profiling provided additional mutants for shade-accelerated flowering. In our condition shade treatment accelerated flowering time about 10% in Col, which is less effective than previously reported (35–40% [42,43,47]). This small acceleration is because our shade treatment started when plants were already close to flowering in long day conditions. Perhaps because of this we observed a strong inverse correlation between flowering time shade response and flowering time in sun (S3H Fig; genotypes that flowered later in sun were more responsive to shade). Given this correlation we defined two categories flowering time shade response mutants. One category (“flowering.time” in Fig 2) consists of mutants whose log2 shade response is different from Col-0. The second category (“flowering.time.resid” in Fig 2) are those mutants whose response is significantly different from that expected based on their flowering time in the sun (see Methods). Even though our experimental conditions were suboptimal for detecting acceleration of flowering time and we could not reproduce reduced responses to shade of pft1, we did find six new flowering time SAS mutants; cryptochrome (cry) 1 cry2 (cry1/2), altered-tryptophan regulation 4 (atr4), reveille 8/lhy-cca1-like 5 (rve8/lcl5), coronatine insensitive 1 (coi1), myc2/jasmonate insensitive1 (jin1), and jasmonate-zim-domain protein 5 (jaz5) (Fig 2, and S3 Table). Details of these mutants will be discussed below.
Separate and overlapping pathways for three different shade avoidance responses
To investigate how many of the genes under study are required for normal shade avoidance response in both the leaf and the hypocotyl, we assayed hypocotyl SAS in the same 59 mutant panel (S3A Fig, Fig 1, and S3 Table). Among the eighteen previously reported hypocotyl SAS mutants tested, we observed altered SAS phenotypes in six mutants (28%). This relatively low validation rate is probably due to differences in growth conditions such as day length, the ratio of red to far-red used for sun and shade, or whether shade was applied throughout the day or simulated by EODFR. In addition to previously described SAS mutants, we discovered seven mutant lines with previously unknown hypocotyl SAS defects (gibberellin 20-oxidase (ga20ox) 1 ga20ox2 (ga20ox1/2), phytochrome rapidly regulated (par) 2–1, indole-3-acetic acid inducible (iaa) 6 iaa19 (iaa6/19), light-regulated zinc finger protein 1 (lzf1), spatula (spt), elongated hypocotyl 5 (hy5), cry1/2, and atr4).
Our mutant phenotypic profiling also revealed that two different indices for one phenotype (“petiole length” and ratio of “petiole blade length ratio” for SAS in petiole; “flowering.time” and “flowering.time.resid” for SAS in flowering time) were clustered together, two elongation phenotypes (hypocotyl elongation and petiole elongation) were clustered while the two elongation phenotypes and flowering time were distinct (top dendrogram in Fig 2). Examining which genes have mutant phenotypes for each trait revealed that there are common and separate pathways for each shade avoidance response (Fig 2 and S3 Table). Auxin-related genes are required for all responses; phytochromes and three TFs (LZF1, HY5, and PIL1) were involved in both hypocotyl and leaf responses; cryptochromes are involved in both hypocotyl and flowering-time response; and JA related pathways were involved in petiole and flowering time responses. Twelve genes were required specifically for petiole response and two genes were required specifically for hypocotyl response (Fig 2 and S2 Table). Our results also showed that our strategy was powerful for identifying new genes required for these shade avoidance responses, including genes in light, auxin, JA, and BR pathways (S2 Table). Details of those genes will be discussed in following sections.
Differences in the sets of genes required for shade avoidance response in hypocotyls and leafs are consistent with GO analysis of shade-responsive genes in hypocotyl or leaf tissue (Table 1, 2, and 4). For example GO:0009753 (response to JA stimulus) is only found in leaf data sets and we found that mutants affecting the JA pathway only affected SAS phenotypes in adult plants. There is a previous report of JA affecting seedling SAS, but this was under extremely low R:FR (0.068) and JA was found to act by modulating phyA signaling [48]. Under the more moderate low R:FR conditions used in this study and for the seedling microarray assays [7], phyA is not involved (Fig 2) and phyB is the major receptor for shade. Thus, under moderate shade conditions, JA is likely to affect adult rather than seedling SAS.
Our findings that pathways of flowering time in response to shade were different from those of hypocotyl are consistent with previous data. For example, altered ARABIDPSIS THALIANA HOMEOBOX PROTEIN 2 expression changed shade-avoidance responses in hypocotyl [49] but not in flowering time [43]. In addition, supressor of phytochrome a-105 (spa1/2/3/4) quadruple mutant and constitutive photomorphogenesis 1 (cop1) show shade-induced acceleration of flowering time, but did not show shade-induced hypocotyl elongation [50].
Updated shade avoidance syndrome pathways
Among 59 tested mutant lines we could detect 33 mutants with defects in at least one shade avoidance response, including 20 new mutant lines (Fig 2). A schematic diagram of SAS signaling pathways is shown in Fig 3. SAS phenotypes with mutants used in this study and known SAS mutants are summarized in S3 Table. Below we discuss details of the pathways corresponding to each mutant category in Fig 1 and then discuss phenotypic clustering of SAS mutants (Fig 2).
Fig. 3. Schematic representation of proposed signal transduction for shade-avoidance syndrome. 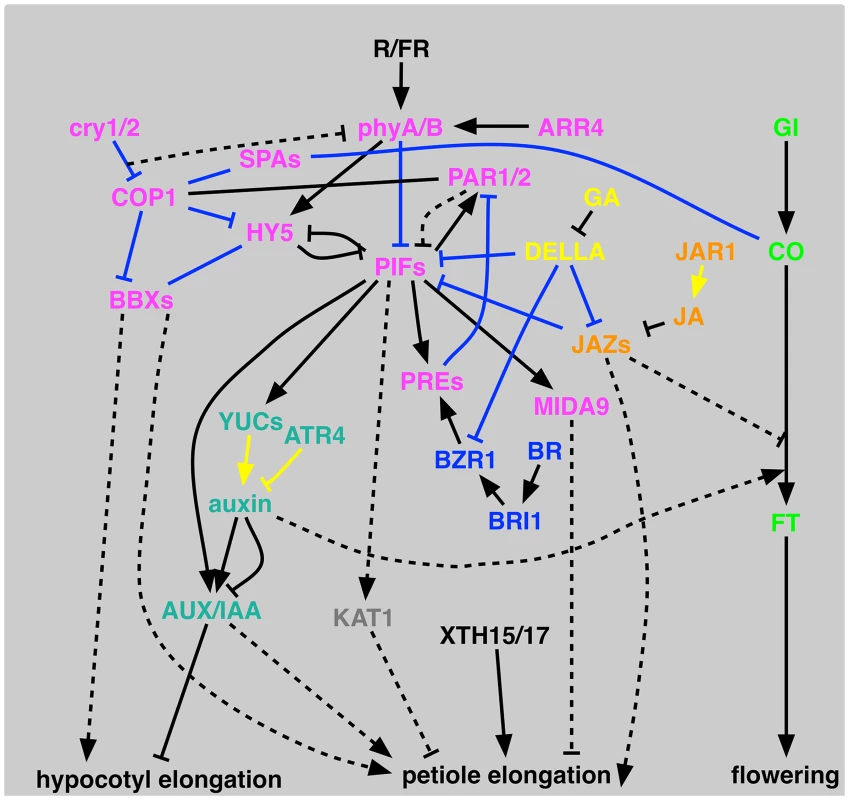
PIFs represent PIF3, PIF4, PIF5, PIF7, PIL1, and SPT. JAZs represents JAZ5 and JAZ10. Black lines represent genetic interactions, blue lines show direct interactions, and yellow lines show hormone biosynthesis/metabolism. Lines without arrowheads are for interactions where directionality is unknown. Dashed lines show hypothetical interactions. SCL13 is omitted because of its unknown function within this context. Light signaling pathway
We simulated canopy shade solely by changing the R/FR ratio, which is perceived by phytochromes. Five PHY genes in Arabidopsis thaliana have partially overlapping functions [51]. Our phenotypic profiling showed PHYB is the major photoreceptor and PHYA has minor function with PHYB in the SAS responses of hypocotyl and leaf in low R/FR condition because phyA mutant phenotypes could be observed only in a phyB mutant background, consistent with previous data [1] (S3 Table, S3A, S3B, and S3F Fig). The situation is different for flowering time where PHYB does not dominate the response, instead it is redundant with PHYD and PHYE [47]. This is consistent with our data that phyB and even phyA/B showed normal shade acceleration of flowering time (Fig 2).
We found that the CRY blue-light photoreceptors were important for proper hypocotyl and flowering time SAS under simulated canopy shade. At first this is surprising because our simulated sun and shade conditions only have altered R/FR ratios but have the same blue irradiance. However, CRY1 and CRY2 blue light photoreceptors of Arabidopsis thaliana are known to interact genetically with phytochrome signaling in other photomorphogenic responses [52,53,54,55,56,57,58,59,60]. There are numerous molecular events known to occur upon phytochrome activation that may explain functional interaction between phytochromes and cryptochromes. One example is light-dependent PHYB-CRY1 interaction [61] and light dependent CRY-COP1 E3 ligase interaction [62]. Recent studies showed COP1 and its interacting proteins SPA1/2/3/4 are also required for SAS in hypocotyl [50,63,64], so that COP1-SPAs system could be a key component of phytochrome-cryptochrome interaction observed in our data. Although the cry1/2 double showed reduced SAS in hypocotyl and reduced acceleration of flowering time, the mode of action for these two traits are different. cry1/2 hypocotyls showed constitutive shade avoidance, while cry1/2 showed constitutive exaggerated “sun” phenotype for flowering time, indicating that the interaction between phytochrome and cryptochrome signaling are different between these two traits. Possible mechanisms of the differences could be related to photoperiod-dependent flowering time regulation by CRY2 [65]. CRY2 interacts with SPA in blue-light dependent manner, which prevents CO degradation by COP1-SPAs [66]. Also CRY activates a subset of bHLH TFs in a blue light dependent manner; these bHLHs in turn activate the flowering time master gene, FT [67]. Since cryptochromes are required to sense depleted blue light under canopy shade [68,69], plants may have evolved a cross-talk system between phytochrome and cryptochrome signaling systems to coordinate shade avoidance syndromes response in natural condition.
Among TFs in light signaling pathways that we tested (Fig 1), our data showed that HY5, LZF1, PIL1, and SPT were required in common between hypocotyl and petiole responses, while PIF1, PIF3, PIF4, PIF5, PAR1, LONG HYPOCOTYL IN FAR-RED (HFR1), SALT TOLERANCE (STO), and SALT TOLERANCE HOMOLOG2/B-BOX DOMAIN PROTEIN 21 (STH2/BBX21) were required in petiole response and PAR2 was required in hypocotyl.
Among them SPT is of particular interest because of its unique spatial expression pattern at the boundary between the leaf blade and petiole, where petiole cell proliferation occurs [70]. Thus the spatial expression pattern of SPT could explain the reduced SAS petiole elongation phenotype of spt-11. Also the severe allele of SPT mutant (spt-11) showed less shade avoidance responses than mild allele (spt-12) (S3 Table), consistent with the reported differences of petiole length and leaf blade area phenotype between spt-11 and spt-12 [70]. SPT regulates gynoecium development by activating genes involved in shade avoidance [71]. Similar mechanisms of SPT regulation of shade avoidance are likely conserved in shade-induced petiole elongation.
It is well recognized that PIFs are growth-promoting proteins whose activities are modulated by a variety of environmental factors [72]. In addition to PIF4 and PIF5, our data showed that both PIF1 and PIF3 are new positive regulators of SAS in petiole (Fig 2). PIF1 is known to regulate hypocotyl growth [72,73] as well as non-growth related processes such as chlorophyll biosynthesis [74]. PIF3 is also known to regulate hypocotyl growth (reviewed in [75]), but the role of either gene in regulating adult plant growth is not known. We confirmed the pif3 result by examining a second allele, pif3-3 [76], and found that pif3-3 also had a significant petiole SAS defect (S4 Fig; p<0.01). In hypocotyl growth, PIF1/3/4/5 regulate overlapping and specific target genes [77], and it seems likely that many of these targets also contribute to petiole SAS.
We found that a knock-out mutant of a PP2C-type phosphatase gene, MISREGULATED IN DARK (MIDA) 9, showed reduced petiole shade response. MIDA9 has been shown to be involved in hook formation and its expression is regulated by PIF3 [23], consistent with our finding that pif3 mutant also showed reduced petiole shade response. We previously found that MIDA9 is induced by PIF4 and/or PIF5 during hypocotyl growing phase [78], suggesting that MIDA9 is involved in growth-control networks. Another link is inactivation of H+-ATPase activity (required for cell wall loosening) via dephosphorylation of H+-ATPase by MIDA9 phosphatase 2C-typeD [79]. It would be interesting to know if other PIFs and SPT controls MIDA9 expression.
Auxin pathways
Previous studies have implicated auxin as being important in hypocotyl and leaf SAS [80]. Here we extend those studies to identify specific auxin signaling components important for SAS in various organs. Consistent with prior studies [4,5,6,7], both our transcriptome data and mutant phenotypic profiling showed auxin pathways were involved in hypocotyl and leaf shade avoidance responses. By assaying many auxin pathway genes we can begin to assign shade avoidance functions to specific components.
Our data showed that components of auxin pathway required for each response are different. For example, two different gene families in one auxin biosynthesis pathway contribute differently in three separate shade avoidance responses. Although TAA1 protein catalyzes auxin biosynthesis in the same pathway as YUC proteins [81,82], TAA1 was required only for hypocotyl shade avoidance response, while YUC2/5/8/9 genes were required for all shade avoidance responses (Fig 2; see Müller-Moulé et al., submitted, for a more detailed analysis). These differences could be explained by lower gene expression level of TAA1 in leaves, in contrast of ubiquitous expression of YUC2/5/8/9 (S5 Fig). Among shade-induced AUX/IAA family genes (see S2 Table), we tested the function of IAA6, and IAA19. The iaa6/19 double showed a defect only in hypocotyl and leaf SAS, but not in acceleration of flowering time. Therefore IAA6/IAA19 are required for response to shade in elongation. Similar to other auxin-related phenotypes, these shade-induced AUX/IAA genes likely function redundantly with other family members in the other two responses [83].
From transcriptome data it is not clear whether the auxin pathway is involved in acceleration of flowering time by shade, but phenotypic profiling indicates its involvement in flowering time control in response to shade. An auxin overproducing mutant (atr4) [84] and reduced auxin level mutant (yuc2/5/8/9 and sav3/taa1) showed opposite flowering responses, supporting a role of auxin in this phenomenon (Fig 2).
How are light signaling and auxin pathways connected? All available data suggested that shade increases active auxin content by inducing auxin biosynthesis genes (YUCs) [80]. Among the PIFs, connection of PIF4/5/7 and SPT to auxin has been reported [4,5,78,85,86,87]. At least some SAS traits are reduced in single or multiple pif4/5/7 mutants (summarized in S3 Table). In our study, yuc2/5/8/9 mutant is defective in all three shade-avoidance responses, which is consistent with upregulation of YUC2/5/8/9 genes by PIF7 [5] and YUC5/8/9 by PIF5 [4]. Interaction of TERMINAL FLOWER 2 (TFL2) protein with IAA6 and IAA19 could be another pathway to link SAS and auxin [88], although we were not able to test TFL2 in this study. Nevertheless, tfl2 showed reduced shade avoidance (EODFR response) in hypocotyl [88].
JA pathway
Evidence showing a link between light and JA pathways has been accumulating for some time (reviewed in [89]). For example JA mediated defense responses are attenuated in shade [11]. This effect is mediated by COI1-JAZ10-dependent, salicylic acid-independent mechanisms [90]. Previous work also implicated JA signaling in phyA mediated responses to very low R:FR [48]. In tomato, stems of plants treated with shade for four days showed reduced JA levels [37]. Our transcriptome ORA with hormone responsive genes also shows shade-attenuation of the JA pathway (Table 3).
While it is known that shade attenuates JA responses, we used JA pathway mutants (Fig 1) to probe which JA components are required for SAS. Because PHYA is not important for the moderately low R/FR used in our study (see “light signaling pathway” section above) this enables us to determine whether JA is important for the typical PHYB mediated shade responses examined here. Our phenotypic profiling of these mutants showed that the JA pathway is involved in shade avoidance responses in leaf and flowering time, but we did not find significant effects on hypocotyl response (Fig 2, and S3 Table). This is consistent with GO analysis of transcriptomes of hypocotyl and juvenile plants (Table 1, 2, and 4) where JA GO categories were over-represented in the leaf/apical region but not in the hypocotyl data set. Reduced JA mediated plant immunity by shade is found in adult plants, but it is not known if this is true for hypocotyl. Comparison of shade-responsive genes between hypocotyl and leaf/apical region and comparison of phenotypic profiling predict that JA mediated plant immunity in hypocotyl is not affected by shade. In conclusion, we found that interaction of JA pathways and SAS network were not unidirectional but bidirectional.
Interestingly, partially different subsets of JA pathway genes are required for normal leaf and flowering shade response: COI1 and JASMONATE RESISTANT 1 (JAR1) for leaf response and COI1 and JAZ5 for flowering time response. What might drive these differences? JAR1 encodes a protein that catalyzes conjugation of Ile to JA to produce active JA, which is required for JA mediated immunity [89]. Therefore one possibility is that JA-Ile is the active component for leaf response while other active JA-related compounds such as OPDA or cis-jasmone [89], could be the active components for flowering time. Another possibility is that JAZ5 acts in a flowering time specific pathway.
It is interesting that JA biosynthesis mutants (allene oxide synthase (aos) and 12-oxophytodienoate-reductase 3 (opr3)) still retained weak shade-avoidance responses although a JA receptor mutant (coi1-16) had a defect of shade avoidance responses in petiole and flowering time (Fig 2). The fact that opr3 mutants retaining JA biosynthesis in certain condition [91] may explain this. Alternatively, native JA related compounds released from neighbor plants in our experiments might partially rescue our JA biosynthesis mutants.
MYC2 is a basic helix-loop-helix (bHLH) TF important for JA mediated immune responses that acts redundantly with its homologs, MYC3 and MYC4 [92]. Mutation in MYC2 slightly reduced SAS in adult plants (Fig 2, and S3 Table), raising the possibility that the MYC2/3/4 redundancy is also true for SAS. PHYTOCHROME AND FLOWERING TIME 1 (PFT1) encodes the conserved MED25 subunit of the mediator complex, which is involved in JA-mediated immune system as well as light-signaling pathways [46,93,94,95]. Our hypothesis that pft1 mutant impairs SAS was not confirmed because pft1 did not show detectable differences compared with Col in any phenotypes we tested.
Our phenotypic profiling data showed less contribution of JA related-genes to shade-induced hypocotyl elongation than expected from previous data. For example, it was previously reported that exogenous MeJA application inhibited hypocotyl growth [96] and that this effect was influenced by PHYB [97]. Another examples is that coi1 mutants showed an increases response to low R/FR [48]. The inconsistency with our results could be due to differences between exogenous JA application in the previous study versus the use of JA pathway mutants in our study, or due to differences in light conditions: previous studies used continuous monochromatic red light or extremely low R/FR (0.068), while our study used low R/FR (0.5). Therefore how light quality influences JA-dependent SAS is of future interest.
How JA pathways modulate petiole elongation is also unclear. Possible mechanism is promotion of PIF activities by binding of JAZ proteins to DELLA proteins that suppress PIF protein activities) [98]. Another link between JA and growth is that exogenous MeJA delays the start of endoreduplication cycle [99]. Interaction of these factors is of future interest.
Gibberellic acid pathways
The GA pathway is involved in shade-induced hypocotyl and petiole elongation as shown by GA biosynthesis inhibitor treatments and a GA deficient mutant (ga requiring 1 (ga1-3)) [8]. The GA signaling DELLA gene family is reported to have a weak effect in hypocotyl SAS and none in petiole SAS [8]. We used another GA deficient mutant line (ga20ox1/2) [78,100] and a quadruple DELLA mutant line (dellaQ) [101] to test if GA is involved in other SAS traits (Fig 2). Similar to ga1-3, ga20ox1/2 showed reduced response to shade in hypocotyl (Fig 2). In contrast to ga1-3, ga20ox1/2 showed shade-induced petiole elongation even though both organs were about 60% of wild-type size (S3A and S3B Fig). In agreement of previous studies, dellaQ showed very weak influence on shade avoidance responses of hypocotyl and petioles in our condition (Fig 2, S3A, and S3B Fig). Both GA deficient and GA signaling mutants showed normal shade response for flowering time (Fig 2 and S3 Table), suggesting that GA pathway is not involved in flowering time acceleration by shade.
Circadian clock pathways
We found that over-expression of RVE8, a circadian clock component, caused moderately exaggerated shade responses in flowering time (Fig 2). This is consistent with the finding that natural variation of ELF3 modulates flowering time response to shade [44]. Recent studies showed direct targets of RVE8 TF [102]. Among them PSEUDO RESPONSE REGULATOR5 (PRR5), another circadian clock component, is of interest because PRR5 is activated by RVE8 [102] and knocking out PRR5 caused exaggerated shade avoidance response in petiole possibly through elevation of PIF4 and PIF5 expression levels [103]. Also it has been shown that circadian modulation of hypocotyl response to shade is mediated by circadian clock regulation of PIL1 expression [30]. However, our shade avoidance assay for flowering time showed pif4/5 and pil1 had normal flowering time shade responses. It would be interesting to test if a RVE8—PRR5 pathway contributes to shade response in flowering time.
Novel component
Our mutant analysis defined a new components of shade avoidance responses; a potassium channel gene (POTASSIUM CHANNEL IN ARABIDOPSIS THALIANA 1, KAT1). The KAT1 gene encodes a potassium channel gene, which is locally expressed in guard cells [104,105]. KAT1 potassium channel is inactivated upon ABA treatment and mediates stomata closure [106]. KAT1 is thought to be important for vegetative growth [107], but its molecular mechanisms are unknown. Recently ABA was shown to be involved in suppression of branching by shade [36]. It is not clear if ABA pathways were involved in shade avoidance responses described in this paper, although ABA responsive genes were enriched in shade responsive genes (see above). Interestingly KAT1 gene expression is induced by auxin [108,109], which may be related to its role in petiole SAS.
Phenotypic-clustered SAS mutants may share common functions
In C. elegans, multiple phenotypic profiling showed that mutants with similar phenotypic profiles function in shared pathways [16]. In our phenotypic clusters (Fig 2), two flowering time mutant lines (gi-2 and co-9) cluster together. These are known from prior studies to act in photoperiodic induction of flowering, and to have reduced shade response for flowering time but not petiole elongation [42] (cluster 7), showing proof of concept. In our data, clusters that consist of SAS mutants are of particular interest (clusters 9, 10, 11, and 12) because they could indicate shared membership in sub-networks of the shade avoidance pathway. Therefore, the following genes likely function in common sub-networks: IAA6, IAA19, PIL1, LZF1, HY5, and SPT in cluster 9 (hyp and pet), and PAR1, KAT1, PIF4, PIF5, MIDA9, PIF3, and JAR1 in cluster 10 (pet only). Opposite effects of mutations were found in cluster 12; mutation caused reduced responses in hypocotyl and petiole, while its caused exaggerated responses in flowering time. Molecular networks within these clusters are of future interests.
Mutants affecting of flowering time regulation under simulated sun
We observed many genotypes that showed altered flowering time in sun condition (S6 Fig). In this section, we will discuss about known components and then novel components in flowering time pathways.
Known components
As reported known knock out of master regulators of flowering time (CO and GI) showed late flowering in our long-day condition [110,111,112]. Light signaling has been known to modulate flowering time [113]. Phytochromes affect flowering time by post-transcriptional regulation and cryptochromes affect flowering time by both transcriptional and post-transcriptional regulation [62]. We observed that phyB, phyA/B, and cry1/2 mutants showed flowering time phenotype as reported. Early flowering phenotype of hy5 hy5 homolog (hyh) is also reported [114]. PIF4 has been known to induce a master regulator of flowering time, FT gene, upon raised temperature [115]. pif4/5 mutant showed late flowering time, so that it is likely that PIF4 and/or PIF5 induce FT gene expression in our condition.
GA is essential for flowering time control. The GA biosynthesis double mutant (ga20ox1/2) showed delayed flowering in our experiment, consistent with earlier reports [100]. However, this mutant showed normal shade effects on flowering time (see above).
The circadian clock is also important for flowering time control. As reported, we found opposite phenotypes of rve8 (early flowering) and RVE8-OX (late flowering) [116].
New components
It is surprising that auxin was involved in flowering time regulation, since it is not incorporated into current models of flowering time pathways (reviewed in [113]). However, exogenous auxin has been reported to delay flowering time, perhaps due to induced damage on plants [117]. This result is consistent with our auxin biosynthesis mutant flowering time data; overproduction delayed flowering time (atr4) while reduced production accelerated flowering time (yuc2/5/8/9 and taa1). These results suggest that it is necessary to reinvestigate the involvement of auxin pathways in flowering time.
It has been shown that coi1 mutants flowered early [48,98]. We observed that other JA mutants also show altered flowering time. Not only JA biosynthesis mutants (aos and opr3) and JA receptor mutant (coi1-16), but also three JA signaling component mutants (JAZ1 RNAi, jaz5-1, myc2) showed early flowering phenotypes under sun condition. Early flowering of myc2 is inconsistent with a previous report showing its late flowering phenotype [118] possibly due to different photoperiods. It will be interesting to investigate how JA pathways regulate flowering time. One possibility is that JA pathways interact with GA pathways in control of flowering, similar to their interaction in growth control [98].
Our data showed that two SPT knock out mutants (spt-11 and spt-12), PAR1 RNAi, sth2, and two KAT1 knock out mutants (kat1-1 and kat1-2) showed early flowering phenotype, while SCL13 anti-sense line 1 (scl13 as1) and hfr1 showed late flowering phenotype. Early flowering of spt-11 has been reported [119] and that was confirmed by another allele (spt-12) in our study. Early flowering phenotype of PAR1 RNAi, sth2, and kat1-1 have not been previously reported and their connections to the flowering time pathway are unknown.
Conclusion
Here we showed that RNA-seq followed by phenotypic profiling is a powerful approach for elucidating complex SAS pathways and discovery of new SAS components. A similar approach was successful in searching new components of de-etiolation of seedlings, a developmental stage with a simple architecture [24]. Our study expanded this approach to show the transcriptome-based discovery of new mutants are also effective for complex syndrome by multiple phonotypic profiling.
After our phenotypic profiling, an additional SAS mutant line has been reported which contained genes that were also in our shade-responsive genes. Specifically, we found that BR ENHANCED EXPRESSION 3 (BEE3), a bHLH TF, was induced by shade (S2 Table). It was recently showed that the bee1 bee2 bee3 triple mutant has altered hypocotyl SAS [120], perhaps due to the altered BR signaling in this triple [121]. This example is additional evidence that our strategy is effective to find novel SAS mutants. Further analysis is needed for elucidating interactions between these genes and/or pathways. Our approaches are straightforward and cost effective, so that these should be applicable to other cases in general.
We found that the effects of some mutations were context-dependent (only found for some organs or developmental stages) whereas others were ubiquitous. Those mutations that affect all organs points to shared mechanisms underlying the SAS in different organs. The mutations that have context-dependent effects could indicate unique genes functioning in the different organs or more quantitative differences in the relative importance of the components in different organs. Regardless the fact that we did find organ-specific effects suggests that we need to be cautious when generalizing conclusions from hypocotyl studies.
Materials and Methods
Light condition
For simulated sun condition, white light (cool-white fluorescent light) was supplemented with far-red light (provided by LEDs (Orbitec, inc) to obtain R/FR = 1.86. For simulated shade condition, white light was supplemented with far-red LEDS to obtain R/FR = 0.52. Both condition had 80–100 μE of Photosynthetically Active Radiation (PAR). Plants were grown under long day condition (16 hour light/8 hour dark) at constant temperature (22°C). For hypocotyl experiments, seedlings were grown under simulated sun (R/FR = 1.3) or simulated shade condition (R/FR = 0.5) with combination of LED lights (Quantum Devices Snap-Lite) [122]. Ambient light spectrum was measured by Black-Comet (StellarNet, Florida).
Plant materials
Arabidopsis seed stocks used in this study are listed in Fig 1. To confirm genotypes of T-DNA insertion lines ordered from Arabidopsis Biological Resource Center (ABRC), genomic DNA was extracted (DNeasy Plant Mini kit, Qiagen) and subject to genomic PCR. cDNA was synthesized by direct mRNA extraction [123] and quantitative PCR (qPCR) was done with homemade SYBR green master mix with the iCycler Multicolor real-time PCR detection system (Bio-Rad). For kat1 mutants, standard RT-PCR was done. Primers used for genomic PCR and (q)RT-PCR and their results were summarized in S4 Table. Arabidopsis seeds were imbibed with water on filter papers and stored them at 4°C for four days. Three days after stratification under sun condition, three germinated seeds were transferred to soil in a well of 5x10 well flat. Fourteen days after stratification, excess seedlings were removed to leave one well-grown plant per pot and the flats were transferred to either sun or shade condition. For hypocotyl growth measurements, seeds were grown on vertical square plates [124] with 1/2 MSMO, 5 mM 2-(N-morpholino)ethanesulfonic acid (MES, pH = 5.8, Sigma), and 0.8% agar (Sigma). Each plate was divided into three rows and two columns and in six spaces five or six seeds of six genotypes were sown. Genotype positions were randomized in repeated sets. 4593 seedling images were taken by a scanner and hypocotyl length was measured by ImageJ (http://rsb.info.nih.gov/ij/) [125].
RNA-seq library preparation and sequencing
For RNA extraction, plants were treated with shade starting at ZT 4 or left in the sun. We prepared two replicates of each sample at 1 hour and 4 hours after sun and shade treatment and five plants were pooled for each replicate. Cotyledons, hypocotyls, and roots were removed from the samples, leaving leaves and apical tissue. Total RNA from the plants was extracted using RNeasy Plant Mini kit (Qiagen) with DNAse treatment (Qiagen). Five μg total RNA was used to construct mRNA library using mRNA-Seq-8 sample Prep kit (Illumina). The resulting cDNA libraries were sequenced by Illumina GAIIx with 40 bp single end mode. Basic statistics of mapping results are given in S1 Table.
Differential expression analysis and over-representation analysis (ORA)
Reads after sorting according to barcodes were subjected to removal of adaptor contamination by custom Perl scripts. Reads were mapped by TopHat [126] to Arabidopsis reference genome using known annotation (TAIR10). Differentially expressed genes were extracted by edgeR package [127] in R statistical environment [128] (FDR <0.001). ORA was done by GOseq package [129] in R statistical environment. GO analysis was done by using GO category database package from Bioconductor (org.At.tair.db and ANNOTATE package). For ORA of hormone responsive genes custom categories were used as defined in Supplemental Table S9 in [130] and Supplemental S1 Table in [131]. GO analysis of shade-responsive genes in hypocotyl [7] was done using the GO Web site (http://amigo.geneontology.org/cgi-bin/amigo/term_enrichment; [132]).
Phenotype measurement and analysis
For scoring leaf phenotypes, 26 day old plants were dissected and leaf images were recorded by a flatbed scanner (Epson, Perfection V700 PHOTO). Scanned images were measured using ImageJ [125] and the LeafJ plugin [25] to determine petiole length, leaf blade length, leaf blade width, and leaf blade area. Days to bolting was scored to measure flowering time. Leaf phenotypes (petiole length, leaf blade length, leaf blade width, leaf blade area) were measured from 10 sets of experiments with 1268 plants in total. For flowering time (days to bolting) measurement, 1950 plants were measured in total. Each phenotype was fitted by mixed effects model, i.e.
where plant is a mutant/overexpressor, treatment is sun or shade condition, plant:treatment is interaction of “plant” and “treatment”, (treatment|set) is the random effect associated with the treatment in set of experiments, and ε is the error. The model was applied to each trait to calculate coefficient (“sun” value). For leaf traits where we measured across multiple leaves (from leaf 3 to leaf 6) for a given trait we treated leaf as a random effect, using the following modelMutants were considered to have a defect in SAS when the plant:treatment term was significant (P<0.05), indicating that the genotype of the plant (mutant versus wild-type) affected the response to shade.
For flowering time, days to bolting was log2 transformed. We found that acceleration of flowering time by shade treatment was strongly correlated with days to bolting in sun condition, i.e., late flowering mutants had more shade-accelerated flowering time than Col (S2G Fig). To address this issue we regressed flowering time shade response on average sun flowering time for each genotype and calculated the residuals from the regression [122]. These residuals represent the amount of flowering time shade response that was not predicted by the sun flowering time. The residuals for shade treated plants were then used in the mixed effects model (S3H Fig).
The lme4 (R package version 1.0–6) [133] and lmerTest [134] packages in R was used for these analyses. All phenotyping data is summarized in S5 Table.
Heatmaps for phenotypic clustering were drawn after scaling each trait data and centered at Col.
All R scripts for this paper and raw data are available at https://bitbucket.org/knozue/sasphenotyping.
Accession numbers
RNA-seq data in this study have been deposited in the NCBI SRA (Study ID PRJNA214254) and the NCBI GEO database (accession GSE66967). Mutants used in this study are listed in Fig 1.
Supporting Information
Zdroje
1. Casal JJ (2013) Photoreceptor signaling networks in plant responses to shade. Annu Rev Plant Biol 64 : 403–427. doi: 10.1146/annurev-arplant-050312-120221 23373700
2. Ballaré CL, Sánchez RA, Scopel AL, Casal JJ, Ghersa CM (1987) Early detection of neighbour plants by phytochrome perception of spectral changes in reflected sunlight. Plant Cell Environ 10 : 551–557.
3. Lorrain S, Allen T, Duek PD, Whitelam GC, Fankhauser C (2008) Phytochrome-mediated inhibition of shade avoidance involves degradation of growth-promoting bHLH transcription factors. Plant J 53 : 312–323. 18047474
4. Hornitschek P, Kohnen MV, Lorrain S, Rougemont J, Ljung K, et al. (2012) Phytochrome interacting factors 4 and 5 control seedling growth in changing light conditions by directly controlling auxin signaling. Plant J 71 : 699–711. doi: 10.1111/j.1365-313X.2012.05033.x 22536829
5. Li L, Ljung K, Breton G, Schmitz RJ, Pruneda-Paz J, et al. (2012) Linking photoreceptor excitation to changes in plant architecture. Genes Dev 26 : 785–790. doi: 10.1101/gad.187849.112 22508725
6. Kozuka T, Kobayashi J, Horiguchi G, Demura T, Sakakibara H, et al. (2010) Involvement of auxin and brassinosteroid in the regulation of petiole elongation under the shade. Plant Physiol 153 : 1608–1618. doi: 10.1104/pp.110.156802 20538889
7. Tao Y, Ferrer JL, Ljung K, Pojer F, Hong F, et al. (2008) Rapid synthesis of auxin via a new tryptophan-dependent pathway is required for shade avoidance in plants. Cell 133 : 164–176. doi: 10.1016/j.cell.2008.01.049 18394996
8. Djakovic-Petrovic T, Wit Md, Voesenek LACJ, Pierik R (2007) DELLA protein function in growth responses to canopy signals. Plant J 51 : 117–126. 17488236
9. Kurepin LV, Emery RJN, Pharis RP, Reid DM (2007) The interaction of light quality and irradiance with gibberellins, cytokinins and auxin in regulating growth of Helianthus annuus hypocotyls. Plant Cell Environ 30 : 147–155. 17238906
10. Carabelli M, Possenti M, Sessa G, Ciolfi A, Sassi M, et al. (2007) Canopy shade causes a rapid and transient arrest in leaf development through auxin-induced cytokinin oxidase activity. Genes Dev 21 : 1863–1868. 17671088
11. Moreno JE, Tao Y, Chory J, Ballare CL (2009) Ecological modulation of plant defense via phytochrome control of jasmonate sensitivity. Proc Natl Acad Sci U S A 106 : 4935–4940. doi: 10.1073/pnas.0900701106 19251652
12. Kegge W, Weldegergis BT, Soler R, Vergeer-Van Eijk M, Dicke M, et al. (2013) Canopy light cues affect emission of constitutive and methyl jasmonate-induced volatile organic compounds in Arabidopsis thaliana. New Phytol 200 : 861–874. doi: 10.1111/nph.12407 23845065
13. Sozzani R, Benfey P (2011) High-throughput phenotyping of multicellular organisms: finding the link between genotype and phenotype. Genome Biol 12 : 219. doi: 10.1186/gb-2011-12-3-219 21457493
14. Winzeler EA, Shoemaker DD, Astromoff A, Liang H, Anderson K, et al. (1999) Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science 285 : 901–906. 10436161
15. Neumann B, Walter T, Heriche JK, Bulkescher J, Erfle H, et al. (2010) Phenotypic profiling of the human genome by time-lapse microscopy reveals cell division genes. Nature 464 : 721–727. doi: 10.1038/nature08869 20360735
16. Green RA, Kao H-L, Audhya A, Arur S, Mayers JR, et al. (2011) A high-resolution C.elegans essential gene network based on phenotypic profiling of a complex tissue. Cell 145 : 470–482. doi: 10.1016/j.cell.2011.03.037 21529718
17. Yanik MF, Rohde CB, Pardo-Martin C (2011) Technologies for micromanipulating, imaging, and phenotyping small invertebrates and vertebrates. Annu Rev Biomed Eng 13 : 185–217. doi: 10.1146/annurev-bioeng-071910-124703 21756142
18. Atwell S, Huang YS, Vilhjalmsson BJ, Willems G, Horton M, et al. (2010) Genome-wide association study of 107 phenotypes in Arabidopsis thaliana inbred lines. Nature 465 : 627–631. doi: 10.1038/nature08800 20336072
19. Berger B, Regt B, Tester M (2012) High-throughput phenotyping of plant shoots. In: Normanly J, editor. High-throughput phenotyping in plants: Humana Press. pp. 9–20.
20. Spalding EP, Miller ND (2013) Image analysis is driving a renaissance in growth measurement. Curr Opin Plant Biol 16 : 100–104. doi: 10.1016/j.pbi.2013.01.001 23352714
21. Dornbusch T, Lorrain S, Kuznetsov D, Fortier A, Liechti R, et al. (2012) Measuring the diurnal pattern of leaf hyponasty and growth in Arabidopsis—a novel phenotyping approach using laser scanning. Funct Plant Biol 39 : 860–869.
22. Bruex A, Kainkaryam RM, Wieckowski Y, Kang YH, Bernhardt C, et al. (2012) A gene regulatory network for root epidermis cell differentiation in Arabidopsis. PLoS Genet 8: e1002446. doi: 10.1371/journal.pgen.1002446 22253603
23. Sentandreu M, Martin G, Gonzalez-Schain N, Leivar P, Soy J, et al. (2011) Functional profiling identifies genes involved in organ-specific branches of the PIF3 regulatory network in Arabidopsis. Plant Cell 23 : 3974–3991. doi: 10.1105/tpc.111.088161 22108407
24. Khanna R, Shen Y, Toledo-Ortiz G, Kikis EA, Johannesson H, et al. (2006) Functional profiling reveals that only a small number of phytochrome-regulated early-response genes in Arabidopsis are necessary for optimal deetiolation. Plant Cell 18 : 2157–2171. 16891401
25. Maloof JN, Nozue K, Mumbach MR, Palmer CM (2013) LeafJ: An ImageJ plugin for semi-automated leaf shape measurement. J Vis Exp 71: e50028.
26. Sessa G, Carabelli M, Sassi M, Ciolfi A, Possenti M, et al. (2005) A dynamic balance between gene activation and repression regulates the shade avoidance response in Arabidopsis. Genes Dev 19 : 2811–2815. 16322556
27. Carabelli M, Morelli G, Whitelam G, Ruberti I (1996) Twilight-zone and canopy shade induction of the Athb-2 homeobox gene in green plants. Proc Natl Acad Sci U S A 93 : 3530–3535. 11607652
28. Roig-Villanova I, Bou-Torrent J, Galstyan A, Carretero-Paulet L, Portoles S, et al. (2007) Interaction of shade avoidance and auxin responses: a role for two novel atypical bHLH proteins. EMBO J 26 : 4756–4767. 17948056
29. Sorin C, Salla-Martret M, Bou-Torrent J, Irma Roig-Villanova I, MartÌnez-GarcÌa J (2009) ATHB4, a regulator of shade avoidance, modulates hormone response in Arabidopsis seedlings. Plant J 59 : 266–277. doi: 10.1111/j.1365-313X.2009.03866.x 19392702
30. Salter MG, Franklin KA, Whitelam GC (2003) Gating of the rapid shade-avoidance response by the circadian clock in plants. Nature 426 : 680–683. 14668869
31. Finlayson SA, Lee IJ, Mullet JE, Morgan PW (1999) The mechanism of rhythmic ethylene production in sorghum. The role of phytochrome B and simulated shading. Plant Physiol 119 : 1083–1089. 10069847
32. Pierik R, Cuppens MLC, Voesenek LACJ, Visser EJW (2004) Interactions between ethylene and gibberellins in phytochrome-mediated shade avoidance responses in tobacco. Plant Physiol 136 : 2928–2936. 15448197
33. Millenaar FF, Cox MC, van Berkel YE, Welschen RA, Pierik R, et al. (2005) Ethylene-induced differential growth of petioles in Arabidopsis. Analyzing natural variation, response kinetics, and regulation. Plant Physiol 137 : 998–1008. 15728343
34. Smalle J, Haegman M, Kurepa J, Van Montagu M, Straeten DVD (1997) Ethylene can stimulate Arabidopsis hypocotyl elongation in the light. Proc Natl Acad Sci U S A 94 : 2756–2761. 11038610
35. Pierik R, Whitelam GC, Voesenek LACJ, de Kroon H, Visser EJW (2004) Canopy studies on ethylene-insensitive tobacco identify ethylene as a novel element in blue light and plant-plant signalling. Plant J 38 : 310–319. 15078333
36. Reddy SK, Holalu SV, Casal JJ, Finlayson SA (2013) Abscisic acid regulates axillary bud outgrowth responses to the ratio of red to far-red light. Plant Physiol 163 : 1047–1058. doi: 10.1104/pp.113.221895 23929720
37. Cagnola J, Ploschuk E, Benech-Arnold T, Finlayson S, Casal JJ (2012) Stem transcriptome reveals mechanisms to reduce the energetic cost of shade-avoidance responses in tomato. Plant Physiol 160 : 1110–1119. doi: 10.1104/pp.112.201921 22872775
38. Nagatani A, Chory J, Furuya M (1991) Phytochrome B is not detectable in the hy3 mutant of Arabidopsis, which Is deficient in responding to end-of-day far-red light treatments. Plant Cell Physiol 32 : 1119–1122.
39. Reed JW, Nagpal P, Poole DS, Furuya M, Chory J (1993) Mutations in the gene for the red/far-red light receptor phytochrome B alter cell elongation and physiological responses throughout Arabidopsis development. Plant Cell 5 : 147–157. 8453299
40. Robson P, Whitelam GC, Smith H (1993) Selected components of the shade-avoidance syndrome are displayed in a normal manner in mutants of Arabidopsis thaliana and Brassica rapa deficient in phytochrome B. Plant Physiol 102 : 1179–1184. 12231894
41. Devlin PF, Robson PR, Patel SR, Goosey L, Sharrock RA, et al. (1999) Phytochrome D acts in the shade-avoidance syndrome in Arabidopsis by controlling elongation growth and flowering time. Plant Physiol 119 : 909–915. 10069829
42. Wollenberg AC, Strasser B, Cerdan PD, Amasino RM (2008) Acceleration of flowering during shade avoidance in Arabidopsis alters the balance between FLOWERING LOCUS C-mediated repression and photoperiodic induction of flowering. Plant Physiol 148 : 1681–1694. doi: 10.1104/pp.108.125468 18790998
43. Kim SY, Yu X, Michaels SD (2008) Regulation of CONSTANS and FLOWERING LOCUS T expression in response to changing light quality. Plant Physiol 148 : 269–279. doi: 10.1104/pp.108.122606 18667727
44. Jimenez-Gomez JM, Wallace AD, Maloof JN (2010) Network analysis identifies ELF3 as a QTL for the shade avoidance response in Arabidopsis. PLoS Genet 6: e1001100. doi: 10.1371/journal.pgen.1001100 20838594
45. Kanyuka K, Praekelt U, Franklin KA, Billingham OE, Hooley R, et al. (2003) Mutations in the huge Arabidopsis gene BIG affect a range of hormone and light responses. Plant J 35 : 57–70. 12834402
46. Cerdan PD, Chory J (2003) Regulation of flowering time by light quality. Nature 423 : 881–885. 12815435
47. Franklin KA, Praekelt U, Stoddart WM, Billingham OE, Halliday KJ, et al. (2003) Phytochromes B, D, and E act redundantly to control multiple physiological responses in Arabidopsis. Plant Physiol 131 : 1340–1346. 12644683
48. Robson F, Okamoto H, Patrick E, Harris SR, Wasternack C, et al. (2010) Jasmonate and phytochrome A signaling in Arabidopsis wound and shade responses are integrated through JAZ1 stability. Plant Cell 22 : 1143–1160. doi: 10.1105/tpc.109.067728 20435902
49. Steindler C, Matteucci A, Sessa G, Weimar T, Ohgishi M, et al. (1999) Shade avoidance responses are mediated by the ATHB-2 HD-zip protein, a negative regulator of gene expression. Development 126 : 4235–4245. 10477292
50. Rolauffs S, Fackendahl P, Sahm J, Fiene G, Hoecker U (2012) Arabidopsis COP1 and SPA genes are essential for plant elongation but not for acceleration of flowering time in response to a low red light to far-red light ratio. Plant Physiol 160 : 2015–2027. doi: 10.1104/pp.112.207233 23093358
51. Franklin KA, Quail PH (2010) Phytochrome functions in Arabidopsis development. J Exp Bot 61 : 11–24. doi: 10.1093/jxb/erp304 19815685
52. Neff MM, Chory J (1998) Genetic interactions between phytochrome A, phytochrome B, and cryptochrome 1 during Arabidopsis development. Plant Physiol 118 : 27–35. 9733523
53. Casal JJ, Mazzella MA (1998) Conditional synergism between cryptochrome 1 and phytochrome B is shown by the analysis of phyA, phyB, and hy4 simple, double, and triple mutants in Arabidopsis. Plant Physiol 118 : 19–25. 9733522
54. Casal JJ, Boccalandro H (1995) Co-action between phytochrome B and HY4 in Arabidopsis thaliana. Planta 197 : 213–218. 8547813
55. Ahmad M, Cashmore AR (1997) The blue-light receptor cryptochrome 1 shows functional dependence on phytochrome A or phytochrome B in Arabidopsis thaliana. Plant J 11 : 421–427. 9107032
56. Hennig L, Funk M, Whitelam GC, Schafer E (1999) Functional interaction of cryptochrome 1 and phytochrome D. Plant J 20 : 289–294. 10571889
57. Castillon A, Shen H, Huq E (2009) Blue light induces degradation of the negative regulator phytochrome interacting factor 1 to promote photomorphogenic development of Arabidopsis seedlings. Genetics 182 : 161–171. doi: 10.1534/genetics.108.099887 19255368
58. Usami T, Matsushita T, Oka Y, Mochizuki N, Nagatani A (2007) Roles for the N - and C-terminal domains of phytochrome B in interactions between phytochrome B and cryptochrome signaling cascades. Plant Cell Physiol 48 : 424–433. 17251203
59. Usami T, Mochizuki N, Kondo M, Nishimura M, Nagatani A (2004) Cryptochromes and phytochromes synergistically regulate Arabidopsis root greening under blue light. Plant Cell Physiol 45 : 1798–1808. 15653798
60. Meijer G, Engelsma G (1965) The synergistic influence of a pre-irradiation on the photoinhibitton of gherkin seedlings. Photochem Photobiol 4 : 251–258.
61. Hughes RM, Vrana JD, Song J, Tucker CL (2012) Light-dependent, dark-promoted interaction between Arabidopsis cryptochrome 1 and phytochrome B proteins. J Biol Chem 287 : 22165–22172. doi: 10.1074/jbc.M112.360545 22577138
62. Liu H, Liu B, Zhao C, Pepper M, Lin C (2011) The action mechanisms of plant cryptochromes. Trends Plant Sci 16 : 684–691. doi: 10.1016/j.tplants.2011.09.002 21983106
63. Crocco CD, Holm M, Yanovsky MJ, Botto JF (2010) AtBBX21 and COP1 genetically interact in the regulation of shade avoidance. Plant J 64 : 551–562. doi: 10.1111/j.1365-313X.2010.04360.x 21070414
64. Pacín M, Legris M, Casal JJ (2013) COP1 re-accumulates in the nucleus under shade. Plant J 75 : 631–641. doi: 10.1111/tpj.12226 23647163
65. Guo H, Yang H, Mockler TC, Lin C (1998) Regulation of flowering time by Arabidopsis photoreceptors. Science 279 : 1360–1363. 9478898
66. Zuo Z, Liu H, Liu B, Liu X, Lin C (2011) Blue light-dependent interaction of CRY2 with SPA1 regulates COP1 activity and floral initiation in Arabidopsis. Curr Biol 21 : 841–847. doi: 10.1016/j.cub.2011.03.048 21514160
67. Liu Y, Li X, Li K, Liu H, Lin C (2013) Multiple bHLH proteins form heterodimers to mediate CRY2-dependent regulation of flowering-time in Arabidopsis. PLoS Genet 9: e1003861. doi: 10.1371/journal.pgen.1003861 24130508
68. Keller MM, Jaillais Y, Pedmale UV, Moreno JE, Chory J, et al. (2011) Cryptochrome 1 and phytochrome B control shade-avoidance responses in Arabidopsis via partially independent hormonal cascades. Plant J 67 : 195–207. doi: 10.1111/j.1365-313X.2011.04598.x 21457375
69. Keuskamp DH, Sasidharan R, Vos I, Peeters AJ, Voesenek LA, et al. (2011) Blue-light-mediated shade avoidance requires combined auxin and brassinosteroid action in Arabidopsis seedlings. Plant J 67 : 208–217. doi: 10.1111/j.1365-313X.2011.04597.x 21457374
70. Ichihashi Y, Horiguchi G, Gleissberg S, Tsukaya H (2010) The bHLH transcription factor SPATULA controls final leaf size in Arabidopsis thaliana. Plant Cell Physiol 51 : 252–261. doi: 10.1093/pcp/pcp184 20040585
71. Reymond MC, Brunoud G, Chauvet A, Martinez-Garcia JF, Martin-Magniette ML, et al. (2012) A light-regulated genetic module was recruited to carpel development in Arabidopsis following a structural change to SPATULA. Plant Cell 24 : 2812–2825. doi: 10.1105/tpc.112.097915 22851763
72. Leivar P, Tepperman JM, Cohn MM, Monte E, Al-Sady B, et al. (2012) Dynamic antagonism between phytochromes and PIF family basic helix-loop-helix factors induces selective reciprocal responses to light and shade in a rapidly responsive transcriptional network in Arabidopsis. Plant Cell 24 : 1398–1419. doi: 10.1105/tpc.112.095711 22517317
73. Soy J, Leivar P, Monte E (2014) PIF1 promotes phytochrome-regulated growth under photoperiodic conditions in Arabidopsis together with PIF3, PIF4, and PIF5. J Exp Bot 65 : 2925–2936. doi: 10.1093/jxb/ert465 24420574
74. Huq E, Al-Sady B, Hudson M, Kim C, Apel K, et al. (2004) Phytochrome-interacting factor 1 is a critical bHLH regulator of chlorophyll biosynthesis. Science 305 : 1937–1941. 15448264
75. Leivar P, Monte E (2014) PIFs: Systems Integrators in Plant Development. The Plant Cell Online 26 : 56–78. doi: 10.1105/tpc.113.120857 24481072
76. Monte E, Tepperman JM, Al-Sady B, Kaczorowski KA, Alonso JM, et al. (2004) The phytochrome-interacting transcription factor, PIF3, acts early, selectively, and positively in light-induced chloroplast development. Proc Natl Acad Sci U S A 101 : 16091–16098. 15505214
77. Zhang Y, Mayba O, Pfeiffer A, Shi H, Tepperman JM, et al. (2013) A quartet of PIF bHLH factors provides a transcriptionally centered signaling hub that regulates seedling morphogenesis through differential expression-patterning of shared target genes in Arabidopsis. PLoS Genet 9: e1003244. doi: 10.1371/journal.pgen.1003244 23382695
78. Nozue K, Harmer SL, Maloof JN (2011) Genomic analysis of circadian clock-, light-, and growth-correlated genes reveals PHYTOCHROME-INTERACTING FACTOR5 as a modulator of auxin signaling in Arabidopsis. Plant Physiol 156 : 357–372. doi: 10.1104/pp.111.172684 21430186
79. Spartz AK, Ren H, Park MY, Grandt KN, Lee SH, et al. (2014) SAUR inhibition of PP2C-D phosphatases activates plasma membrane H+-ATPases to promote cell expansion in Arabidopsis. Plant Cell 26 : 2129–2142. 24858935
80. de Wit M, Lorrain S, Fankhauser C (2014) Auxin-mediated plant architectural changes in response to shade and high temperature. Physiol Plant 151 : 13–24. doi: 10.1111/ppl.12099 24011166
81. Mashiguchi K, Tanaka K, Sakai T, Sugawara S, Kawaide H, et al. (2011) The main auxin biosynthesis pathway in Arabidopsis. Proc Natl Acad Sci U S A 108 : 18512–18517. doi: 10.1073/pnas.1108434108 22025724
82. Won C, Shen X, Mashiguchi K, Zheng Z, Dai X, et al. (2011) Conversion of tryptophan to indole-3-acetic acid by TRYPTOPHAN AMINOTRANSFERASES OF ARABIDOPSIS and YUCCAs in Arabidopsis. Proc Natl Acad Sci U S A 108 : 18518–18523. doi: 10.1073/pnas.1108436108 22025721
83. Overvoorde PJ, Okushima Y, Alonso JM, Chan A, Chang C, et al. (2005) Functional genomic analysis of the AUXIN/INDOLE-3-ACETIC ACID gene family members in Arabidopsis thaliana. Plant Cell 17 : 3282–3300. 16284307
84. Delarue M, Prinsen E, Va H, Onckelen, Caboche M, et al. (1998) Sur2 mutations of Arabidopsis thaliana define a new locus involved in the control of auxin homeostasis. Plant J 14 : 603–611. 9675903
85. Franklin KA, Lee SH, Patel D, Kumar SV, Spartz AK, et al. (2011) Phytochrome-interacting factor 4 (PIF4) regulates auxin biosynthesis at high temperature. Proc Natl Acad Sci U S A 108 : 20231–20235. doi: 10.1073/pnas.1110682108 22123947
86. Nemhauser JL, Feldman LJ, Zambryski PC (2000) Auxin and ETTIN in Arabidopsis gynoecium morphogenesis. Development 127 : 3877–3888. 10952886
87. Foreman J, White J, Graham I, Halliday K, Josse EM (2011) Shedding light on flower development: phytochrome B regulates gynoecium formation in association with the transcription factor SPATULA. Plant Signal Behav 6 : 471–476. 21364315
88. Valdes AE, Rizzardi K, Johannesson H, Para A, Sundas-Larsson A, et al. (2011) Arabidopsis thaliana TERMINAL FLOWER2 is involved in light-controlled signalling during seedling photomorphogenesis. Plant Cell Environ 35 : 1013–1025.
89. Wasternack C, Hause B (2013) Jasmonates: biosynthesis, perception, signal transduction and action in plant stress response, growth and development. An update to the 2007 review in Annals of Botany. Ann Bot 111 : 1021–1058. doi: 10.1093/aob/mct067 23558912
90. Cerrudo I, Keller MM, Cargnel MD, Demkura PV, de Wit M, et al. (2012) Low red/far-red ratios reduce Arabidopsis resistance to Botrytis cinerea and jasmonate responses via a COI1-JAZ10-dependent, salicylic acid-independent mechanism. Plant Physiol 158 : 2042–2052. doi: 10.1104/pp.112.193359 22371506
91. Chehab EW, Kim S, Savchenko T, Kliebenstein D, Dehesh K, et al. (2011) Intronic T-DNA insertion renders Arabidopsis opr3 a conditional jasmonic acid-producing mutant. Plant Physiol 156 : 770–778. doi: 10.1104/pp.111.174169 21487047
92. Fernandez-Calvo P, Chini A, Fernandez-Barbero G, Chico JM, Gimenez-Ibanez S, et al. (2011) The Arabidopsis bHLH transcription factors MYC3 and MYC4 are targets of JAZ repressors and act additively with MYC2 in the activation of jasmonate responses. Plant Cell 23 : 701–715. doi: 10.1105/tpc.110.080788 21335373
93. Cevik V, Kidd BN, Zhang P, Hill C, Kiddle S, et al. (2012) MEDIATOR25 acts as an integrative hub for the regulation of jasmonate-responsive gene expression in Arabidopsis. Plant Physiol 160 : 541–555. doi: 10.1104/pp.112.202697 22822211
94. Chen R, Jiang H, Li L, Zhai Q, Qi L, et al. (2012) The Arabidopsis mediator subunit MED25 differentially regulates jasmonate and abscisic acid signaling through interacting with the MYC2 and ABI5 transcription factors. Plant Cell 24 : 2898–2916. doi: 10.1105/tpc.112.098277 22822206
95. Klose C, Buche C, Fernandez AP, Schafer E, Zwick E, et al. (2012) The mediator complex subunit PFT1 interferes with COP1 and HY5 in the regulation of Arabidopsis light signaling. Plant Physiol 160 : 289–307. doi: 10.1104/pp.112.197319 22760208
96. Hou X, Lee LY, Xia K, Yan Y, Yu H (2010) DELLAs modulate jasmonate signaling via competitive binding to JAZs. Dev Cell 19 : 884–894. doi: 10.1016/j.devcel.2010.10.024 21145503
97. Chen J, Sonobe K, Ogawa N, Masuda S, Nagatani A, et al. (2013) Inhibition of arabidopsis hypocotyl elongation by jasmonates is enhanced under red light in phytochrome B dependent manner. J Plant Res 126 : 161–168. doi: 10.1007/s10265-012-0509-3 22825635
98. Yang D-L, Yao J, Mei C-S, Tong X-H, Zeng L-J, et al. (2012) Plant hormone jasmonate prioritizes defense over growth by interfering with gibberellin signaling cascad. Proc Natl Acad Sci U S A 109: E1192–E1200. doi: 10.1073/pnas.1201616109 22529386
99. Noir S, Bomer M, Takahashi N, Ishida T, Tsui TL, et al. (2013) Jasmonate controls leaf growth by repressing cell proliferation and the onset of endoreduplication while maintaining a potential stand-by mode. Plant Physiol 161 : 1930–1951. doi: 10.1104/pp.113.214908 23439917
100. Rieu I, Ruiz-Rivero O, Fernandez-Garcia N, Griffiths J, Powers SJ, et al. (2008) The gibberellin biosynthetic genes AtGA20ox1 and AtGA20ox2 act, partially redundantly, to promote growth and development throughout the Arabidopsis life cycle. Plant J 53 : 488–504. 18069939
101. Achard P, Cheng H, De Grauwe L, Decat J, Schoutteten H, et al. (2006) Integration of plant responses to environmentally activated phytohormonal signals. Science 311 : 91–94. 16400150
102. Hsu PY, Devisetty UK, Harmer SL (2013) Accurate timekeeping is controlled by a cycling activator in Arabidopsis. eLife 2: e00473. doi: 10.7554/eLife.00473 23638299
103. Takase M, Mizoguchi T, Kozuka T, Tsukaya H (2013) The unique function of the Arabidopsis circadian clock gene PRR5 in the regulation of shade avoidance response. Plant Signal Behav 8: e23534. doi: 10.4161/psb.23534 23333981
104. Schachtman D, Schroeder J, Lucas W, Anderson J, Gaber R (1992) Expression of an inward-rectifying potassium channel by the Arabidopsis KAT1 cDNA. Science 258 : 1654–1658. 8966547
105. Nakamura RL, McKendree WL Jr., Hirsch RE, Sedbrook JC, Gaber RF, et al. (1995) Expression of an Arabidopsis potassium channel gene in guard cells. Plant Physiol 109 : 371–374. 7480337
106. Sutter J-U, Sieben C, Hartel A, Eisenach C, Thiel G, et al. (2007) Abscisic acid triggers the endocytosis of the Arabidopsis KAT1 K+ channel and its recycling to the plasma membrane. Current Biol 17 : 1396–1402. 17683934
107. Eisenach C, Chen Z-H, Grefen C, Blatt MR (2011) The trafficking protein SYP121 of Arabidopsis connects programmed stomatal closure and K+ channel activity with vegetative growth. Plant J 69 : 241–251. doi: 10.1111/j.1365-313X.2011.04786.x 21914010
108. Abel S, Nguyen MD, Theologis A (1995) The PS-IAA4/5-like family of early auxin-inducible mRNAs in Arabidopsis thaliana. J Mol Biol 251 : 533–549. 7658471
109. Philippar K, Ivashikina N, Ache P, Christian M, Luthen H, et al. (2004) Auxin activates KAT1 and KAT2, two K+-channel genes expressed in seedlings of Arabidopsis thaliana. Plant J 37 : 815–827. 14996216
110. Putterill J, Robson F, Lee K, Simon R, Coupland G (1995) The CONSTANS gene of Arabidopsis promotes flowering and encodes a protein showing similarities to zinc finger transcription factors. Cell 80 : 847–857. 7697715
111. Park DH, Somers DE, Kim YS, Choy YH, Lim HK, et al. (1999) Control of circadian rhythms and photoperiodic flowering by the Arabidopsis GIGANTEA gene. Science 285 : 1579–1582. 10477524
112. Fowler S, Lee K, Onouchi H, Samach A, Richardson K, et al. (1999) GIGANTEA: a circadian clock-controlled gene that regulates photoperiodic flowering in Arabidopsis and encodes a protein with several possible membrane-spanning domains. EMBO J 18 : 4679–4688. 10469647
113. Song YH, Ito S, Imaizumi T (2013) Flowering time regulation: photoperiod - and temperature-sensing in leaves. Trends Plant Sci 18 : 575–583. doi: 10.1016/j.tplants.2013.05.003 23790253
114. Holm M, Ma LG, Qu LJ, Deng XW (2002) Two interacting bZIP proteins are direct targets of COP1-mediated control of light-dependent gene expression in Arabidopsis. Genes Dev 16 : 1247–1259. 12023303
115. Kumar SV, Lucyshyn D, Jaeger KE, Alos E, Alvey E, et al. (2012) Transcription factor PIF4 controls the thermosensory activation of flowering. Nature 484 : 242–245. doi: 10.1038/nature10928 22437497
116. Rawat R, Takahashi N, Hsu PY, Jones MA, Schwartz J, et al. (2011) REVEILLE8 and PSEUDO-REPONSE REGULATOR5 form a negative feedback loop within the Arabidopsis circadian clock. PLoS Genet 7: e1001350. doi: 10.1371/journal.pgen.1001350 21483796
117. Leopold AC (1958) Auxin uses in the control of flowering and fruiting. Annu Rev Plant Physiol 9 : 281–310.
118. Gangappa SN, Chattopadhyay S (2010) MYC2, a bHLH transcription factor, modulates the adult phenotype of SPA1. Plant Signal Behav 5 : 1650–1652. doi: 10.4161/psb.5.12.13981 21512327
119. Makkena S, Lamb RS (2013) The bHLH transcription factor SPATULA is a key regulator of organ size in Arabidopsis thaliana. Plant Signal Behav 8: e24140. doi: 10.4161/psb.24140 23470719
120. Cifuentes-Esquivel N, Bou-Torrent J, Galstyan A, Galllemi M, Sessa G, et al. (2013) The bHLH proteins BEE and BIM positively modulate the shade avoidance syndrome in Arabidopsis seedlings. Plant J 75 : 989–1002. doi: 10.1111/tpj.12264 23763263
121. Friedrichsen DM, Nemhauser J, Muramitsu T, Maloof JN, Alonso J, et al. (2002) Three redundant brassinosteroid early response genes encode putative bHLH transcription factors required for normal growth. Genetics 162 : 1445–1456. 12454087
122. Filiault DL, Maloof JN (2012) A genome-wide association study identifies variants underlying the Arabidopsis thaliana shade avoidance response. PLoS Genet 8: e1002589. doi: 10.1371/journal.pgen.1002589 22438834
123. Kumar R, Ichihashi Y, Kimura S, Chitwood DH, Headland LR, et al. (2012) A high-throughput method for Illumina RNA-Seq library preparation. Front Plant Sci 3 : 202. doi: 10.3389/fpls.2012.00202 22973283
124. Nozue K, Covington MF, Duek PD, Lorrain S, Fankhauser C, et al. (2007) Rhythmic growth explained by coincidence between internal and external cues. Nature 448 : 358–361. 17589502
125. Abramoff MD, Magalhaes PJ, Ram SJ (2004) Image processing with ImageJ. Biophotonics International 11 : 36–42.
126. Trapnell C, Pachter L, Salzberg S (2009) TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25 : 1105–1111. doi: 10.1093/bioinformatics/btp120 19289445
127. Robinson MD, McCarthy DJ, Smyth GK (2010) edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26 : 139–140. doi: 10.1093/bioinformatics/btp616 19910308
128. R_Development_Core_Team (2005) R: A language and environment for statistical computing. R Foundation for Statistical Computing. Vienna, Austria.
129. Young MD, Wakefield MJ, Smyth GK, Oshlack A (2010) Gene ontology analysis for RNA-seq: accounting for selection bias. Genome Biol 11: R14. doi: 10.1186/gb-2010-11-2-r14 20132535
130. Nemhauser JL, Hong F, Chory J (2006) Different plant hormones regulate similar processes through largely nonoverlapping transcriptional responses. Cell 126 : 467. 16901781
131. Zentella R, Zhang Z-L, Park M, Thomas SG, Endo A, et al. (2007) Global analysis of DELLA direct targets in early gibberellin signaling in Arabidopsis. Plant Cell 19 : 3037–3057. 17933900
132. Carbon S, Ireland A, Mungall CJ, Shu S, Marshall B, et al. (2009) AmiGO: online access to ontology and annotation data. Bioinformatics 25 : 288–289. doi: 10.1093/bioinformatics/btn615 19033274
133. Bates D, Maechler M, Bolker B, Walker S (2014) lme4: Linear mixed-effects models using Eigen and S4. http://CRAN.R-project.org/package=lme4.
134. Kuznetsova A, Brockhoff PB, Christensen RHB (2014) lmerTest: Tests for random and fixed effects for linear mixed effect models (lmer objects of lme4 package). http://CRAN.R-project.org/package=lmerTest.
135. Benjamini Y, Hochberg Y (1995) Controlling the false discovery rate—a practical and powerful approach to multiple testing. J Roy Statist Soc Ser B 57 : 289–300.
136. Winter D, Vinegar B, Nahal H, Ammar R, Wilson GV, et al. (2007) An "Electronic Fluorescent Pictograph Browser" for exploring and analyzing large-scale biological data sets. PLoS ONE 2: e718. 17684564
Štítky
Genetika Reprodukční medicína
Článek Retraction: Astakine 2—the Dark Knight Linking Melatonin to Circadian Regulation in CrustaceansČlánek Adventures in WonderlandČlánek Genomic Location of the Major Ribosomal Protein Gene Locus Determines Global Growth and InfectivityČlánek Spatial Fluctuations in Expression of the Heterocyst Differentiation Regulatory Gene in FilamentsČlánek Genome-Wide Negative Feedback Drives Transgenerational DNA Methylation Dynamics in ArabidopsisČlánek Systematic Dissection of the Sequence Determinants of Gene 3’ End Mediated Expression ControlČlánek The Chromatin Remodeler CHD8 Is Required for Activation of Progesterone Receptor-Dependent EnhancersČlánek Selection against Heteroplasmy Explains the Evolution of Uniparental Inheritance of MitochondriaČlánek The DNA Helicase Recql4 Is Required for Normal Osteoblast Expansion and Osteosarcoma FormationČlánek Dual-Specificity Anti-sigma Factor Reinforces Control of Cell-Type Specific Gene Expression in
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2015 Číslo 4- Růst a vývoj dětí narozených pomocí IVF
- Délka menstruačního cyklu jako marker ženské plodnosti
- Vztah užívání alkoholu a mužské fertility
- Intrauterinní inseminace a její úspěšnost
- Šanci na úspěšný průběh těhotenství snižují nevhodné hladiny progesteronu vznikající při umělém oplodnění
-
Všechny články tohoto čísla
- Retraction: Astakine 2—the Dark Knight Linking Melatonin to Circadian Regulation in Crustaceans
- Adventures in Wonderland
- Experimental Swap of 's Assortative Mating Preferences Demonstrates Key Role of X-Chromosome Divergence Island in Incipient Sympatric Speciation
- Chromosome Replacement and Deletion Lead to Clonal Polymorphism of Berry Color in Grapevine
- The Protein Quality Control Machinery Regulates Its Misassembled Proteasome Subunits
- Genome-Wide Association Study Identifies as a Critical Gene for Susceptibility to Noise-Induced Hearing Loss
- Genomic Location of the Major Ribosomal Protein Gene Locus Determines Global Growth and Infectivity
- Viable Neuronopathic Gaucher Disease Model in Medaka () Displays Axonal Accumulation of Alpha-Synuclein
- Multi-locus Analysis of Genomic Time Series Data from Experimental Evolution
- The Genetic Legacy of the Expansion of Turkic-Speaking Nomads across Eurasia
- Lack of GDAP1 Induces Neuronal Calcium and Mitochondrial Defects in a Knockout Mouse Model of Charcot-Marie-Tooth Neuropathy
- The Pif1 Helicase, a Negative Regulator of Telomerase, Acts Preferentially at Long Telomeres
- Inhibiting K63 Polyubiquitination Abolishes No-Go Type Stalled Translation Surveillance in
- SYD-1C, UNC-40 (DCC) and SAX-3 (Robo) Function Interdependently to Promote Axon Guidance by Regulating the MIG-2 GTPase
- Spatial Fluctuations in Expression of the Heterocyst Differentiation Regulatory Gene in Filaments
- Synergistic and Independent Actions of Multiple Terminal Nucleotidyl Transferases in the 3’ Tailing of Small RNAs in Arabidopsis
- Host Genetic Variation Influences Gene Expression Response to Rhinovirus Infection
- Contribution of Large Region Joint Associations to Complex Traits Genetics
- Volatility of Mutator Phenotypes at Single Cell Resolution
- Proteolysis of Virulence Regulator ToxR Is Associated with Entry of into a Dormant State
- Genome-Wide Negative Feedback Drives Transgenerational DNA Methylation Dynamics in Arabidopsis
- A Multi-layered Protein Network Stabilizes the FtsZ-ring and Modulates Constriction Dynamics
- Systematic Dissection of the Sequence Determinants of Gene 3’ End Mediated Expression Control
- Genome Sequencing of the Perciform Fish Provides Insights into Molecular and Genetic Mechanisms of Stress Adaptation
- Natural Variant E610G Is a Semi-dominant Suppressor of IAP-Induced RNA Processing Defects
- The Alkaline Response Pathway: Identification of a Novel Rim Pathway Activator
- Transgenerational Inheritance of Diet-Induced Genome Rearrangements in Drosophila
- A Single Nucleotide Polymorphism Uncovers a Novel Function for the Transcription Factor Ace2 during Hyphal Development
- DNA Damage Response and Spindle Assembly Checkpoint Function throughout the Cell Cycle to Ensure Genomic Integrity
- The Functional Interplay Between the t(9;22)-Associated Fusion Proteins BCR/ABL and ABL/BCR in Philadelphia Chromosome-Positive Acute Lymphatic Leukemia
- Extreme Recombination Frequencies Shape Genome Variation and Evolution in the Honeybee,
- Beyond Glycolysis: GAPDHs Are Multi-functional Enzymes Involved in Regulation of ROS, Autophagy, and Plant Immune Responses
- Comprehensive Profiling of Amino Acid Response Uncovers Unique Methionine-Deprived Response Dependent on Intact Creatine Biosynthesis
- Windpipe Controls Intestinal Homeostasis by Regulating JAK/STAT Pathway via Promoting Receptor Endocytosis and Lysosomal Degradation
- Ataxin-2 Regulates Translation in a New BAC-SCA2 Transgenic Mouse Model
- Cross-Population Joint Analysis of eQTLs: Fine Mapping and Functional Annotation
- The Power of Gene-Based Rare Variant Methods to Detect Disease-Associated Variation and Test Hypotheses About Complex Disease
- The Chromatin Remodeler CHD8 Is Required for Activation of Progesterone Receptor-Dependent Enhancers
- Competition between VanU Repressor and VanR Activator Leads to Rheostatic Control of Vancomycin Resistance Operon Expression
- A Missense Change in the Gene Links Aberrant Autophagy to a Neurodegenerative Vacuolar Storage Disease
- Simultaneous Discovery, Estimation and Prediction Analysis of Complex Traits Using a Bayesian Mixture Model
- Selection against Heteroplasmy Explains the Evolution of Uniparental Inheritance of Mitochondria
- Genome-Destabilizing Effects Associated with Top1 Loss or Accumulation of Top1 Cleavage Complexes in Yeast
- Imputation-Based Population Genetics Analysis of Malaria Parasites
- Heterozygosity for a Hypomorphic Polβ Mutation Reduces the Expansion Frequency in a Mouse Model of the Fragile X-Related Disorders
- Neto-Mediated Intracellular Interactions Shape Postsynaptic Composition at the Neuromuscular Junction
- Ndd1 Turnover by SCF Is Inhibited by the DNA Damage Checkpoint in
- Frameshift Variant Associated with Novel Hoof Specific Phenotype in Connemara Ponies
- The DNA Helicase Recql4 Is Required for Normal Osteoblast Expansion and Osteosarcoma Formation
- Spastin Binds to Lipid Droplets and Affects Lipid Metabolism
- Maintenance of Glia in the Optic Lamina Is Mediated by EGFR Signaling by Photoreceptors in Adult Drosophila
- Auxin Influx Carriers Control Vascular Patterning and Xylem Differentiation in
- Dual-Specificity Anti-sigma Factor Reinforces Control of Cell-Type Specific Gene Expression in
- The Lowe Syndrome Protein OCRL1 Is Required for Endocytosis in the Zebrafish Pronephric Tubule
- Postnatal Loss of Hap1 Reduces Hippocampal Neurogenesis and Causes Adult Depressive-Like Behavior in Mice
- CAPER Is Vital for Energy and Redox Homeostasis by Integrating Glucose-Induced Mitochondrial Functions via ERR-α-Gabpa and Stress-Induced Adaptive Responses via NF-κB-cMYC
- Distinct and Cooperative Activities of HESO1 and URT1 Nucleotidyl Transferases in MicroRNA Turnover in
- The Evolutionary Origination and Diversification of a Dimorphic Gene Regulatory Network through Parallel Innovations in and
- MAPK Signaling Pathway Alters Expression of Midgut ALP and ABCC Genes and Causes Resistance to Cry1Ac Toxin in Diamondback Moth
- Spatio-temporal Remodeling of Functional Membrane Microdomains Organizes the Signaling Networks of a Bacterium
- Asymmetric Transcript Discovery by RNA-seq in . Blastomeres Identifies , a Gene Important for Anterior Morphogenesis
- A Stress-Induced Small RNA Modulates Alpha-Rhizobial Cell Cycle Progression
- Systematic Profiling of Poly(A)+ Transcripts Modulated by Core 3’ End Processing and Splicing Factors Reveals Regulatory Rules of Alternative Cleavage and Polyadenylation
- The UPR Branch IRE1- in Plants Plays an Essential Role in Viral Infection and Is Complementary to the Only UPR Pathway in Yeast
- A Non-canonical RNA Silencing Pathway Promotes mRNA Degradation in Basal Fungi
- Co-chaperone p23 Regulates . Lifespan in Response to Temperature
- Re-replication of a Centromere Induces Chromosomal Instability and Aneuploidy
- Shade Avoidance Components and Pathways in Adult Plants Revealed by Phenotypic Profiling
- Lipid-Induced Epigenomic Changes in Human Macrophages Identify a Coronary Artery Disease-Associated Variant that Regulates Expression through Altered C/EBP-Beta Binding
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Lack of GDAP1 Induces Neuronal Calcium and Mitochondrial Defects in a Knockout Mouse Model of Charcot-Marie-Tooth Neuropathy
- Proteolysis of Virulence Regulator ToxR Is Associated with Entry of into a Dormant State
- Frameshift Variant Associated with Novel Hoof Specific Phenotype in Connemara Ponies
- Ataxin-2 Regulates Translation in a New BAC-SCA2 Transgenic Mouse Model
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání






