-
Články
- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Experimental Evolution of a Novel Sexually Antagonistic Allele
Evolutionary conflict permeates biological systems. In sexually reproducing organisms, sex-specific optima mean that the same allele can have sexually antagonistic expression, i.e. beneficial in one sex and detrimental in the other, a phenomenon known as intralocus sexual conflict. Intralocus sexual conflict is emerging as a potentially fundamental factor for the genetic architecture of fitness, with important consequences for evolutionary processes. However, no study to date has directly experimentally tested the evolutionary fate of a sexually antagonistic allele. Using genetic constructs to manipulate female fecundity and male mating success, we engineered a novel sexually antagonistic allele (SAA) in Drosophila melanogaster. The SAA is nearly twice as costly to females as it is beneficial to males, but the harmful effects to females are recessive and X-linked, and thus are rarely expressed when SAA occurs at low frequency. We experimentally show how the evolutionary dynamics of the novel SAA are qualitatively consistent with the predictions of population genetic models: SAA frequency decreases when common, but increases when rare, converging toward an equilibrium frequency of ∼8%. Furthermore, we show that persistence of the SAA requires the mating advantage it provides to males: the SAA frequency declines towards extinction when the male advantage is experimentally abolished. Our results empirically demonstrate the dynamics underlying the evolutionary fate of a sexually antagonistic allele, validating a central assumption of intralocus sexual conflict theory: that variation in fitness-related traits within populations can be maintained via sex-linked sexually antagonistic loci.
Published in the journal: . PLoS Genet 8(8): e32767. doi:10.1371/journal.pgen.1002917
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1002917Summary
Evolutionary conflict permeates biological systems. In sexually reproducing organisms, sex-specific optima mean that the same allele can have sexually antagonistic expression, i.e. beneficial in one sex and detrimental in the other, a phenomenon known as intralocus sexual conflict. Intralocus sexual conflict is emerging as a potentially fundamental factor for the genetic architecture of fitness, with important consequences for evolutionary processes. However, no study to date has directly experimentally tested the evolutionary fate of a sexually antagonistic allele. Using genetic constructs to manipulate female fecundity and male mating success, we engineered a novel sexually antagonistic allele (SAA) in Drosophila melanogaster. The SAA is nearly twice as costly to females as it is beneficial to males, but the harmful effects to females are recessive and X-linked, and thus are rarely expressed when SAA occurs at low frequency. We experimentally show how the evolutionary dynamics of the novel SAA are qualitatively consistent with the predictions of population genetic models: SAA frequency decreases when common, but increases when rare, converging toward an equilibrium frequency of ∼8%. Furthermore, we show that persistence of the SAA requires the mating advantage it provides to males: the SAA frequency declines towards extinction when the male advantage is experimentally abolished. Our results empirically demonstrate the dynamics underlying the evolutionary fate of a sexually antagonistic allele, validating a central assumption of intralocus sexual conflict theory: that variation in fitness-related traits within populations can be maintained via sex-linked sexually antagonistic loci.
Introduction
Understanding the mechanisms that promote variation in fitness-related traits within populations presents an enduring challenge in evolutionary biology [1], [2]: intralocus sexual conflict is predicted to be one such mechanism [3]–[6]. Intralocus conflict occurs when the same allele at a single locus provides net fitness benefits when expressed in one sex but net fitness costs when expressed in the other [7]. Although this conflict can potentially be resolved by the evolution of sexual dimorphism [8], a growing body of studies provide evidence that substantial sexually antagonistic variation occurs in both natural [9], [10] and laboratory-adapted populations [11]–[18]. To date, the main approaches used to identify the presence of intralocus sexual conflict have been the detection of negative genetic correlations for fitness between males and females [9]–[17] and experimental evolution using sex-limited selection [14], [19]. These studies have highlighted the extent to which sexually antagonistic selection affects fitness-related traits, and have identified candidate sexually antagonistic genes. However, no previous empirical studies have characterized the evolutionary dynamics of a specific sexually antagonistic allele.
We aimed to validate predictions made by intra-locus sexually antagonistic theory by experimentally engineering a novel sexually antagonistic X-linked allele. We empirically explored a fundamental principle of intralocus sexual conflict theory: that a recessive allele that benefits the heterogametic sex but harms the homogametic sex can invade a population, even when the cost exceeds the benefit, if the locus is located on the homogametic sex-chromosome [6]. This prediction arises because at low population frequency the costly effects of the allele for the homogametic sex are limited to homozygotes, which are rare, whereas the benefits are always expressed in the hemizygous sex. Consequently, such an allele could theoretically invade and reach an equilibrium frequency [6]. This makes the X-chromosome a potential hot spot for such sexually antagonistic genetic variation [20] and thus an ideal target for intralocus sexual conflict research.
We first used genetic manipulations to generate a putative sexually antagonistic allele on the X-chromosome of Drosophila melanogaster. We then tested: a) the magnitude of the cost to females (in terms of offspring production) and benefits to males (in terms of mating success), b) whether the allele could invade and persist in a population and how the invasion dynamics compared to predictions derived from theoretical models, and c) whether the evolutionary persistence of the allele was dependent upon the benefit provided to males.
Results/Discussion
Generation of a Novel Sexually Antagonistic Locus
To create a novel sexually antagonistic allele on the D. melanogaster X chromosome, we used two genetic constructs: 1) Df(1)Exel6234, a genetic deficiency which covers the sex-peptide receptor gene and 4 other genes of unknown function [21] and 2) w1118, a loss of function allele for the white gene which determines eye color [22]. Both Df(1)Exel6234 and w1118 are located on the X-chromosome. Homozygous Df(1)Exel6234 females fail to react to the male seminal protein, sex peptide [23], and show reduced levels of sex-peptide-induced post-mating responses. For example, Df(1)Exel6234 females lay significantly fewer eggs after mating than wild-type females [21]. Flies lacking white have white eyes, and white-eyed males suffer from impaired vision and reduced mating success compared to wild-type males (which have red eyes) in photophase (i.e., the light) [24], but not in the scotophase (i.e., the dark) [25]. In contrast, females lacking white suffer no obvious reduction in adult fitness (i.e., lifespan, fecundity or fertility) under standard laboratory conditions [26]. The Df(1)Exel6234 deficiency carries a white+ transgene [27], which provides a partial rescue of white mutations (i.e., red eyes and improved vision). Tight linkage between the Df(1)Exel6234 deficiency and the white+ transgene ensures that recombination between them is negligible. Thus, in a w1118 background, male hemizygote and female homozygote carriers of Df(1)Exel6234 possess red eyes, whilst heterozygote females possess orange eyes (Figure 1).
Fig. 1. Summary of fly genotypes and phenotypes, and the predicted fitness consequence for males and females expressing the X-linked SAA (sexually antagonistic allele) relative to controls and heterozygotes. 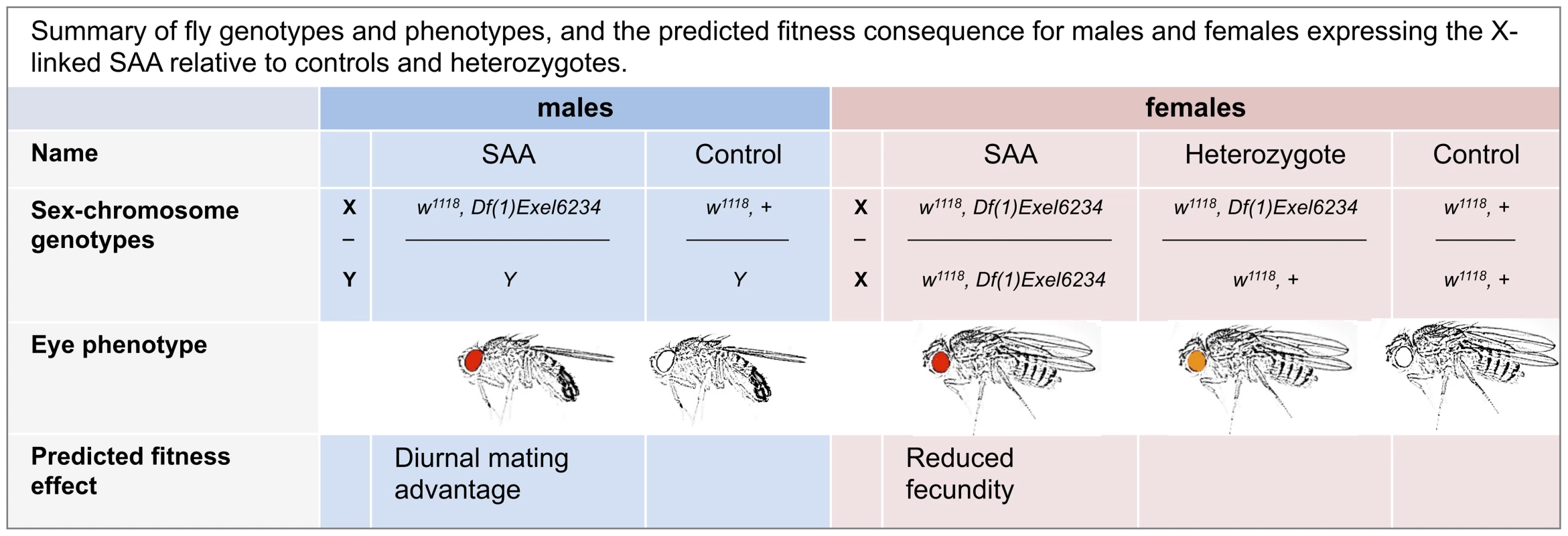
We confirmed that red-eyed Df(1)Exel6234 bearing males have increased competitive mating success relative to w1118 white-eyed males in photophase, presumably due to improved vision. In direct, one-on-one, male-male competition, Df(1)Exel6234 bearing males were significantly more likely to achieve the first mating with a single virgin female in photophase (26/28 trials, binomial test, p<0.0001) but not in scotophase (winning 14/28 trials, binomial test, p = 0.57). We also tested whether the SAA has an effect on male post-copulatory competitive ability. Female D. melanogaster mate multiply [28] resulting in sperm competition [29], [30], and variation in sperm competitive ability can potentially have major impacts on male fitness [31], [32]. However, we found no significant differences in the sperm defense (P1) or sperm offense (P2) abilities of SAA and control males (P1 assay, Z = 1.145, P = 0.252; P2 assay, Z = 0.247, P = 0.805; Figure S1A and S1B).
As expected, homozygous Df(1)Exel6234 females suffer significant reproductive costs compared to heterozygote and control females (Figure 2a, Table S1). Thus, in a w1118 background population, Df(1)Exel6234 fits the conditions required for an X-linked sexually antagonistic allele: it benefits one sex but harms the other. Moreover, the costs of Df(1)Exel6234 to females are recessive: we detected no significant fecundity cost to heterozygote females (Figure 2a, Tables S1, S2). We hereafter refer to individuals carrying the deficiency Df(1)Exel6234 as the SAA (sexually antagonistic allele) flies and non-carriers as controls (Figure 1). All experimental flies carry w1118. We predicted that selection favouring the SAA males should drive the SAA allele to higher frequency in populations when it is rare, whilst selection against the SAA homozygote females should drive the SAA frequency down when it is common.
Fig. 2. Reproductive success of male and female genotypes. 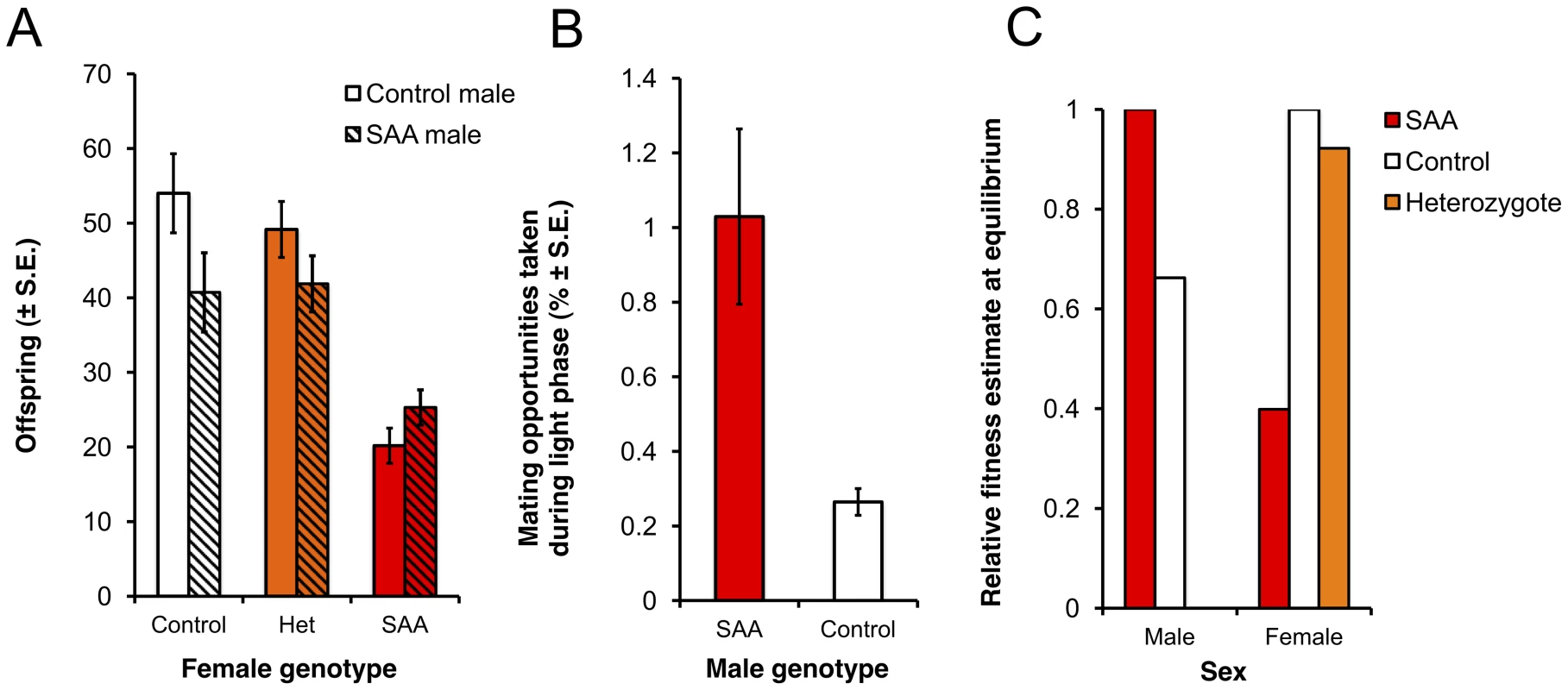
(a) Homozygote sexually antagonistic allele (SAA) females suffer reproductive success costs compared to control and heterozygote females (F2,168 = 55.4, p<0.0001). Furthermore, reproducing with control males rather than SAA-males exacerbates the relative cost to SAA-female reproductive success (male*female: F2,168 = 5.27, p = 0.07). (b) SAA-males have a photophase mating advantage over control males in P4-P4 (χ21 = 35.58, p<0.0001) (c) Estimates of relative fitness at the SAA equilibrium frequency (12.6%) for males and females of different genotypes. Relative fitness is calculated from the population genetic model for a 12∶12 light∶dark cycle. Note that the relative fitness of males is adjusted for scotophase, during which time the mating success of SAA and control males is equal. Therefore, the overall advantage to SAA males is lower than in photophase only (as shown in b) and the predicted fitness cost of SAA to homozygote females exceeds the predicted fitness benefit of SAA to males. Experimental Evolution and Modeling of a Novel Sexually Antagonistic Locus
To test the evolutionary fate of the male-beneficial, female-detrimental SAA, we simultaneously set up four replicate experimental populations (P1–P4) containing a mixture of SAA and control individuals. We initiated the populations with a SAA frequency of 3% and tracked the frequency of SAA for 16 generations in P1–P4, and a further 7 generations in two of these populations that we randomly selected (P1 and P2). Populations were maintained on a 12∶12 light dark cycle, and thus for 50% of the time (during the photophase), SAA males were predicted to possess a mating advantage (D. melanogaster mating activity occurs slightly more frequently in the dark [33], [34] when the mating advantage of SAA males is absent). We observed matings in P1–P4 during photophase over multiple generations, allowing us to estimate the relative mating fitness of SAA - versus control males in the population cage environment. We found that, as expected, SAA-males possessed a significant mating advantage in P1–P4 during photophase (Figure 2b).
Using these male mating frequency estimates (and assuming equal mating success between SAA and control males during scotophase), together with the expected mating rates during light vs dark phases [33], [34] and the genotype-specific frequencies of offspring produced from each type of cross (Table S1), we generated quantitative predictions for the spread and equilibrium of the SAA based on Rice's population genetic model [6]. Parameterizing the model with these data leads to the prediction that, over evolutionary time, the SAA should reach an equilibrium frequency at which the fitness cost to homozygote SAA females will exceed the fitness benefits to SAA-males (Figure 2c).
As predicted, average SAA frequency in P1–P4 significantly increased from the 3% starting frequency and appeared to reach a plateau at an equilibrium frequency. Initially, the frequency increased more rapidly than predicted by the model but thereafter stabilized around 8% (Figure 3a), which broadly agrees with the model predictions over the first 23 generations (Figure 3b). The model predicts an ultimate equilibrium of 12.6% (0.05–0.20 95% CI) after 700 generations, suggesting that over the 23 generations we measured, the SAA may not have reached its final equilibrium frequency.
Fig. 3. Experimental evolution of the SAA. 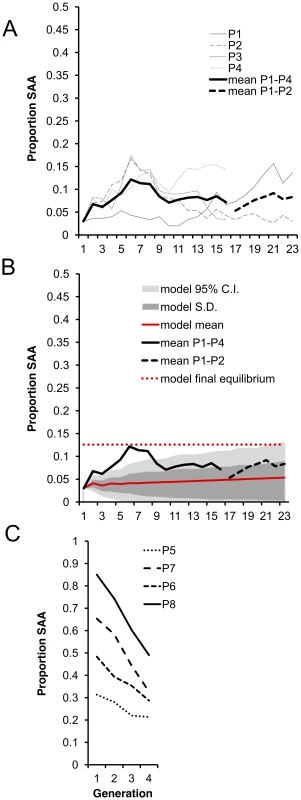
(a) Mean SAA frequency changed significantly over the 16 experimental generations in a log-linear manner (ln linear term, χ21 = 94.1, p<0.0001, linear term, χ21 = 0.30, p = 0.58). SAA frequency increased significantly from the 1st to 2nd generation (χ21 = 6.07, p = 0.014), indicating that the SAA bearing males had high fitness relative to controls (SAA was present only in males in the 1st generation) and confirming that SAA frequency increases when rare. Segmented regressions showed that mean SAA frequency continued to increase until generation 6 (change from generation 1–6, χ21 = 6.71, p = 0.0096) reaching ∼12%. SAA frequency then underwent a significant decline to ∼8% at generation 10 (change from generation 6–10, χ21 = 5.14, p = 0.023) and thereafter did not change significantly in frequency (change from generation 10–16, χ21 = 0.013, p = 0.91). Populations P1 and P2 only were maintained for generations 17–23. (b) The model (red solid line) predicted a steady increase in SAA frequency until an equilibrium frequency of 12.6% after 700 generations (red dashed line). The range of values expected from the model is shown by the 95% confidence limits (light grey area). (c) SAA frequency declined over 4 generations for each of the P5–8 populations (χ21 = 10.89, p = 0.001). There was a significant interaction between the initial SAA frequency and generation, showing that the higher the initial SAA frequency, the further it declined (χ21 = 11.049, p = 0.0009). To test the prediction that, due to the harmful effects on female fecundity, the SAA frequency should decline if the SAA is common, we set up a further 4 populations (P5–P8) with a range of higher initial SAA frequencies (31% to 85%) and measured SAA frequency over 3 subsequent generations. As expected, SAA frequency significantly declined in P5–P8. Moreover, the steepness of the decline was significantly greater in populations with higher initial frequencies (Figure 3c), confirming that SAA cannot be maintained at high frequencies, and suggesting that – regardless of the original frequency – SAA tends to converge towards a single stable equilibrium.
SAA Persistence Is Dependent upon the Male Mating Advantage
A central assumption of our hypothesis is that the SAA invades, and is maintained in the population, as a result of the mating advantage it provides males during photophase. Without this advantage, we expect a decline in the SAA and eventual extinction due to the costs imposed upon SAA females. To test this prediction we set up replicate populations of P1 and P2 at generation 16 (in which the SAA frequencies were 0.073 and 0.033, respectively) and maintained adults in these populations in permanent dark (P1 dark, P2 dark) conditions, under which SAA males should posses no mating advantage. To control for the disruption to circadian rhythm we set up replicate control populations maintained in permanent light (P1 light, P2 light). We measured SAA frequency over 6 subsequent generations in the dark and light populations. As expected, within each replicate SAA frequency significantly decreased in the dark population relative to the light population (Figure 4a and 4b) indicating that the SAA male mating advantage in photophase is essential for the maintenance of SAA. Surprisingly, SAA did not increase in light populations, suggesting that additional hours of light did not provide significant additional fitness benefits to SAA males over the standard 12∶12 light∶dark conditions. Male Drosophila require scotophases to initiate courtship efficiently [35], therefore courtship and mating in SAA males might have been negatively affected by permanent light. Additionally, there may be constraints on male courtship rates, mating rates or ejaculate production that set an upper limit to SAA male reproductive capacity. Nevertheless, the results provide support for the hypothesis that SAA persists in populations as a result of the mating advantage it provides males during photophase.
Fig. 4. Changes in SAA frequency in the light and dark populations. 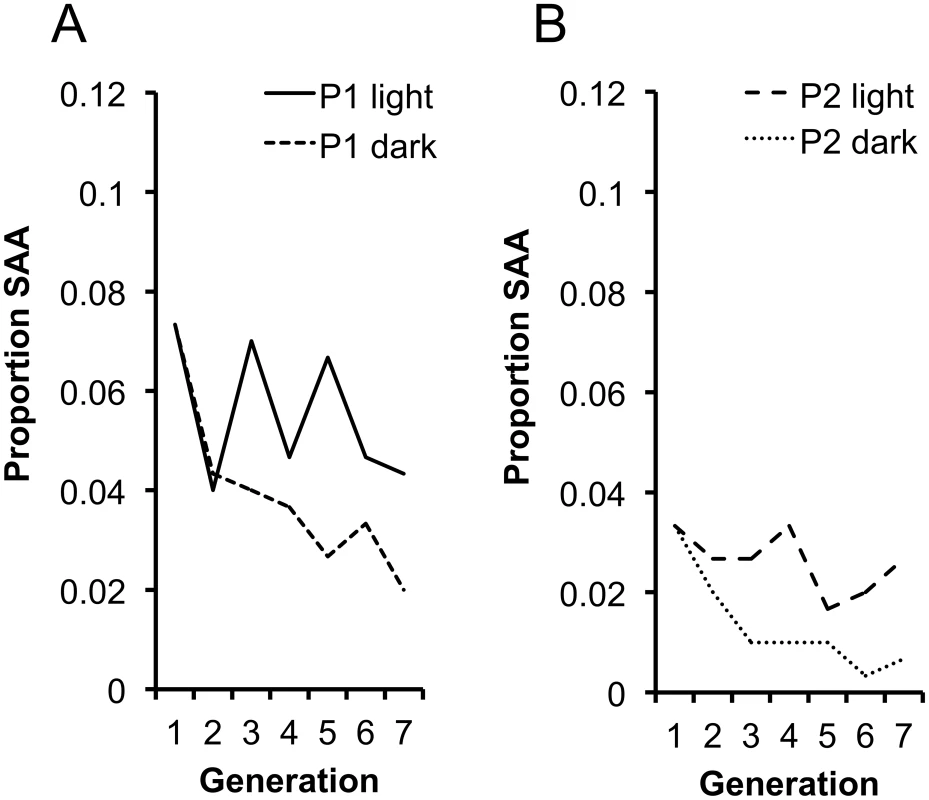
SAA frequency was affected by the manipulation of light/dark regimes (χ21 = 18.82, p<0.0001) across (a) P1 light and dark populations and (b) P2 light and dark populations. There was a significant interaction between light treatment and generation (χ21 = 4.54, p = 0.033) showing that SAA frequency significantly diverged between the continuous light and continuous dark populations. SAA frequency did not significantly change in light populations (χ21 = 2.97, p = 0.085) but significantly declined in dark populations (χ21 = 4.81, p = 0.028). Experimental Support for Intralocus Sexual Conflict Theory
Our experimental data indicate that 1) SAA frequency declines when it is common, because there is a large negative impact on the fecundity of homozygous females 2) SAA persists in populations because of the mating benefit it provides males in photophase, and SAA frequency declines towards extinction if the mating advantage of SAA males is abolished and 3) SAA has a single equilibrium frequency that is of broadly similar magnitude to that predicted by models based on intra-locus sexual conflict theory. Quantitative discrepancies between the model and our empirical data – for example, the surprisingly rapid increase in SAA frequency in the P1–4 lines – may derive from a range of factors. For example, any potential subtle effects of the Df(1)Exel6234 deficiency that have not been characterized – on development time, ejaculate depletion rates or other traits that might impact male or female fitness – might contribute to differences between model predictions and our observed SAA frequencies. Nevertheless, our results provide robust qualitative support for sexually antagonistic evolution.
Conclusion
Previous empirical evidence for intralocus sexual conflict derives from studies that demonstrate negative intersexual correlations for fitness, sexually antagonistic selection on phenotypes, or changes in sexually dimorphic traits under sex-limited evolution (reviewed in reference [4]). Here we provide direct experimental support for the idea that that sexually antagonistic alleles can invade and persist in populations. Thus, our work provides a novel demonstration that – as predicted by theory – evolution can maintain fitness variation within populations via sex chromosome-linked sexually antagonistic alleles.
Materials and Methods
General Fly Methods
The control, white-eyed whiteDahomey, stock [36] was generated by repeatedly backcrossing w1118 into the Dahomey wild-type background (>7 generations). Df(1)Exel6234 [21] was backcrossed for 5 generations into whiteDahomey to generate SAA flies. Thus, all flies were in the same genetic background before experiments began. All stocks and experimental flies were maintained in plastic vials or bottles on sugar-yeast-molasses medium with ad libitum live yeast granules at 25°C on a 12∶12 hr light dark cycle (except where specified). We used a standard density method to rear flies. First instar larvae were picked from petri dishes containing an agar-grape-juice laying medium and placed in batches of 150 into plastic bottles containing 50 mL of food.
Reproductive Success of SAA and Control Males and Females
We measured male mating success by introducing a single virgin wild-type female (N = 28) into a vial containing a virgin control male and a virgin SAA male of matched age. Experiments were conducted in light or in dark under red-light (D. melanogaster cannot see red light). We recorded which male mated first. To assay the post-copulatory competitive ability of SAA and control males, we conducted tests of sperm defense (P1, the paternity share of the first male to mate with a female) and sperm offense (P2, the paternity share of the second male to mate with a female). The competitor males and the females were homozygous for the sparklingpoliert (spapol) mutation [37]. spapol homozygotes posses a distinct eye phenotype which allows for easy visual determination of paternity. All flies were 3–5 days post-eclosion at the time of first mating. To assay P1, single virgin spapol females were first mated to either a SAA or control male, and then mated to a single spapol male 24 hours after this initial mating. Females were then allowed to oviposit individually in vials for 24 hours. Offspring from these vials were assayed for paternity (SAA, N vials = 23; control, N vials = 27). The P2 assay was identical except that the matings were reversed: the first mating was conducted with spapol males, and the second mating with either a SAA or control male (SAA, N = 21; control, N = 16). To measure offspring production of females we placed 5 3-day old virgin SAA, heterozygote or control females in vials with 5 virgin SAA or control males of the same age (i.e., 6 cross combinations). Flies were transferred to fresh vials every 2 or 3 days until day 10 when they were separated into pairs of 1 male and 1 female and transferred to fresh vials for 24 hrs. Eggs oviposited over the 24 hrs were counted. 14 days later the eclosed offspring were counted and scored for eye colour.
Experimental Evolution Populations
Flies for the 1st generation P1–P8 populations were virgins generated from crosses between heterozygote females and SAA and control males. P1–P4 initially contained 9 SAA and 81 control males, and 100 control females (i.e., 3% SAA bearing X-chromosomes, 97% control X-chromosomes). Initial numbers of SAA and control males, and SAA, heterozygote and control females were, respectively, P5) 44, 56, 4, 42, 54 (i.e., 31% SAA X-chromosomes); P6) 65, 35, 12, 56, 31 (i.e., 48% SAA); P7) 81, 19, 29, 57, 14 (i.e., 65% SAA); P8) 94, 6, 64, 33, 2 (i.e., 85% SAA). These proportions were calculated based on selection at Hardy-Weinberg equilibrium using rudimentary fitness estimates (calculated when P5–P8 were set up) for each genotype (1 for SAA and 0.55 for control males, 0.388 for SAA females, 0.9 for heterozygote females, and 1 for control females).
Adult flies were placed in a 4.5 L plastic cage containing a food bottle, which was replaced every 2 or 3 days. After 8 days eggs were collected for propagation of the subsequent generation. 13 days later (i.e., typically 2–3 days after the majority of flies had eclosed, allowing ample time for development), offspring were counted and eye colour recorded to determine genotypes. The proportions of genotypes were calculated and the next generation of 100 males and 100 females was established for each population based on these proportions, rounded to the nearest integer. During photophase we made a total of 62 spot-check mating observations on P1–P4 – over generations 1, 3–7, 9, 11, 12 and 15 – to estimate the relative mating success of SAA and control males in the population cage environment.
Mathematical Modeling
We modeled the spread and maintenance of the SAA using a standard population genetic approach. We consider a population of SAA and control genotypes. At each generation the number of matings between males and females of each genotype combination was calculated based on the frequency of each male and female genotype in the population and the empirically-derived advantage for the SAA allele in males. This SAA male advantage was calculated by taking the mean mating success of males during light phases in the experimental environment (Figure 2b), and adjusting it for the hours of light in the light-cycle (e.g. 12∶12) and the proportion of matings expected to occur in light vs dark (0.402∶0.598, light∶dark, calculated from references [33], [34]). The frequencies of each male and female genotype for the following generation were then calculated based on the mean number of surviving offspring of each genotype produced by each type of mating (i.e., male-female genotype combination) observed in our experiments (Table S1). We set the initial genotype frequencies at generation 1 to be the initial frequencies used in the experiment and determined the equilibrium SAA frequency after 1000 generations.
To generate confidence intervals around the predicted equilibria, we introduced the random selection of 300 offspring genotypes from all those generated to make up the next generation. This step mirrors the experimental procedure, in which 300 larvae were taken each generation from all those available. The total number of offspring generated (from which 300 were selected) varied with each generation and with the parameter values used, and was typically 2500–5400. Each run of this simulation model generated new frequencies of the SAA at each generation. We performed 100 runs of the model with each set of parameter values and then calculated at each generation the mean, standard deviation, and 95% confidence interval for SAA frequency.
Statistical Analysis
Data were analysed using R and JMP v9. SAA male mating advantage was calculated using chi square tests on the total number of observed SAA-male and control-male mating opportunities taken as a proportion of the total number of potential mating opportunities (i.e., a product of the frequency of SAA in each generation and the total number of mating observations each generation). P1 and P2 data for the sperm competitive ability assays could not be satisfactorily normalized and so were analyzed using Wilcoxon signed ranks tests. Analyses using parametric methods (i.e., t-tests on data that was Box-Cox transformed) produced qualitatively similar (i.e., non-significant) results. Female fitness costs of bearing the SAA were analyzed using a generalized linear model (GLM) with Poisson error distribution on the total number of offspring resulting from each of the six combinations of parental crosses. Father (2 level factor), mother (3 level factor) and their interaction were specified as fixed effects. SAA frequency data in P1–P8 and in the light/dark lines were analyzed with generalized linear mixed-effects (GLMM) models. To account for replicate lines and for repeated measures across generations, line within generation was specified as a random effect in all GLMM models. Generation and, where appropriate, ln generation, light manipulation or initial SAA frequency were specified as fixed effects. To analyze the change in SAA frequency in P1–4 in more detail we conducted a segmented regression. We partitioned the data based on the observation that the change in SAA frequency appeared to follows 3 distinct phases of increase, decrease, and plateau. Thus, we tested for changes in SAA frequency between generations 1–6, 6–10, and 10–16.
Supporting Information
Zdroje
1. EllegrenH, SheldonBC (2008) Genetic basis of fitness differences in natural populations. Nature 452 : 169–175.
2. TurelliM, BartonNH (2004) Polygenic variation maintained by balancing selection: pleiotropy, sex-dependent allelic effects and G×E interactions. Genetics 166 : 1053–1079.
3. ParkerGA, PartridgeL (1998) Sexual conflict and speciation. Phil Trans R Soc Lond B 353 : 261–274.
4. BondurianskyR, ChenowethSF (2009) Intralocus sexual conflict. Trends in Ecology & Evolution 24 : 280–288.
5. ConnallonT, ClarkAG (2012) A general population genetic framework for antagonistic selection that accounts for demography and recurrent mutation. Genetics 190 : 1477–1489.
6. RiceWR (1984) Sex chromosomes and the evolution of sexual dimorphism. Evolution 38 : 735–742.
7. LandeR (1980) Sexual dimorphism, sexual selection, and adaptation in polygenic characters. Evolution 34 : 292–305.
8. StewartAD, PischeddaA, RiceWR (2010) Resolving intralocus sexual conflict: genetic mechanisms and time frame. Journal of Heredity 101: S94–S99.
9. FoersterK, CoulsonT, SheldonBC, PembertonJM, Clutton-BrockTH, et al. (2007) Sexually antagonistic genetic variation for fitness in red deer. Nature 447 : 1107.
10. MokkonenM, KokkoH, KoskelaE, LehtonenJ, MappesT, et al. (2011) Negative frequency-dependent selection of sexually antagonistic alleles in Myodes glareolus. Science 334 : 972–974.
11. ChippindaleAK, GibsonJR, RiceWR (2001) Negative genetic correlation for adult fitness between sexes reveals ontogenetic conflict in Drosophila. Proc Natl Acad Sci USA 98 : 1671–1675.
12. FedorkaKM, MousseauTA (2004) Female mating bias results in conflicting sex-specific offspring fitness. Nature 429 : 65–67.
13. PischeddaA, ChippindaleAK (2006) Intralocus sexual conflict diminishes the benefits of sexual selection. PLoS Biol 4: e356 doi:10.1371/journal.pbio.0040356.
14. PrasadN, BedhommeS, DayT, ChippindaleA (2007) An evolutionary cost of separate genders revealed by male-limited evolution. The American Naturalist 169 : 29–37.
15. BildeT, FogedA, SchillingN, ArnqvistG (2009) Postmating sexual selection favors males that sire offspring with low fitness. Science 324 : 1705–1706.
16. DelcourtM, BlowsMW, RundleHD (2009) Sexually antagonistic genetic variance for fitness in an ancestral and a novel environment. Proceedings of the Royal Society B: Biological Sciences 276 : 2009–2014.
17. InnocentiP, MorrowEH (2010) The sexually antagonistic genes of Drosophila melanogaster. PLoS Biol 8: e1000335 doi:10.1371/journal.pbio.1000335.
18. Van DoornGS (2009) Intralocus sexual conflict. Annals of the New York Academy of Sciences 1168 : 52–71.
19. MorrowEH, StewartAD, RiceWR (2008) Assessing the extent of genome-wide intralocus sexual conflict via experimentally enforced gender-limited selection. Journal of Evolutionary Biology 21 : 1046–1054.
20. GibsonJR, ChippindaleAK, RiceWR (2002) The X chromosome is a hot spot for sexually antagonistic fitness variation. Proc R Soc London Ser B 269 : 499–505.
21. YapiciN, KimY-J, RibeiroC, DicksonBJ (2008) A receptor that mediates the post-mating switch in Drosophila reproductive behaviour. Nature 451 : 33–37.
22. RabinowL, BirchlerJA (1989) A dosage-sensitive modifier of retrotransposon-induced alleles of the Drosophila white locus. The EMBO Journal 8 : 879–889.
23. ChenPS, Stumm-ZollingerE, AigakiT, BalmerJ, BienzM, et al. (1988) A male accessory gland peptide that regulates reproductive behavior of female D. melanogaster. Cell 54 : 291–298.
24. ReedSC, ReedEW (1950) Natural selection in laboratory populations of Drosophila. II. Competition between a white-eye gene and its wild type allele. Evolution 4 : 34–42.
25. BurnetB, ConnollyK (1973) The visual component in the courtship of Drosophila melanogaster. Cellular and Molecular Life Sciences 29 : 488–489.
26. GrandisonRC, WongR, BassTM, PartridgeL, PiperMDW (2009) Effect of a standardised dietary restriction protocol on multiple laboratory strains of Drosophila melanogaster. PLoS ONE 4: e4067 doi:10.1371/journal.pone.0004067.
27. ParksAL, CookKR, BelvinM, DompeNA, FawcettR, et al. (2004) Systematic generation of high-resolution deletion coverage of the Drosophila melanogaster genome. Nat Genet 36 : 288–292.
28. ImhofM, HarrB, BremG, SchlöttererC (1998) Multiple mating in wild Drosophila melanogaster revisited by microsatellite analysis. Mol Ecol 7 : 915–917.
29. CivettaA (1999) Direct visualization of sperm competition and sperm storage in Drosophila. Curr Biol 9 : 841–844.
30. ManierMK, BeloteJM, BerbenKS, NovikovD, StuartWT, et al. (2010) Resolving Mechanisms of Competitive Fertilization Success in Drosophila melanogaster. Science 328 : 354–357.
31. ClarkAG, BegunDJ, ProutT (1999) Female×male interactions in Drosophila sperm competition. Science 283 : 217–220.
32. BretmanA, FrickeC, ChapmanT (2009) Plastic responses of male Drosophila melanogaster to the level of spermcompetition increase male reproductive fitness. Proceedings of the Royal Society B: Biological Sciences 276 : 1705–1711.
33. TauberE, RoeH, CostaR, HennessyJM, KyriacouCP (2003) Temporal mating isolation driven by a behavioral gene in Drosophila. Current Biology 13 : 140–145.
34. FujiiS, KrishnanP, HardinP, AmreinH (2007) Nocturnal male sex drive in Drosophila. Current Biology 17 : 244–251.
35. HardelandR, StangeG (1971) Einflüsse von geschlecht und alter auf die lokomotorische aktivität von Drosophila. Journal of Insect Physiology 17 : 427–434.
36. BroughtonSJ, PiperMDW, IkeyaT, BassTM, JacobsonJ, et al. (2005) Longer lifespan, altered metabolism, and stress resistance in Drosophila from ablation of cells making insulin-like ligands. Proceedings of the National Academy of Sciences of the United States of America 102 : 3105–3110.
37. FrickeC, WigbyS, HobbsR, ChapmanT (2009) The benefits of male ejaculate sex peptide transfer in Drosophila melanogaster. Journal of Evolutionary Biology 22 : 275–286.
Štítky
Genetika Reprodukční medicína
Článek Mutational Signatures of De-Differentiation in Functional Non-Coding Regions of Melanoma GenomesČlánek Rescuing Alu: Recovery of Inserts Shows LINE-1 Preserves Alu Activity through A-Tail ExpansionČlánek Genetics and Regulatory Impact of Alternative Polyadenylation in Human B-Lymphoblastoid CellsČlánek Retrovolution: HIV–Driven Evolution of Cellular Genes and Improvement of Anticancer Drug ActivationČlánek The Mi-2 Chromatin-Remodeling Factor Regulates Higher-Order Chromatin Structure and Cohesin DynamicsČlánek Identification of Human Proteins That Modify Misfolding and Proteotoxicity of Pathogenic Ataxin-1
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2012 Číslo 8- Akutní intermitentní porfyrie
- Souvislost haplotypu M2 genu pro annexin A5 s opakovanými reprodukčními ztrátami
- Vztah užívání alkoholu a mužské fertility
- Doc. Miloš Kubánek: Nemocní se srdeční amyloidózou jsou často skryti a sledováni pod jinými diagnózami
- Hysteroskopická resekce děložního septa zlepšuje šanci na graviditu žen s jinak nevysvětlenou infertilitou
-
Všechny články tohoto čísla
- Mutational Signatures of De-Differentiation in Functional Non-Coding Regions of Melanoma Genomes
- Rescuing Alu: Recovery of Inserts Shows LINE-1 Preserves Alu Activity through A-Tail Expansion
- Genetics and Regulatory Impact of Alternative Polyadenylation in Human B-Lymphoblastoid Cells
- Chromosome Territories Meet a Condensin
- It's All in the Timing: Too Much E2F Is a Bad Thing
- Fine-Mapping and Initial Characterization of QT Interval Loci in African Americans
- Genome Patterns of Selection and Introgression of Haplotypes in Natural Populations of the House Mouse ()
- A Combinatorial Amino Acid Code for RNA Recognition by Pentatricopeptide Repeat Proteins
- Advances in Quantitative Trait Analysis in Yeast
- Experimental Evolution of a Novel Sexually Antagonistic Allele
- Variation of Contributes to Dog Breed Skull Diversity
- , a Gene Involved in Axonal Pathfinding, Is Mutated in Patients with Kallmann Syndrome
- A Single Origin for Nymphalid Butterfly Eyespots Followed by Widespread Loss of Associated Gene Expression
- Cryptocephal, the ATF4, Is a Specific Coactivator for Ecdysone Receptor Isoform B2
- Retrovolution: HIV–Driven Evolution of Cellular Genes and Improvement of Anticancer Drug Activation
- The PARN Deadenylase Targets a Discrete Set of mRNAs for Decay and Regulates Cell Motility in Mouse Myoblasts
- A Sexual Ornament in Chickens Is Affected by Pleiotropic Alleles at and , Selected during Domestication
- Use of Allele-Specific FAIRE to Determine Functional Regulatory Polymorphism Using Large-Scale Genotyping Arrays
- Novel Loci for Metabolic Networks and Multi-Tissue Expression Studies Reveal Genes for Atherosclerosis
- The Genetic Basis of Pollinator Adaptation in a Sexually Deceptive Orchid
- Uncovering the Genome-Wide Transcriptional Responses of the Filamentous Fungus to Lignocellulose Using RNA Sequencing
- Inheritance Beyond Plain Heritability: Variance-Controlling Genes in
- The Metabochip, a Custom Genotyping Array for Genetic Studies of Metabolic, Cardiovascular, and Anthropometric Traits
- Reprogramming to Pluripotency Can Conceal Somatic Cell Chromosomal Instability
- Condensin II Promotes the Formation of Chromosome Territories by Inducing Axial Compaction of Polyploid Interphase Chromosomes
- PTEN Negatively Regulates MAPK Signaling during Vulval Development
- A Dynamic Response Regulator Protein Modulates G-Protein–Dependent Polarity in the Bacterium
- Population Genomics of the Facultatively Mutualistic Bacteria and
- Components of a Fanconi-Like Pathway Control Pso2-Independent DNA Interstrand Crosslink Repair in Yeast
- Polysome Profiling in Liver Identifies Dynamic Regulation of Endoplasmic Reticulum Translatome by Obesity and Fasting
- Stromal Liver Kinase B1 [STK11] Signaling Loss Induces Oviductal Adenomas and Endometrial Cancer by Activating Mammalian Target of Rapamycin Complex 1
- Reprogramming of H3K27me3 Is Critical for Acquisition of Pluripotency from Cultured Tissues
- Transgene Induced Co-Suppression during Vegetative Growth in
- Hox and Sex-Determination Genes Control Segment Elimination through EGFR and Activity
- A Quantitative Comparison of the Similarity between Genes and Geography in Worldwide Human Populations
- Minibrain/Dyrk1a Regulates Food Intake through the Sir2-FOXO-sNPF/NPY Pathway in and Mammals
- Comparative Analysis of Regulatory Elements between and by Genome-Wide Transcription Start Site Profiling
- Simple Methods for Generating and Detecting Locus-Specific Mutations Induced with TALENs in the Zebrafish Genome
- S Phase–Coupled E2f1 Destruction Ensures Homeostasis in Proliferating Tissues
- Cell-Nonautonomous Signaling of FOXO/DAF-16 to the Stem Cells of
- The Mi-2 Chromatin-Remodeling Factor Regulates Higher-Order Chromatin Structure and Cohesin Dynamics
- Comparative Analysis of the Genomes of Two Field Isolates of the Rice Blast Fungus
- Role of Mex67-Mtr2 in the Nuclear Export of 40S Pre-Ribosomes
- Genetic Modulation of Lipid Profiles following Lifestyle Modification or Metformin Treatment: The Diabetes Prevention Program
- HAL-2 Promotes Homologous Pairing during Meiosis by Antagonizing Inhibitory Effects of Synaptonemal Complex Precursors
- SLX-1 Is Required for Maintaining Genomic Integrity and Promoting Meiotic Noncrossovers in the Germline
- Phylogenetic and Transcriptomic Analysis of Chemosensory Receptors in a Pair of Divergent Ant Species Reveals Sex-Specific Signatures of Odor Coding
- Reduced Prostasin (CAP1/PRSS8) Activity Eliminates HAI-1 and HAI-2 Deficiency–Associated Developmental Defects by Preventing Matriptase Activation
- Dissecting the Gene Network of Dietary Restriction to Identify Evolutionarily Conserved Pathways and New Functional Genes
- Identification of Human Proteins That Modify Misfolding and Proteotoxicity of Pathogenic Ataxin-1
- and Link Transcription of Phospholipid Biosynthetic Genes to ER Stress and the UPR
- CDK9 and H2B Monoubiquitination: A Well-Choreographed Dance
- Rare Copy Number Variations in Adults with Tetralogy of Fallot Implicate Novel Risk Gene Pathways
- Ccdc94 Protects Cells from Ionizing Radiation by Inhibiting the Expression of
- NOL11, Implicated in the Pathogenesis of North American Indian Childhood Cirrhosis, Is Required for Pre-rRNA Transcription and Processing
- Human Developmental Enhancers Conserved between Deuterostomes and Protostomes
- A Luminal Glycoprotein Drives Dose-Dependent Diameter Expansion of the Hindgut Tube
- Melanophore Migration and Survival during Zebrafish Adult Pigment Stripe Development Require the Immunoglobulin Superfamily Adhesion Molecule Igsf11
- Dynamic Distribution of Linker Histone H1.5 in Cellular Differentiation
- Combining Comparative Proteomics and Molecular Genetics Uncovers Regulators of Synaptic and Axonal Stability and Degeneration
- Chemical Genetics Reveals a Specific Requirement for Cdk2 Activity in the DNA Damage Response and Identifies Nbs1 as a Cdk2 Substrate in Human Cells
- Experimental Relocation of the Mitochondrial Gene to the Nucleus Reveals Forces Underlying Mitochondrial Genome Evolution
- Rates of Gyrase Supercoiling and Transcription Elongation Control Supercoil Density in a Bacterial Chromosome
- Mutations in a P-Type ATPase Gene Cause Axonal Degeneration
- A General G1/S-Phase Cell-Cycle Control Module in the Flowering Plant
- Multiple Roles and Interactions of and in Development of the Respiratory System
- UNC-40/DCC, SAX-3/Robo, and VAB-1/Eph Polarize F-Actin during Embryonic Morphogenesis by Regulating the WAVE/SCAR Actin Nucleation Complex
- Epigenetic Remodeling of Meiotic Crossover Frequency in DNA Methyltransferase Mutants
- Modulating the Strength and Threshold of NOTCH Oncogenic Signals by
- Loss of Axonal Mitochondria Promotes Tau-Mediated Neurodegeneration and Alzheimer's Disease–Related Tau Phosphorylation Via PAR-1
- Acetyl-CoA-Carboxylase Sustains a Fatty Acid–Dependent Remote Signal to Waterproof the Respiratory System
- ATXN2-CAG42 Sequesters PABPC1 into Insolubility and Induces FBXW8 in Cerebellum of Old Ataxic Knock-In Mice
- Cohesin Rings Devoid of Scc3 and Pds5 Maintain Their Stable Association with the DNA
- The MicroRNA Inhibits Calcium Signaling by Targeting the TIR-1/Sarm1 Adaptor Protein to Control Stochastic L/R Neuronal Asymmetry in
- Rapid-Throughput Skeletal Phenotyping of 100 Knockout Mice Identifies 9 New Genes That Determine Bone Strength
- The Genes Define Unique Classes of Two-Partner Secretion and Contact Dependent Growth Inhibition Systems
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Dissecting the Gene Network of Dietary Restriction to Identify Evolutionarily Conserved Pathways and New Functional Genes
- It's All in the Timing: Too Much E2F Is a Bad Thing
- Variation of Contributes to Dog Breed Skull Diversity
- The PARN Deadenylase Targets a Discrete Set of mRNAs for Decay and Regulates Cell Motility in Mouse Myoblasts
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



