-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaCandidate Gene Screen in the Red Flour Beetle Reveals as Ancient Regulator of Anterior Median Head and Central Complex Development
Several highly conserved genes play a role in anterior neural plate patterning of vertebrates and in head and brain patterning of insects. However, head involution in Drosophila has impeded a systematic identification of genes required for insect head formation. Therefore, we use the red flour beetle Tribolium castaneum in order to comprehensively test the function of orthologs of vertebrate neural plate patterning genes for a function in insect head development. RNAi analysis reveals that most of these genes are indeed required for insect head capsule patterning, and we also identified several genes that had not been implicated in this process before. Furthermore, we show that Tc-six3/optix acts upstream of Tc-wingless, Tc-orthodenticle1, and Tc-eyeless to control anterior median development. Finally, we demonstrate that Tc-six3/optix is the first gene known to be required for the embryonic formation of the central complex, a midline-spanning brain part connected to the neuroendocrine pars intercerebralis. These functions are very likely conserved among bilaterians since vertebrate six3 is required for neuroendocrine and median brain development with certain mutations leading to holoprosencephaly.
Published in the journal: . PLoS Genet 7(12): e32767. doi:10.1371/journal.pgen.1002416
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1002416Summary
Several highly conserved genes play a role in anterior neural plate patterning of vertebrates and in head and brain patterning of insects. However, head involution in Drosophila has impeded a systematic identification of genes required for insect head formation. Therefore, we use the red flour beetle Tribolium castaneum in order to comprehensively test the function of orthologs of vertebrate neural plate patterning genes for a function in insect head development. RNAi analysis reveals that most of these genes are indeed required for insect head capsule patterning, and we also identified several genes that had not been implicated in this process before. Furthermore, we show that Tc-six3/optix acts upstream of Tc-wingless, Tc-orthodenticle1, and Tc-eyeless to control anterior median development. Finally, we demonstrate that Tc-six3/optix is the first gene known to be required for the embryonic formation of the central complex, a midline-spanning brain part connected to the neuroendocrine pars intercerebralis. These functions are very likely conserved among bilaterians since vertebrate six3 is required for neuroendocrine and median brain development with certain mutations leading to holoprosencephaly.
Introduction
The insect head is composed of several fused segments, the number of which remains disputed (e.g. [1]–[6]). The posterior labial, maxillary and mandibular segments are patterned by the well-studied trunk segmentation cascade. In contrast, the patterning of the procephalic region (intercalary, antennal, ocular segments and anterior non-segmental region) is less well understood. It has been suggested in the fruit fly Drosophila melanogaster that the head gap-like genes orthodenticle (otd), empty-spiracles (ems), buttonhead (btd) and sloppy-paired (slp) activate segment polarity genes directly or via second order regulators [7]–[9] but an instructive role could not be confirmed for btd and ems [10], [11]. Moreover, the segment polarity interactions of head segments differ from those in the trunk. For example, hedgehog (hh) expression in the intercalary segment is driven by its own unique enhancer element [12], [13] (see [4] and [6] for further details).
However, the development of the larval head of Drosophila is highly derived. During late stages of embryogenesis, the head gets turned outside into the thorax (head involution) and consequently cuticular head structures are highly reduced [14]–[16]. Also the emergence of the everted adult head from imaginal discs is derived within insects [17], [18]. These morphological differences correlate with changes in embryonic pattern formation. Comparisons with other insects have revealed that the upstream levels of head formation differ profoundly (e.g. no bicoid in most insects [19], no torso signaling in head development [20], different input of decapentaplegic (dpp) on head development [21]) while some degree of conservation is found on lower levels like the head gap like genes, second order regulators and segment polarity genes ([4] and references therein). Notably, the head expression of wingless (wg) appears to be reduced in Drosophila correlating with its derived head development. Hence, the evolution of the Drosophila head involved both structural changes and alterations of the gene regulatory network [4], [6], [14], [22].
Intriguingly, orthologs of several genes required for Drosophila head patterning play a role in vertebrate neural plate patterning (e.g. otd/otx, ems/emx, slp/bf1, tailless(tll)/tlx). These data indicated that anterior patterning in insects and vertebrates relies on a strongly overlapping gene set [2], [7], [8], [13], [23]–[36]. Indeed, additional similarities between vertebrate and insect head and brain patterning have subsequently been revealed (e.g. [37]–[47]). Furthermore, an urbilaterian origin of anterior brain patterning has also been supported by similar data in an annelid [48]–[52].
The red flour beetle Tribolium castaneum has recently been established as a model for insect head development because it has an insect typical non-involuted head developing from a ventral-posterior region of the blastoderm, reflecting the ancestral situation (reviewed in [4], [53]). Several orthologs of Drosophila head patterning genes have been studied in Tribolium revealing differences with respect to the head gap-like genes [54] and knirps [55] but also a number of similarities with respect to second order regulators [56], [57]. Furthermore, genes required for vertebrate placode development were found to be active at similar locations in the Tribolium embryonic head [44].
Tc-six3 is a member of the six type homeobox gene family, which has three members in insects [58] while two paralogs of each family are found in mouse (six3/six6, six1/2 and six4/5). It is required for the formation of the labrum, an appendage of the non-segmental part of the head [3]. The Drosophila ortholog, called optix, is required for eye development [59]–[61], and for maxillary and clypeolabral structures of the larval head [62]. However, genetic interactions of this gene in the context of head development have not been analyzed in insects so far. Suggestively, the vertebrate six3 gene is essential for eye development [63]–[70] and anterior neural plate patterning [71]–[75]. Furthermore, vertebrate six3 and its paralog six6 are involved in the development of the neuroendocrine pituitary and hypothalamus [76]–[80]. As the six3 expression domain is anterior to the otd domain in arthropods, annelids and vertebrates, it is likely that this was also a feature of the last common ancestor of bilaterian animals [81].
In order to identify novel insect head patterning genes and with the high degree of conservation in mind, we comprehensively tested Tribolium orthologs of vertebrate neural plate patterning genes for a function in the insect head. Indeed, we find that many of them are required for head development. Closer examination of Tc - six3 reveals that it acts upstream of Tc-wingless (Tc-wg), Tc-otd and Tc-eyeless (Tc-ey) in anterior median head patterning. Further, we find that Tc-six3 is required for median brain development with a specific role in central body formation.
Results
18 out of 24 Vertebrate Neural Plate Patterning Genes Are Expressed in the Tribolium Head
From the literature we identified 24 genes involved in early vertebrate neural plate patterning (see Table S1) [32], [33], [41], [45], [74], [80], [82]–[127]. Three genes do not possess orthologs in either the Tribolium or Drosophila genome (Dmbx1/Atx, Vax1, Hesx1/Rpx; see phylogenetic trees in Figure S1). Tc-FGF8 does not cluster unequivocally with mouse FGF8 but with the Drosophila Pyramus and Thisbe proteins which have previously been identified as FGF8 orthologs [128], [129]. Of the 21 orthologs, Tc-BarH, Tc-Wnt11 and Tc-munster/arx are not expressed in the head anlagen (not shown), while the remaining 18 genes are active in the embryonic head. For these genes, we determined the expression pattern at several stages (determined by Tc-wg counter stain) in order to reveal their dynamics during head development. Genes that had not been described before in Tribolium are shown in Figure 1. In order to produce a comprehensive dataset with comparable staging, we also included previously described genes (Figures S2 and S3) (Tc-otd [130], Tc-ems [54], Tc-tailless (Tc-tll) [131], Tc-six3 [3], Tc-hedgehog (Tc-hh) [132], [133], Tc-cubitus-interruptus (Tc-ci) [132], Tc-wg [134], Tc-fgf8 [128], Tc-sloppy paired (Tc-slp) [135], Tc-eyeless (Tc-ey) [136], Tc-ptx [44], Tc-irx [137]).
Fig. 1. Novel genes with anterior but not segmental expression. 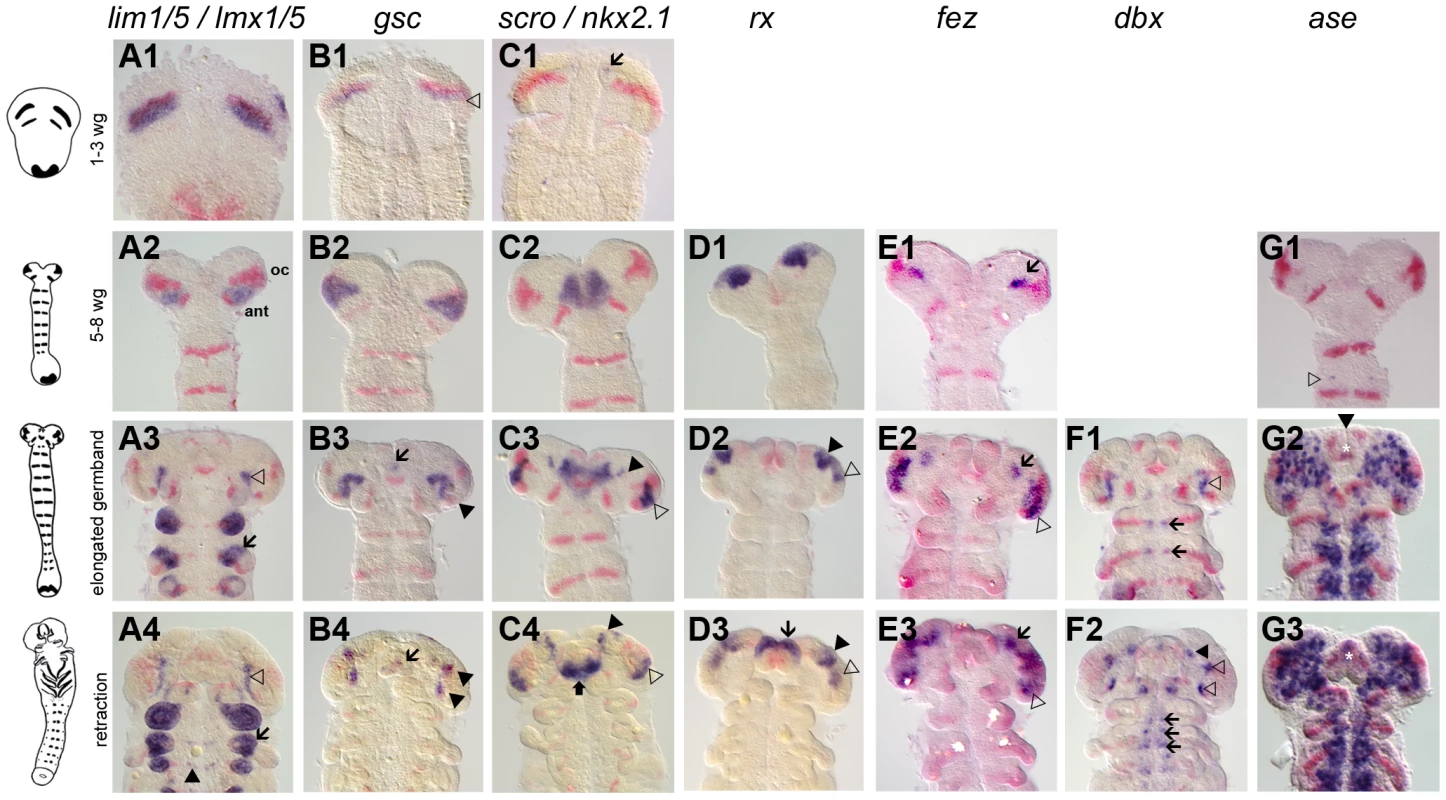
Gene expression depicted in blue with Tc-wg as red counterstain. Germband stages are oriented with anterior to the top. The embryos in one row are staged according to their Tc-wg pattern and shape. Stages prior to onset of expression are omitted. (A) Tc-lim1/5 expression starts in parallel to the ocular Tc-wg domain (A1) before it covers parts of the antennal segment (A2). Later, this domain shrinks (open arrowhead in A3) and expression in limb buds (arrow in A3–4) and in some cells in the CNS starts (black arrowhead in A4). (B) Tc-gsc expression starts in parallel to the ocular Tc-wg domain (open arrowhead in B1) before covering parts of the antennal segment (B2). Some cells close to the eye anlagen express Tc-gsc (black arrowheads in B3–4) and eventually stomodeal expression starts (arrow in B3–4). (C) Tc-scro expression starts as bilateral median dots (arrow in C1) and expands to cover an anterior median region with the stomodeum in the center (C2). Additional expression is found in the eye anlagen (open arrowhead in C3–4), in lateral/anterior cells (black arrowhead in C3–4) and in the labrum (black arrow in C4). (D) Tc-rx expression starts in elongating embryos in two bilateral patches, which are later subdivided (black and open arrowheads in D2–3). Still later, an expression at the base of the labrum arises (arrow in D3). (E) Initial Tc-fez expression covers a very restricted domain (arrow in E1–3). Later, an additional domain in the eye anlagen develops (open arrowheads in E2–3). (F) In addition to segmental dots (arrows in F1–2), strong Tc-dbx expression is found close to the ocular Tc-wg domain (open arrowhead in F1) which later splits (open arrowheads in F2). A more anterior domain becomes visible at later stages (black arrowhead in F2). (G) Tc-ase is used as marker for neuroectoderm because it is expressed in recently delaminated neural stem cells (neuroblasts). Expression starts in the 5–8 wg-stripe stage embryo (open arrowhead in G1). The neuroectoderm consists of a salt and pepper pattern of neural and epidermal precursors (G2–3). Note the stomodeal neuroblasts (white star in G2–3) in an otherwise non-neural anterior median region. Interestingly, a number of these genes are expressed in the head but not segmentally reiterated in the trunk (Tc-otd, Tc-six3, Tc-tll, Tc-lim1, Tc-gsc, Tc-scro, Tc-rx, Tc-fez1) supporting the notion that the anterior patterning system differs from the one of the trunk. However, other genes do have segmentally reiterated expression in addition to anterior expression (Tc-hh, Tc-wg, Tc-ci, Tc-irx/mirr, Tc-fgf8, Tc-slp, Tc-ems, Tc-ey, Tc-dbx, Tc-ptx), linking these two systems.
All Genes But One Are Involved in Head Epidermis Patterning
The embryonic preantennal region gives rise to the lateral and dorsal head capsule (compare white area anterior to the dark grey shaded antennal segment in a flattened germband in Figure 2D with a non-flattened embryo depicted in E) [4], [44]. This region is marked by an invariant bristle pattern in the first instar larval cuticle [54] (see Figure 2F for most prominent bristles and Figure S4 for more details), which allows the localization of cuticle defects, which arose in pre-antennal tissues. Unfortunately, previously published RNAi phenotypes had not been scored for the head bristle pattern except for Tc-otd and Tc-ems [54] and Tc-ey/toy, where a small subset of three setae was scored [136]. Therefore, we performed RNAi for all novel genes as well for those where the head capsule defects had not been described previously. We excluded Tc-hh and Tc-wg because RNAi for these genes induces severe generalized embryonic defects, which impede the analysis of the bristle pattern (data not shown and [132], [138]).
Fig. 2. Fate map and morphogenesis of the head. 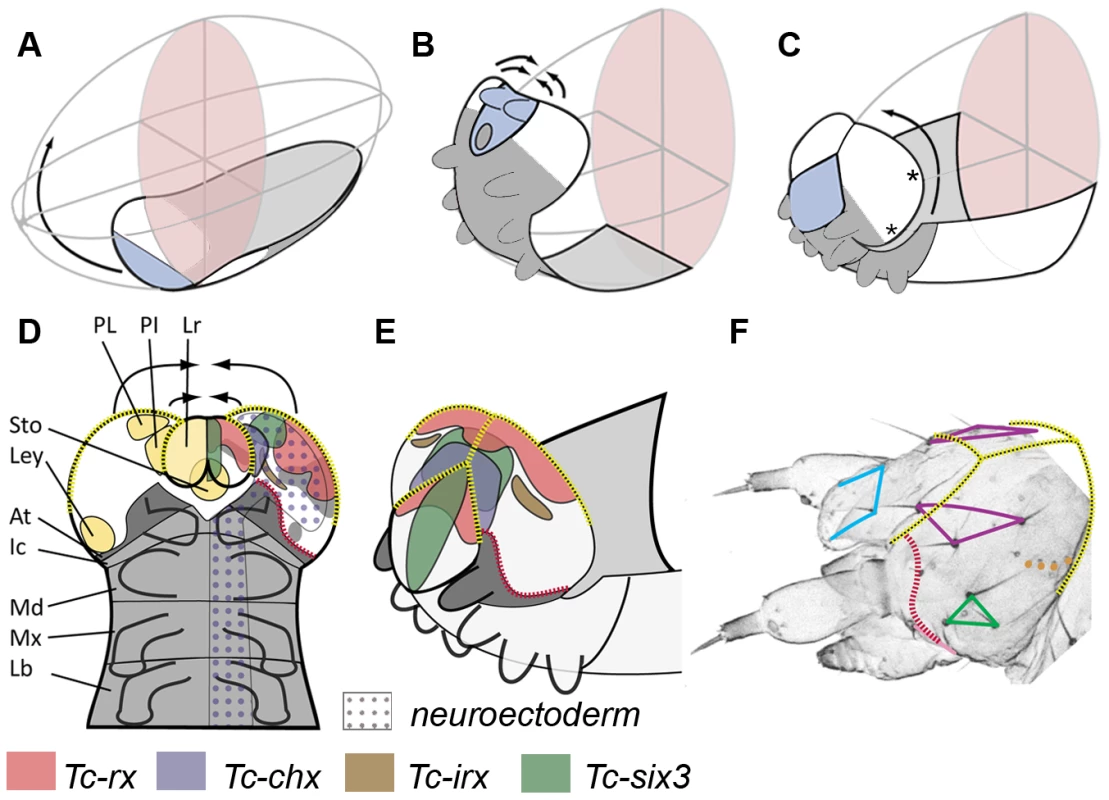
(A–C) Our recently proposed “bend and zipper” model of head development describes the movement of the ventral head anlagen (A) towards the anterior, their upward bending (B) and their zippering together at the dorsal midline (C) (after [137]). Hence, the anterior lateral tissue (white in A–C) forms the dorsal and lateral head capsule. Many of the genes shown in Figure 1 and Figures S2 and S3 are expressed in the preantennal region, hence, cuticle phenotypes are expected to affect the head capsule (see text for further details). (D) Schematic of a retracting germband embryo with segments shown in grey (At = antennal (dark gray); Ic = intercalary; Md = mandibular; Mx = maxillary; Lb = labial). Light yellow shaded domains in the left head lobe indicate the approximate location of the neuroendocrine anlagen pars lateralis (PL, marked by Tc-fas2) and pars intercerebralis (PI, marked by Tc-chx; both markers established in Drosophila by [141]). Further, the stomodeum with stomatogastric nervous system anlagen (Sto), and the larval eye anlagen (Ley, based on [136]) are shown. The neuroectoderm is marked by the dotted field in the right half of the embryo (based on the neuroblast marker Tc-asense). Expression patterns of head markers in flat preparations of retracting embryos are indicated in different colors in the right head lobe (compare to Figure 1 and Figures S2 and S3). (E) Expression of the same genes is shown in a non-flattened embryo, in which the head is zippering together at the dorsal midline (based on Figure S6). (F) Head cuticle of a first instar larva with the location of the most prominent head bristles marked by the corners of the colored shapes (see Figure S4 for complete pattern). The yellow broken lines in D–F help to relate the tissue boundaries of a retracting embryo with the L1 head. The red broken line indicates the adjacent expression of ocular Tc-hh and Tc-wg during early embryogenesis (see Figure S3A) and by Tc-tll and Tc-ems at later stages (see Figure S2 and S3). This fate map remains an approximation because it assumes that cells continue to express a certain gene and that the relative locations of cell groups do not change during the development from retracted germband stage to the hatchling. Tc-ci RNAi interfered with segmentation of the entire embryo as described [132]. Head defects ranged from the total loss of the head (9.1%, n = 11) to the loss and malformation of gnathal segments (90.9%; Figure 3C). Where accessible, the head bristle pattern was analyzed, revealing mainly a disruption of the vertex setae marking the dorsal tissue (Figure 3C′; the numbers for this and other bristle pattern defects are given in Table S2, the names of the setae and bristles are given in Figure S4). Knock down of the pair rule gene Tc-slp resulted in a pair rule phenotype [135] (70%, n = 10; Figure 3D). We found additional head defects in the median part of the vertex, the bell row and the maxilla escort bristles (Figure 3D′). Tc-six3 knock down leads to loss of the labrum and clypeolabral parts of the anterior head capsule [3], [62] (100%, n = 16; Figure 3E). In line with the loss of anterior median cuticle, the anterior vertex seta and the anterior vertex bristle were missing (Figure 3E′), while the median part of the dorsal head cuticle often displayed an irregular pattern of additional bristles and setae. Tc-ey and Tc-toy have been shown to act synergistically in eye formation, while the respective analysis on the head bristle pattern was restricted to 3 bristles [136]. Re-analysis of single and double RNAi revealed more extensive defects than previously described (Figure 3F, 3F′). In comparison with single RNAi experiments, double RNAi revealed a 1,4 - to 6-fold increase of penetrance of bristle pattern defects using the same final concentration of dsRNA (Table S2) confirming that the two Tribolium pax6 orthologs also act synergistically in epidermis development. The head of Tc-lim1/5 RNAi larvae was compacted and shortened (16.7%, n = 12; Figure 3G). Head appendages were present but mostly malformed (41.7%). The anterior and median maxilla escort bristles failed to form (Figure 3G′), while in 20.8% no bell row was observed (Figure 3G′). In Tc-scro RNAi larvae the labrum failed to fuse either completely (60%; n = 15) (black arrowheads in Figure 3H) or partially (13.3%; not shown). The bristle pattern remained largely unaltered except for the anterior vertex bristle (Figure 3H′). Interestingly, the labrum quartet bristles were present even on unfused labra. In Tc-rx knock down larvae, the labrum was narrower than in wild type leading to a widened space between the labrum and the antennae (25%, n = 8; arrow in Figure 3I) and the adjacent clypeus bristles of the labrum quartet were lost in more than half of the analyzed RNAi larvae (Figure 3I′). Additionally, the antenna basis bristle and the median maxilla escort seta were sensitive to Tc-rx knock down (Figure 3I′).
Fig. 3. Genes whose knock down result in severe defects in larval cuticles. 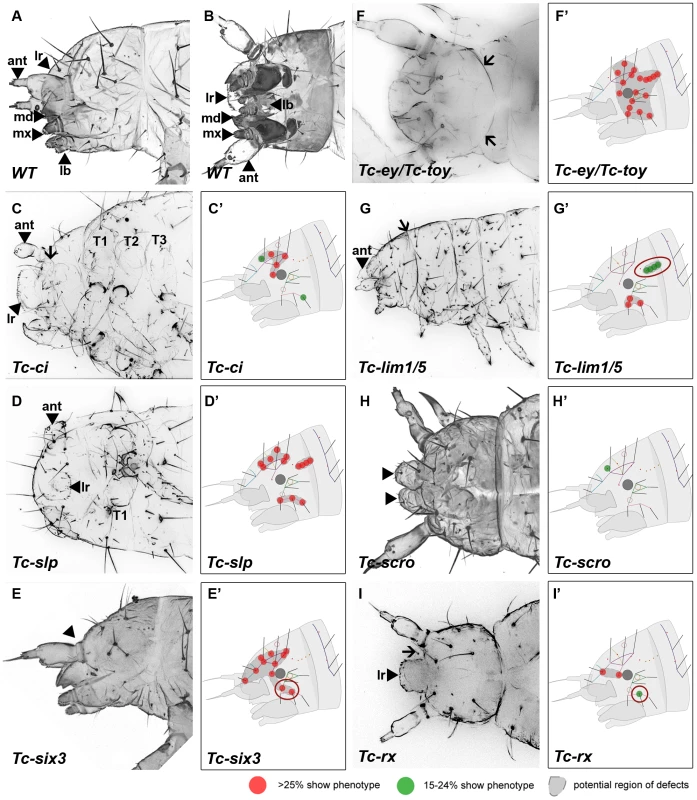
Larval head cuticles and schematic representations of respective head bristle pattern defects. All larval heads are oriented with the anterior to the left. See Figure 2D–2F for fate map. The wildtype bristle pattern is shown in Figure S4 (based on Schinko et al. 2008 but extended with the dorsal ridge row). Red dots indicate disturbance in >25%, green dots in 15–24% of scored cuticles (see Table S2 for quantification). Assuming that tissue surrounding a given bristle is affected as well, a putative region of defects is marked in dark grey (C′–I′). The location of the cuticle defects generally correspond to the embryonic expression domains of the respective genes – exceptions are highlighted with red circles (G′, E′, I′). (A–B) Lateral and ventral view of a wild type larval head cuticle with head bristle pattern, labrum (lr), antenna (ant), mandibles (md), maxillae (mx) and labium (lb). (C) Tc-ci RNAi affects gnathal segments and antennae probably due to its role in segmental Hedgehog signaling. In addition, some dorsal head bristles are strongly affected. (D) Tc-slp RNAi strongly affects gnathal segments in line with being a pair rule gene. Widespread dorsal bristle defects confirm the function in anterior patterning. (E) Tc-six3 RNAi leads to loss of the labrum (black arrowhead in E) and adjacent head bristles. (F) Tc-ey/toy double RNAi leads to strong reduction of head size and disturbance of the “neck” (arrows). In addition, almost all head bristles are strongly affected (see Figure 4 for respective single RNAi). (G) Gnathal appendages are malformed in Tc-lim1/5 RNAi, consistent with its expression in that domain. Strong defects in the lateral head and overall shortening of the head capsule are observed. (H) The labrum is not fused in Tc-scro RNAi embryos (black arrowheads) and one head bristle is affected to some extent. (I) Tc-rx RNAi results in reduction of the labrum and an increased distance between the labrum and the antennae (arrow) while two nearby bristles are affected frequently. RNAi against the remaining genes did not elicit large deletions but alterations of the head bristle pattern (Figure 4 and Table S2). Lateral defects were found after RNAi against Tc-dbx, Tc-ey single RNAi and Tc-gsc (Figure 4A–4C). Tc-ptx and Tc-irx led to dorsal defects (Figure 4D, 4E) while the bristle defects of Tc-toy, Tc-fez and Tc-tll were more widespread (Figure 4F–4H). No bristles were missing in Tc-fgf8 RNAi (n = 29), although lethality of most larvae and a bent flagellum phenotype of the antenna in 41.5% (n = 53, not shown) confirmed the RNAi effect. In summary, we showed that all vertebrate neural plate patterning genes investigated here (except for Tc-fgf8) are indeed involved in anterior head epidermis patterning in Tribolium. By and large, the cuticle defects correspond well with the location of the expression domain, but we also find some indication for indirect defects (see red circles in Figure 3 and Figure 4 and discussion for details).
Fig. 4. Genes whose knock down results in minor defects in larval cuticles. 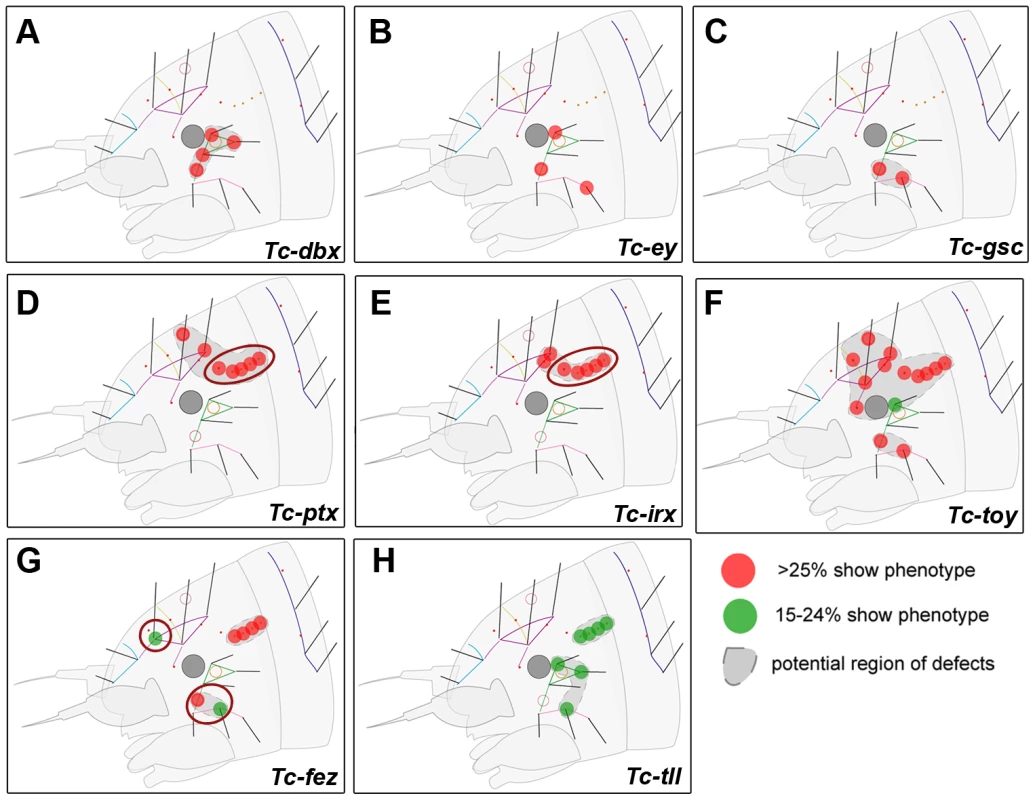
RNAi against these genes did not lead to strong head capsule or head appendage defects (not shown). Hence, only schematic representations of bristle pattern defects are depicted (see Table S2 for quantification). Cuticle defects, which do not correspond to the embryonic expression domains of the respective genes are marked by red circles (D, E, G). (A–C) The knock down of Tc-dbx, Tc-ey/pax6 or Tc-gsc affects the formation of lateral bristles. (D–E) More dorsal defects are found after RNAi against Tc-ptx and Tc- irx. (F) Tc-toy single RNAi leads to widespread disturbance of head bristles while the paralog Tc-ey has a more restricted effect (compare with B). Only double RNAi leads to strong head capsule defects, indicating a synergistic function (Figure 3F). (G–H) Tc-fez and Tc-tll RNAi results in rather widespread defects. Tc-six3 Is a Repressor of Tc-wg and Tc-otd1
Considering its early expression and severe RNAi phenotype, Tc-six3 was likely to play a central role in insect head patterning. Therefore, we centered our subsequent analysis on the epidermal and neural function of this gene. We tested the effect of Tc-six3 RNAi on genes that-based on our expression and RNAi data-were likely to interact. Indeed, the protocerebral neuroectodermal expression domain of Tc-wg (pne) [133] (also called the ocular Tc-wg domain or the head blob in Drosophila [2], [133]) expanded medially and anteriorly in early elongating RNAi embryos (black arrowheads in Figure 5B), resulting in massive median misexpression in fully elongated germbands (Figure 5D). The lateral aspects of Tc-wg expression appeared largely unchanged (open arrows in Figure 5C–5D). Moreover, loss of median embryonic tissue was apparent (white outline in Figure 5C–5D) including the stomodeum (see also Figure 5U) and labrum anlagen (white and black stars in Figure 5C, respectively). As a consequence, the head lobes were not bent outwards and the antennal Tc-wg stripes became perpendicular to the body axis instead of being twisted outwards as in wildtype (compare arrows in Figure 5D with 5C; see Figure S5L–S50 for phenotypes of more advanced stages where the assignment of the antennal stripe becomes obvious). The expression of Tc-otd1 was strongly expanded towards anterior and median tissue (compare Figure 5F, 5H with Figure 5E, 5G) while the lateral aspects appeared normal (open arrowheads in Figure 5G–5H). Despite being partially coexpressed (Tc-tll and Tc-scro) or expressed adjacent to the Tc-six3 domain (Tc-rx), the expression domains of these genes remained unchanged after Tc - six3 RNAi (not shown).
Fig. 5. Tc-six3 is an upstream regulator of the anterior median region of the embryo. 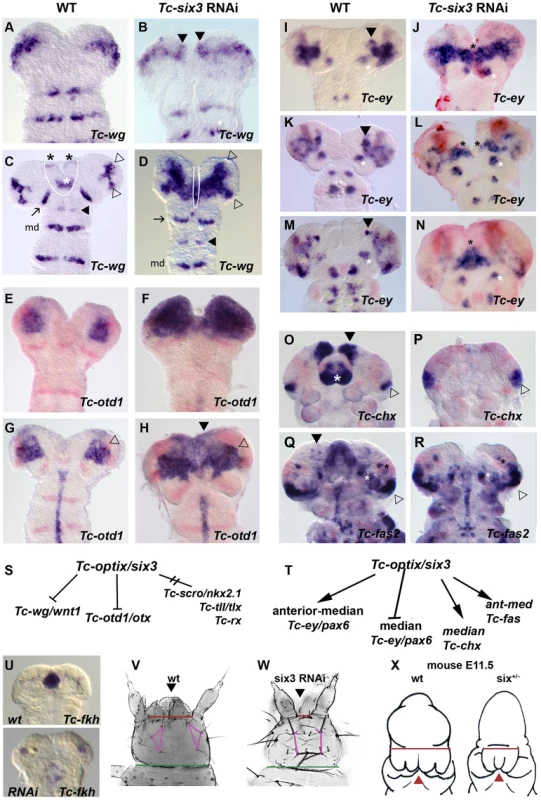
Heads are oriented with anterior to the top. Tc-wg expression is shown in red in E–R. (A–D) Upon Tc-six3 RNAi, Tc-wg is derepressed in the median head (black arrowheads in B, strong medial domain in D) while the lateral parts appear unchanged (compare open arrowheads in D with C). Moreover, an anterior median region including the labrum and stomodeum (black and white asterisks in C, respectively) is lost (compare region outlined by white line in C with D). Intriguingly, the loss of median tissue leads to antennal Tc-wg stripes oriented perpendicular to the body axis (like all trunk stripes) in RNAi embryos. In wildtype, in contrast, they are turned outwards by the intervening tissue (compare distance and orientation of antennal stripes marked by arrow in D with C). Antennal, intercalary and mandibular domains are marked with black arrow, black arrowhead and md in both panels, respectively. (E–H) Upon Tc-six3 RNAi, Tc-otd1 is derepressed medially and anteriorly (e.g. black arrowhead in H) while the lateral expression appears normal (open arrowhead in G and H). (I–N) The effect of Tc-six3 RNAi is different with respect to the various domains of Tc-ey. The major ocular domain is expanded medially (J) and ectopic medial expression can remain paired (black stars in L) or fused at the midline (black star in N). The mushroom body domain of Tc-ey (black arrowhead in I,K,M) is lost in Tc-six3 RNAi embryos while the antennal domains are not altered (white asterisks in I–N). (O–R) Tc-six3 RNAi leads to loss of neuroendocrine markers. (O–P) The pars intercerebralis anlagen marked by Tc-chx (black arrowhead) and the labrum (white asterisk) are lost in Tc-six3 RNAi embryos while the Tc-chx eye domain remains present (open arrowhead in O,P). (Q–R) Tc-fas2 marks neuroendocrine cells of the pars lateralis (black arrowhead in Q), which are lost upon Tc-six3 RNAi, while more lateral aspects including eye expression remain unchanged (black and white stars and open arrowhead). (S–T) Summary of genetic interactions of Tc-six3 identified in this work. Tc-scro, Tc-tll and Tc-rx are not altered after Tc-six3 RNAi (not shown). (U–W) Tc-six3 RNAi leads to the loss of anterior median structures like the stomodeum as shown by the loss of Tc-fkh expression (lower panel of U). (V–W) In dorsal views of head cuticles, loss of the labrum (black arrowhead) and the anterior vertex bristle, and a reduced distance between the antennae (red bar) and the head bristles (pink lines) are evident while more posterior aspects like the width of the neck (green bar) remain unchanged. (X) Strikingly, loss of anterior median head and brain tissue (holoprosencephaly phenotype) is also found in hypomorphic six3 mutants in mouse embryos, where the distance between the eyes is reduced (red bar) and median structures are lost or fused (red arrowhead). Embryos are redrawn from Geng et al. 2008 [170]. Early Ocular Tc-ey/Pax6 Is Repressed by Tc-six3 While More Anterior Expression Domains Require Tc-six3 Function
The effect of Tc-six3 RNAi was different with respect to the various domains of Tc-ey (Figure 5I–5N). Tc-ey expression starts in a prominent ocular domain (open arrowheads in Figure S3 H2–3) before an additional anterior median expression arises (black arrowheads in Figure 5I, 5K, 5M and Figure S3 H4–5). We found coexpression of Tc-dachshund with parts of the anterior median domain (white arrowheads in Figure S5C–S5C″), making it possible that it marks mushroom body anlagen as in Drosophila [139], [140]. In Tc-six3 RNAi embryos, the early ocular domain was strongly expanded towards the midline (compare Figure 5J with Figure 5I). Later, a domain remained visible at the midline (black star in Figure 5N). The anterior median domain did not develop in Tc-six3 RNAi embryos (Figure 5J, 5L, 5N). Again, the lateral aspects of the ocular domain as well as the segmental domains appeared unaffected.
Tc-six3 Is Essential for the Expression of Neuroendocrine Marker Genes
six3 has been implicated in neuroendocrine development in vertebrates [80] and protostomes [81]; and in Drosophila, it is coexpressed with the neuroendocrine markers fas2 and chx [81], [141]. The functional relevance of this co-expression remained unknown. We confirmed co-expression in Tribolium (Figure S5D–S5G, S5H–S5K) and found that in Tc-six3 RNAi embryos the anterior median domains of Tc-fas2 and Tc-chx were absent (compare domains marked by black arrowheads in Figure 5O, 5Q to Figure 5P, 5R).
Tc-six3 Is Required for Development of the Central Body and the Median Brain
In insects, epidermal and neural precursor cells are intermingled in the neuroectoderm. The neural stem cells receive spatial patterning cues before they delaminate from the neuroectoderm and contribute to the central nervous system in a cell autonomous way. The remaining epidermal cells eventually secrete the cuticle [142], [143]. This explains why mutations in segment polarity genes elicit both cuticular and CNS defects. With this in mind, it was likely that Tc-six3 knock down would induce brain defects. First, we determined that eight Tc-ase marked neuroblasts are found within the Tc-six3 marked neuroectoderm until 24 hours of development (extended germband stage, white stars and arrows in Figure 6A). Later, in 42–48 hour old embryos, Tc-six3 is expressed in the developing brain lateral and anterior to the stomodeum (Figure 6C, stomodeum marked by black asterisk). Additionally, staining is evident in the overlaying dorsal epidermis (Figure 6B), the stomodeal roof (Figure 6D) and the labrum (Figure 6E).
Fig. 6. Tc-six3 is required for central body development. 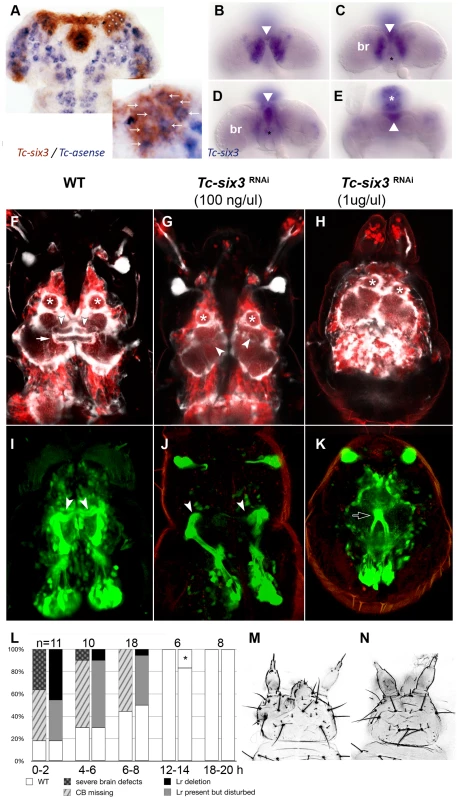
(A) Fully elongated embryo stained for the neuroblast marker Tc-asense (blue) and Tc-six3 (red). Eight neuroblasts arise from the lateral Tc-six3 expression domain by 24 hours of development. (B–E) Staining of Tc-six3 in 42–48 hour old embryos shown in different optical sections taken from dorsal. Expression is observed in the dorsal epidermis (B), in the median brain (br) lateral to the stomodeum (C), the stomodeal roof (D) and the labrum (white asterisk in E). The stomodeum is marked by a black asterisk, the labrum with a white asterisk. Note additional staining of Tc-six3 in median cells (white arrowheads in B–E). (F–K) First instar larval brains are imaged from dorsal within the head cuticle. Anterior is up in all panels; white asterisks mark antennal lobes. (F,I) Wildtype brain with neural cells marked in red, glia shown in white (F) and mushroom bodies marked in green (I). The central body is a neuropil ensheathed with glia, which spans the midline (white arrow). It is located posterior to the median lobes of the mushroom bodies (white arrowheads in F,I). (G,J) After Tc-six3 RNAi with low doses, the central body is not detectable in an otherwise normal brain. The mushroom bodies are intact but their median lobes do not meet at the midline (white arrowheads in G,J). (H,K) In strong RNAi phenotypes, the separation of the brain hemispheres is lost (H) and the mushroom bodies come into close apposition and are reduced in size (black arrow in K). (L) The strength of brain and epidermal phenotypes correlates. Injection of dsRNA against Tc-six3 at different points in time after egg deposition lead to decreasing strength of the cuticle phenotype (right bars) and concomitant decrease of brain phenotype strength (left bars). The black star indicates that in one embryo, a strong cuticle defect was observed, which was not part of the phenotypic series of Tc-six3. As this was likely a background effect or injection artifact, this embryo was not considered as Tc-six3 phenotype. (M) Weak cuticle phenotype with labrum reduced but present. (N) Strong phenotype – note the loss of the labrum and the reduced distance between the antennae. In order to test the hypothesis of a neural function of Tc-six3, we generated transgenic imaging lines marking neural cells, glia and mushroom bodies (Koniszewski, Kollmann, Averof, in preparation) and we identified the central body at the L1 larval stage (white arrow in Figure 6F). Indeed, Tc-six3 RNAi at low doses led to the loss of the central body in an otherwise normal brain (Figure 6G). Higher doses additionally reduced the distance between the two brain hemispheres (Figure 6H). In weak phenotypes the orientation of the median lobes of the mushroom bodies towards the midline was lost (compare white arrowheads in Figure 6J with Figure 6I) while in strong phenotypes, upon convergence of the brain hemispheres, the mushroom bodies approached each other and were reduced in size (see black arrow in Figure 6K).
In the light of the Tc-six3 expression profile, these brain defects could be due to an early neuroectodermal function of Tc-six3 (see neuroectodermal expression in Figure S2B) or to a later function in developing neural cells (see expression in the brain in Figure 6B–6E). In the first case, epidermal and neural phenotypes would be elicited at the same time and, hence, be tightly linked. In the second case, knockdown at late embryonic stages (when epidermal patterning is already finished) would lead to the induction of neural phenotypes in otherwise unaffected heads. To test this, we injected 1 ug/ul Tc-six3 dsRNA in embryos at 0–2, 4–6, 6–8, 12–14 and 18–20 hours post egg laying and scored the resulting L1 larvae for both cuticle and brain phenotypes (Figure 6L). Indeed, the severity of neural and epidermal phenotypes correlated strongly and we never observed brain phenotypes in embryos without cuticle defects. This indicates that both cuticle and brain phenotype are outcomes of the same early neuroectodermal patterning events.
Discussion
Identification of Novel Insect Head Patterning Genes
With our candidate gene approach we identified five genes that had not been implicated in insect head epidermis patterning before (goosecoid, scarecrow, fez1, dbx, ptx). For four additional genes, we show involvement in embryonic head capsule development while a role in adult Drosophila head patterning had been described previously (ci, Drx, lim1, mirror) [144]–[147]. Based on our fate map, the cuticle defects generally correspond well with the head expression of the respective gene. However, the bell row and the setae of the maxilla escort appear to be sensitive to indirect effects because they were affected in several RNAi experiments with genes, which-based on our fatemap-do not show expression in the respective regions (red circles in Figure 3 Tc-lim1/5, Tc-ptx, Tc-irx and Figure 4 Tc-six3, Tc-rx, Tc-fez). Both regions are located where the head lobes are predicted to fuse either with gnathal segments (maxilla escort) or the trunk (bell row; see black stars in Figure 2C). Hence, primary defects of a gene knockdown in head lobe morphology or size could lead to the observed secondary defects.
A Novel Regulator of Central Complex Formation
We show that Tc-six3 is required for proper formation of the central body, which is a midline spanning neuropile and part of the central complex. To our knowledge, this is the first gene known to be required for embryonic central complex development. Our data are in line with cell lineage tracing experiments in grasshopper embryos, where neuroblasts at a corresponding anterior median position contribute to the central complex [148]. Further, expression of optix/six3 in corresponding neuroblasts was also shown in Drosophila [81]. Together, these data are consistent with the hypothesis that Tc-six3 is required in the neuroectoderm for specifying the identity of central body neuroblasts. However, tools to genetically trace the offspring of these neuroblasts [149] are needed to prove this link.
In hemimetabolous insects, which represent the ancestral mode of embryogenesis, all neuropils of the central complex are formed during embryogenesis. In Drosophila, in contrast, the central complex develops during late larval stages [150]–[153]. Tribolium takes an intermediate position by forming a subset of central complex neuropils during embryogenesis, a situation apparently conserved with another tenebrionid beetle [154]. With the newly available brain imaging lines and its amenability to functional genomics Tribolium is an excellent model to investigate the genetics of embryonic central complex development.
Anterior-Median Development in Insects
We showed that Tc-six3 is expressed in an anterior median domain from earliest stages on and that it acts as an upstream component of anterior median patterning. Drosophila optix/six3 is expressed in an anterior blastodermal ring anterior to otd, which persists at the dorsal side [58], [70], [81] and is required for labral and maxillary structures [62]. Its ring like expression does not support an involvement in median patterning but relevant genetic interactions remain to be studied. The later expression in the labrum and in bilateral dorsal domains, however, is similar in both species.
Interestingly, aspects of dorsal median head patterning are controlled by dpp in Drosophila. Shortly before gastrulation, the action of dpp and its downstream target zen at the dorsal midline separate the neuroectoderm into paired anlagen by medial repression of genes and by promoting median cell death. This results in the establishment of bilateral expression of marker genes of the respective brain parts (e.g. Dchx (pars intercerebralis); Fas2 and Drx (pars lateralis); sine oculis and eyes-absent (visual system)) [46], [141], [155], [156]. Actually, many other anterior patterning genes initiate their expression as unpaired domains across the dorsal midline that are subsequently medially subdivided in Drosophila (e.g. otd [157], tll [158], fezf [159], Dsix4 [58]). In contrast, the Tribolium orthologs of most of these genes are initiated as separate bilateral domains (Tc-rx and Tc-fez (Figure 1E and 1D), Tc-chx and Tc-Fas2 (Figure S5), Tc-tll [131]), Tc-six4 [44], Tc-sine oculis, Tc-eyes-absent [160]). Tc-otd1 starts out with ubiquitous expression related to axis formation [130], [161], [162] but then resolves into paired head lobe domains which are separate as with the aforementioned genes.
Due to differences in topology of the head anlagen (see below), median repression of anterior patterning genes by Tc-dpp is not required in Tribolium. Nevertheless, it is expressed along the rim of the head anlagen at blastoderm stages, some parts of which will become the site of dorsal fusion [163], [164]. However, Tc-Dpp activity (detected by antibodies against pMad) does not occur at the site of expression and is clearly distant from the arising Tc-rx, Tc-chx, Tc-six4, Tc-sine oculis or Tc-fas2 domains [21]. Also the Tc-dpp RNAi phenotypes differ from Drosophila mutants in that the head anlagen are expanded and appear to have lost their dorso-ventral orientation (shown by expansion of Tc-otd1 and the proneural gene Tc-ASH) in an overall ventralized embryo [21]. Hence, the early expression of dpp at the future dorsal midline might be ancestral, but its function with respect to medially repressing gene expression has probably evolved in Drosophila.
Tribolium Probably Reflects the More Ancestral Situation
The difference in generation of paired dorsal domains in these two insect species reflects the different location of the head anlagen. In the long germ insect Drosophila, extraembryonic tissues are reduced to the dorsally located amnioserosa while the head anlagen are situated in the anterior dorsal blastoderm from earliest stages on [165], [166]. Consequently, the head lobes are never separated along the midline. In contrast, in the short germ insect Tribolium, the anterior blastoderm gives rise to extraembryonic amnion and serosa, which eventually ensheath the embryo [166]. In contrast to Drosophila, the Tribolium head anlagen are located in the ventral median blastoderm from where they move towards anteriorly and bend dorsally. The head lobes are separate from the beginning but fuse at late stages at the dorsal midline forming the dorsal head (bend and zipper model, see Figure 2A–2C and [4], [137] for more details). During these morphogenetic movements, the initially separate expression domains of the head lobes eventually come into close proximity at the dorsal midline like in Drosophila (Figure 2D–2F). Both the anterior dorsal location of extraembryonic tissue anlagen and the ventral location of the head anlagen are found in most insects [166] and in the hemimetabolous milkweed bug Oncopeltus fasciatus, gene expression data show a clear separated origin of the head lobes in the blastoderm [167]. Hence, Tribolium is likely to represent the ancestral state in insects.
In striking analogy to Drosophila, the expressions of vertebrate eye field patterning genes start out as one midline spanning domain (e.g. Rx and Pax6 [65], [168], [169]). Later, these domains split medially, which is the prerequisite for the formation of bilateral eye anlagen. shh as well as six3 are involved in medial repression of Pax6 and Rx2 [65], [169] with six3 acting upstream of shh [170]. This appears to be more similar to the derived Drosophila situation than to the ancestral split of head lobe anlagen (see above). However, the molecules involved in median split are different (dpp in Drosophila versus six3 and shh in vertebrates) and we find involvement of Tribolium six3 but not dpp in median patterning. Hence, the molecular data actually suggest a higher degree of conservation between Tribolium and vertebrates and convergent evolution of the similarity between Drosophila and vertebrates.
Conserved Function of six3 in Neuroendocrine and Anterior Median Patterning of Bilaterians
Regarding the likely difference to Drosophila, it is striking that the role of vertebrate six3 in median separation of anterior expression domains is similar to what we find in Tribolium. In vertebrates, six3 represses midbrain derived Wnt signaling [72], [73], which we also find in Tribolium. In vertebrates, six3 and its paralog six6 are involved in pituitary and hypothalamus development [76]–[80]. Based on its expression, six3 has been predicted to contribute to neuroendocrine brain parts in annelids and Drosophila [81]. More generally, the similarity of bilaterian neuroendocrine systems and their common origin from placode like precursors have been noted [44], [47], [171]. Here, we have added functional data showing that Tc-six3 is indeed required for the expression of neuroendocrine markers for the pars intercerebralis (Tc-chx) and pars lateralis (Tc-fas2) [141] placing it high in the hierarchy of neuroendocrine development in bilaterians.
In mouse embryos with reduced levels of six3 and shh expression, median head and brain structures are affected (e.g. median nasal prominence) or absent (e.g. nasal septum, the septum, corpus callosum)(see Figure 5X) [170]. Such holoprosencephaly phenotypes are also seen in some human six3 mutations [172]. Very similarly, we see loss of median brain structures in Tribolium after RNAi for Tc-six3 Overall, these similarities functionally confirm that the ancestral role of six3 orthologs was in the anterior median patterning of the Urbilateria (Figure 5V–5X) [81].
Materials and Methods
Animals
Most experiments were performed using the wild type Tribolium castaneum strain San Bernardino. For brain imaging, a transgenic line for 6XP3-ECFP (marking glia) and elongation factor1-alpha regulatory region-DsRedEx (EF1-B; marking neural cells) and the enhancer trap line Gö-11410 (marking mushroom bodies with EGFP; identified in the GEKU screen [173]) were used (Koniszewski, Kollmann, Averof, in preparation).
Identification of Candidate Genes in Tribolium
Mouse protein sequences of the candidate genes (see Table S1) were obtained from the NCBI database (www.ncbi.nlm.nih.gov/). Tribolium orthologs were identified by BLAST at the Beetle Base server (beetlebase.org/). Test of orthology: Tribolium sequences were blasted against the NCBI protein database and the top 5–15 hits from insects, vertebrate and selected other groups were retrieved, as well as the three most similar Tribolium genes. These were BLASTed against the entire NCBI nucleotide database and the first three hits were retrieved. All these sequences were aligned using the ClustalW algorithm of Mega 4 [174],[175]. Phylogenetic trees were calculated in Mega 4 using the Neighbor-Joining method [176] (bootstrap consensus tree inferred from 10.000 replicates [177]; evolutionary distances computed using the Poisson correction method [178]; all positions containing gaps and missing data were eliminated from the dataset (complete deletion option)). See Figure S1 for phylogenetic trees. Phylogenetic relationships for the following genes were already published: Tc-wnt11 and Tc-wg/wnt1 [179], Tc-otd1/otx and Tc-ems/emx [54], Tc-ey/pax6 and Tc-toy/pax6 [136], Tc-eya [160].
Cloning of Candidate Genes
mRNA of 0–48 h embryos was isolated using the MicroPoly(A)Purist Kit (Ambion) and cDNA was synthesized by using the SMART PCR cDNA Synthesis Kit (ClonTech). Gene fragments obtained by PCR with gene specific primers (see Table S3) were cloned into the pCRII vector using the TA Cloning Dual Promotor Kit (Invitrogen) and their sequence was confirmed.
Whole-Mount In Situ Hybridization
Single (NBT/BCIP) and double in situ stainings (NBT/BCIP & FastRed or INT/BCIP) were performed and documented as described [180], [181].
Knock Down of Gene Function by RNA Interference (RNAi)
RNAi was performed by injection of dsRNA into pupae (pRNAi), adults (aRNAi) or embryos (eRNAi) as described [182]–[184]. Lengths of gene fragments and mode of injection are listed in Table S1. Concentrations used in pupal RNAi and adult RNAi: 2–4 µg/µl; in embryonic RNAi: 1–2 µg/µl. A negative control for pupal RNAi was performed by pricking pupae with the needle or injecting water, injection buffer or dsRNA against tGFP. These controls did not show significant developmental effects in the offspring (not shown). In order to identify potential off-target regions, sequences were BLASTed against the Tribolium genome (BLASTn) at Beetle Base. For five genes, sequence similarity of 21 or more successive identical nucleotides was found to hit other gene predictions. For those, RNAi analysis was repeated by another person using subfragments that did not contain the potentially off-target sequences. The phenotypic effects were very similar with respect to the cuticle phenotype (not shown) and the head bristle pattern (Table S2). See Table S3 for primers for the subfragments. Because Tc-six3 was investigated in more detail, two non-overlapping fragments were cloned, injected and analyzed separately by another person. Staining of Tc-six3 in Tc-six3 RNAi embryos confirmed strongly reduced or imperceptible expression in knock down embryos. The cuticle phenotype (not shown) and the head bristle pattern (Table S2) was very similar, confirming specificity.
Epifluorescent and Confocal Imaging
Image stacks of cleared first instar cuticles were gathered by using a Zeiss LSM 510 or a Zeiss Axioplan 2 microscope and projections were calculated as described previously [44], [181]. Brain imaging was performed using a Zeiss LSM 510.
Supporting Information
Zdroje
1. Schmidt-OttUGonzalez-GaitanMJackleHTechnauGM 1994 Number, identity, and sequence of the Drosophila head segments as revealed by neural elements and their deletion patterns in mutants. Proc Natl Acad Sci U S A 91 8363 8367
2. Schmidt-OttUTechnauGM 1992 Expression of en and wg in the embryonic head and brain of Drosophila indicates a refolded band of seven segment remnants. Development 116 111 125
3. PosnienNBashasabFBucherG 2009 The insect upper lip (labrum) is a nonsegmental appendage-like structure. Evol Dev 11 479 487
4. PosnienNSchinkoJBKittelmannSBucherG 2010 Genetics, development and composition of the insect head - A beetle's view. Arthropod Struct Dev E pub ahead of print
5. ScholtzGEdgecombeGD 2006 The evolution of arthropod heads: reconciling morphological, developmental and palaeontological evidence. Dev Genes Evol 216 395 415
6. RogersBTKaufmanTC 1997 Structure of the insect head in ontogeny and phylogeny: a view from Drosophila. Int Rev Cytol 174 1 84
7. CohenSMJurgensG 1990 Mediation of Drosophila head development by gap-like segmentation genes. Nature 346 482 485
8. GrossniklausUPearsonRKGehringWJ 1992 The Drosophila sloppy paired locus encodes two proteins involved in segmentation that show homology to mammalian transcription factors. Genes Dev 6 1030 1051
9. CrozatierMValleDDuboisLIbnsoudaSVincentA 1996 Collier, a novel regulator of Drosophila head development, is expressed in a single mitotic domain. Curr Biol 6 707 718
10. SchockFReischlJWimmerETaubertHPurnellBA 2000 Phenotypic suppression of empty spiracles is prevented by buttonhead. Nature 405 351 354
11. WimmerEACohenSMJackleHDesplanC 1997 buttonhead does not contribute to a combinatorial code proposed for Drosophila head development. Development 124 1509 1517
12. NtiniEWimmerEA 2011 Unique establishment of procephalic head segments is supported by the identification of cis-regulatory elements driving segment-specific segment polarity gene expression in Drosophila. Dev Genes Evol 221 1 16
13. Gallitano-MendelAFinkelsteinR 1997 Novel segment polarity gene interactions during embryonic head development in Drosophila. Dev Biol 192 599 613
14. SnodgrassRE 1935 Principles of Insect Morphology New York McGRaw Hill
15. Campos-OrtegaJAHartensteinV 1985 The Embryonic Development of Drosophila melanogaster New York Springer-Verlag
16. TurnerFRMahowaldAP 1979 Scanning electron microscopy of Drosophila melanogaster embryogenesis. III. Formation of the head and caudal segments. Dev Biol 68 96 109
17. SnodgrassRE 1953 The metamorphosis of a fly's head. Smithsonian Miscellaneous Collections 122 1 25
18. SnodgrassRE 1954 Insect Metamorphosis. Washington
19. StauberMJackleHSchmidt-OttU 1999 The anterior determinant bicoid of Drosophila is a derived Hox class 3 gene. Proc Natl Acad Sci U S A 96 3786 3789
20. SchoppmeierMSchröderR 2005 Maternal torso signaling controls body axis elongation in a short germ insect. Curr Biol 15 2131 2136
21. van der ZeeMStockhammerOvon LevetzowCNunes da FonsecaRRothS 2006 Sog/Chordin is required for ventral-to-dorsal Dpp/BMP transport and head formation in a short germ insect. Proc Natl Acad Sci U S A 103 16307 16312
22. CohenSJurgensG 1991 Drosophila headlines. Trends Genet 7 267 272
23. GrossniklausUCadiganKMGehringWJ 1994 Three maternal coordinate systems cooperate in the patterning of the Drosophila head. Development 120 3155 3171
24. StreckerTRKongsuwanKLengyelJAMerriamJR 1986 The zygotic mutant tailless affects the anterior and posterior ectodermal regions of the Drosophila embryo. Dev Biol 113 64 76
25. MohlerJ 1995 Spatial regulation of segment polarity gene expression in the anterior terminal region of the Drosophila blastoderm embryo. Mech Dev 50 151 161
26. Younossi-HartensteinAGreenPLiawGJRudolphKLengyelJ 1997 Control of early neurogenesis of the Drosophila brain by the head gap genes tll, otd, ems, and btd. Dev Biol 182 270 283
27. SimeoneAAcamporaDGulisanoMStornaiuoloABoncinelliE 1992 Nested expression domains of four homeobox genes in developing rostral brain. Nature 358 687 690
28. SimeoneAGulisanoMAcamporaDStornaiuoloARambaldiM 1992 Two vertebrate homeobox genes related to the Drosophila empty spiracles gene are expressed in the embryonic cerebral cortex. Embo J 11 2541 2550
29. LeuzingerSHirthFGerlichDAcamporaDSimeoneA 1998 Equivalence of the fly orthodenticle gene and the human OTX genes in embryonic brain development of Drosophila. Development 125 1703 1710
30. HirthFReichertH 1999 Conserved genetic programs in insect and mammalian brain development. Bioessays 21 677 684
31. HollandPInghamPKraussS 1992 Development and evolution. Mice and flies head to head. Nature 358 627 628
32. YuRTMcKeownMEvansRMUmesonoK 1994 Relationship between Drosophila gap gene tailless and a vertebrate nuclear receptor Tlx. Nature 370 375 379
33. MonaghanAPGrauEBockDSchutzG 1995 The mouse homolog of the orphan nuclear receptor tailless is expressed in the developing forebrain. Development 121 839 853
34. AcamporaDAvantaggiatoVTuortoFBaronePReichertH 1998 Murine Otx1 and Drosophila otd genes share conserved genetic functions required in invertebrate and vertebrate brain development. Development 125 1691 1702
35. HirthFKammermeierLFreiEWalldorfUNollM 2003 An urbilaterian origin of the tripartite brain: developmental genetic insights from Drosophila. Development 130 2365 2373
36. ReichertH 2005 A tripartite organization of the urbilaterian brain: developmental genetic evidence from Drosophila. Brain Res Bull 66 491 494
37. De VelascoBShenJGoSHartensteinV 2004 Embryonic development of the Drosophila corpus cardiacum, a neuroendocrine gland with similarity to the vertebrate pituitary, is controlled by sine oculis and glass. Dev Biol 274 280 294
38. ErclikTHartensteinVLipshitzHDMcInnesRR 2008 Conserved role of the Vsx genes supports a monophyletic origin for bilaterian visual systems. Curr Biol 18 1278 1287
39. EggertTHauckBHildebrandtNGehringWJWalldorfU 1998 Isolation of a Drosophila homolog of the vertebrate homeobox gene Rx and its possible role in brain and eye development. Proc Natl Acad Sci U S A 95 2343 2348
40. DavisRJShenWHeanueTAMardonG 1999 Mouse Dach, a homologue of Drosophila dachshund, is expressed in the developing retina, brain and limbs. Dev Genes Evol 209 526 536
41. Gomez-SkarmetaJLGlavicAde la Calle-MustienesEModolellJMayorR 1998 Xiro, a Xenopus homolog of the Drosophila Iroquois complex genes, controls development at the neural plate. Embo J 17 181 190
42. GorielyAStellaMCoffinierCKesslerDMailhosC 1996 A functional homologue of goosecoid in Drosophila. Development 122 1641 1650
43. UrbachR 2007 A procephalic territory in Drosophila exhibiting similarities and dissimilarities compared to the vertebrate midbrain/hindbrain boundary region. Neural Develop 2 23
44. PosnienNKoniszewskiNBucherG 2011 Insect Tc-six4 marks a unit with similarity to vertebrate placodes. Dev Biol
45. CavodeassiFModolellJGomez-SkarmetaJL 2001 The Iroquois family of genes: from body building to neural patterning. Development 128 2847 2855
46. HartensteinVRehTA 2002 Homologies between vertebrate and invertebrate eyes. MosesK Drosophila eye development Berlin Heidelberg Springer-Verlag
47. HartensteinV 2006 The neuroendocrine system of invertebrates: a developmental and evolutionary perspective. J Endocrinol 190 555 570
48. TomerRDenesASTessmar-RaibleKArendtD 2010 Profiling by image registration reveals common origin of annelid mushroom bodies and vertebrate pallium. Cell 142 800 809
49. DenesASJekelyGSteinmetzPRRaibleFSnymanH 2007 Molecular architecture of annelid nerve cord supports common origin of nervous system centralization in bilateria. Cell 129 277 288
50. ArendtD 2005 Genes and homology in nervous system evolution: comparing gene functions, expression patterns, and cell type molecular fingerprints. Theory Biosci 124 185 197
51. Tessmar-RaibleKRaibleFChristodoulouFGuyKRemboldM 2007 Conserved sensory-neurosecretory cell types in annelid and fish forebrain: insights into hypothalamus evolution. Cell 129 1389 1400
52. ArendtDDenesASJekelyGTessmar-RaibleK 2008 The evolution of nervous system centralization. Philos Trans R Soc Lond B Biol Sci 363 1523 1528
53. BucherGWimmerEA 2005 Beetle a-head. BIF Futura 20 164 169
54. SchinkoJBKreuzerNOffenNPosnienNWimmerEA 2008 Divergent functions of orthodenticle, empty spiracles and buttonhead in early head patterning of the beetle Tribolium castaneum (Coleoptera). Dev Biol 317 600 613
55. CernyACGrossmannDBucherGKlinglerM 2008 The Tribolium ortholog of knirps and knirps-related is crucial for head segmentation but plays a minor role during abdominal patterning. Dev Biol 321 284 294
56. SchaeperNDPechmannMDamenWGPrpicNMWimmerEA 2010 Evolutionary plasticity of collier function in head development of diverse arthropods. Dev Biol 344 363 376
57. EconomouADTelfordMJ 2009 Comparative gene expression in the heads of Drosophila melanogaster and Tribolium castaneum and the segmental affinity of the Drosophila hypopharyngeal lobes. Evol Dev 11 88 96
58. SeoHCCurtissJMlodzikMFjoseA 1999 Six class homeobox genes in drosophila belong to three distinct families and are involved in head development. Mech Dev 83 127 139
59. CheyetteBNGreenPJMartinKGarrenHHartensteinV 1994 The Drosophila sine oculis locus encodes a homeodomain-containing protein required for the development of the entire visual system. Neuron 12 977 996
60. SerikakuMAO'TousaJE 1994 sine oculis is a homeobox gene required for Drosophila visual system development. Genetics 138 1137 1150
61. PignoniFHuBZavitzKHXiaoJGarrityPA 1997 The eye-specification proteins So and Eya form a complex and regulate multiple steps in Drosophila eye development. Cell 91 881 891
62. CoiffierDCharrouxBKerridgeS 2008 Common functions of central and posterior Hox genes for the repression of head in the trunk of Drosophila. Development 135 291 300
63. LiuWLagutinOSwindellEJamrichMOliverG 2010 Neuroretina specification in mouse embryos requires Six3-mediated suppression of Wnt8b in the anterior neural plate. J Clin Invest 120 3568 3577
64. Del BeneFTessmar-RaibleKWittbrodtJ 2004 Direct interaction of geminin and Six3 in eye development. Nature 427 745 749
65. CarlMLoosliFWittbrodtJ 2002 Six3 inactivation reveals its essential role for the formation and patterning of the vertebrate eye. Development 129 4057 4063
66. GoudreauGPetrouPRenekerLWGrawJLosterJ 2002 Mutually regulated expression of Pax6 and Six3 and its implications for the Pax6 haploinsufficient lens phenotype. Proc Natl Acad Sci U S A 99 8719 8724
67. LagutinOZhuCCFurutaYRowitchDHMcMahonAP 2001 Six3 promotes the formation of ectopic optic vesicle-like structures in mouse embryos. Dev Dyn 221 342 349
68. LoosliFWinklerSWittbrodtJ 1999 Six3 overexpression initiates the formation of ectopic retina. Genes Dev 13 649 654
69. ToyJYangJMLeppertGSSundinOH 1998 The optx2 homeobox gene is expressed in early precursors of the eye and activates retina-specific genes. Proc Natl Acad Sci U S A 95 10643 10648
70. SeimiyaMGehringWJ 2000 The Drosophila homeobox gene optix is capable of inducing ectopic eyes by an eyeless-independent mechanism. Development 127 1879 1886
71. GestriGCarlMAppolloniIWilsonSWBarsacchiG 2005 Six3 functions in anterior neural plate specification by promoting cell proliferation and inhibiting Bmp4 expression. Development 132 2401 2413
72. LagutinOVZhuCCKobayashiDTopczewskiJShimamuraK 2003 Six3 repression of Wnt signaling in the anterior neuroectoderm is essential for vertebrate forebrain development. Genes Dev 17 368 379
73. LavadoALagutinOVOliverG 2008 Six3 inactivation causes progressive caudalization and aberrant patterning of the mammalian diencephalon. Development 135 441 450
74. LoosliFKosterRWCarlMKroneAWittbrodtJ 1998 Six3, a medaka homologue of the Drosophila homeobox gene sine oculis is expressed in the anterior embryonic shield and the developing eye. Mech Dev 74 159 164
75. KobayashiMToyamaRTakedaHDawidIBKawakamiK 1998 Overexpression of the forebrain-specific homeobox gene six3 induces rostral forebrain enlargement in zebrafish. Development 125 2973 2982
76. Gaston-MassuetCAndoniadouCLSignoreMSajediEBirdS 2008 Genetic interaction between the homeobox transcription factors HESX1 and SIX3 is required for normal pituitary development. Dev Biol 324 322 333
77. JeanDBernierGGrussP 1999 Six6 (Optx2) is a novel murine Six3-related homeobox gene that demarcates the presumptive pituitary/hypothalamic axis and the ventral optic stalk. Mech Dev 84 31 40
78. GhanbariHSeoHCFjoseABrandliAW 2001 Molecular cloning and embryonic expression of Xenopus Six homeobox genes. Mech Dev 101 271 277
79. GallardoMELopez-RiosJFernaud-EspinosaIGranadinoBSanzR 1999 Genomic cloning and characterization of the human homeobox gene SIX6 reveals a cluster of SIX genes in chromosome 14 and associates SIX6 hemizygosity with bilateral anophthalmia and pituitary anomalies. Genomics 61 82 91
80. OliverGMailhosAWehrRCopelandNGJenkinsNA 1995 Six3, a murine homologue of the sine oculis gene, demarcates the most anterior border of the developing neural plate and is expressed during eye development. Development 121 4045 4055
81. SteinmetzPRUrbachRPosnienNErikssonJKostyuchenkoRP 2010 Six3 demarcates the anterior-most developing brain region in bilaterian animals. Evodevo 1 14
82. BoncinelliEGulisanoMBroccoliV 1993 Emx and Otx homeobox genes in the developing mouse brain. J Neurobiol 24 1356 1366
83. RubensteinJLShimamuraKMartinezSPuellesL 1998 Regionalization of the prosencephalic neural plate. Annu Rev Neurosci 21 445 477
84. RubensteinJLRShimamuraK 1997 Regulation of patterning and differentiation in the embryonic vertebrate forebrain. CowanWMJessellTMZipurskySL Molecular and Cellular Approaches to Neural Development New York Oxford Oxford University Press 356 390
85. TakahashiMOsumiN 2008 Expression study of cadherin7 and cadherin20 in the embryonic and adult rat central nervous system. BMC Dev Biol 8 87
86. HollemannTBellefroidEPielerT 1998 The Xenopus homologue of the Drosophila gene tailless has a function in early eye development. Development 125 2425 2432
87. KitambiSSHauptmannG 2007 The zebrafish orphan nuclear receptor genes nr2e1 and nr2e3 are expressed in developing eye and forebrain. Gene Expr Patterns 7 521 528
88. ShengHZBertuzziSChiangCShawlotWTairaM 1997 Expression of murine Lhx5 suggests a role in specifying the forebrain. Dev Dyn 208 266 277
89. CamusADavidsonBPBilliardsSKhooPRivera-PerezJA 2000 The morphogenetic role of midline mesendoderm and ectoderm in the development of the forebrain and the midbrain of the mouse embryo. Development 127 1799 1813
90. LemaireLRoeserTIzpisua-BelmonteJCKesselM 1997 Segregating expression domains of two goosecoid genes during the transition from gastrulation to neurulation in chick embryos. Development 124 1443 1452
91. ChuangJCMathersPHRaymondPA 1999 Expression of three Rx homeobox genes in embryonic and adult zebrafish. Mech Dev 84 195 198
92. DeschetKBourratFRistoratoreFChourroutDJolyJS 1999 Expression of the medaka (Oryzias latipes) Ol-Rx3 paired-like gene in two diencephalic derivatives, the eye and the hypothalamus. Mech Dev 83 179 182
93. MathersPHGrinbergAMahonKAJamrichM 1997 The Rx homeobox gene is essential for vertebrate eye development. Nature 387 603 607
94. MeijlinkFBeverdamABrouwerAOosterveenTCBergeDT 1999 Vertebrate aristaless-related genes. Int J Dev Biol 43 651 663
95. HashimotoHYabeTHirataTShimizuTBaeY 2000 Expression of the zinc finger gene fez-like in zebrafish forebrain. Mech Dev 97 191 195
96. HirataTNakazawaMMuraokaONakayamaRSudaY 2006 Zinc-finger genes Fez and Fez-like function in the establishment of diencephalon subdivisions. Development 133 3993 4004
97. HirataTSudaYNakaoKNarimatsuMHiranoT 2004 Zinc finger gene fez-like functions in the formation of subplate neurons and thalamocortical axons. Dev Dyn 230 546 556
98. JeongJYEinhornZMathurPChenLLeeS 2007 Patterning the zebrafish diencephalon by the conserved zinc-finger protein Fezl. Development 134 127 136
99. MatsuoISudaYYoshidaMUekiTKimuraC 1997 Otx and Emx functions in patterning of the vertebrate rostral head. Cold Spring Harb Symp Quant Biol 62 545 553
100. McMahonAPJoynerALBradleyAMcMahonJA 1992 The midbrain-hindbrain phenotype of Wnt-1-/Wnt-1 - mice results from stepwise deletion of engrailed-expressing cells by 9.5 days postcoitum. Cell 69 581 595
101. AotoKNishimuraTEtoKMotoyamaJ 2002 Mouse GLI3 regulates Fgf8 expression and apoptosis in the developing neural tube, face, and limb bud. Dev Biol 251 320 332
102. HebertJMFishellG 2008 The genetics of early telencephalon patterning: some assembly required. Nat Rev Neurosci
103. GlavicAGomez-SkarmetaJLMayorR 2002 The homeoprotein Xiro1 is required for midbrain-hindbrain boundary formation. Development 129 1609 1621
104. ScholppSLohsCBrandM 2003 Engrailed and Fgf8 act synergistically to maintain the boundary between diencephalon and mesencephalon. Development 130 4881 4893
105. ShimamuraKRubensteinJL 1997 Inductive interactions direct early regionalization of the mouse forebrain. Development 124 2709 2718
106. FjoseAIzpisua-BelmonteJCFromental-RamainCDubouleD 1994 Expression of the zebrafish gene hlx-1 in the prechordal plate and during CNS development. Development 120 71 81
107. LuSBogaradLDMurthaMTRuddleFH 1992 Expression pattern of a murine homeobox gene, Dbx, displays extreme spatial restriction in embryonic forebrain and spinal cord. Proc Natl Acad Sci U S A 89 8053 8057
108. GershonAARudnickJKalamLZimmermanK 2000 The homeodomain-containing gene Xdbx inhibits neuronal differentiation in the developing embryo. Development 127 2945 2954
109. LuSWiseTLRuddleFH 1994 Mouse homeobox gene Dbx: sequence, gene structure and expression pattern during mid-gestation. Mech Dev 47 187 195
110. ShojiHItoTWakamatsuYHayasakaNOhsakiK 1996 Regionalized expression of the Dbx family homeobox genes in the embryonic CNS of the mouse. Mech Dev 56 25 39
111. DickinsonASiveH 2007 Positioning the extreme anterior in Xenopus: cement gland, primary mouth and anterior pituitary. Semin Cell Dev Biol 18 525 533
112. DuttaSDietrichJEAspockGBurdineRDSchierA 2005 pitx3 defines an equivalence domain for lens and anterior pituitary placode. Development 132 1579 1590
113. SchweickertASteinbeisserHBlumM 2001 Differential gene expression of Xenopus Pitx1, Pitx2b and Pitx2c during cement gland, stomodeum and pituitary development. Mech Dev 107 191 194
114. ZilinskiCAShahRLaneMEJamrichM 2005 Modulation of zebrafish pitx3 expression in the primordia of the pituitary, lens, olfactory epithelium and cranial ganglia by hedgehog and nodal signaling. Genesis 41 33 40
115. CrossleyPHMartinGR 1995 The mouse Fgf8 gene encodes a family of polypeptides and is expressed in regions that direct outgrowth and patterning in the developing embryo. Development 121 439 451
116. ColomboEGalliRCossuGGeczJBroccoliV 2004 Mouse orthologue of ARX, a gene mutated in several X-linked forms of mental retardation and epilepsy, is a marker of adult neural stem cells and forebrain GABAergic neurons. Dev Dyn 231 631 639
117. El-HodiriHMQiXLSeufertDW 2003 The Xenopus arx gene is expressed in the developing rostral forebrain. Dev Genes Evol 212 608 612
118. FriocourtGPoirierKRakicSParnavelasJGChellyJ 2006 The role of ARX in cortical development. Eur J Neurosci 23 869 876
119. SeufertDWPrescottNLEl-HodiriHM 2005 Xenopus aristaless-related homeobox (xARX) gene product functions as both a transcriptional activator and repressor in forebrain development. Dev Dyn 232 313 324
120. CavodeassiFCarreira-BarbosaFYoungRMConchaMLAllendeML 2005 Early stages of zebrafish eye formation require the coordinated activity of Wnt11, Fz5, and the Wnt/beta-catenin pathway. Neuron 47 43 56
121. BroccoliVColomboECossuG 2002 Dmbx1 is a paired-box containing gene specifically expressed in the caudal most brain structures. Mech Dev 114 219 223
122. GogoiRNSchubertFRMartinez-BarberaJPAcamporaDSimeoneA 2002 The paired-type homeobox gene Dmbx1 marks the midbrain and pretectum. Mech Dev 114 213 217
123. HallonetMHollemannTWehrRJenkinsNACopelandNG 1998 Vax1 is a novel homeobox-containing gene expressed in the developing anterior ventral forebrain. Development 125 2599 2610
124. JonesFSKioussiCCopertinoDWKallunkiPHolstBD 1997 Barx2, a new homeobox gene of the Bar class, is expressed in neural and craniofacial structures during development. Proc Natl Acad Sci U S A 94 2632 2637
125. KawaharaAChienCBDawidIB 2002 The homeobox gene mbx is involved in eye and tectum development. Dev Biol 248 107 117
126. MartynovaNEroshkinFErmakovaGBayramovAGrayJ 2004 Patterning the forebrain: FoxA4a/Pintallavis and Xvent2 determine the posterior limit of Xanf1 expression in the neural plate. Development 131 2329 2338
127. ThomasPBeddingtonR 1996 Anterior primitive endoderm may be responsible for patterning the anterior neural plate in the mouse embryo. Curr Biol 6 1487 1496
128. BeermannASchroderR 2008 Sites of Fgf signalling and perception during embryogenesis of the beetle Tribolium castaneum. Dev Genes Evol 218 153 167
129. StathopoulosATamBRonshaugenMFraschMLevineM 2004 pyramus and thisbe: FGF genes that pattern the mesoderm of Drosophila embryos. Genes Dev 18 687 699
130. LiYBrownSJHausdorfBTautzDDenellRE 1996 Two orthodenticle-related genes in the short germ beetle Tribolium castaneum. Dev Genes Evol 206 35 45
131. SchröderREckertCWolffCTautzD 2000 Conserved and divergent aspects of terminal patterning in the beetle Tribolium castaneum. Proc Natl Acad Sci U S A 97 6591 6596
132. FarzanaLBrownSJ 2008 Hedgehog signaling pathway function conserved in Tribolium segmentation. Dev Genes Evol 218 181 192
133. LiuZYangXDongYFriedrichM 2006 Tracking down the “head blob”: comparative analysis of wingless expression in the developing insect procephalon reveals progressive reduction of embryonic visual system patterning in higher insects. Arthropod Struct Dev 35 341 356
134. NagyLMCarrollS 1994 Conservation of wingless patterning functions in the short-germ embryos of Tribolium castaneum. Nature 367 460 463
135. ChoeCPBrownSJ 2007 Evolutionary flexibility of pair-rule patterning revealed by functional analysis of secondary pair-rule genes, paired and sloppy-paired in the short-germ insect, Tribolium castaneum. Dev Biol 302 281 294
136. YangXWeberMZarinkamarNPosnienNFriedrichF 2009 Probing the Drosophila retinal determination gene network in Tribolium (II): The Pax6 genes eyeless and twin of eyeless. Dev Biol
137. PosnienNBucherG 2010 Formation of the insect head involves lateral contribution of the intercalary segment, which depends on Tc-labial function. Dev Biol 338 107 116
138. BolognesiRFarzanaLFischerTDBrownSJ 2008 Multiple Wnt genes are required for segmentation in the short-germ embryo of Tribolium castaneum. Curr Biol 18 1624 1629
139. NoveenADanielAHartensteinV 2000 Early development of the Drosophila mushroom body: the roles of eyeless and dachshund. Development 127 3475 3488
140. KurusuMNagaoTWalldorfUFlisterSGehringWJ 2000 Genetic control of development of the mushroom bodies, the associative learning centers in the Drosophila brain, by the eyeless, twin of eyeless, and Dachshund genes. Proc Natl Acad Sci U S A 97 2140 2144
141. de VelascoBErclikTShyDSclafaniJLipshitzH 2007 Specification and development of the pars intercerebralis and pars lateralis, neuroendocrine command centers in the Drosophila brain. Dev Biol 302 309 323
142. TechnauGMBergerCUrbachR 2006 Generation of cell diversity and segmental pattern in the embryonic central nervous system of Drosophila. Dev Dyn 235 861 869
143. SkeathJBThorS 2003 Genetic control of Drosophila nerve cord development. Curr Opin Neurobiol 13 8 15
144. AminALiYFinkelsteinR 1999 Hedgehog activates the EGF receptor pathway during Drosophila head development. Development 126 2623 2630
145. DavisRJTavsanliBCDittrichCWalldorfUMardonG 2003 Drosophila retinal homeobox (drx) is not required for establishment of the visual system, but is required for brain and clypeus development. Dev Biol 259 272 287
146. RoignantJYLegentKJanodyFTreismanJE 2010 The transcriptional co-factor Chip acts with LIM-homeodomain proteins to set the boundary of the eye field in Drosophila. Development 137 273 281
147. CavodeassiFModolellJCampuzanoS 2000 The Iroquois homeobox genes function as dorsal selectors in the Drosophila head. Development 127 1921 1929
148. WilliamsJLBoyanGS 2008 Building the central complex of the grasshopper Schistocerca gregaria: axons pioneering the w, x, y, z tracts project onto the primary commissural fascicle of the brain. Arthropod Struct Dev 37 129 140
149. EvansCJOlsonJMNgoKTKimELeeNE 2009 G-TRACE: rapid Gal4-based cell lineage analysis in Drosophila. Nat Methods 6 603 605
150. RennSCArmstrongJDYangMWangZAnX 1999 Genetic analysis of the Drosophila ellipsoid body neuropil: organization and development of the central complex. J Neurobiol 41 189 207
151. PereanuWYounossi-HartensteinALovickJSpindlerSHartensteinV 2011 Lineage-based analysis of the development of the central complex of the Drosophila brain. J Comp Neurol 519 661 689
152. YoungJMArmstrongJD Building the central complex in Drosophila: the generation and development of distinct neural subsets. J Comp Neurol 518 1525 1541
153. WilliamsJLD 1975 Anatomical studies of the insect central nervous system: a ground plan of the midbrain and an introduction to the central complex in the locust, Schistocerca gregaria (Orthoptera). Journal of Zoology, London 176 67 86
154. WegerhoffRBreidbachO 1992 Structure and development of the larval central complex in a holometabolous insect, the beetle Tenebrio molitor. Cell Tissue Res 268 341 358
155. ChangTMazottaJDumstreiKDumitrescuAHartensteinV 2001 Dpp and Hh signaling in the Drosophila embryonic eye field. Development 128 4691 4704
156. ChangTShyDHartensteinV 2003 Antagonistic relationship between Dpp and EGFR signaling in Drosophila head patterning. Dev Biol 263 103 113
157. FinkelsteinRSmouseDCapaciTMSpradlingACPerrimonN 1990 The orthodenticle gene encodes a novel homeo domain protein involved in the development of the Drosophila nervous system and ocellar visual structures. Genes Dev 4 1516 1527
158. PignoniFBaldarelliRMSteingrimssonEDiazRJPatapoutianA 1990 The Drosophila gene tailless is expressed at the embryonic termini and is a member of the steroid receptor superfamily. Cell 62 151 163
159. IrimiaMPineiroCMaesoIGomez-SkarmetaJLCasaresF 2010 Conserved developmental expression of Fezf in chordates and Drosophila and the origin of the Zona Limitans Intrathalamica (ZLI) brain organizer. Evodevo 1 7
160. YangXZarinkamarNBaoRFriedrichM 2009 Probing the Drosophila retinal determination gene network in Tribolium (I): The early retinal genes dachshund, eyes absent and sine oculis. Dev Biol 333 202 214
161. KotkampKKlinglerMSchoppmeierM 2010 Apparent role of Tribolium orthodenticle in anteroposterior blastoderm patterning largely reflects novel functions in dorsoventral axis formation and cell survival. Development 137 1853 1862
162. SchröderR 2003 The genes orthodenticle and hunchback substitute for bicoid in the beetle Tribolium. Nature 422 621 625
163. ChenGHandelKRothS 2000 The maternal NF-kappaB/dorsal gradient of Tribolium castaneum: dynamics of early dorsoventral patterning in a short-germ beetle. Development 127 5145 5156
164. Sanchez-SalazarJPletcherMTBennettRBrownSJDandamudiTJ 1996 The Tribolium decapentaplegic gene is similar in sequence, structure, and expression to the Drosophila dpp gene. Dev Genes Evol 206 237 246
165. TechnauGMCampos-OrtegaJA 1985 Fate-mapping in wildtype Drosophila melanogaster. II. Injections of horseradish peroxidase in cells of the early gastrula stage. Roux's Arch Dev Biol 194 196 212
166. RothS 2004 Gastrulation in other insects. SternCD Gastrulation: From cells to Embryo New York Cold Spring Harbor Laboratory Press 105 121
167. BirkanMSchaeperNDChipmanAD 2011 Early patterning and blastodermal fate map of the head in the milkweed bug Oncopeltus fasciatus. Evol Dev 13 436 447
168. ZuberMEGestriGViczianASBarsacchiGHarrisWA 2003 Specification of the vertebrate eye by a network of eye field transcription factors. Development 130 5155 5167
169. MacdonaldRBarthKAXuQHolderNMikkolaI 1995 Midline signalling is required for Pax gene regulation and patterning of the eyes. Development 121 3267 3278
170. GengXSpeirsCLagutinOInbalALiuW 2008 Haploinsufficiency of Six3 fails to activate Sonic hedgehog expression in the ventral forebrain and causes holoprosencephaly. Dev Cell 15 236 247
171. Tessmar-RaibleK 2007 The evolution of neurosecretory centers in bilaterian forebrains: insights from protostomes. Semin Cell Dev Biol 18 492 501
172. CohenMMJr 2006 Holoprosencephaly: clinical, anatomic, and molecular dimensions. Birth Defects Res A Clin Mol Teratol 76 658 673
173. TraunerJSchinkoJLorenzenMDShippyTDWimmerEA 2009 Large-scale insertional mutagenesis of a coleopteran stored grain pest, the red flour beetle Tribolium castaneum, identifies embryonic lethal mutations and enhancer traps. BMC Biol 7 73
174. KumarSNeiMDudleyJTamuraK 2008 MEGA: a biologist-centric software for evolutionary analysis of DNA and protein sequences. Brief Bioinform 9 299 306
175. TamuraKDudleyJNeiMKumarS 2007 MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol 24 1596 1599
176. SaitouNNeiM 1987 The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4 406 425
177. FelsensteinJ 1985 Confidence limits on phylogenies: An approach using the bootstrap. Evolution 39 783 791
178. ZuckerkandlEPaulingL 1965 Evolutionary divergence and convergence in proteins. BrysonVVogelHJ Evolving Genes and Proteins New York Academic Press 97 166
179. BolognesiRBeermannAFarzanaLWittkoppNLutzR 2008 Tribolium Wnts: evidence for a larger repertoire in insects with overlapping expression patterns that suggest multiple redundant functions in embryogenesis. Dev Genes Evol 218 193 202
180. SchinkoJPosnienNKittelmannSKoniszewskiNBucherG 2009 Single and Double Whole-Mount In Situ Hybridization in Red Flour Beetle (Tribolium) Embryos. Cold Spring Harbor Protocols 2009 pdb.prot5258-
181. WohlfromHSchinkoJBKlinglerMBucherG 2006 Maintenance of segment and appendage primordia by the Tribolium gene knodel. Mech Dev 123 430 439
182. BrownSJMahaffeyJPLorenzenMDDenellREMahaffeyJW 1999 Using RNAi to investigate orthologous homeotic gene function during development of distantly related insects. Evol Dev 1 11 15
183. BucherGScholtenJKlinglerM 2002 Parental RNAi in Tribolium (Coleoptera). Curr Biol 12 R85 86
184. PosnienNSchinkoJGrossmannDShippyTDKonopovaB 2009 RNAi in the red flour beetle (Tribolium). Cold Spring Harb Protoc 2009 pdb prot5256
185. BeermannASchroderR 2004 Functional stability of the aristaless gene in appendage tip formation during evolution. Dev Genes Evol 214 303 308
Štítky
Genetika Reprodukční medicína
Článek A Complex Genomic Rearrangement Involving the Locus Causes Dermal Hyperpigmentation in the ChickenČlánek Genome Instability and Transcription Elongation Impairment in Human Cells Depleted of THO/TREXČlánek A Population Genetics-Phylogenetics Approach to Inferring Natural Selection in Coding SequencesČlánek Interspecific Sex in Grass Smuts and the Genetic Diversity of Their Pheromone-Receptor SystemČlánek Genomic Distribution and Inter-Sample Variation of Non-CpG Methylation across Human Cell Types
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2011 Číslo 12- Akutní intermitentní porfyrie
- Souvislost haplotypu M2 genu pro annexin A5 s opakovanými reprodukčními ztrátami
- Příjem alkoholu a menstruační cyklus
- Doporučení pro diagnostiku a léčbu akutních jaterních porfyrií
- Doc. Miloš Kubánek: Nemocní se srdeční amyloidózou jsou často skryti a sledováni pod jinými diagnózami
-
Všechny články tohoto čísla
- The Connection between Space and Thinking: An Interview with Rafael Viñoly
- An Assessment of the Individual and Collective Effects of Variants on Height Using Twins and a Developmentally Informative Study Design
- Widespread Cotranslational Formation of Protein Complexes
- Genomes Reveal Transition of Bacteria from Aquatic to Terrestrial Environments
- A Complex Genomic Rearrangement Involving the Locus Causes Dermal Hyperpigmentation in the Chicken
- Plasticity of BRCA2 Function in Homologous Recombination: Genetic Interactions of the PALB2 and DNA Binding Domains
- Transcription Is Required to Establish Maternal Imprinting at the Prader-Willi Syndrome and Angelman Syndrome Locus
- Substitutions in the Amino-Terminal Tail of Neurospora Histone H3 Have Varied Effects on DNA Methylation
- MAPK/ERK Signaling Regulates Insulin Sensitivity to Control Glucose Metabolism in
- A Comprehensive Analysis of Shared Loci between Systemic Lupus Erythematosus (SLE) and Sixteen Autoimmune Diseases Reveals Limited Genetic Overlap
- Genome Instability and Transcription Elongation Impairment in Human Cells Depleted of THO/TREX
- Genome-Wide Meta-Analysis of Five Asian Cohorts Identifies as a Susceptibility Locus for Corneal Astigmatism
- A Population Genetics-Phylogenetics Approach to Inferring Natural Selection in Coding Sequences
- HIF-1 Regulates Iron Homeostasis in by Activation and Inhibition of Genes Involved in Iron Uptake and Storage
- Ror2 Enhances Polarity and Directional Migration of Primordial Germ Cells
- DNA Methylation of the Gonadal Aromatase () Promoter Is Involved in Temperature-Dependent Sex Ratio Shifts in the European Sea Bass
- A Genetic Screening Strategy Identifies Novel Regulators of the Proteostasis Network
- Interspecific Sex in Grass Smuts and the Genetic Diversity of Their Pheromone-Receptor System
- The Synthetic Multivulva Genes Prevent Ras Pathway Activation by Tightly Repressing Global Ectopic Expression of EGF
- Mining the Allelic Spectrum Reveals the Contribution of Rare and Common Regulatory Variants to HDL Cholesterol
- Identification of a Genomic Reservoir for New Genes in Primate Genomes
- Genomic Distribution and Inter-Sample Variation of Non-CpG Methylation across Human Cell Types
- Identification of Evolutionarily Conserved Exons as Regulated Targets for the Splicing Activator Tra2β in Development
- Acute Multiple Organ Failure in Adult Mice Deleted for the Developmental Regulator Wt1
- Age-Related Neuronal Degeneration: Complementary Roles of Nucleotide Excision Repair and Transcription-Coupled Repair in Preventing Neuropathology
- Target Site Recognition by a Diversity-Generating Retroelement
- Ancestral Components of Admixed Genomes in a Mexican Cohort
- Targeted Proteolysis of Plectin Isoform 1a Accounts for Hemidesmosome Dysfunction in Mice Mimicking the Dominant Skin Blistering Disease EBS-Ogna
- Autosomal Recessive Dilated Cardiomyopathy due to Mutations Results from Abnormal Dystroglycan O-Mannosylation
- SREBP Coordinates Iron and Ergosterol Homeostasis to Mediate Triazole Drug and Hypoxia Responses in the Human Fungal Pathogen
- The RNA Silencing Enzyme RNA Polymerase V Is Required for Plant Immunity
- An Anti-Checkpoint Activity for Rif1
- The FGFR4-G388R Polymorphism Promotes Mitochondrial STAT3 Serine Phosphorylation to Facilitate Pituitary Growth Hormone Cell Tumorigenesis
- Common Variants Show Predicted Polygenic Effects on Height in the Tails of the Distribution, Except in Extremely Short Individuals
- The Fission Yeast Stress-Responsive MAPK Pathway Promotes Meiosis via the Phosphorylation of Pol II CTD in Response to Environmental and Feedback Cues
- Integrating Genome-Wide Genetic Variations and Monocyte Expression Data Reveals -Regulated Gene Modules in Humans
- Repetitive Elements May Comprise Over Two-Thirds of the Human Genome
- A Novel Checkpoint and RPA Inhibitory Pathway Regulated by Rif1
- Hierarchical Generalized Linear Models for Multiple Groups of Rare and Common Variants: Jointly Estimating Group and Individual-Variant Effects
- The Major Roles of DNA Polymerases Epsilon and Delta at the Eukaryotic Replication Fork Are Evolutionarily Conserved
- A High-Resolution Whole-Genome Map of Key Chromatin Modifications in the Adult
- A Densely Interconnected Genome-Wide Network of MicroRNAs and Oncogenic Pathways Revealed Using Gene Expression Signatures
- A Functional Phylogenomic View of the Seed Plants
- Histone H3K9 Trimethylase Eggless Controls Germline Stem Cell Maintenance and Differentiation
- Ribosomal Protein Mutants Control Tissue Growth Non-Autonomously via Effects on the Prothoracic Gland and Ecdysone
- , , and Are Required to Activate or Delimit the Spread of the Transcriptional Response to Epidermal Wounds in
- Mechanisms Establishing TLR4-Responsive Activation States of Inflammatory Response Genes
- Candidate Gene Screen in the Red Flour Beetle Reveals as Ancient Regulator of Anterior Median Head and Central Complex Development
- Charcot-Marie-Tooth–Linked Mutant GARS Is Toxic to Peripheral Neurons Independent of Wild-Type GARS Levels
- The RNA–Methyltransferase Misu (NSun2) Poises Epidermal Stem Cells to Differentiate
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Targeted Proteolysis of Plectin Isoform 1a Accounts for Hemidesmosome Dysfunction in Mice Mimicking the Dominant Skin Blistering Disease EBS-Ogna
- The RNA Silencing Enzyme RNA Polymerase V Is Required for Plant Immunity
- The FGFR4-G388R Polymorphism Promotes Mitochondrial STAT3 Serine Phosphorylation to Facilitate Pituitary Growth Hormone Cell Tumorigenesis
- Target Site Recognition by a Diversity-Generating Retroelement
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



