-
Články
- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
The Arabidopsis Silencing Pathway Modulates PAMP- and Effector-Triggered Immunity through the Post-transcriptional Control of Disease Resistance Genes
RNA-DEPENDENT RNA POLYMERASE 6 (RDR6) is a key RNA silencing factor initially characterized in transgene silencing and virus resistance. This enzyme also contributes to the biosynthesis of endogenous short interfering RNAs (siRNAs) from non-coding RNAs, transposable elements and protein-coding transcripts. One class of protein-coding transcripts that have recently emerged as major sources of RDR6-dependent siRNAs are nucleotide-binding leucine-rich repeat (NB-LRR) proteins, a family of immune-receptors that perceive specific pathogen effector proteins and mount Effector-Triggered Immunity (ETI). Nevertheless, the dynamic post-transcriptional control of NB-LRR transcripts during the plant immune response and the functional relevance of NB-LRRs in signaling events triggered by Pathogen-Associated Molecular Patterns (PAMPs) remain elusive. Here, we show that PTI is constitutive and sensitized in the Arabidopsis rdr6 loss-of-function mutant, implicating RDR6 as a novel negative regulator of PTI. Accordingly, rdr6 mutant exhibits enhanced basal resistance towards a virulent Pseudomonas syringae strain. We further provide evidence that dozens of CC-NB-LRRs (CNLs), including the functionally characterized RPS5 gene, are post-transcriptionally controlled by RDR6 both constitutively and during PTI. These CNL transcripts are also regulated by the Arabidopsis microRNA miR472 and knock-down of this miRNA recapitulates the PTI and basal resistance phenotypes observed in the rdr6 mutant background. Furthermore, both miR472 and rdr6 mutants were more resistant to Pto DC3000 expressing AvrPphB, a bacterial effector recognized by the disease resistance protein RPS5, whereas transgenic plants overexpressing miR472 were more susceptible to this bacterial strain. Finally, we show that the enhanced basal and RPS5-mediated resistance phenotypes observed in the rdr6 mutant are dependent on the proper chaperoning of NB-LRR proteins, and might therefore be due to the enhanced accumulation of CNL proteins whose cognate mRNAs are no longer controlled by RDR6-dependent siRNAs. Altogether, this study supports a model whereby the miR472 - and RDR6-mediated silencing pathway represents a key regulatory checkpoint modulating both PTI and ETI responses through the post-transcriptional control of disease resistance genes.
Published in the journal: . PLoS Pathog 10(1): e32767. doi:10.1371/journal.ppat.1003883
Category: Research Article
doi: https://doi.org/10.1371/journal.ppat.1003883Summary
RNA-DEPENDENT RNA POLYMERASE 6 (RDR6) is a key RNA silencing factor initially characterized in transgene silencing and virus resistance. This enzyme also contributes to the biosynthesis of endogenous short interfering RNAs (siRNAs) from non-coding RNAs, transposable elements and protein-coding transcripts. One class of protein-coding transcripts that have recently emerged as major sources of RDR6-dependent siRNAs are nucleotide-binding leucine-rich repeat (NB-LRR) proteins, a family of immune-receptors that perceive specific pathogen effector proteins and mount Effector-Triggered Immunity (ETI). Nevertheless, the dynamic post-transcriptional control of NB-LRR transcripts during the plant immune response and the functional relevance of NB-LRRs in signaling events triggered by Pathogen-Associated Molecular Patterns (PAMPs) remain elusive. Here, we show that PTI is constitutive and sensitized in the Arabidopsis rdr6 loss-of-function mutant, implicating RDR6 as a novel negative regulator of PTI. Accordingly, rdr6 mutant exhibits enhanced basal resistance towards a virulent Pseudomonas syringae strain. We further provide evidence that dozens of CC-NB-LRRs (CNLs), including the functionally characterized RPS5 gene, are post-transcriptionally controlled by RDR6 both constitutively and during PTI. These CNL transcripts are also regulated by the Arabidopsis microRNA miR472 and knock-down of this miRNA recapitulates the PTI and basal resistance phenotypes observed in the rdr6 mutant background. Furthermore, both miR472 and rdr6 mutants were more resistant to Pto DC3000 expressing AvrPphB, a bacterial effector recognized by the disease resistance protein RPS5, whereas transgenic plants overexpressing miR472 were more susceptible to this bacterial strain. Finally, we show that the enhanced basal and RPS5-mediated resistance phenotypes observed in the rdr6 mutant are dependent on the proper chaperoning of NB-LRR proteins, and might therefore be due to the enhanced accumulation of CNL proteins whose cognate mRNAs are no longer controlled by RDR6-dependent siRNAs. Altogether, this study supports a model whereby the miR472 - and RDR6-mediated silencing pathway represents a key regulatory checkpoint modulating both PTI and ETI responses through the post-transcriptional control of disease resistance genes.
Introduction
To defend themselves against pathogens, plants have evolved potent inducible immune responses. The first line of active defense relies on the recognition of common features of microbial pathogens, such as flagellin (the major protein of bacterial flagellum), lipopolysaccharides, glycoproteins and chitin [1]. These microbial determinants are referred to as Pathogen - or Microbe - Associated Molecular Patterns (PAMPs/MAMPs) and are sensed by host-encoded Pattern-Recognition Receptors (PRRs) or surface receptors, which encode transmembrane receptor-like kinases. Upon PAMP detection, PRRs trigger a series of immune responses including, for instance, MAPK (mitogen-activated protein kinase) activation, reactive oxygen species (ROS) production, differential expression of genes, callose (β-1->3 glucose polymer) deposition and stomatal closure, which ultimately leads to basal immunity or PAMP-Triggered Immunity (PTI) [2]–[5]. To enable disease, pathogens produce a large array of divergent virulent determinants known as pathogen effectors that suppress different steps of PTI, resulting in disease susceptibility [6], [7]. As a counter-counter defense strategy, plants have evolved a repertoire of immune receptors, called disease resistance (R) proteins that can sense effector proteins and establish effector-triggered-immunity (ETI) [1]. The largest class of R proteins is composed of intracellular receptors that share structural homologies with mammalian innate immune receptors, such as NUCLEOTIDE-BINDING OLIGOMERIZATION DOMAIN-CONTAINING PROTEIN 1 (NOD1) and NOD2, which perceive bacterial PAMPs [8]. Plant NOD-like receptors (NLRs) are composed of nucleotide-binding (NB) and leucine-rich repeat (LRR) domains. They additionally contain an N-terminal domain that is composed of either a Toll/interleukin1 receptor (TIR) or a coiled-coil (CC) module, and are thus referred to as TNLs or CNLs, respectively [9]. These R proteins can directly sense pathogen effectors [1], however, in most cases they recognize indirectly these virulent determinants by detecting their effects on plant target proteins called ‘guardees’ [10]. Upon pathogen effector recognition, R proteins trigger a series of immune responses that significantly overlap with PTI responses, albeit with a stronger amplitude, and often result in a form of programmed cell death known as the hypersensitive response (HR) [1]. Importantly, constitutive expression or activation of R proteins often leads to constitutive cell death as well as severe developmental defects in the absence of pathogen [11]–[16], indicating that R genes and their products must be under tight negative control in unchallenged conditions. Consistent with this idea, transcriptional regulation, RNA processing, protein modifications, protein stability, and nucleocytoplasmic trafficking were shown to play a critical role in controlling R-mediated autoimmune responses [17].
More recently, RNA silencing has also emerged as a key regulatory mechanism that negatively regulates R gene expression [18]–[24]. RNA silencing is an ancestral gene regulatory mechanism that controls gene expression at the transcriptional (TGS, Transcriptional Gene Silencing) and post-transcriptional (PTGS, Post-transcriptional Gene Silencing) levels. The core mechanism of RNA silencing starts with the production of double stranded RNAs (dsRNAs) that are processed by RNase-III enzymes DICERs into 20–24 nt small RNA duplexes. One selected strand is subsequently incorporated into an RNA-induced silencing complex (RISC) containing an argonaute (AGO) protein, and guides these complexes onto sequence complementary RNA/DNA targets. The plant model Arabidopsis thaliana encodes 4 DICER-like proteins and 10 AGOs. DCL1 processes miRNA precursors into mature microRNAs that are mostly incorporated into the AGO1-RISC that guides mRNA degradation and/or translation inhibition of sequence complementary mRNA targets. DCL2, DCL3 and DCL4 are involved in the biogenesis of short interfering RNAs (siRNAs) from extensive dsRNAs produced from read through, convergent or overlapping transcription, endogenous hairpins as well as some miRNA precursors [25], [26]. As an example, overlapping sense and antisense transcripts that are produced at a functionally relevant disease resistance gene cluster, were found to be processed into siRNAs, leading to the down-regulation of several disease resistance gene transcripts within this cluster [18]. In addition, a large proportion of dsRNAs are produced by RNA-dependent RNA polymerases (RDRs) that convert single stranded RNAs into dsRNAs. RDR6, which is one out of six Arabidopsis RDRs, produces dsRNAs from viral and transgene transcripts as well as some endogenous transcripts [27]. These dsRNAs are processed in part by DCL4 into 21 nt siRNAs that direct PTGS of endogenous mRNA targets or exogenous RNAs derived from sense-transgenes or viral RNAs [28]–[31]. In addition to the biogenesis of primary siRNAs, plants have evolved the production of secondary siRNAs as a feed-forward amplification of silencing signals. These siRNAs are produced by the combined action of primary siRNA/miRNA-directed transcript cleavage and the activity of RDRs that use the target transcripts as template to generate dsRNAs [32]. In plants, the best-characterized endogenous secondary siRNAs are termed trans-acting siRNAs (tasiRNAs) [33], [34]. The biogenesis of these small RNA molecules is initiated by 22 nt long miRNAs that direct AGO1-mediated cleavage of a non-coding TAS primary transcript [35], [36]. One of the cleavage products is then converted by RDR6 into dsRNAs, which are processed by DCL4 into 21-nt phased siRNA duplexes. These secondary siRNAs guide an AGO protein to silence sequence complementary mRNA targets in trans. Importantly, this phenomenon is not restricted to non-coding transcripts but also targets protein-coding transcripts and both TNLs and CNLs have emerged as major targets of this silencing pathway [20], [21], [22]. For example, two 22 nt long miRNAs that initiate the production of RDR6-dependent secondary siRNAs, were found to directly control the tobacco disease resistance gene N, which recognizes the C-terminal helicase domain of the Tobacco Mosaic Virus (TMV) replicase protein [22]. These miRNAs play a functional role in N-regulation because their overexpression was shown to compromise N-mediated resistance to TMV [22]. Another recent study conducted in Solanum lycopersicum showed that miR482, a 22 nt long conserved miRNA that targets dozen of CNLs, was down-regulated in response to unrelated viruses as well as to a bacterium that encode RNA silencing suppressors [21]. Interestingly, this phenomenon was associated with the derepression of some CNLs that are targeted by miR482, suggesting that pathogen-triggered suppression of RNA silencing likely derepresses a whole repertoire of immune receptors during infection that might contribute to plant immunity [21].
Recent findings have thus revealed a critical role of miRNA-directed phased siRNA production in controlling the expression of R gene transcripts in the context of pathogen infection. Nevertheless, the interplay between the dynamic regulation of the RNA silencing machinery involved in miRNA-directed secondary siRNA production and the post-transcriptional regulation of R gene transcripts that are targeted by these small RNA species remains unknown. In addition, whereas some intracellular immune-receptors have recently been characterized in basal defense as well as plant defense against a disarmed bacterium very little is known on the functional relevance of plant NLRs in PTI [21]. The present study addresses some of these important issues by studying the regulation of RDR6 during antibacterial defense and the role of this silencing factor in the control of CNLs that are targeted by the Arabidopsis miR472, a miRNA related to miR482.
Results
Arabidopsis RDR6 negatively regulates PTI responses
Although ARGONAUTE 1 (AGO1) and DICER-LIKE 1 (DCL1) were previously shown to contribute to PTI [37], [38], their regulation during the plant innate immune response has not been determined. To get a first insight into the regulation of components of PTGS during plant defense, we examined the expression levels of well-characterized PTGS factors in multiple conditions known to trigger PTI responses (Genevestigator database: https://www.genevestigator.com). Results from this analysis revealed that RDR6, AGO1 and SUPPRESSOR OF GENE SILENCING 3 (SGS3) mRNAs [39] were all down-regulated, with RDR6 showing the highest difference (consistently more than 2-fold in the various conditions analyzed) (Figures S1A, S1B). Accordingly, Reverse-Transcriptase Quantitative Polymerase chain reaction (RT-qPCR) analyses revealed a significant decrease in RDR6 mRNA levels in Arabidopsis leaves and seedlings treated with the flagellin-derived peptide flg22 (Figure 1), with a decrease in RDR6 transcripts starting at 10 min in Arabidopsis elicited seedlings (Figure S1C). A similar effect was observed with the type-three secretion (TTS) defective mutant Pto DC3000 hrcC−, which can elicit, but not suppress, PTI responses due to its inability to inject effector proteins within host cells (figure S1C). The PAMP-triggered dynamic regulation of RDR6 transcripts therefore suggested a potential role for RDR6 in orchestrating PTI responses. To test this idea, we first monitored the effect of the rdr6-15 loss-of-function mutation on the production of reactive oxygen species (ROS), one of the earliest cellular responses following PAMP perception, which is known to orchestrate the establishment of different defensive barriers against biotrophic pathogens [40]. We observed a more pronounced flg22-triggered oxidative burst in the rdr6 mutant as compared to WT-elicited plants (Figure 2A). However, given that the kinetics of flg22-triggered ROS production precedes the down-regulation of RDR6 transcripts in wild-type treated plants (Figure 1), these results suggest that the repression of RDR6 mRNAs is unlikely causative for this early PTI response. We also monitored the expression of PTI marker genes and found a primed induction of Flg22 RECEPTOR KINASE 1 (FRK1) in the rdr6-elicited mutant (Figure 2B, [41]). Of note, induction of FRK1 as well as the two other early PTI marker genes WRKY22 and WRKY29 was also moderately sensitized upon syringe infiltration of water in rdr6 - versus WT-leaves (Figure 2B), suggesting that RDR6 may additionally repress a wounding response caused by mechanical stress.
Fig. 1. RDR6 and AGO1 mRNA levels rapidly decrease after flg22 treatment. 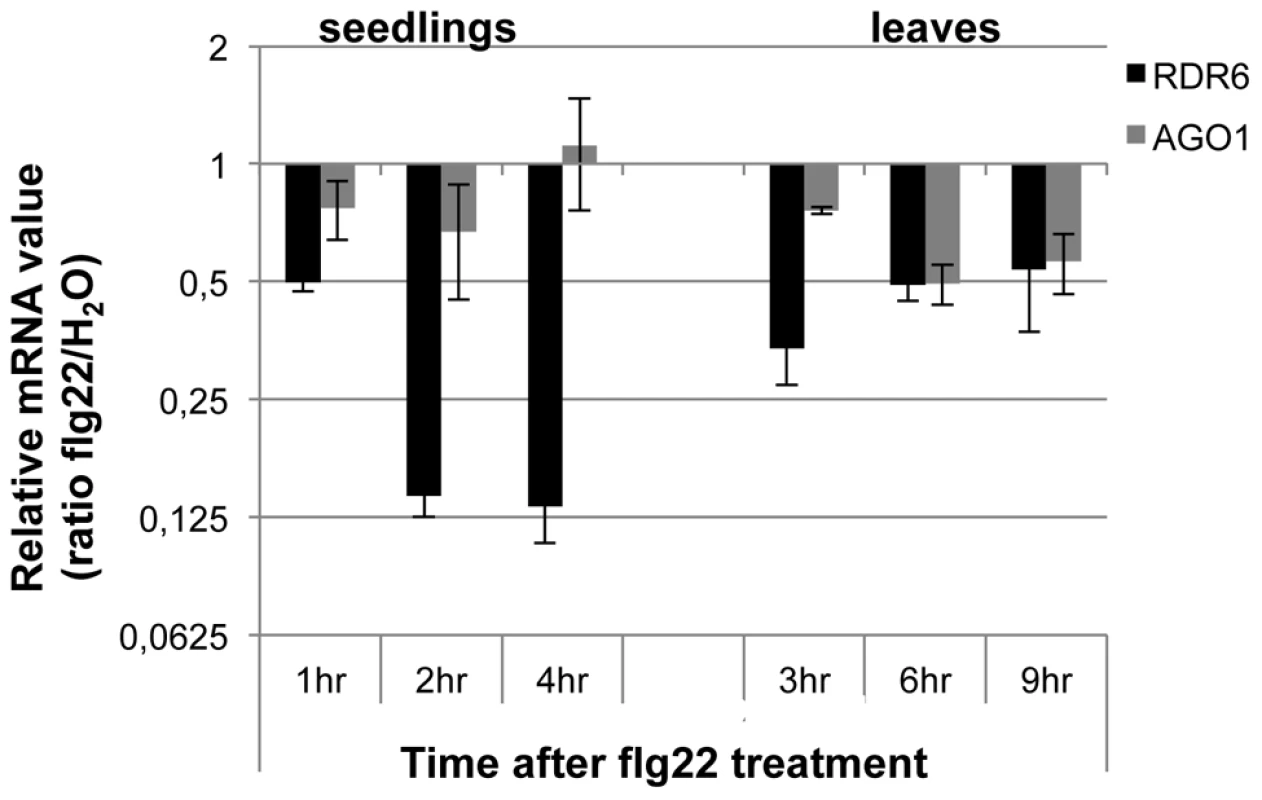
Seedlings were treated for 1, 2 and 4-qPCR. Expression levels are relative to three reference genes (At2g36060; At4g29130; At5g13440). The Log2 of mRNA values are normalized to that of control WT seedlings or leaves plants treated or infiltrated with water. Error bars indicate standard deviation from technical repeats. Similar results were obtained in two independent experiments. Fig. 2. RDR6 negatively regulates PTI responses. 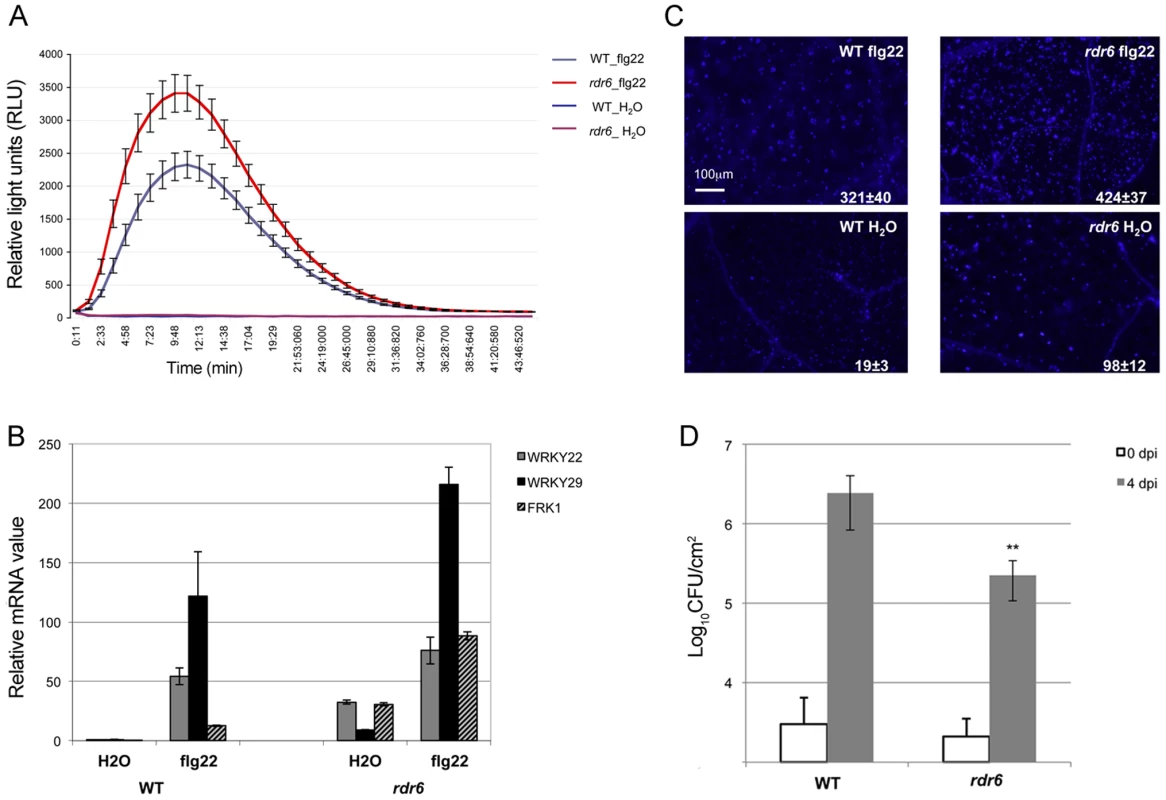
(A) H2O2-dependent luminescence upon H2O or flg22 (100 nM) treatment in WT and rdr6-15 leaf discs. (B) Expression levels of PAMP-responsive mRNAs, FRK1, WRKY22 and WRKY29 detected by RT-qPCR in leaves treated with 100 nM flg22 or water for 4 hours. (C) Callose deposition upon H2O or flg22 (100 nM) treatment in WT and rdr6-15 leaves blade at stage 7. Values are average ± se (standard error) with n = 25 to 30. (D) Bacterial growth in five- to six-week-old plants (WT or rdr6) 4 days after being sprayed (108 CFU mL−1) with Pto DC3000. Values are average ± se of four leaf discs (n = 8). Wilcoxon test was performed to determine the significant differences between rdr6 and WT plants. Asterisk “**” indicates statistically significant differences (P<0.01). Experiments were performed in two independent biological replicates with similar results. We further monitored the flg22-triggered formation of cell wall depositions of callose, a late PTI response that plays a critical role in the establishment of basal immunity [42], [43]. An increase in flg22-induced callose depositions was observed in the rdr6-15 mutant as compared to WT plants, reinforcing a role for RDR6 in repressing this late PTI response (Figure 2C). It is noteworthy that a higher number of callose deposits were also observed in mock-treated rdr6-15 mutant versus WT plants, but not in untreated rdr6-15 mutant leaves (data not shown), suggesting that RDR6 may additionally prevent callose deposition upon wounding caused by syringe infiltration.
Natural surface openings, such as stomata, are important entry sites for bacterial plant pathogens such as Pto DC3000 and previous studies have shown that stomata closure plays an active role in limiting bacterial invasion as part of PTI responses [4]. Furthermore, fls2 mutants were found to be more susceptible to Pto DC3000 upon spray inoculation, although no discernible phenotype was observed using classical syringe infiltration assay, which bypassed basal immunity present at the leaf surface [44]. Given that the rdr6-15 mutant was sensitized for multiple flg22-triggered PTI responses, we reasoned that such silencing-deficient mutant might display enhanced resistance to Pto DC3000 upon spray inoculation. Consistent with this hypothesis, we found ∼10 times lower bacterial titer on rdr6-15 mutant as compared to WT plants spray inoculated with Pto DC3000 (Figure 2D). Collectively, these data provide evidence that the RNA silencing factor RDR6 acts as a negative regulator of basal immunity. These results also suggest that some positive regulators of plant defense are likely to be directly controlled by RDR6-dependent siRNAs.
RDR6-dependent secondary siRNAs target a subset of mRNAs encoding CNL proteins
Besides generating siRNAs directed against viral-, transgene - and transposon-derived RNAs, RDR6 is known to produce secondary 21 nt siRNAs from several endogenous loci including TAS genes [25], [26]. We thus searched for candidate defense gene transcripts that would be directly controlled by RDR6-dependent siRNAs. We used publicly available small RNA libraries derived from WT and rdr6 mutant leaves and selected candidate genes with a significant reduced amount of 21 nt siRNAs in the rdr6 as compared to the WT background. Using such criterion, we identified 75 loci that were likely targeted by RDR6-dependent siRNAs. Among those, 27 were previously annotated as TAS genes or tasiRNA targets. The remaining 48 protein-coding genes were enriched in GO categories ‘response to stress’ (http://bar.utoronto.ca/welcome.htm), (Figure S2), and include well-characterized RDR6-dependent targets such as AGO1, which is targeted by miR168-directed secondary siRNAs [45]. In addition, thirteen other candidate genes were annotated as miRNA targets and include multiple disease resistance gene transcripts that were previously identified as targets of miR472 (Figures S3, S4), a 22 nt long miRNA that is at least in part loaded into AGO1-RISC [35], [46]. These R genes are phylogenetically related to the functionally relevant disease resistance gene RPS5, which was previously characterized in ETI [47].
RDR6-dependent siRNAs negatively regulate a subset of CNL transcripts both constitutively and during flg22 elicitation
Given that At1g51480 and At5g43730 were among the CNL transcripts with the most matching secondary siRNAs (Figure S4), we decided to further characterize their regulation by RDR6 in both naïve and flg22-challenged conditions. These candidate genes are referred to here as Resistance Silenced Gene 1 (RSG1, At1g51480) and Resistance Silenced Gene 2 (RSG2, At5g43730). We also included RPS5 in this analysis, which was previously validated as miR472 target in Parallel Analysis of RNA Ends (PARE) datasets. We found a mild enhanced accumulation of these three candidate transcripts in unchallenged rdr6 mutant as compared to non-treated WT seedlings (Figure 3A), suggesting that these mRNAs are weakly controlled by RDR6-dependent siRNAs in naïve conditions, presumably due to their low basal transcriptional level in unchallenged conditions as previously observed for several disease resistance genes [17], [48]. We next monitored the levels of these mRNAs upon flg22 treatment in both WT and rdr6 mutant backgrounds. Whereas a mild increased induction of these transcripts was found in WT-elicited background, as observed in publicly available datasets (Figure S5), a 10 - to 20-fold enhanced accumulation of these transcripts was obtained in the rdr6-elicited mutant seedlings, indicating cell priming in the absence of RDR6-dependent siRNAs (Figure 3B). These results therefore indicate that RDR6-dependent secondary siRNAs negatively regulate these CNL transcripts and that this post-transcriptional regulatory control is particularly relevant during PTI, when these disease resistance genes are presumably transcriptionally activated.
Fig. 3. RDR6-dependent siRNAs negatively regulate a subset of CNL transcripts both constitutively and during flg22 elicitation. 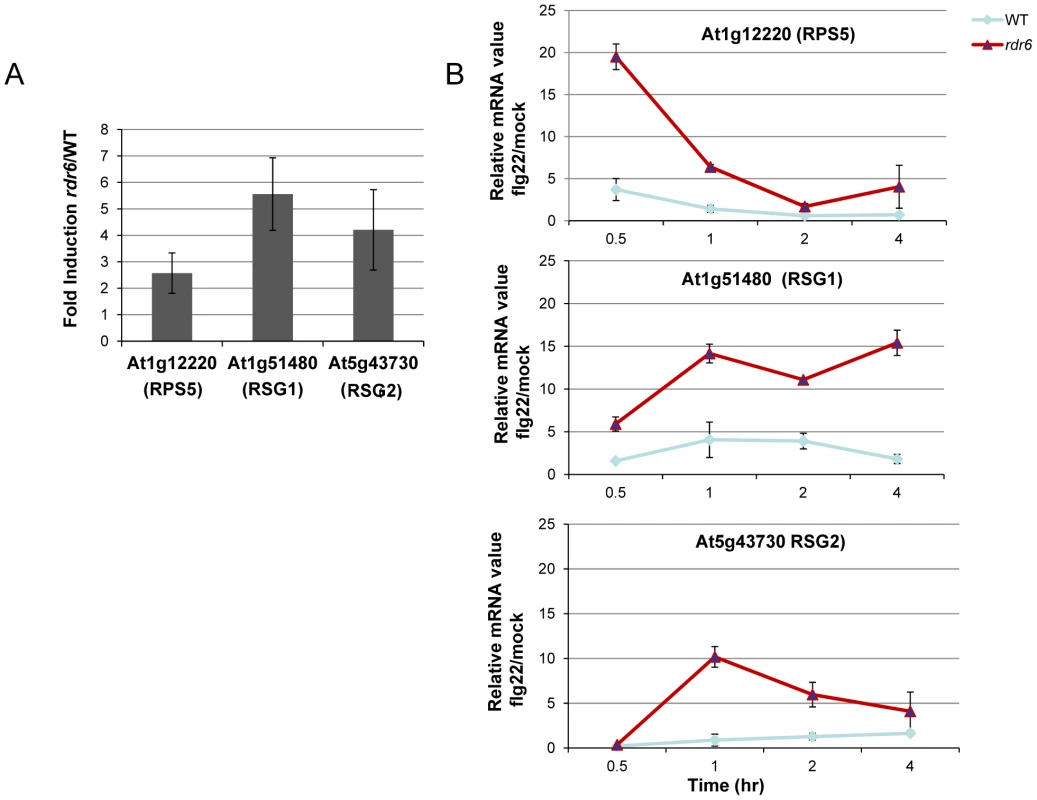
(A) The transcript levels of At1g12220 (RPS5), At1g51480 (RSG1) and At5g43730 (RSG2) were detected by RT-qPCR in untreated seedlings. Results of expression represent the ratio rdr6/WT. (B) Transcript levels of RPS5, RSG1 and RSG2 by RT-qPCR in seedling treated or not with flg22 (100 nM) at different time-points. Results represent the ratio of the values between samples treated with flg22 relative to H2O (mock) for rdr6 and WT seedlings. Expression levels are always normalized to the same internal controls At2g36060, At4g29130, and At5g13440. Error bars indicate standard deviation from technical repeats. These experiments were performed in two biological replicates with similar results. MiR472 negatively regulates PTI responses and resistance against virulent Pto DC3000
Given that miR472 was shown to target the above CNL mRNAs and to initiate the production of RDR6-dependent secondary siRNAs at these loci [20], [21], [22], we next characterized the role of this particular miRNA in the regulation of these candidate CNL transcripts as well as other orphan targets. For this purpose, we first transformed Arabidopsis with a construct containing AtmiR472 driven by the strong Cauliflower Mosaic Virus (CaMV) 35S promoter and selected a reference line (referred to as miR472OE line) exhibiting high miR472 accumulation compared to WT (Figure 4A). This line displayed a 25% and 30% reduction in the accumulation of RPS5 and RSG1 transcripts, respectively (Figure S6), providing further evidence that miR472 targets these CNL mRNAs in unchallenged conditions. Furthermore, genome-wide small RNA deep sequencing analyses revealed a drastic enhanced accumulation of secondary siRNAs at the 3′ ends of miR472 target sites for RPS5, RSG1 and RSG2 mRNAs as well as for 16 other CNL transcripts in the miR472OE line as compared to wild type seedlings (Figures 4B, 4C, S7). It is noteworthy that no siRNA were identified upstream the miR472 target site, which is in agreement with the rapid degradation of this region after miRNA-guided cleavage [49]. Furthermore, normal levels of tasiRNAs were identified in miR472OE as compared to WT seedlings (Figure S8), proving evidence that the enhanced accumulation of CNL-derived secondary siRNAs are not due to a general activation of the RDR6-dependent pathway in this transgenic line. Collectively, these results strongly reinforce a role for miR472 in initiating the biosynthesis of RDR6-dependent secondary siRNAs at our candidate CNL transcripts and revealed additional CNLs that are directly targeted by this regulatory process including the other functionally relevant disease resistance gene SUMM2 (Figure S9) [50].
Fig. 4. Overexpression of miR472 drastically enhances the accumulation of secondary siRNAs at multiple CNL transcripts. 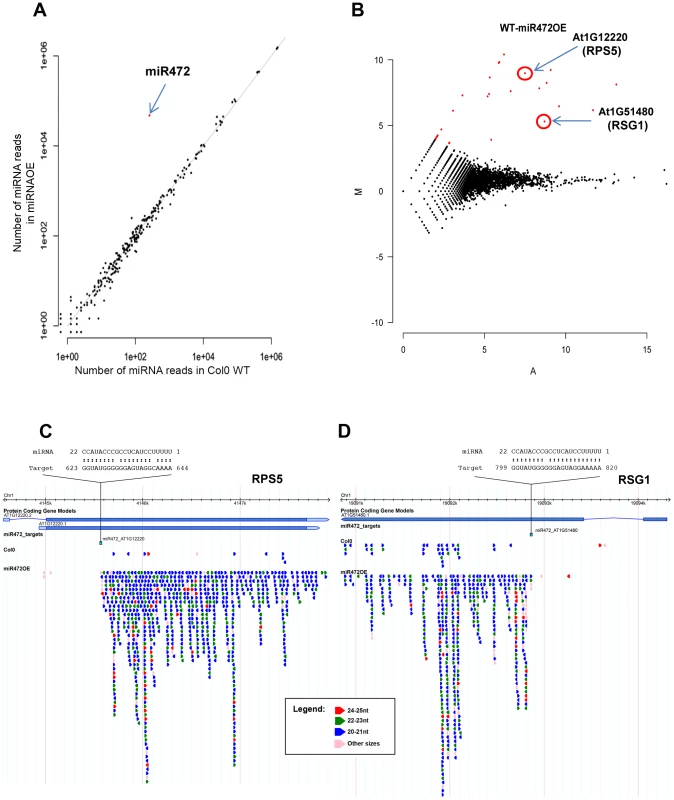
(A) Scatter plot representation of the number of reads corresponding to miRNA stem-loop loci (miRBase release 19) in WT and miR472OE mutant sRNA libraries. The number of reads was library size normalized. The red dot corresponds to miR472. (B) MA plot representation of the results obtained after differential analysis of 20–22 nt small RNAs accumulation from genes, between WT and miR472OE mutant. The y axis represents the log ratio (log10) of the library size normalized number of reads between the 2 datasets and the x axis the average number of reads in the two libraries. Genes with a significantly higher sRNAs accumulation in miR472OE library are shown in red. (C) Example of sRNAs accumulation in WT and miR4720E libraries along 2 CNL genes, RPS5 (AT1g12220) and RSG1 (AT1G51480). Genome browser representation of sRNA reads along. Each arrow corresponds to a specific sRNA sequence with a colour code corresponding to it length as indicated in the legend. Position and alignment of miR472 recognition sites are indicated. It is remarkable that the accumulation of siRNAs is observed downstream the cleavage site of miR472. We next analyzed the mRNA accumulation of two candidate CNLs in the miR472OE line challenged with flg22. Flg22-triggered induction of RPS5 and RSG1 mRNAs was significantly impaired in miR472OE line as compared to WT-elicited control (Figure 5A), supporting a role for miR472 in regulating the accumulation of these targets during flg22 elicitation. We also examined different PTI features in the miR472OE reference line by monitoring ROS production and callose deposition upon flg22 treatment. While this transgenic line displayed a normal flg22-triggered ROS production as compared to WT-elicited control (Figure 5B), we found a reduced number of flg22-induced callose deposits relative to WT-treated plants (Figure 5C), indicating that the miR472OE reference line is altered in the latter PTI response. It is noteworthy that similar PTI phenotypes were observed in another independent transgenic line overexpressing miR472 (Figure S10).
Fig. 5. MiR472 negatively regulates PTI responses and resistance against virulent Pto DC3000. 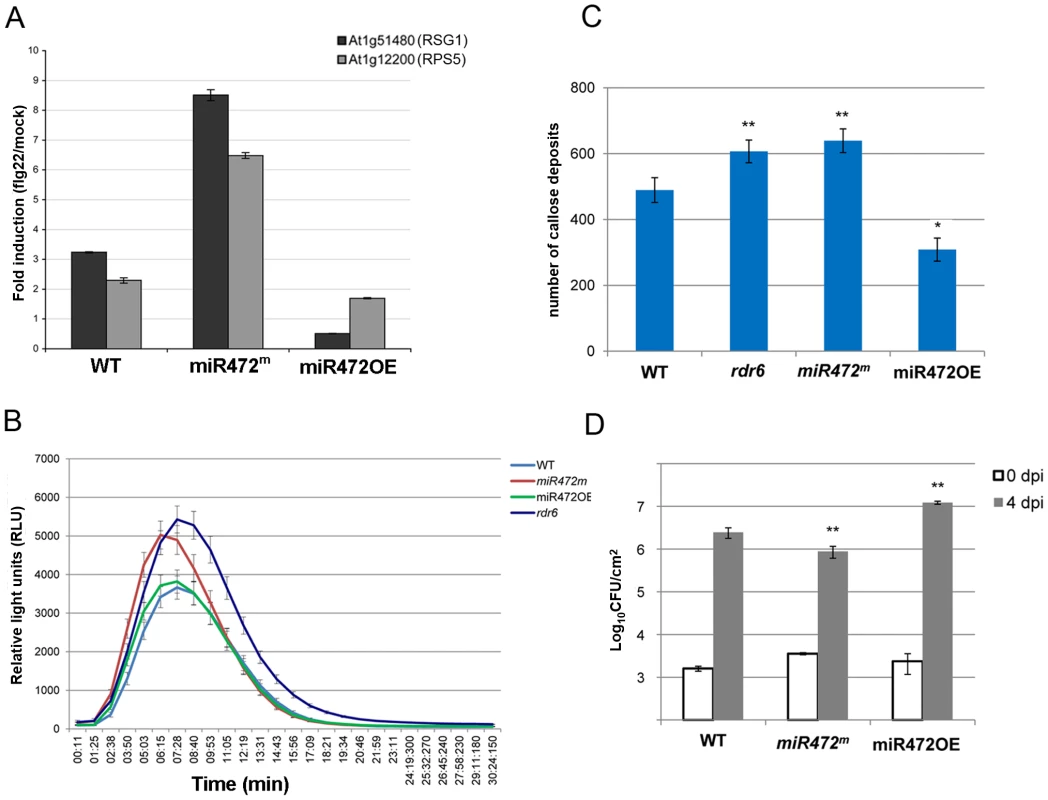
(A) Expression levels of At1g12220 and At1g51480 detected by RT-qPCR in WT, miR472OE (overexpressor) and miR472m (mutant) seedling treated with either H2O or flg22 (100 nM) for 2 hours. (B) H2O2-dependent luminescence induced by flg22 (100 nM) in WT, miR472OE and miR472m leaf discs. (C) Callose deposition induced by flg22 (100 nM) in WT, miR472OE and miR472m leaves. (D) Bacterial growth in five- to six-week-old plants from WT, miR472OE and miR472m infiltrated with Pto DC3000 (2×105 CFU mL−1). For C and D values are average ± se of four leaf discs (n = 8). Wilcoxon test was performed to determine the significant differences as compared to rdr6 plants. Asterisks “*” and “**” indicate statistically significant differences at a P value<0.05 and <0.01 respectively. These experiments were performed in two biological replicates with similar results. To get further insights into the role of miR472 in the regulation of CNL transcripts and PTI responses, we further characterized a transgenic line carrying a T-DNA insertion within the promoter of the AtmiR472 locus (Salk_087945, referred to as miR472m). This line displayed a drastic decrease in the accumulation of the mature form of miR472 relative to the levels of this miRNA in WT background (Figure S11). Furthermore, a primed induction of RPS5 and RSG1 transcripts was found in the miR472m line relative to WT background treated with flg22 (Figure 5A), supporting a role for miR472 in repressing mRNA accumulation of these CNL mRNAs during flg22 elicitation. Further phenotypic analyses in this line revealed a more pronounced flg22-induced ROS production and callose deposition, thereby mimicking the primed PTI responses observed in the rdr6-elicited mutant (Figures 5B, 5C). We thus conclude that miR472 and RDR6-dependent secondary siRNAs regulate PTI responses likely by targeting a whole repertoire of CNL transcripts.
Finally to determine the role of miR472 in basal resistance, we inoculated the virulent Pto DC3000 strain on miR472OE and miR472m lines and monitored bacterial titers in these genetic backgrounds as compared to WT-infected control. We found an increased Pto DC3000 titer in the miR472OE line, and, conversely, a reduced growth of this bacterium in the miR472m line as compared to WT-infected control (Figures 5D, S10). These results indicate that miR472 not only represses PTI responses but also negatively regulates basal resistance against Pto DC3000. These results also suggest that a subset of CNLs, which are targeted by miR472 and RDR6-dependent secondary siRNAs, may control basal resistance against Pto DC3000.
MiR472 and RDR6 negatively regulate RPS5-mediated resistance
The effective targeting of RPS5 mRNAs by miR472 and RDR6-dependent secondary siRNAs (Figure 4B, 4C), together with the well-characterized role of RPS5 in recognizing the bacterial effector AvrPphB and mounting ETI [47], prompted us to investigate the role of miR472 and RDR6 in RPS5-mediated resistance. For this purpose, the rdr6-15 and miR472m lines were first inoculated with a Pto DC3000 strain carrying AvrPphB and bacterial titers were monitored at 4 days post-inoculation. Results from these analyses indicated a significant enhanced RPS5-mediated resistance in both rdr6 and miR472m as revealed by lower bacterial titers in these mutants as compared to WT-infected plants (Figure 6A), which is consistent with the enhanced accumulation of RPS5 transcripts in these mutant backgrounds (Figures 3A, 3B, 5A). Of note, this phenomenon was specific to RPS5-mediated resistance, because no phenotype was observed upon inoculation of rdr6 and miR472m lines with Pto DC3000 expressing AvrRpt2, a bacterial effector that is recognized by another CNL that is not targeted by miR472 (Figure S12, [13]). We next inoculated the Pto DC3000 (AvrPphB) strain on the miR472OE reference line and monitored bacterial titers as well as disease symptoms at 4 days post-inoculation. Interestingly, we found a significant enhanced Pto DC3000 (AvrPphB) titer in the miR472OE line as compared to WT-infected plants (Figure 6B), which was associated with a rescue of both chlorotic and necrotic disease symptoms in this transgenic plants (data not shown), thereby mimicking the phenotypes observed in rps5 loss-of-function mutants (Figure 6A, [47]). We conclude that overexpression of miR472 is sufficient to compromise RPS5-mediated resistance, which is consistent with the reduced levels of RPS5 mRNAs in this transgenic line (Figures 5A, S6). Collectively, these results indicate that miR472 and RDR6 negatively regulate not only PTI but also RPS5-mediated resistance, suggesting a critical role for RPS5 and other CNLs in basal and race-specific immunity.
Fig. 6. MiR472m and rdr6 are more resistant to Pto DC3000 (AvrPphB). 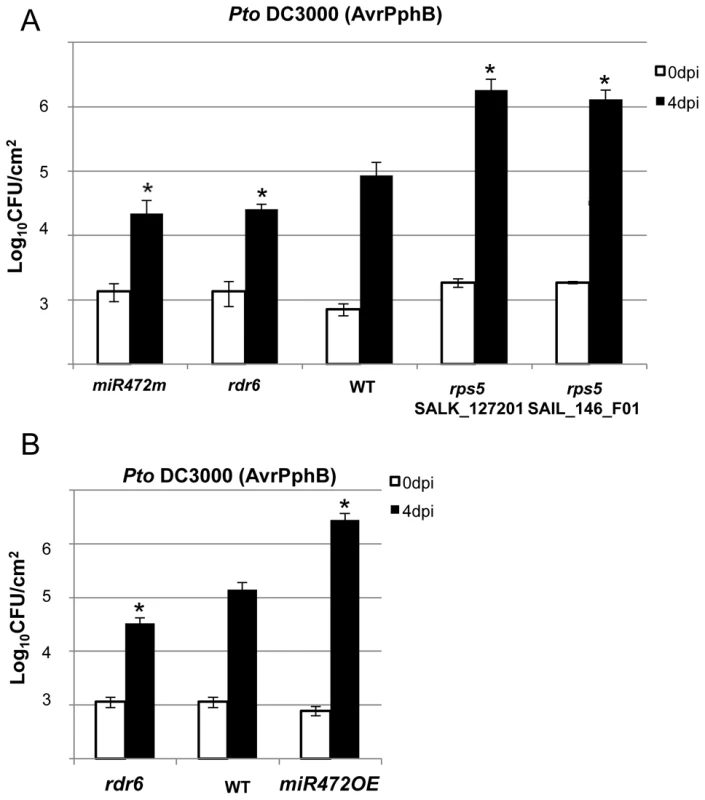
(A) Bacterial growth in five- to six-week-old plants from WT, rdr6 and miR472m syringe-infiltrated with Pto DC3000 AvrPphB (2×105 CFU mL−1). Values are average ± se of four leaf discs (n = 8). Wilcoxon test was performed to determine the significant differences as compared to WT plants. As a positive control the susceptible mutant rps5 (two independent mutant alleles SalK_127201 and SAIL_146_F01) has been used as control. Asterisk “*” indicates statistically significant differences at a P value<0.05. (B) Bacterial growth in five- to six-week-old plants from WT and miR472OE lines syringe-infiltrated with Pto DC3000 AvrPphB (2×105 CFU mL−1). Values are average ± se of four leaf discs (n = 8). Wilcoxon test was performed to determine the significant differences as compared to WT plants. These experiments were performed in two biological replicates with similar results. Proper chaperoning of NB-LRRs is required for the enhanced basal immunity and RPS5-mediated resistance phenotypes observed in the rdr6 mutant
The predicted target site of miR472 is embedded within a region encoding the P-loop domain, which is highly conserved in a large repertoire of CNL disease resistance proteins [9]. It is therefore likely that multiple CNLs are controlled by this particular miRNA and, in agreement, 19 CNL transcripts were experimentally validated as miR472 targets in Arabidopsis seedlings overexpressing miR472 (Figures 4, S7). This suggests that the enhanced basal resistance phenotype observed in the rdr6 and miR472m mutants might not only be due to the constitutive expression and/or primed induction of the few CNLs that have been characterized in these mutant backgrounds (e.g. RPS5), but also likely to multiple other relatives that are targeted by these small RNAs, rendering the functional characterization of these CNLs challenging. To circumvent this issue, we first introduced, in the rdr6 mutant background, mutations that abolish CNL-mediated signaling, and subsequently monitored Pto DC3000 titer in these double mutant backgrounds. Since several CNLs are known to trigger SA-signaling/biosynthesis [51], including RPS5 [52], we hypothesized that the SA-dependent defense response might be constitutive in the rdr6 mutant background. Consistent with this idea, we found a constitutive expression of the SA-dependent marker gene PATHOGENESIS-RELATED 1 (PR1) and the ISOCHORISMATE SYNTHASE1 (ICS1) (Figures 7A, 7B) [51], [53], as well as an enhanced resistance to Pto DC3000, in the rdr6 mutant as compared to WT control (Figures 7C, 7D, 7E, 7F). Importantly, this increased resistance to Pto DC3000 was abolished by introducing mutations that compromise SA-biosynthesis (the sid2-2 mutation, [53]) or SA-signaling (the npr1-1 mutation, [54]) in the rdr6-15 mutant background (Figures 7C, 7D). These results therefore indicate that the enhanced basal resistance achieved in the rdr6 mutant relies on the constitutive activation of the SA-dependent defense response, which might be initially triggered by the enhanced accumulation of CNLs that are no longer controlled by RDR6-dependent secondary siRNAs in this mutant background.
Fig. 7. Enhanced basal resistance towards Pto DC3000 observed in the rdr6 mutant requires SA and proper chaperoning of NLRs. 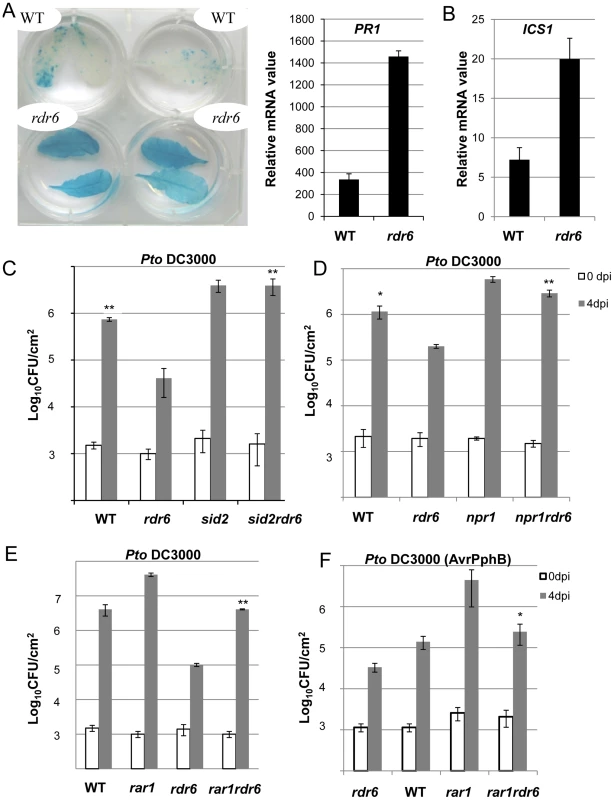
(A) β-glucuronidase (GUS) activity in plants PR1p:GUS and rdr6 PR1p:GUS plants reporting PR1 transcriptional activity in WT and rdr6-15 mutant, respectively. (B) The transcript level of PR1 and ICS1 were detected by RT-qPCR. Error bars indicate standard deviation from technical repeats. Expression levels are normalized to the same internal controls At2g36060, At4g29130, and At5g13440. (C) Bacterial growth in five- to six-week-old plants from WT, single rdr6-15 and sid2-2 mutants or double rdr6-sid2 mutant infiltrated with Pto DC3000 (2×105 CFU mL−1). (D) Bacterial growth in five- to six-week-old plants from WT, simple rdr6-15 and npr1-1 mutants or double rdr6-npr1 mutant infiltrated with Pto DC3000 (2×105 CFU mL−1). (E) Bacterial growth in five- to six-week-old plants from WT, single (rdr6-15, rar1-21) or double mutant (rdr6-rar1) infiltrated with Pto DC3000 (2×105 CFU mL1). F) Bacterial growth in five- to six-week-old plants from WT, single (rdr6-15, rar1-21) or double mutant (rdr6-rar1) infiltrated with Pto DC3000 (AvrPphB) (2 105 CFU mL1). For C, D, E and F values are average ± se of four leaf discs (n = 8). Wilcoxon test was performed to determine the significant differences between rdr6 and double mutant plants. Asterisks “**” and “*” indicate statistically significant differences at a P value<0.01 and <0.05 respectively. Experiments were performed in two independent biological replicates with similar results. To get further insights into the role of these CNLs in the enhanced basal resistance phenotype observed in the rdr6 mutant, we took advantage of the property of the REQUIRED FOR MLA12 RESISTANCE (RAR1) protein. RAR1 is part of a molecular chaperone complex, containing HEAT SHOCK PROTEIN 90 (HSP90) and SUPPRESSOR OF G-TWO ALLELE OF SKP1 (SGT1), and plays a major role in NLR protein stability and activity [55]–[65]. Importantly, the steady-state accumulation of several CNL proteins, including RPS5, was shown to be dramatically impaired in rar1 loss-of-function mutants [58], [64]–[68]. We thus reasoned that by introducing a rar1 loss-of-function mutation in the rdr6 mutant background, we would destabilize CNL proteins whose cognate mRNAs are targeted by RDR6-dependent siRNAs, and therefore potentially restore disease susceptibility. Consistent with this hypothesis, we found that the increased resistance achieved in the rdr6 mutant was abolished in the rdr6-rar1 double mutant (Figure 7E). It is noteworthy that an enhanced Pto DC3000 titer was also found in the single rar1 and double rdr6-rar1 mutants as compared to WT control, indicating that RAR1 contributes to basal resistance as previously reported [64]. Given that RDR6 was found to negatively regulate RPS5-mediated resistance (Figure 6), we also monitored Pto DC3000 (AvrPphB) titer in the single rdr6 mutant as compared to the rdr6-rar1 double mutant. Results from these analyses indicated that the enhanced RPS5-mediated resistance observed in rdr6 mutants was partially compromised in the rdr6-rar1 mutant (Figure 7F). Collectively, these results indicate that the increased basal and specific resistance observed in the rdr6 mutant is dependent on the proper chaperoning of CNL proteins (e.g. RPS5), and might therefore be due to the enhanced accumulation of CNL proteins whose cognate mRNAs are no longer controlled by endogenous secondary siRNAs in this silencing-defective mutant.
Discussion
Arabidopsis RDR6 negatively regulates PTI, basal resistance, SA-dependent defense and RPS5-mediated resistance
RDR6 has been clearly implicated as a positive regulator of virus and viroid resistance. Indeed silencing of RDR6 in Nicotiana benthamiana results in hyper-susceptibility to some viruses and viroids [29], [30]. Moreover, in situ hybridization shows that viruses and viroids invade floral and vegetative meristems of N. benthamiana rdr6 RNAi plants [69], [70]. Here, by combining microbiological, genetic, genomic and molecular techniques, we demonstrate that RDR6 also acts as a negative regulator of PTI, basal defense as well as RPS5-mediated resistance. Indeed, we first showed that knock-out of RDR6 renders the plants more resistant to the hemibiotrophic pathogen Pto DC3000 and to the avirulent Pto DC3000 (AvrPphB) strain (Figures 2D, 6A, 7C, 7D, 7E). Furthermore, classical PTI responses such as ROS production, mRNA accumulation of PAMP-response genes as well as callose deposition were increased in rdr6 plants as compared to WT plants upon flg22 treatment (Figures 2A, 2B, 2C) [71]. Our results are thus in sharp contrast with the previously reported PTI phenotypes observed in ago1 loss-of-function mutants [38]. Why is there such a discrepancy between these PTGS-defective mutant phenotypes during PTI? One would argue that AGO1 is not only involved in the siRNA pathway but also in the canonical miRNA pathway. AGO1 impairment has thus additional consequences on the action of several miRNAs necessary for PTI [38], [72], thereby leading to the previously reported compromised PTI responses in ago1 loss-of-function mutants such as in other miRNA-defective mutants [37], [38]. It is also possible that RDR6-derived siRNAs that target disease resistance genes may not only be loaded into AGO1-RISC but also into other as-yet unknown AGO-RISCs, thereby contributing in part to the post-transcriptional regulation of CNLs in an AGO1-independent manner.
We also observed a constitutive activation of the SA defense marker gene PR1 and an enhanced expression of ICS1 in the rdr6 loss-of function mutant (Figures 7A, 7B). To examine the involvement of the SA-dependent defense in the enhanced disease resistance phenotype observed in rdr6 mutant, the rdr6-15 mutation was combined with the sid2-2, a loss-of-function mutation in ICS1 also referred to as SID2 [53]. Inactivation of ISC1/SID2 abolishes rdr6 resistance to Pto DC3000 and similar results were obtained in the npr1 mutant, which is impaired in SA signaling [54] (Figures 7C, 7D). Therefore, the SA-dependent defense pathway plays a critical role in the enhanced basal resistance phenotype observed in the rdr6 mutant. Such constitutive SA-dependent defense response might result from a derepression of a subset of CNL transcripts (e.g. RPS5 mRNAs) that are no longer regulated by secondary siRNAs in this silencing-defective mutant. Additionally, it may result from the post-translational activation of R proteins that would be constitutively present in a protein complex with RDR6 and active in the absence of this silencing factor, as observed in classical ‘guardee’ mutants [10]. Further investigations will be necessary to address these possibilities. Moreover, additional experiments will be required to determine whether the constitutive SA-dependent defense response observed in the rdr6 mutant is linked with the mild constitutive PTI responses in this silencing-defective mutant or whether both processes remain independent.
We observed a higher expression of PAMP-response marker genes in unchallenged rdr6 mutant as compared to WT seedlings and a significant hyper-induction of FRK1 in the rdr6-elicited mutant (Figure 2B). Furthermore, a more pronounced callose deposition as well as ROS production were observed in the rdr6 mutant challenged with flg22 as compared to WT-elicited seedlings (Figures 2A, 2C), indicating that this silencing-defective mutant is in a physiological situation known as “primed” state [73]. Those results also indicate that RDR6 encodes a novel negative regulator of PTI and further reinforce the idea that PTI is under a tight negative regulatory control as previously reported [2], [74], [75], [76]. Interestingly, an analogous RNA silencing-dependent regulatory phenomenon has been recently described in the transcriptional control of a disease resistance gene during PTI [24]. In this case, flg22 was shown to trigger the repression of a subset of RNA-directed DNA methylation factors and this process was associated with TGS release and with the transcriptional activation of this immune receptor, which is targeted by siRNA-directed DNA methylation in its promoter region [24]. Although RDR6 mRNAs were down-regulated in response to flg22 (Figure 1), it remains to be tested whether this molecular effect could be accompanied with a decrease in RDR6 protein levels as well as an eventual global release of RDR6-silencing as part of PTI responses.
Arabidopsis RDR6 and miR472 repress basal resistance and RPS5-mediated resistance likely by controlling a subset of CNLs at the post-transcriptional level
How does RDR6 repress PTI, basal resistance and RPS5-mediated resistance? A first in silico analysis of small RNA populations derived from rdr6 mutant as compared to wild-type leaf samples allowed us to identify R gene mRNA candidates that are targeted by RDR6-dependent secondary siRNAs (Figure S4). However, the low abundance of secondary siRNAs in the majority of cases limited the identification of such miR472/RDR6 targets. By contrast, the use of our transgenic line overexpressing miR472 was instrumental in identifying with confidence 19 bona fide CNL target transcripts that contain the miR472 recognition sites as well as a large number of secondary siRNAs located downstream of their miR472-guided cleavage site (Figures 4, S9). Among these candidates, we have identified RPS5 and SUMM2 transcripts, which encode functionally relevant disease resistance proteins with well-characterized role in ETI [47], [50], [56]. These results therefore suggested that the miR472/RDR6-silencing pathway inhibits the accumulation not only of disease resistance gene transcripts encoding R proteins required for PTI and basal resistance but also of transcripts encoding immune receptors required for ETI. This implicates miR472 and RDR6 in a central regulatory pathway that modulates both ETI and PTI responses. Consistent with this, we found that RDR6 and miR472 act not only as negative regulators of PTI and basal immunity but also as repressors of RPS5-mediated resistance (Figure 6). In addition, the use of the rar1 mutant, which destabilizes disease resistance proteins including RPS5 [58], [59] was useful to provide genetic evidence that the enhanced disease resistance phenotypes observed in the rdr6 mutant is likely the result of a higher accumulation of NB-LRR proteins in this silencing-defective mutant (Figure 7).
The present work also provides genetic evidence that miR472 - and RDR6-dependent secondary siRNAs efficiently control the steady state levels of three CNL transcripts. Indeed, we first showed that RPS5, RSG1 and RSG2 mRNAs were moderately up-regulated in untreated rdr6 mutant and significantly hyper-induced in this silencing-defective mutant challenged with flg22 (Figure 3B). Accordingly, a lower level of RPS5 and RSG1 mRNAs was detected in the miR472OE line (Figure S6) and a compromised induction of these CNL transcripts was also observed in this transgenic line challenged with flg22 (Figures 5A, 5B, 5C). Conversely, the miR472 knock-down line displayed higher accumulation of CNL mRNAs, which was associated with increased PTI responses (Figure 5), therefore mimicking the phenotypes observed in the rdr6 mutant (Figure 2). Collectively, these results indicate that both Arabidopsis RDR6 and miR472 negatively regulate the steady state levels of these candidate CNL transcripts in normal growth conditions and during PTI, although these effects appear more pronounced during the elicitation possibly due to the concomitant transcriptional activation of these R genes as previously demonstrated for other biotic stress responsive disease resistance genes [17], [18], [48].
Based on these results, we propose a model, which integrates the contribution of the miR472/RDR6-dependent PTGS pathway in plant immunity (Figure 8). In unchallenged conditions, both miR472 and RDR6 are constitutively expressed and negatively regulate a subset of CNL mRNAs at the post-transcriptional level (Figure 8). MiR472 guides cleavage of RPS5, RSG1, RSG2 and at least 16 other CNL transcripts that carry miR472 recognition sites and RDR6 uses 3′ cleavage products as substrates to generate dsRNAs that are presumably processed by DCL4 into 21 nt siRNAs (Figure 8). These secondary siRNAs can act in cis by guiding mRNA degradation of the CNL transcripts from which they are produced, but also likely in trans presumably by targeting CNLs as well as unrelated mRNAs that display sequence complementary to these small RNA species as was recently suggested in tomato ([21], Figure S7).
Fig. 8. Schematic representation illustrating the relationship between miR472/RDR6 through CNL regulation during Arabidopsis immunity. 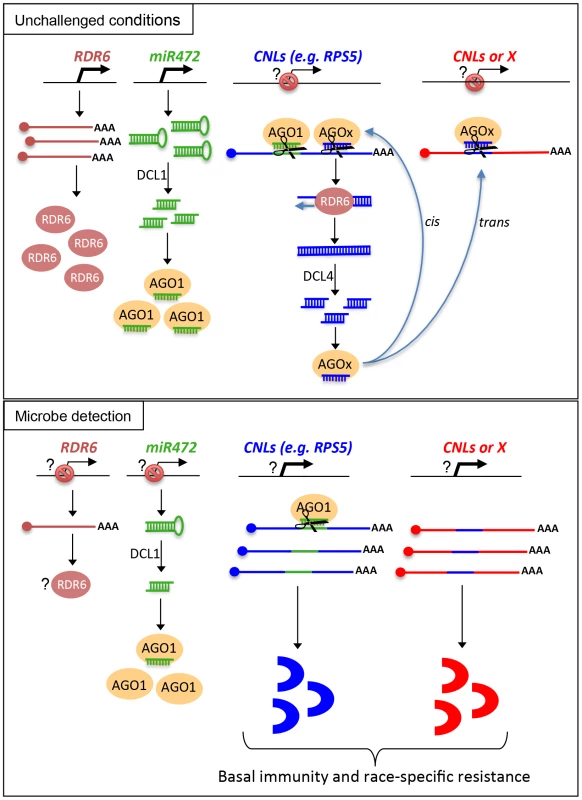
It is also likely that the genes encoding the above immune receptors remain at a transcriptionally inactive state in unchallenged conditions as demonstrated for several other disease resistance genes [17], [48]. In this case, the concomitant low basal transcriptional expression of CNLs and the miR472/RDR6-dependent post-transcriptional regulatory process would effectively deplete immune receptor mRNAs in the absence of pathogens, thus preventing an autoimmune response that would have detrimental consequences on plant fitness [1], [77]. This is reminiscent of recent findings on other 22 nt miRNAs/secondary siRNAs that target NLR transcripts in different plant species [20], [21], [22], as well as with the observation that the production of siRNAs at the disease resistance RPP4 cluster repress basal expression of several R gene transcripts within this cluster and likewise prevent constitutive activation of the SA-dependent defense pathway [18]. Our model also suggests that the mature form of miR472 is down-regulated during PTI, as a 4-fold decrease in the accumulation of this microRNA was observed in small RNA libraries generated by Li et al [38] upon flg22 treatment, which was confirmed in Arabidopsis leaves and seedlings treated with flg22 (Figure S13). We thus propose that upon pathogen detection, and perhaps also perception of non-adapted microbes, microbe-associated molecular patterns trigger the down-regulation of miR472, which in concert with the eventual transcriptional activation of CNLs, may contribute to the transient enhanced accumulation of CNL mRNAs/proteins at an early phase of the elicitation (Figure 8). This gene regulatory mechanism may also be reinforced by the down-regulation of RDR6-dependent silencing pathway as suggested by the rapid repression of RDR6 mRNAs during PTI (Figure 1). At a later phase of the elicitation, we propose that this double post-transcriptional layer of regulation mediated by miR472 and RDR6 likely trigger a robust resilencing of these CNL transcripts to prevent a sustained activation of the plant immune response.
Functional relevance of disease resistance proteins in both ETI and PTI responses
Although R proteins have been extensively characterized in ETI [10], there is increasing evidence that these immune receptors can also contribute to basal defense as well as PTI responses in plants [78]. For example, a compromised basal resistance to virulent Pto DC3000 was previously reported in a rar1 loss-of-function mutant [64], and confirmed in the present study (Figure 7E), suggesting that plant NLRs contribute to basal immunity. More recently, a subclade of CNL proteins, characterized as ‘helper NB-LRR’, where not only required for ETI but also for basal resistance and this process was independent of their P-loop motifs [79]. Importantly, these CNLs additionally regulate PAMP-triggered SA-accumulation in response to a disarmed P. syringae strain, which provides evidence that plant NLRs contribute to PTI [79]. Nevertheless, these CNLs do not control early events of PTI responses triggered by flg22 or the elongation factor-derived peptide elf18, indicating that these immune receptors likely act downstream or independently of these early PTI signaling events [79]. In the present work, we showed that another subclade of CNLs, which are targeted by miR472 and RDR6-dependent siRNAs, possibly contribute to multiple PTI signaling events, including potentially flg22-triggered callose deposition and ROS production (Figure 3B). Interestingly, the product of one if this mRNA target, the RPS5 protein, was previously shown to reside in the same protein complex as the PTI receptor FLS2 [80], further supporting a molecular link between ETI and PTI components.
Conclusion
In conclusion we have established a direct link between miR472/RDR6-dependent PTGS and plant immunity. We showed that both miR472 and RDR6 act as negative regulators of PTI and ETI, presumably by repressing a subset of CNLs at the post-transcriptional level. Our data therefore sustain previous anticipations suggesting that in addition to their role in specific resistance, R proteins contribute to PTI [10], [64], [78], [81], [82]. Furthermore, given that flg22 as well as disarmed bacteria were shown to trigger Systemic Acquired Resistance (SAR), such as in response to pathogens expressing Avr products [83]–[86], we speculate that a potential release of miR472 - and eventually of RDR6-dependent PTGS may also occur in distal tissues, and thereby might contribute to the transient derepression of a whole repertoire of disease resistance genes as part of the SAR response.
Materials and Methods
Plants and bacterial strains
Arabidopsis thaliana seeds from the Col-0 accession were used as wild-type, the rdr6-15 T-DNA insertion line has been previously described in Xi et al [87]. We also used sid2-2, npr1-1 and rar1-21 mutant alleles. Plants were genotyped with the following primers and conditions: rdr6 (RDR6_LP:TGAATCCATTCCTGAACAAGC; RDR6_RP: CAATGCAACCTCATCTTGGATG; LB3: TAGCATCTGAATTTCATAACCAATCTCGATACAC), npr1(1g64280_F: AGGGGATATACGGTGCTTCAT; 1g64280_R: GAGCAGCGTCATCTTCAATTC); sid2 (sid2_F:CAGTCCGAAAGACGACCTCGAGTT;sid2_R:CTCATCATCTTCCTTCGTAAGTCTCC); rar1 (5g51700_F: AAGCAGGGAGTAAGTCAAATTTAC; 5g51700_R CAAACTGAAATCATGACTTCTTTG). All plants were grown in short days conditions subjected to a cycle of 8 h and 16 h of light and darkness, respectively, at a day/night temperature of 22.5/18.5° with 50–60% humidity for about 5–6 weeks. The plants were watered 16 h before inoculation to promote stomatal opening, thereby facilitating inoculation.
Pseudomonas syringae pv. tomato DC3000 (Pto DC3000) was grown at 28°C on NYGB medium (5 g L−1 bactopeptone, 3 g L−1 yeast extract, 20 ml L−1 glycerol) containing kanamycin (50 mg mL−1) and rifampicin (25 mg mL−1) for selection.
Plant inoculations and bacterial counting
Pto DC3000, Pto DC3000 AvrPphB and Pto DC3000 AvrRpt2 from overnight culture were collected, washed once and resuspended in 10 mM MgCl2 at a concentration of 5×105 colony-forming units (CFU) mL−1. A. thaliana leaves were infiltrated with bacterial suspensions using a needleless syringe. Leaves were harvested immediately (0 dpi) or after 4 days. Two leaf discs (d = 0.4 mm) from two different leaves were washed in 10 mM MgCl2 and then ground with a Microfuge pestle. After grinding of the tissue, the samples were diluted 1∶10 serially. Samples were plated on NYGA solid medium (NYGB with 10 g L−1 agar) supplemented with antibiotics. Plates were placed at 28°C for 4 days and the CFU were counted. For spray inoculation bacteria were resuspended in 10 mM MgCl2 at OD600 of 0.2 (108 CFU/mL) and Silwet was added to a final concentration of 0.04%. All experiments presented were repeated three times and statistical differences were detected with a Wilcoxon test (*, P<0.05; **, P<0.01).
ROS measurements and callose staining
Reactive oxygen species released by leaf discs were assayed by H2O2-dependent luminescence of luminal [88]. Leaf discs were deposed into 96-well plate and incubated overnight in 200 µL H2O in a growth chamber. The next morning, 100 µL H2O containing 20 µM luminol and 1 µg horseradish peroxidase (Sigma) with or without 100 nM flg22 were added. Luminescence was immediately measured for 45 min using a Tristar LB 941 plate reader (Berthold technologies, Thoiry). At least 25 to 30 discs were tested by conditions.
For callose detection, leaves were infiltrated with 100 nM flg22 or water using a needleless syringe. After 15 h, about ten leaves from at least four independent plants were cleared by immersion in an alcoholic lactophenol solution by the method of Shipton and Brown [89] modified by Adam and Sommerville [90]. They were rinsed in 50% ethanol, then in water. Callose was detected by staining for 30 min in 150 mM K2HPO4 (pH 9.5) buffer containing 0.01% aniline blue (Sigma-Aldrich). After staining each leaf was mounted in 50% glycerol and examined with an Olympus Macro Zoom System Microscope MVX10 fluorescent microscope (excitation filter 365 nm and barrier filter 420 nm). Representative pictures are shown. The number of callose deposits per picture was determined using ImageJ (National Institutes of Health, Bethesda, MD, U.S.A.) and compared using a Wilcoxon test (P<0.05). We analyzed 25 to 30 pictures corresponding to more than five independent leaves for each treatment.
RNA extraction and RT-qPCR analyses
For RNA extraction, leaves or seedlings were collected, immediately frozen in liquid nitrogen, and then stored at −80°C. Total RNA was prepared by TRIzol (Invitrogen) extraction as recommended by the supplier (Invitrogen). For RT-PCR analysis, first-strand cDNA was synthesized using Superscript reverse transcriptase (Invitrogen,) from 1 µg of RNase-free DNaseI-treated (Promega) total RNA in a 20 µl reaction volume. Quantitative PCR reactions were performed on 1/40 of cDNA, 300 nM final concentration of each primer pair and LightCycler 480 SYBR Green I Master 2× conc. (Roche). PCR was performed in 384-well optical reaction plates heated at 95°C for 10 min, followed by 45 cycles of denaturation at 95°C for 15 s and annealing and elongation at 60°C for 30 s. A melting curve was performed at the end of the amplification by steps of 1°C (from 95°C to 50°C). Each experiment was repeated two to three times. Transcript levels were normalized to that of At2G36060, At4G29130 and At5G13440 genes. These reference genes display invariant expression over hundreds of publicly available microarray experiments. The gene-specific primers used in this analysis were listed in Figure S14.
For miR472 quantification, total RNA was isolated from plants using TRIzol reagent (Invitrogen) and treated with RNase-free DNaseI (Promega). Small RNAs were polyadenylated with ATP by poly(A) polymerase following the manufacturer's directions for the Poly(A) Tailing Kit (Ambion). After phenol-chloroform extraction and ethanol precipitation, the RNAs were reverse-transcribed with 200 U SuperScript III Reverse Transcriptase (Invitrogen) and 0.5 µg poly(T) adapter (Figure S14) according to the manufacturer's protocols (Invitrogen). The cDNAs were used for qPCR with miR472 as one primer and the reverse primer as described by Shi and Chiang [91]. 5,8S ribosomal RNA gene was used as internal control as previously described [91]. Sequences of miR472, reverse primer, poly(T) adapter and 5S primers are listed in Figure S14.
Deep-sequencing
Total cellular RNA (5 µg), extracted using TRIzol reagent (Invitrogen) was processed into sequencing libraries using adapted Illumina protocols and sequenced at Fasteris (http://www.fasteris.com, Switzerland) using the Hi-seq 2000 sequencer. All next-generation sequencing data have been deposited to the NCBI Gene Expression Omnibus (GEO).
Bioinformatic analyses
We took advantage of publicly available sRNA libraries from leaf tissue [92]. These data correspond to 2 replicates of WT and rdr6 sRNA sequenced using Illumina Genome Analyser technology. Replicates were pooled and sequence reads were matched against the Arabidopsis thaliana genome (TAIR10) using MUMmer v3.0 [93]. Only 15 to 30-nt long sRNAs reads with perfect match over their entire length were analysed further (2 434 780 and 1 753 064 for WT and rdr6 respectively). The number of 20–22 nt reads matching TAIR10 annotated protein coding genes locus or tasiRNA were then compared between WT and rdr6 libraries by differential analysis with NOISeq [94] using the parameters indicated below: k = NULL, norm = “rpkm”, long = 1000, q = 0.90, pnr = 0.5, nss = 1000, v = 0.02, lc = 1.
The WT and miR472OE sRNA libraries, containing 17 828 872 and 30 869 878 sRNA reads respectively, were processed using the same methods. Over those reads, 88.9% are 15 to 30-nt long and can be perfectly aligned to Arabidopsis genome. The miR472OE line was validated by comparing the number of reads mapping to all miRNA stem-loop loci (miRBase release 19; [95]–[98]) between WT and mutant sRNA libraries. Differential analysis of 20–22 nt reads in genes has then been done as described in the previous paragraph.
Supporting Information
Zdroje
1. DanglJL, JonesJD (2001) Plant pathogens and integrated defense responses to infection. Nature 411 : 826–833.
2. NavarroL, ZipfelC, RowlandO, KellerI, RobatzekS, et al. (2004) The transcriptional innate immune response to flg22. Interplay and overlap with Avr gene-dependent defense responses and bacterial pathogenesis. Plant Physiol 135 : 1113–1128.
3. ZipfelC, RobatzekS, NavarroL, OakeleyEJ, JonesJD, et al. (2004) Bacterial disease resistance in Arabidopsis through flagellin perception. Nature 428 : 764–767.
4. MelottoM, UnderwoodW, KoczanJ, NomuraK, HeSY (2006) Plant stomata function in innate immunity against bacterial invasion. Cell 126 : 969–980.
5. SchwessingerB, ZipfelC (2008) News from the frontline: recent insights into PAMP-triggered immunity in plants. Curr Opin Plant Biol 11 : 389–395.
6. GöhreV, RobatzekS (2008) Breaking the barriers: microbial effector molecules subvert plant immunity. Annu Rev Phytopathol 46 : 189–215.
7. LindebergM, CunnacS, CollmerA (2009) The evolution of Pseudomonas syringae host specificity and type III effector repertoires. Mol Plant Pathol 10 : 767–75.
8. KuferTA, SansonettiPJ (2011) NLR functions beyond pathogen recognition. Nat Immunol 12 : 121–8.
9. MeyersBC, KozikA, GriegoA, KuangH, MichelmoreRW (2003) Genome-wide analysis of NBS-LRR-encoding genes in Arabidopsis. Plant Cell 15 : 809–34.
10. JonesJD, DanglJL (2006) The plant immune system. Nature 444 : 323–329.
11. OldroydGE, StaskawiczBJ (1998) Genetically engineered broad-spectrum disease resistance in tomato. Proc Natl Acad Sci USA 95 : 10300–10305.
12. BendahmaneA, Kanyuka1K, BaulcombeDC (1999) The Rx Gene from Potato Controls Separate Virus Resistance and Cell Death Responses. Plant Cell 11 : 781–792.
13. TaoY, YuanF, LeisterRT, AusubelFM, KatagiriF (2000) Mutational analysis of the Arabidopsis nucleotide binding site-leucine-rich repeat resistance gene RPS2. Plant Cell 12 : 2541–2554.
14. ZhangY, GoritschnigS, DongX, LiX (2003) A gain-of-function mutation in a plant disease resistance gene leads to constitutive activation of downstream signal transduction pathways in suppressor of npr1-1, constitutive 1. Plant Cell 15 : 2636–46.
15. ZhangY, DoreyS, SwiderskiM, JonesJD (2004) Expression of RPS4 in tobacco induces an AvrRps4-independent HR that requires EDS1, SGT1 and HSP90. Plant J 40 : 213–24.
16. ZhouF, MosherS, TianM, SassiG, ParkerJ, KlessigDF (2008) The Arabidopsis gain-of-function mutant ssi4 requires RAR1 and SGT1b differentially for defense activation and morphological alterations. Mol Plant Microbe Interact 21 : 40–9.
17. JohnsonKCM, DongOX, HuangY, LiX (2012) A Rolling Stone Gathers No Moss, but Resistant Plants Must Gather Their MOSes. Cold Spring Harb Symp Quant Biol 77 : 259–68.
18. YiH, RichardsEJ (2007) A cluster of disease resistance genes in Arabidopsis is coordinately regulated by transcriptional activation and RNA silencing. Plant Cell 19 : 2929–2939.
19. Ruiz-FerrerV, VoinnetO (2009) Roles of plant small RNAs in biotic stress responses. Annu Rev Plant Biol 60 : 485–510.
20. ZhaiJ, JeongDH, De PaoliE, ParkS, RosenBD, et al. (2011) MicroRNAs as master regulators of the plant NB-LRR defense gene family via the production of phased trans-acting siRNAs. Genes Dev 25 : 2540–2553.
21. ShivaprasadPV, ChenHM, PatelK, BondDM, SantosBA, BaulcombeDC (2012) A MicroRNA Superfamily Regulates Nucleotide Binding Site-Leucine-Rich Repeats and other mRNAs. Plant Cell 24 : 859–874.
22. LiF, PignattaD, BendixC, BrunkardJO, CohnMM, et al. (2012) MicroRNA regulation of plant innate immune receptors. Proc Natl Acad Sci USA 109 : 1790–1795.
23. DowenRH, PelizzolaM, SchmitzRJ, ListerR, DowenJM, et al. (2012) Widespread dynamic DNA methylation in response to biotic stress. Proc Natl Acad Sci USA 109: E2183–91.
24. YuA, LepèreG, JayF, WangJY, BapaumeL, et al. (2013) Dynamics and biological relevance of DNA demethylation in Arabidopsis antibacterial defense. Proc Natl Acad Sci USA 110 : 2389–94.
25. VaucheretH (2006) Post-transcriptional small RNA pathways in plants: mechanisms and regulations. Genes Dev 20 : 759–71.
26. BrodersenP, VoinnetO (2006) The diversity of RNA silencing pathways in plants. Trends Genet 22 : 268–80.
27. BaulcombeD (2004) RNA silencing in plants. Nature 2431 : 356–63.
28. MourrainP, BéclinC, ElmayanT, FeuerbachF, GodonC, et al. (2000) Arabidopsis SGS2 and SGS3 genes are required for posttranscriptional gene silencing and natural virus resistance. Cell 101 : 533–42.
29. DalmayT, HamiltonA, RuddS, AngellS, BaulcombeDC (2000) An RNA-dependent RNA polymerase gene in Arabidopsis is required for posttranscriptional gene silencing mediated by a transgene but not by a virus. Cell 101 : 543–53.
30. QuF, YeX, HouG, SatoS, ClementeTE, MorrisTJ (2005) RDR6 has a broad-spectrum but temperature-dependent antiviral defense role in Nicotiana benthamiana. J Virol 79 : 15209–15217.
31. SchwachF, VaistijFE, JonesL, BaulcombeDC (2005) An RNA-dependent RNA polymerase prevents meristem invasion by potato virus X and is required for the activity but not the production of a systemic silencing signal. Plant Physiol 138 : 1842–1852.
32. Katiyar-AgarwalS, JinH (2010) Role of small RNAs in host-microbe interactions. Annu Rev Phytopathol 48 : 225–46.
33. VazquezF, VaucheretH, RajagopalanR, LepersC, GasciolliV, et al. (2004) Endogenous trans-acting siRNAs regulate the accumulation of Arabidopsis mRNAs. Mol Cell 16 : 69–79.
34. YoshikawaM, PeragineA, ParkMY, PoethigRS (2005) A pathway for the biogenesis of trans-acting siRNAs in Arabidopsis. Genes Dev 19 : 2164–75.
35. CuperusJT, CarbonellA, FahlgrenN, Garcia-RuizH, BurkeRT, et al. (2010) Unique functionality of 22-nt miRNAs in triggering RDR6-dependent siRNA biogenesis from target transcripts in Arabidopsis. Nat Struct Mol Biol 17 : 997–1003.
36. ManavellaPA, KoenigD, WeigelD (2012) Plant secondary siRNA production determined by microRNA-duplex structure. Proc Natl Acad Sci USA 109 : 2461–6.
37. NavarroL, JayF, NomuraK, HeSY, VoinnetO (2008) Suppression of the MicroRNA Pathway by Bacterial Effector Proteins. Science 321 : 964–967.
38. LiY, ZhangQ, ZhangJ, WuL, QiY, et al. (2010) Identification of microRNAs involved in pathogen-associated molecular pattern-triggered plant innate immunity. Plant Physiol 152 : 2222–2231.
39. PeragineA, YoshikawaM, WuG, AlbrechtHL, PoethigRS (2004) SGS3 and SGS2/SDE1/RDR6 are required for juvenile development and the production of trans-acting siRNAs in Arabidopsis. Genes Dev 18 : 2368–79.
40. TorresMA, JonesJD, DanglJL (2006) Reactive oxygen species signaling in response to pathogens. Plant Physiol 141 : 373–8.
41. AsaiT, TenaG, PlotnikovaJ, WillmannMR, ChiuWL, et al. (2002) MAP kinase signalling cascade in Arabidopsis innate immunity. Nature 415 : 977–983.
42. HauckP, ThilmonyR, HeSY (2003) A Pseudomonas syringae type III effector suppresses cell wall-based extracellular defense in susceptible Arabidopsis plants. Proc Natl Acad Sci USA 100 : 8577–8582.
43. DebRoyS, ThilmonyR, KwackYB, NomuraK, HeSY (2004) A family of conserved bacterial effectors inhibits salicylic acid-mediated basal immunity and promotes disease necrosis in plants. Proc Natl Acad Sci USA 101 : 9927–9932.
44. ZengW, HeSY (2010) A prominent role of the flagellin receptor FLAGELLIN-SENSING2 in mediating stomatal response to Pseudomonas syringae pv tomato DC3000 in Arabidopsis. Plant Physiol 153 : 1188–98.
45. MalloryAC, VaucheretH (2009) ARGONAUTE 1 homeostasis invokes the coordinate action of the microRNA and siRNA pathways. EMBO Rep 10 : 521–526.
46. LuC, KulkarniK, SouretFF, MuthuValliappanR, TejSS, et al. (2006) MicroRNAs and other small RNAs enriched in the Arabidopsis RNA-dependent RNA polymerase-2 mutant. Genome Res 16 : 1276–88.
47. SimonichMT, InnesRW (1995) A disease resistance gene in Arabidopsis with specificity for the avrPph3 gene of Pseudomonas syringae pv. phaseolicola. Mol Plant Microbe Interact 8 : 637–640.
48. HuangX, LiJ, BaoF, ZhangX, YangS (2010) A gain-of-function mutation in the Arabidopsis disease resistance gene RPP4 confers sensitivity to low temperature. Plant Physiol 154 : 796–809.
49. LlaveC, XieZ, KasschauKD, CarringtonJC (2002) Cleavage of Scarecrow-like mRNA Targets Directed by a Class of Arabidopsis miRNA. Science 297 : 2053–2056.
50. ZhangZ, WuY, GaoM, ZhangJ, KongQ, et al. (2012) Disruption of PAMP-induced MAP kinase cascade by a Pseudomonas syringae effector activates plant immunity mediated by the NB-LRR protein SUMM2. Cell Host Microbe 11 : 253–63.
51. PieterseCM, Leon-ReyesA, Van der EntS, Van WeesSC (2009) Networking by small-molecule hormones in plant immunity. Nat Chem Biol 5 : 308–316.
52. ShapiroAD, ZhangC (2001) The Role of NDR1 in Avirulence Gene-Directed Signaling and Control of Programmed Cell Death in Arabidopsis. Plant Physiol 127 : 1089–1101.
53. WildermuthMC, DewdneyJ, WuG, AusubelF (2001) Isochorismate synthase is required to synthetize salicylic acid for plant defense. Nature 414 : 562–565.
54. CaoH, GlazebrookJ, ClarkeJD, VolkoS, DongX (1997) The Arabidopsis NPR1 gene that controls systemic acquired resistance encodes a novel protein containing ankyrin repeats. Cell 88 : 57–63.
55. ShirasuK, LahayeT, TanMW, ZhouF, AzevedoC, Schulze-LefertP (1999) A novel class of eukaryotic zinc-binding proteins is required for disease resistance signaling in barley and development in C. elegans. Cell 99 : 355–366.
56. WarrenRF, MerrittPM, HolubE, InnesRW (1999) Identification of three putative signal transduction genes involved in R gene-specified disease resistance in Arabidopsis. Genetics 152 : 401–12.
57. AustinMJ, MuskettPJ, KahnK, FeysBJ, JonesJDG, et al. (2002) Regulatory role of SGT1 in early R-mediated plant defenses. Science 295 : 2077–2080.
58. MuskettPR, KahnK, AustinMJ, MoisanLJ, SadanandomA, et al. (2002) Arabidopsis RAR1 exerts rate-limiting control of R gene-mediated defenses against multiple pathogens. Plant Cell 14 : 979–992.
59. TorneroP, MerrittP, SadanandomA, ShirasuK, InnesRW, et al. (2002) RAR1 and NDR1 contribute quantitatively to disease resistance in Arabidopsis and their relative contributions are dependent on the R gene assayed. Plant Cell 14 : 1005–1015.
60. AzevedoC, SadanandomA, KitigawaK, FreialdenhovenA, ShirasuK, et al. (2002) The RAR1 interactor SGT1 is an essential component of R-gene triggered disease resistance. Science 295 : 2073–2076.
61. TörM, GordonP, CuzickA, EulgemT, SinapidouE, et al. (2002) Arabidopsis SGT1b is required for defense signaling conferred by several downy mildew (Peronospora parasitica) resistance genes. Plant Cell 14 : 993–1003.
62. TakahashiA, CasaisC, IchimuraK, ShirasuK (2003) HSP90 interacts with RAR1 and SGT1 and is essential for RPS2-mediated disease resistance in Arabidopsis. Proc Natl Acad Sci USA 100 : 11777–11782.
63. HubertDA, TorneroP, BelkhadirY, KrishnaP, TakahashiA, et al. (2003) Cytosolic HSP90 associates with and modulates the Arabidopsis RPM1 disease resistance protein. EMBO J 22 : 5679–5689.
64. HoltBF, BelkhadirY, DanglJL (2005) Antagonistic control of disease resistance protein stability in the plant immune system. Science 309 : 929–932.
65. VenugopalSC, JeongRD, MandalMK, ZhuS, Chandra-ShekaraAC, et al. (2009) Enhanced disease susceptibility 1 and salicylic acid act redundantly to regulate resistance gene-mediated signaling. PLoS Genet 5: e1000545.
66. BoyesDC, NamJ, DanglJL (1998) The Arabidopsis thaliana RPM1 disease resistance gene product is a peripheral plasma membrane protein that is degraded coincident with the hypersensitive response. Proc Natl Acad Sci USA 95 : 15849–15854.
67. AxtellMJ, StaskawiczBJ (2003) Initiation of RPS2-specified disease resistance in Arabidopsis is coupled to the AvrRpt2-directed elimination of RIN4. Cell 112 : 369–377.
68. BelkhadirY, NimchukZ, HubertDA, MackeyD, DanglJL (2004) Arabidopsis RIN4 negatively regulates disease resistance mediated by RPS2 and RPM1 downstream or independent of the NDR1 signal modulator, and is not required for the virulence functions of bacterial type III effectors AvrRpt2 or AvrRpm1. Plant Cell 16 : 2822–2835.
69. VaistijFE, JonesL (2009) Compromised virus-induced gene silencing in RDR6-deficient plants. Plant Physiol 149 : 1399–1407.
70. Di SerioF, Martínez de AlbaAE, NavarroB, GiselA, FloresR (2010) RNA-dependent RNA polymerase 6 delays accumulation and precludes meristem invasion of a viroid that replicates in the nucleus. J Virol 84 : 2477–89.
71. NicaiseV, RouxM, ZipfelC (2009) Recent Advances in PAMP-Triggered Immunity against Bacteria: Pattern Recognition Receptors Watch over and Raise the Alarm. Plant Physiol 150 : 1638–1647.
72. NavarroL, DunoyerP, JayF, ArnoldB, DharmasiriN, et al. (2006) A plant miRNA contributes to antibacterial resistance by repressing auxin signaling. Science 312 : 436–439.
73. ConrathU (2011) Molecular aspects of defense priming. Trends Plant Sci 16 : 524–531.
74. ShenQH, SaijoY, MauchS, BiskupC, BieriS, et al. (2007) Nuclear activity of MLA immune receptors links isolate-specific and basal disease-resistance responses. Science 315 : 1098–1103.
75. TrujilloM, IchimuraK, CasaisC, ShirasuK (2008) Negative regulation of PAMP-triggered immunity by E3 ubitquitin ligase triplet in Arabidopsis. Curr Biol 18 : 1396–1401.
76. LuD, LinW, GaoX, WuS, ChengC, et al. (2011) Direct ubiquitination of pattern recognition receptor FLS2 attenuates plant innate immunity. Science 332 : 1439–42.
77. BombliesK, LempeJ, EppleP, WarthmannN, LanzC, et al. (2007) Autoimmune Response as a Mechanism for a Dobzhansky-Muller-Type Incompatibility Syndrome in Plants. PLoS Biol 5: e236.
78. TsudaK, KatagiriF (2010) Comparing signaling mechanisms engaged in pattern-triggered and effector-triggered immunity. Curr Opin Plant Biol 13 : 459–65.
79. BonardiV, TangS, StallmannA, RobertsM, CherkisK, et al. (2011) Expanded functions for a family of plant intracellular immune receptors beyond specific recognition of pathogen effectors. Proc Natl Acad Sci U S A 108 : 16463–8.
80. ZhangZ, WuY, GaoM, QiY, TsudaK, et al. (2011) Physical association of pattern-triggered immunity (PTI) and effector-triggered immunity (ETI) immune receptors in Arabidopsis. Mol Plant Pathol 12 : 702–8.
81. TsudaK, SatoM, StoddardT, GlazebrookJ, KatagiriF (2009) Network properties of robust immunity in plants. PLoS Genet 5: e1000772.
82. ThommaBP, NürnbergerT, JoostenMH (2011) Of PAMPs and effectors: the blurred PTI-ETI dichotomy. Plant Cell 23 : 4–15.
83. RossAF (1961) Systemic acquired resistance induced by localized virus infections in plants. Virology 14 : 340–358.
84. DurrantWE, DongX (2004) Systemic acquired resistance. Annu Rev Phytopathol 42 : 185–209.
85. MishinaTE, ZeierJ (2007) Pathogen-associated molecular pattern recognition rather than development of tissue necrosis contributes to bacterial induction of systemic acquired resistance in Arabidopsis. Plant J 50 : 500–513.
86. SpoelSH, DongX (2012) How do plants achieve immunity? Defence without specialized immune cells. Nat Rev Immunol 12 : 89–100.
87. XieZ, JohansenLK, GustafsonAM, KasschauKD, LellisAD, et al. (2004) Genetic and functional diversification of small RNA pathways in plants. PLoS Biol 2: e104.
88. SchwackeR, HagerA (1992) Fungal elicitors induce a transient release of active oxygen species from cultured spruce cells that is dependent on Ca2+ and protein-kinase activity. Planta 187 : 136–141.
89. ShiptonWA, BrownJF (1962) A whole-leaf clearing and staining technique to demonstrate host-pathogen relationships of wheat stem rust. Phytopathology 52 : 1313–1318.
90. AdamL, SomervilleSC (1996) Genetic characterization of five powdery mildew disease resistance loci in Arabidopsis thaliana. Plant J 9 : 341–356.
91. ShiR, ChiangVL (2005) Facile means for quantifying microRNA expression by real-time PCR. BioTechniques 39 : 519–525.
92. HardcastleTJ, KellyKA (2010) baySeq: empirical Bayesian methods for identifying differential expression in sequence count data. BMC Bioinformatics 11 : 422.
93. KurtzS, PhillippyA, DelcherAL, SmootM, ShumwayM, et al. (2004) Versatile and open software for comparing large genomes. Genome Biology 5: R12.
94. TarazonaS, García-AlcaldeF, DopazoJ, FerrerA, ConesaA (2011) Differential expression in RNA-seq: a matter of depth. Genome Res 21 : 2213–23.
95. Griffiths-JonesS (2004) The miRNA Registry. Nucleic Acids Res 32: D109–D111.
96. Griffiths-JonesS, GrocockRJ, van DongenS, BatemanA, EnrightAJ (2006) miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res 34: D140–D144.
97. Griffiths-JonesS, SainiHK, van DongenS, EnrightAJ (2008) miRBase: tools for microRNA genomics. Nucleic Acids Res 36: D154–D158.
98. KozomaraA, Griffiths-JonesS (2011) miRBase: integrating microRNA annotation and deep-sequencing data. Nucleic Acids Res 2011 39: D152–D157.
Štítky
Hygiena a epidemiologie Infekční lékařství Laboratoř
Článek Serotonin Signaling in : A Serotonin–Activated G Protein-Coupled Receptor Controls Parasite MovementČlánek Regulators of Cell Cycle Progression and Differentiation Identified Using a Kinome-Wide RNAi ScreenČlánek IFNγ/IL-10 Co-producing Cells Dominate the CD4 Response to Malaria in Highly Exposed ChildrenČlánek Functions of CPSF6 for HIV-1 as Revealed by HIV-1 Capsid Evolution in HLA-B27-Positive SubjectsČlánek Decreases in Colonic and Systemic Inflammation in Chronic HIV Infection after IL-7 Administration
Článek vyšel v časopisePLOS Pathogens
Nejčtenější tento týden
2014 Číslo 1- Stillova choroba: vzácné a závažné systémové onemocnění
- Diagnostický algoritmus při podezření na syndrom periodické horečky
- Diagnostika virových hepatitid v kostce – zorientujte se (nejen) v sérologii
- Perorální antivirotika jako vysoce efektivní nástroj prevence hospitalizací kvůli COVID-19 − otázky a odpovědi pro praxi
- Infekční komplikace virových respiračních infekcí – sekundární bakteriální a aspergilové pneumonie
-
Všechny články tohoto čísla
- HIV-1 Accessory Proteins Adapt Cellular Adaptors to Facilitate Immune Evasion
- Ranaviruses: Not Just for Frogs
- Effectors and Effector Delivery in
- Plasmacytoid Dendritic Cell Dynamics Tune Interferon-Alfa Production in SIV-Infected Cynomolgus Macaques
- Lu/BCAM Adhesion Glycoprotein Is a Receptor for Cytotoxic Necrotizing Factor 1 (CNF1)
- A Substrate-Fusion Protein Is Trapped inside the Type III Secretion System Channel in
- Parvovirus-Induced Depletion of Cyclin B1 Prevents Mitotic Entry of Infected Cells
- Red Blood Cell Invasion by : Structural Basis for DBP Engagement of DARC
- NsrR, GadE, and GadX Interplay in Repressing Expression of the O157:H7 LEE Pathogenicity Island in Response to Nitric Oxide
- Loss of Circulating CD4 T Cells with B Cell Helper Function during Chronic HIV Infection
- TREM-1 Deficiency Can Attenuate Disease Severity without Affecting Pathogen Clearance
- Origin, Migration Routes and Worldwide Population Genetic Structure of the Wheat Yellow Rust Pathogen f.sp.
- Glutamate Utilization Couples Oxidative Stress Defense and the Tricarboxylic Acid Cycle in Phagosomal Escape
- Serotonin Signaling in : A Serotonin–Activated G Protein-Coupled Receptor Controls Parasite Movement
- Recovery of an Antiviral Antibody Response following Attrition Caused by Unrelated Infection
- Regulators of Cell Cycle Progression and Differentiation Identified Using a Kinome-Wide RNAi Screen
- Absence of Intestinal PPARγ Aggravates Acute Infectious Colitis in Mice through a Lipocalin-2–Dependent Pathway
- Induction of a Stringent Metabolic Response in Intracellular Stages of Leads to Increased Dependence on Mitochondrial Metabolism
- CTCF and Rad21 Act as Host Cell Restriction Factors for Kaposi's Sarcoma-Associated Herpesvirus (KSHV) Lytic Replication by Modulating Viral Gene Transcription
- Gammaherpesviral Gene Expression and Virion Composition Are Broadly Controlled by Accelerated mRNA Degradation
- The Arabidopsis Silencing Pathway Modulates PAMP- and Effector-Triggered Immunity through the Post-transcriptional Control of Disease Resistance Genes
- Inflammatory Stimuli Reprogram Macrophage Phagocytosis to Macropinocytosis for the Rapid Elimination of Pathogens
- Alphavirus Mutator Variants Present Host-Specific Defects and Attenuation in Mammalian and Insect Models
- Phosphopyruvate Carboxylase Identified as a Key Enzyme in Erythrocytic Carbon Metabolism
- IFNγ/IL-10 Co-producing Cells Dominate the CD4 Response to Malaria in Highly Exposed Children
- Electron Tomography of HIV-1 Infection in Gut-Associated Lymphoid Tissue
- Characterisation of a Multi-ligand Binding Chemoreceptor CcmL (Tlp3) of
- Single Cell Stochastic Regulation of Pilus Phase Variation by an Attenuation-like Mechanism
- Cell Tropism Predicts Long-term Nucleotide Substitution Rates of Mammalian RNA Viruses
- Functions of CPSF6 for HIV-1 as Revealed by HIV-1 Capsid Evolution in HLA-B27-Positive Subjects
- RNA-seq Analysis of Host and Viral Gene Expression Highlights Interaction between Varicella Zoster Virus and Keratinocyte Differentiation
- Kaposi's Sarcoma Associated Herpesvirus Tegument Protein ORF75 Is Essential for Viral Lytic Replication and Plays a Critical Role in the Antagonization of ND10-Instituted Intrinsic Immunity
- DAMP Molecule S100A9 Acts as a Molecular Pattern to Enhance Inflammation during Influenza A Virus Infection: Role of DDX21-TRIF-TLR4-MyD88 Pathway
- Variable Suites of Non-effector Genes Are Co-regulated in the Type III Secretion Virulence Regulon across the Phylogeny
- Reengineering Redox Sensitive GFP to Measure Mycothiol Redox Potential of during Infection
- Preservation of Tetherin and CD4 Counter-Activities in Circulating Alleles despite Extensive Sequence Variation within HIV-1 Infected Individuals
- KSHV 2.0: A Comprehensive Annotation of the Kaposi's Sarcoma-Associated Herpesvirus Genome Using Next-Generation Sequencing Reveals Novel Genomic and Functional Features
- Nutrient Limitation Governs Metabolism and Niche Adaptation in the Human Nose
- Decreases in Colonic and Systemic Inflammation in Chronic HIV Infection after IL-7 Administration
- Investigation of Acetylcholine Receptor Diversity in a Nematode Parasite Leads to Characterization of Tribendimidine- and Derquantel-Sensitive nAChRs
- Intranasal Vaccination Promotes Detrimental Th17-Mediated Immunity against Influenza Infection
- -Mediated Inhibition of Iron Export Promotes Parasite Replication in Macrophages
- Variation in RNA Virus Mutation Rates across Host Cells
- A Single Amino Acid in the Stalk Region of the H1N1pdm Influenza Virus HA Protein Affects Viral Fusion, Stability and Infectivity
- Group B Engages an Inhibitory Siglec through Sialic Acid Mimicry to Blunt Innate Immune and Inflammatory Responses
- Synthesis and Biological Properties of Fungal Glucosylceramide
- HIV Protective KIR3DL1/S1-HLA-B Genotypes Influence NK Cell-Mediated Inhibition of HIV Replication in Autologous CD4 Targets
- Recruitment of PfSET2 by RNA Polymerase II to Variant Antigen Encoding Loci Contributes to Antigenic Variation in
- Human and Plant Fungal Pathogens: The Role of Secondary Metabolites
- Lyme Disease: Call for a “Manhattan Project” to Combat the Epidemic
- Enhancing Virus-Specific Immunity by Combining Therapeutic Vaccination and PD-L1 Blockade in Chronic Hepadnaviral Infection
- Suppression of Interferon Lambda Signaling by SOCS-1 Results in Their Excessive Production during Influenza Virus Infection
- Inflammation Fuels Colicin Ib-Dependent Competition of Serovar Typhimurium and in Blooms
- Host-Specific Enzyme-Substrate Interactions in SPM-1 Metallo-β-Lactamase Are Modulated by Second Sphere Residues
- STING-Dependent Type I IFN Production Inhibits Cell-Mediated Immunity to
- From Scourge to Cure: Tumour-Selective Viral Pathogenesis as a New Strategy against Cancer
- Lysine Acetyltransferase GCN5b Interacts with AP2 Factors and Is Required for Proliferation
- Narrow Bottlenecks Affect Populations during Vertical Seed Transmission but not during Leaf Colonization
- Targeted Cytotoxic Therapy Kills Persisting HIV Infected Cells During ART
- Murine Gammaherpesvirus M2 Protein Induction of IRF4 via the NFAT Pathway Leads to IL-10 Expression in B Cells
- iNKT Cell Production of GM-CSF Controls
- Malaria-Induced NLRP12/NLRP3-Dependent Caspase-1 Activation Mediates Inflammation and Hypersensitivity to Bacterial Superinfection
- Detection of Host-Derived Sphingosine by Is Important for Survival in the Murine Lung
- PLOS Pathogens
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Lyme Disease: Call for a “Manhattan Project” to Combat the Epidemic
- Origin, Migration Routes and Worldwide Population Genetic Structure of the Wheat Yellow Rust Pathogen f.sp.
- IFNγ/IL-10 Co-producing Cells Dominate the CD4 Response to Malaria in Highly Exposed Children
- Human and Plant Fungal Pathogens: The Role of Secondary Metabolites
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



