-
Články
- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Murine Gamma-Herpesvirus 68 Hijacks MAVS and IKKβ to Initiate Lytic Replication
Upon viral infection, the mitochondrial antiviral signaling (MAVS)-IKKβ pathway is activated to restrict viral replication. Manipulation of immune signaling events by pathogens has been an outstanding theme of host-pathogen interaction. Here we report that the loss of MAVS or IKKβ impaired the lytic replication of gamma-herpesvirus 68 (γHV68), a model herpesvirus for human Kaposi's sarcoma-associated herpesvirus and Epstein-Barr virus. γHV68 infection activated IKKβ in a MAVS-dependent manner; however, IKKβ phosphorylated and promoted the transcriptional activation of the γHV68 replication and transcription activator (RTA). Mutational analyses identified IKKβ phosphorylation sites, through which RTA-mediated transcription was increased by IKKβ, within the transactivation domain of RTA. Moreover, the lytic replication of recombinant γHV68 carrying mutations within the IKKβ phosphorylation sites was greatly impaired. These findings support the conclusion that γHV68 hijacks the antiviral MAVS-IKKβ pathway to promote viral transcription and lytic infection, representing an example whereby viral replication is coupled to host immune activation.
Published in the journal: . PLoS Pathog 6(7): e32767. doi:10.1371/journal.ppat.1001001
Category: Research Article
doi: https://doi.org/10.1371/journal.ppat.1001001Summary
Upon viral infection, the mitochondrial antiviral signaling (MAVS)-IKKβ pathway is activated to restrict viral replication. Manipulation of immune signaling events by pathogens has been an outstanding theme of host-pathogen interaction. Here we report that the loss of MAVS or IKKβ impaired the lytic replication of gamma-herpesvirus 68 (γHV68), a model herpesvirus for human Kaposi's sarcoma-associated herpesvirus and Epstein-Barr virus. γHV68 infection activated IKKβ in a MAVS-dependent manner; however, IKKβ phosphorylated and promoted the transcriptional activation of the γHV68 replication and transcription activator (RTA). Mutational analyses identified IKKβ phosphorylation sites, through which RTA-mediated transcription was increased by IKKβ, within the transactivation domain of RTA. Moreover, the lytic replication of recombinant γHV68 carrying mutations within the IKKβ phosphorylation sites was greatly impaired. These findings support the conclusion that γHV68 hijacks the antiviral MAVS-IKKβ pathway to promote viral transcription and lytic infection, representing an example whereby viral replication is coupled to host immune activation.
Introduction
Host cells activate innate immune signaling pathways to defend against invading pathogens. Pattern recognition receptors, including Toll-like receptors and cytosolic sensors (such as NOD-like receptors and RIG-I-like receptors), recognize pathogen-associated structural components and initiate signal transduction that leads to the biosynthesis and secretion of pro-inflammatory cytokines and interferons, thereby mounting a potent host immune response [1], [2]. To survive within an infected host, viruses have evolved intricate strategies to counteract host immune responses. Herpesviruses and poxviruses have large genomes and therefore have the capacity to encode numerous proteins that modulate host immune responses.
Mitochonrial antiviral signaling (MAVS, also known as IPS-1, VISA, and CARDIF) protein serves as an adaptor to activate both the NFκB and interferon regulatory factor (IRF) pathways [3], [4], [5], [6]. MAVS relays signals from RIG-I and MDA-5, cytosolic sensors that recognize viral dsRNA or ssRNA bearing 5′-triphosphate [7], [8], to the IKKα/β/γ and TBK-1/IKKε (also known as IKKi) kinase complexes [4], [6]. IKKα/β, together with the scaffold protein IKKγ, phosphorylates the inhibitor of NFκB (IκB) and promotes its subsequent ubiquitination and degradation by the proteasome, thereby unleashing NFκB that translocates into the nucleus to activate gene expression of pro-inflammatory cytokines [9], [10]. By contrast, TBK-1 and IKKε directly phosphorylate a serine/threonine-rich sequence within the carboxyl termini of IRF3 and IRF7, leading to the dimerization and nuclear translocation of these transcription factors [11], [12]. Together with NFκB and c-Jun/ATF-2, IRF3 and IRF7 bind to the interferon (IFN)-β enhancer and initiate the transcription of IFN-β [13], [14]. Ultimately, these signaling events promote cytokine and interferon production, establishing an antiviral state in infected cells. Although it is not clear how MAVS activates these immune kinases, recent findings have established the vital roles of MAVS in host antiviral innate immunity [15]. Interestingly, the mitochondrial localization of MAVS is critical for its ability to activate downstream signaling events. As such, various RNA viruses, exemplified by human hepatitis C virus (HCV), encode proteases that cleave MAVS from the outer membrane of the mitochondrion, thereby disarming MAVS-dependent signaling cascades and the host antiviral innate immunity [6], [16], [17], [18].
Murine gamma-herpesvirus 68 (γHV68 or MHV-68) is closely related to human Kaposi's sarcoma-associated herpesvirus (KSHV) and Epstein-Barr virus (EBV) [19]. KSHV and EBV are lymphotropic DNA viruses that are causally linked to malignancies of lymphoid or endothelial/epithelial origin, including lymphoma, nasopharyngeal carcinoma, and Kaposi's sarcoma [20], [21]. Persisting within host immune cells, KSHV and EBV are known to evade, manipulate, and exploit host immune pathways [22], [23]. Emerging studies suggest that γ-herpesviruses may usurp host innate immune responses for their infection [24], [25], [26]. However, it is not known how human KSHV and EBV manipulate innate immune pathways in vivo. Such investigations are greatly hampered by the lack of permissive cell lines and animal models for both KSHV and EBV. By contrast, γHV68 infection in laboratory mice leads to a robust acute infection in the lung and a long-term latent infection in the spleen. For murine γHV68 and human KSHV, the replication and transcription activator (RTA, encoded by ORF50) is necessary and sufficient to initiate lytic replication from latently-infected cells, supporting the notion that RTA integrates diverse signaling pathways to initiate lytic replication [27], [28], [29]. Using γHV68 as a surrogate for human KSHV and EBV, we have unexpectedly discovered that γHV68 activated IKKβ to phosphorylate RTA and promote RTA transcriptional activation, thereby increasing viral gene transcription and lytic replication. As such, RTA phosphorylation by IKKβ couples γHV68 gene expression and lytic replication to host innate immune activation, representing the first example whereby a virus hijacks the antiviral MAVS-IKKβ pathway to promote its lytic replication.
Results
MAVS is Necessary for Efficient γHV68 Lytic Replication
To investigate the roles of MAVS in γHV68 infection, wild-type (MAVS+/+), heterozygous (MAVS+/−), and knockout (MAVS−/−) mice were intranasally (i.n.) infected with 40 plaque-forming unit (PFU) γHV68. γHV68 acute infection in the lung was measured by plaque assays at 4, 7, 10, 13, and 16 days post-infection (d.p.i.). In MAVS+/+ mice, γHV68 titers peaked at 7 d.p.i. with approximately 500 PFU/lung and declined to 100 PFU/lung at 10 d.p.i. Viral load was undetectable by 13 d.p.i., indicating that γHV68 acute infection in the lung had been cleared (Figure 1A). Similar viral loads in the lungs of heterozygous mice (MAVS+/−) were observed (data not shown). Surprisingly, although viral loads at 7 d.p.i. in the lungs of MAVS−/− mice were comparable to those in the lungs of MAVS+/+ mice, γHV68 was nearly undetectable at 10 d.p.i. (Figure 1A). By contrast, γHV68 latent infection as characterized by viral genome frequency, persistent lytic replication, and reactivation was similar in splenocytes of MAVS+/+ and MAVS−/− mice at 16 and 45 d.p.i. (Figure S1). These observations suggest that MAVS plays a specific role(s) in γHV68 acute infection.
Fig. 1. MAVS deficiency reduces γHV68 lytic replication. 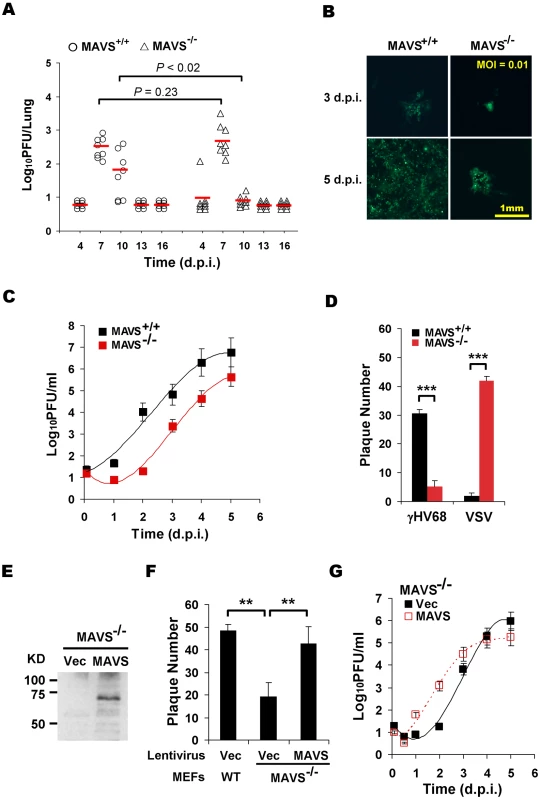
MAVS wild-type (MAVS+/+) or knockout (MAVS−/−) mice were intranasally (i.n.) infected with 40 PFU γHV68. The lungs were harvested at 4, 7, 10, 13, and 16 days post-infection (d.p.i.) and viral titer was determined by a plaque assay. (B) Mouse embryonic fibroblasts (MEFs) were infected with a GFP-marked γHV68 K3/GFP at multiplicities of infection (MOI) of 0.01. Viral replication in MAVS+/+ and MAVS−/− MEFs were photographed. (C) MEFs were infected with γHV68 K3/GFP as in (B) and viral titers were determined by a plaque assay. Data represent three independent experiments and error bars denote standard error of the mean (SEM). (D) MAVS+/+ and MAVS−/− MEFs were infected with 120 PFU γHV68 K3/GFP or 5 PFU vesicular stomatitis virus (VSV), and plaques were counted. Data represent the mean ± SEM of three independent experiments. (E to G) MAVS−/− MEFs were respectively infected with control lentivirus (Vec) or lentivirus containing the Flag-tagged human MAVS (MAVS), and selected with puromycin. (E) MAVS expression was confirmed by immunoprecipitation and immunoblot with anti-Flag antibody. (F) γHV68 plaque assays were performed as in (D). (G) Reconstituted MAVS−/− MEFs as indicated were infected with γHV68 K3/GFP (MOI = 0.01), and viral multi-step growth was determined by a plaque assay. Statistical significance in (A), (D), and (F): *, P<0.05; **, P<0.02; ***, P<0.005. To determine whether γHV68 infection altered MAVS expression, we infected BL/6 mice intranasally with a high dose (1×105 PFU) of γHV68, presumably permitting synchronized and maximal infection of lung epithelial cells. MAVS mRNA levels were determined by quantitative real-time PCR (qRT-PCR). The levels of MAVS mRNA were transiently increased at 2.5 and 5 d.p.i. in the lung and spleen, respectively (Figure S2A). Interestingly, the up-regulation of MAVS mRNA preceded that of viral RTA mRNA (Figure S2A and S2B), and that higher viral RTA mRNA levels tightly correlated with higher MAVS mRNA levels at 2.5 and 5 d.p.i., when MAVS mRNA levels peaked in the lung and spleen (Figure S2C). Together with the reduced viral load in the lungs of MAVS−/− mice (Figure 1A), these results suggest that MAVS is necessary for efficient lytic replication in mice and that the transiently induced MAVS expression by γHV68 infection may facilitate viral lytic replication in vivo.
To investigate the roles of MAVS in γHV68 infection, we then assessed the effects of MAVS-deficiency on γHV68 lytic replication ex vivo. Mouse embryonic fibroblasts (MEFs) were infected with a GFP-marked recombinant γHV68 (γHV68 K3/GFP) and viral replication was examined by fluorescence microscopy and plaque assays. Surprisingly, γHV68 displayed delayed replication kinetics in MAVS−/− MEFs compared to MAVS+/+ MEFs at multiplicities of infection (MOI) of 0.01 and 0.1 (Figure 1B, 1C and S3). To quantitatively determine the effect of MAVS on γHV68 lytic infection, we examined γHV68 lytic replication in MAVS+/+ and MAVS−/− MEFs by plaque assays. In fact, γHV68 formed approximately four-fold more plaques in MAVS+/+ MEFs than those in MAVS−/− MEFs, indicative of reduced initiation of lytic replication in MAVS-deficient MEFs (Figure 1D, S4A, and S4B). Interestingly, the plaque size of γHV68 was equivalent in MAVS+/+ and MAVS−/− MEFs (Figure S4C and S4D). To test whether MAVS−/− MEFs are defective in supporting viral lytic replication in general, we examined the lytic replication of vesicular stomatitis virus (VSV), a prototype RNA virus, with a plaque assay. Consistent with an antiviral activity of MAVS against RNA viruses, VSV formed 10-fold more plaques in MAVS−/− MEFs than those in MAVS+/+ MEFs (Figure 1D). The diminished lytic replication of γHV68 in MAVS-deficient MEFs is consistent with the reduced acute infection observed in the lung. To test whether exogenously expressed MAVS is able to restore γHV68 lytic replication, we generated lentivirus in 293T cells and MEFs stably expressing human MAVS (hMAVS) was established with puromycin selection (Figure 1E). As shown in Figure 1F and 1G, exogenous hMAVS restored γHV68 lytic replication by a plaque assay and multi-step growth curves. Nevertheless, these results together support the conclusion that MAVS is necessary for efficient γHV68 lytic replication in vivo and ex vivo.
The IKKβ/γ Complex is an Effector Downstream of MAVS in γHV68 Lytic Replication
Two known pathways, the IKKα/β/γ-NFκB and TBK-1/IKKε-IRF3/7 pathway, have been characterized downstream of MAVS (Figure 2A) [3], [4]. We therefore used MEFs deficient in key components of aforementioned pathways to identify downstream effectors of MAVS that are critical for γHV68 lytic infection. Plaque assays and multi-step growth curves of γHV68 lytic infection showed that deficiency in TRAF6, IKKγ, and IKKβ, but not deficiency in the closely related IKKα, recapitulated phenotypes of MAVS deficiency (Figure 2B and 2C). Notably, TRAF6 is necessary for MAVS to activate IKKβ that requires IKKγ, a scaffold protein for both IKKα and IKKβ [5]. By contrast, deficiency in type I IFN receptor (IFNAR) and double deficiency in IRF3 and IRF7 had no discernable effect on the plaque numbers of γHV68 in MEFs, indicating that the IRF-IFN signaling pathway is not critical for the initiation of γHV68 lytic replication (Figure 2B). Furthermore, the exogenous IKKβ expression reconstituted by lentivirus restored the lytic replication of γHV68 as determined by a plaque assay and multi-step growth curves (Figure 2D, 2E, and 2F). Interestingly, the expression of IKKβ in MAVS−/− did not increase γHV68 lytic replication by a plaque assay (Figure 2E), suggesting that the MAVS-dependent activation of IKKβ, rather than the absolute expression level of IKKβ, is crucial for efficient γHV68 lytic replication. Additionally, exogenous IKKβ did not increase γHV68 plaque numbers in MAVS+/+ MEFs (Figure 2E), implying that endogenous IKKβ is sufficient to support efficient γHV68 lytic replication. Of note, lentivirus infection reduces the difference of γHV68 plaque forming capacity in wild-type MEFs and in MEFs deficient in MAVS and IKKβ (Figure 1F and 2D). Collectively, these data indicate that the MAVS-dependent IKKβ activation is critical for efficient γHV68 lytic replication.
Fig. 2. The MAVS-IKKβ pathway is necessary for efficient γHV68 lytic replication ex vivo. 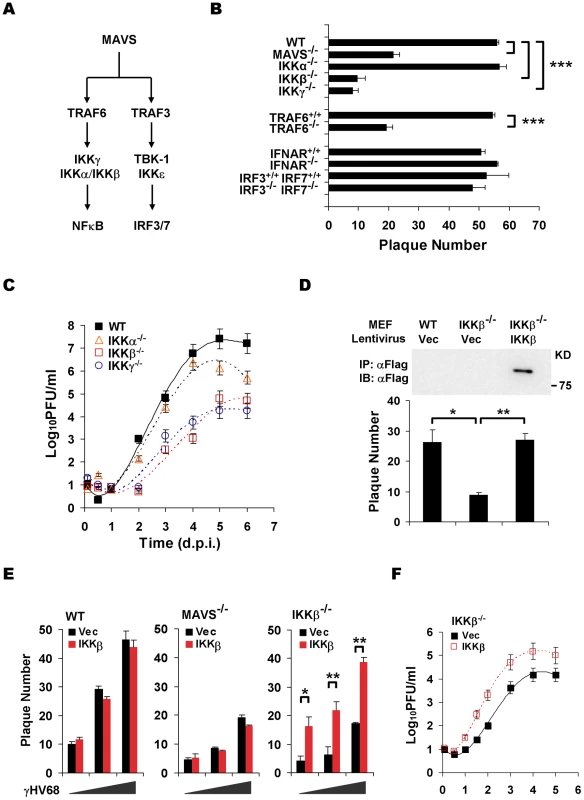
(A) Two known pathways, the IKKα/β/γ-NFκB and TBK-1/IKKε-IRF pathways, downstream of MAVS. (B) The initiation of γHV68 lytic replication in wild-type (WT) MEFs and MAVS−/−, IKKα−/−, IKKβ−/−, IKKγ−/−, TRAF6−/−, IFNAR−/−, and IRF3−/−IRF7−/− (double knockout) MEFs was assessed by a plaque assay. Data represent the mean ± SEM of three independent experiments. (C) Multi-step growth properties of γHV68 (MOI = 0.01) in wild-type MEFs and IKKβ−/−, IKKγ−/−, and IKKα−/− MEFs were examined by plaque assays. Data represents three independent experiments. (D to F) Wild-type, MAVS−/−, and IKKβ−/− MEFs were respectively infected with control lentivirus (Vec) or lentivirus containing the Flag-tagged IKKβ (IKKβ), and selected with puromycin. (D) IKKβ expression was confirmed by immunoprecipitation and immunoblot with anti-Flag antibody (top). γHV68 plaque assays were performed as in (B). (E) Reconstituted MEFs of indicated genotypes were used for γHV68 plaque assays as in (B) with increasing doses of γHV68. Data represent the mean ± SEM of three independent experiments. (F) Reconstituted IKKβ−/− MEFs as indicated were infected with γHV68 K3/GFP (MOI = 0.01), and viral multi-step growth was determined by a plaque assay. Statistical significance (P-value) in (B), (D), and (E) was calculated with two-tailed unpaired Student's t-test: *, P<0.05; **, P<0.02; ***, P<0.005. To assess whether the kinase activity of IKKβ is important for γHV68 lytic infection, we performed plaque assays with or without the specific IKKβ inhibitor, Bay11-7082 (Bay11). This experiment revealed that Bay11 reduced the plaque number of γHV68 in a dose-dependent manner (Figure 3A). Whereas treatment with 1 µM of Bay11 at 0.5 h before infection reduced γHV68 plaque number by 52%, the same treatment at 7 h post-infection (h.p.i.) reduced the plaque number by 29%, emphasizing the important roles of IKKβ during early γHV68 infection (Figure 3A). We further examined IKKβ activity by an in vitro kinase assay with IKKβ precipitated from MAVS+/+ and MAVS−/− MEFs infected with γHV68. The IKKβ kinase activity was transiently and moderately increased in MAVS+/+ MEFs, however, it was drastically diminished in MAVS−/− MEFs after γHV68 infection (Figure 3B). The activation of IKKβ was further supported by the rapid degradation of IκBα concurrent to IKKβ activation by γHV68 infection in MAVS+/+ MEFs, but not in MAVS−/− MEFs (Figure 3C). To test whether UV-inactivated virus is able to trigger IKKβ activation, we examined the levels of IKKβ kinase activity and IκBα in MAVS+/+ MEFs by in vitro kinase and immunoblot assays, respectively. Interestingly, UV-inactivated γHV68 activated IKKβ and reduced IκBα protein levels, although less efficiently than live γHV68 (Figure 3D and 3E). This observation suggests that γHV68 lytic replication is necessary to activate the MAVS-IKKβ pathway. Alternatively, UV treatment may damage or disrupt viral structural components whose integrity is necessary to activate the MAVS-IKKβ pathway.
Fig. 3. γHV68 infection activates IKKβ in a MAVS-dependent manner. 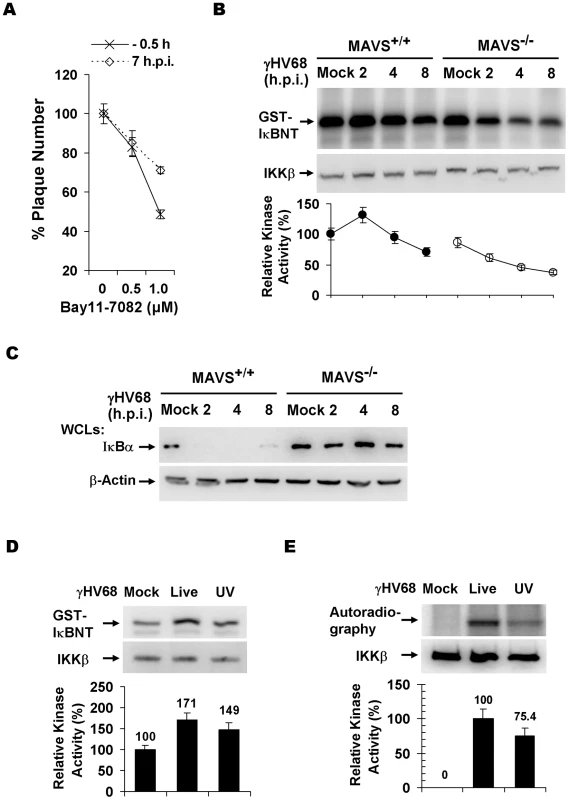
(A) Wild-type MEFs were treated with the IKKβ inhibitor, Bay11-7082, for 30 min at 0.5 h before infection or 7 h post-infection (h.p.i.) with γHV68. Cells were washed with medium and incubated for plaque formation. Plaques formed at 6 d.p.i. were counted. Data represent the mean ± SEM. (B) MEFs were infected with γHV68 (MOI = 10) and whole cell lysates of MEFs at indicated time points after γHV68 infection were precipitated with anti-IKKβ antibody. One half of IKKβ was used for an in vitro kinase assay with GST-IκBNT (amino terminal 50 amino acids of IκBα) (top) or analyzed by immunoblot (middle). Relative intensity of phosphorylated GST-IκBNT was normalized to IKKβ protein (bottom). (C) γHV68 infection was carried out as in (B) and whole cell lysates were analyzed by immunoblot with anti-IκBα (top) and β-actin (bottom). (D and E) Equal amount of live (MOI = 10) or UV-inactivated (UV) γHV68 was used to infect wild-type MEFs. The IKKβ kinase activity was assessed as in (B) and whole cell lysates were analyzed by immunoblot as in (C) for IκBα and β-actin. Graphs at the bottom show normalized IKKβ kinase activity (D) and IκBα protein (E). MAVS activation by RNA viruses is known to increase the expression of pro-inflammatory cytokines and interferons. However, γHV68 appears to be a poor inducer for these antiviral molecules, suggesting that γHV68 evades signaling events downstream of the MAVS adaptor. Indeed, γHV68 infection failed to up-regulate the expression of IFN-β (Figure S5A). In agreement with this observation, γHV68 RTA, similar to KSHV RTA [30], is sufficient to reduce IRF3 expression (Figure S5B). Meanwhile, it was previously shown that γHV68 infection did not significantly activate NFκB during early infection [31], suggesting that γHV68 uncouples NFκB activation from activated IKKβ. Taken together, these results support the conclusion that γHV68 infection selectively activates IKKβ to promote viral lytic replication.
The MAVS-IKKβ Pathway is Implicated in γHV68 Transcriptional Activation
To discern the molecular mechanisms underlying the requirement of the MAVS-IKKβ pathway in γHV68 lytic infection, levels of γHV68 genomic DNA and mRNA were assessed by PCR or reverse transcription followed by real-time PCR analyses, respectively. At a low MOI (0.01), analyses by PCR (Figure 4A) and real-time PCR (Figure 4B) revealed comparable levels of viral genomes in MAVS+/+ and MAVS−/− MEFs early after de novo infection, suggesting comparable viral entry into MAVS+/+ and MAVS−/− MEFs. Interestingly, levels of viral mRNA transcripts representing immediate early (RTA, ORF73, and ORF57) and early (ORF60 and ORF9) gene products in MAVS+/+ MEFs were higher than those in MAVS−/− MEFs as determined by reverse-transcriptase PCR (Figure 4C). Real-time PCR analyses with cDNA showed approximately 4 - to 16-fold higher levels of γHV68 mRNA transcripts in MAVS+/+ MEFs compared to those in MAVS−/− MEFs at 2 and 3 d.p.i. (Figure 4D). It has been shown that TRAF6 is necessary for MAVS to activate IKKβ [5] and exogenous TRAF6 is sufficient to activate IKKβ. To further examine the effects of the MAVS-IKKβ pathway on levels of γHV68 mRNA transcripts, a bacterial artificial chromosome (BAC) containing the γHV68 genome and a plasmid expressing TRAF6 were transfected into 293T cells. The effects of exogenous TRAF6 (that activates IKKβ) on viral transcription were determined by reverse transcription and real-time PCR. At 28 h post-transfection, a time point when immediate early and early genes are transcribed, exogenous TRAF6 efficiently increased the mRNA levels of γHV68 RTA, ORF57, ORF60, and ORF73, without discernable effect on levels of viral genomic DNA (Figure 4E and 4F). These results, obtained under conditions of loss of function (MAVS−/− MEFs) and gain of function (TRAF6 expression), indicate that the activated IKKβ increases the levels of γHV68 mRNA transcripts.
Fig. 4. The MAVS-IKKβ pathway is important for γHV68 mRNA production. 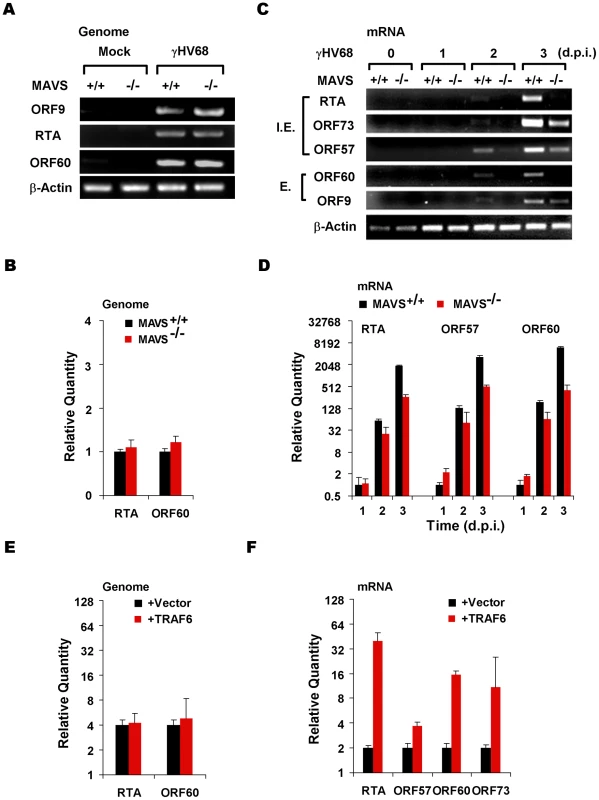
(A and B) MAVS+/+ and MAVS−/− MEFs were infected with γHV68 (MOI = 0.01) and cells were harvested at two hours post-infection. Total DNA was extracted and viral genomes were analyzed by PCR and agarose gel electrophoresis (A) or quantitative real-time PCR (qRT-PCR) (B). (C and D) MEFs were infected with γHV68 as in (A). Total RNA was extracted and levels of γHV68 mRNA transcripts were examined by reverse transcription and PCR (C) or qRT-PCR (D) using primers specific for viral genes as indicated. I.E., immediate early; E, early. (E and F) 293T cells were transfected with the γHV68 BAC and a plasmid containing TRAF6. At 28 h post-transfection, levels of the γHV68 genome were determined by qRT-PCR (E) and levels of various γHV68 gene transcripts as indicated were determined by reverse transcription and qRT-PCR analyses (F). Data represent the mean ± SEM. IKKβ Phosphorylates γHV68 RTA and Promotes γHV68 Transcription
MAVS is an adaptor that activates IKKβ and the MAVS-dependent IKKβ increases γHV68 mRNA levels. We thus postulated that MAVS influences γHV68 transcription via its downstream IKKβ on RTA, because RTA, the master transcription activator, is critical for γHV68 lytic replication. To test this hypothesis, we examined whether IKKβ phosphorylates γHV68 RTA. IKKβ was purified from 293T cells and bacterial GST fusion proteins containing the RTA internal region (RTA-M, aa 335–466) or the RTA C-terminal transactivation domain (RTA-C, aa 457–583) were purified from E.coli (Figure 5A). In the presence of [32P]γATP, IKKβ efficiently transferred the phosphate group to GST-RTA-C. By contrast, GST was not phosphorylated and GST-RTA-M was weakly phosphorylated by IKKβ. Furthermore, the kinase domain deletion variant of IKKβ (IKKβΔKD) failed to phosphorylate GST-RTA-C and GST-RTA-M (Figure 5A), and IKKα had only residual kinase activity toward RTA-C (Figure S6). To confirm the MAVS - and IKKβ-dependent phosphorylation of RTA, RTA phosphorylation in γHV68-infected cells was analyzed by autoradiography and immunoblot. We found that MAVS - and IKKβ deficiency reduced RTA phosphorylation by 50% and 85%, respectively, while reconstituted IKKβ expression restored RTA phosphorylation to that of RTA in MAVS+/+ MEFs (Figure 5B). To assess the roles of phosphorylation of RTA in transcription regulation, luciferase reporter assays were carried out with plasmids containing RTA-responsive promoters of RTA, ORF57, and M3. As shown in Figure 5C, the transcription activity of RTA on all three promoters was significantly increased by exogenous TRAF6 and IKKβ, but not by the kinase dead variant IKKβΔKD, supporting the notion that IKKβ promotes RTA transcription activation via phosphorylation. When expressed to similar levels of IKKβ, IKKβΔKD had no significant effect on RTA transcriptional activation (Figure S7). Given that RTA is a substrate for IKKβ, we sought to examine whether RTA can physically associate with the IKKα/β/γ complex. However, we were unable to detect interaction between RTA and any of the three subunits of IKKα/β/γ by co-immunoprecipitation (data not shown), suggesting that the RTA interaction with the IKKα/β/γ complex is transient or mediated via additional cellular proteins.
Fig. 5. IKKβ phosphorylates and potentiates the transcription activity of γHV68 RTA. 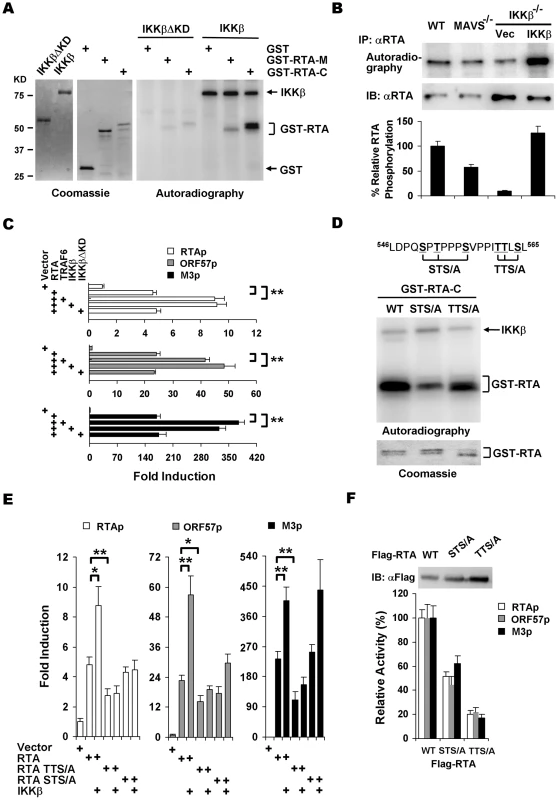
(A) IKKβ or IKKβΔKD purified from 293T cells (left) were incubated with [32P]γATP and bacterial GST fusion proteins containing RTA fragments (middle), and examined by autoradiography (right). (B) MEFs of indicated genotype were infected with γHV68 (MOI = 2) for 4 h, labeled with [32P]-orthophosphoric acid for 8 h. Whole cell lysates were precipitated with anti-RTA antibody and analyzed by autoradiography (top) or immunoblot with anti-RTA antibody (bottom). (C) 293T cells were transfected with reporter plasmids and plasmids containing RTA, TRAF6, IKKβ, and IKKβΔKD. Luciferase activity normalized against β-galactosidase activity was shown. (D) Phosphorylation of GST fusion proteins, containing the C-terminal 127 amino acids of wild-type RTA, STS/A, or TTS/A variants, by IKKβ was analyzed similarly as in (A). (E) 293T transfection and luciferase reporter assays were carried out as in (C). (F) Whole cell lysates of 293T transfected with plasmids containing wild-type RTA, STS/A, or TTS/A variants were analyzed by immunoblot (top) and used to normalize the basal transcriptional activity of wild-type RTA, the STS/A and TTS/A variants (bottom). Data in (C), (E), and (F) represent the mean ± SEM with indicated P values (*, P<0.05; **, P<0.02) of at least three independent experiments. To identify IKKβ phosphorylation sites, series of truncations from the C-terminus of RTA were constructed and purified as GST fusion proteins for in vitro kinase assays with IKKβ. These experiments demonstrated that the IKKβ phosphorylation sites were located within the region containing residues 540 through 567 (Figure S8). Given that IKKβ is a serine/threonine kinase, clusters of various serine/threonine residues were changed to alanines and RTA phosphorylation was assessed similarly. Two clusters of mutations, replacement of S550T552S556 (STS/A) and T561T562S564 (TTS/A) by alanines, reduced the phosphorylation levels of RTA-C by approximately 72% and 45%, respectively (Figure 5D and S8). These results indicate that the STS and TTS sequences represent two major IKKβ phosphorylation sites within the transactivation domain of RTA.
To further examine the roles of IKKβ phosphorylation in regulating RTA transcription activity, reporter assays with plasmids containing wild-type RTA, the STS/A and TTS/A variants were carried out with exogenously expressed IKKβ. The STS/A and TTS/A variants had lower basal activity to activate promoters of RTA, ORF57, and M3. Moreover, exogenous IKKβ failed to further stimulate the transcription activities of the STS/A and TTS/A variants to activate promoters of RTA and ORF57 (Figure 5E). Interestingly, the STS/A variant activated M3 promoter to the level of wild-type RTA with or without IKKβ, indicating that the STS site is dispensable for IKKβ to promote RTA transcriptional activity on the M3 promoter (Figure 5E). It is noteworthy that the STS/A and TTS/A variants were expressed at higher levels than wild-type RTA, the transcription activities of the STS/A and TTS/A variants were approximately 50% and 20% of that of wild-type RTA, respectively, when luciferase activity was normalized against protein levels (Figure 5F). Collectively, these results demonstrated that IKKβ promotes RTA transcriptional activation via phosphorylation of the TTS and STS sites within the transactivation domain.
Impaired Lytic Replication of Recombinant γHV68 Carrying Mutations within the IKKβ Phosphorylation Sites
To further investigate the roles of RTA phosphorylation, we assessed the effects of the STS/A and TTS/A mutations on γHV68 lytic replication. Taking advantage of the γHV68-containing BAC with a transposon insertion that inactivates RTA (ORF50 Null) [32], a recombination-based strategy [33] was employed to generate viruses carrying wild-type RTA (Null Rescued, designated NR), the STS/A allele, or the TTS/A allele (Figure 6A). Whereas we easily obtained recombinant γHV68 containing wild-type RTA (γHV68.NR) or the TTS/A allele (γHV68.TTS/A), the STS/A variant failed to support γHV68 recombination in multiple independent experiments. This observation suggests an essential role for the phosphorylated STS sequence in γHV68 lytic replication. To confirm the integrity of viral genomic DNA, we performed restriction digestion with KpnI and EcoRI, and analyzed with agarose gel electrophoresis. As expected, the removal of the Kanamycin cassette within RTA alleles reduced the 9-kb fragment to 7.5-kb counterpart released by KpnI digestion (Figure 6B), and abolished an EcoRI site within the Kanamycin cassette (Figure 6C). To assess the transcriptional activity of RTA derived from BAC DNA, BAC DNA and the M3p luciferase reporter plasmid were transfected into 293T cells and RTA transcriptional activity was assessed by luciferase reporter assay. The activity of wild-type RTA to activate M3 promoter was approximately 6-fold higher than that of the TTS/A mutant (Figure 6D). Using 293T cells transfected with the γHV68 BAC containing the TTS/A allele and a plasmid expressing TRAF6, we assessed the effects of TRAF6 (that activates IKKβ) on γHV68 gene expression. In contrast to what was observed for the γHV68 BAC containing wild-type RTA (Figure 4F), exogenous TRAF6 had marginal effects on the levels of viral mRNAs transcribed from γHV68 BAC containing the TTS/A allele (Figure 6E). These findings are consistent with the observation that IKKβ failed to further promote the transcription of the TTS/A variant (Figure 5E), supporting the conclusion that the TTS residues constitute an IKKβ phosphorylation sequence by which RTA-dependent transcription is positively regulated.
Fig. 6. Generation and characterization of γHV68 BAC carrying the TTS/A mutation. 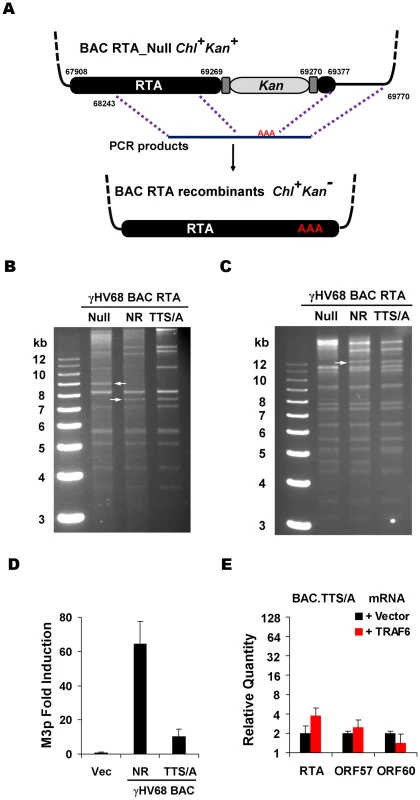
(A) Diagram of the strategy to generate recombinant γHV68. Briefly, wild-type RTA or the STS/A and TTS/A alleles were PCR amplified with overlapping PCR primers. Purified PCR products were transfected into NIH3T3 cells, together with the BAC clone containing a transposon within the transactivation domain of RTA. Recombinant viruses in the supernatant were used to infect NIH3T3 cells. Circular BAC DNA was purified and electroporated into DH10B cells. Chl, chloramphenicol; Kan, kanamycin. (B and C) BACs containing γHV68 genome were analyzed by KpnI (B) or EcoRI (C) digestion, and resolved on 0.8% agarose gels stained with ethidium bromide. The white arrows indicate the specific fragment shift caused by homologous recombination within the RTA locus. NR, RTA-null rescued. (D) 293T cells were transfected with γHV68 M3 reporter plasmid and BAC.NR or BAC.TTS/A. At 28 h post-transfection, luciferase activity and β-galactosidase activity were determined and M3 transcriptional activation by RTA was shown. (E) Transfection of 293T cells with the BAC.TTS/A DNA and a plasmid containing TRAF6, and qRT-PCR were carried out as in Figure 4F. Next, we examined whether recombinant γHV68.TTS/A recapitulates the defects of wild-type γHV68 lytic replication in MEFs deficient in MAVS and IKKβ (plaque assays and multi-step growth curves). To assess the effects of the TTS/A mutation on γHV68 transcription activation, we normalized viral genomes immediately after γHV68 de novo infection of MEFs by qRT-PCR. With equal number of viral genomes, γHV68.NR displayed approximately 32-fold higher of RTA mRNA than recombinant γHV68.TTS/A in MAVS+/+ MEFs at 30 h.p.i. (Figure 7A). This is consistent with the observation that RTA activates its own promoter to facilitate viral lytic replication (Figure 5C and 5E). Furthermore, multi-step growth curves (at an MOI of 0.01) demonstrated that γHV68.TTS/A had delayed replication kinetics and produced >3 orders of magnitude less virion progeny in MAVS+/+ MEFs (Figure 7B). To test whether RTA phosphorylation and the MAVS-IKKβ pathway are functionally redundant, we examined the replication kinetics of recombinant γHV68.NR and γHV68.TTS/A in wild-type, MAVS−/−, and IKKβ−/− MEFs. Consistent with our previous observations (Figure 1C, 2C, and S3B), γHV68.NR showed delayed lytic replication in MAVS−/− and IKKβ−/− MEFs (Figure 7B and 7C). Remarkably, γHV68.TTS/A replicated with similar kinetics in wild-type, MAVS−/−, and IKKβ−/− MEFs, suggesting that the MAVS-IKKβ pathway functions on RTA to promote viral lytic replication (Figure 7B and 7C). However, these replication defects of recombinant γHV68 carrying the TTS/A mutation are much more pronounced than the phenotypes of wild-type γHV68 in MAVS−/− and IKKβ−/− MEFs, implying that additional kinases may influence RTA transcriptional activation via phosphorylation of the TTS site. Taken together, we conclude that the TTS site of RTA is likely phosphorylated by IKKβ and is crucially important for γHV68 lytic replication.
Fig. 7. Impaired lytic replication of recombinant γHV68 carrying the TTS/A mutation. 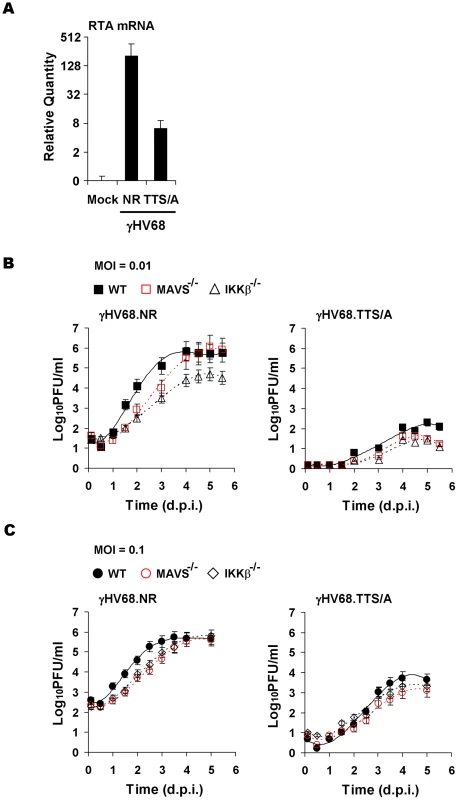
(A) Wild-type MEFs were infected with equal number of genomes, measured by qRT-PCR, of recombinant γHV68.NR (MOI = 0.1) or γHV68.TTS/A. At 30 h post-infection, the RTA mRNA levels were determined by reverse transcription and qRT-PCR. (B) Wild-type, MAVS−/−, and IKKβ−/− MEFs were infected with recombinant γHV68.NR (left) and γHV68.TTS/A (right) (MOI = 0.01) and the multi-step growth curves were determined by a plaque assay. Data represent three independent experiments. (C) The lytic replication of recombinant γHV68.NR and γHV68.TTS/A was examined similarly as in (B) with an MOI of 0.1 (γHV68.NR). Discussion
Here we provide evidence that murine γHV68 hijacks the antiviral MAVS-IKKβ pathway to promote its lytic replication. The MAVS adaptor is important for host defense against invading pathogens, including various DNA and RNA viruses. For example, mice lacking MAVS were severely compromised in innate immune defense against VSV infection, leading to an elevated peak viral load and prolonged acute viral infection [34]. The antiviral effects of MAVS have been observed against the infection of a number of RNA and DNA pathogens [35], [36], [37]. To our surprise, γHV68 viral load in the lungs of MAVS−/− mice was significantly lower than that in the lungs of MAVS+/+ mice at 10 d.p.i. The reduced viral load of γHV68 in MAVS−/− mice is counter-intuitive to the presumed antiviral function of the MAVS adaptor in promoting innate immune responses. Although type I interferons in γHV68-infected mice were undetectable [38], mice deficient in type I IFN receptor had higher viral loads and succumbed to γHV68 infection [39]. We surmise that the effects of MAVS deficiency on γHV68 acute infection is likely under-estimated, providing that MAVS is critical for interferon production in response to viral infection. Thus, the viral load of γHV68 acute infection in MAVS−/− mice likely represents a “neutralized” phenotype, in which reduced γHV68 lytic replication is compensated by the lack of type I interferon inhibition. Moreover, the observation that viral RTA mRNA levels correlates tightly with the MAVS mRNA levels during early γHV68 acute infection suggests that MAVS is necessary for γHV68 lytic replication (Figure S2). Although we have not formally excluded the contribution of host immune responses against γHV68 infection to the reduced viral load at 10 d.p.i. in MAVS−/− mice, our experiments with γHV68 replication ex vivo demonstrated critical roles of the MAVS-IKKβ pathway in facilitating γHV68 lytic infection.
During early stages of viral infection, γHV68 activated IKKβ in a MAVS-dependent manner, a signaling event that is likely triggered by a variety of pathogens. The MAVS-dependent activation was supported by elevated IKKβ kinase activity and accelerated IκBα degradation, signature signaling events downstream of the MAVS adaptor. Although the up-regulation of IKKβ kianse activity appears modest, γHV68 may direct IKKβ kinase activity to efficiently modify cellular and viral components that are critical for γHV68 infection, such as RTA. Consequently, γHV68 can harness activated IKKβ without inducing NFκB activation that may be resulted from massive IKKβ activation. Indeed, it was reported that γHV68 infection does not induce NFκB activation during early infection [40], suggesting that modest IKKβ activation is beneficial for γHV68 infection and that γHV68 may uncouple NFκB activation from IKKβ activation. Interestingly, γHV68 appears to block the interferon limb of the MAVS-dependent innate immune pathway. In fact, we found that γHV68 infection failed to induce the expression of IFN-β (Figure S5A). Consistent with this observation, γHV68 RTA, similar to KSHV RTA [30], is sufficient to reduce IRF3 protein (Figure S5B), potentially abrogating the production of interferons that otherwise would potently thwart γHV68 replication. Moreover, ORF36 was reported to deregulate the phosphorylated form of IRF3 and inhibit interferon production [41]. These observations suggest that γHV68 selectively activates the MAVS-IKKβ pathway to promote viral lytic replication.
Within this report, we have identified one requisite role of the MAVS-IKKβ pathway in γHV68 lytic replication with MEFs deficient in key components of this pathway. Phenotypically, γHV68 displayed similar replication defects in MEFs deficient in MAVS, IKKβ, and IKKγ, although the replication defects in IKKβ−/− and IKKγ−/− MEFs were more pronounced than those in MAVS−/− MEFs (Figure 1C, 2B, and 2C). This result supports the corollary that IKKβ, with the scaffold protein IKKγ, functions downstream of MAVS and likely integrates additional signaling emanating from other innate immune pathways including Toll-like receptors. It is worthy to point out that our result does not exclude the antiviral activity of the IRF-IFN pathway in γHV68 lytic replication, although deficiency of IRF3 and IRF7 or IFNAR did not appear to impact the initiation of γHV68 lytic infection as assessed by plaque assays (Figure 2B). It is possible that the IRF-IFN pathway may inhibit molecular events other than the initiation of lytic replication and reduce viral yield during γHV68 infection. Mechanistically, we identified γHV68 RTA, the master viral replication transactivator, as one of the IKKβ kinase substrates. Phosphorylation of RTA by IKKβ increases RTA transcriptional activity and consequently viral mRNA production. Indeed, γHV68 had lower levels of various mRNA transcripts that correlated with reduced lytic replication in MAVS−/− MEFs (Figure 1 and 4). Conversely, exogenous TRAF6 potentiated RTA transcriptional activity and substantially increased the levels of viral mRNA transcripts (Figure 4F and 5C). Additionally, exogenously reconstituted expression of MAVS and IKKβ restored RTA phosphorylation (Figure 5B) and restored γHV68 lytic replication (Figure 1 and 2). Moreover, lytic replication of recombinant γHV68 viruses carrying mutations within the IKKβ phosphorylation sites was greatly impaired, displaying phenotypes that are more pronounced than those of wild-type γHV68 in MEFs deficient in components of the MAVS-IKKβ pathway. Conceivably, other kinases and signaling pathways may converge to modulate RTA transcriptional activation via phosphorylation within these identified IKKβ sites. For example, virus-encoded kinases, such as the functionally conserved ORF36, may amplify the phosphorylation cascade that is initiated by the MAVS-IKKβ pathway [42]. Most importantly, RTA auto-activates its own promoter and increases RTA protein that, in turn, up-regulates the expression of numerous immediate early and early genes during γHV68 infection. Thus, the 50–80% reduction in RTA transcriptional activity of the STS/A and TTS/A variants (Figure 5F) likely translates into, through the aforementioned amplification cascades, the viral yields that are less than 0.1% of the recombinant γHV68.NR (Figure 7B). Finally, it is noteworthy that deficiency in MAVS and IKKβ and mutations within RTA exhibited distinct phenotypes (such as peak viral titers of multi-step growth curves), in addition to the shared reduction of γHV68 lytic replication. These differing effects on γHV68 infection are likely due to their unique hierarchical position within the MAVS-IKKβ-RTA signaling axis. In essence, these experiments identified novel phosphorylation sites within RTA that couples γHV68 lytic replication to the antiviral IKKβ kinase. These findings collectively demonstrate that the MAVS-dependent IKKβ kinase activity is critical for RTA transcriptional activation and γHV68 lytic replication. Interestingly, Gwack et al. reported that phosphorylation of the internal serine/threonine-rich region of KSHV and γHV68 RTA inhibited RTA transcriptional activity and suppressed viral lytic replication [43]. Together with our findings, these results indicate that site-specific phosphorylation determines the transcriptional activity, and likely the promoter-specificity, of gamma-herpesvirus RTA.
Although it is well accepted that the NFκB pathway is crucial for gamma-herpesvirus latent infection [44], the roles of this pathway in gamma-herpesvirus lytic replication appear to be inconsistent. Particularly, Krug et al. reported that the recombinant γHV68 expressing the IκBα super suppressor replicated indistinguishably compared to wild type γHV68 [31]. Thus, the authors concluded that the NFκB pathway is dispensable for γHV68 lytic replication. By contrast, it was shown that RelA, the p65 subunit of an NFκB transcription dimer, inhibits γHV68 lytic replication through suppressing RTA transcription activity in 293T cells [45]. Finally, our current report indicates that the MAVS-IKKβ pathway is necessary for efficient γHV68 lytic replication. However, the seemingly paradox can be explained by the differential effects of three distinct components of the NFκB pathway on γHV68 lytic replication. Although the IκBα super suppressor is commonly employed to inhibit the activation of the NFκB transcription factors, it is important to note that no significant NFκB activation was observed during early γHV68 infection (within the first 6 hours post-infection) [40], temporal phase in which the critical roles of IKKβ was indentified by our genetic and biochemical experiments. Conceivably, the unphosphorylatable IκBα super suppressor may not impact IKKβ kinase activity. By contrast, we have focused on the IKKβ kinase and our study indicated that the ability of IKKβ to promote viral lytic replication largely stems from IKKβ kinase activity to phosphorylate RTA and increase RTA transcriptional activation. Apparently, neither IκBα, nor RelA can do so in replace of IKKβ function. On the other hand, although RelA was shown to suppress γHV68 lytic replication [45], the lack of NFκB activation during early γHV68 infection implies that γHV68 uncouples NFκB activation from IKKβ activation, which are otherwise tightly correlated. As such, γHV68 infection may selectively activate the IKKβ kinase, while sparing the inhibition by preventing NFκB activation. Therefore, a scenario that potentially accommodates all three reports is that nuclear activated RelA is necessary to inhibit γHV68 lytic replication and γHV68 is capable of preventing RelA activation in an IκBα-independent manner. Crucial to this hypothesis is the mechanisms that γHV68 has evolved to thwart NFκB activation and future experiments are necessary to address this possibility.
It was previously reported that γHV68 was impaired for latency establishment and reactivation in MyD88-deficient mice, although the lytic replication of γHV68 appeared to be normal in these mice [26]. Moreover, agonists specific for TLR7/8, which activate downstream signaling events through MyD88, induced KSHV lytic gene expression and reactivated KSHV replication from latently-infected B cells [25]. The specific roles of MAVS in lytic replication and MyD88 in latent infection are consistent with their distinct functions in innate immune responses of epithelial cells and immune cells, respectively. Given that MyD88 also activates the IKKα/β kinase complex, it is possible that IKKβ-dependent activation of RTA may contribute to γHV68 and KSHV latent infection as well. Finally, reduced lytic replication of human KSHV and cytomegalovirus has been observed under experimental conditions in which IKKβ was inhibited by Bay11, implying that human KSHV and cytomegalovirus have evolved similar molecular mechanisms to facilitate lytic replication [46], [47], [48]. Taken together, the mechanism whereby an antiviral innate immune signaling pathway is exploited to promote viral lytic replication may be applied to other herpesviruses and viral reactivation from latency. This study thus has uncovered an intricate interplay between the viral replication transactivator, RTA, and the MAVS-IKKβ pathway. To our best knowledge, this is the first example that illustrates how a virus hijacks an antiviral signaling pathway, downstream of cytosolic sensors, to initiate its lytic replication. Perhaps, co-evolution between the persistent herpesviruses and their hosts has selected viruses that exploited the inevitable innate immune activation by viral infection. Although our current study delineates the key signaling events downstream of MAVS and IKKβ, it remains unknown what viral components and cellular factors activate the MAVS-IKKβ pathway and whether these mechanisms are shared by the oncogenic KSHV and EBV to promote lytic replication or reactivation.
Materials and Methods
Plasmids
For protein expression in mammalian cells, all genes were cloned into pcDNA5/FRT/TO (Invitrogen) unless specified. For protein expression and purification in E.coli, the internal region (RTA-M, aa 335–466) and C-terminal transactivation domain (RTA-C, aa 457–583) of RTA were cloned into pGEX-4T-1 (Promega) with BamHI and XhoI sites.
Cells and Viruses
NIH3T3 cells, HEK293T (293T) cells and mouse embryonic fibroblasts (MEFs) were maintained in DMEM (Mediatech) with 8% newborn calf serum (NCS) or fetal bovine serum (FBS), respectively. MAVS+/+, MAVS−/−, IKKβ−/−, IKKγ−/− and TRAF6−/− MEFs were described previously [4], [34]. IKKα−/− MEFs were kindly provided by Dr. Amyn A. Habib (Neurology, UT Southwestern). IFNAR+/+ and IFNAR−/− MEFs were kindly provided by Dr. Michael Gale (Immunology, University of Washington). IRF3+/+IRF7+/+ and IRF3−/−IRF7−/− MEFs were kindly provided by Dr. Jae Jung (Microbiology, University of Southern California). γHV68 K3/GFP was kindly provided by Dr. Philip Stevenson (Cambridge University, UK). Wild-type γHV68 and γHV68 K3/GFP were amplified in NIH3T3 cells, and VSV-GFP virus was amplified in BHK-21 cells. Viral titer was determined by a plaque assay with NIH3T3 cells.
Mice and Infections
All animal experiments were performed in accordance to NIH guidelines, the Animal Welfare Act, and US federal law. The experimental protocol (entitled: Innate immune pathways in γHV68 infection) were approved by the Institutional Animal Care and Use Committee (IACUC). All animals were housed in a centralized research animal facility that is accredited by the Association of Assessment and Accreditation of Laboratory Animal Care International, and that is fully staffed with trained husbandry, technical, and veterinary personnel.
Wild-type (MAVS+/+), heterozygous (MAVS+/−), and knockout (MAVS−/−) mice were described previously [34]. Gender-matched, 6 - to 8-week old littermate mice were intranasally (i.n.) inoculated with 40 plaque-forming unit (PFU) wild-type γHV68. To assess MAVS expression in the lung and spleen, BL/6 mice were intranasally infected with 1×105 PFU γHV68. The lungs and spleens were harvested and homogenized in DMEM.
Plaque Assay
Viral titer of mice tissues or cell lysates was assessed by a plaque assay on NIH3T3 monolayers. After three rounds of freezing and thawing, 10-fold serially-diluted virus supernatants were added onto NIH3T3 cells and incubated for 2 hours at 37°C. Then, DMEM containing 2% NCS and 0.75% methylcellulose (Sigma) was added after removing the supernatant. Plaques were counted at day 6 post-infection. The detection limit for this assay is 5 PFU. To assess the infectivity of γHV68 on various MEFs, a similar plaque assay was carried out with the initial cell density of 5000 cells/cm2. In Bay11-7082 treatment assay, 0.5µM or 1µM Bay11-7082 was added at 0.5 h before infection or 7 h post-infection. Supernatant was removed after 30 min incubation at 37°C, and cells were washed with medium and incubated for plaque formation.
Protein Expression and Purification
Glutathione-S-transferase (GST) and GST fusion proteins containing the internal region and the transactivation domain of RTA were expressed with IPTG induction and purified with glutathione-sepharose as previously described [33]. Eluted proteins were re-suspended in 25% glycerol and stored at −20°C for kinase assays. To purify IKKβ and IKKβΔKD, 293T cells were transfected with pcDNA3 containing Flag-IKKβ and Flag-IKKβΔKD. At 48 h post-transfection, cells were lysed with kinase purification buffer (150 mM NaCl, 20 mM Tris.HCl pH7.4, 10% Glycerol, 0.5% Triton X-100, 0.5 mM DTT) and subject to one-step affinity purification with anti-Flag M2-conjugated agarose (Sigma). Proteins were eluted with 0.2 mg/ml Flag peptide in kinase buffer (50 mM KCl, 2 mM MgCl2, 2 mM MnCl2, 1 mM DTT, 10 mM NaF, 25 mM HEPES, pH7.5) and stored in 25% glycerol at −80°C.
Antibodies
Commercial antibodies used in this study include: anti-Flag (Sigma), anti-GFP (Covance), anti-IKKβ (H4), anti-IκBα (C20) (Santa Cruz Biotech.), anti-actin (Abcam.). To generate antibody to γHV68 RTA, the mixture of GST fusion proteins containing the RTA-M and RTA-C was used to immunize a rabbit and polyclonal antibodies were tested for the specificity with pre-immune serum as control.
In Vitro Kinase Assay
Endogenous IKKβ or exogenously expressed IKKβ and IKKβΔKD were used for in vitro kinase assays. The kinase reaction includes 0.5 µg GST or GST fusion proteins, 100 µCi [32P]γATP, and approximately 250 ng kinase in 20 µl of kinase buffer. Reaction was incubated at room temperature for 25 min and denatured proteins were analyzed by SDS-PAGE and autoradiography.
Reverse Transcription (RT)-PCR and Quantitative Real-Time PCR (qRT-PCR) Analysis
To determine the relative levels of viral transcripts, total RNA was extracted from MEFs or mice tissues using TRIzol reagent (Invitrogen). To remove genomic DNA, total RNA was treated with RNase-free DNase I (New England Biolab) at 37°C for 1 hour. After heat inactivation, total RNA was re-purified with TRIzol reagent. cDNA was prepared with 1.5 µg total RNA and reverse transcriptase (Invitrogen). RNA was then removed by incubation with RNase H (Epicentre). Abundance of viral transcripts was assessed by qRT-PCR. Mouse β-actin was used as an internal control. Primers used in this study were summarized in Table S1.
Limiting-Dilution Ex Vivo Reactivation Analyses
Bulk splenocytes were re-suspended in DMEM, and plated onto primary MEF monolayers in 96-well plates in 2-fold serial dilutions (from 105 to 48 cells/well) as previously described [49]. Twelve wells were plated every dilution. Reactivation percentage was scored for cytopathic effects (CPE) positive wells on day 6. In order to measure preformed infectious virus, disrupted cells were plated onto primary MEF monolayers. This procedure destroys over 99% of the cells, but has minimal effect on preformed infectious virus, thus allowing distinction between reactivation from latency and persistent infection.
Limiting-Dilution Nested PCR (LDPCR) Detection of γHV68 Genome-positive Cells
The frequency of splenocytes harboring wild-type γHV68 genome was assessed by a single-copy-sensitive nested PCR analysis of serial dilutions of splenocytes as previously described [49]. Briefly, mice spleens were homogenized and re-suspended in isotonic buffer and subjected to 3-fold serial dilutions (from 104 to 41 cells/well) in a background of uninfected RAW 264.7 cells, with a total of 104 cells per well. Twelve replicates were plated for each cell dilution. After being plated, cells were subjected to lysis by proteinase K at 56°C for 8 hours. After inactivating the enzyme for 30 minutes at 85°C, samples were subjected to nested PCR using primers specific for γHV68 ORF72. Positive controls of 10, 1, and 0.1 copies of viral DNA and negative controls of uninfected RAW 264.7 cells alone were included on each plate. Reaction products were separated using 2.5% UltraPure agarose (Invitrogen) gels and visualized by ethidium bromide staining.
Statistical Analyses
Reactivation and LDPCR results were analyzed using GraphPad Prism software (GraphPad Software, San Diego, CA). The frequencies of genome-positive cells were statistically analyzed using the paired Student's t-test. The frequencies of viral genome-positive cells were determined from a nonlinear regression analysis of sigmoidal dose-response best-fit curve data. Based on a Poisson distribution, the frequency at which at least one event is present in a given population occurs at the point at which the regression analysis line intersects 63.2%. Pooled data of at least three independent experiments were used to calculate P values with the two-tailed, unpaired Student's t-test.
Luciferase Reporter Assay
293T cells (2×105 cells/well) were seeded in 24-well plates 16 hours before transfection. A total of 377 ng of plasmid DNA per well was co-transfected by the calcium phosphate method (Clontech). The plasmid cocktail includes 75 ng of luciferase plasmid (RTAp_luc, ORF57p_Luc or M3p_luc), 200 ng of pCMV-β-galactosidase plasmid, 2 ng of pcDNA5_RTA and 100 ng of pcDNA5 containing TRAF6, IKKβ or IKKβΔKD. At 21 hours post-transfection, whole cell lysates were used to measure the firefly luciferase activity and β-galactosidase activity.
Generating Recombinant γHV68
The bacterial artificial chromosome (BAC) system was used to generate recombinant γHV68 similarly to what was described previously [33]. Briefly, wild-type RTA or the STS/A and TTS/A alleles were PCR amplified with overlapping PCR primers. Purified PCR products, along with the BAC clone 5.15 [32] containing a transposon within the transactivation domain of RTA (between nucleotide 69269 and 69270, according to accession number U97553), were transfected into NIH3T3 cells with Lipofectamine 2000 (Invitrogen). Virus in the supernatant was further amplified with NIH3T3 cells. To isolate circular BAC DNA, NIH3T3 cells were infected with recombinant γHV68 and DNA was extracted according to Hirt's protocol [50] and electroporated into ElectroMAX DH10B cells (Invitrogen). BAC DNA containing γHV68 genome was digested with EcoRI and KpnI to rule out chromosome rearrangement. Meanwhile, the RTA alleles were amplified by PCR and sequenced to confirm desired mutations. Selected clones were transfected into NIH3T3 cells and recombinant γHV68 was amplified for subsequent experiments.
NCBI Entrez Gene ID List
RIG-I, 230073; MDA-5, 71586; MAVS, 228607; TBK-1, 56480; IKKε, 56489; IRF3, 54131; IRF7, 54123; c-Jun, 16476; ATF-2, 11909; IFNβ, 15977; IFNAR, 15975; TRAF3, 22031; TRAF6, 22034; IKKγ, 16151; IKKα, 12675; IKKβ, 16150; IκBα, 18035; NFκB1, 18033; RelA, 19697; MyD88, 35956; TLR7, 170743; TLR8, 170744.
Supporting Information
Zdroje
1. AkiraS
UematsuS
TakeuchiO
2006 Pathogen recognition and innate immunity. Cell 124 783 801
2. MedzhitovR
2007 Recognition of microorganisms and activation of the immune response. Nature 449 819 826
3. KawaiT
TakahashiK
SatoS
CobanC
KumarH
2005 IPS-1, an adaptor triggering RIG-I - and Mda5-mediated type I interferon induction. Nat Immunol 6 981 988
4. SethRB
SunL
EaCK
ChenZJ
2005 Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-kappaB and IRF 3. Cell 122 669 682
5. XuLG
WangYY
HanKJ
LiLY
ZhaiZ
2005 VISA is an adapter protein required for virus-triggered IFN-beta signaling. Mol Cell 19 727 740
6. MeylanE
CurranJ
HofmannK
MoradpourD
BinderM
2005 Cardif is an adaptor protein in the RIG-I antiviral pathway and is targeted by hepatitis C virus. Nature 437 1167 1172
7. HornungV
EllegastJ
KimS
BrzozkaK
JungA
2006 5′-Triphosphate RNA is the ligand for RIG-I. Science 314 994 997
8. PichlmairA
SchulzO
TanCP
NaslundTI
LiljestromP
2006 RIG-I-mediated antiviral responses to single-stranded RNA bearing 5′-phosphates. Science 314 997 1001
9. MercurioF
ZhuH
MurrayBW
ShevchenkoA
BennettBL
1997 IKK-1 and IKK-2: cytokine-activated IkappaB kinases essential for NF-kappaB activation. Science 278 860 866
10. ChenZJ
ParentL
ManiatisT
1996 Site-specific phosphorylation of IkappaBalpha by a novel ubiquitination-dependent protein kinase activity. Cell 84 853 862
11. FitzgeraldKA
McWhirterSM
FaiaKL
RoweDC
LatzE
2003 IKKepsilon and TBK1 are essential components of the IRF3 signaling pathway. Nat Immunol 4 491 496
12. SharmaS
tenOeverBR
GrandvauxN
ZhouGP
LinR
2003 Triggering the interferon antiviral response through an IKK-related pathway. Science 300 1148 1151
13. PanneD
ManiatisT
HarrisonSC
2007 An atomic model of the interferon-beta enhanceosome. Cell 129 1111 1123
14. ThanosD
ManiatisT
1995 Virus induction of human IFN beta gene expression requires the assembly of an enhanceosome. Cell 83 1091 1100
15. JohnsonCL
GaleMJr
2006 CARD games between virus and host get a new player. Trends Immunol 27 1 4
16. LiK
FoyE
FerreonJC
NakamuraM
FerreonAC
2005 Immune evasion by hepatitis C virus NS3/4A protease-mediated cleavage of the Toll-like receptor 3 adaptor protein TRIF. Proc Natl Acad Sci U S A 102 2992 2997
17. LiXD
SunL
SethRB
PinedaG
ChenZJ
2005 Hepatitis C virus protease NS3/4A cleaves mitochondrial antiviral signaling protein off the mitochondria to evade innate immunity. Proc Natl Acad Sci U S A 102 17717 17722
18. YangY
LiangY
QuL
ChenZ
YiM
2007 Disruption of innate immunity due to mitochondrial targeting of a picornaviral protease precursor. Proc Natl Acad Sci U S A 104 7253 7258
19. VirginHWt
LatreilleP
WamsleyP
HallsworthK
WeckKE
1997 Complete sequence and genomic analysis of murine gammaherpesvirus 68. J Virol 71 5894 5904
20. BoshoffC
WeissRA
2001 Epidemiology and pathogenesis of Kaposi's sarcoma-associated herpesvirus. Philos Trans R Soc Lond B Biol Sci 356 517 534
21. CarboneA
CesarmanE
SpinaM
GloghiniA
SchulzTF
2009 HIV-associated lymphomas and gamma-herpesviruses. Blood 113 1213 1224
22. MoorePS
ChangY
2003 Kaposi's sarcoma-associated herpesvirus immunoevasion and tumorigenesis: two sides of the same coin? Annu Rev Microbiol 57 609 639
23. LiangC
LeeJS
JungJU
2008 Immune evasion in Kaposi's sarcoma-associated herpes virus associated oncogenesis. Semin Cancer Biol 18 423 436
24. StevensonPG
EfstathiouS
2005 Immune mechanisms in murine gammaherpesvirus-68 infection. Viral Immunol 18 445 456
25. GregorySM
WestJA
DillonPJ
HilscherC
DittmerDP
2009 Toll-like receptor signaling controls reactivation of KSHV from latency. Proc Natl Acad Sci U S A 106 11725 11730
26. GarganoLM
MoserJM
SpeckSH
2008 Role for MyD88 signaling in murine gammaherpesvirus 68 latency. J Virol 82 3853 3863
27. StaudtMR
DittmerDP
2007 The Rta/Orf50 transactivator proteins of the gamma-herpesviridae. Curr Top Microbiol Immunol 312 71 100
28. HairJR
LyonsPA
SmithKG
EfstathiouS
2007 Control of Rta expression critically determines transcription of viral and cellular genes following gammaherpesvirus infection. J Gen Virol 88 1689 1697
29. WuTT
TongL
RickabaughT
SpeckS
SunR
2001 Function of Rta is essential for lytic replication of murine gammaherpesvirus 68. J Virol 75 9262 9273
30. YuY
WangSE
HaywardGS
2005 The KSHV immediate-early transcription factor RTA encodes ubiquitin E3 ligase activity that targets IRF7 for proteosome-mediated degradation. Immunity 22 59 70
31. KrugLT
MoserJM
DickersonSM
SpeckSH
2007 Inhibition of NF-kappaB activation in vivo impairs establishment of gammaherpesvirus latency. PLoS Pathog 3 e11
32. SongMJ
HwangS
WongWH
WuTT
LeeS
2005 Identification of viral genes essential for replication of murine gamma-herpesvirus 68 using signature-tagged mutagenesis. Proc Natl Acad Sci U S A 102 3805 3810
33. FengP
LiangC
ShinYC
XiaofeiE
ZhangW
2007 A novel inhibitory mechanism of mitochondrion-dependent apoptosis by a herpesviral protein. PLoS Pathog 3 e174
34. SunQ
SunL
LiuHH
ChenX
SethRB
2006 The specific and essential role of MAVS in antiviral innate immune responses. Immunity 24 633 642
35. KawaiT
AkiraS
2006 Innate immune recognition of viral infection. Nat Immunol 7 131 137
36. ScottI
2009 Mitochondrial factors in the regulation of innate immunity. Microbes Infect 11 729 736
37. ArnoultD
CarneiroL
TattoliI
GirardinSE
2009 The role of mitochondria in cellular defense against microbial infection. Semin Immunol 21 223 232
38. Weslow-SchmidtJL
JewellNA
MertzSE
SimasJP
DurbinJE
2007 Type I interferon inhibition and dendritic cell activation during gammaherpesvirus respiratory infection. J Virol 81 9778 9789
39. BartonES
LutzkeML
RochfordR
VirginHWt
2005 Alpha/beta interferons regulate murine gammaherpesvirus latent gene expression and reactivation from latency. J Virol 79 14149 14160
40. KrugLT
CollinsCM
GarganoLM
SpeckSH
2009 NF-kappaB p50 plays distinct roles in the establishment and control of murine gammaherpesvirus 68 latency. J Virol 83 4732 4748
41. HwangS
KimKS
FlanoE
WuTT
TongLM
2009 Conserved herpesviral kinase promotes viral persistence by inhibiting the IRF-3-mediated type I interferon response. Cell Host Microbe 5 166 178
42. TarakanovaVL
Leung-PinedaV
HwangS
YangCW
MatatallK
2007 Gamma-herpesvirus kinase actively initiates a DNA damage response by inducing phosphorylation of H2AX to foster viral replication. Cell Host Microbe 1 275 286
43. GwackY
NakamuraH
LeeSH
SouvlisJ
YusteinJT
2003 Poly(ADP-ribose) polymerase 1 and Ste20-like kinase hKFC act as transcriptional repressors for gamma-2 herpesvirus lytic replication. Mol Cell Biol 23 8282 8294
44. HiscottJ
NguyenTL
ArguelloM
NakhaeiP
PazS
2006 Manipulation of the nuclear factor-kappaB pathway and the innate immune response by viruses. Oncogene 25 6844 6867
45. BrownHJ
SongMJ
DengH
WuTT
ChengG
2003 NF-kappaB inhibits gammaherpesvirus lytic replication. J Virol 77 8532 8540
46. GrossmannC
GanemD
2008 Effects of NFkappaB activation on KSHV latency and lytic reactivation are complex and context-dependent. Virology 375 94 102
47. SadagopanS
Sharma-WaliaN
VeettilMV
RaghuH
SivakumarR
2007 Kaposi's sarcoma-associated herpesvirus induces sustained NF-kappaB activation during de novo infection of primary human dermal microvascular endothelial cells that is essential for viral gene expression. J Virol 81 3949 3968
48. CaposioP
MussoT
LuganiniA
InoueH
GariglioM
2007 Targeting the NF-kappaB pathway through pharmacological inhibition of IKK2 prevents human cytomegalovirus replication and virus-induced inflammatory response in infected endothelial cells. Antiviral Res 73 175 184
49. TibbettsSA
LohJ
Van BerkelV
McClellanJS
JacobyMA
2003 Establishment and maintenance of gammaherpesvirus latency are independent of infective dose and route of infection. J Virol 77 7696 7701
50. HirtB
1967 Selective extraction of polyoma DNA from infected mouse cell cultures. J Mol Biol 26 365 369
Štítky
Hygiena a epidemiologie Infekční lékařství Laboratoř
Článek The Role of Coupled Positive Feedback in the Expression of the SPI1 Type Three Secretion System inČlánek A Systems Immunology Approach to Plasmacytoid Dendritic Cell Function in Cytopathic Virus Infections
Článek vyšel v časopisePLOS Pathogens
Nejčtenější tento týden
2010 Číslo 7- Stillova choroba: vzácné a závažné systémové onemocnění
- Diagnostika virových hepatitid v kostce – zorientujte se (nejen) v sérologii
- Perorální antivirotika jako vysoce efektivní nástroj prevence hospitalizací kvůli COVID-19 − otázky a odpovědi pro praxi
- Jak souvisí postcovidový syndrom s poškozením mozku?
- Infekční komplikace virových respiračních infekcí – sekundární bakteriální a aspergilové pneumonie
-
Všechny články tohoto čísla
- The Mouse Resistance Protein Irgm1 (LRG-47): A Regulator or an Effector of Pathogen Defense?
- Leprosy and the Adaptation of Human Toll-Like Receptor 1
- Intergenomic Arms Races: Detection of a Nuclear Rescue Gene of Male-Killing in a Ladybird
- The Role of Chemokines during Viral Infection of the CNS
- Bottlenecks and the Maintenance of Minor Genotypes during the Life Cycle of
- DNA Damage Triggers Genetic Exchange in
- The Role of Coupled Positive Feedback in the Expression of the SPI1 Type Three Secretion System in
- Uropathogenic Modulates Immune Responses and Its Curli Fimbriae Interact with the Antimicrobial Peptide LL-37
- Biogenesis of the Inner Membrane Complex Is Dependent on Vesicular Transport by the Alveolate Specific GTPase Rab11B
- A Spatio-Temporal Analysis of Matrix Protein and Nucleocapsid Trafficking during Vesicular Stomatitis Virus Uncoating
- Hepatitis B Virus Polymerase Blocks Pattern Recognition Receptor Signaling via Interaction with DDX3: Implications for Immune Evasion
- Quasispecies Theory and the Behavior of RNA Viruses
- Bid Regulates the Pathogenesis of Neurotropic Reovirus
- Distinct Roles for Dectin-1 and TLR4 in the Pathogenesis of Keratitis
- Unexpected Inheritance: Multiple Integrations of Ancient Bornavirus and Ebolavirus/Marburgvirus Sequences in Vertebrate Genomes
- Balanced Nuclear and Cytoplasmic Activities of EDS1 Are Required for a Complete Plant Innate Immune Response
- Adaptation of Hepatitis C Virus to Mouse CD81 Permits Infection of Mouse Cells in the Absence of Human Entry Factors
- An Outer Membrane Receptor of Involved in Zinc Acquisition with Vaccine Potential
- Inositol Hexakisphosphate-Induced Autoprocessing of Large Bacterial Protein Toxins
- Plus- and Minus-End Directed Microtubule Motors Bind Simultaneously to Herpes Simplex Virus Capsids Using Different Inner Tegument Structures
- Distinct Pathogenesis and Host Responses during Infection of by and
- Can Bacteria Evolve Resistance to Quorum Sensing Disruption?
- RNA Virus Replication Complexes
- PPARγ and LXR Signaling Inhibit Dendritic Cell-Mediated HIV-1 Capture and -Infection
- The Virulence Protein SopD2 Regulates Membrane Dynamics of -Containing Vacuoles
- Genome-Wide Mutagenesis Reveals That ORF7 Is a Novel VZV Skin-Tropic Factor
- Adaptive Evolution of Includes Retroviral Insertion and Positive Selection at Two Clusters of Residues Flanking the Substrate Groove
- A Systems Immunology Approach to Plasmacytoid Dendritic Cell Function in Cytopathic Virus Infections
- Transduction of Human T Cells with a Novel T-Cell Receptor Confers Anti-HCV Reactivity
- Identification of GBV-D, a Novel GB-like Flavivirus from Old World Frugivorous Bats () in Bangladesh
- Virus-Infection or 5′ppp-RNA Activates Antiviral Signal through Redistribution of IPS-1 Mediated by MFN1
- HIV gp41 Engages gC1qR on CD4+ T Cells to Induce the Expression of an NK Ligand through the PIP3/H2O2 Pathway
- Oxidation of Helix-3 Methionines Precedes the Formation of PK Resistant PrP
- Protection from the 2009 H1N1 Pandemic Influenza by an Antibody from Combinatorial Survivor-Based Libraries
- Murine Gamma-Herpesvirus 68 Hijacks MAVS and IKKβ to Initiate Lytic Replication
- Viral Protein Inhibits RISC Activity by Argonaute Binding through Conserved WG/GW Motifs
- TOPO3α Influences Antigenic Variation by Monitoring Expression-Site-Associated Switching in
- Functional Genetic Diversity among Complex Clinical Isolates: Delineation of Conserved Core and Lineage-Specific Transcriptomes during Intracellular Survival
- The Meningococcal Vaccine Candidate Neisserial Surface Protein A (NspA) Binds to Factor H and Enhances Meningococcal Resistance to Complement
- Network Modeling Reveals Prevalent Negative Regulatory Relationships between Signaling Sectors in Arabidopsis Immune Signaling
- Endothelial Galectin-1 Binds to Specific Glycans on Nipah Virus Fusion Protein and Inhibits Maturation, Mobility, and Function to Block Syncytia Formation
- Extreme CD8 T Cell Requirements for Anti-Malarial Liver-Stage Immunity following Immunization with Radiation Attenuated Sporozoites
- Integration Preferences of Wildtype AAV-2 for Consensus Rep-Binding Sites at Numerous Loci in the Human Genome
- Epigenetic Analysis of KSHV Latent and Lytic Genomes
- Vaccinia Virus–Encoded Ribonucleotide Reductase Subunits Are Differentially Required for Replication and Pathogenesis
- PLOS Pathogens
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- RNA Virus Replication Complexes
- Virus-Infection or 5′ppp-RNA Activates Antiviral Signal through Redistribution of IPS-1 Mediated by MFN1
- Functional Genetic Diversity among Complex Clinical Isolates: Delineation of Conserved Core and Lineage-Specific Transcriptomes during Intracellular Survival
- Extreme CD8 T Cell Requirements for Anti-Malarial Liver-Stage Immunity following Immunization with Radiation Attenuated Sporozoites
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



