-
Články
- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Production of an Attenuated Phenol-Soluble Modulin Variant Unique to the MRSA Clonal Complex 30 Increases Severity of Bloodstream Infection
Methicillin-resistant Staphylococcus aureus (MRSA) is a major cause of morbidity and mortality and a great concern for public health. The CC30 MRSA lineage is especially notorious for causing bloodstream infections with complications such as seeding into organs. In our study, we show that this lineage produces an attenuated form of a key S. aureus toxin with decreased pro-inflammatory features. Our results suggest that attenuation of this toxin allows the bacteria to evade recognition and subsequent elimination by host defenses, thereby increasing pathogen success during blood infection.
Published in the journal: . PLoS Pathog 10(8): e32767. doi:10.1371/journal.ppat.1004298
Category: Research Article
doi: https://doi.org/10.1371/journal.ppat.1004298Summary
Methicillin-resistant Staphylococcus aureus (MRSA) is a major cause of morbidity and mortality and a great concern for public health. The CC30 MRSA lineage is especially notorious for causing bloodstream infections with complications such as seeding into organs. In our study, we show that this lineage produces an attenuated form of a key S. aureus toxin with decreased pro-inflammatory features. Our results suggest that attenuation of this toxin allows the bacteria to evade recognition and subsequent elimination by host defenses, thereby increasing pathogen success during blood infection.
Introduction
Staphylococcus aureus is a dangerous human pathogen that is responsible for thousands of deaths annually in the U. S. alone [1]. Virulence of S. aureus is due to a large repertoire of virulence factors, including immune evasion factors and aggressive cytolytic toxins [2]. S. aureus infections become particularly dangerous when they are caused by strains that are resistant to commonly used antibiotics. Methicillin-resistant S. aureus (MRSA) is of especially great concern, as identification of MRSA eliminates the therapeutic use of most beta-lactam antibiotics, which are antibiotics of first choice against pathogenic staphylococci. Many countries report high rates of methicillin resistance among hospital-associated infections caused by S. aureus [3]. In addition, community-associated strains of MRSA (CA-MRSA) have emerged over the last two decades that have the capacity to infect healthy individuals outside of hospital settings [4].
MRSA strains belonging to clonal complex (CC) 30 are a major cause of hospital-associated infections in the U. S., Europe, and elsewhere [5]–[7]. Infections with CC30 MRSA present predominantly as bloodstream infections with complications such as hematogenous seeding [8]. Historical methicillin-susceptible CC30 strains (phage type 80/81) caused serious, in part community-associated infections of the skin and lungs in addition to blood infections. In contrast to contemporary CC30 isolates, many historical phage type 80/81 clones had genes encoding the Panton-Valentine leukocidin (PVL) [7]. Furthermore, contemporary CC30 clones contain mutations in the global virulence regulator Agr (agrC gene, non-synonymous mutation, G55R) and the gene encoding α-toxin (hla, STOP mutation) [9]. The resulting overall lower expression of cytolytic toxins in contemporary compared to historic CC30 clones has been linked to the fact that contemporary CC30 clones predominantly cause hospital-associated infections [9].
PSMs are short, amphipathic, α-helical peptides with a major impact on S. aureus virulence [10], [11]. The PSMα peptides of S. aureus in particular cause lysis of a variety of cell types, including neutrophils (or polymorphonuclear leukocytes, PMNs), monocytes, erythrocytes, and osteoblasts [11]–[13]. The rather low target specificity of PSM-mediated cytolysis is due to the fact that lysis is believed to be receptor-independent [14], [15], which is reflected by the capacity of PSMs to lyse artificial vesicles [14]. In addition, PSMs have pro-inflammatory capacities that are receptor-dependent, leading for example to neutrophil chemotaxis and activation [11], [14]. Similar to other S. aureus toxins, these immune-stimulatory activities are observed at sublytic concentrations [14], [16]. PSMα3 is the by far most pro-inflammatory and cytolytic PSM of S. aureus [11]. Notably, the capacity of PSMα3 to elicit chemotaxis by neutrophils by far exceeds that of any other PSM of S. aureus [11].
Except for a variation in the PSM δ-toxin sequence (serine substitution for glycine at position 10 in some strains), whose effect on peptide function has not yet been analyzed, naturally occurring variants of PSM peptides have not yet been reported; and in general, the consequences of non-synonymous variations in psm genes are not understood. Here, we report an allelic variation in the PSMα3-encoding gene that is characteristic of CC30 strains and leads to significantly lower cytolytic and chemotactic activity, and increased hematogenous seeding in a bacteremia model. For the first time, our study describes an allelic variant of a psm gene that has key biological consequences and whose appearance is strongly correlated with a specific MRSA lineage. Furthermore, our findings reveal a molecular mechanism supporting the notion that MRSA strains such as those of the CC30 lineage evolved to cause specific infections, in which reduction of the expression of immune-stimulatory toxins allows to evade recognition and elimination by host defenses.
Results
CC30 strains contain a unique non-synonymous mutation in the psmα3 gene
When analyzing CC30 strains using an HPLC/MS-based PSM analysis method [17], we realized that CC30 strains lacked the m/z values typical for PSMα3, but instead produced a peptide with a changed mass (Fig. 1A,B). The mass of that peptide was 49 Da higher than that of PSMα3. Sequencing the psmα3 gene in selected CC30 strains and comparing to the published genome of the CC30 strain MRSA252 (EMRSA16) [18], we identified the cause for that difference as a non-synonymous mutation in the psmα3 gene in the codon coding for the C-terminal amino acid position (TAC instead of AAC, leading to an exchange of asparagine with tyrosine). Notably, we found the mass corresponding to the PSMα3N22Y peptide in all CC30 strains that we analyzed, containing contemporary and historic strains (Fig. 1C) [9], but never in any of the large number of S. aureus strains of different genetic backgrounds that we had analyzed over the last ∼5 years. Some CC30 strains did not produce any PSM peptide (9 among the 41 analyzed). Lack of PSM production is commonly found in about a quarter of strains in staphylococcal strain collections and due to non-functionality of the Agr system, which strictly regulates PSM production [19], [20]. Furthermore, a BLASTP search only detected the PSMα3N22Y peptide in the CC30 strain MRSA252, but not in any other sequenced S. aureus genome. Therefore, we conclude that the N22Y variant of PSMα3 is characteristic of the CC30 lineage.
Fig. 1. CC30 strains produce a variant of PSMα3. 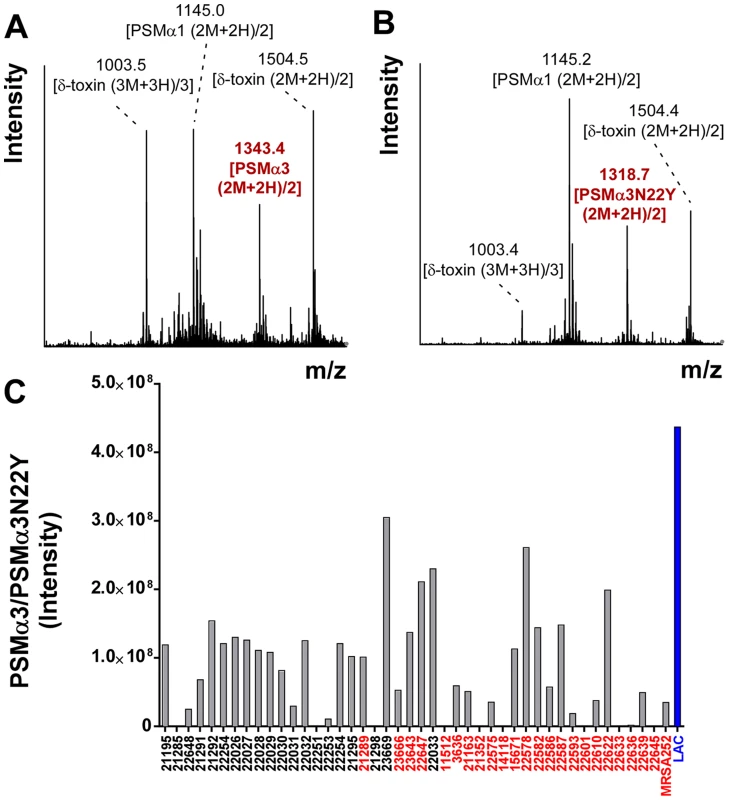
(A,B) Mass spectrogram of the region where PSMα3 elutes during RP-HPLC/MS of the culture filtrate of a non-CC30 strain (USA300) (A) or a CC30 strain (B), respectively. The peak corresponding to PSMα3 is marked in red. Other peaks are from co-eluting PSM peptides. (C) PSMα3N22Y production in a collection of historic and contemporary CC30 strains. Strains shown in red contain a mutation in agrC that is characteristic of contemporary CC30 strains. Production of PSMα3 in strain LAC (USA300) is shown as comparison. Production of α-toxin, but not PSM production and Agr functionality, is significantly different in contemporary and historic CC30 strains
Contemporary CC30 MRSA strains contain two key mutations, one in the hla (α-toxin) and one in the agrC gene [9]. When introduced in a historic CC30 strain (strain 22030), both mutations caused a significant reduction in virulence in a mouse infection model; and the agrC mutation led to reduced production of RNAIII [9], the intracellular effector molecule of the Agr system [21]. However, we did not detect a significant difference in the average PSMα3N22Y production comparing historic and contemporary CC30 strains from that previous study [9] (Fig. 2). Given that PSM production is a direct readout of Agr functionality [19], our present results indicate that the agrC mutation in contemporary CC30 strains does not cause a significant reduction of Agr functionality in average in the contemporary CC30 strain population. We confirmed the results obtained with PSMα3 by analyzing production of δ-toxin, which is the translational product of RNAIII. In both PSMα3 and δ-toxin analyses, contemporary CC30 strains only showed a slightly reduced average production (PSMα3, 0.99×107 versus 1.16×108 intensity units; δ-toxin 3.18×108 versus 3.81×108); both differences were not significant (Fig. 2). In contrast, production of α-toxin as measured by densitometry of Western blot signals was strongly different between the historical CC30 strains and the contemporary strains bearing a stop codon mutation in the α-toxin gene (Fig. 2). As expected, no α-toxin could be detected in culture filtrates of the latter. These findings attribute a greatly more important impact of the hla than the agrC mutation to causing overall lower aggressive virulence of contemporary as compared to historic CC30 strains.
Fig. 2. PSM and α-toxin production in historic and contemporary CC30 MRSA strains. 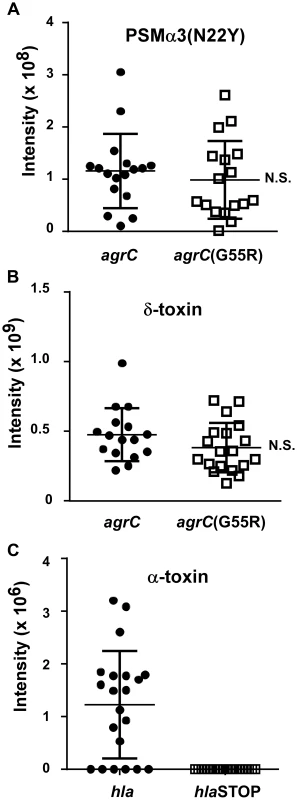
(A,B) Production of PSMα3 and δ-toxin in culture filtrates of a CC30 strain collection (see Fig. 1C). PSMs were measured by HPLC/MS. (C) Production of α-toxin, which was measured by Western blots and densitometry of α-toxin bands in the same culture filtrates. N.S., not significant. For the comparison of α-toxin production, no significance could be computed, as all contemporary strains with hla mutations completely lacked α-toxin production. One strain in the collection contains only the agrC, but not the hla mutation, and was grouped according to presence/absence of those mutations. PSMα3N22Y has decreased chemotactic activity
Neutrophil chemotaxis is one of the most important pro-inflammatory activities exerted by PSMs via their interaction with formyl peptide receptors (FPRs) [10], [14]. PSMs activate FPRs at nanomolar amounts, with activation of FPR2 (also termed FPRL1) being ∼10 times stronger than that of FPR1 and FPR3 [14]. We analyzed the capacities of PSMα3 to cause neutrophil chemotaxis in comparison to PSMα3N22Y. PSMα3 showed a capacity to cause neutrophil chemotaxis that was about twice higher than that caused by PSMα3N22Y at all tested concentrations (Fig. 3A).
Fig. 3. PSMα3N22Y has decreased chemotactic activity toward human neutrophils. 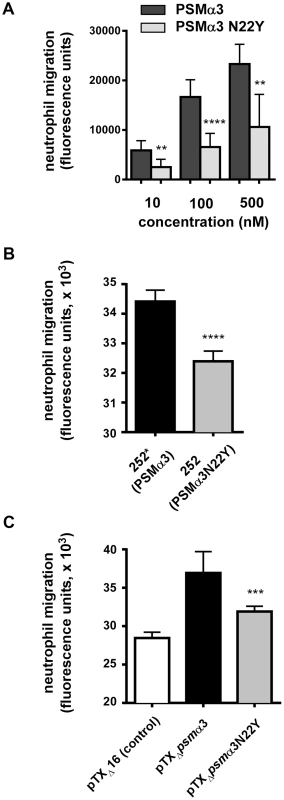
(A) Chemotaxis of human neutrophils (neutrophil migration) was measured using a transwell system with synthetic peptides at the indicated concentrations. Neutrophils were from three donors and the measurements were performed in quadruplicate. **, p<0.01; ****, p<0.0001. (B) Chemotaxis of human neutrophils tested with culture filtrates (diluted 1∶100) of strains MRSA252 (252, expressing PSMα3N22Y) and 252* (MRSA252 altered to express PSMα3); ****, p<0.0001. Data are from two donors with triplicate measurements. (C) Chemotaxis of human neutrophils tested with culture filtrates (diluted 1∶100) of strains expressing PSMα3 or PSMα3N22Y constitutively from plasmids; pTXΔ16 is the empty control plasmid. ***, p<0.001 (versus pTXΔpsmα3). Data are from two donors with triplicate measurements. To test the impact of the psmα3 gene mutation in CC30 strains on chemotactic activity in the natural strain background, we changed the corresponding, last codon in the psmα3 coding sequence in the genome of strain MRSA252 by an allelic replacement-based strategy to the sequence found in other S. aureus strains (from TAC to AAC, tyrosine to asparagine codon). Furthermore, we analyzed the impact of PSMα3 and PSMα3N22Y expressed constitutively from a plasmid in a PSM-negative strain background (strain USA300 with all psm genes deleted) [22]. PSMα3N22Y and PSMα3 expression was verified by HPLC/MS and found to be highly similar in the respective strain pairs (Supplemental Fig. S1). Chemotaxis of human neutrophils was significantly higher with the MRSA252 strain that was altered to express non-CC30 PSMα3 (strain 252*) compared to the wild-type strain MRSA252 expressing PSMα3N22Y (Fig. 3B). Similarly, chemotaxis was significantly higher in the PSM-free strain expressing PSMα3 from a plasmid compared to the isogenic strain expressing PSMα3N22Y (Fig. 3C). These results demonstrate that PSMα3N22Y causes decreased chemotaxis of human neutrophils compared to PSMα3, also when expressed in its natural strain background.
Diminished activation of FPR1 plays contributes significantly to the differences in pro-inflammatory activities of PSMα3N22Y versus PSMα
Next, we analyzed Ca2+ flux as a general measure to test for neutrophil activation, as used in previous studies [14]. HL60 cells stably transfected with different FPR receptor genes [14] were used to determine which FPR receptors are differentially activated by PSMα3 versus PSMα3N22Y (Fig. 4A). Somewhat surprisingly, only FPR1 was activated differentially by the two peptides in this assay, but not FPR2 and FPR3. As expected, both peptides activated FPR2 much stronger than FPR1 and FPR3, but we detected no difference in the FPR2-activating potential between the two peptides. Using a one site-binding model, we obtained Kd values for FPR1 of 615.6±101.2 nM (PSMα3) versus 3039±682 nM (PSMα3N22Y), which represents a significant difference with a factor of ∼5. In contrast, Kd values were not significantly different for FPR2 (PSMα3, 11.90±1.90 nM; PSMα3N22Y, 10.81±2.38 nM) and FPR3 (PSMα3, 1377±285 nM; PSMα3N22Y, 953.0±253.9). However, in human neutrophil chemotaxis experiments using specific inhibitors of FPR1 (chemotaxis inhibitory protein of S. aureus, CHIPS) and FPR2 (FPRL1 inhibitory protein FLIPr) [23], [24], we found that both FPR1 and FPR2 appear to be responsible for the observed differences between PSMα3 and PSMα3N22Y (Fig. 4B). At a peptide concentration of 100 nM, we observed a difference of ∼factor 2 in chemotactic activity comparing the two peptides when FPR1 recognition was inhibited by CHIPS, likely attributable to FPR2-mediated recognition. Owing to the overall ∼10 times lower capacity to stimulate FPR receptors other than FPR2, we observed no chemotaxis at that peptide concentration with addition of the FPR2 blocking agent FLIPr. However, at a 10 times higher peptide concentration of 1 µM, there was a very strong difference in chemotactic potential when FPR2-mediated recognition was blocked (Fig. 4B), which is in agreement with the results of the transfected HL60 experiments and according to our results mainly attributable to differences in activation of FPR1. Together, these results indicate that the differences in the pro-inflammatory potentials of PSMα3 and PSMα3N22Y are due to a considerable extent to differential activation of FPR1, while the overall stronger activation of FPR2 does not differ that much between the two peptides.
Fig. 4. PSMα3N22Y shows decreased activation of FPR1. 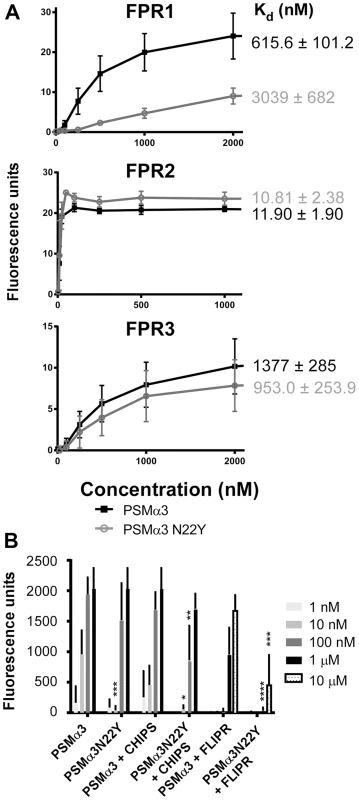
(A) Differential activation of FPRs by PSMα3 versus PSMα3N22Y was measured using synthetic peptides and Ca2+ flux observed in HL60 cells stably transfected with the corresponding FPR genes as readout, using three biological and two technical replicates. The calculated Kd values are shown on the right. The obtained curves were significantly different for PSMα3 versus PSMα3N22Y only in the case of FPR1. (B) Ca2+ flux in human neutrophils upon addition of PSMα3 or PSMα3N22Y and inhibition by FLIPr (FPR2 inhibitor) or CHIPS (FPR1 inhibitor). Neutrophils were from three donors and the measurements were performed in quadruplicate. *, p<0.05; **, p<0.01; ***, p<0.001; ****, p<0.0001 (comparing values obtained with PSMα3 versus corresponding values obtained with PSMα3N22Y). PSMα3N22Y has reduced cytolytic activity toward human neutrophils and erythrocytes compared to PSMα3
PSMα peptides are strongly cytolytic to a variety of cell types, which is believed to be a major determinant of their impact on disease progression observed in several manifestations of S. aureus disease, in particular infections of the skin [10]. PSM-mediated cytolysis is assumed to occur independently of receptor interaction by promoting disintegration of eukaryotic plasma membranes [14], [15], [25].
To compare the cytolytic capacities of PSMα3 and PSMα3N22Y, we first analyzed the degree to which synthetic peptides lysed human erythrocytes and neutrophils. PSMα3 showed significantly higher cytolytic capacities toward human neutrophils and erythrocytes compared to PSMα3N22Y (Fig. 5A,B). Then, we analyzed the impact of PSMα3 and PSMα3N22Y on cytolysis when expressed from plasmids and the genome (see above). In accordance with the results achieved with the synthetic peptides, culture filtrates of the PSMα3-expressing strain had significantly higher capacities to lyse human erythrocytes and neutrophils (Fig. 5C–F). These findings show that the CC30 psmα3 gene mutation leads to significantly decreased cytolytic activity toward human neutrophils and erythrocytes, including in its original strain background.
Fig. 5. PSMα3N22Y has diminished cytolytic activities. 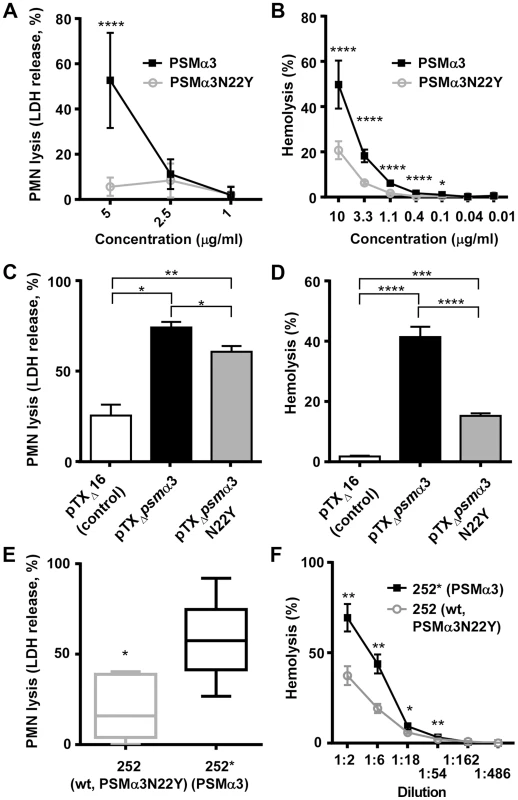
(A) PMN lysis of synthetic PSMα3N22Y versus PSMα3. Data are from 2 donors, measured in triplicate each after incubation at 37°C for 1 h. At concentrations of 10 µg/ml or higher, both peptides completely lysed the neutrophils. (B) Hemolysis of synthetic PSMα3N22Y versus PSMα3. Data are from 2 donors, measured in triplicate, after incubation at 37°C for 1 h. (C, D) PMN lysis and hemolysis, respectively, of culture filtrates from PSMα3N22Y- versus PSMα3-producing strains when expressed on plasmids. pTXΔ16 is the empty control vector. The background strain in all constructs is a PSM-free USA300 strain (LACΔpsmαΔpsmβΔhld). Culture filtrates were diluted 1∶20 for the PMN lysis and 1∶2 for the hemolysis measurements. PMN lysis results are from two and hemolysis data from one donor, with three replicates measured each. (E) PMN lysis of culture filtrates from MRSA252 (252) versus 252*, in which the psmα3 gene (expressing PSMα3N22Y) on the genome was changed to express PSMα3 as present in all non-CC30 strains. The graph shows the results from six different donors. Culture filtrates were diluted 1∶10. *, p<0.05 (paired t-test). (F) Hemolysis of culture filtrates from the same strains as in (E) at different dilutions. (A–F), *, p<0.05; **, p<0.01; ***, p<0.001; ****, p<0.0001. The psmα3 non-synonymous mutation in CC30 isolates leads to increased virulence during bacteremia
The CC30 MRSA lineage is a leading cause of sepsis and subsequent hematogenous complications in hospitals. Therefore, we used a mouse bacteremia (renal abscess) model with assessment of seeding into and abscess formation in the kidneys to evaluate the impact of the psmα3 mutation on CC30 pathogenic success. Both the contemporary CC30 MRSA strain 252 and the historic strain 22030 showed significantly increased seeding into kidneys and kidney abscess formation compared to the respective isogenic mutants in which the psmα3 gene was altered to the non-CC30 form (Fig. 6A, B). Considerable bacterial numbers (>1000/kidney) were found in 70% of kidneys in mice infected with MRSA252, while never in kidneys infected with the isogenic mutant expressing the non-CC30 form of PSMα3 (Fig. 6A). Similar results were found for the 22030 strain pair (65% versus 20%) (Fig. 6B). Histological analyses of several kidneys from mice infected with MRSA252 showed clear signs of infection: they had developed tubulointerstitial nephritis that was associated with colonies of cocci (Fig. 6 C, D). This was not seen in the kidneys of mice infected with the isogenic strain expressing the non-CC30 form of PSMα3 (Fig. 6E). These results indicate that the specific features of the CC30 mutant of PSMα3 (PSMα3N22Y) result in an increased capacity to cause sepsis and subsequent complications. This effect appeared to be stronger in the contemporary clone MRSA252 than in the historic strain 22030, possibly because the latter strongly expresses a series of other pro-inflammatory toxins that to a certain extent overshadow the effect of the PSMα3 mutation.
Fig. 6. The PSMα3 variation significantly impacts virulence of CC30 MRSA in experimental blood infection. 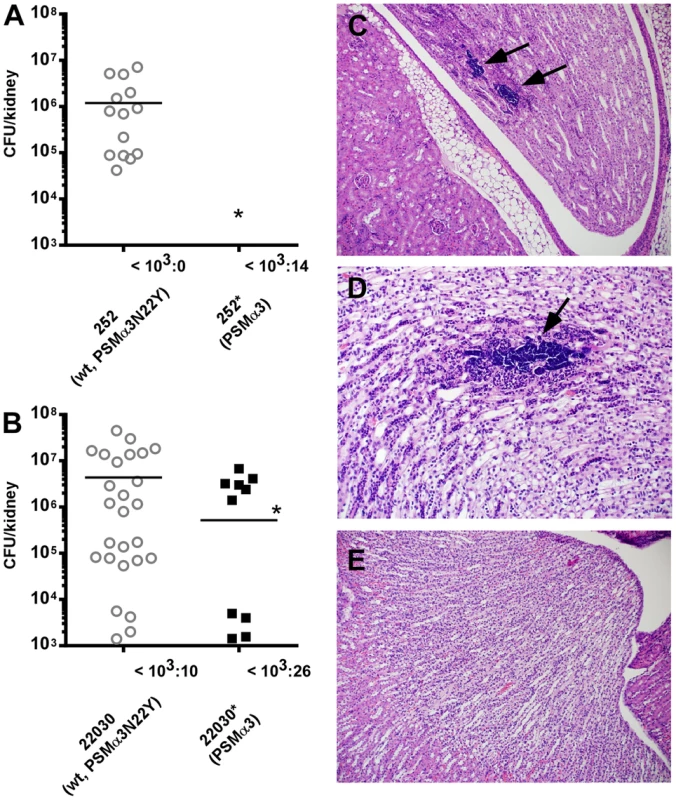
(A) The MRSA252 strain (252, expressing PSMα3N22Y) was compared to a strain (252*) in which the psmα3 gene was changed to express non-CC30 PSMα3 in a mouse model of blood infection. Hematogenous seeding into the kidneys was measured 4 days after infecting mice via the tail vein with 1×107 CFU. Results from kidneys with considerable bacterial numbers (>103 CFU) are shown in the graph; the number of kidneys with lower or no bacteria is shown at the bottom. Analysis of significance was performed with the numbers obtained from all kidneys; *, p<0.05. Number of mice used per strain: 10. Two counts per analyzed kidney were performed. (B) Experiment as in (A) with strain 22030 expressing PSMα3N22Y and the isogenic 22030* strain expressing PSMα3. Number of mice used per strain: 20; two counts per analyzed kidney are included; inoculum: 1×106. *, p<0.05. (C, D) Tubulointerstitial nephritis with large colonies of bacteria (arrows) in kidneys infected with strain 252, magnification 100× (C) and 200× (D). (E) Absence of signs of infection in kidneys infected with strain 252* (magnification 100×). PSMs of the α-type are known to have a strong impact on acute skin infections [11], [26]. This is believed to be due primarily to their cytolytic activities, which they exert for example on neutrophils after phagocytosis [27]. We found that the CC30 form of PSMα3 (PSMα3N22Y) in strains MRSA252 and 22030 did not cause increased virulence in skin infections as measured by abscess sizes. Rather, abscess sizes were in general slightly larger in the MRSA252 strain expressing non-CC30 PSMα3 compared to the MRSA252 wild-type strain, but differences were not significant. No differences were detectable with the 22030 strain pair (Supplemental Fig. S2). This indicates that in contrast to bacteremia, the altered features of the CC30-type PSMα3 (PSMα3N22Y) do not significantly impact the outcome of skin infections.
Discussion
In the present study, we report an allelic variant in a psm gene that is characteristic for a specific S. aureus lineage, namely CC30, and shows reduced immune-stimulatory and cytolytic activities. Our results indicate that the increased capacity of CC30 strains to cause hematogenous complications is at least in part due to the production of this attenuated form of PSMα3. Furthermore, the strongly different impact of that form of PSMα3 on bacteremia versus skin infection underscores that the two main features of PSMs, i.e. cytolytic and pro-inflammatory capacity, may have a strongly different influence on the development of distinct disease types.
Given that a reduction in cytolytic activity can hardly explain increased virulence and hematogenous seeding, the differences that we detected in the bacteremia model are very likely due to the attenuated pro-inflammatory features of the attenuated form of PSMα3 in CC30 strains. These findings are in accordance with the notion that the innate immune system makes use of S. aureus toxins for pathogen recognition and evading that recognition is of benefit for bacterial survival in specific types of disease [28]. This notion is founded on several previous observations: first, S. aureus produces a series of molecules such as CHIPS or FLIPr that block toxin recognition by receptors on innate host defense cells, including the FPRs that recognize PSMs [14], [23], [24]; second, it was found that the pro-inflammatory properties of PVL [16], [29] enhanced clearing of pneumonia and anti-PVL antibodies led to increased severity of infection [30]–[32]. Finally, persistent bacteremia has been reported to be associated with Agr dysfunction [33]. While the latter has been explained by an increased fitness cost associated with the production of RNAIII [34], this correlation may also be explained by the lower expression of pro-inflammatory molecules in agr mutants (such as PSMs and other toxins). All these observations indicate that toxin production is a double-edged sword for the bacteria: toxins not only serve to eliminate immune cells but also trigger the launch of host defenses, representing a sort of pathogen-associated molecular pattern. Importantly, our study is the first to provide an example in which a staphylococcal toxin variant appears to have evolved to circumvent pathogen recognition.
The psmα3 allelic variant is present in all CC30 strains, including historic and contemporary strains. Our findings therefore do not answer the much-debated question which specific mutations are linked to the fact that the latter are predominantly restricted to the hospital setting [7], [9]. However, our findings suggest that the hla and agrC mutations in contemporary CC30 clones may add to immune evasion by lowering pathogen recognition in a similar fashion as the PSMα3 attenuation, because they also result in an attenuation of the expression of pro-inflammatory toxins. These mutations, which are believed to make contemporary strains of CC30 better adapted to long-term colonization and persistence in the human host, may thus not decrease virulence in a general fashion as has been suggested [9]. They may rather shift the disease spectrum to types of infection in which the production of aggressive toxins that alert the immune system is counterproductive for the establishment and progression of infection. According to our results, the hla mutation plays a much greater role in that adaptation of contemporary CC30 clones to hospital-associated infections than the agrC mutation, Altogether, these findings attribute a key role to point mutations in major S. aureus toxins for virulence and correlation with specific disease types.
Results that we previously obtained using an alanine exchange peptide library screen of PSMα3 indicated that the C-terminal amino acids are critical for interaction with FPR2 [35]. In general, the difference in receptor interaction between PSMα3 and PSMα3N22Y as determined in the present study supports this concept. However, exchange of the C-terminal asparagine with tyrosine (CC30 PSMα3N22Y) leads to a different change in receptor interaction as compared to an exchange with alanine (PSMα3N22A), inasmuch as the former more strongly affects interaction with FPR1. This indicates that the molecular details of PSM – FPR interaction differ between the different subtypes of FPRs.
In conclusion, we here report an allelic variation of a key member of the PSM toxin family that significantly impacts in-vitro virulence phenotypes and in-vivo virulence of strains of the CC30 lineage. Our findings lend support to the general idea that mutations leading to lower pro-inflammatory potential are linked to the involvement in specific types of disease.
Materials and Methods
Ethics statement
The used animal protocol (LHBP1E) was reviewed and approved by the Animal Use Committee at NIAID, NIH, according to the animal welfare act of the United States (7 U.S.C. 2131 et. seq.). Human neutrophils were isolated from venous blood of healthy volunteers in accordance with protocols approved by the Institutional Review Board for Human Subjects, NIAID, and the University of Tübingen, Germany. Informed written consent was obtained from all volunteers.
Bacterial strains and culture conditions
The bacterial strains used in this study are contemporary and historic CC30 strains as published in a previous study [9]. MRSA252 is a widespread contemporary CC30 isolate and a frequent cause of hospital-associated infections [18]. LAC is a CA-MRSA strain of the pulsed-field type USA300. Strains were grown in tryptic soy broth (TSB) with addition of tetracycline at 12.5 µg/ml when appropriate. Culture filtrates for all experiments were obtained from 50-ml cultures grown for 18 h in 100-ml flasks with shaking at 180 rpm. They were filtered with PES filters (0.2 µm pore size, Millipore) and used fresh or stored at −20°C for further use. Only for the analysis of the CC30 strain collection (Fig. 1C), strains were grown as 1-ml cultures in 5-ml tubes, with all other conditions being the same.
Peptides
Synthetic PSM peptides were obtained from commercial vendors at a purity of >95%. They all carry the N-terminal N-formyl methionine as common for PSMs, which are secreted into the medium without a signal peptide [36]. Peptide stock solutions were made in dimethyl sulfoxide at 10 mg/ml, which were diluted in RPMI 1640 (Gibco) for PMN or Dulbecco's phosphate-buffered saline (DPBS) (Sigma) for hemolysis assays.
Allelic replacement and plasmid construction
Allelic replacement was performed as previously described [11] using the plasmid pKOR1-based method [37] to change the psmα3 gene in MRSA252 and 22030 (encoding PSMα3N22Y) to non-CC30 psmα3. Template DNA was from strain LAC (USA300). Oligonucleotides used are shown in Table 1. Plasmids for PSM expression are based on plasmid pTXΔ, which is a derivative of the xylose-inducible plasmid pTX15 [38], in which inducible was changed to constitutive expression by deletion of the xylR repressor gene [11]. Oligonucleotides for construction of the pTXΔpsmα3N22Y plasmid were used as reported [11] to amplify the psmα3 gene from genomic DNA of strain MRSA252. Fidelity of the replacement was ascertained by DNA sequencing of the psmα operon and HPLC/MS of PSMs showing the exchanged mass of PSMα3 versus PSMα3N22Y. Additionally, production of PSMs was verified in the culture filtrates and cultures of strains used for all experiments prior to performing the experiment, to ensure that PSMα3 and PSMα3N22Y production was at the same level and no agr or other mutation impacting PSM levels had occurred (see Supplemental Fig. S1).
Tab. 1. Oligonucleotides used in this study. 
Measurement of PSM production by high-pressure liquid chromatography/mass spectrometry (HPLC/MS)
PSM concentrations in culture filtrates grown for 16 h in TSB were measured using HPLC/MS as described [17].
Measurement of α-toxin by western blot
Staphylococcal strains were grown in TSB at 37°C overnight. 10 µl of these cultures were inoculated into 1 ml of TSB and grown for 8 h. Culture supernatants were loaded onto 15% SDS-PAGE gels and run at 150 V for 1 h. Proteins in the gels were blotted on nitrocellulose membranes using an iBlot Western blotting system (Life Technologies, Grand Island, NY). Blotted membranes were incubated with Odyssey blocking buffer (LI-COR, Lincoln, NE) for 1 h at room temperature. Anti-staphylococcal α-toxin rabbit serum (1∶2,000, Sigma-Aldrich, St. Louis, MO) was added to the blocking buffer and incubated for another hour at room temperature. Membranes were washed five times with washing buffer (Tris-buffered saline containing 0.1% Tween-20, pH 7.4) and incubated with 1∶10,000 diluted Cy5-labeled goat anti-rabbit IgG (Life Technologies, Grand Island, NY) in Odyssey blocking buffer in dark for 1 h at room temperature. Membranes were washed five times with the washing buffer and scanned with a Typhoon TRIO+ Variable Mode Imager (GE Healthcare, Piscataway, NJ). The amount of α-toxin in the scanned image was quantified using ImageQuant TL software (GE Healthcare, Piscataway, NJ).
Measurement of neutrophil chemotaxis, lysis, and calcium flux
Human neutrophils were isolated from venous blood of healthy donors as described [24], [39]. Neutrophil chemotaxis and Ca2+ flux were determined as previously described [11], [24]. Briefly, neutrophils were subjected to a brief hypotonic shock with pyrogen-free water, washed, and suspended at 5×106 cells/ml. Chemotaxis of neutrophils was determined using a transwell system (Costar) with analysis of neutrophil migration using fluorescence labeling. Calcium fluxes were monitored as a surrogate marker for chemotaxis since it can be measured more robustly and accurately than chemotaxis. For measurement of Ca2+ fluxes, 5×106 neutrophils/ml were labeled with a fluorescent dye and analyzed with a FACScalibur (Becton Dickinson) as described [24]. For measuring the influence of CHIPS or FLIPr, 1×106 cells/ml were pre-incubated with CHIPS or FLIPr at final concentrations of 1.4 µg/ml or 0.5 µg/ml, respectively, for 20 min at room temperature under agitation. The CHIPS and FLIPr proteins (a kind gift from K. van Kessel) were prepared as described [23], [24]. Measurements of 2,000 events were performed and Ca2+ flux was expressed as relative fluorescence corrected for buffer controls. For the measurement of neutrophil lysis, synthetic PSMs or (diluted) culture filtrates were added to wells of a 96-well tissue culture plate containing 106 PMNs and plates were incubated at 37°C. At the desired times, PMN lysis was determined by release of lactate dehydrogenase (LDH) (Cytotoxicity Detection Kit, Roche Applied Sciences).
Hemolysis assays
Hemolysis assays were performed according to Wang et al. [11] Briefly, whole blood from humans was washed twice with ice-cold DPBS. The final percentage of blood was 2% (v/v) in ice-cold DPBS. Equal volumes of diluted synthetic PSMs or dilutions of culture filtrates were incubated with the 2% erythrocyte suspension in a total volume of 200 µl in 96 round-bottomed plates. After incubation at 37°C for 1 h, the plates were centrifuged at 233×g at 4°C for 5 min, supernatants were collected, and the optical density was measured at 540 nm.
Mouse model of skin infection
The mouse skin infection model was performed as described [11]. Briefly, six to eight week-old female Crl∶SKH1-hrBR hairless mice (Charles River Laboratories) were injected subcutaneously with ∼4×107 CFU of S. aureus strains 252 or 252*, or ∼5×106 CFU of S. aureus strains 22030 or 22030* in 50 µl of PBS in the left flank of the mouse. The length (L) and width (W) of the abscess or lesion caused by the bacterial infection was measured with an electronic caliper daily for 14 d post infection and calculated using the formula L×W. Typically, strain MRSA252 caused closed abscesses and strain 22030 open lesions. All animals were euthanized after completion of the entire procedure. All mouse experiments were performed blinded at the animal care facility of the NIAID, Building 33, in compliance with the guidelines of the NIAID/NIH Institutional Animal Care and Use Committee.
Mouse bacteremia (renal abscess) model
For the renal abscess model, S. aureus strains were inoculated from a pre-culture and grown to mid-exponential growth phase (∼2 h), harvested, washed, and diluted with sterile PBS. Six to eight week-old female CD-1 mice (Charles River Laboratories) were infected with 50 µl of bacteria in PBS via the tail vein. Mice received ∼1×107 CFU of S. aureus strains 252 or 252*, or ∼1×106 CFU of S. aureus strains 22030 or 22030*. Four days post infection terminal cardiac bleeds were performed. There were no bacteria found in the blood. All mice were euthanized by CO2 inhalation and kidneys were collected. One kidney of each mouse was placed into a 2-ml tube containing 1 ml of sterile PBS with 500 mg of 2 mm borosilicate glass beads (Sigma). The kidney was homogenized in a Fast Prep bead beater (Thermo Savant) at 6 m/s for 20 s. The homogenates were diluted in PBS, plated onto TSB plates, and incubated overnight at 37°C for CFU counting. The other kidney was placed in 10% formalin (Sigma) for subsequent histopathological examination.
Statistics
Statistical analysis was performed using Graph Pad Prism version 6.02. For the comparison of two groups, t-tests were used (unpaired unless otherwise noted), for three or more, 1-way or 2-way ANOVA, as appropriate. All error bars depict the standard deviation.
Supporting Information
Zdroje
1. LowyFD (1998) Staphylococcus aureus infections. N Engl J Med 339 : 520–532.
2. FosterTJ (2005) Immune evasion by staphylococci. Nat Rev Microbiol 3 : 948–958.
3. OttoM (2012) MRSA virulence and spread. Cell Microbiol 14 : 1513–1521.
4. OttoM (2013) Community-associated MRSA: what makes them special? Int J Med Microbiol 303 : 324–330.
5. CoxRA, ConquestC, MallaghanC, MarplesRR (1995) A major outbreak of methicillin-resistant Staphylococcus aureus caused by a new phage-type (EMRSA-16). J Hosp Infect 29 : 87–106.
6. JohnsonAP, AuckenHM, CavendishS, GannerM, WaleMC, et al. (2001) Dominance of EMRSA-15 and -16 among MRSA causing nosocomial bacteraemia in the UK: analysis of isolates from the European Antimicrobial Resistance Surveillance System (EARSS). J Antimicrob Chemother 48 : 143–144.
7. McAdamPR, TempletonKE, EdwardsGF, HoldenMT, FeilEJ, et al. (2012) Molecular tracing of the emergence, adaptation, and transmission of hospital-associated methicillin-resistant Staphylococcus aureus. Proc Natl Acad Sci U S A 109 : 9107–9112.
8. FowlerVGJr, NelsonCL, McIntyreLM, KreiswirthBN, MonkA, et al. (2007) Potential associations between hematogenous complications and bacterial genotype in Staphylococcus aureus infection. J Infect Dis 196 : 738–747.
9. DeLeoFR, KennedyAD, ChenL, Bubeck WardenburgJ, KobayashiSD, et al. (2011) Molecular differentiation of historic phage-type 80/81 and contemporary epidemic Staphylococcus aureus. Proc Natl Acad Sci U S A 108 : 18091–18096.
10. PeschelA, OttoM (2013) Phenol-soluble modulins and staphylococcal infection. Nat Rev Microbiol 11 : 667–673.
11. WangR, BraughtonKR, KretschmerD, BachTH, QueckSY, et al. (2007) Identification of novel cytolytic peptides as key virulence determinants for community-associated MRSA. Nat Med 13 : 1510–1514.
12. CheungGY, DuongAC, OttoM (2012) Direct and synergistic hemolysis caused by Staphylococcus phenol-soluble modulins: implications for diagnosis and pathogenesis. Microbes Infect 14 : 380–386.
13. RasigadeJP, Trouillet-AssantS, FerryT, DiepBA, SapinA, et al. (2013) PSMs of hypervirulent Staphylococcus aureus act as intracellular toxins that kill infected osteoblasts. PLoS One 8: e63176.
14. KretschmerD, GleskeAK, RautenbergM, WangR, KoberleM, et al. (2010) Human formyl peptide receptor 2 senses highly pathogenic Staphylococcus aureus. Cell Host Microbe 7 : 463–473.
15. CheungGY, JooHS, ChatterjeeSS, OttoM (2013) Phenol-soluble modulins - critical determinants of staphylococcal virulence. FEMS Microbiol Rev doi: 10.1111/1574-6976.12057. [Epub ahead of print]
16. GravesSF, KobayashiSD, BraughtonKR, WhitneyAR, SturdevantDE, et al. (2012) Sublytic concentrations of Staphylococcus aureus Panton-Valentine leukocidin alter human PMN gene expression and enhance bactericidal capacity. J Leukoc Biol 92 : 361–374.
17. JooHS, OttoM (2014) The isolation and analysis of phenol-soluble modulins of Staphylococcus epidermidis. Methods Mol Biol 1106 : 93–100.
18. HoldenMT, FeilEJ, LindsayJA, PeacockSJ, DayNP, et al. (2004) Complete genomes of two clinical Staphylococcus aureus strains: evidence for the rapid evolution of virulence and drug resistance. Proc Natl Acad Sci U S A 101 : 9786–9791.
19. QueckSY, Jameson-LeeM, VillaruzAE, BachTH, KhanBA, et al. (2008) RNAIII-independent target gene control by the agr quorum-sensing system: insight into the evolution of virulence regulation in Staphylococcus aureus. Mol Cell 32 : 150–158.
20. TraberKE, LeeE, BensonS, CorriganR, CanteraM, et al. (2008) agr function in clinical Staphylococcus aureus isolates. Microbiology 154 : 2265–2274.
21. NovickRP, RossHF, ProjanSJ, KornblumJ, KreiswirthB, et al. (1993) Synthesis of staphylococcal virulence factors is controlled by a regulatory RNA molecule. EMBO J 12 : 3967–3975.
22. JooHS, CheungGY, OttoM (2011) Antimicrobial activity of community-associated methicillin-resistant Staphylococcus aureus is caused by phenol-soluble modulin derivatives. J Biol Chem 286 : 8933–8940.
23. PratC, BestebroerJ, de HaasCJ, van StrijpJA, van KesselKP (2006) A new staphylococcal anti-inflammatory protein that antagonizes the formyl peptide receptor-like 1. J Immunol 177 : 8017–8026.
24. de HaasCJ, VeldkampKE, PeschelA, WeerkampF, Van WamelWJ, et al. (2004) Chemotaxis inhibitory protein of Staphylococcus aureus, a bacterial antiinflammatory agent. J Exp Med 199 : 687–695.
25. DuongAC, CheungGY, OttoM (2012) Interaction of phenol-soluble modulins with phosphatidylcholine vesicles. Pathogens 1 : 3–11.
26. KobayashiSD, MalachowaN, WhitneyAR, BraughtonKR, GardnerDJ, et al. (2011) Comparative analysis of USA300 virulence determinants in a rabbit model of skin and soft tissue infection. J Infect Dis 204 : 937–941.
27. SurewaardBG, de HaasCJ, VervoortF, RigbyKM, DeLeoFR, et al. (2013) Staphylococcal alpha-phenol soluble modulins contribute to neutrophil lysis after phagocytosis. Cell Microbiol 15 : 1427–1437.
28. SpaanAN, SurewaardBG, NijlandR, van StrijpJA (2013) Neutrophils versus Staphylococcus aureus: a biological tug of war. Annu Rev Microbiol 67 : 629–650.
29. HolzingerD, GieldonL, MysoreV, NippeN, TaxmanDJ, et al. (2012) Staphylococcus aureus Panton-Valentine leukocidin induces an inflammatory response in human phagocytes via the NLRP3 inflammasome. J Leukoc Biol 92 : 1069–1081.
30. YoongP, PierGB (2012) Immune-activating properties of Panton-Valentine leukocidin improve the outcome in a model of methicillin-resistant Staphylococcus aureus pneumonia. Infect Immun 80 : 2894–2904.
31. HermosCR, YoongP, PierGB (2010) High levels of antibody to Panton-Valentine leukocidin are not associated with resistance to Staphylococcus aureus-associated skin and soft-tissue infection. Clin Infect Dis 51 : 1138–1146.
32. YoongP, PierGB (2010) Antibody-mediated enhancement of community-acquired methicillin-resistant Staphylococcus aureus infection. Proc Natl Acad Sci U S A 107 : 2241–2246.
33. FowlerVGJr, SakoulasG, McIntyreLM, MekaVG, ArbeitRD, et al. (2004) Persistent bacteremia due to methicillin-resistant Staphylococcus aureus infection is associated with agr dysfunction and low-level in vitro resistance to thrombin-induced platelet microbicidal protein. J Infect Dis 190 : 1140–1149.
34. PaulanderW, Nissen VarmingA, BaekKT, HaaberJ, FreesD, et al. (2013) Antibiotic-mediated selection of quorum-sensing-negative Staphylococcus aureus. mBio 3: e00459-00412.
35. CheungGY, KretschmerD, QueckSY, JooHS, WangR, et al. (2014) Insight into structure-function relationship in phenol-soluble modulins using an alanine screen of the phenol-soluble modulin (PSM) alpha3 peptide. FASEB J 28 : 153–161.
36. ChatterjeeSS, JooHS, DuongAC, DieringerTD, TanVY, et al. (2013) Essential Staphylococcus aureus toxin export system. Nat Med 19 : 364–367.
37. BaeT, SchneewindO (2006) Allelic replacement in Staphylococcus aureus with inducible counter-selection. Plasmid 55 : 58–63.
38. PeschelA, OttenwalderB, GotzF (1996) Inducible production and cellular location of the epidermin biosynthetic enzyme EpiB using an improved staphylococcal expression system. FEMS Microbiol Lett 137 : 279–284.
39. VoyichJM, OttoM, MathemaB, BraughtonKR, WhitneyAR, et al. (2006) Is Panton-Valentine leukocidin the major virulence determinant in community-associated methicillin-resistant Staphylococcus aureus disease? J Infect Dis 194 : 1761–1770.
Štítky
Hygiena a epidemiologie Infekční lékařství Laboratoř
Článek Disruption of Fas-Fas Ligand Signaling, Apoptosis, and Innate Immunity by Bacterial PathogensČlánek A Tick Gut Protein with Fibronectin III Domains Aids Congregation to the Gut during TransmissionČlánek The Vi Capsular Polysaccharide Enables Serovar Typhi to Evade Microbe-Guided Neutrophil ChemotaxisČlánek Structure of CfaA Suggests a New Family of Chaperones Essential for Assembly of Class 5 Fimbriae
Článek vyšel v časopisePLOS Pathogens
Nejčtenější tento týden
2014 Číslo 8- Stillova choroba: vzácné a závažné systémové onemocnění
- Jak souvisí postcovidový syndrom s poškozením mozku?
- Perorální antivirotika jako vysoce efektivní nástroj prevence hospitalizací kvůli COVID-19 − otázky a odpovědi pro praxi
- Diagnostika virových hepatitid v kostce – zorientujte se (nejen) v sérologii
- Infekční komplikace virových respiračních infekcí – sekundární bakteriální a aspergilové pneumonie
-
Všechny články tohoto čísla
- Regulatory RNAs Involved in Bacterial Antibiotic Resistance
- From Dandruff to Deep-Sea Vents: -like Fungi Are Ecologically Hyper-diverse
- Pathogenicity and Epithelial Immunity
- Mother–Infant HIV Transmission: Do Maternal HIV-Specific Antibodies Protect the Infant?
- Hell's BELs: acterial 3 igases That Exploit the Eukaryotic Ubiquitin Machinery
- Disruption of Fas-Fas Ligand Signaling, Apoptosis, and Innate Immunity by Bacterial Pathogens
- Primary Seronegative but Molecularly Evident Hepadnaviral Infection Engages Liver and Induces Hepatocarcinoma in the Woodchuck Model of Hepatitis B
- TLR2 Signaling Decreases Transmission of by Limiting Bacterial Shedding in an Infant Mouse Influenza A Co-infection Model
- Production of an Attenuated Phenol-Soluble Modulin Variant Unique to the MRSA Clonal Complex 30 Increases Severity of Bloodstream Infection
- Inhibition of the TRAIL Death Receptor by CMV Reveals Its Importance in NK Cell-Mediated Antiviral Defense
- Early Mucosal Sensing of SIV Infection by Paneth Cells Induces IL-1β Production and Initiates Gut Epithelial Disruption
- Limited HIV Infection of Central Memory and Stem Cell Memory CD4+ T Cells Is Associated with Lack of Progression in Viremic Individuals
- Virus-Specific Regulatory T Cells Ameliorate Encephalitis by Repressing Effector T Cell Functions from Priming to Effector Stages
- A Tick Gut Protein with Fibronectin III Domains Aids Congregation to the Gut during Transmission
- The HIV-1 Envelope Transmembrane Domain Binds TLR2 through a Distinct Dimerization Motif and Inhibits TLR2-Mediated Responses
- Infection with MERS-CoV Causes Lethal Pneumonia in the Common Marmoset
- VGIII Isolates Causing Infections in HIV/AIDS Patients in Southern California: Identification of the Local Environmental Source as Arboreal
- Diverse Host-Seeking Behaviors of Skin-Penetrating Nematodes
- Capsid Protein VP4 of Human Rhinovirus Induces Membrane Permeability by the Formation of a Size-Selective Multimeric Pore
- The Murine Gammaherpesvirus Immediate-Early Rta Synergizes with IRF4, Targeting Expression of the Viral M1 Superantigen to Plasma Cells
- Characterization of an Insecticidal Toxin and Pathogenicity of against Insects
- The Vi Capsular Polysaccharide Enables Serovar Typhi to Evade Microbe-Guided Neutrophil Chemotaxis
- Histone Deacetylase Inhibitors Impair the Elimination of HIV-Infected Cells by Cytotoxic T-Lymphocytes
- A Locus Encompassing the Epstein-Barr Virus Kinase Regulates Expression of Genes Encoding Viral Structural Proteins
- Distinct APC Subtypes Drive Spatially Segregated CD4 and CD8 T-Cell Effector Activity during Skin Infection with HSV-1
- Structure of CfaA Suggests a New Family of Chaperones Essential for Assembly of Class 5 Fimbriae
- Adoptive Transfer of EBV Specific CD8 T Cell Clones Can Transiently Control EBV Infection in Humanized Mice
- Schistosome Feeding and Regurgitation
- EVM005: An Ectromelia-Encoded Protein with Dual Roles in NF-κB Inhibition and Virulence
- Rabies Virus Hijacks and Accelerates the p75NTR Retrograde Axonal Transport Machinery
- Why HIV Virions Have Low Numbers of Envelope Spikes: Implications for Vaccine Development
- Identification of Anti-virulence Compounds That Disrupt Quorum-Sensing Regulated Acute and Persistent Pathogenicity
- HIV-1 Receptor Binding Site-Directed Antibodies Using a VH1-2 Gene Segment Orthologue Are Activated by Env Trimer Immunization
- Cooperation between Epstein-Barr Virus Immune Evasion Proteins Spreads Protection from CD8 T Cell Recognition across All Three Phases of the Lytic Cycle
- Parasite Extracellular Vesicles: Mediators of Intercellular Communication
- RC1339/APRc from Is a Novel Aspartic Protease with Properties of Retropepsin-Like Enzymes
- Cyclic di-GMP-dependent Signaling Pathways in the Pathogenic Firmicute
- Non-random Escape Pathways from a Broadly Neutralizing Human Monoclonal Antibody Map to a Highly Conserved Region on the Hepatitis C Virus E2 Glycoprotein Encompassing Amino Acids 412–423
- Neutrophil Elastase Causes Tissue Damage That Decreases Host Tolerance to Lung Infection with Species
- Ly6C Monocyte Recruitment Is Responsible for Th2 Associated Host-Protective Macrophage Accumulation in Liver Inflammation due to Schistosomiasis
- SGNH Hydrolase-Like Proteins AlgJ and AlgX Have Similar Topology but Separate and Distinct Roles in Alginate Acetylation
- Why Sexually Transmitted Infections Tend to Cause Infertility: An Evolutionary Hypothesis
- Late Engagement of CD86 after Influenza Virus Clearance Promotes Recovery in a FoxP3 Regulatory T Cell Dependent Manner
- Determinants of Influenza Transmission in South East Asia: Insights from a Household Cohort Study in Vietnam
- A Novel Signal Transduction Pathway that Modulates Quorum Sensing and Bacterial Virulence in
- Host Responses to Group A Streptococcus: Cell Death and Inflammation
- A Cysteine Protease Inhibitor of Is Essential for Exo-erythrocytic Development
- EBNA3C Augments Pim-1 Mediated Phosphorylation and Degradation of p21 to Promote B-Cell Proliferation
- On the Front Line: Quantitative Virus Dynamics in Honeybee ( L.) Colonies along a New Expansion Front of the Parasite
- Assembly and Architecture of the EBV B Cell Entry Triggering Complex
- NLR-Associating Transcription Factor bHLH84 and Its Paralogs Function Redundantly in Plant Immunity
- The PDZ-Binding Motif of Severe Acute Respiratory Syndrome Coronavirus Envelope Protein Is a Determinant of Viral Pathogenesis
- Strain-Specific Properties and T Cells Regulate the Susceptibility to Papilloma Induction by Papillomavirus 1
- Human Cytomegalovirus pUL79 Is an Elongation Factor of RNA Polymerase II for Viral Gene Transcription
- The GAP Activity of Type III Effector YopE Triggers Killing of in Macrophages
- PLOS Pathogens
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Disruption of Fas-Fas Ligand Signaling, Apoptosis, and Innate Immunity by Bacterial Pathogens
- Ly6C Monocyte Recruitment Is Responsible for Th2 Associated Host-Protective Macrophage Accumulation in Liver Inflammation due to Schistosomiasis
- Host Responses to Group A Streptococcus: Cell Death and Inflammation
- Pathogenicity and Epithelial Immunity
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



