-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaGenetic Dissection of Gut Epithelial Responses to
In malaria vector mosquitoes, the presence of bacteria and malaria parasites is tightly linked. Bacteria that are part of the mosquito gut ecosystem are critical modulators of the immune response elicited during infection with malaria parasites. Furthermore, responses against oral bacterial infections can affect malaria parasites. Here, we combined mosquito gut infections with the enterobacterium Serratia marcescens with genome-wide discovery and phenotypic analysis of genes involved in antibacterial responses to characterize molecular processes that control gut bacterial infections thus possibly affecting the mosquito susceptibility to infection by malaria parasites. Our data reveal complex genetic networks controlling the gut bacterial infection load and ecosystem homeostasis. These networks appear to exhibit much higher specificity toward specific classes of bacteria than previously thought and include behavioral response circuits involved in antibacterial immunity.
Published in the journal: . PLoS Pathog 10(3): e32767. doi:10.1371/journal.ppat.1003897
Category: Research Article
doi: https://doi.org/10.1371/journal.ppat.1003897Summary
In malaria vector mosquitoes, the presence of bacteria and malaria parasites is tightly linked. Bacteria that are part of the mosquito gut ecosystem are critical modulators of the immune response elicited during infection with malaria parasites. Furthermore, responses against oral bacterial infections can affect malaria parasites. Here, we combined mosquito gut infections with the enterobacterium Serratia marcescens with genome-wide discovery and phenotypic analysis of genes involved in antibacterial responses to characterize molecular processes that control gut bacterial infections thus possibly affecting the mosquito susceptibility to infection by malaria parasites. Our data reveal complex genetic networks controlling the gut bacterial infection load and ecosystem homeostasis. These networks appear to exhibit much higher specificity toward specific classes of bacteria than previously thought and include behavioral response circuits involved in antibacterial immunity.
Introduction
Genetic variation within populations of the An. gambiae mosquito, especially with regard to genes encoding immune factors, is believed to play an important role in the mosquito susceptibility to infection by the malaria parasite Plasmodium falciparum [1]–[3]. Many immune factors exhibit both anti-Plasmodium and antibacterial activities, such as those involved in the IMD/REL2 pathway, which is triggered by bacteria through the peptidoglycan recognition receptor PGRPLC [4], [5]. Bacterial infections can affect mosquito survival [6] and are thought to constitute a major evolutionary drive [7] as opposed to Plasmodium infections the impact of which on mosquito fitness is unclear [8]. An example is the segregation of TEP1 alleles between the M and S molecular forms of An. gambiae in west Africa, which differentially affect Plasmodium infections, and is thought to be largely driven by bacterial pathogen pressure in larval habitats [2]. Therefore, genetic associations related to the outcome of bacterial infections may, directly or indirectly, influence mosquito vectorial capacity.
The adult mosquito gut harbors a wide spectrum of bacterial populations, mainly Gram-negative enterobacteria [9]–[11]. The broad variation in gut microbiota composition observed both at the individual and population levels is probably the result of an interplay between the environmental bacterial diversity and the mosquito genetic makeup [12]–[14]. Moreover, a precipitous bacterial increase after a blood meal, whose peak coincides with midgut invasion by Plasmodium [15], can affect the Plasmodium infection load both indirectly, by triggering PGRPLC-mediated mosquito immune responses [5], [16] or through generation of immune memory [17], and directly, through the generation of reactive oxygen species by specific enterobacteria that compromise malaria parasites [18].
Epithelial responses against Gram-negative bacteria have been extensively studied in Drosophila [19]. They involve recognition of peptidoglycan [20], [21] that triggers a finely-tuned immune response mainly through the Imd pathway, resulting in the expression of antimicrobial peptides that limit bacterial populations [22], [23]. Production of reactive oxygen species, which target bacteria, through the Dual Oxidase (DUOX) pathway, has also been reported [24]. Gut stem cell proliferation and epithelial cell renewal following tissue damage due to bacterial infection are regulated by the EGFR and JAK/STAT pathways [25]–[27]. However, the mechanisms involved in achieving gut homeostasis remain poorly understood. It has been suggested that regulation of Imd responses can influence the microbiota composition in Drosophila [28]. Further discrimination between commensal and pathogenic bacteria can be provided by recognition of pathogen-derived uracil, most likely by unidentified G protein-coupled receptors (GPCRs), which triggers the DUOX pathway [29], but the possibility of more specific responses that shape the gut microbiota remains open.
One unexplored aspect of antibacterial immunity is the behavioral immune responses that limit or disrupt the intake of pathogens, thus making an infection more controllable by the immune system. Feeding behavior in Drosophila is known to be finely regulated through an interplay between allatostatin A and neuropeptide F (NPF) [30] while feeding suppression is shown to occur following an immune challenge [31]–[33]. Gustatory receptors are shown to modulate feeding behavior by acting as nutrient sensors [34] and may also be involved in aversion circuits [35] or antibacterial responses through recognition of bacterial-derived metabolites as in mammalian chemoattractant receptors [36].
Here we set out to examine the genetic basis of bacterial infection in the mosquito gut using An. gambiae infections with the Gram-negative enterobacterium Serratia marcescens that is prevalent in both lab-reared and field collected mosquitoes and is shown to affect the Plasmodium infection load [10], [12], [37]. To achieve this, we used an Affymetrix 400 k SNP genotyping array to identify genetic variation associated with the outcome of oral S. marcescens infection in a recently established M form An. gambiae colony. The results identify 138 genes associated with the outcome of infection, including the gene encoding the major IMD/REL2 receptor PGRPLC and the epidermal growth factor receptor EGFR, and further suggest that epithelial immune responses against gut bacteria are more complex than previously thought. We identify a set of three type III fibronectins that modulate homeostasis of the gut microbiota with specificity mainly against Enterobacteriaceae. We also present evidence that behavioral responses following S. marcescens infection can modulate the bacterial load. These data could be further exploited in mosquito microbiota-based interventions aiming to limit malaria transmission.
Results
S. marcescens infection of the mosquito gut
An. gambiae female adults were treated with antibiotics to reduce their natural gut microbiota load (Figure S1) and subsequently fed with fluorescently labeled S. marcescens (Db11-GFP) added to the sugar meal. The bacterial levels in the gut of sugar-fed mosquitoes (henceforth referred to as infection) were monitored from day 2 to 6 post infection and showed considerable variation including highly and lowly infected mosquitoes as well as mosquitoes that despite ingesting bacteria-containing sugar showed no sign of fluorescence in their gut (Figure 1A). While the proportion of lowly infected mosquitoes remained rather constant at approximately 50% throughout the course of the experiment, the relative proportions of highly and non-infected mosquitoes changed between days 2 and 3 in favor of highly infected mosquitoes and remained stable thereafter until day 5 (Figure 1B). At day 6, highly infected mosquitoes decreased by ca. 15% with a parallel increase of non-infected mosquitoes.
Fig. 1. Gut infection with S. marcescens varies between individual An. gambiae mosquitoes. 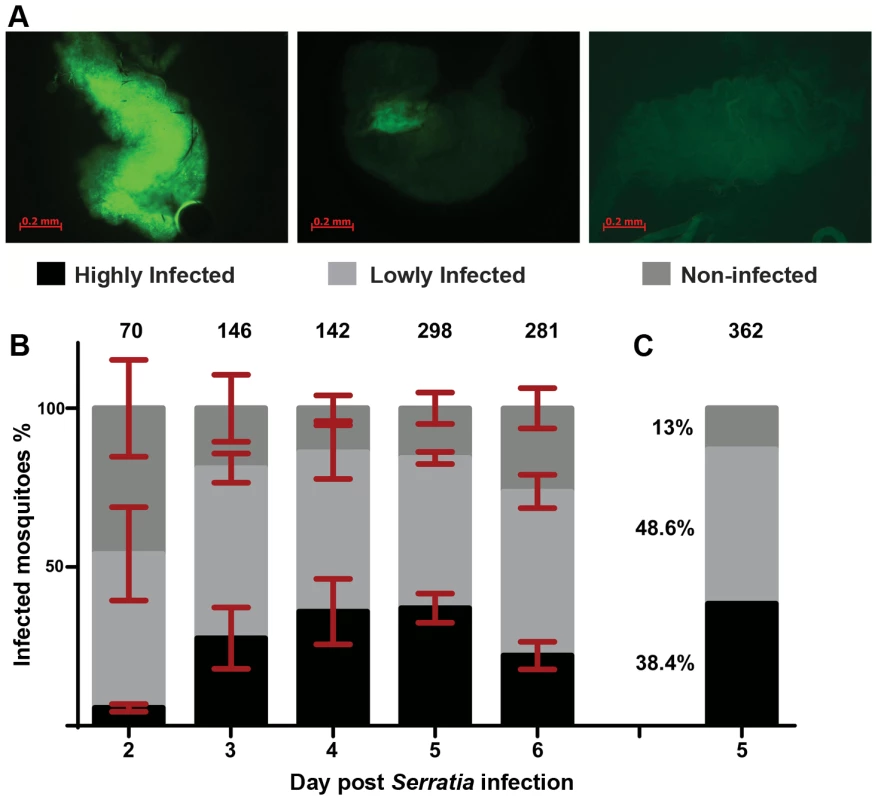
Mosquitoes were antibiotic treated for 5 days and subsequently fed on sugar containing the Db11-GFP strain of S. marcescens. Bacteria-fed mosquitoes were selected 2 days post infection and the prevalence of fluorescent bacteria in their gut was monitored from day 2 to 6 post infection. 1A: The level of S. marcescens infection in the mosquito gut showed considerable variation: mosquitoes with intense fluorescence in most of the gut were characterized as highly infected (left panel), mosquitoes in which fluorescence was evident but confined to a part of the gut were characterized as lowly infected (middle panel) and mosquitoes with no sign of fluorescence were characterized as non-infected (right panel). 1B: S. marcescens infected mosquitoes were dissected each day, from day 2 to 6 post infection, and the proportions of highly, lowly and non-infected mosquitoes were determined over 4 independent infections. The average percentage ±SEM for each level of infection is indicated for each day post infection, with the total number of mosquitoes dissected each day in all 4 infections shown over each bar. 1C: In 2 independent infections used for SNP genotyping, mosquitoes were dissected 5 days post infection and the percentage of highly, lowly and non-infected mosquitoes, pooled from both infections, can be seen beside the respective part of the bar representing each level of infection. Identification of SNP divergence associated with the outcome of S. marcescens infection
To investigate whether genetic variation could partly explain the observed S. marcescens infection phenotype, single nucleotide polymorphism (SNP) divergence between the highly and non-infected phenotypic pools was interrogated using a 400 k SNP genotyping array. Mosquitoes were orally infected with S. marcescens, and gut infection levels were determined at day 5 post infection. The results were similar to those obtained in the previous replicate experiments: 38.4% of mosquitoes could be classified as highly infected, 48.6% lowly infected and 13% non-infected (Figure 1C). Pools of equimolar amounts of genomic DNA (gDNA) prepared from carcasses of 15 highly infected and 15 non-infected mosquitoes out of 139 and 47 mosquitoes in each phenotypic group, respectively, were hybridized onto two Affymetrix SNP genotyping arrays. These SNP chips interrogate genetic variation at ∼400,000 variable positions in the An. gambiae genome (Table S1) [38], and were previously shown to provide useful quantitative information regarding divergence between pooled mosquito samples [39].
Allele calls for each SNP locus were used to determine the minor allele frequency (MAF) differences between highly and non-infected gDNA pools. Two approaches were used to assess genotypic association with the S. marcescens infection phenotype. The first included MAF difference at a SNP locus between highly infected and non-infected pools >0.5, suggesting a preponderance of different genotypes between the two pools for the respective locus. The second involved a permutation analysis in which the average MAF difference of 10 adjacent SNP loci (SNPs) was compared with that of 10 random SNPs. Statistical significance was assessed for each of the ∼40,000 non-overlapping 10-SNP windows (Table S2) and those showing a p-value<10−5, following a Bonferroni correction for the number of tests conducted, were considered as being associated with the S. marcescens infection phenotype.
The two approaches detected 140 SNPs with MAF difference >0.5 and 44 10-SNP windows with significant p-values, respectively. As shown in Figure 2, these SNPs and 10-SNP windows together formed distinctive clusters along the An. gambiae genome that were designated as peaks so that they are discerned from each other, although assessed association was limited to genes within a 5 kb radius of highlighted SNPs or within genomic areas delineated by significant 10-SNP windows. Overall, 118 genes were found to reside within a 5 kb radius of highlighted SNPs (Table S3), while 27 genes fell within significant 10-SNP windows (Table S4). The two approaches combined detected 138 genes (Table S5), as there was an overlap of 7 genes between the two sets, including the highly relevant CLIPE6 and EGFR as discussed below.
Fig. 2. Mapping of An. gambiae genetic variation associated with the S. marcescens infection phenotype. 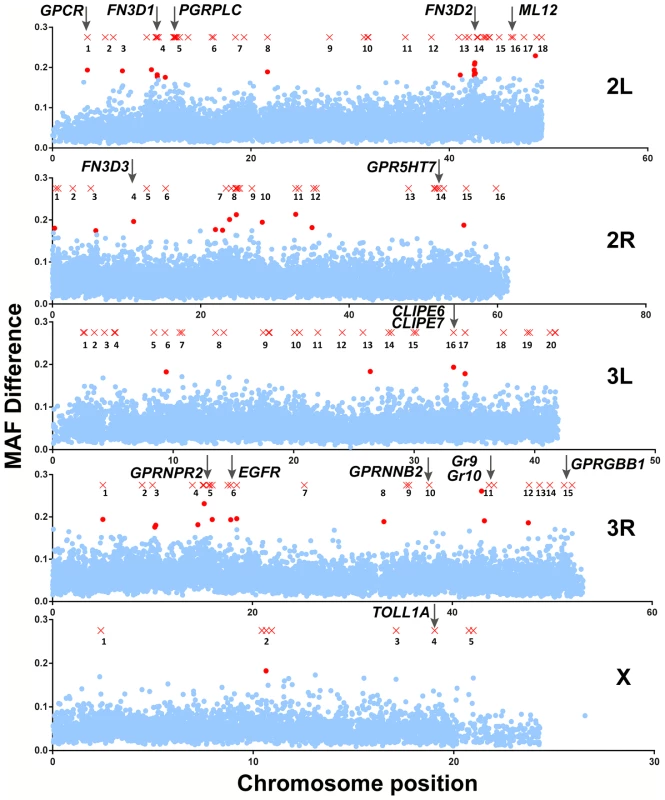
SNPs with MAF difference >0.5 and 10-SNP windows with Bonferroni-corrected significance (p-value<10−5) are shown in their respective chromosomal position as red X crosses and dots, respectively. Non-significant 10-SNP windows are shown as blue dots. Genomic areas with highlighted SNPs and/or significant 10-SNP windows in close proximity are referred to as peaks and are numbered. Each peak is referred to using the chromosomal arms it resides on and its respective assigned number. The genomic positions of genes of interest found within a 5 kb radius of highlighted SNPs or within genomic areas delineated by 10-SNP windows with a significant p-value are indicated by vertical arrows. In peak 2L-5 (chromosome 2L, peak 5), the gene encoding PGRPLC is found within a 5 kb radius of a highlighted SNP. PGRPLC recognizes peptidoglycan and activates the IMD/REL2 NF-kappaB signaling pathway, thus eliciting antibacterial responses [21], [40], [41]. This pathway is constitutively triggered by mosquito gut bacteria maintaining an elevated level of antimicrobial peptide production [5], [42]. The association of PGRPLC with the S. marcescens infection phenotype suggests that genetic variation within the mosquito population may influence the ability to mount an antibacterial response via the IMD/REL2 pathway. Adjacent to PGRPLC, in peak 2L-5, is another peptidoglycan recognition protein encoding gene, PGRPLA.
Of the remaining genes, several exhibit homologies suggesting involvement in antibacterial immune responses, especially in recognition of pathogen or host derived signals as well as in signal transduction and regulation of immune responses (Table 1). The permutation analysis revealed 3 genes, out of a total of 27, encoding proteins with type III fibronectin domains (FN3D) in different peaks: FN3D1 in peak 2L-4, which was also in the proximity of a highlighted SNP, FN3D2 in 2L-14 and FN3D3 in 2R-4. A total of 65 An. gambiae genes contain FN3 domains, including the hypervariable pattern recognition receptor AgDscam, the insulin receptor INR and the JAK/STAT receptor DOME. FN3D2 and FN3D3 additionally possess immunoglobulin and putative transmembrane domains, while FN3D2 is an ortholog of Drosophila Dscam4. Drosophila Dscam is shown to bind bacteria and influence the efficiency of phagocytosis [43], while its An. gambiae ortholog, AgDscam, is also shown to bind bacteria and mediate antibacterial and anti-Plasmodium responses [44]. Importantly, Dscam genes in various organisms generate a diverse repertoire of isoforms, suggestive of challenge-specific pattern recognition through alternative splicing [43], [45], [46], with particular AgDscam isoforms specifically targeting P. berghei, P. falciparum or commensal bacteria [47], [48].
Tab. 1. Genes of interest associated with the S. marcescens infection phenotype. 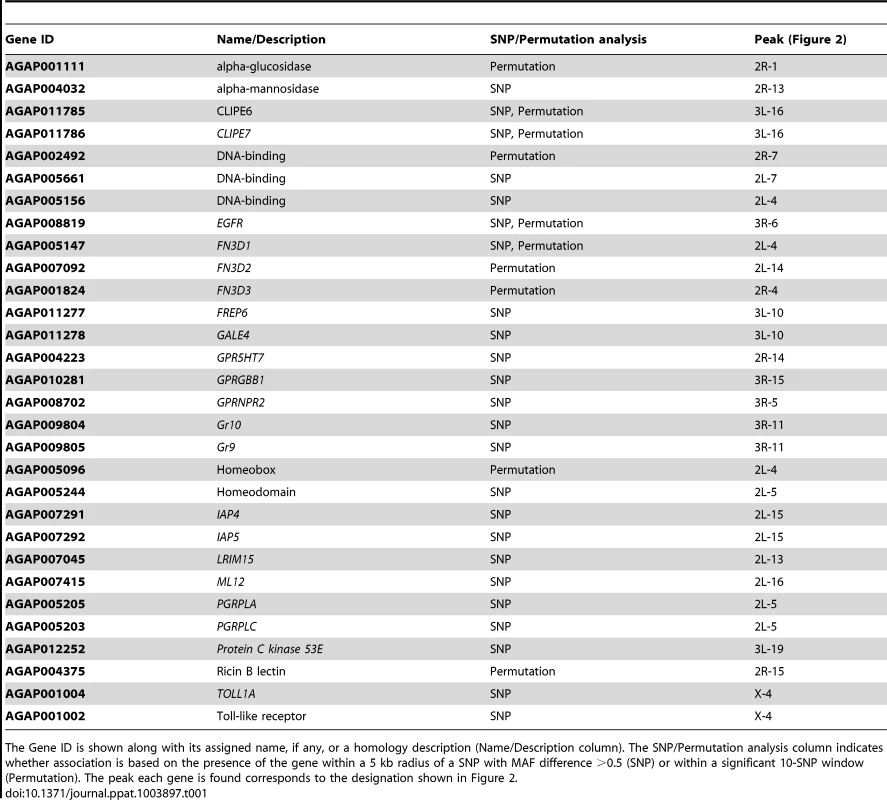
The Gene ID is shown along with its assigned name, if any, or a homology description (Name/Description column). The SNP/Permutation analysis column indicates whether association is based on the presence of the gene within a 5 kb radius of a SNP with MAF difference >0.5 (SNP) or within a significant 10-SNP window (Permutation). The peak each gene is found corresponds to the designation shown in Figure 2. Several putative transcription factors with homeobox-like or DNA-binding domains were found in the identified peaks. AGAP005096 in 2L-4 and AGAP005244 in 2L-5 (together with PGRPLC and PGRPLA) encode homeodomains. The homeobox gene, Caudal, has been previously implicated in the regulation of epithelial immune responses and shown to influence the gut bacterial population structure in Drosophila [28], while its mosquito homolog has been shown to regulate the IMD/REL2 pathway [49]. Thus, these putative transcription factors could play similar regulatory roles. AGAP002492 in peak 2R-7 encodes a DNA-binding domain, while its Drosophila ortholog ewg is involved in the Wnt/Wingless pathway [50]. AGAP005156, in peak 2L-4 encodes an ARID/BRIGHT DNA-binding domain, with its Drosophila ortholog, retained, is involved in behavioral modulations and repression of male courtship [51], [52]. AGAP005661, in peak 2L-7, a putative ligand-regulated transcription factor, is an ortholog of the Drosophila nuclear receptor FTZ-F1, involved in juvenile hormone mediated gene expression [53].
Genes encoding alpha-glucosidase and alpha-mannosidase homologs were detected in peaks 2R-1 and 2R-13, respectively. These genes possess glycoside hydrolase domains that are also present in the conserved chitinase gene family [54], involved in bacterial clearance and host tolerance [55].
The gene encoding the epidermal growth factor receptor, EGFR, was identified in the prominent peak 3R-6 both by both the permutation and the individual SNP analysis. The Drosophila EGFR pathway has been implicated in gut remodeling following oral bacterial infection [27], suggesting that the EGFR pathway may influence the outcome of S. marcescens infection in Anopheles, possibly through synergistic functions in gut homeostasis.
CLIPE6 and CLIPE7, found in peak 3L-16, belong to the non-catalytic E sub-family of CLIP-type serine proteases, a family known to participate in proteolytic cascades in antibacterial and anti-Plasmodium responses [56], [57], with SPCLIP1, another E sub-family member, involved in anti-Plasmodium responses by regulating complement recruitment [58], [59]. Several leucine-rich repeat containing genes were also detected, including LRIM15 (peak 2L-13), a transmembrane member of the LRIM family of immune proteins [60]. LRIMs have also been implicated in complement anti-Plasmodium responses [61]–[63].
Two Toll-like receptors, TOLL1A and a previously uncharacterized paralog of TOLL5B, were found in peak X-4. Little is known about the role of Toll-like receptors in Anopheles immunity, however, cross-talk between the REL1 and REL2 signaling pathways in the yellow fever mosquito Aedes aegypti [64] and synergistic interactions between the Toll and Imd pathway in Drosophila [65], leave open the possibility for involvement of Toll-like receptors in defenses against Gram-negative bacteria, also in Anopheles [66].
A gene encoding a protein with a ricin B lectin domain was found in peak 2R-15. Lectins bind oligosaccharides and have been shown to modulate mosquito immune responses [6], [63], while mammalian lectins modulate host and gut microbiota interactions [67]. Genes belonging to other families of putative pattern recognition receptors were also found to be associated with the S. marcescens infection phenotype, including a fibrinogen-related protein (FBN or FREP) and a galectin in peak 3L-10 and an MD2-like receptor in 2L-16 [59], [68], [69].
Five annotated or putative GPCRs were found to be associated with the S. marcescens infection phenotype, including three putative neurotransmitter-triggered receptors: the serotonin receptor GPR5HT7 in peak 2R-14, the GABA-B family receptor GPRGBB1 in peak 3R-15 and the neuropeptide receptor GPRNPR2 in 3R-5. GPCRs have been previously implicated in modulation of P. falciparum infection in An. gambiae [70], but the mechanism by which this is accomplished remains unclear. NPR-1, a neurotransmitter-triggered GPCR of Caenorhabditis elegans, has been shown to modulate antibacterial defenses in a behavior dependent or independent manner, and NPR-1 genetic polymorphisms are suggested to be major determinants of bacterial susceptibility [71], [72]. Serotonin is a major modulator of mammalian intestinal inflammation [73], [74], in an interplay between the nervous and immune system [75]. The Drosophila ortholog of GPR5HT7 is involved in various behavioral processes [76], [77], including aggressive behavior, a process also modulated by NPF [78]. Interestingly, the Drosophila ortholog of GPRGBB1 has been implicated in behavioral responses to alcohol sensitivity [79], a process in which NPF is also a major modulator [80], [81].
Two gustatory receptor genes, Gr9 and Gr10, encoding 7-transmembrane chemoreceptor domains, were associated with the outcome of S. marcescens infection (Figure 2, peak 3R-11). Gr9 and Gr10 are paralogs and show co-orthologous relationships with the Drosophila Gr32a, Gr39a and Gr68a [82]. Gr32a and Gr68a act as pheromone receptors in modulating mating behavior [83], [84], while Gr39a has been implicated, through 4 splice variants, in sustaining courtship behavior [85]. Gr32a is also involved in regulating aggressive behavior through recognition of small non-volatile hydrocarbons [86], or feeding suppression triggered by DEET or other antifeedants [87].
Gustatory receptor family members have also been implicated in aversive taste [35], [88], CO2 responses [89], [90] and sugar recognition [91]–[94]. A Drosophila gustatory receptor, Gr43a, has been shown to recognize fructose and act as a nutrient sensor, promoting or suppressing feeding [34]. Since enhanced or suppressed feeding of bacteria-containing sugar can decisively influence the abundance of S. marcescens that the mosquito takes in and its immune system can handle, it is possible that Gr9 or Gr10 variants linked to altered mosquito feeding behavior can affect the outcome of infection. Furthermore, GPR43, a mammalian chemoattractant receptor, has been shown to recognize short-chain fatty acids of bacterial origin and participate in antibacterial responses [36], while other mammalian chemoattractant receptors regulate inflammatory responses by recognizing endogenous factors [95]. Recognition of bacterial-derived uracil has recently been shown to modulate Drosophila antibacterial responses through the DUOX pathway [29]. Therefore, another possibility is that Gr9 or Gr10 recognize bacterial-derived metabolites or infection-induced mosquito molecules and mediate antibacterial responses.
Several other genes with no known or unrelated to immune responses homologies were also associated with the S. marcescens infection phenotype such as AGAP013684 in peak 2R-8, encoding a putative miRNA. MiRNAs are known to modulate gene regulation in processes that include epithelial immunity [96], [97]. AGAP006405 in peak 2L-10 encodes a tyrosine protein kinase, while its Drosophila ortholog, derailed2, is involved in Wnt5 signaling and establishment of olfactory circuits [98]. In peak 2L-15 the inhibitors of apoptosis IAP4 and IAP5 were found. The Drosophila IAP2 is known to regulate Imd signaling [99], suggesting that the An. gambiae IAP4 or IAP5 may also play similar roles. AGAP012252, in peak 3L-19, encodes the ortholog of Drosophila PKC53E, implicated in NPF-mediated alcohol sensitivity [100], [101]. AGAP011363, in peak 3L-11, encodes the ortholog of Drosophila rab6, implicated in phagocytosis [102] but also trafficking of Grk, the EGFR ligand [103], [104]. AGAP010503, in peak 3L-4, encodes the ortholog of the Drosophila SK channel, implicated in behavioral courtship memory [105]. AGAP005216, in peak 2L-5, encodes the ortholog of Drosophila fab1, involved in autophagy but also the lysosomal degradation of necrotic, a modulator of the Toll pathway [106]–[109].
Candidate gene prioritization for further phenotypic analysis was based on homologies with genes known to be involved in species-specific antibacterial responses, e.g. FN3D2 and Dscam [43] or demonstrably regulating the response to gut microbiota in other systems, e.g. Gr9 and the mammalian chemoattractant receptor GPR43 [36], with the aim of the identification of novel functions of genes or gene families in antibacterial responses.
Serratia infection phenotypic analysis of FN3D1-3
The involvement of the three FN3D genes in shaping the outcome of An. gambiae gut infection with S. marcescens was investigated by RNAi-mediated gene silencing (Figure 3). Antibiotic treated mosquitoes were orally infected with S. marcescens following knockdown (kd) of each of the FN3Ds (Figure S2). The bacterial load in mosquito guts was determined 5 days post infection by quantitative RT-PCR (qRT-PCR), using both broad range bacterial 16S and Serratia-specific primers. Highly significant and robust increase of the S. marcescens load was observed after silencing any of the three genes compared to dsLacZ-treated controls: 21 to 53-fold in FN3D1 (Figure 3A), 41 to 60-fold in FN3D2 (Figure 3B) and 13 to 29-fold in FN3D3 kd (Figure 3C).
Fig. 3. Silencing of FN3D1–3 increases Serratia levels in orally infected mosquitoes or mosquitoes retaining their natural gut microbiota. 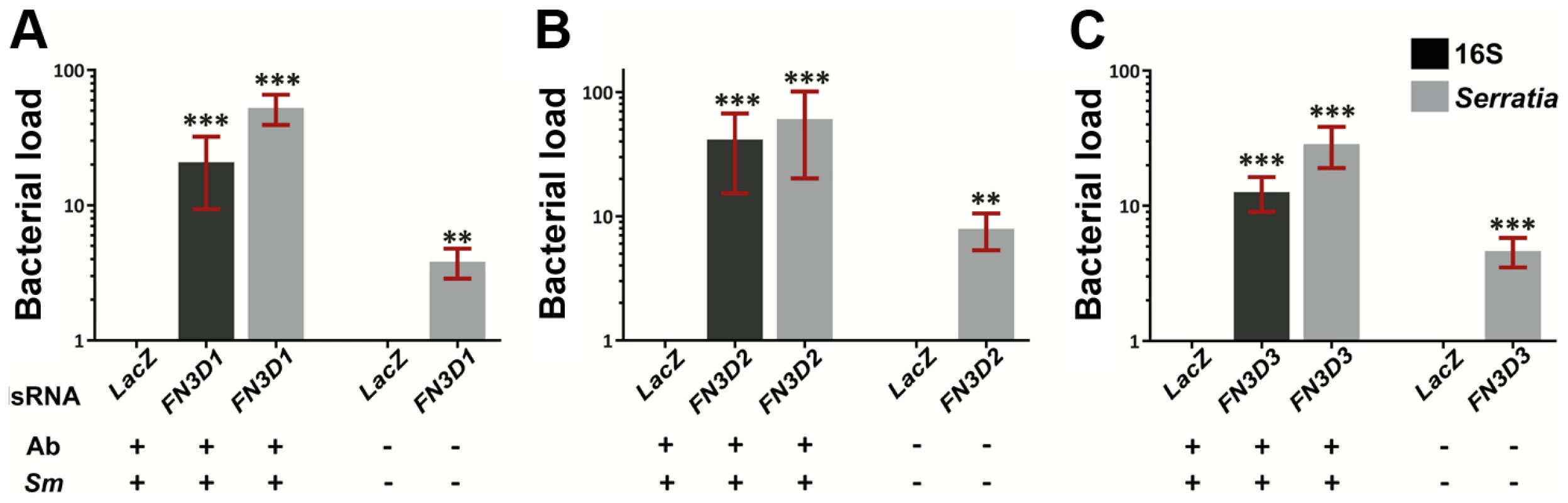
Antibiotic treated and subsequently orally infected with S. marcescens (Ab+Sm+) or non-treated mosquitoes retaining their natural midgut microbiota (Ab−Sm−), were dsRNA treated to silence FN3D1 (3A), FN3D2 (3B) or FN3D3 (3C) or treated with the LacZ dsRNA control. The bacterial load in the midguts of surface sterilized mosquitoes was determined 5 days post S. marcescens infection for Ab+Sm+ mosquitoes, or 5 days post dsRNA treatment for Ab−Sm− mosquitoes. Bacterial load was determined using broad range bacterial 16S or Serratia-specific primers using qRT-PCR, in which relative to the endogenous AgS7 control bacterial abundance was determined for each sample and then normalized to the relative abundance of the dsLacZ treated control. For Ab+Sm+ mosquitoes, the average ±SEM of the fold-change in bacterial load is shown as determined over 7 independent infections for FN3D1 (3A) and FN3D2 (3B), or 8 independent infections for FN3D3 (3C), with the qRT-PCR in each infection replicated at least twice. For Ab−Sm− mosquitoes, the average ±SEM of the fold-change in bacterial load is shown as determined over 4 independent assays for FN3D1 and FN3D3, or 5 independent assays for FN3D2. Asterisks indicate significance in an one-sample t-test against zero using the log2-transformed fold-change values so that zero corresponds to no difference from dsLacZ treatment. Two asterisks indicate a p-value<0.005 while three asterisks indicate a p-value<0.0005. We also assessed the role of FN3Ds in shaping the load of Serratia naturally found in the mosquito gut. Mosquitoes reared in standard conditions, without antibiotic treatment or infection with S. marcescens, were treated with dsRNA against each of FN3D1–3 and the level of commensal Serratia was determined 5 days later (Figure 3 A–C, last bar in each panel). Silencing any of the three genes resulted in a significant 4 to 8-fold increase in the levels of commensal Serratia compared to dsLacZ-treated controls. These data indicate the involvement of FN3D1–3 in constitutive antibacterial effects that shape the load and composition of the mosquito natural gut microbiota.
FN3D1–3 kd alters the gut microbiota composition in favor of Enterobacteriaceae
When the effect of FN3D1–3 kd was assessed on the total bacterial load in the gut of mosquitoes that retained their natural gut microbiota, a non-uniform effect was observed between 4 independent replicate assays (Figure S3). In some cases, FN3D silencing resulted in moderate increases of both Serratia and total bacterial load, while in other cases the total bacterial load showed no or marginal increase while Serratia showed a strong increase. This variability suggested that the FN3D effect on total bacteria may depend on the initial Serratia load and that FN3Ds may function in shaping the population structure of the gut microbiota by affecting a subset of bacteria inhabiting the mosquito gut, including Serratia.
To further investigate these hypotheses, we carried out a microbiome analysis using 454 pyrosequencing of samples from two of the replicate assays in which FN3D1–3 kd increased Serratia but not total bacteria abundance (Figure 4A–B) and from a replicate assay in which FN3D3 kd increased both Serratia and total bacterial load (Figure 4C). The resulting sequence reads were assigned to their respective bacterial family. Reads aligning to Serratia reference sequences were categorized separately from other Enterobacteriaceae (Table S6).
Fig. 4. FN3D1–3 silencing changes the composition of the mosquito gut microbiota in favor of Enterobacteriaceae. 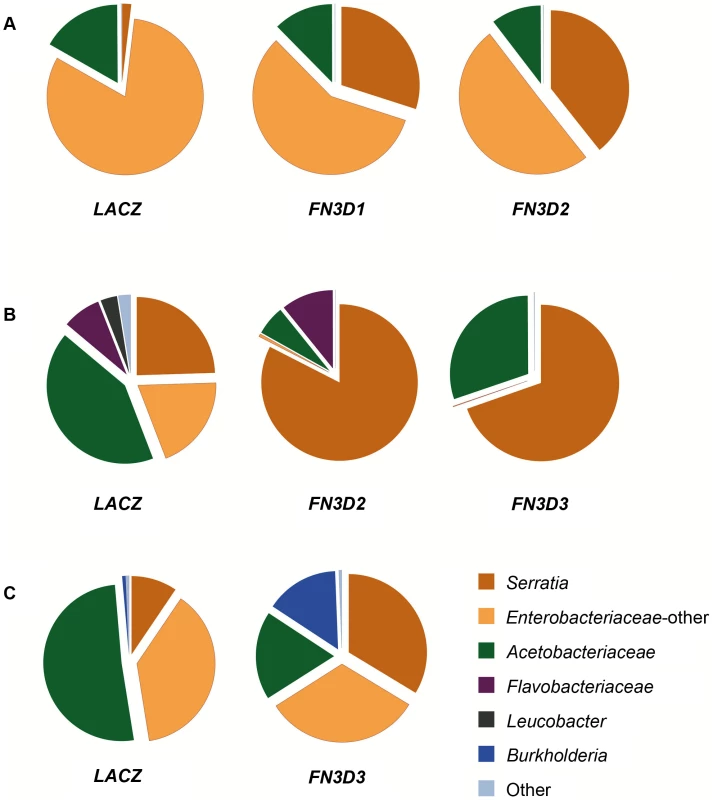
The 16S V4–V6 hypervariable regions of gut bacterial populations from mosquitoes retaining their natural gut microbiota without antibiotic treatment or S. marcescens infection (Ab−Sm−, Figure S3) were sequenced using 454 pyrosequencing (Table S6). cDNA pools from guts of FN3D1–3 dsRNA treated mosquitoes or dsLacZ treated controls, surface sterilized and dissected 5 days post dsRNA treatment, were PCR amplified and sequenced over 3 independent assays (panels A to C). The gut microbiota composition of the FN3D1–3 dsRNA treated pools or the dsLacZ-treated control in each independent assay can be seen in the respective pie charts, with the dsRNA treatment indicated below each pie chart. The color legend indicates the bacterial family corresponding to each pie chart color. Modulation of total bacteria or Serratia abundance can be seen for each sequenced pool in Figure S3, with FN3D1 kd corresponding to replicate 1 in panel 4A, FN3D2 kd corresponding to replicate 1 in panel 4A and 3 in panel 4B and FN3D3 kd corresponding to replicate 2 in panel 4B and 4 in panel 4C. Considerable variation in bacterial composition was observed in control gut pools between the three assays (Figure 4). This variation is consistent with previously reported metagenomic analyses in lab-reared and field-collected mosquitoes, which revealed extensive gut microbiota diversity at both the individual and population levels [12]–[14]. Total Enterobacteriaceae (Serratia and other Enterobacteriaceae) were highly prevalent in all pools corresponding to 83.2%, 44.2% and 47.5% of total reads, respectively, while significant variation was observed in the specific representation of Serratia that corresponded to 1.9%, 24.5% and 9.5% of total sequence reads, respectively. This natural Serratia variation is consistent with the variation observed following oral infection with Db11-GFP S. marcescens (see Figure 1) and may be related to the underlying genetic variation. Acetobacteriaceae was a prominent family in all assays, while Flavobacteriaceae was the prevailing family in the second assay.
In the first assay, FN3D1 or FN3D2 kd increased the representation of total Enterobacteriaceae to 87.6% and 89.6%, respectively (Figure 4A). Remarkably, silencing FN3D1 or FN3D2 resulted in a dramatic increase in Serratia representation from 1.9% in the dsLacZ-treated control to 30% and 39.3% of total sequence reads, respectively, in agreement with the qRT-PCR analysis of the same samples (Figure S3). Similar results were obtained in the second assay whereby silencing FN3D2 or FN3D3 resulted in an increase in total Enterobacteriaceae representation, from 44.2% to 83.1% and 69.7%, respectively (Figure 4B). In both cases, Serratia representation showed a precipitous increase from an initial intermediate level of 24.5%, to almost all Enterobacteriaceae sequence reads aligning to Serratia reference sequences, again in consistence with the qRT-PCR analysis (Figure S3). Although non-Enterobacteriaceae representation decreased in both FN3D2 and FN3D3 kd, Flavobacteriaceae persisted following FN3D2 kd but were completely eliminated following FN3D3 kd, indicating a difference in the effect between the two FN3Ds related to non-Enterobacteriaceae strains.
Taken together, these data indicate that FN3Ds indeed play a major role in shaping the population structure of the mosquito gut microbiota, as silencing any of FN3D1–3 led to increased Serratia abundance but also shifted the composition of the mosquito gut microbiota in favor of Enterobacteriaceae, mainly Serratia or strains that show similarity to Serratia reference sequences. This shift may be a result of a specific FN3D function against Serratia or a subset of gut bacteria. Alternatively, bacterial interactions or differential growth potential of different bacterial strains may account for the observed shift following a uniform FN3D antibacterial effect.
We tested this hypothesis by examining whether FN3D1–3 silencing could affect the levels of gut infection with non-Enterobacteriaceae. Antibiotic treated dsLacZ treated controls and FN3D1–3 kd An. gambiae mosquitoes were orally infected with bacteria of the genus Asaia, a member of the Acetobacteriaceae family, common in both field and laboratory-reared An. gambiae [9]–[11] and present in all of our sequenced samples. FN3D1–3 silencing resulted in moderate, non-significant increases in bacterial load, compared to controls (Figure S4), distinguishably lower than following oral S. marcescens infection (Figure 3). These data suggest that the observed FN3D antibacterial effect is not uniform across all Gram-negative bacteria and may be specific to a subset of the gut bacterial population including Enterobacteriaceae.
The observed shift in favor of Enterobacteriaceae representation when both Serratia and total bacterial abundance increased following FN3D3 kd was also confirmed by microbiome sequencing that showed an increase of Serratia from 9.5% to 33.7% of total sequence reads and of total Enterobacteriaceae from 47.5% to 66% (Figure 4C). Remarkably, FN3D3 kd also increased the representation of bacteria of the genus Burkholderia, from an initial 0.71% to 15.1% of total reads (Figure 4C). Burkholderia were not traced in the dsLacZ treated control pool in which the effect of FN3D3 kd was also assayed (Figure 4B). These data suggest that FN3D3 limits a subset of the mosquito gut bacterial community including Enterobacteriaceae but also bacteria of the genus Burkholderia.
Gr9 modulates S. marcescens infection levels
The genomic area encompassing genes encoding the gustatory receptors Gr9 and Gr10 was associated with the outcome of S. marcescens infection. As alternative splicing of Gr9 has been previously suggested [82], with Gr9 possessing 13 splice variants compared to one for the adjacent Gr10, we considered Gr9 genetic variation more likely to influence the outcome of S. marcescens infection, leading to the observed SNP divergence. Gr9 has shown significant upregulation compared to other tissues in the midgut of blood-fed adult mosquitoes [110] and also in the midgut of adult mosquito tissues [111]. The Gr9 midgut expression was also confirmed here (Figure S2). Furthermore, comparison of transcription profiles between antennae or maxillary palps and whole body transcriptomes in female mosquitoes has previously shown a non-significant upregulation of Gr9 in those two tissues (1.42 for antennae and 1.15 for maxillary palps) [112].
We carried out RNAi-mediated silencing of Gr9 in adult mosquitoes and examined the outcome of oral S. marcescens Db11-GFP infection. Gr9 knockdown resulted in a precipitous 36 to 48-fold increase in S. marcescens levels compared to dsLacZ treated controls, as determined using both broad range 16S and Serratia-specific primers (Figure 5A). These data suggest that Gr9 exerts an antibacterial effect that influences the outcome of S. marcescens infection.
Fig. 5. Gr9 silencing increases S. marcescens levels in orally infected mosquitoes. 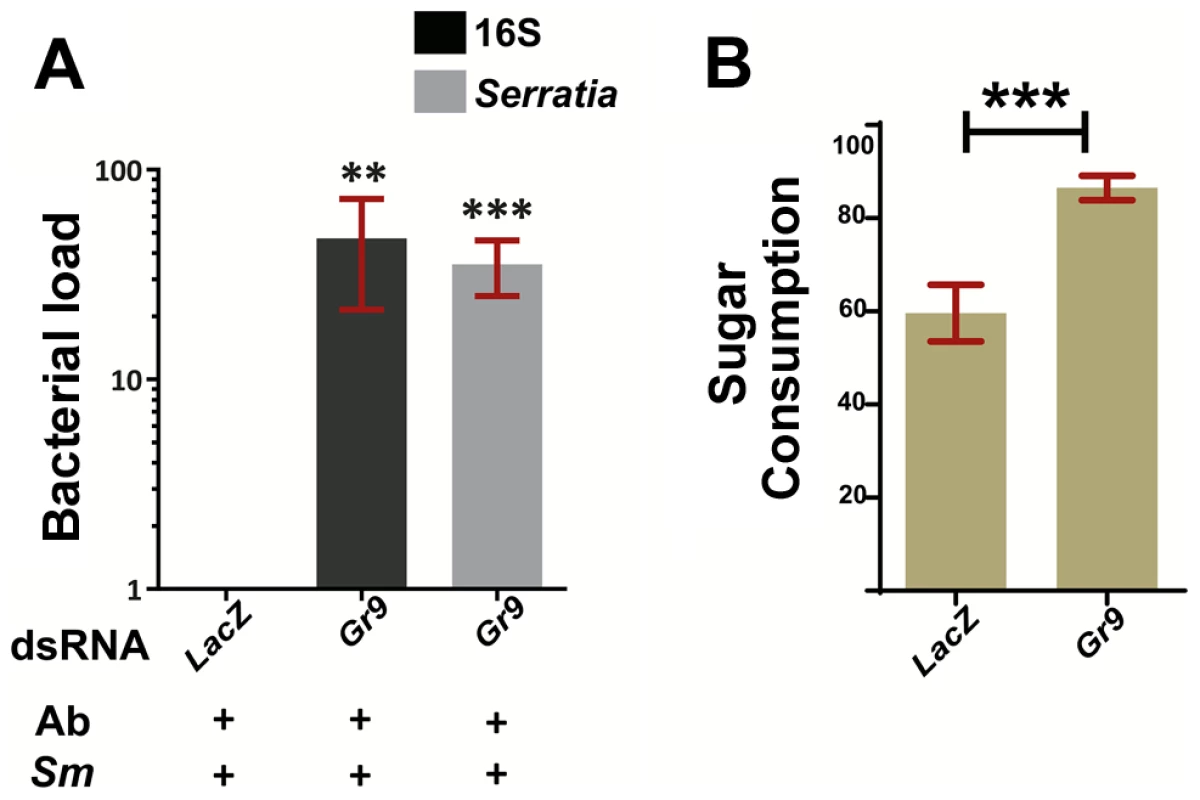
5A: The bacterial load of antibiotic treated mosquitoes orally infected with S. marcescens (Ab+Sm+), treated either with Gr9 dsRNA or the dsLacZ control was determined at day 5 post infection either with broad range 16S or Serratia-specific primers. The average ±SEM of the bacterial fold-increase is shown, compared to dsLacZ treated mosquitoes over 5 independent infections, with the qRT-PCR performed at least twice for each infection. 5B: Antibiotic treated mosquitoes were starved overnight and then offered a sugar meal through a 5 µl capillary. Sugar meal consumption was determined 16 hours later for 38 LacZ and 55 Gr9 dsRNA treated mosquitoes. The average ±SEM percentage of sugar consumption through the capillary for each mosquito is shown for LacZ and Gr9 dsRNA treatments. In panel 5A, asterisks indicate significance in a one-sample t-test against zero using the log2-transformed fold-change values while in panel 5B asterisks indicate significance in the non-parametric Mann-Whitney test. Two asterisks indicate a p-value<0.005 while three asterisks indicate a p-value<0.0005. We next examined the possibility that the Gr9 antibacterial effect is related to changes in mosquito feeding behavior. Based on the Gr9 many-to-many orthologous relationship with the Drosophila Gr32a, Gr39a and Gr68a, it would be more likely that any Gr9 effects on mosquito behavior would be exerted through the recognition of mosquito-induced or bacterial-derived molecules rather than nutrient sensing [83]–[85], [87].Therefore we first examined the possibility that Gr9 mediates aversion to bacteria-containing sugar thus limiting sugar meal uptake upon oral S. marcescens infection. A two-choice preference assay, in which mosquitoes were offered to feed from a capillary that contained S. marcescens and another that contained only sugar, indicated that there was no significant difference due to Gr9 silencing in consumption between the two capillaries (Figure S5).
Another possibility that could explain the Gr9 antibacterial effect is that Gr9 modulates meal size irrespective of the presence of bacteria. Antibiotic treated mosquitoes were starved and then offered a sugar meal. Consumption was determined 16 hours later in LacZ or Gr9 dsRNA treated mosquitoes (Figure 5B). Indeed, Gr9 silencing resulted in a significant 1.45-fold increase in meal size compared to dsLacZ treated mosquitoes. An increased meal size could result in higher S. marcescens uptake following oral infection, thus contributing to the precipitous increase in S. marcescens load following Gr9 silencing. Our data suggest that Gr9 influences feeding behavior by triggers that do not rely on the presence of bacteria. As the presence of S. marcescens does not seem to affect sugar uptake following Gr9 silencing, there is no reason to assume that the presence of S. marcescens influences the Gr9 effect on meal size, although Gr9-independent aversion circuits could conceivably taper overall consumption.
Transcriptional responses following S. marcescens infection
To examine the relationship between genes identified in the population genetics analysis to be associated with the S. marcescens infection outcome and infection-induced transcriptional responses, we used DNA microarrays to monitor the transcriptional profile of mosquito guts 3 days post infection with S. marcescens added to the sugar meal. Uninfected mosquitoes, which were also treated with antibiotics, were used as controls. Three independent replicate infections were performed. Overall, 55 and 44 transcripts were found to be up and down regulated by at least 1.75-fold, respectively, with 38 and 28 respective up or down regulated transcripts yielding a significant p-value in a t-test against zero, where zero corresponds to no transcriptional regulation (Figure 6A and Table S7). Functional classification of all 97 differentially regulated genes, accounting for multiple transcripts of the same gene, identified serine-type endopeptidases and protein/receptor binding as the most represented classes (Figure 6B). The protein/receptor binding functional class comprised 12 members, including several up or downregulated FREPs, zinc finger containing proteins, PGRPLC and the complement factor regulator LRIM1, which has been previously shown to be regulated by the IMD/REL2 pathway [113]. The oxidoreductase class comprised 7 members, including two P450 cytochromes, possibly involved in detoxification [114], the hydrolase class included a glycoside hydrolase and the nucleotide metabolic process class included 5 heat shock proteins, likely to be involved in stress responses [115]. The antimicrobial peptide LYSC2, showing the highest 3.44-fold upregulation of all genes, has been previously shown to be upregulated following a bacterial challenge [116].
Fig. 6. Transcriptional regulation following S. marcescens infection using DNA microarrays. 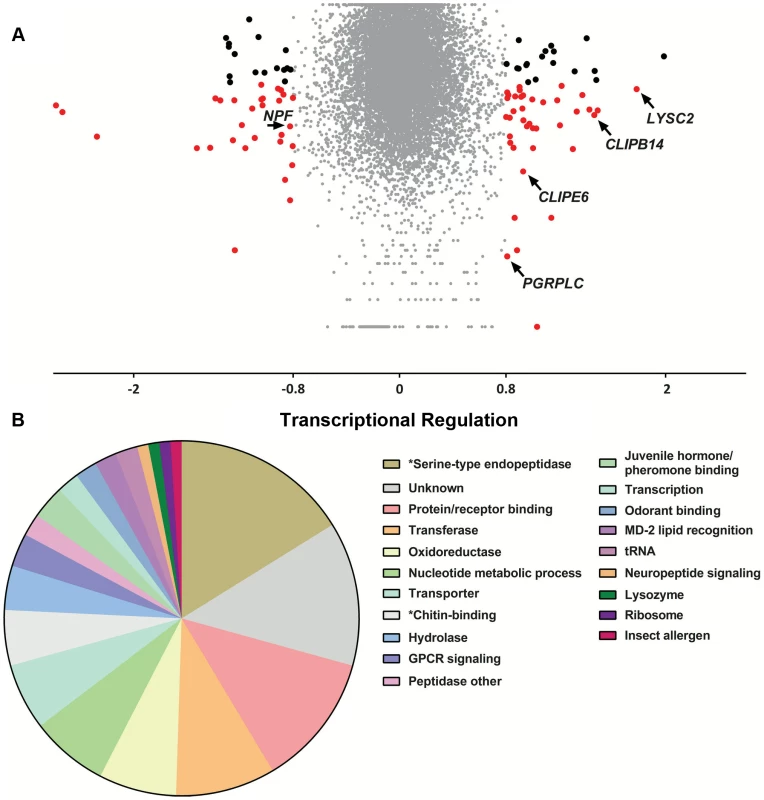
Antibiotic treated mosquitoes were orally infected with S. marcescens and, 3 days post infection, transcriptional regulation in the gut of bacteria-fed mosquitoes was determined using DNA microarrays, compared to uninfected mosquitoes further antibiotic treated for 3 days. 6A: Volcano plot of transcriptional regulation as determined over 3 independent infections. The log2-transformed fold-change values for each transcript, as determined by two probes for each of the three arrays, were used for a one-sample t-test against zero, where zero corresponds to no regulation. Transcripts with more than 1.75-fold regulation are indicated either by black dots if the p-value of the t-test is >0.05 or red dots if the p-value is <0.05. Transcripts corresponding to LYSC2, PGRPLC, CLIPE6, CLIPB14 and NPF are indicated by arrows. 6B: Functional classification of more than 1.75-fold regulated genes. The 97 genes with more than 1.75-fold regulation were assigned to a functional class based on assigned GO terms, InterPro-predicted domains or Drosophila orthologs. The pie chart shows the proportion of genes assigned to each functional class. Functional classes corresponding to significantly overrepresented GO terms are indicated by asterisks. A hypergeometric test followed by Benjamini-Hochberg correction was used to determine enriched GO terms in the set of 97 genes. The results identified 16 GO terms that were significantly overrepresented, most of which were related to just two functional classes: serine-type endopeptidases and chitin-binding genes (Figure S6 and Table S8). In total, 16 serine-type endopeptidase genes were differentially regulated, including CLIPE6, which was also associated with the outcome of infection, CLIPB14 that has been implicated in defense against Gram-negative bacteria [57], [117], CLIPB17 and CLIPB20. The group of chitin-binding genes comprised 5 members, including the gene encoding the scavenger receptor SCRASP1, previously shown to be upregulated following bacterial infection and bind chitin [117], [118] and two downregulated peritrophic matrix components identified by a previous proteomic analysis [119]. Chitin-binding genes are upregulated following oral bacterial infection in Drosophila [26], with one member participating in barrier formation that protects against oral S. marcescens infection [120], while their suggested role in mosquitoes is recognition of danger signals following tissue remodeling due to a bacterial infection [118].
Several transcriptionally regulated genes suggested a mosquito behavioral response following S. marcescens infection (Table 2). Among these genes, NPF was downregulated after infection. NPF is expressed in the midgut of Drosophila [121] and Aedes aegypti [122], and has been implicated in modulation of feeding behavior in Drosophila [30], aversion to noxious food [123] as well as in alcohol sensitivity [80] and regulation of reward systems [81]. It has been also linked to food signaling by integrating sugar gustatory stimuli [124] and behavioral immune responses against endoparasitoid wasps, by mediating oviposition behavior [125].
Tab. 2. Transcripts related to mosquito behavior regulated more than 1.75-fold following S. marcescens oral infection. 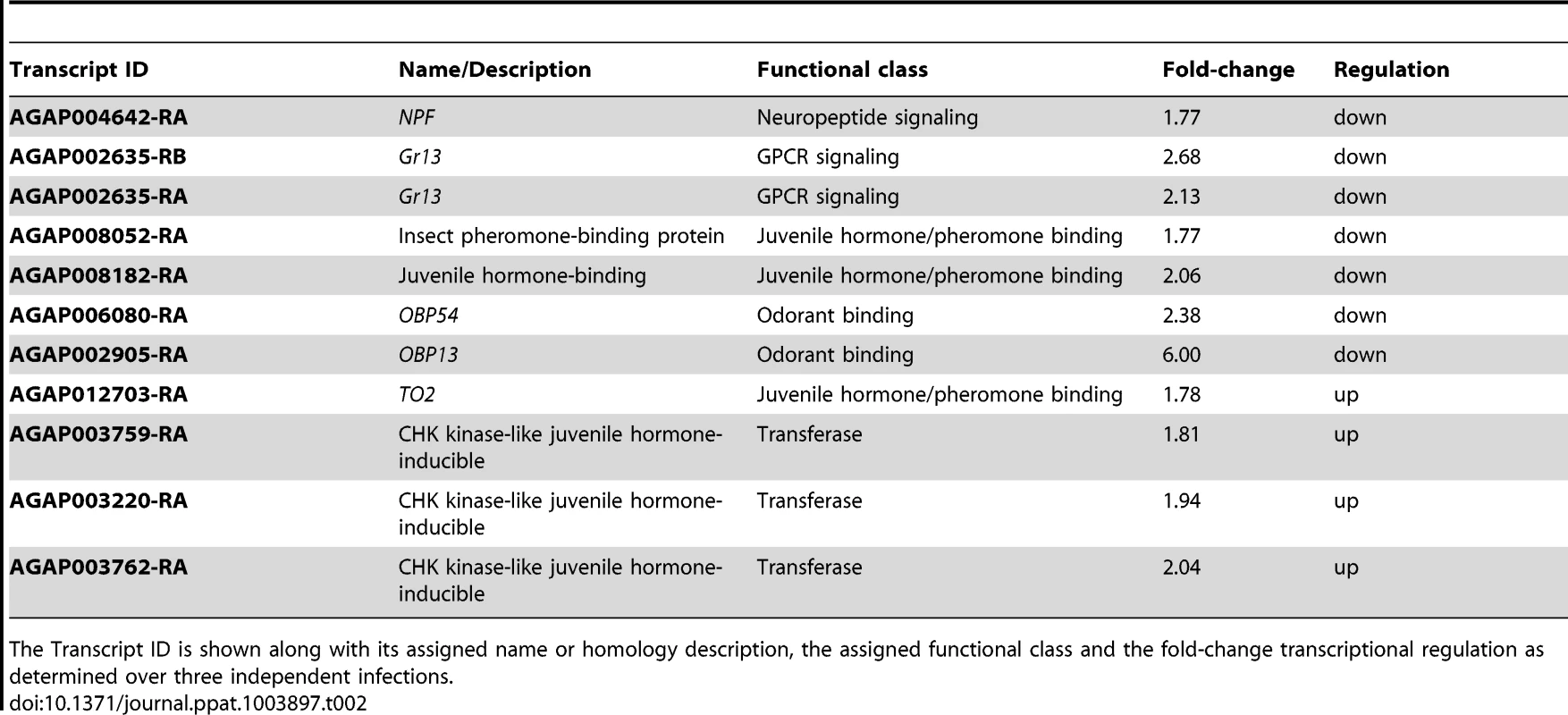
The Transcript ID is shown along with its assigned name or homology description, the assigned functional class and the fold-change transcriptional regulation as determined over three independent infections. Additional behavior-related genes that were transcriptionally regulated following S. marcescens infection included the gustatory receptor Gr13 with two downregulated transcripts, three upregulated juvenile hormone-inducible kinases, two downregulated genes encoding a pheromone and a juvenile hormone binding protein and the downregulated odorant binding protein genes OBP13 and OBP54. Juvenile hormone circuits are known to affect gustatory perception and feeding behavior in various organisms including Ae. aegypti [126]–[129], while pheromone and olfaction circuits are also known to affect mosquito behavior [130], [131]. The gene encoding the juvenile hormone binding protein TO2 (takeout2), was also upregulated following S. marcescens infection and may also participate in behavioral responses, as its Drosophila homolog, takeout, is known to regulate feeding behavior [132], [133].
Gr9 modulates S. marcescens infection via NPF
We examined a possible link between the observed downregulation of the known modulator of feeding behavior, NPF, following S. marcescens infection and the role of Gr9 in modulating the S. marcescens infection outcome. The Gr9 ortholog Gr39a has been previously shown to be expressed in the Drosophila midgut and co-localize with NPF in enteroendocrine cells [134], raising the possibility of a functional link between these two genes. In mosquitoes orally infected with S. marcescens, NPF expression showed a significant 1.8-fold increase following Gr9 silencing compared to dsLacZ treated controls (Figure 7A). This modulation of NPF expression following Gr9 silencing suggested that NPF expression may mediate the observed Gr9 antibacterial effect. Therefore, we further examined whether silencing NPF can affect the increase of S. marcescens observed in Gr9 kd mosquitoes. Indeed, concomitant silencing of Gr9 and NPF resulted in a significant 10-fold decrease in infection load, compared to Gr9 silencing alone (Figure 7B).
Fig. 7. The Gr9 antibacterial effect mostly relies on changes in NPF expression. 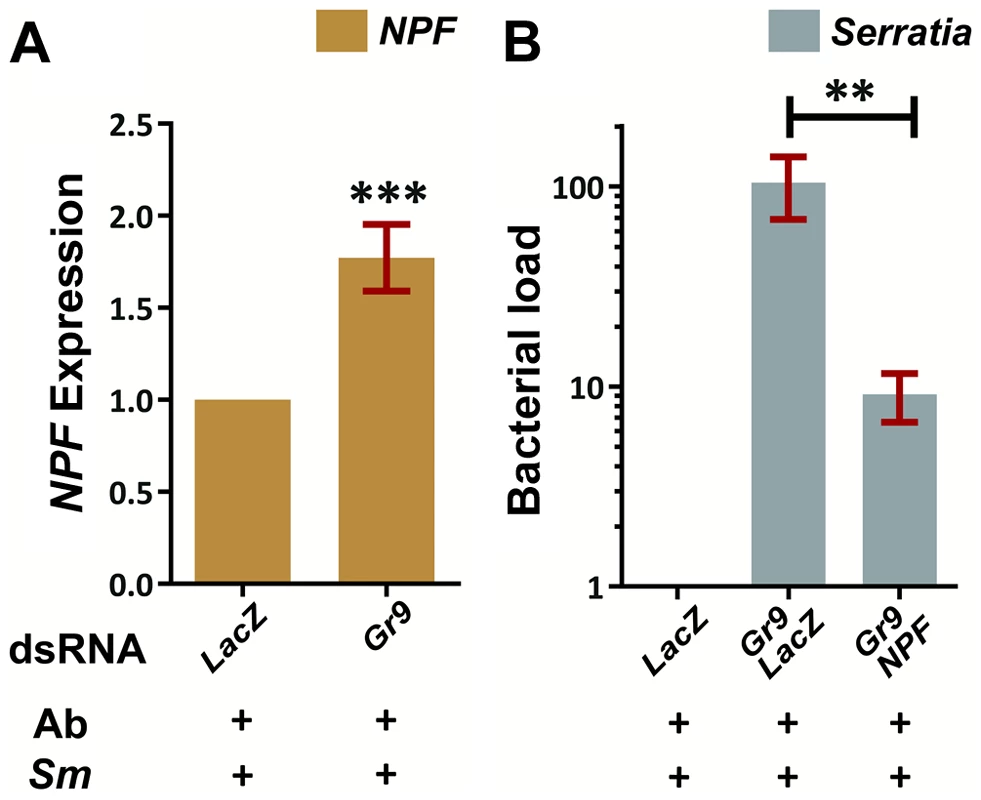
7A: Antibiotic treated mosquitoes orally infected with S. marcescens (Ab+Sm+) were dissected 5 days post infection and the NPF levels of mosquitoes treated either with LacZ or Gr9 dsRNA were determined in their guts. The average ±SEM of the NPF fold-increase as determined over 8 independent infections, with the qRT-PCR performed at least twice for each infection, can be seen. 7B: The bacterial load of antibiotic treated mosquitoes orally infected with S. marcescens (Ab+Sm+) treated either with LacZ dsRNA or with a 50–50% mix of either Gr9 and LacZ or Gr9 and NPF dsRNA was determined and normalized to the levels of dsLacZ treated mosquitoes. The average ±SEM of the bacterial fold-increase as determined over 3 independent infections can be seen, with the qRT-PCR performed at least twice for each infection. In panel 7A asterisks indicate significance in a one-sample t-test against zero using the log2-transformed fold-change values while in panel 7B asterisks indicate significance in the non-parametric Mann-Whitney test. Two asterisks indicate a p-value<0.005 while three asterisks indicate a p-value<0.0005. Taken together, our data suggest a behavioral immune response involving Gr9, mostly relying on changes of NPF expression. One hypothesis is that Gr9 activation in the midgut, most likely through a mosquito-induced cue, tapers the expression of NPF, resulting in feeding suppression that limits the mosquito meal size and thus the abundance of ingested S. marcescens. Gr9 variants that influence the efficiency of this suppression may lead to enhanced feeding which, depending on the efficiency of the epithelial response to handle the infection, can influence the outcome of S. marcescens infection, thus explaining the observed Gr9 association with the S. marcescens infection phenotype.
Discussion
The rapidly evolving and adapting mosquito species have become tractable systems for genetic association studies that could yield important information about vector/parasite interactions leading to malaria transmission [135]. Previous studies have focused on the outcome of Plasmodium infections, using laboratory or field mosquitoes and genetic tools such as microsatellite markers and targeted SNP loci genotyping [1], [3], [136]. These studies have not considered the effect of gut bacteria on the outcome of Plasmodium infections, which has been revealed recently [5], [16]–[18]. Furthermore, the influence of associated complement factors on natural P. falciparum infections remains questionable [137]. Indeed, the presence of Enterobacteriaceae, such as S. marcescens, a common member of the mosquito gut flora, has been correlated with P. falciparum susceptibility in field mosquito populations [12], while intraspecific variation within S. marcescens populations also is shown to affect the Plasmodium infection load [37]. Therefore, genome-wide studies to determine factors that modulate the levels of mosquito gut bacteria can provide novel insights into how midgut bacteria affect the outcome of Plasmodium infection and hence malaria transmission.
The unprecedented level of detail achieved in the population genetics analysis presented here in identifying SNPs associated with the outcome of S. marcescens infection is a result of the strong evolutionary drive exerted by gut bacteria on mosquito genetic variation, the use of a high-resolution SNP genotyping array and the use of a recently established laboratory colony of An. gambiae which retains genetic variation found in field populations but also shows elevated linkage disequilibrium due to colonization bottlenecks. This population homogeneity can facilitate gene discovery as shown in human genome-wide association studies in isolated populations [138], [139].
A dual implication can be inferred for genes associated with the S. marcescens infection phenotype; they are putatively involved in shaping the infection outcome, while their level of involvement may also be affected by genetic variation within the mosquito population. It is possible that identified associations are the result of causal polymorphisms such as gain or loss of function mutations in coding or regulatory sequences or the result of allele combination in several genetic loci which shapes the outcome of infection through synergism, epistatic interactions or redundant function. In any of the latter cases, a reverse genetics approach may not be capable of capturing such interactions.
The involvement of the three FN3Ds in the outcome of Serratia infection reveals a novel function of this family in modulating the load and composition of the mosquito gut microbiota and opens new avenues in investigating the complexity of such responses and possible synergisms with known antibacterial pathways such as the IMD/REL2. The three FN3D genes identified here emerge as major modulators of the bacterial population structure in the mosquito gut, limiting the representation of Enterobacteriaceae, mainly Serratia or strains with similarity to Serratia reference sequences, but also, for FN3D3, bacteria of the genus Burkholderia. As shifts in gut microbiota population structure can elicit gut pathology [28], [140], while Serratia can influence the outcome of Plasmodium infection [37], FN3Ds can play critical roles in gut homeostatic interactions and malaria transmission dynamics.
Further insights into the FN3D mode of action remain to be determined. Our data showing that the knockdown effects of FN3Ds may be limited to Serratia or to a fraction of the microbiota raise intriguing questions about the specificity of bacterial recognition in the mosquito gut. The homology of FN3D2 with the hypervariable pattern recognition receptor Dscam opens the possibility that the specific pathogen recognition shown for AgDscam [47] concerns a broader family of FN3Ds, equipping mosquitoes with the capacity for specific recognition resembling that of animals possessing adaptive immune systems. The phylogenetically unrelated FN3D2 and FN3D3 share a similar domain architecture comprising immunoglobulin and FN3 domains, as is the case with Dscam. The identification of FN3D2 and FN3D3 as being both associated with the outcome of S. marcescens infection and exhibiting discrete but similar phenotypic characteristics in modulating the bacterial population structure in the mosquito gut, parallels the discrete but similar functions of the phylogenetically unrelated Dscam, Frazzled and Roundabout in Drosophila axon guidance, with all three receptors sharing immunoglobulin and FN3 domains [141]–[144]. FN3D1 has a distinct domain architecture with an FN3 domain, while its orthologous relationship with Drosophila windei [145] and sequence similarity with the activating transcription factor 7 - interacting protein [146], suggest a role in regulating gene expression.
The identification of An. gambiae genes involved in immune responses against bacteria and/or Plasmodium has been largely based to date on studies that combine bioinformatic identification of known immunity gene homologs and transcriptional profiling of genes following a pathogen challenge. This approach, however, has the limitation of the a priori assumption that genes of interest show significant change in transcriptional regulation, mostly induction, which is true for most effectors, but not all genes, for example pattern recognition receptors or transcription factors. In addition, it is possible that even strong changes in transcriptional regulation are the consequence of the infection rather than part of the response. Especially for quantitative traits within mosquito populations, such as Plasmodium infection intensity, different infection intensities can correlate with variable transcriptional responses [70], while the underlying genetic variation further complicates the observed transcriptional regulation.
The microarray approach adopted here has identified a limited set of 99 differentially regulated transcripts following oral S. marcescens infection. The number of regulated transcripts is consistent with that of a previous microarray-based comparison of antibiotic treated and untreated mosquitoes, which showed differential expression for 185 transcripts [16], attributing this limited transcriptional regulation to symbiotic relationships that have led to adaptation of commensal bacteria. A much broader set of differentially expressed genes has been identified following oral bacterial infections in Drosophila [26], [32]. This is most likely due to differences in gene pool diversity between the genetically homogeneous fly lines and the recently established mosquito laboratory colony used here, which retains considerable genetic variation thus enabling the SNP genotyping analysis. The different levels of infection seen between mosquitoes (high, low and no infection), which are largely attributed to genetic variation within the colony population, are most likely linked to differences in the mosquito transcription profiles that are averaged out in our study design. Therefore, our analysis identifies transcripts with the most pronounced and consistent differential expression, comprising the core response to S. marcescens infection. Future studies investigating the transcription profile of highly, lowly or non-infected mosquitoes are most likely to reveal components of transcriptional regulation that lead to the respective outcome of infection. Indeed, genes identified to show prominent differential expression after bacterial challenge in previous studies also showed transcriptional regulation following oral S. marcescens infection, including CLIPB14 [117], LRIM1 [63], [113], LYSC2 [116] and SCRASP1 [117], [118].
The identification of diverse transcriptional responses to different bacteria in Drosophila [32] along with the specificity of mosquito responses to a subset of bacteria, as suggested by the SNP genotyping analysis presented here, may explain the surprisingly little overlap between differentially expressed genes following S. marcescens infection and antibiotic treated vs. untreated mosquitoes [16]. Remarkably, however, consistency is seen in gene families present in both datasets, including CLIPs, chitin-binding genes, homeobox genes, PGRPs and FREPs, suggesting that similar defense strategies are employed, which are customized for each type of infection through utilization of different gene family members.
The approach we adopted here to identify genes involved in mosquito gut infection with S. marcescens combines transcriptional profiling of infected guts with the identification of SNPs segregating between phenotypic pools, whereby an association implies contribution to the outcome of infection, while the study design incorporates variation that leads to different observed phenotypes. This approach addresses some of the aforementioned shortcomings but introduces others, as it cannot capture genes with redundant functions, genes with additional housekeeping functions or a role during development, of which variants are eliminated from the population, or genes with rare variants that are not in the variation pool of our colony. Furthermore, an association may be the result of a selective sweep in the proximity of the gene that creates linkage disequilibrium and leads to SNP divergence between the phenotypic pools. Therefore, although each of the approaches cannot provide by itself a complete picture, the combination of the two can provide novel insights into the mosquito gut responses to S. marcescens.
The comparison between the datasets of transcriptionally regulated genes and genes associated with the outcome of S. marcescens infection shows limited overlap, with only PGRPLC and CLIPE6 found in both datasets. Again, considerable overlap is detected in identified gene families, which are represented by different members in each dataset. These include acyl-transferase, glycoside hydrolase, kinase, GPCR, LRIM, homeobox, zinc-finger, PGRP, peptidase, FREP, MD2-like and chitin-binding genes. Interestingly, a previous study investigating differential expression following a bacterial challenge in mosquito immunoglobulin-containing genes failed to identify significant regulation for FN3D2 or FN3D3 [147], strengthening the case for the complementarity of the SNP genotyping and expression analysis approaches. The specific role of gene family members, especially those showing considerable expansion in Anopheles, e.g. FREPs [68], [148], remains unclear. Therefore, SNP genotyping reveals a different set of candidate genes involved in antibacterial immunity while at the same time it is intriguing to postulate whether this divergence between associated and differentially expressed genes within each gene family constitutes a functional divergence between them.
A novel finding stemming from this combinatorial approach is a mosquito behavioral response to S. marcescens infection that involves Gr9 signaling and is mediated by changes of NPF expression. Although Gr9 orthologs in Drosophila recognize chemosensory cues and mediate aversive behaviors [83], [84], [87], surprisingly, Gr9 appears to suppress feeding irrespective of the presence of bacteria. One explanation is that the Gr9 antibacterial effect relies on its expression in the midgut rather than external sensory organs, where the role of its Drosophila counterparts has been studied. The role of gustatory receptor midgut expression [134] remains poorly understood and could involve detection of nutrients or host-derived molecules that triggers downstream responses. The role of NPF midgut expression [121], [122] also remains poorly understood. NPF downregulation following S. marcescens infection implies its involvement in an aversion circuit triggered by the presence of S. marcescens, with a possible NPF role in integrating aversion and satiation signals that lead to feeding suppression. Such NPF involvement remains to be further investigated, in conjunction with the involvement of other genes related to mosquito behavior which were either associated with the outcome of S. marcescens infection or were transcriptionally regulated following infection. These include Gr13, downregulated following S. marcescens infection but also three neurotransmitter-triggered GPCRs, associated with the outcome of infection, pointing to complex behavioral circuits involved in antibacterial responses, which are yet to be revealed.
The identification of FN3Ds as well as Gr9 and NPF in responses affecting the outcome of S. marcescens infection, in addition to known responses including the IMD/REL2 and DUOX pathways, suggests that the response to gut infection is the result of a complex molecular interplay. Both the SNP genotyping and expression analysis suggest that the mosquito response to oral S. marcescens infection involves two discrete but inextricably linked modes of defense, a behavioral and an epithelial immune response. A behavioral immune response involving Gr9 and NPF can limit or disrupt pathogen intake, a defense conceptually similar to barrier responses that inhibit pathogen contact with triggers of epithelial or systemic immune responses. An impaired behavioral response, e.g. due to Gr9 variants that affect feeding behavior, can decisively influence the efficacy of the epithelial response and thus the infection outcome. This implies a threshold after which epithelial immunity cannot efficiently handle the pathogen load, an aspect of immunity that remains poorly understood. Nevertheless, in mosquito infections with Plasmodium parasites, the intensity of infection has been correlated with the efficacy of different components of the IMD/REL2 pathway, suggesting that different effectors may be deployed in low, mid or high intensity parasite infections [4].
As pathogen abundance most likely relies on feeding behavior, the interplay between behavioral and epithelial immunity can shape both responses. Our implementation of a model of natural bacterial infections through the oral route integrated both behavioral and epithelial responses and not only revealed the previously unknown behavioral component but also allowed the study of aspects of epithelial immunity that, by being infection intensity dependent, possibly rely on the behavioral component. This integrative approach to behavioral and epithelial immunity can be further employed to reveal aspects of this interplay that may involve regulation of behavioral responses by host-derived factors induced by the epithelial component. This implies that the study of behavioral immunity alone may be insufficient to uncover some aspects of its biological consequences.
In Drosophila, a balance between immune response and tolerance, achieved by various Imd regulators, largely shapes the gut microbiota population structure, although the only known elicitor of such responses is DAP-type peptidoglycan, common to all Gram-negative bacteria [28], [149], [150]. A similar mechanism has been suggested for mosquitoes through alternative splicing of the modular IMD/REL2 pathway receptor PGRPLC that leads to production of positive and negative pathway modulators [5], [42]. Indeed, utilization of alternative splicing as a mechanism to derive new immune functions and increase the specificity of pathogen recognition by the mosquito innate immune system has been described for the FN3D2 homolog, Dscam [43], [44], [47]. Whether the Enterobacteriaceae-specific effect of FN3D2 knockdown is due to specific recognition and activation of highly specialized or targeted effector reactions remains to be investigated. Furthermore, the significance of alternative splicing suggested for Gr9 [82] remains to be determined along with the cue that triggers its antibacterial effect, and could also involve recognition of, most likely, host-derived signals. In addition, recognition of differentially produced metabolites after infection as shown for the DUOX pathway [29] could further increase the specificity in antibacterial responses. Whether PAMPs (bacterial-derived) or DAMPs (host-derived), such metabolites can be recognized by gustatory receptors triggering specific antibacterial responses, which together with FN3Ds and the rather generalist response of the IMD/REL2 pathway can shape the load and composition of the mosquito gut microbiota. In conclusion, our findings suggest that mosquitoes can mount a much more complex and specific antibacterial response than previously thought, which not only contributes to fending off intestinal bacterial infections but also to achieving homeostasis of the complex gut ecosystem.
Materials and Methods
Ethics statement
This study was carried out in strict accordance with the United Kingdom Animals (Scientific Procedures) Act 1986. The protocols for maintenance of mosquitoes by blood feeding were approved and carried out under the UK Home Office License PPL70/7170. The procedures are of mild to moderate severity and the numbers of animals used are minimized by incorporation of the most economical protocols. Opportunities for reduction, refinement and replacement of animal experiments are constantly monitored and new protocols are implemented following approval by the Imperial College Ethical Review Committee.
Mosquito rearing and maintenance
The N'gousso strain of An. gambiae was used. This is an M form strain colonized in 2006 [1] and kept in large numbers to retain genetic variation. Rearing and maintenance of the strain was performed as described previously [151]. Mosquitoes were collected after emergence and kept on a cocktail of 25 µg/ml gentamicin, 10 µg/ml penicillin and 10 units/ml streptomycin, diluted in 10% D-(-)-Fructose (Sigma). This antibiotic treatment regime was used for 5 days, with the antibiotic solution refreshed every 24 hours. At day 5 post emergence, the antibiotic solution was replaced by dH2O and mosquitoes were starved overnight prior to oral bacterial infection.
Mosquito oral infection with S. marcescens or Asaia
We used the S. marcescens Db11-GFP strain, modified to be GFP-fluorescent and resistant to tetracycline and carbenicillin [152]. S. marcescens glycerol stock was grown in 5 ml LB cultures containing 50 µg/ml tetracycline and carbenicillin (Sigma) at 37°C. Following overnight incubation, the cultures were expanded to 100 ml and further incubated overnight at 37°C. OD600 and GFP fluorescence (excitation/emission at 485/520 nm) were then measured to ensure cultures maintained GFP fluorescence, using the Fluostar Omega spectrophotometer (BMG Labtech). Bacterial pellets following centrifugation at 2500 rpm for 5 minutes were washed twice with PBS and resuspended in such volume of 10% D-(-)-Fructose, so that 1 ml of the bacteria-containing sugar solution corresponded to OD600 = 0.1 of the initial 100 ml culture. The sugar solution was further diluted 1∶12 in a 10% D-(-)-Fructose solution that contained tetracycline and carbenicillin at 50 µg/ml and 5% v/v of scarlet dye (Langdale). Mosquitoes were fed with this solution for 2 days. Subsequently, mosquitoes fed with bacteria-containing sugar were separated based on the presence of the dye in their gut and kept on 10% D-(-)-Fructose containing tetracycline and carbenicillin at 50 µg/ml.
Oral infections with Asaia were conducted in a similar manner. The Asaia SF2.1 (GFP) strain was used, grown as previously described [153] and maintained in 50 µg/ml kanamycin (Sigma).
Fluorescence microscopy
The levels of S. marcescens infection were determined by microscopic observation of dissected midguts immersed in Vectashield mounting medium (Vecta), immediately after dissection. The Zeiss Axiophot fluorescence microscope was used, equipped with light and GFP filters while photos of observed midguts were taken with the Axiocam HRc and Axiovision software (Zeiss).
SNP genotyping arrays
All carcasses corresponding to midguts of S. marcescens infected mosquitoes were kept numbered in 96-well plates immersed in 75% ethanol at −80°C. Carcasses from selected midguts were used for gDNA extraction using the QIAquick Blood and Tissue kit (QIAGEN). Subsequently, gDNA concentrations were determined using the Picogreen dsDNA kit (Invitrogen) and equimolar gDNA quantities from each mosquito were pooled. The design and validation of the SNP genotyping array used along with the treatment of gDNA pools, hybridization, calling of SNP genotypes and measurement of differentiation in each pooled hybridization between allele A and B have been described previously [39], [154]. The frequency of designated allele A was considered as the minor allele frequency and was used to measure the difference between pooled hybridizations. The permutation analysis used has been described previously [39], with a modified length of non-overlapping 10-SNP windows. Determination of genes residing in identified genomic areas and homology analysis was performed using Biomart 0.7 and the AgamP3.7 An. gambiae gene annotation [155]. The SNP genotyping array datasets have been deposited to ArrayExpress under the experiment name Serratia_SNP1 and accession number E-MEXP-3951.
DNA microarrays
Total RNA was extracted from midguts using the Trizol reagent (Invitrogen), and treated with Turbo DNAse I (Ambion). Samples were further purified using the RNeasy kit (QIAGEN). Quantification was performed using the Nanodrop 1000 spectrophotometer (Thermo Scientific) and RNA integrity was assessed using the RNA 6000 Pico Chip kit (Agilent). Labeling and hybridization were performed using the Low Input Quick Amp Labeling kit for two-color microarray based expression analysis (Agilent). We used Agilent custom 4×44 k gene expression microarrays. The microarray design Pfalcip_Agamb2009 (A-MEXP-2324) comprises oligonucleotide probes encompassing all An. gambiae annotated transcripts of the AgamP3.6 release along with P. falciparum probes, with each probe represented in duplicate. Slides were scanned using the Genepix 4000B scanner equipped with the Genepix Pro 6.1 software (Axon instruments).
All dataset files were normalized using the Genespring 11.0 GX software (Agilent). The Lowess normalization method was used while the threshold of raw signals was set to 5, which was sufficient to eliminate background regulation of P. falciparum probes. Further analysis of transcriptionally regulated genes and GO analysis was performed using the Genespring 11.0 GX software. For GO analysis, GO accession numbers for all An. gambiae transcripts were obtained using Biomart 0.7 and a hypergeometric test with Benjamini-Hochberg correction was performed on the set of more than 1.75-fold regulated genes. The corrected p-value for testing multiple GO accession numbers for their significance was set to 0.1. The log2-transformed transcriptional regulation for each transcript was extracted from the normalized datasets for each of the two probes corresponding to each transcript and the obtained values from all three independent infections were used in a t-test against zero, with a p-value cut-off of 0.05. The DNA microarray datasets have been deposited to ArrayExpress under the experiment name Serratia_infections and accession number E-MEXP-3952.
Bacterial load following RNAi-mediated silencing
Mosquitoes were treated with the respective dsRNA at the day of emergence, as described previously [156]. For each targeted gene or the dsLacZ control, dsRNA was synthesized using the T7 Megascript kit (Invitrogen) and further purified using the RNeasy kit (QIAGEN) to a concentration of 3 µg/µl. For each T7 Megascript reaction, 1 µg of purified PCR product was added, derived using the T7 primer sets shown in Table S9, using An. gambiae cDNA as template.
Mosquitoes were surface sterilized by immersing them in 70% ethanol for 30 seconds and washing them twice in PBS and midguts were dissected in RNA later (Invitrogen). Total RNA from mosquito midguts was extracted after homogenization with a pestle motor in RNA later using the RNeasy kit (QIAGEN). cDNA was synthesized from total RNA using the QuantiTect Reverse Transcription kit (QIAGEN).
Quantification of bacterial load or the efficiency of RNAi-mediated silencing was performed using qRT-PCR with the respective primers shown in Table S9. In a 20 µl reaction of Fast SYBR Green Master Mix (Applied Biosystems), 1 µl of cDNA template and 2 µl of each respective primer at a 0.5 to 9 µM concentration, optimized for each primer set, were added. The 7500 Real-Time PCR System (Applied Biosystems) was used with its respective software to perform the reaction and any further analysis. The relative abundance of each sample was determined using the standard curve method as described in User Bulletin #2 for the ABI Prism 7700 Sequence Detection system (Applied Biosystems) in which the housekeeping AgS7 gene was used as an endogenous control.
454 pyrosequencing
cDNA pools were amplified with the GO Taq DNA polymerase (Promega) using the 16S V4–V6 primers shown in Table S9 and suitable barcode sequences and purified using PCR purification and Gel Extraction kits (QIAGEN). PCR products were sequenced by Beckman Genomics (Grenoble, France) using the Roche 454 GS FLX+ and standard procedures. The resulting FASTA files were filtered to a minimum read length of 250 bp using Galaxy [157] and blasted against the NCBI 16SMicrobial database using BLAST+ and prfectBLAST [158] with standard blastn algorithm settings and 10 maximum target sequences. Further analysis was performed using MEGAN4 [159].
Meal size and two-choice preference assays
Sugar meal size was determined through a modified capillary feeder assay [160]. Mosquitoes treated with LacZ or Gr9 dsRNA were antibiotic treated for 5 days, starved overnight and, subsequently, individual mosquitoes were fed on a 5 µl glass capillary (VWR) containing 10% D-(-)-Fructose and 5% v/v scarlet dye. For alive mosquitoes, sugar consumption was determined 16 hours later through the reduction of sugar solution in each capillary. The two-choice preference assay was also conducted based on a previously described capillary feeder assay [160]. Mosquitoes treated with LacZ or Gr9 dsRNA were antibiotic treated for 5 days, starved overnight and placed in pools of 8–11 mosquitoes. Mosquitoes were offered to feed from two capillaries, one containing a sugar solution as above and one also containing S. marcescens, prepared as described above for oral infection. Water-containing cotton pads were also used and pools with mosquito mortality were disregarded. 16 hours later, consumption for each capillary was determined based on the reduction of the sugar solution.
Supporting Information
Zdroje
1. HarrisC, LambrechtsL, RoussetF, AbateL, NsangoSE, et al. (2010) Polymorphisms in Anopheles gambiae immune genes associated with natural resistance to Plasmodium falciparum. PLoS Pathog 6: e1001112.
2. WhiteBJ, LawniczakMK, ChengC, CoulibalyMB, WilsonMD, et al. (2011) Adaptive divergence between incipient species of Anopheles gambiae increases resistance to Plasmodium. Proc Natl Acad Sci U S A 108 : 244–249.
3. RiehleMM, MarkianosK, NiareO, XuJ, LiJ, et al. (2006) Natural malaria infection in Anopheles gambiae is regulated by a single genomic control region. Science 312 : 577–579.
4. GarverLS, BahiaAC, DasS, Souza-NetoJA, ShiaoJ, et al. (2012) Anopheles imd pathway factors and effectors in infection intensity-dependent anti-Plasmodium action. PLoS Pathog 8: e1002737.
5. MeisterS, AgianianB, TurlureF, RelogioA, MorlaisI, et al. (2009) Anopheles gambiae PGRPLC-mediated defense against bacteria modulates infections with malaria parasites. PLoS Pathog 5: e1000542.
6. SchnitgerAK, YassineH, KafatosFC, OstaMA (2009) Two C-type lectins cooperate to defend Anopheles gambiae against Gram-negative bacteria. J Biol Chem 284 : 17616–17624.
7. GrossmanSR, AndersenKG, ShlyakhterI, TabriziS, WinnickiS, et al. (2013) Identifying recent adaptations in large-scale genomic data. Cell 152 : 703–713.
8. SangareI, MichalakisY, YameogoB, DabireR, MorlaisI, et al. (2013) Studying fitness cost of Plasmodium falciparum infection in malaria vectors: validation of an appropriate negative control. Malar J 12 : 2.
9. RaniA, SharmaA, RajagopalR, AdakT, BhatnagarRK (2009) Bacterial diversity analysis of larvae and adult midgut microflora using culture-dependent and culture-independent methods in lab-reared and field-collected Anopheles stephensi-an Asian malarial vector. BMC Microbiol 9 : 96.
10. Gonzalez-CeronL, SantillanF, RodriguezMH, MendezD, Hernandez-AvilaJE (2003) Bacteria in midguts of field-collected Anopheles albimanus block Plasmodium vivax sporogonic development. J Med Entomol 40 : 371–374.
11. Gendrin M, Christophides GK (2013) The Anopheles Mosquito Microbiota and Their Impact on Pathogen Transmission. In Manguin S, Ed. Anopheles mosquitoes - New insights into malaria vectors
12. BoissièreA, TchioffoMT, BacharD, AbateL, MarieA, et al. (2012) Midgut Microbiota of the Malaria Mosquito Vector Anopheles gambiae and Interactions with Plasmodium falciparum Infection. PLoS Pathogens 8: e1002742.
13. WangY, GilbreathTM3rd, KukutlaP, YanG, XuJ (2011) Dynamic Gut Microbiome across Life History of the Malaria Mosquito Anopheles gambiae in Kenya. PLoS One 6: e24767.
14. Osei-PokuJ, MbogoCM, PalmerWJ, JigginsFM (2012) Deep sequencing reveals extensive variation in the gut microbiota of wild mosquitoes from Kenya. Mol Ecol 21 : 5138–5150.
15. KumarS, Molina-CruzA, GuptaL, RodriguesJ, Barillas-MuryC (2010) A peroxidase/dual oxidase system modulates midgut epithelial immunity in Anopheles gambiae. Science 327 : 1644–1648.
16. DongY, ManfrediniF, DimopoulosG (2009) Implication of the mosquito midgut microbiota in the defense against malaria parasites. PLoS Pathog 5: e1000423.
17. RodriguesJ, BraynerFA, AlvesLC, DixitR, Barillas-MuryC (2010) Hemocyte differentiation mediates innate immune memory in Anopheles gambiae mosquitoes. Science 329 : 1353–1355.
18. CirimotichCM, DongY, ClaytonAM, SandifordSL, Souza-NetoJA, et al. (2011) Natural microbe-mediated refractoriness to Plasmodium infection in Anopheles gambiae. Science 332 : 855–858.
19. LemaitreB, HoffmannJ (2007) The host defense of Drosophila melanogaster. Annu Rev Immunol 25 : 697–743.
20. KanekoT, GoldmanWE, MellrothP, SteinerH, FukaseK, et al. (2004) Monomeric and polymeric gram-negative peptidoglycan but not purified LPS stimulate the Drosophila IMD pathway. Immunity 20 : 637–649.
21. ChoeKM, LeeH, AndersonKV (2005) Drosophila peptidoglycan recognition protein LC (PGRP-LC) acts as a signal-transducing innate immune receptor. Proc Natl Acad Sci U S A 102 : 1122–1126.
22. MailletF, BischoffV, VignalC, HoffmannJ, RoyetJ (2008) The Drosophila peptidoglycan recognition protein PGRP-LF blocks PGRP-LC and IMD/JNK pathway activation. Cell Host Microbe 3 : 293–303.
23. LhocineN, RibeiroPS, BuchonN, WepfA, WilsonR, et al. (2008) PIMS modulates immune tolerance by negatively regulating Drosophila innate immune signaling. Cell Host Microbe 4 : 147–158.
24. HaEM, OhCT, BaeYS, LeeWJ (2005) A direct role for dual oxidase in Drosophila gut immunity. Science 310 : 847–850.
25. CroninSJ, NehmeNT, LimmerS, LiegeoisS, PospisilikJA, et al. (2009) Genome-wide RNAi screen identifies genes involved in intestinal pathogenic bacterial infection. Science 325 : 340–343.
26. BuchonN, BroderickNA, PoidevinM, PradervandS, LemaitreB (2009) Drosophila intestinal response to bacterial infection: activation of host defense and stem cell proliferation. Cell Host Microbe 5 : 200–211.
27. BuchonN, BroderickNA, KuraishiT, LemaitreB (2010) Drosophila EGFR pathway coordinates stem cell proliferation and gut remodeling following infection. BMC Biol 8 : 152.
28. RyuJH, KimSH, LeeHY, BaiJY, NamYD, et al. (2008) Innate immune homeostasis by the homeobox gene caudal and commensal-gut mutualism in Drosophila. Science 319 : 777–782.
29. LeeKA, KimSH, KimEK, HaEM, YouH, et al. (2013) Bacterial-derived uracil as a modulator of mucosal immunity and gut-microbe homeostasis in Drosophila. Cell 153 : 797–811.
30. HergardenAC, TaylerTD, AndersonDJ (2012) Allatostatin-A neurons inhibit feeding behavior in adult Drosophila. Proc Natl Acad Sci U S A 109 : 3967–3972.
31. AdamoSA (2005) Parasitic suppression of feeding in the tobacco hornworm, Manduca sexta: parallels with feeding depression after an immune challenge. Arch Insect Biochem Physiol 60 : 185–197.
32. ChakrabartiS, LiehlP, BuchonN, LemaitreB (2012) Infection-induced host translational blockage inhibits immune responses and epithelial renewal in the Drosophila gut. Cell Host Microbe 12 : 60–70.
33. LiehlP, BlightM, VodovarN, BoccardF, LemaitreB (2006) Prevalence of local immune response against oral infection in a Drosophila/Pseudomonas infection model. PLoS Pathog 2: e56.
34. MiyamotoT, SloneJ, SongX, AmreinH (2012) A Fructose Receptor Functions as a Nutrient Sensor in the Drosophila Brain. Cell 151 : 1113–1125.
35. MoonSJ, LeeY, JiaoY, MontellC (2009) A Drosophila gustatory receptor essential for aversive taste and inhibiting male-to-male courtship. Curr Biol 19 : 1623–1627.
36. MaslowskiKM, VieiraAT, NgA, KranichJ, SierroF, et al. (2009) Regulation of inflammatory responses by gut microbiota and chemoattractant receptor GPR43. Nature 461 : 1282–1286.
37. BandoH, OkadoK, GuelbeogoWM, BadoloA, AonumaH, et al. (2013) Intra-specific diversity of Serratia marcescens in Anopheles mosquito midgut defines Plasmodium transmission capacity. Sci Rep 3 : 1641.
38. LawniczakMK, EmrichSJ, HollowayAK, RegierAP, OlsonM, et al. (2010) Widespread divergence between incipient Anopheles gambiae species revealed by whole genome sequences. Science 330 : 512–514.
39. NeafseyDE, LawniczakMK, ParkDJ, RedmondSN, CoulibalyMB, et al. (2010) SNP genotyping defines complex gene-flow boundaries among African malaria vector mosquitoes. Science 330 : 514–517.
40. GottarM, GobertV, MichelT, BelvinM, DuykG, et al. (2002) The Drosophila immune response against Gram-negative bacteria is mediated by a peptidoglycan recognition protein. Nature 416 : 640–644.
41. ChoeKM, WernerT, StovenS, HultmarkD, AndersonKV (2002) Requirement for a peptidoglycan recognition protein (PGRP) in Relish activation and antibacterial immune responses in Drosophila. Science 296 : 359–362.
42. LinH, ZhangL, LunaC, HoNT, ZhengL (2007) A splice variant of PGRP-LC required for expression of antimicrobial peptides in Anopheles gambiae. Insect Science 14 : 185–192.
43. WatsonFL, Puttmann-HolgadoR, ThomasF, LamarDL, HughesM, et al. (2005) Extensive diversity of Ig-superfamily proteins in the immune system of insects. Science 309 : 1874–1878.
44. DongY, TaylorHE, DimopoulosG (2006) AgDscam, a hypervariable immunoglobulin domain-containing receptor of the Anopheles gambiae innate immune system. PLoS Biol 4: e229.
45. WatthanasurorotA, JiravanichpaisalP, LiuH, SoderhallI, SoderhallK (2011) Bacteria-Induced Dscam Isoforms of the Crustacean, Pacifastacus leniusculus. PLoS Pathog 7: e1002062.
46. WojtowiczWM, WuW, AndreI, QianB, BakerD, et al. (2007) A vast repertoire of Dscam binding specificities arises from modular interactions of variable Ig domains. Cell 130 : 1134–1145.
47. DongY, CirimotichCM, PikeA, ChandraR, DimopoulosG (2012) Anopheles NF-kappaB-Regulated Splicing Factors Direct Pathogen-Specific Repertoires of the Hypervariable Pattern Recognition Receptor AgDscam. Cell Host Microbe 12 : 521–530.
48. SmithPH, MwangiJM, AfraneYA, YanG, ObbardDJ, et al. (2011) Alternative splicing of the Anopheles gambiae Dscam gene in diverse Plasmodium falciparum infections. Malar J 10 : 156.
49. ClaytonAM, CirimotichCM, DongY, DimopoulosG (2012) Caudal is a negative regulator of the Anopheles IMD Pathway that controls resistance to P. falciparum infection. Dev Comp Immunol 39 (4) 323–32.
50. XinN, BenchabaneH, TianA, NguyenK, KlofasL, et al. (2011) Erect Wing facilitates context-dependent Wnt/Wingless signaling by recruiting the cell-specific Armadillo-TCF adaptor Earthbound to chromatin. Development 138 : 4955–4967.
51. DitchLM, ShirangiT, PitmanJL, LathamKL, FinleyKD, et al. (2005) Drosophila retained/dead ringer is necessary for neuronal pathfinding, female receptivity and repression of fruitless independent male courtship behaviors. Development 132 : 155–164.
52. ShirangiTR, TaylorBJ, McKeownM (2006) A double-switch system regulates male courtship behavior in male and female Drosophila melanogaster. Nat Genet 38 : 1435–1439.
53. DubrovskyEB, DubrovskayaVA, BernardoT, OtteV, DiFilippoR, et al. (2011) The Drosophila FTZ-F1 nuclear receptor mediates juvenile hormone activation of E75A gene expression through an intracellular pathway. J Biol Chem 286 : 33689–33700.
54. FunkhouserJD, AronsonNNJr (2007) Chitinase family GH18: evolutionary insights from the genomic history of a diverse protein family. BMC Evol Biol 7 : 96.
55. Dela CruzCS, LiuW, HeCH, JacobyA, GornitzkyA, et al. (2012) Chitinase 3-like-1 Promotes Streptococcus pneumoniae Killing and Augments Host Tolerance to Lung Antibacterial Responses. Cell Host Microbe 12 : 34–46.
56. VolzJ, MullerHM, ZdanowiczA, KafatosFC, OstaMA (2006) A genetic module regulates the melanization response of Anopheles to Plasmodium. Cell Microbiol 8 : 1392–1405.
57. VolzJ, OstaMA, KafatosFC, MullerHM (2005) The roles of two clip domain serine proteases in innate immune responses of the malaria vector Anopheles gambiae. J Biol Chem 280 : 40161–40168.
58. PovelonesM, BhagavatulaL, YassineH, TanLA, UptonLM, et al. (2013) The CLIP-Domain Serine Protease Homolog SPCLIP1 Regulates Complement Recruitment to Microbial Surfaces in the Malaria Mosquito Anopheles gambiae. PLoS Pathog 9: e1003623.
59. DongY, AguilarR, XiZ, WarrE, MonginE, et al. (2006) Anopheles gambiae immune responses to human and rodent Plasmodium parasite species. PLoS Pathog 2: e52.
60. WaterhouseRM, PovelonesM, ChristophidesGK (2010) Sequence-structure-function relations of the mosquito leucine-rich repeat immune proteins. BMC Genomics 11 : 531.
61. PovelonesM, UptonLM, SalaKA, ChristophidesGK (2011) Structure-Function Analysis of the Anopheles gambiae LRIM1/APL1C Complex and its Interaction with Complement C3-Like Protein TEP1. PLoS Pathog 7: e1002023.
62. PovelonesM, WaterhouseRM, KafatosFC, ChristophidesGK (2009) Leucine-rich repeat protein complex activates mosquito complement in defense against Plasmodium parasites. Science 324 : 258–261.
63. OstaMA, ChristophidesGK, KafatosFC (2004) Effects of mosquito genes on Plasmodium development. Science 303 : 2030–2032.
64. KazuraJW, ZouZ, Souza-NetoJ, XiZ, KokozaV, et al. (2011) Transcriptome Analysis of Aedes aegypti Transgenic Mosquitoes with Altered Immunity. PLoS Pathogens 7: e1002394.
65. ValanneS, WangJH, RametM (2011) The Drosophila Toll signaling pathway. J Immunol 186 : 649–656.
66. GarverLS, DongY, DimopoulosG (2009) Caspar controls resistance to Plasmodium falciparum in diverse anopheline species. PLoS Pathog 5: e1000335.
67. VaishnavaS, YamamotoM, SeversonKM, RuhnKA, YuX, et al. (2011) The antibacterial lectin RegIIIgamma promotes the spatial segregation of microbiota and host in the intestine. Science 334 : 255–258.
68. DongY, DimopoulosG (2009) Anopheles fibrinogen-related proteins provide expanded pattern recognition capacity against bacteria and malaria parasites. J Biol Chem 284 : 9835–9844.
69. PaceKE, BaumLG (2004) Insect galectins: roles in immunity and development. Glycoconj J 19 : 607–614.
70. MendesAM, Awono-AmbenePH, NsangoSE, CohuetA, FontenilleD, et al. (2011) Infection intensity dependent responses of Anopheles gambiae to African malaria parasites Plasmodium falciparum. Infect Immun 79 (11) 4708–15.
71. StyerKL, SinghV, MacoskoE, SteeleSE, BargmannCI, et al. (2008) Innate immunity in Caenorhabditis elegans is regulated by neurons expressing NPR-1/GPCR. Science 322 : 460–464.
72. ReddyKC, AndersenEC, KruglyakL, KimDH (2009) A polymorphism in npr-1 is a behavioral determinant of pathogen susceptibility in C. elegans. Science 323 : 382–384.
73. CirilloC, Vanden BergheP, TackJ (2011) Role of serotonin in gastrointestinal physiology and pathology. Minerva Endocrinol 36 : 311–324.
74. KhanWI, GhiaJE (2010) Gut hormones: emerging role in immune activation and inflammation. Clin Exp Immunol 161 : 19–27.
75. BaganzNL, BlakelyRD (2013) A dialogue between the immune system and brain, spoken in the language of serotonin. ACS Chem Neurosci 4 : 48–63.
76. BecnelJ, JohnsonO, LuoJ, NasselDR, NicholsCD (2011) The serotonin 5-HT7Dro receptor is expressed in the brain of Drosophila, and is essential for normal courtship and mating. PLoS One 6: e20800.
77. JohnsonO, BecnelJ, NicholsCD (2011) Serotonin receptor activity is necessary for olfactory learning and memory in Drosophila melanogaster. Neuroscience 192 : 372–381.
78. DierickHA, GreenspanRJ (2007) Serotonin and neuropeptide F have opposite modulatory effects on fly aggression. Nat Genet 39 : 678–682.
79. DzitoyevaS, DimitrijevicN, ManevH (2003) Gamma-aminobutyric acid B receptor 1 mediates behavior-impairing actions of alcohol in Drosophila: adult RNA interference and pharmacological evidence. Proc Natl Acad Sci U S A 100 : 5485–5490.
80. WenT, ParrishCA, XuD, WuQ, ShenP (2005) Drosophila neuropeptide F and its receptor, NPFR1, define a signaling pathway that acutely modulates alcohol sensitivity. Proc Natl Acad Sci U S A 102 : 2141–2146.
81. Shohat-OphirG, KaunKR, AzanchiR, HeberleinU (2012) Sexual deprivation increases ethanol intake in Drosophila. Science 335 : 1351–1355.
82. HillCA, FoxAN, PittsRJ, KentLB, TanPL, et al. (2002) G protein-coupled receptors in Anopheles gambiae. Science 298 : 176–178.
83. BrayS, AmreinH (2003) A putative Drosophila pheromone receptor expressed in male-specific taste neurons is required for efficient courtship. Neuron 39 : 1019–1029.
84. MiyamotoT, AmreinH (2008) Suppression of male courtship by a Drosophila pheromone receptor. Nat Neurosci 11 : 874–876.
85. WatanabeK, TobaG, KoganezawaM, YamamotoD (2011) Gr39a, a highly diversified gustatory receptor in Drosophila, has a role in sexual behavior. Behav Genet 41 : 746–753.
86. WangL, HanX, MehrenJ, HiroiM, BilleterJC, et al. (2011) Hierarchical chemosensory regulation of male-male social interactions in Drosophila. Nat Neurosci 14 : 757–762.
87. LeeY, KimSH, MontellC (2010) Avoiding DEET through insect gustatory receptors. Neuron 67 : 555–561.
88. MoonSJ, KottgenM, JiaoY, XuH, MontellC (2006) A taste receptor required for the caffeine response in vivo. Curr Biol 16 : 1812–1817.
89. JonesWD, CayirliogluP, KadowIG, VosshallLB (2007) Two chemosensory receptors together mediate carbon dioxide detection in Drosophila. Nature 445 : 86–90.
90. ErdelyanCN, MahoodTH, BaderTS, WhyardS (2011) Functional validation of the carbon dioxide receptor genes in Aedes aegypti mosquitoes using RNA interference. Insect Mol Biol
91. ChybS, DahanukarA, WickensA, CarlsonJR (2003) Drosophila Gr5a encodes a taste receptor tuned to trehalose. Proc Natl Acad Sci U S A 100 Suppl 2 : 14526–14530.
92. UenoK, OhtaM, MoritaH, MikuniY, NakajimaS, et al. (2001) Trehalose sensitivity in Drosophila correlates with mutations in and expression of the gustatory receptor gene Gr5a. Curr Biol 11 : 1451–1455.
93. DahanukarA, FosterK, van der Goes van NatersWM, CarlsonJR (2001) A Gr receptor is required for response to the sugar trehalose in taste neurons of Drosophila. Nat Neurosci 4 : 1182–1186.
94. JiaoY, MoonSJ, MontellC (2007) A Drosophila gustatory receptor required for the responses to sucrose, glucose, and maltose identified by mRNA tagging. Proc Natl Acad Sci U S A 104 : 14110–14115.
95. SerhanCN, ChiangN, Van DykeTE (2008) Resolving inflammation: dual anti-inflammatory and pro-resolution lipid mediators. Nat Rev Immunol 8 : 349–361.
96. LucasKJ, MylesKM, RaikhelAS (2013) Small RNAs: a new frontier in mosquito biology. Trends Parasitol 29 : 295–303.
97. ZhouR, HuG, LiuJ, GongAY, DrescherKM, et al. (2009) NF-kappaB p65-dependent transactivation of miRNA genes following Cryptosporidium parvum infection stimulates epithelial cell immune responses. PLoS Pathog 5: e1000681.
98. SakuraiM, AokiT, YoshikawaS, SantschiLA, SaitoH, et al. (2009) Differentially expressed Drl and Drl-2 play opposing roles in Wnt5 signaling during Drosophila olfactory system development. J Neurosci 29 : 4972–4980.
99. GesellchenV, KuttenkeulerD, SteckelM, PelteN, BoutrosM (2005) An RNA interference screen identifies Inhibitor of Apoptosis Protein 2 as a regulator of innate immune signalling in Drosophila. EMBO Rep 6 : 979–984.
100. ChenJ, ZhangY, ShenP (2008) A protein kinase C activity localized to neuropeptide Y-like neurons mediates ethanol intoxication in Drosophila melanogaster. Neuroscience 156 : 42–47.
101. ChenJ, ZhangY, ShenP (2010) Protein kinase C deficiency-induced alcohol insensitivity and underlying cellular targets in Drosophila. Neuroscience 166 : 34–39.
102. YeT, TangW, ZhangX (2012) Involvement of Rab6 in the regulation of phagocytosis against virus infection in invertebrates. J Proteome Res 11 : 4834–4846.
103. CoutelisJB, EphrussiA (2007) Rab6 mediates membrane organization and determinant localization during Drosophila oogenesis. Development 134 : 1419–1430.
104. TianAG, TamoriY, HuangYC, MelendezNT, DengWM (2013) Efficient EGFR signaling and dorsal-ventral axis patterning requires syntaxin dependent Gurken trafficking. Dev Biol 373 : 349–358.
105. Abou TayounAN, PikielnyC, DolphPJ (2012) Roles of the Drosophila SK channel (dSK) in courtship memory. PLoS One 7: e34665.
106. RustenTE, VaccariT, LindmoK, RodahlLM, NezisIP, et al. (2007) ESCRTs and Fab1 regulate distinct steps of autophagy. Curr Biol 17 : 1817–1825.
107. SoukupSF, CuliJ, GubbD (2009) Uptake of the necrotic serpin in Drosophila melanogaster via the lipophorin receptor-1. PLoS Genet 5: e1000532.
108. LevashinaEA (1999) Constitutive Activation of Toll-Mediated Antifungal Defense in Serpin-Deficient Drosophila. Science 285 : 1917–1919.
109. PelteN, RobertsonAS, ZouZ, BelorgeyD, DaffornTR, et al. (2006) Immune challenge induces N-terminal cleavage of the Drosophila serpin Necrotic. Insect Biochem Mol Biol 36 : 37–46.
110. MarinottiO, CalvoE, NguyenQK, DissanayakeS, RibeiroJM, et al. (2006) Genome-wide analysis of gene expression in adult Anopheles gambiae. Insect Mol Biol 15 : 1–12.
111. BakerDA, NolanT, FischerB, PinderA, CrisantiA, et al. (2011) A comprehensive gene expression atlas of sex - and tissue-specificity in the malaria vector, Anopheles gambiae. BMC Genomics 12 : 296.
112. PittsRJ, RinkerDC, JonesPL, RokasA, ZwiebelLJ (2011) Transcriptome profiling of chemosensory appendages in the malaria vector Anopheles gambiae reveals tissue - and sex-specific signatures of odor coding. BMC Genomics 12 : 271.
113. MeisterS, KanzokSM, ZhengXL, LunaC, LiTR, et al. (2005) Immune signaling pathways regulating bacterial and malaria parasite infection of the mosquito Anopheles gambiae. Proc Natl Acad Sci U S A 102 : 11420–11425.
114. FelixRC, SilveiraH (2011) The Interplay between Tubulins and P450 Cytochromes during Plasmodium berghei Invasion of Anopheles gambiae Midgut. PLoS One 6: e24181.
115. BenoitJB, Lopez-MartinezG, PatrickKR, PhillipsZP, KrauseTB, et al. (2011) From the Cover: Drinking a hot blood meal elicits a protective heat shock response in mosquitoes. Proc Natl Acad Sci U S A 108 : 8026–8029.
116. LiB, CalvoE, MarinottiO, JamesAA, PaskewitzSM (2005) Characterization of the c-type lysozyme gene family in Anopheles gambiae. Gene 360 : 131–139.
117. ChristophidesGK, ZdobnovE, Barillas-MuryC, BirneyE, BlandinS, et al. (2002) Immunity-related genes and gene families in Anopheles gambiae. Science 298 : 159–165.
118. DanielliA, LoukerisTG, LagueuxM, MullerHM, RichmanA, et al. (2000) A modular chitin-binding protease associated with hemocytes and hemolymph in the mosquito Anopheles gambiae. Proc Natl Acad Sci U S A 97 : 7136–7141.
119. DinglasanRR, DevenportM, FlorensL, JohnsonJR, McHughCA, et al. (2009) The Anopheles gambiae adult midgut peritrophic matrix proteome. Insect Biochem Mol Biol 39 : 125–134.
120. KuraishiT, BinggeliO, OpotaO, BuchonN, LemaitreB (2011) Genetic evidence for a protective role of the peritrophic matrix against intestinal bacterial infection in Drosophila melanogaster. Proc Natl Acad Sci U S A 108 : 15966–15971.
121. BrownMR, CrimJW, ArataRC, CaiHN, ChunC, et al. (1999) Identification of a Drosophila brain-gut peptide related to the neuropeptide Y family. Peptides 20 : 1035–1042.
122. StanekDM, PohlJ, CrimJW, BrownMR (2002) Neuropeptide F and its expression in the yellow fever mosquito, Aedes aegypti. Peptides 23 : 1367–1378.
123. WuQ, ZhaoZ, ShenP (2005) Regulation of aversion to noxious food by Drosophila neuropeptide Y - and insulin-like systems. Nat Neurosci 8 : 1350–1355.
124. ShenP, CaiHN (2001) Drosophila neuropeptide F mediates integration of chemosensory stimulation and conditioning of the nervous system by food. J Neurobiol 47 : 16–25.
125. KacsohBZ, LynchZR, MortimerNT, SchlenkeTA (2013) Fruit flies medicate offspring after seeing parasites. Science 339 : 947–950.
126. WangY, BrentCS, FennernE, AmdamGV (2012) Gustatory perception and fat body energy metabolism are jointly affected by vitellogenin and juvenile hormone in honey bees. PLoS Genet 8: e1002779.
127. XuJ, ShengZ, PalliSR (2013) Juvenile hormone and insulin regulate trehalose homeostasis in the red flour beetle, Tribolium castaneum. PLoS Genet 9: e1003535.
128. NoriegaFG (2004) Nutritional regulation of JH synthesis: a mechanism to control reproductive maturation in mosquitoes? Insect Biochem Mol Biol 34 : 687–693.
129. Perez-HedoM, Rivera-PerezC, NoriegaFG (2013) The insulin/TOR signal transduction pathway is involved in the nutritional regulation of juvenile hormone synthesis in Aedes aegypti. Insect Biochem Mol Biol 43 (6) 495–500.
130. SharonG, SegalD, RingoJM, HefetzA, Zilber-RosenbergI, et al. (2010) Commensal bacteria play a role in mating preference of Drosophila melanogaster. Proc Natl Acad Sci U S A 107 : 20051–20056.
131. CareyAF, WangG, SuCY, ZwiebelLJ, CarlsonJR (2010) Odorant reception in the malaria mosquito Anopheles gambiae. Nature 464 : 66–71.
132. MeunierN, BelgacemYH, MartinJR (2007) Regulation of feeding behaviour and locomotor activity by takeout in Drosophila. J Exp Biol 210 : 1424–1434.
133. VanaphanN, DauwalderB, ZufallRA (2012) Diversification of takeout, a male-biased gene family in Drosophila. Gene 491 : 142–148.
134. ParkJH, KwonJY (2011) Heterogeneous expression of Drosophila gustatory receptors in enteroendocrine cells. PLoS One 6: e29022.
135. NiareO, MarkianosK, VolzJ, OduolF, ToureA, et al. (2002) Genetic loci affecting resistance to human malaria parasites in a West African mosquito vector population. Science 298 : 213–216.
136. BlandinSA, Wang-SattlerR, LamacchiaM, GagneurJ, LycettG, et al. (2009) Dissecting the genetic basis of resistance to malaria parasites in Anopheles gambiae. Science 326 : 147–150.
137. Molina-CruzA, GarverLS, AlabasterA, BangioloL, HaileA, et al. (2013) The human malaria parasite Pfs47 gene mediates evasion of the mosquito immune system. Science 340 : 984–987.
138. HeutinkP, OostraBA (2002) Gene finding in genetically isolated populations. Hum Mol Genet 11 : 2507–2515.
139. GudmundssonJ, SulemP, GudbjartssonDF, MassonG, AgnarssonBA, et al. (2012) A study based on whole-genome sequencing yields a rare variant at 8q24 associated with prostate cancer. Nat Genet 44 : 1326–1329.
140. LeeK-A, KimS-H, KimE-K, HaE-M, YouH, et al. (2013) Bacterial-Derived Uracil as a Modulator of Mucosal Immunity and Gut-Microbe Homeostasis in Drosophila. Cell 153 : 797–811.
141. MeijersR, Puettmann-HolgadoR, SkiniotisG, LiuJH, WalzT, et al. (2007) Structural basis of Dscam isoform specificity. Nature 449 : 487–491.
142. KolodziejPA, TimpeLC, MitchellKJ, FriedSR, GoodmanCS, et al. (1996) frazzled encodes a Drosophila member of the DCC immunoglobulin subfamily and is required for CNS and motor axon guidance. Cell 87 : 197–204.
143. KiddT, BroseK, MitchellKJ, FetterRD, Tessier-LavigneM, et al. (1998) Roundabout controls axon crossing of the CNS midline and defines a novel subfamily of evolutionarily conserved guidance receptors. Cell 92 : 205–215.
144. EvansTA, BashawGJ (2010) Axon guidance at the midline: of mice and flies. Curr Opin Neurobiol 20 : 79–85.
145. KochCM, Honemann-CapitoM, Egger-AdamD, WodarzA (2009) Windei, the Drosophila homolog of mAM/MCAF1, is an essential cofactor of the H3K9 methyl transferase dSETDB1/Eggless in germ line development. PLoS Genet 5: e1000644.
146. LiuL, IshiharaK, IchimuraT, FujitaN, HinoS, et al. (2009) MCAF1/AM is involved in Sp1-mediated maintenance of cancer-associated telomerase activity. J Biol Chem 284 : 5165–5174.
147. GarverLS, XiZ, DimopoulosG (2008) Immunoglobulin superfamily members play an important role in the mosquito immune system. Dev Comp Immunol 32 : 519–531.
148. ZdobnovEM, von MeringC, LetunicI, TorrentsD, SuyamaM, et al. (2002) Comparative genome and proteome analysis of Anopheles gambiae and Drosophila melanogaster. Science 298 : 149–159.
149. Bosco-DrayonV, PoidevinM, BonecaIG, Narbonne-ReveauK, RoyetJ, et al. (2012) Peptidoglycan Sensing by the Receptor PGRP-LE in the Drosophila Gut Induces Immune Responses to Infectious Bacteria and Tolerance to Microbiota. Cell Host Microbe 12 : 153–165.
150. ParedesJC, WelchmanDP, PoidevinM, LemaitreB (2011) Negative Regulation by Amidase PGRPs Shapes the Drosophila Antibacterial Response and Protects the Fly from Innocuous Infection. Immunity 35 : 770–779.
151. Crampton JM, Beard CB, Louis C, World Health Organization. (1997) The molecular biology of insect disease vectors : a methods manual. London: Chapman & Hall. xxv: , 578 p.
152. NehmeNT, LiegeoisS, KeleB, GiammarinaroP, PradelE, et al. (2007) A model of bacterial intestinal infections in Drosophila melanogaster. PLoS Pathog 3: e173.
153. FaviaG, RicciI, DamianiC, RaddadiN, CrottiE, et al. (2007) Bacteria of the genus Asaia stably associate with Anopheles stephensi, an Asian malarial mosquito vector. Proc Natl Acad Sci U S A 104 : 9047–9051.
154. ReidenbachKR, NeafseyDE, CostantiniC, SagnonN, SimardF, et al. (2012) Patterns of genomic differentiation between ecologically differentiated M and S forms of Anopheles gambiae in West and Central Africa. Genome Biol Evol 4 (12) 1202–12.
155. LawsonD, ArensburgerP, AtkinsonP, BesanskyNJ, BruggnerRV, et al. (2009) VectorBase: a data resource for invertebrate vector genomics. Nucleic Acids Res 37: D583–587.
156. BlandinS, MoitaLF, KocherT, WilmM, KafatosFC, et al. (2002) Reverse genetics in the mosquito Anopheles gambiae: targeted disruption of the Defensin gene. EMBO Rep 3 : 852–856.
157. GoecksJ, NekrutenkoA, TaylorJ, GalaxyT (2010) Galaxy: a comprehensive approach for supporting accessible, reproducible, and transparent computational research in the life sciences. Genome Biol 11: R86.
158. Santiago-SoteloP, Ramirez-PradoJH (2012) prfectBLAST: a platform-independent portable front end for the command terminal BLAST+ stand-alone suite. Biotechniques 53 : 299–300.
159. HusonDH, MitraS, RuscheweyhHJ, WeberN, SchusterSC (2011) Integrative analysis of environmental sequences using MEGAN4. Genome Res 21 : 1552–1560.
160. JaWW, CarvalhoGB, MakEM, de la RosaNN, FangAY, et al. (2007) Prandiology of Drosophila and the CAFE assay. Proc Natl Acad Sci U S A 104 : 8253–8256.
Štítky
Hygiena a epidemiologie Infekční lékařství Laboratoř
Článek DHX36 Enhances RIG-I Signaling by Facilitating PKR-Mediated Antiviral Stress Granule FormationČlánek Oral Bacteria and CancerČlánek A Non-Coding RNA Promotes Bacterial Persistence and Decreases Virulence by Regulating a Regulator in
Článek vyšel v časopisePLOS Pathogens
Nejčtenější tento týden
2014 Číslo 3- Stillova choroba: vzácné a závažné systémové onemocnění
- Diagnostika virových hepatitid v kostce – zorientujte se (nejen) v sérologii
- Diagnostický algoritmus při podezření na syndrom periodické horečky
- Perorální antivirotika jako vysoce efektivní nástroj prevence hospitalizací kvůli COVID-19 − otázky a odpovědi pro praxi
- Infekční komplikace virových respiračních infekcí – sekundární bakteriální a aspergilové pneumonie
-
Všechny články tohoto čísla
- Conflicting Interests in the Pathogen–Host Tug of War: Fungal Micronutrient Scavenging Versus Mammalian Nutritional Immunity
- Putting Fungi to Work: Harvesting a Cornucopia of Drugs, Toxins, and Antibiotics
- Mycobacteriophages: Windows into Tuberculosis
- Human African Trypanosomiasis and Immunological Memory: Effect on Phenotypic Lymphocyte Profiles and Humoral Immunity
- Five Things to Know about Genetically Modified (GM) Insects for Vector Control
- A Missing Dimension in Measures of Vaccination Impacts
- Eosinophils Are Important for Protection, Immunoregulation and Pathology during Infection with Nematode Microfilariae
- Clonality of HTLV-2 in Natural Infection
- Production, Fate and Pathogenicity of Plasma Microparticles in Murine Cerebral Malaria
- Group B Streptococcal Infection of the Choriodecidua Induces Dysfunction of the Cytokeratin Network in Amniotic Epithelium: A Pathway to Membrane Weakening
- New Insights into How Adapts to Its Mammalian Host during Bubonic Plague
- Foodborne Transmission of Nipah Virus in Syrian Hamsters
- A Polysaccharide Virulence Factor from Elicits Anti-inflammatory Effects through Induction of Interleukin-1 Receptor Antagonist
- Structural and Functional Characterization of a Complex between the Acidic Transactivation Domain of EBNA2 and the Tfb1/p62 Subunit of TFIIH
- Adaptive Gene Amplification As an Intermediate Step in the Expansion of Virus Host Range
- DHX36 Enhances RIG-I Signaling by Facilitating PKR-Mediated Antiviral Stress Granule Formation
- Hepatitis B Virus Infection and Immunopathogenesis in a Humanized Mouse Model: Induction of Human-Specific Liver Fibrosis and M2-Like Macrophages
- Crk Adaptors Negatively Regulate Actin Polymerization in Pedestals Formed by Enteropathogenic (EPEC) by Binding to Tir Effector
- Fatty Acid Biosynthesis Contributes Significantly to Establishment of a Bioenergetically Favorable Environment for Vaccinia Virus Infection
- A Cytosolic Chaperone Complexes with Dynamic Membrane J-Proteins and Mobilizes a Nonenveloped Virus out of the Endoplasmic Reticulum
- Intracellular Promote Invasive Cell Motility through Kinase Regulation of the Host Actin Cytoskeleton
- MAVS-MKK7-JNK2 Defines a Novel Apoptotic Signaling Pathway during Viral Infection
- RON5 Is Critical for Organization and Function of the Moving Junction Complex
- Immune Suppression by Neutrophils in HIV-1 Infection: Role of PD-L1/PD-1 Pathway
- and Exhibit Metabolic Symbioses
- The Herpes Virus Fc Receptor gE-gI Mediates Antibody Bipolar Bridging to Clear Viral Antigens from the Cell Surface
- Target Cell Availability, Rather than Breast Milk Factors, Dictates Mother-to-Infant Transmission of SIV in Sooty Mangabeys and Rhesus Macaques
- Evolution of the Retroviral Restriction Gene : Inhibition of Non-MLV Retroviruses
- Infection of Adult Thymus with Murine Retrovirus Induces Virus-Specific Central Tolerance That Prevents Functional Memory CD8 T Cell Differentiation
- Fha Interaction with Phosphothreonine of TssL Activates Type VI Secretion in
- In Vivo Administration of a JAK3 Inhibitor during Acute SIV Infection Leads to Significant Increases in Viral Load during Chronic Infection
- Lack of Detectable HIV-1 Molecular Evolution during Suppressive Antiretroviral Therapy
- Activation of HIV-1 from Latent Infection via Synergy of RUNX1 Inhibitor Ro5-3335 and SAHA
- A Compact, Multifunctional Fusion Module Directs Cholesterol-Dependent Homomultimerization and Syncytiogenic Efficiency of Reovirus p10 FAST Proteins
- The Role of Host and Microbial Factors in the Pathogenesis of Pneumococcal Bacteraemia Arising from a Single Bacterial Cell Bottleneck
- Genetic Dissection of Gut Epithelial Responses to
- Two-Component System Cross-Regulation Integrates Response to Heme and Cell Envelope Stress
- Oral Mycobiome Analysis of HIV-Infected Patients: Identification of as an Antagonist of Opportunistic Fungi
- A Model System for Studying the Transcriptomic and Physiological Changes Associated with Mammalian Host-Adaptation by Serovar Copenhageni
- Inflammasome Sensor NLRP1 Controls Rat Macrophage Susceptibility to
- ChIP-Seq and RNA-Seq Reveal an AmrZ-Mediated Mechanism for Cyclic di-GMP Synthesis and Biofilm Development by
- The Hypervariable Amino-Terminus of P1 Protease Modulates Potyviral Replication and Host Defense Responses
- Caspase-1-Dependent and -Independent Cell Death Pathways in Infection of Macrophages
- The Effect of Cell Growth Phase on the Regulatory Cross-Talk between Flagellar and Spi1 Virulence Gene Expression
- Different Mutagenic Potential of HIV-1 Restriction Factors APOBEC3G and APOBEC3F Is Determined by Distinct Single-Stranded DNA Scanning Mechanisms
- Oral Bacteria and Cancer
- Identification of OmpA, a Protein Involved in Host Cell Invasion, by Multi-Phenotypic High-Content Screening
- Transovarial Transmission of a Plant Virus Is Mediated by Vitellogenin of Its Insect Vector
- VE-Cadherin Cleavage by LasB Protease from Facilitates Type III Secretion System Toxicity in Endothelial Cells
- Dimerization of VirD2 Binding Protein Is Essential for Induced Tumor Formation in Plants
- Crystal Structure of the Vaccinia Virus DNA Polymerase Holoenzyme Subunit D4 in Complex with the A20 N-Terminal Domain
- Post-Translational Regulation via Clp Protease Is Critical for Survival of
- Modulation of Phagosomal pH by Promotes Hyphal Morphogenesis and Requires Stp2p, a Regulator of Amino Acid Transport
- Rotavirus Activates Lymphocytes from Non-Obese Diabetic Mice by Triggering Toll-Like Receptor 7 Signaling and Interferon Production in Plasmacytoid Dendritic Cells
- Cytomegalovirus m154 Hinders CD48 Cell-Surface Expression and Promotes Viral Escape from Host Natural Killer Cell Control
- Interferon Regulatory Factor-1 Protects from Fatal Neurotropic Infection with Vesicular Stomatitis Virus by Specific Inhibition of Viral Replication in Neurons
- HMGB1-Promoted and TLR2/4-Dependent NK Cell Maturation and Activation Take Part in Rotavirus-Induced Murine Biliary Atresia
- An Immunomics Approach to Schistosome Antigen Discovery: Antibody Signatures of Naturally Resistant and Chronically Infected Individuals from Endemic Areas
- PPARγ Agonists Improve Survival and Neurocognitive Outcomes in Experimental Cerebral Malaria and Induce Neuroprotective Pathways in Human Malaria
- A Non-Coding RNA Promotes Bacterial Persistence and Decreases Virulence by Regulating a Regulator in
- Viral OTU Deubiquitinases: A Structural and Functional Comparison
- Heterogeneity and Breadth of Host Antibody Response to KSHV Infection Demonstrated by Systematic Analysis of the KSHV Proteome
- Influenza A Virus Assembly Intermediates Fuse in the Cytoplasm
- Broadly Reactive Human CD8 T Cells that Recognize an Epitope Conserved between VZV, HSV and EBV
- Oncogenic Human Papillomaviruses Activate the Tumor-Associated Lens Epithelial-Derived Growth Factor (LEDGF) Gene
- Erythrocyte Invasion: Combining Function with Immune Evasion
- IL-1α and Complement Cooperate in Triggering Local Neutrophilic Inflammation in Response to Adenovirus and Eliminating Virus-Containing Cells
- Chronic Exposure to Type-I IFN under Lymphopenic Conditions Alters CD4 T Cell Homeostasis
- PLOS Pathogens
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Cytomegalovirus m154 Hinders CD48 Cell-Surface Expression and Promotes Viral Escape from Host Natural Killer Cell Control
- Human African Trypanosomiasis and Immunological Memory: Effect on Phenotypic Lymphocyte Profiles and Humoral Immunity
- DHX36 Enhances RIG-I Signaling by Facilitating PKR-Mediated Antiviral Stress Granule Formation
- Conflicting Interests in the Pathogen–Host Tug of War: Fungal Micronutrient Scavenging Versus Mammalian Nutritional Immunity
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



