-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaNew Insights into How Adapts to Its Mammalian Host during Bubonic Plague
In order to understand and combat infectious diseases, it is essential to characterize the full set of genes required by pathogenic bacteria to overcome the many immunological and physiological challenges encountered during infection. Here, we used a genome-scale approach to identify genes required by the bacterium Yersinia pestis in the production of bubonic plague (a fatal, flea-borne zoonosis). Our results suggest that when colonizing the mammalian host, the bacterium (i) relies on carbohydrates as its carbon source, (ii) shifts to anaerobic respiration or fermentation and (iii) experiences and resists several (but not all) types of stress generated by the host's innate immune system. Strikingly, only a small set of genes (including horizontally acquired and uncharacterized sequences) are required for these infectious processes. Further investigations of the ypmt1,66c gene provided evidence to suggest that accretion of genetic material via horizontal transfer has played a key role in Yersinia pestis' ability to successfully initiate infection after the dermal fleabite. Lastly, we believe that (i) application of our approach to other pathogens and (ii) additional studies of selected Yersinia pestis genes important for plague pathogenesis (some of which are shared with other pathogens) will provide a better understanding of bacterial pathogenesis in general.
Published in the journal: . PLoS Pathog 10(3): e32767. doi:10.1371/journal.ppat.1004029
Category: Research Article
doi: https://doi.org/10.1371/journal.ppat.1004029Summary
In order to understand and combat infectious diseases, it is essential to characterize the full set of genes required by pathogenic bacteria to overcome the many immunological and physiological challenges encountered during infection. Here, we used a genome-scale approach to identify genes required by the bacterium Yersinia pestis in the production of bubonic plague (a fatal, flea-borne zoonosis). Our results suggest that when colonizing the mammalian host, the bacterium (i) relies on carbohydrates as its carbon source, (ii) shifts to anaerobic respiration or fermentation and (iii) experiences and resists several (but not all) types of stress generated by the host's innate immune system. Strikingly, only a small set of genes (including horizontally acquired and uncharacterized sequences) are required for these infectious processes. Further investigations of the ypmt1,66c gene provided evidence to suggest that accretion of genetic material via horizontal transfer has played a key role in Yersinia pestis' ability to successfully initiate infection after the dermal fleabite. Lastly, we believe that (i) application of our approach to other pathogens and (ii) additional studies of selected Yersinia pestis genes important for plague pathogenesis (some of which are shared with other pathogens) will provide a better understanding of bacterial pathogenesis in general.
Introduction
Plague (caused by the bacterium Yersinia pestis) is one of the diseases that has most greatly affected the course of history [1], [2] and is still an international public health concern. The WHO has even categorized this disease as re-emergent in some parts of the world. This problem is heightened by the emergence of multidrug-resistant strains and the potential use of plague as a bioweapon [3], [4]. Plague is commonly acquired through the bite of an infected flea, when Y. pestis is regurgitated into the dermis during the flea blood meal [5]. It is thought that Y. pestis is then taken up by macrophages, within which it replicates and produces antiphagocytic factors [6]. Next, bacteria released from dying macrophages spread into the draining lymph node, where they actively replicate, circumvent phagocytosis by polymorphonuclear neutrophils (PMNs) and confront a number of toxic effectors released by degranulating or lysing PMNs [7], [8]. These events lead to a hemorrhagic, edematous, swollen and painful lymph node (the bubo), which is the defining clinical feature in the bubonic form of plague [9], [10]. Overwhelmed, the lymph node allows the bacteria to disseminate into the blood stream; this produces a fulminant, systemic infection and fatal septicemia. In a very low proportion of infections, septicemia causes pneumonia, which in turn enables Y. pestis to be transmitted from person to person via contaminated aerosols. However, we lack important data on the above-mentioned mechanisms. For instance, genes dedicated to the intracellular step have not been identified. Furthermore, recent work suggests that transit through the flea vector enhances Y. pestis' ability to resist phagocytosis by the macrophage [11]. Very few pathways for resisting innate immune effectors have been elucidated [12]. Lastly, the metabolic pathways that enable Y. pestis to replicate during infection are incompletely understood.
Regurgitation of Y. pestis into the skin does not always cause bubonic plague and bubonic plague does not always progress consistently to septicemic plague. This indicates that the skin and the lymph node are the two decisive arenas in which Y. pestis must deploy its arsenal to produce a successful infection. Better knowledge of this arsenal will undoubtedly improve our understanding of the pathogenesis of plague and should help us to develop better countermeasures against this disease. To this end, we previously determined the transcriptome of Y. pestis in the most relevant context of infection: the rat bubo [7]. Although the study results were valuable, they were not predictive of the requirements for virulence and merely provided clues about putative biological roles of upregulated genes in vivo that will have to be confirmed using traditional experimental approaches. We therefore pursued our efforts by devising a means of screening a mutant Y. pestis library in which genes previously identified as being up-regulated in vivo had been deleted. This method enabled us to draw up a hierarchical table ranking genes in order of their importance in disease production. The results of further investigations suggest that horizontal gene acquisition had a key role in establishing Y. pestis' ability to successfully initiate infection after the dermal fleabite.
Methods
Ethics statement
Animals were housed at the Institut Pasteur de Lille animal facility (accredited by the French authorities for the performance of experiments on live rodents (#B59-350009) and which complies with French and European regulations on the care and protection of laboratory animals (EC Directive 86/609 and the French Act #2001-486, issued on June 6, 2001)). At the time some experiments were performed, regulations did not require ethical approval. However, all animal experiments had been authorized by the French veterinary authorities (#59-350218) and were performed in compliance with the terms of NIH Animal Welfare Assurance #A5476-01 (issued on July 2, 2007). Since 2013, animal experiments were approved by the Nord-Pas-de-Calais Ethics board committee (#CEEA 23012). Human serum samples were obtained after the provision of written, informed consent by the donors. The use of human samples has been declared and approved by the MESR and CPP boards (#DC-2013-1765).
The strains, the mutant library and the plasmids
The bacterial strains and plasmids used in this study are presented in Table S1 and Text S1. Mutant Y. pestis strains were generated using allelic exchange methods (based on lambda Red recombinase or the pCVD442 suicide vector [13], [14]) and with the primer sets and plasmids described in Tables S1 and S2. The coding sequence of interest (apart from the first and last 50 base pairs) was replaced by a kanamycin, zeocin or trimethoprim resistance gene that had been amplified from the suicide vectors listed in Table S1. Each mutant's genotype was confirmed by PCR assays using primer sets that were complementary to the target flanking sequence not involved in the allelic exchange (Table S2). Mutant strains were complemented with the recombinant pCRII plasmid (Life technologies, Saint Aubin, France) containing a wild-type (WT) copy of the gene of interest (including the latter's putative promoter sequence).
Determination of bacterial virulence in rodent models of bubonic plague
Virulence attenuation was evaluated in either pools of mutants or individual mutants. Wild-type and mutant strains were cultured in Luria broth (LB) at 21°C, quantified (via the optical density at 600 nm), diluted to the desired cell density in sterile phosphate-buffered saline (PBS) and mixed with equal colony-forming unit (CFU) counts of other mutants if required. Fifty microliters of the bacterial suspension were intradermally inoculated (as previously described [9]) into 8-week-old female Brown-Norway rats (Janvier, France) or OF-1 female mice (Charles River, France). For individual screening, rats and mice were inoculated with 20 and 10 CFUs respectively. For per-pool screening, rats were injected with a total of 100 CFUs (20 CFUs of each of five mutants). Groups of eight and ten animals were respectively used for individual and per-pool screening experiments. For per-pool screening, animals were euthanized when signs of terminal plague appeared. The inguinal lymph nodes proximal to the inoculation site and the spleens were immediately collected after euthanasia and then triturated. Serial dilutions of the triturated organs were plated on blood agar containing the antibiotics of interest. The CFUs were counted after a 48-hour incubation at 28°C. The virulence attenuation of Y. pestis was measured by calculating the average relative competitive index (ARCI), as follows: where is the CFU count of the tested mutant, is the count for the most abundant mutant (i.e. the strain that acts like a WT strain in the animal of interest), is the animal of interest, is the number of animals used in the experiment, and . Groups of five 8-week-old female Brown-Norway rats (inoculated as above) were used to study the time course of lymph node, blood and spleen colonization by bacteria. Lastly, in order to study the role of PMNs cells during infection, groups of 8-week-old OF-1 mice were inoculated intraperitoneally with 100 µg of anti-GR1+ (RB6-8C5) antibody (as described above) on the day before the intradermal challenge [8].
Growth in human serum
Y. pestis grown overnight in LB at 21°C were centrifuged, washed and adjusted to 2×107 bacteria/mL in PBS. Twenty microliters of the bacterial suspension were added to 180 µl of fresh human serum in the wells of a 96-well plate. After different incubation times at 37°C, serial dilutions of the serum were plated on blood agar and the CFUs were subsequently quantified.
Bacterial survival in macrophages
Macrophages were cultured in high-glucose (4.5 g/L) Dulbecco's Modified Eagle Medium (DMEM) supplemented with 5% heat-inactivated fetal bovine serum. All incubations were performed at 37°C in a 5% CO2 atmosphere. In the intracellular survival assay, cells were infected as previously described [15]. The day before interaction with Y. pestis, 1.4×105 RAW macrophages in the incubation medium were allowed to seed in the wells of a 24-well plate. The macrophages were washed three times in incubation medium prior to addition of Y. pestis that had been grown overnight at 21°C in LB, washed and suspended in DMEM. The multiplicity of infection was 10. Immediately after mixture of the bacterial suspension and the macrophages, the plates were centrifuged (at 18 g for 5 minutes) and then incubated. After 30 minutes, the cells were washed three times. Fresh medium supplemented with 8 µg/mL gentamicin was then added. One hour later, the wells were washed twice and fresh medium containing 2 µg/mL of gentamicin was added. At various time points after contact, macrophages were lysed by incubation in 500 µl of cold water for 10 minutes on ice. The lysate was plated on blood agar and the number of CFUs was counted.
Statistical analysis
A mutant was considered to be virulence-affected when the survival curves for animals infected with the mutant and the wild-type strain were found to be significantly different (p<0.05) in a Gehan-Breslow-Wilcoxon test. Two-way analysis of variance was also used to compare data obtained from in vitro experiments; a p<0.05 was considered significant.
Results
More than 15% of the Y. pestis loci up-regulated in vivo are required for virulence
We previously reported that 526 Y. pestis genes (other than previously characterized virulence factors) were up-regulated more than two-fold in the rat bubo, relative to Y. pestis cultures grown at 21 or 37°C [7]. Seventy-nine of these are highly likely to encode factors important for plague pathogenesis because they are (i) up-regulated in both the rat bubo and the mouse lung or (ii) thought to be involved in the bacterium's defensive response against nitric oxide, oxidative stress and iron stress generated by the immune system during bubonic plague in rat [7], [16]. We therefore generated a library of Y. pestis mutants in which one or more of these 79 genes had been deleted and then tested the individual mutant's virulence in mouse or rat models of bubonic plague. In order to reduce the number of mutants and the number of animals to be sacrificed, we deleted blocks of neighboring genes (whenever possible and regardless of their genetic organization and relationship). Hence, a total of 33 mutants (rather than 79) were produced. The mutant lacking the functional pyruvate dehydrogenase AceEF [17] was the only one to show a serious growth defect on LB agar. As expected, this mutant did not kill any mice 15 days after inoculation with a dose (∼20 CFUs) that usually kills all the animals (Table S3) - indicating that the mutant was virulence-attenuated. Screening of the library for virulence revealed five attenuated mutants (Table S3). The strains lacking the homoserine O-succinyltransferase gene metA, the ribonucleoside reductase operon nrdHIEF or the uncharacterized gene ypo0426 required significantly more time to cause fatal plague (median survival ≥5 days vs 4 days for the mutants and the wild-type strain respectively). The Δypo0988, ΔptsG and ΔznuABC ypo2062 mutants (respectively lacking the genes for an uncharacterized protein, the glucose transport PtsG and both the Zinc (Zn) transporter ZnuABC and the putative zinc murein DD-endopeptidase YebA [18]) caused plague but at a lower incidence (survival rate >60% vs ≤12.5 for the mutants and the wild-type respectively). Further virulence assays showed that deletion of yebA (but not znu) was responsible for the loss of virulence in the ΔznuABC ypo2062 mutant (Figure 1).
Fig. 1. Incidence of plague in mice infected with Y. pestis lacking the zinc transporter Znu and/or the putative zinc murein DD-endopeptidase YebA (YPO2062). 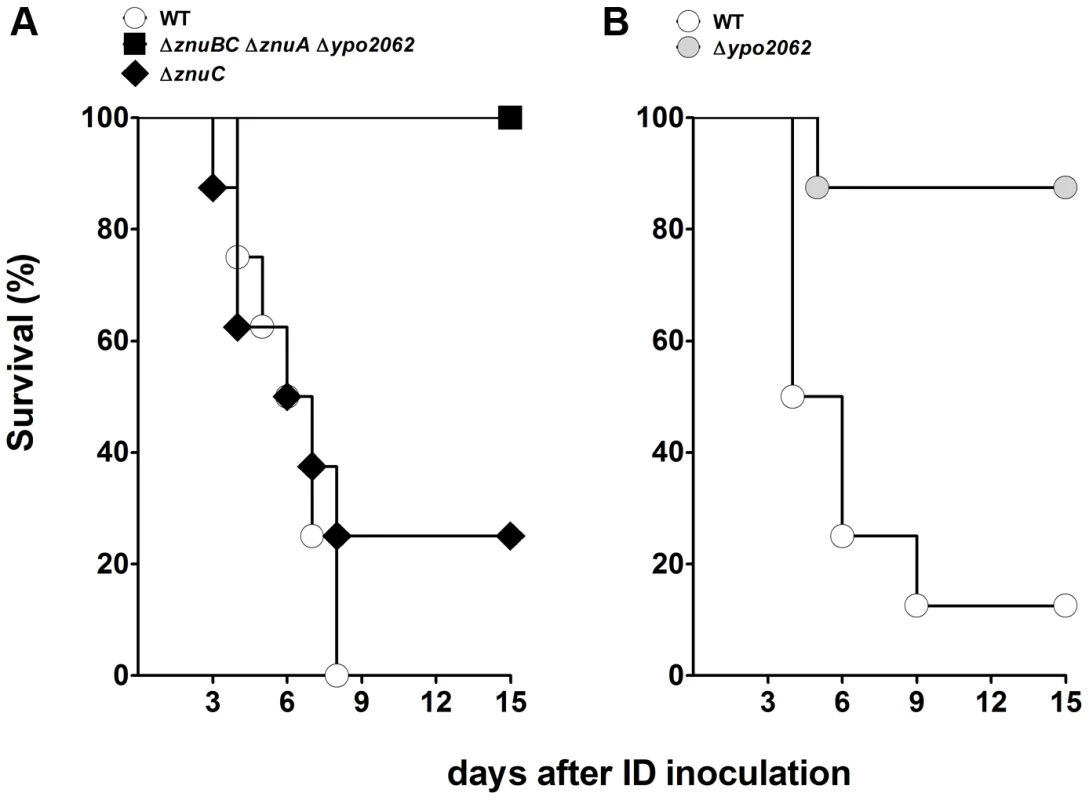
(A), mice were injected intradermally with ∼10 Y. pestis WT (white circles) and ΔznuABC ypo2062 (black squares) or ΔznuC (black diamonds). (B), mice were inoculated with as in (A) with WT (white circles) or Δypo2062 (grey circles) mutants. Only the loss of YPO2062 significantly decreased the virulence of Y. pestis (p<0.05, relative to WT). Groups of 8 mice were used. Data obtained with the ΔznuABC ypo2062 mutant are representative of two experiments. Data obtained with the ΔznuC and Δypo2062 mutants come from one experiment each. Next, we sought to establish which of the remaining 447 up-regulated genes were required for plague progression in rats. To this end, we used the approach described above to build a library of 137 mutants and designed a per-pool screening method for application in the most relevant model of infection. Hence, rats were intradermally inoculated with about the dose of Y. pestis (the input pool) that is regurgitated by the rat flea Xenopsylla cheopis (i.e 100 CFUs) [19]. In order to reduce the selection of false positives and false negatives, each mutant in the pool was given at the lowest dose that consistently killed all the rats in experiments with the WT (20 CFUs). In other words, each pool comprised five mutants. A wild-type strain was not included in each pool because (i) we postulated that at least one mutant in the pool would have a wild-type like phenotype (i.e. colonizing and killing the animals as efficiently as the wild type) and (ii) we expected a bottleneck restricting bacterial dissemination from the skin to the other tissues [20]. To overcome bias due to bottlenecking, we were therefore obliged to evaluate each pool in several animals (see below). To identify mutants within a pool, we provided each one with a specific antibiotic resistance marker (kanamycin, trimethoprim or zeocin, denoted respectively as K, T and Z). The mutants were generated using a virulent strain bearing (or not) a zeocin resistance gene at the only neutral att Tn7 chromosomal site (Figure S1). Hence, five groups of tagged mutants were produced (K, T, Z, ZK, and ZT). Each mutant was counted in the lymph nodes and the spleen (the output pool), in order to (i) identify genes that might be necessary during early or late stages of infection and (ii) reduce the likelihood of missing false negatives (i.e. attenuated mutants that might artificially appear to be virulent as a result of transcomplementation of the mutant's defects by other strains in the pool or the production of an abnormal environment that facilitates the growth of an otherwise virulence-attenuated mutant. We expected rescued mutants to be more outcompeted in the spleen than in the lymph node. Hence, we did not expect a virulence-attenuated mutant to be fully rescued in the final “bacterial read” in the spleen - even when it may have been transcomplemented in the pool. Lastly, the bacterial loads for each mutant were determined in animals displaying terminal clinical signs of plague; at this stage of the disease, (i) the bacterial load recovered from an organ (>107 CFU) is high enough to ensure reproducible recovery of each mutant, and (ii) all the mutants would have experienced similar in vivo growth conditions because the organs from terminally ill animals described in the literature (and certainly from our experience) were all histologically similar [9], [21]. Using this strategy, a total of 24 pools comprising five mutants and one pool comprising four mutants (i.e. 124 mutants in total) was tested. Due to technical issues, the remaining 13 mutants had to be individually evaluated for virulence. One of the 13 had a longer incubation period in animals and was found to lack the prophage ypo1089-ypo1091 locus (Table S3).
Terminal signs of plague occurred 2 to 5 days after inoculation of the pools. The mean time to sacrifice was 2.6±0.4 days (Figure S2). At the time of sacrifice, the lymph nodes and the spleens from all rats were heavily colonized, with a median bacterial count of >108 CFU (Figure S3). These data suggested that each pool behaved collectively like a WT strain and that at least one mutant within each pool was fully virulent because i) the time course of infection of animals infected with either ∼20 or ∼100 wild-type CFU did not differ significantly (data not shown) and ii) organs from animals showing terminal signs of plague reportedly contain more than 107 CFU [9], [22], [23]. Analysis of the CFU counts in the output pools revealed that there was a bottleneck that affected bacterial dissemination from the skin to the other tissues. The proportions of clones recovered from the lymph nodes and the spleens did not appear to follow a particular trend; a clone that completely dominated colonization in one animal could be totally outcompeted in another (Figure S4). This variable “dominant effect” hampers the identification of genes needed for microbial pathogenesis [24], [25]. However, we considered that the “dominant effect” observed in individual animals would be balanced by an analysis of the group of animals (notably by averaging the data obtained from the different replicates). Hence, we determined the relative virulence of each mutant for a specific organ by calculating an average relative competitive index (ARCI) for each mutant by using the equation described in the Methods section. Following inspection of the data, we divided the mutants into three virulence classes with either a high to moderate likelihood of being virulence-attenuated (ARCI≤10), a moderate to low likelihood (10<ARCI<20) or no likelihood (ARCI≥20) (Table S4). The first two categories comprised about 40% of the mutants (48 out of 124) (Table 1). Fifteen of the 48 mutants lacked metabolic genes, 5 lacked stress response genes, 2 lacked fimbriae/adhesin genes and 26 lacked uncharacterized genes. Interestingly, some mutants were more outcompeted in the lymph node than in the spleen (or vice-versa), suggesting that specific factors are required to colonize each organ. To confirm the validity of our hierarchical ranking, the individual virulence of a number of mutants from each of the three different ARCI classes (11, 5 and 4 mutants, respectively) was evaluated in the rat. Eight mutants (variously lacking Pap fimbriae, Ail or uncharacterized proteins) were found to be significantly less virulent (Table S4) and all had an ARCI<20. Furthermore, 65% and 20% of the attenuated mutants were from the “high ARCI” and “low ARCI” categories, respectively; this suggests the existence of a correlation between the ARCI category and loss of virulence.
Tab. 1. List of mutants selected using the per-pool screening method. 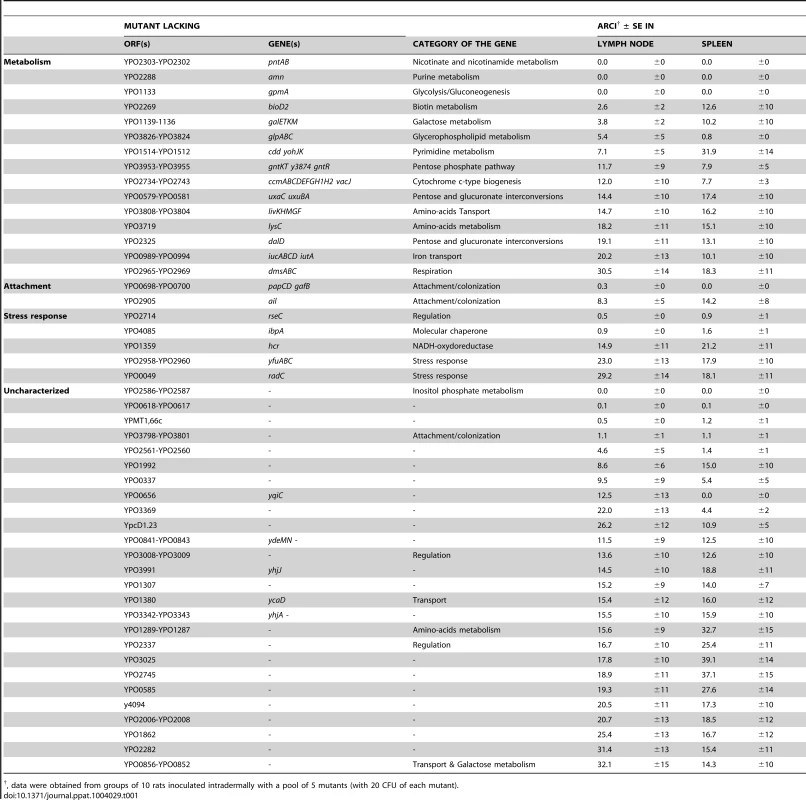
, data were obtained from groups of 10 rats inoculated intradermally with a pool of 5 mutants (with 20 CFU of each mutant). The attenuation of the mutants described above might conceivably result from a secondary mutation generated during the construction process. To test this hypothesis, we compared the virulence of the wild-type strain and several mutants (Δypo0988, Δypo2062, Δypo0656, Δypo2560-61, Δypo3369 and Δypo3991) harboring (or not) a wild-type copy of the gene of interest on a plasmid (Figure S5). In accordance with the results of the above-described infection experiments, all mutants were significantly affected in virulence (p<0.05 in a Gehan-Breslow-Wilcoxon test). The virulence of the complemented mutants Δypo0988, Δypo2062 and Δypo0656 was fully restored. In contrast, the virulence of the complemented mutants Δypo2560-61, Δypo3369 and Δypo3391 was not restored - indicating that these mutants carry a secondary mutation or that the methodology used for complementation was inadequate.
Y. pestis requires a limited number of metal acquisition systems to counter metal starvation in vivo
Iron, manganese and zinc play a crucial role in many biological processes - including bacterial virulence. Hence, it is not surprising that a marked number of metal acquisition systems are up-regulated by Y. pestis in vivo [7], [16]. Notably, Y. pestis overexpresses 12 of the 15 proven or potential iron transport systems (Table S5 and Text S1). It has previously been shown that the absence of the Yersiniabactin (Ybt) siderophore-dependent system or the Yfe ATP-binding cassette (ABC) iron transporter (but not that of the heme transporter Hmu, the hemin storage system Hms or the ABC iron transporters Yiu and Yfu) attenuates the virulence of Y. pestis in a mouse model of bubonic plague [26], [27], [28], [29], [30]. Our present data (obtained in a different Y. pestis strain and a different rodent species and strain) confirmed that Hmu, Hms, Yiu and Yfu are not required for bubonic plague (Table S5). Furthermore, we found that three siderophore-based systems, two ABC ion transporters and two iron permeases were not necessary for disease production, despite upregulation of the corresponding genes in vivo; this is presumably because some of these transport systems are not functional in Y. pestis due to frameshift mutations or gene disruption by an insertion sequence [31].
In vivo, Y. pestis also overexpresses the ZnuABC zinc transporter and the MntH manganese transporter (which are also the predominant zinc and manganese importers in vitro). However, mutants lacking Znu and MntH were found to be fully virulent (Figure 1 and Table S5), which agrees with recent findings in a different Y. pestis strain and a different animal model [32], [33].
Yersinia pestis requires carbohydrate metabolism when colonizing the mammalian host
Inspection of the expression patterns for genes involved in metabolic pathways has suggested that carbohydrates constitute Y. pestis' main carbon and energy source during successful colonization of the mammalian host [7]. The results of our virulence tests indicated that the uptake of glucose, gluconate and, to a lesser extent, maltose (but not that of fructose, mannose or fructuronate) was important for plague production (Figure 2 and Table S3 and S4). Glucose is known to be metabolized via glycolysis; however, the latter did not appear to be essential for plague production because deletion of the first two enzymes in this pathway (Pgi and PfkA) did not affect Y. pestis' virulence. Although the presence of a complete glycolysis pathway was not essential for host colonization, its terminal part appeared to be essential because a gpmA mutant was completely outcompeted in vivo. The end product of glycolysis (pyruvate) is an important source of acetyl CoA, some of which enters the tricarboxylic acid (TCA) cycle via reaction with citrate synthase. However, this step (and perhaps the TCA cycle as a whole) was not essential for virulence. Thus, Y. pestis relies on carbohydrate metabolism to produce disease but carbohydrates such as glucose are not necessarily directed towards the expected metabolic pathways.
Fig. 2. Carbohydrate uptake and metabolism by Y. pestis in its mammalian host. 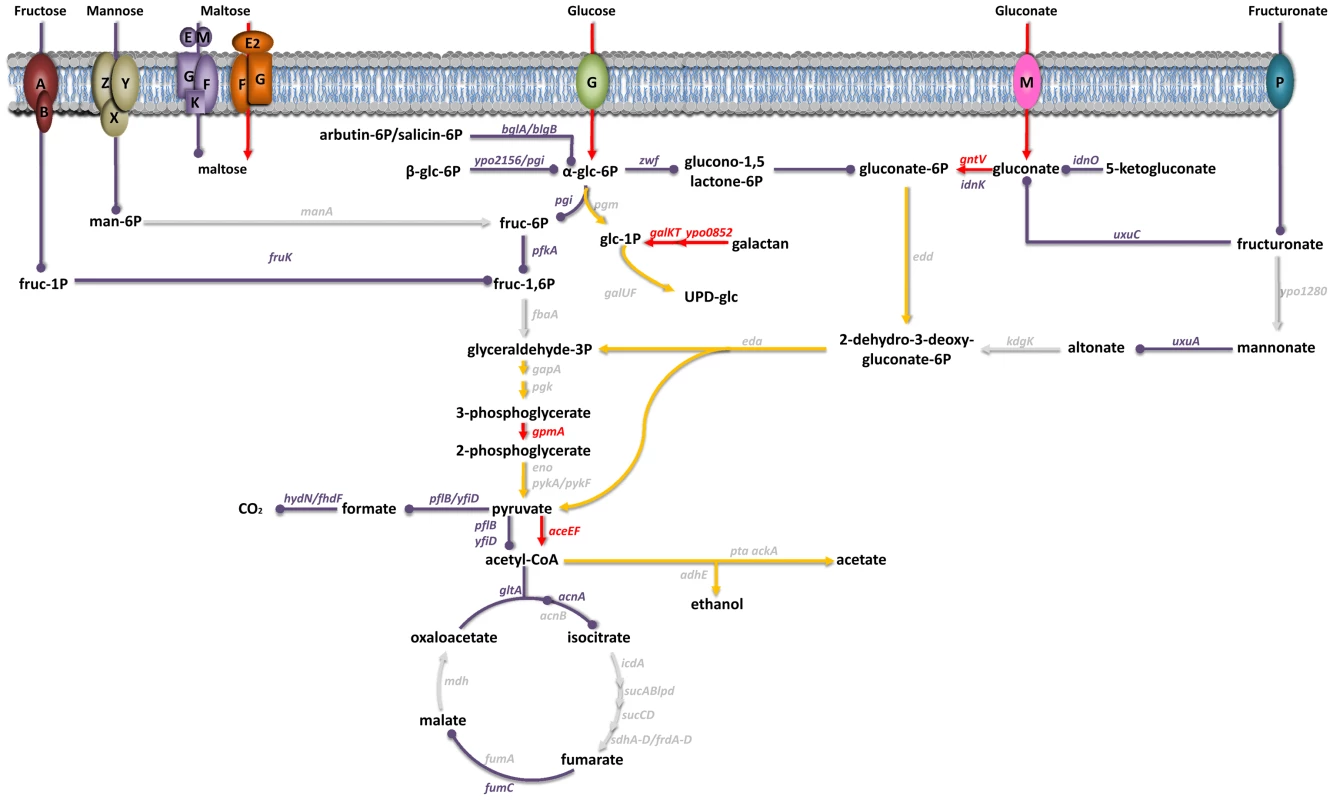
Genes encoding transporters for fructose (FruAB), mannose (ManZYX), maltose (MalEMGKF and MalE2FG), glucose (PtsG), gluconate (GntM) and fructuronate (GntP) and the carbohydrate metabolism genes shown in red or purple were all overexpressed in vivo [7]. In virulence tests, mutants lacking a gene highlighted in red were virulence-attenuated, whereas those lacking a gene highlighted in purple were not. Genes in grey were not upregulated in vivo and their role in virulence has yet to be elucidated; hence, the importance of the metabolic fluxes involving the corresponding enzymes in virulence is unknown (grey arrows). Red and purple arrows respectively indicate the metabolic fluxes considered to be important and non-essential for virulence. Yellow arrowheads represent the metabolic pathways that glucose and gluconate might follow. A limited number of genes enable Y. pestis to resist nitrosative stress in the bubo
During infection, Y. pestis is in close contact with PMNs that produce inducible nitric oxide synthase. The bacterium uses the flavoglobin Hmp to detoxify reactive nitrogen species (RNS) released by the PMNs [7]. Of the Y. pestis genes that are up-regulated in the rat bubo and for which homologs in other bacterial species are involved in the response to RNS, aceE that we found to be necessary for virulence was also found to be required for optimal bacterial growth in vitro (Table S6 and Text S1). Therefore, one can hypothesize that aceE is essential for virulence but is not necessarily required to counter the host PMNs' production of nitric oxide. Of the 8 other loci thought to protect Y. pestis against RNS in vivo, only nrdHIEF (the ribonucleotide reductase operon) appeared to contribute moderately to virulence; deletion of this operon was associated with a slightly longer time to disease onset (Table S6).
Further evidence to suggest that Y. pestis may not encounter oxidative stress in vivo or may require a limited number of genes to resist oxidative stress
The absence of induction in the rat bubo of (i) the OxyR oxidative stress regulator regulon and (ii) the genes known to protect bacteria against peroxynitrite (i.e. the product of the reaction between hydrogen peroxide and nitric oxide) suggested that Y. pestis is not exposed to reactive oxygen species (ROS) in the mammalian host [7]. However, a limited number of genes involved in bacterial resistance against ROS are known to be activated in the rat bubo, and they may encode the major detoxifying proteins. Furthermore, several genes involved in the detoxification of ROS (which are downregulated or at least not upregulated in the rat bubo) were shown to be upregulated in Y. pestis replicating inside a macrophage-cell line in vitro [34]; macrophage infection is thought to be important in the initial stages of infection [6]. Of the genes known to protect bacteria against oxidative stress, Y. pestis overexpresses mntH, fumC, yfiD (grcA) and ibpA in vivo. As mentioned above, MntH is required for manganese import (which in turn is involved in protection against ROS), whereas FumC and GrcA rescue enzyme reactions when FumA and PflB are damaged by ROS [35], [36], [37]. IbpA protects several metabolic enzymes against oxidation [38]. Deletion of the ibpA gene was associated with attenuation of virulence because the mutant was outcompeted in per-pool infection. However, IbpA is also involved in resistance to other forms of stress, such as high temperatures [38], [39]. In contrast, deletion of fumC, yfiD or mntH did not affect the virulence of Y. pestis (Table S6). Thus, our data provide further evidence for the hypothesis whereby ROS production may not be a significant host defense factor in plague.
Horizontal genetic transfer had an important role in acquisition of Y. pestis capacity for intracellular survival
Some of the Y. pestis genes overexpressed in the rat bubo and required for virulence in the present study have been acquired horizontally. These genes include the P4-like prophage genes (ypo1089 to ypo1091) and the ypmt1.66c gene (which encodes a putative DNA binding protein and is located on the pMT1 plasmid). A Δypmt1.66c mutant was outcompeted in a per-pool assay (Table S4). Furthermore, the absence of the ypmt1.66c gene decreased the incidence of plague in both rats and mice (Figure 3). The virulence of the mutant expressing a WT copy of ypmt1.66c under the control of its own promoter from a high copy number plasmid (pCR2) was almost identical to that of the wild-type strain. The complemented mutant killed 100% of animals but caused fatal plague with a slight lag (a median survival time of 4 days, vs. 3.5 for the WT). Even though this slight difference in virulence was statistically significant (P = 0.038), this result indicated that loss of virulence was certainly due to deletion of ypmt1.66c. Interestingly, animals which survived inoculation with the Δypmt1.66c mutant had enlarged, purulent lymph nodes but not severe splenomegaly - suggesting that the mutant was impaired in its ability to initiate skin colonization and/or to escape from the draining lymph node. Consistently, the times to colonization of the lymph node, blood and spleen were significantly longer in rats infected with the mutant than in rats infected with the WT strain (Figure 4). Taken as a whole, our data indicate that a mutant's virulence is attenuated because the host manages to contain the infection during the initial skin colonization step, for which intracellular replication within macrophages is thought to be important [6]. In accordance with this model, the Δypmt1.66c mutant was more susceptible to macrophages than the WT and the complemented mutant (Figure 5A). The mutant's failure to initiate infection at the fleabite site could also result from its inability to use available nutrients or to survive contact with PMNs once released from lysing macrophages. The mutant showed a slight but statistically significant growth defect in serum (used here as model of nutrient acquisition in the skin), when compared with the WT and the complemented strain (Figure 5B). Furthermore, the mutant's respective levels of virulence in neutropenic and immunocompetent mice did not differ significantly (Figure 3C); this indicates that YPMT1.66c is involved in a biological process situated upstream of interaction with PMNs. Taken as a whole, our data suggest that YPMT1.66c's role in survival within macrophages is important in the production of plague.
Fig. 3. Incidence of plague in immunocompetent rats (A), immunocompetent mice (B), and neutropenic mice (C) injected intradermally with ∼20 WT Y. pestis (white circles), Δypmt1.66c (black circles) or complemented mutant (grey circles). 
The mutant was significantly (p<0.05) less virulent than the WT strain in immunocompetent rats (A), immunocompetent mice (B and C) and neutropenic mice (C). The survival curves of mice infected with the complemented mutant harboring the ypmt1.66c gene on a high copy number plasmid (pCR2) and the wild-type (B) were significantly different (p<0.05). The virulence of Y. pestis lacking ympt1.66c was no greater in neutropenic mice than it was in immunocompetent mice (dashed lines) (p>0.05). Data were obtained from groups of 8 or 9 animals. P values were determined using the Gehan-Breslow-Wilcoxon test. Fig. 4. The change over time in colonization and bacterial load in rats inoculated intradermally with ∼20 WT Y. pestis (white circles) or Δypmt1.66c (black circles). 
N.D: not determined. Each circle represents one animal. The times to colonization were significantly longer in rats infected with the mutant than in rats infected with the WT strain (p<0.05). P values were determined using 2-way analysis of variance. Fig. 5. YPMT1.66c is required for intracellular survival in macrophages (A) and for optimal growth in serum (B). 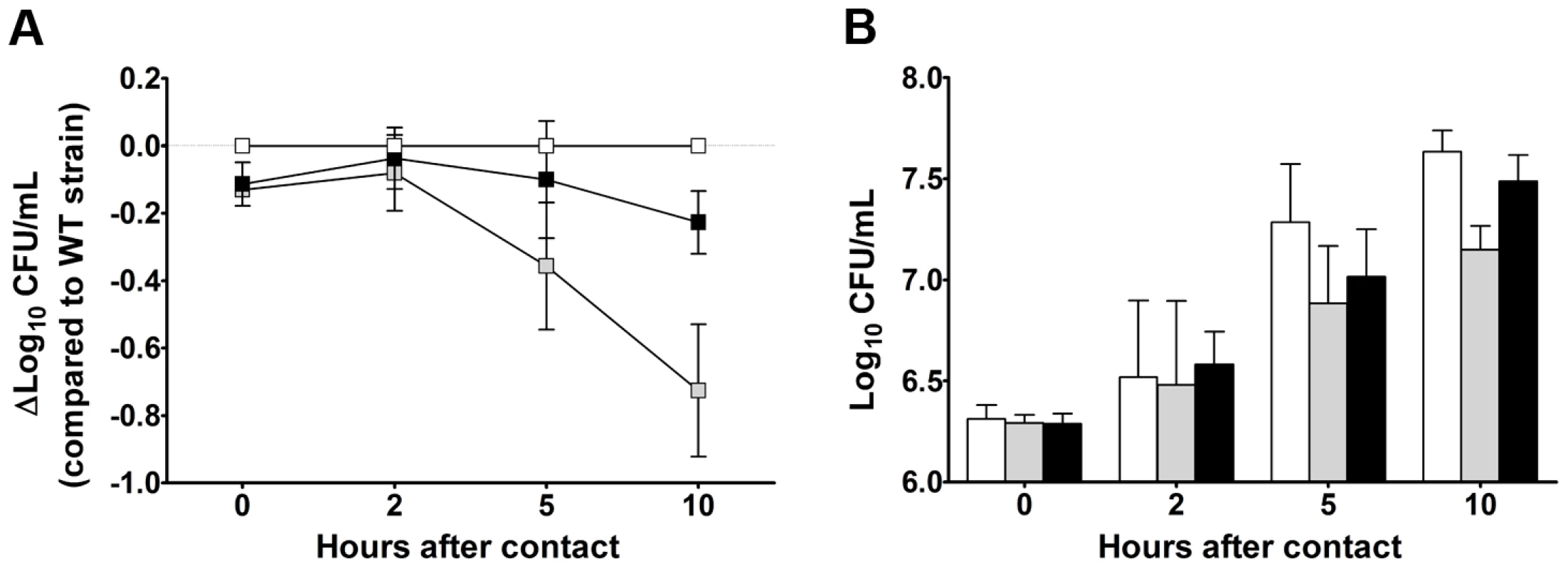
Shown are the data obtained with the wild-type (white squares and bars), the Δypmt1.66c (grey squares and bars) and the complemented mutant (black squares and bars) strains. Data are quoted as the mean (SD) from three independent experiments using macrophages (A) and 5 independent experiments using serum from 5 different healthy donors (B). The mutant's survival (A) and growth curves (B) differed significantly from those of the WT and the complemented strain (p<0.05 in a two-way analysis of variance). The complemented mutant's and the WT's survival (A) and growth curves (B) did not differ significantly (p>0.05 in a two-way analysis of variance). Stress, metabolic and uncharacterized genes are required for resistance and/or growth in serum
Approximately 40% of the mutants found to be virulence-attenuated in individual tests and classified in the high-to-low likelihood class for virulence attenuation (i.e. ARCI≤10 and 10<ARCI<20) are known to encode proteins related to metabolic pathways. The remaining 60% of mutants lack factors considered to be involved in stress resistance (e.g. IbpA or RseC), bacterial attachment or serum resistance (e.g. Ail). Indeed, most of the remaining mutants are involved in hypothetical or uncharacterized processes. To determine whether the selected mutants were impaired for bacterial growth in vivo rather than for resistance to the host immune system, we measured their ability to grow in fresh serum (Figure 6). However, the mutants with a 10<ARCI<20 were not evaluated because we expected few strains in this ARCI category to be truly virulence-affected (see discussion) and therefore expected even fewer mutants to be serum-sensitive. Furthermore, the ΔaceEF mutant (identified in an individual virulence test) was not evaluated because its serious growth defect in LB led us to expect a growth defect in serum. Lastly, the ARCI≤10 mutants lacking genes that encode fimbriae or genes found to be non-essential for plague in individual tests were not evaluated because (i) we did not expect fimbriae to be involved in serum resistance and (ii) mutants that were found to be attenuated in per-pool screening but not in individual tests were considered to be false positives. Hence, a total of 23 mutants were evaluated for serum resistance. Ten of them differed significantly from the wild-type strain. In particular, the ΔrseC, Δypo0337 and ΔgpmA mutants were sensitive to serum (presumably because of the bactericidal effect of the complement), and the ΔibpA, Δypo3369, Δypo0988, Δamn, Δypo0617-0618, Δypo2586-2587 and Δypo0426 mutants had significant a growth defect (either because they confer a metabolic advantage or effective complement tolerance).
Fig. 6. Stress, metabolic and uncharacterized genes are needed for resistance and/or growth in serum. 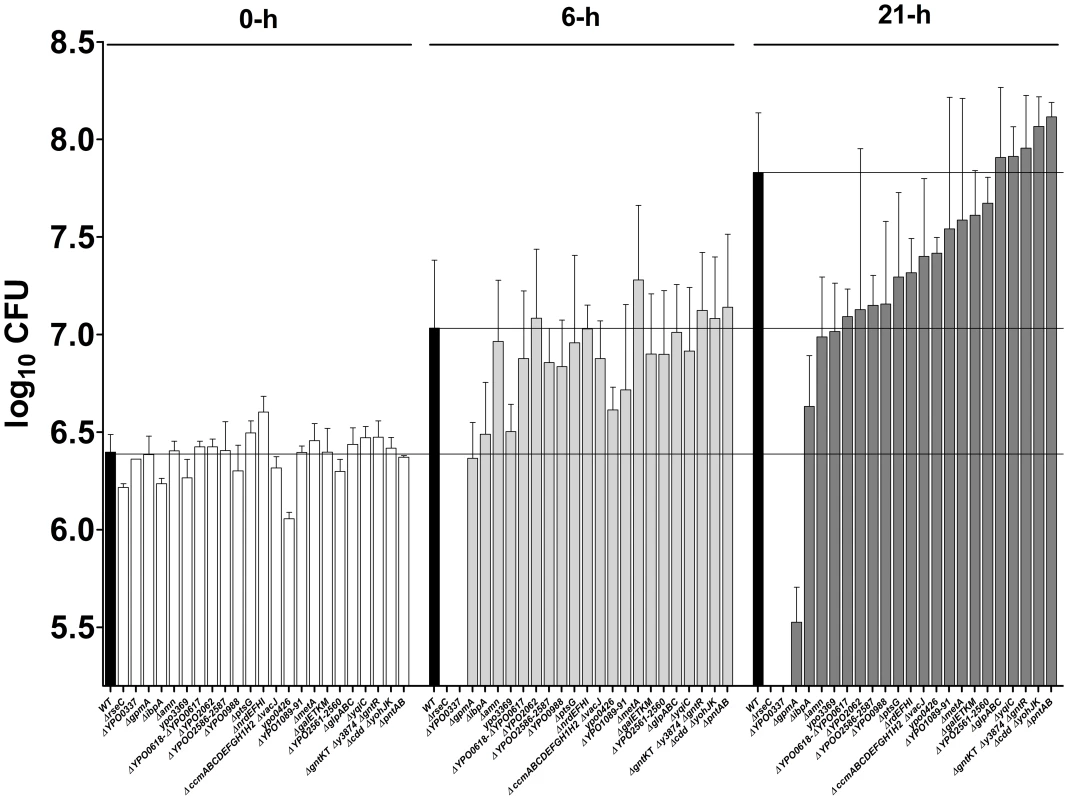
The ability of selected mutants to grow in fresh serum was evaluated after 6 and 21ΔrseC, Δypo0337, ΔgpmA, ΔibpA, Δypo3369, Δypo0988, Δamn, Δypo0617-0618, Δypo2586-2587 and Δypo0426 mutants differed significantly from that of the wild-type strain (p<0.05 in a two-way analysis of variance). Discussion
Here, we presented a novel experimental approach for establishing a hierarchical table of Y. pestis genes previously identified (in a microarray analysis) as being up-regulated in vivo. The genes' ranking in terms of their importance in virulence should provide new insights into the pathogenesis of plague. Our approach consisted of (i) testing the virulence of individual mutants each lacking a locus likely to encode factors important for plague pathogenesis and (ii) per-pool screening of a library of mutants composed of mutants strains each lacking a locus with no obvious role in pathogenesis and of which ∼60% of the strains lacked genes that were not directly involved in any known metabolic pathway. Our per-pool screening method is based on signature-tagged mutagenesis (STM) [40] and is closely related to “array-based analysis of cistron under selection” (ABACUS) technology [41]. We first generated a library of non-polar, tagged deletion mutants by using lambda-Red technology [13] (as in ABACUS) rather than a transposon mutant library (as in STM). Each mutant in the pool was tagged with a unique antibiotic resistance cassette, which ultimately allowed the numbers of viable, tagged mutants in infected tissues to be quantified by CFU counting on selective media. This aspect contrasts with the ABACUS method, in which (i) a T7 RNA polymerase promoter is inserted into the target gene and (ii) mutants are monitored with a microarray using labeled RNAs synthesized in vitro from the T7 promoter and mutant DNA that has been fragmented, poly(A)-tailed and amplified prior to transcription. Hence, in contrast to ABACUS and other methodologies based on microarrays or deep sequencing (such as STM-like or transposon-directed insertion-site sequencing), our method does not require nucleic acid purification, RNA synthesis, PCRs, microarray hybridization or sequencing [41], [42], [43]. It therefore minimizes the likelihood of introducing bias related to sample preparation, hybridization and/or sequencing. One could argue that our “low complexity”, per-pool approach is limited by the number of antibiotic tags available. However, various fluorescent tags (such as green or red fluorescent protein), bioluminescent tags (blue or red) and enzymatic tags (such as LacZ) can be used to increase the number of mutants in the pool. Hence, it would be easy to match the pool to the desired complexity and, ultimately, to the most relevant infection conditions. In fact, the tag approach enabled us to generate a low-complexity pool that corresponded to inoculation of a physiological dose of bacteria, i.e. that delivered by a flea during a blood meal.
By injecting pools of mutants under the most relevant infection conditions, we confirmed previous suggestions whereby Y. pestis encounters a bottleneck during dissemination from the skin (Figure S4) [20], [44], [45]. This dissemination bottleneck has been observed for several pathogens (including Bacillus anthracis and Francisella tularensis) and was previously considered to prevent the use of per-pool screening methodologies other than when very high doses were inoculated [25], [41]. Our results do not support the latter hypothesis. Our calculation of the ARCI enabled us to address the bottleneck problem and thus draw up a hierarchical table of Y. pestis genes in terms of their importance in plague. We then validated this hierarchical table by testing the virulence of several mutants individually. With the exclusion of ail (which was identified as needed for plague pathogenesis by other researchers while we were performing our experiments [46]), 16 new loci were found here to be important for virulence in mice and/or rats (Tables S3 and S4). Among them, the uncharacterized ypmt1.66c and ypo0988 genes were required for optimal growth in serum (Figures 5B and 6). Furthermore, the uncharacterized ypo0337 gene (previously identified by signature-tagged mutagenesis in Yersinia pseudotuberculosis [45], the recent ancestor of Y. pestis [47]) was essential for serum resistance (Figure 6); this finding suggested that YPO0337 is a new, uncharacterized virulence factor. Thirty-two mutants identified as being virulence-attenuated in per-pool screening were not reassessed individually in the present study (Table S4). Eleven and 20 of these mutants belonged to the ARCI≤10 and ARCI<20 classes respectively. Based on the virulence of individual mutants in the different ARCI classes, we estimate that 65% of the mutants with an ARCI<10 and 20% of those with an ARCI<20 might truly be virulence-attenuated. Hence, at least 12 other loci can conceivably be added to the current list of 16 new loci found to be important in plague pathogenesis. Accordingly, several genes were required for survival or optimal growth in serum (Figure 6). However, we cannot rule out the possibility that some of the mutants identified here are defective in virulence because they harbor a secondary mutation (Figure S5). The apparent absence of complementation might also result from use of an inadequate method or other, uncharacterized features.
Our virulence testing results suggest that in the mammalian host, Y. pestis (i) relies on carbohydrates (i.e. glucose, galactans, gluconate and perhaps maltose) as its carbon source and (ii) shifts to anaerobic respiration (maybe using dimethyl sulfoxide (DMSO) or glycerol) and/or fermentation (Figure 2 and Tables S4, S5, S6). The data indicated that glucose (imported via the PtsG transporter) and galactans were channeled towards the synthesis of UDP-glucose rather than to glycolysis. Firstly, Y. pestis does not encode a functional glucose-6-P dehydrogenase (Zwf, the enzyme that catalyzes the first step in the metabolism of hexose via the pentose phosphate pathway [48]. Secondly, deletion of the first two enzymes in the glycolysis pathway (Pgi and PfkA) did not impact virulence (Figure 2 and Table S3). Whereas carbohydrates do not feed into the upper part of glycolysis, gluconate (taken by the GntM transporter) can potentially supply glyceraldehyde-3-phosphate (G3P) to the lower part of the pathway. The latter metabolite appeared to be essential for plague production, since a gpmA mutant was completely outcompeted in vivo (Figure 2 and Table S4). Glyceraldehyde-3-phosphate is an important carbon source in hexose and amino-acid anabolism and pyruvate production [49]. However, we do not have any evidence to suggest that G3P is involved in hexose and amino-acids anabolism in Y. pestis. In particular, SerA (YPO0914, which catalyzes the first step in the biosynthesis of serine from glycerate-3P that is produced from G3P) was not necessary for plague. Whereas G3P's role in hexose and amino-acid anabolism in Y. pestis has yet to be confirmed, this compound is likely to be crucial for the production of pyruvate in vivo because a mutant lacking the pyruvate dehydrogenase AceEF was unable to produce fatal plague when injected at a dose known to kill most animals. Lastly, the facts that (i) the tricarboxylic acid cycle (TCA) is down-regulated in vivo [7], (ii) the mutant lacking the acotinase A (part of the TCA cycle) is fully virulent (unpublished data) and (iii) the mutant lacking the heat-stable iron-independent fumarate enzyme (FumC, part of the TCA cycle) was not outcompeted in per-pool virulence screening in contrast to the mutants lacking DMSO reductase (DmsABC, YPO2965-2969) and glycerol-3P dehydrogenase (GlpABC) – both involved in anaerobic respiration - suggested that Y. pestis relies on anaerobic respiration to produce plague.
Our virulence testing results also suggest that in the mammalian host, Y. pestis experiences iron, RNS and temperature stresses but not ROS stress (Figure 2 and Tables S4, S5, S6). This supports the hypothesizes generated on the basis of in vivo gene expression profiling [7]. However, we tested a small number of genes involved in bacterial resistance against ROS (Table S6) because few oxidative stress response genes are activated in the rat bubo [7]. Several of the genes involved in the detoxification of ROS which are not upregulated or even downregulated in vivo are upregulated in Y. pestis replicating inside macrophage-cell line [34] - a step suggested to be important in the initial stages of infection [6]. The difference in expression levels found in rat bubo and in macrophages cell-line in vitro may indicate that Y. pestis encounters ROS during the initial stage of infection but not later on. However, the detoxifying hydroperoxidase katG gene (the expression of which is upregulated ∼10 fold during replication within the macrophage) was found to be dispensable for plague pathogenesis [50]. Hence, the role of ROS during plague infection remains to be accurately established by systematic deletion of the ROS resistance/responding genes and characterization of the resulting mutants.
Our screening results also suggested that Y. pestis only needs a limited number of genes to successfully infect its mammalian host. This might be attributable to redundant genes that constitute a “spare wheel” system. For instance, serine biosynthesis enzyme SerA (YPO0914) was not required for virulence but its loss could be rescued by a second putative SerA (YPO1288) enzyme. Alternatively, the requirement for a limited number of genes may be because only one of several genes with overlapping functions are needed in a given context. For example, Y. pestis upregulates both idnK and gntV encoding gluconate kinases but only gntV was required for virulence (Figure 2) [51]. One can hypothesize that IndK is needed to establish an infection in the flea. Lastly, the requirement for just a limited number of genes might indicate ancestral needs for colonizing other environments. Hence, one would expect at least some of the redundant genes to be inactivated and/or deleted from the genome as the bacterium retains only the most appropriate system for its current lifestyle. This is consistent with the suggestion that Y. pestis is undergoing genome decay [52]. Data on the role of iron acquisition systems in plague provide support for this decay scenario. Of the four siderophore-based uptake systems overexpressed by Y. pestis in vivo (yersiniabactin, pseudochelin, aerobactin and alcaligin) and whose homologs in other bacteria are known to have a role in lifestyle, only the Yersiniabactin system is crucial for plague - presumably because the aerobactin and alcaligin systems are not functional in Y. pestis (Table S5) [7], [26], [53], [54].
In addition to genome decay, the emergence of Y. pestis has been characterized by the horizontal acquisition of the plasmids pPst and pMT1/pFra [52]. The two plasmids have had a crucial role in the emergence of flea-borne plague [55], [56], [57], [58], [59], [60], [61]. pPst encodes a plasminogen activator that is thought to prevent the host from containing the Y. pestis infection at the dermal fleabite site. pFra encodes the pseudocapsule F1, whose antiphagocytic activity and/or ability to interfere with interleukin-1 signaling increases the incidence of plague. pFra harbors also several uncharacterized regions (located near the caf operon and elsewhere) which may encode virulence factors [62]. None of these regions contained the uncharacterized gene ypmt1.66c (encoding a protein that is conserved among Y. pestis strains). Here, we found that that ypmt1.66c is also important for maximum disease production at the dermal infection site (Figures 3 and 4). Our observation provides a new example of the importance of horizontal gene acquisition in the emergence of flea-borne plague.
The deletion of ypmt1.66c reduced Y. pestis' ability to resist macrophages, and slightly hampers the bacterium's ability to grow in normal serum (Figure 5) - suggesting that ympt1.66c is partially needed for resisting complement and/or to acquiring nutrients from the serum (and perhaps also at the fleabite site). We did not test whether YPMT1.66c is needed to confront the PMNs that are recruited en masse in infected tissues. Even if this is the case, YPMT1.66c's role as an intracellular survival factor appears to be a prerequisite for resistance to PMNs because the virulence of a mutant lacking YPMT1.66c was not restored in neutropenic mice (Figure 3C). Taken as a whole, our data provide support for the scenario in which Y. pestis regurgitated at the fleabite site is taken up by macrophages and indicate that YPMT166c is required for replication within the phagocyte. This scenario is consistent with the current model of plague, in which the conditions for intracellular replication at the initial stage of infection (i.e. at the dermal flea bite site) are critical for the subsequent production of a bubonic infection [6], [9].
Conclusion
We leveraged our previously collected in vivo gene expression profiling data on Y. pestis [7] to generate and screen a non-polar tagged deletion mutant library composed of bacteria lacking genes known to be upregulated in vivo. This approach constituted the first integrated attempt to identify the molecular mechanisms of the pathogenesis of plague in the most relevant biological model of infection - the mammalian host. Our approach could be transposed to other pathogens and notably those for which a dissemination bottleneck has been described [24], [25]. Our data provided further insights into the pathogenesis of plague by identifying new genes important for plague pathogenesis (including several previously uncharacterized sequences). The fact that these genes are shared with several other pathogens suggests that they belong to a core set of bacterial pathogenesis genes. Confirmation of these virulence results (by individually testing the virulence of mutants selected from the pools and by performing complementation experiments) and further characterization of these new identified genes will be necessary. Future studies should therefore improve our understanding of pathogenesis and, in particular, yield putative antibiotic targets for several diseases.
Supporting Information
Zdroje
1. Sherman IW (2007) Twelve diseases that changed our world. Washington DC: ASM Press: 68.
2. YersinA (1894) La peste bubonique à Hong-Kong. Ann Inst Pasteur 8 : 662–667.
3. GuiyouleA, GerbaudG, BuchrieserC, GalimandM, RahalisonL, et al. (2001) Transferable plasmid-mediated resistance to streptomycin in a clinical isolate of Yersinia pestis. Emerg Infect Dis 7 : 43–48.
4. JoskoD (2004) Yersinia pestis: still a plague in the 21st century. Clin Lab Sci 17 : 25–29.
5. SimondPL (1898) La propagation de la peste. Ann Inst Pasteur 12 : 626–686.
6. PujolC, BliskaJB (2005) Turning Yersinia pathogenesis outside in: subversion of macrophage function by intracellular yersiniae. Clin Immunol 114 : 216–226.
7. SebbaneF, LemaitreN, SturdevantDE, RebeilR, VirtanevaK, et al. (2006) Adaptive response of Yersinia pestis to extracellular effectors of innate immunity during bubonic plague. Proc Natl Acad Sci U S A 103 : 11766–11771.
8. DerbiseA, PierreF, MerchezM, PradelE, LaouamiS, et al. (2013) Inheritance of the lysozyme inhibitor Ivy was an important evolutionary step by Yersinia pestis to avoid the host innate immune response. J Infect Dis 207 : 1535–1543.
9. SebbaneF, GardnerD, LongD, GowenBB, HinnebuschBJ (2005) Kinetics of disease progression and host response in a rat model of bubonic plague. Am J Pathol 166 : 1427–1439.
10. FlexnerS (1901) The pathology of bubonic plague. Am J Med Sci 122 : 396–416.
11. VadyvalooV, JarrettC, SturdevantDE, SebbaneF, HinnebuschBJ (2010) Transit through the flea vector induces a pretransmission innate immunity resistance phenotype in Yersinia pestis. PLoS Pathog 6: e1000783.
12. Hinnebusch BJ (2012) Yersinia. Systems Biology and Control. Caister Academic Press.
13. DatsenkoKA, WannerBL (2000) One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A 97 : 6640–6645.
14. DonnenbergMS, KaperJB (1991) Construction of an eae deletion mutant of enteropathogenic Escherichia coli by using a positive-selection suicide vector. Infect Immun 59 : 4310–4317.
15. PujolC, BliskaJB (2003) The ability to replicate in macrophages is conserved between Yersinia pestis and Yersinia pseudotuberculosis. Infect Immun 71 : 5892–5899.
16. LathemWW, CrosbySD, MillerVL, GoldmanWE (2005) Progression of primary pneumonic plague: a mouse model of infection, pathology, and bacterial transcriptional activity. Proc Natl Acad Sci U S A 102 : 17786–17791.
17. AngelidesKJ, AkiyamaSK, HammesGG (1979) Subunit stoichiometry and molecular weight of the pyruvate dehydrogenase multienzyme complex from Escherichia coli. Proc Natl Acad Sci U S A 76 : 3279–3283.
18. UeharaT, DinhT, BernhardtTG (2009) LytM-domain factors are required for daughter cell separation and rapid ampicillin-induced lysis in Escherichia coli. J Bacteriol 191 : 5094–5107.
19. LorangeEA, RaceBL, SebbaneF, Joseph HinnebuschB (2005) Poor vector competence of fleas and the evolution of hypervirulence in Yersinia pestis. J Infect Dis 191 : 1907–1912.
20. SebbaneF, DevalckenaereA, FoulonJ, CarnielE, SimonetM (2001) Silencing and reactivation of urease in Yersinia pestis is determined by one G residue at a specific position in the ureD gene. Infect Immun 69 : 170–176.
21. LemaitreN, SebbaneF, LongD, HinnebuschBJ (2006) Yersinia pestis YopJ suppresses tumor necrosis factor alpha induction and contributes to apoptosis of immune cells in the lymph node but is not required for virulence in a rat model of bubonic plague. Infect Immun 74 : 5126–5131.
22. ComerJE, SturdevantDE, CarmodyAB, VirtanevaK, GardnerD, et al. (2010) Transcriptomic and innate immune responses to Yersinia pestis in the lymph node during bubonic plague. Infect Immun 78 : 5086–5098.
23. BosioCF, JarrettCO, GardnerD, HinnebuschBJ (2012) Kinetics of innate immune response to Yersinia pestis after intradermal infection in a mouse model. Infect Immun 80 : 4034–4045.
24. WeissDS, BrotckeA, HenryT, MargolisJJ, ChanK, et al. (2007) In vivo negative selection screen identifies genes required for Francisella virulence. Proc Natl Acad Sci U S A 104 : 6037–6042.
25. PlautRD, KellyVK, LeeGM, StibitzS, MerkelTJ (2012) A dissemination bottleneck in murine inhalational anthrax. Infect Immun 80 : 3189–3193.
26. ItemanI, GuiyouleA, de AlmeidaAM, GuilvoutI, BarantonG, et al. (1993) Relationship between loss of pigmentation and deletion of the chromosomal iron-regulated irp2 gene in Yersinia pestis: evidence for separate but related events. Infect Immun 61 : 2717–2722.
27. BeardenSW, PerryRD (1999) The Yfe system of Yersinia pestis transports iron and manganese and is required for full virulence of plague. Mol Microbiol 32 : 403–414.
28. RossiMS, FetherstonJD, LetoffeS, CarnielE, PerryRD, et al. (2001) Identification and characterization of the hemophore-dependent heme acquisition system of Yersinia pestis. Infect Immun 69 : 6707–6717.
29. LillardJWJr, BeardenSW, FetherstonJD, PerryRD (1999) The haemin storage (Hms+) phenotype of Yersinia pestis is not essential for the pathogenesis of bubonic plague in mammals. Microbiology 145(Pt 1): 197–209.
30. KirillinaO, BobrovAG, FetherstonJD, PerryRD (2006) Hierarchy of iron uptake systems: Yfu and Yiu are functional in Yersinia pestis. Infect Immun 74 : 6171–6178.
31. Perry RD (2004) in Iron Transport in Bacteria, eds Crosa, J H, Mey, A R & Payne, S M (Am Soc Microbiol, Washington, DC): 219–240.
32. DesrosiersDC, BeardenSW, MierIJr, AbneyJ, PaulleyJT, et al. (2010) Znu is the predominant zinc importer in Yersinia pestis during in vitro growth but is not essential for virulence. Infect Immun 78 : 5163–5177.
33. PerryRD, CraigSK, AbneyJ, BobrovAG, KirillinaO, et al. (2012) Manganese transporters Yfe and MntH are Fur-regulated and important for the virulence of Yersinia pestis. Microbiology 158 : 804–815.
34. FukutoHS, SvetlanovA, PalmerLE, KarzaiAW, BliskaJB (2010) Global gene expression profiling of Yersinia pestis replicating inside macrophages reveals the roles of a putative stress-induced operon in regulating type III secretion and intracellular cell division. Infect Immun 78 : 3700–3715.
35. AnjemA, VargheseS, ImlayJA (2009) Manganese import is a key element of the OxyR response to hydrogen peroxide in Escherichia coli. Mol Microbiol 72 : 844–858.
36. TsengCP, YuCC, LinHH, ChangCY, KuoJT (2001) Oxygen - and growth rate-dependent regulation of Escherichia coli fumarase (FumA, FumB, and FumC) activity. J Bacteriol 183 : 461–467.
37. WagnerAF, SchultzS, BomkeJ, PilsT, LehmannWD, et al. (2001) YfiD of Escherichia coli and Y06I of bacteriophage T4 as autonomous glycyl radical cofactors reconstituting the catalytic center of oxygen-fragmented pyruvate formate-lyase. Biochem Biophys Res Commun 285 : 456–462.
38. KitagawaM, MiyakawaM, MatsumuraY, TsuchidoT (2002) Escherichia coli small heat shock proteins, IbpA and IbpB, protect enzymes from inactivation by heat and oxidants. Eur J Biochem 269 : 2907–2917.
39. ThomasJG, BaneyxF (1998) Roles of the Escherichia coli small heat shock proteins IbpA and IbpB in thermal stress management: comparison with ClpA, ClpB, and HtpG In vivo. J Bacteriol 180 : 5165–5172.
40. MahanMJ, SlauchJM, MekalanosJJ (1993) Selection of bacterial virulence genes that are specifically induced in host tissues. Science 259 : 686–688.
41. SantiviagoCA, ReynoldsMM, PorwollikS, ChoiSH, LongF, et al. (2009) Analysis of pools of targeted Salmonella deletion mutants identifies novel genes affecting fitness during competitive infection in mice. PLoS Pathog 5: e1000477.
42. MecsasJ (2002) Use of signature-tagged mutagenesis in pathogenesis studies. Curr Opin Microbiol 5 : 33–37.
43. LangridgeGC, PhanMD, TurnerDJ, PerkinsTT, PartsL, et al. (2009) Simultaneous assay of every Salmonella Typhi gene using one million transposon mutants. Genome Res 19 : 2308–2316.
44. OystonPC, KarlyshevAV, WrenBW, TitballRW (2003) Signature-tagged mutagenesis of Yersinia pestis. Adv Exp Med Biol 529 : 39–41.
45. KarlyshevAV, OystonPC, WilliamsK, ClarkGC, TitballRW, et al. (2001) Application of high-density array-based signature-tagged mutagenesis to discover novel Yersinia virulence-associated genes. Infect Immun 69 : 7810–7819.
46. HinnebuschBJ, JarrettCO, CallisonJA, GardnerD, BuchananSK, et al. (2011) Role of the Yersinia pestis Ail protein in preventing a protective polymorphonuclear leukocyte response during bubonic plague. Infect Immun 79 : 4984–4989.
47. AchtmanM, ZurthK, MorelliG, TorreaG, GuiyouleA, et al. (1999) Yersinia pestis, the cause of plague, is a recently emerged clone of Yersinia pseudotuberculosis. Proc Natl Acad Sci U S A 96 : 14043–14048.
48. MortlockRP (1962) Gluconate metabolism of Pasteurella pestis. J Bacteriol 84 : 53–59.
49. AtkinsonDE (1969) Regulation of enzyme function. Annu Rev Microbiol 23 : 47–68.
50. HanY, GengJ, QiuY, GuoZ, ZhouD, et al. (2008) Physiological and regulatory characterization of KatA and KatY in Yersinia pestis. DNA Cell Biol 27 : 453–462.
51. IsturizT, PalmeroE, Vitelli-FloresJ (1986) Mutations affecting gluconate catabolism in Escherichia coli. Genetic mapping of the locus for the thermosensitive gluconokinase. J Gen Microbiol 132 : 3209–3219.
52. ChainPS, CarnielE, LarimerFW, LamerdinJ, StoutlandPO, et al. (2004) Insights into the evolution of Yersinia pestis through whole-genome comparison with Yersinia pseudotuberculosis. Proc Natl Acad Sci U S A 101 : 13826–13831.
53. GarciaEC, BrumbaughAR, MobleyHL (2011) Redundancy and specificity of Escherichia coli iron acquisition systems during urinary tract infection. Infect Immun 79 : 1225–1235.
54. BrickmanTJ, CummingsCA, LiewSY, RelmanDA, ArmstrongSK (2012) Transcriptional profiling of the iron starvation response in Bordetella pertussis provides new insights into siderophore utilization and virulence gene expression. J Bacteriol 193 : 4798–4812.
55. SodeindeOA, SubrahmanyamYV, StarkK, QuanT, BaoY, et al. (1992) A surface protease and the invasive character of plague. Science 258 : 1004–1007.
56. SebbaneF, JarrettCO, GardnerD, LongD, HinnebuschBJ (2006) Role of the Yersinia pestis plasminogen activator in the incidence of distinct septicemic and bubonic forms of flea-borne plague. Proc Natl Acad Sci U S A 103 : 5526–5530.
57. SebbaneF, JarrettC, GardnerD, LongD, HinnebuschBJ (2009) The Yersinia pestis caf1M1A1 fimbrial capsule operon promotes transmission by flea bite in a mouse model of bubonic plague. Infect Immun 77 : 1222–1229.
58. DuY, RosqvistR, ForsbergA (2002) Role of fraction 1 antigen of Yersinia pestis in inhibition of phagocytosis. Infect Immun 70 : 1453–1460.
59. Zav'yalovVP, ChernovskayaTV, NavolotskayaEV, KarlyshevAV, MacIntyreS, et al. (1995) Specific high affinity binding of human interleukin 1 beta by Caf1A usher protein of Yersinia pestis. FEBS Lett 371 : 65–68.
60. HinnebuschBJ, RudolphAE, CherepanovP, DixonJE, SchwanTG, et al. (2002) Role of Yersinia murine toxin in survival of Yersinia pestis in the midgut of the flea vector. Science 296 : 733–735.
61. WeeningEH, CathelynJS, KaufmanG, LawrenzMB, PriceP, et al. (2011) The dependence of the Yersinia pestis capsule on pathogenesis is influenced by the mouse background. Infect Immun 79 : 644–652.
62. WelkosSL, AndrewsGP, LindlerLE, SnellingsNJ, StrachanSD (2004) Mu dI1(Ap lac) mutagenesis of Yersinia pestis plasmid pFra and identification of temperature-regulated loci associated with virulence. Plasmid 51 : 1–11.
Štítky
Hygiena a epidemiologie Infekční lékařství Laboratoř
Článek DHX36 Enhances RIG-I Signaling by Facilitating PKR-Mediated Antiviral Stress Granule FormationČlánek Oral Bacteria and CancerČlánek A Non-Coding RNA Promotes Bacterial Persistence and Decreases Virulence by Regulating a Regulator in
Článek vyšel v časopisePLOS Pathogens
Nejčtenější tento týden
2014 Číslo 3- Stillova choroba: vzácné a závažné systémové onemocnění
- Diagnostika virových hepatitid v kostce – zorientujte se (nejen) v sérologii
- Diagnostický algoritmus při podezření na syndrom periodické horečky
- Perorální antivirotika jako vysoce efektivní nástroj prevence hospitalizací kvůli COVID-19 − otázky a odpovědi pro praxi
- Infekční komplikace virových respiračních infekcí – sekundární bakteriální a aspergilové pneumonie
-
Všechny články tohoto čísla
- Conflicting Interests in the Pathogen–Host Tug of War: Fungal Micronutrient Scavenging Versus Mammalian Nutritional Immunity
- Putting Fungi to Work: Harvesting a Cornucopia of Drugs, Toxins, and Antibiotics
- Mycobacteriophages: Windows into Tuberculosis
- Human African Trypanosomiasis and Immunological Memory: Effect on Phenotypic Lymphocyte Profiles and Humoral Immunity
- Five Things to Know about Genetically Modified (GM) Insects for Vector Control
- A Missing Dimension in Measures of Vaccination Impacts
- Eosinophils Are Important for Protection, Immunoregulation and Pathology during Infection with Nematode Microfilariae
- Clonality of HTLV-2 in Natural Infection
- Production, Fate and Pathogenicity of Plasma Microparticles in Murine Cerebral Malaria
- Group B Streptococcal Infection of the Choriodecidua Induces Dysfunction of the Cytokeratin Network in Amniotic Epithelium: A Pathway to Membrane Weakening
- New Insights into How Adapts to Its Mammalian Host during Bubonic Plague
- Foodborne Transmission of Nipah Virus in Syrian Hamsters
- A Polysaccharide Virulence Factor from Elicits Anti-inflammatory Effects through Induction of Interleukin-1 Receptor Antagonist
- Structural and Functional Characterization of a Complex between the Acidic Transactivation Domain of EBNA2 and the Tfb1/p62 Subunit of TFIIH
- Adaptive Gene Amplification As an Intermediate Step in the Expansion of Virus Host Range
- DHX36 Enhances RIG-I Signaling by Facilitating PKR-Mediated Antiviral Stress Granule Formation
- Hepatitis B Virus Infection and Immunopathogenesis in a Humanized Mouse Model: Induction of Human-Specific Liver Fibrosis and M2-Like Macrophages
- Crk Adaptors Negatively Regulate Actin Polymerization in Pedestals Formed by Enteropathogenic (EPEC) by Binding to Tir Effector
- Fatty Acid Biosynthesis Contributes Significantly to Establishment of a Bioenergetically Favorable Environment for Vaccinia Virus Infection
- A Cytosolic Chaperone Complexes with Dynamic Membrane J-Proteins and Mobilizes a Nonenveloped Virus out of the Endoplasmic Reticulum
- Intracellular Promote Invasive Cell Motility through Kinase Regulation of the Host Actin Cytoskeleton
- MAVS-MKK7-JNK2 Defines a Novel Apoptotic Signaling Pathway during Viral Infection
- RON5 Is Critical for Organization and Function of the Moving Junction Complex
- Immune Suppression by Neutrophils in HIV-1 Infection: Role of PD-L1/PD-1 Pathway
- and Exhibit Metabolic Symbioses
- The Herpes Virus Fc Receptor gE-gI Mediates Antibody Bipolar Bridging to Clear Viral Antigens from the Cell Surface
- Target Cell Availability, Rather than Breast Milk Factors, Dictates Mother-to-Infant Transmission of SIV in Sooty Mangabeys and Rhesus Macaques
- Evolution of the Retroviral Restriction Gene : Inhibition of Non-MLV Retroviruses
- Infection of Adult Thymus with Murine Retrovirus Induces Virus-Specific Central Tolerance That Prevents Functional Memory CD8 T Cell Differentiation
- Fha Interaction with Phosphothreonine of TssL Activates Type VI Secretion in
- In Vivo Administration of a JAK3 Inhibitor during Acute SIV Infection Leads to Significant Increases in Viral Load during Chronic Infection
- Lack of Detectable HIV-1 Molecular Evolution during Suppressive Antiretroviral Therapy
- Activation of HIV-1 from Latent Infection via Synergy of RUNX1 Inhibitor Ro5-3335 and SAHA
- A Compact, Multifunctional Fusion Module Directs Cholesterol-Dependent Homomultimerization and Syncytiogenic Efficiency of Reovirus p10 FAST Proteins
- The Role of Host and Microbial Factors in the Pathogenesis of Pneumococcal Bacteraemia Arising from a Single Bacterial Cell Bottleneck
- Genetic Dissection of Gut Epithelial Responses to
- Two-Component System Cross-Regulation Integrates Response to Heme and Cell Envelope Stress
- Oral Mycobiome Analysis of HIV-Infected Patients: Identification of as an Antagonist of Opportunistic Fungi
- A Model System for Studying the Transcriptomic and Physiological Changes Associated with Mammalian Host-Adaptation by Serovar Copenhageni
- Inflammasome Sensor NLRP1 Controls Rat Macrophage Susceptibility to
- ChIP-Seq and RNA-Seq Reveal an AmrZ-Mediated Mechanism for Cyclic di-GMP Synthesis and Biofilm Development by
- The Hypervariable Amino-Terminus of P1 Protease Modulates Potyviral Replication and Host Defense Responses
- Caspase-1-Dependent and -Independent Cell Death Pathways in Infection of Macrophages
- The Effect of Cell Growth Phase on the Regulatory Cross-Talk between Flagellar and Spi1 Virulence Gene Expression
- Different Mutagenic Potential of HIV-1 Restriction Factors APOBEC3G and APOBEC3F Is Determined by Distinct Single-Stranded DNA Scanning Mechanisms
- Oral Bacteria and Cancer
- Identification of OmpA, a Protein Involved in Host Cell Invasion, by Multi-Phenotypic High-Content Screening
- Transovarial Transmission of a Plant Virus Is Mediated by Vitellogenin of Its Insect Vector
- VE-Cadherin Cleavage by LasB Protease from Facilitates Type III Secretion System Toxicity in Endothelial Cells
- Dimerization of VirD2 Binding Protein Is Essential for Induced Tumor Formation in Plants
- Crystal Structure of the Vaccinia Virus DNA Polymerase Holoenzyme Subunit D4 in Complex with the A20 N-Terminal Domain
- Post-Translational Regulation via Clp Protease Is Critical for Survival of
- Modulation of Phagosomal pH by Promotes Hyphal Morphogenesis and Requires Stp2p, a Regulator of Amino Acid Transport
- Rotavirus Activates Lymphocytes from Non-Obese Diabetic Mice by Triggering Toll-Like Receptor 7 Signaling and Interferon Production in Plasmacytoid Dendritic Cells
- Cytomegalovirus m154 Hinders CD48 Cell-Surface Expression and Promotes Viral Escape from Host Natural Killer Cell Control
- Interferon Regulatory Factor-1 Protects from Fatal Neurotropic Infection with Vesicular Stomatitis Virus by Specific Inhibition of Viral Replication in Neurons
- HMGB1-Promoted and TLR2/4-Dependent NK Cell Maturation and Activation Take Part in Rotavirus-Induced Murine Biliary Atresia
- An Immunomics Approach to Schistosome Antigen Discovery: Antibody Signatures of Naturally Resistant and Chronically Infected Individuals from Endemic Areas
- PPARγ Agonists Improve Survival and Neurocognitive Outcomes in Experimental Cerebral Malaria and Induce Neuroprotective Pathways in Human Malaria
- A Non-Coding RNA Promotes Bacterial Persistence and Decreases Virulence by Regulating a Regulator in
- Viral OTU Deubiquitinases: A Structural and Functional Comparison
- Heterogeneity and Breadth of Host Antibody Response to KSHV Infection Demonstrated by Systematic Analysis of the KSHV Proteome
- Influenza A Virus Assembly Intermediates Fuse in the Cytoplasm
- Broadly Reactive Human CD8 T Cells that Recognize an Epitope Conserved between VZV, HSV and EBV
- Oncogenic Human Papillomaviruses Activate the Tumor-Associated Lens Epithelial-Derived Growth Factor (LEDGF) Gene
- Erythrocyte Invasion: Combining Function with Immune Evasion
- IL-1α and Complement Cooperate in Triggering Local Neutrophilic Inflammation in Response to Adenovirus and Eliminating Virus-Containing Cells
- Chronic Exposure to Type-I IFN under Lymphopenic Conditions Alters CD4 T Cell Homeostasis
- PLOS Pathogens
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Cytomegalovirus m154 Hinders CD48 Cell-Surface Expression and Promotes Viral Escape from Host Natural Killer Cell Control
- Human African Trypanosomiasis and Immunological Memory: Effect on Phenotypic Lymphocyte Profiles and Humoral Immunity
- DHX36 Enhances RIG-I Signaling by Facilitating PKR-Mediated Antiviral Stress Granule Formation
- Conflicting Interests in the Pathogen–Host Tug of War: Fungal Micronutrient Scavenging Versus Mammalian Nutritional Immunity
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



