-
Články
- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
The ISWI Chromatin Remodeler Organizes the hsrω ncRNA–Containing Omega Speckle Nuclear Compartments
The complexity in composition and function of the eukaryotic nucleus is achieved through its organization in specialized nuclear compartments. The Drosophila chromatin remodeling ATPase ISWI plays evolutionarily conserved roles in chromatin organization. Interestingly, ISWI genetically interacts with the hsrω gene, encoding multiple non-coding RNAs (ncRNA) essential, among other functions, for the assembly and organization of the omega speckles. The nucleoplasmic omega speckles play important functions in RNA metabolism, in normal and stressed cells, by regulating availability of hnRNPs and some other RNA processing proteins. Chromatin remodelers, as well as nuclear speckles and their associated ncRNAs, are emerging as important components of gene regulatory networks, although their functional connections have remained poorly defined. Here we provide multiple lines of evidence showing that the hsrω ncRNA interacts in vivo and in vitro with ISWI, regulating its ATPase activity. Remarkably, we found that the organization of nucleoplasmic omega speckles depends on ISWI function. Our findings highlight a novel role for chromatin remodelers in organization of nucleoplasmic compartments, providing the first example of interaction between an ATP-dependent chromatin remodeler and a large ncRNA.
Published in the journal: . PLoS Genet 7(5): e32767. doi:10.1371/journal.pgen.1002096
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1002096Summary
The complexity in composition and function of the eukaryotic nucleus is achieved through its organization in specialized nuclear compartments. The Drosophila chromatin remodeling ATPase ISWI plays evolutionarily conserved roles in chromatin organization. Interestingly, ISWI genetically interacts with the hsrω gene, encoding multiple non-coding RNAs (ncRNA) essential, among other functions, for the assembly and organization of the omega speckles. The nucleoplasmic omega speckles play important functions in RNA metabolism, in normal and stressed cells, by regulating availability of hnRNPs and some other RNA processing proteins. Chromatin remodelers, as well as nuclear speckles and their associated ncRNAs, are emerging as important components of gene regulatory networks, although their functional connections have remained poorly defined. Here we provide multiple lines of evidence showing that the hsrω ncRNA interacts in vivo and in vitro with ISWI, regulating its ATPase activity. Remarkably, we found that the organization of nucleoplasmic omega speckles depends on ISWI function. Our findings highlight a novel role for chromatin remodelers in organization of nucleoplasmic compartments, providing the first example of interaction between an ATP-dependent chromatin remodeler and a large ncRNA.
Introduction
ISWI, the catalytic subunit of several ATP-dependent chromatin remodeling complexes, is highly conserved during evolution and is essential for cell viability [1]. ISWI-containing complexes play central roles in DNA replication, gene expression and chromosome organization [2]. ISWI uses ATP hydrolysis to catalyze nucleosome spacing and sliding reactions [1]. Loss of ISWI function in Drosophila causes global transcription defects and dramatic alterations in higher-order chromatin structure, including decondensation of chromosomes [3], [4]. In vitro and in vivo studies in several model organisms have also shown the involvement of ISWI-containing complexes in other nuclear functions like telomere silencing, stem cell renewal, neural morphogenesis and epigenetic reprogramming during nuclear transfer in animal cloning [2], [5], [6]. The diverse functions associated with ISWI depend upon the ability of other cellular and nuclear factors to regulate its ATP-dependent chromatin remodeling activity [7]–[9]. In order to identify new regulators of ISWI function, we developed in vivo assays to identify genetic interactors of ISWI in D.melanogaster [10], [11]. Using an eye-based assay to identify factors antagonizing ISWI activity, we recovered, among other genes, a genetic interaction between ISWI and hsrω [10]. The hsrω gene is developmentally expressed in almost all cells types and is one of the most strongly induced heat shock genes in flies [12]–[14]. The hsrω locus encodes multiple non-coding RNAs (ncRNA), of which the large >10 kb nuclear species (hsrω-n) is essential for the assembly and organization of the hnRNP-containing omega speckles [14]. These specialized nuclear compartments are distinct from other nuclear speckles and are localized in the nucleoplasm close to chromatin edges [14]. Omega speckles play essential roles in storage and sequestration of members of the hnRNP family and other proteins involved in RNA processing and maturation in normal as well as environmentally or genotoxically stressed cells (for a list of hsrω interactors see [13]–[15]. Here we show that the hsrω ncRNA interacts in vivo and in vitro with ISWI to directly regulate its ATPase activity. Additionally, we provide in vivo data showing that omega speckle nuclear organization depends on ISWI function. Our work thus suggests that ISWI and the omega speckle associated hsrω ncRNAs interact and reciprocally affect each other's activities. Our findings highlight a novel role for chromatin remodelers in organization of nuclear speckles.
Results
ISWI Genetically Interacts with hsrω
Loss of hsrω function by RNAi [15] results in a striking amelioration of morphological defects in eyes exclusively composed of ISWI-null mitotic clones (Figure 1A, 1B and Figure S1A–S1D, S1J; Table S1A). Mutations in the sqd gene, which encodes the Squid hnRNP, a component of omega speckles, also suppresses ISWI mutant eye defects (Figure S1F–S1I and S1K; Table S1A) [10]. Absence of ISWI in larval salivary gland cells causes a dramatic decondensation of the male X polytene chromosome [4]. Remarkably, hsrω-RNAi suppresses the ISWI null male X chromosome condensation defects as well (Figure 1C, 1D). Tissue-specific mis-expression of the catalytically inactive ISWIK159R transgene also produces eye phenotypes and global chromosome decondensation [3], [4], [11]. Silencing of hsrω-n activity not only suppresses ISWIK159R eye phenotypes (Figure 1E–1H) but also restores normal polytene chromosome condensation (Figure 1I, 1J). In agreement with the above observations, the larval lethality of ISWI-null mutants is also partially suppressed by hsrω-RNAi (Figure 1K; Table S1B), strongly indicating that reduction of hsrω-n transcripts improves ISWI activity. On the other hand, over-expression of hsrω through the hsrωEP93D allele [15] antagonizes ISWI activity, resulting in enhanced chromosome condensation defects and eye phenotypes in ISWI-null background (Figure 1L and Figure S1E; Table S1A).
Fig. 1. Loss of the hsrω ncRNA suppresses ISWI mutant defects. 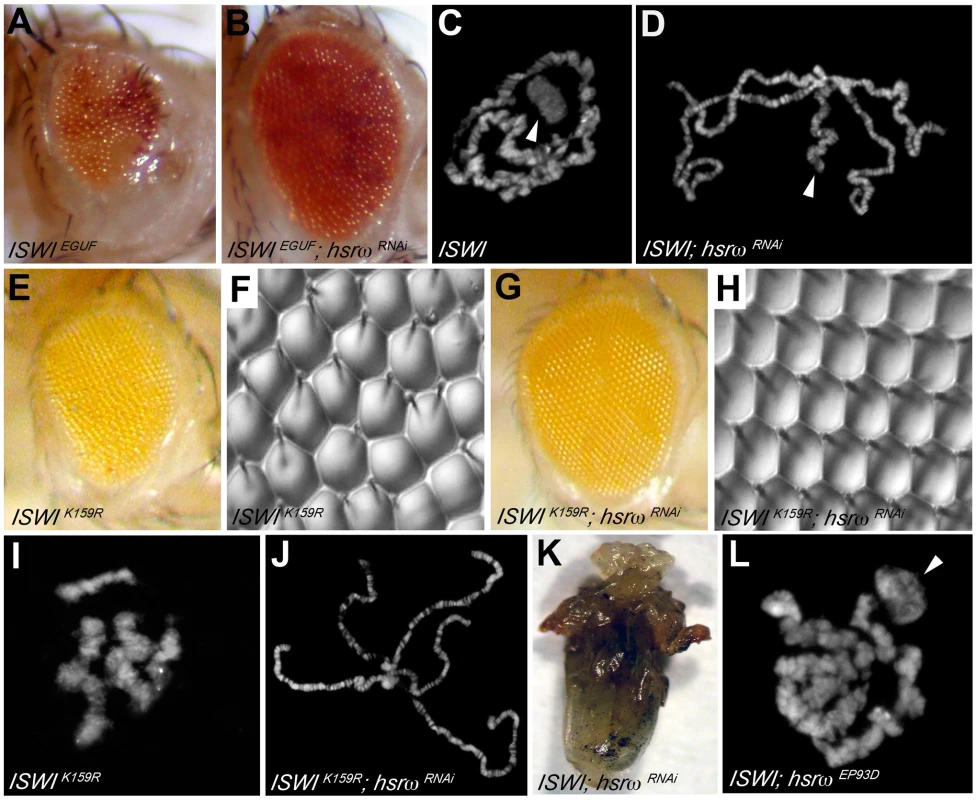
(A) The EGUF technique allows generation of adult eyes homozygous for a specific mutation in an otherwise heterozygous adult fly [34]. The ISWIEGUF eye is composed exclusively of clones that have lost ISWI activity, and is characterized by roughness and reduced size caused by loss of ommatidial boundaries or their orientation and reduced number of photoreceptors [11]. (B) Eye specific loss of hsrω function by RNAi (ey-GAL4 driven hsrω-RNAi3) [15] can suppress ISWIEGUF eye phenotypes with 100% penetrance. (C) Salivary gland polytene chromosomes from ISWI1/ISWI2 trans-heterozygous ISWI null male larvae [4] (ISWI) are characterized by condensation defects, particularly of the X chromosome (arrowhead). (D) ey-GAL4 driven hsrω-RNAi3 suppresses the ISWI mutant male X chromosome condensation defects (arrowhead) with a 100% penetrance. In addition to the eye imaginal discs, the eyeless promoter is known to efficiently drive the expression of GAL4 also in salivary glands also [15]. (E) ey-GAL4 driven mis-expression of the catalytically inactive ISWIK159R transgene produces reduced and rough eyes [4], [11]. (F) Nail polish imprint of eye mis-expressing ISWIK159R highlights ommatidial roughness. (G) Knock down of hsrω-n RNA by RNAi suppresses the ISWIK159R eye defects with a 100% penetrance. (H) Nail polish imprint of eye co-expressing ISWIK159R and hsrω -RNAi3 transgenes confirms suppression of ISWIK159R ommatidial roughness. (I) ey-GAL4 driven expression of ISWIK159R in salivary glands causes global decondensation of polytene chromosomes [3]. (J) hsrω-RNAi completely suppresses polytene chromosome condensation defects caused by ISWIK159R mis-expression. (K) The larval lethality associated with ISWI-null condition is partially rescued by the simultaneous down regulation of hsrω-n transcripts, as seen in this ventral view of a pharate dissected from a ISWI1/ISWI2; Act5C-GAL4/hsrω RNAi3 dead pupa (also see Table S1B). (L) Act5c-GAL4 driven EP93D [15] mediated over-expression of hsrω transcripts exaggerates the chromosome condensation defects of ISWI-null mutants (arrowhead points to the putative X chromosome). The suppression of chromosome condensation and eye defects in ISWI nulls by hsrω-RNAi is not due to a reduction in the efficiency of the GAL4/UAS driving system used to produce the ISWI-null and ISWIK159R mutant phenotypes (Figure S2A, S2B). Furthermore, the effect of hsrω-RNAi is highly specific for the loss of ISWI function (Figure S2C, S2D). Given the role played by omega speckles in nuclear RNA processing [13], we also checked if the levels of ISWI or ISWIK159R proteins and their corresponding mRNA were affected by hsrω-RNAi, which could account for the suppression of ISWI-null or ISWIK159R defects. However, depletion of hsrω transcripts by RNAi does not detectably affect ISWI protein or mRNA levels in either of these cases (Figure S3).
Loss of ISWI Causes Global Defects in Omega Speckle Organization
In order to understand the molecular basis of the specific suppression of ISWI phenotypes by hsrω-RNAi, we examined the distribution and organization of omega speckles in the ISWI mutant third instar larval Malpighian tubule nuclei, which show abundant omega speckles using either RNA∶RNA in situ hybridization to hsrω-n ncRNA or immunostaining for some of the omega speckle associated hnRNPs [14]. Interestingly, the organization and distribution of omega speckles in ISWI mutants is profoundly altered when compared with wild type cells. Instead of typical speckles, the hsrω-n transcripts form “trail”-like structures in ISWI-null mutant nucleoplasm, indicating a severe defect in their maturation or organization (Figure 2A, 2B). Interestingly, Squid, NonA and other omega speckle associated hnRNPs also form “trail”-like structures in ISWI mutants (Figure 2C–2F, Figure 3A–3D, and Figure S4), which shows that distribution of not only the hsrω-n ncRNA but also of the omega speckle-associated hnRNPs is compromised in ISWI mutant nuclei. As shown earlier [15], the omega speckles do not form in the absence of hsrω-n transcripts and the omega speckle-associated hnRNPs remain diffused in the nucleoplasm (Figure 3E–3F). Interestingly, when the ISWI as well as hsrω-n ncRNA are absent, omega “trails” are not formed (Figure 3G–3H), strongly indicating that ISWI mutant specific omega “trails” are dependent on the presence of the hsrω-n ncRNA.
Fig. 2. Loss of ISWI causes global defects in organization of omega speckles. 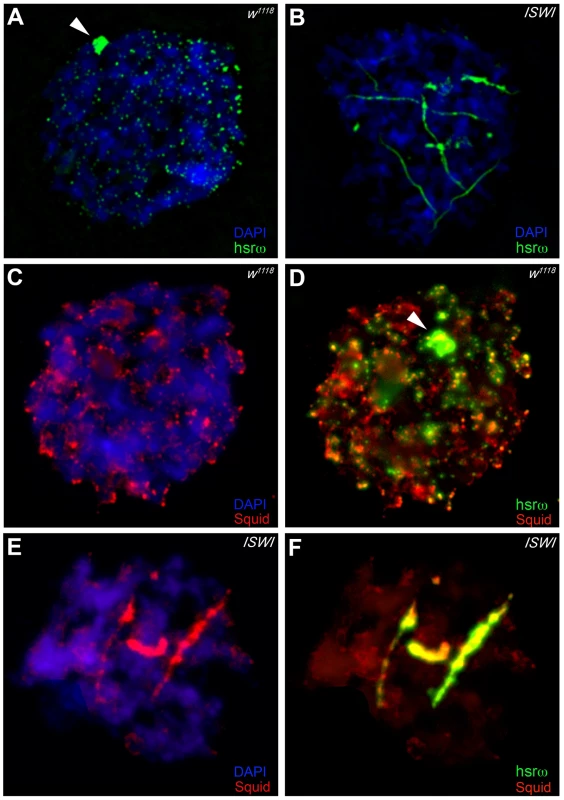
(A) Confocal projection image of wild type (w1118) Malpighian tubule whole nucleus following fluorescent RNA in situ hybridization (FRISH) using the 280b tandem repeat riboprobe specific for the nuclear hsrω–n ncRNA [32]. The hsrω-n RNA localizes in omega speckles in the nucleoplasm mainly in the nuclear space not occupied by chromosomes, and at the 93D cytogenetic region corresponding to its site of transcription (arrowhead) [14]. (B) FRISH on ISWI1/ISWI2 mutant (ISWI) larval Malpighian tubule nucleus reveals a dramatic change in the classic punctate pattern of omega speckles with a penentrance of 100%. The omega speckles in ISWI mutant cells are totally disorganized and form “trail”-like structures. DAPI stained DNA is shown in blue and hsrω–n RNA in green. (C) and (D) Besides its chromatin localization, the Squid protein largely colocalizes with the hsrω-n ncRNA [14] in omega speckles as seen after immunostaining for the Squid protein (red) and FRISH for hsrω-n RNA (green) on w1118 Malpighian tubule whole nucleus. The 93D cytologenetic location is indicated by arrowhead. DNA was counterstained with DAPI (blue). (E) and (F) Immuno-FRISH of Squid (red) and hsrω-n RNA (green) on ISWI1/ISWI2 mutant (ISWI) Malpighian tubule nucleus shows that besides the chromatin associated Squid, its nucleoplasmic fraction localizes in “trail”-like structures in ISWI mutant nuclei, instead of in the characteristic nucleoplasmic speckles. The Squid “trails” completely overlap with the hsrω signal, strongly indicating that loss of ISWI causes profound alteration in the organization not only of the hsrω-n ncRNA component but also of the associated Squid hnRNP. DNA was counterstained with DAPI (blue). Fig. 3. Defects in omega speckles in ISWI mutant cells are dependent on the presence of the hsrω ncRNA. 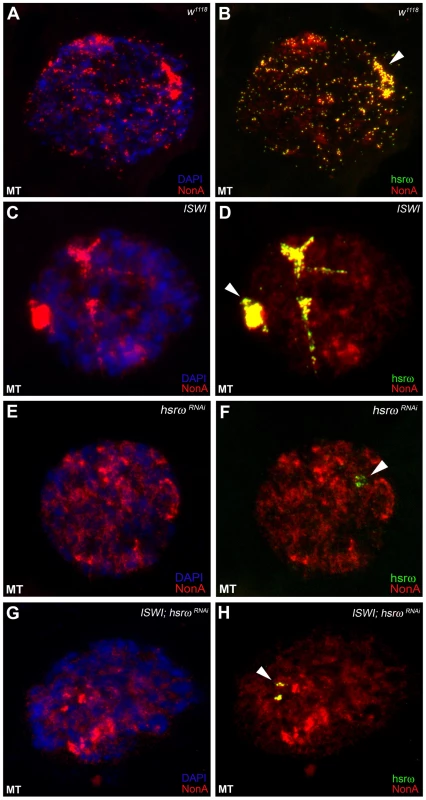
(A) and (B) The NonA protein colocalizes with the hsrω-n ncRNA [14] in omega speckles as seen after immunostaining for the NonA protein (red) and FRISH for hsrω-n RNA (green) on w1118 Malpighian tubule whole nucleus [14]. The 93D cytologenetic locus is indicated by the arrowhead. DNA was counterstained with DAPI (blue). (C) and (D) Immuno-FRISH of NonA (red) and hsrω-n RNA (green) on ISWI1/ISWI2 mutant (ISWI) Malpighian tubule nucleus shows that the nucleoplasmic fraction of NonA also localizes in “trail”-like structures in ISWI mutant nuclei. The NonA “trails” completely overlap with the hsrω signal, strongly indicating that loss of ISWI causes profound alteration in the organization not only of the hsrω-n ncRNA component but also of omega speckle associated hnRNPs like Squid (see Figure 2C–2F) and Hrb87F (see Figure S4). (E) and (F) Down regulation of hsrω-n transcripts causes the omega speckle associated hnRNP components to diffuse in the nucleoplasm together with the absence of classic omega speckles. (G) and (H) ISWI null mutant condition does not result in formation of omega “trails” when the hsrω-n ncRNA is down regulated by RNAi. Analysis of live cells expressing a SquidGFP transgene [16] clearly identifies the GFP-positive “trails” in live ISWI mutant cells similar to those observed in fixed cells (Figure S5). This shows that the ISWI omega “trails” are not a fixation artifact. Significantly, comparable hsrω RNA “trails” were not seen (Figure S6) in the presence of other mutants like jil1, ada2 and gcn5 which also display chromosome condensation defects similar to those observed in the ISWI mutants [17], [18]. This excludes the possibility that the omega “trails” in ISWI mutant nuclei result from a “squeezing” effect of the nucleoplasm due to a massive “fallout” of chromatin associated proteins following global chromosome decondensation.
Studies in several model organisms have shown that ISWI plays a global role in transcriptional activation as well as repression [1], [3], [4]. Therefore, we examined if ISWI mutation altered the levels of hsrω-n ncRNA or the omega speckle-associated proteins. However, no significant difference in their levels was found between ISWI mutant and wild type cells (Figure S7). The >10 Kb hsrω-n ncRNA that organizes the omega speckles contains a small 0.7 Kb intron [14], [19]. It has been recently observed [20] that a spliced form of the hsrω-n transcript is also associated with the omega speckles. Therefore, we checked if the ISWI mutant condition affects splicing of this RNA which may result in the “trail”-like organization. RT-PCR and Northern blot analyses clearly indicate that ISWI mutation does not affect splicing of the hsrω-n ncRNA (Figure S8A–S8C).
In light of the significant role played by ISWI in gene expression, we checked whether an engulfment of the nuclear RNA export machinery in ISWI mutants affected RNA transport from nucleus, which in turn could modify the omega speckles into “trails”. In situ hybridization to cellular RNA with poly-dT probe did not reveal any difference in the nuclear vs cytoplasmic distribution of poly-A RNAs between wild type and ISWI mutant cells (Figure S8D). Thus, ISWI mutant nuclei do not seem to have a general RNA export defect, which could have been responsible for the observed omega “trails”.
Omega speckles are thought to provide a dynamic system to sequester and release specific RNA processing factors in normal as well as stressed cells [13]. Following heat shock, hsrω is one of the most highly transcribed genes [13], [21] and omega speckles coalesce into bigger growing clusters that finally get restricted to the hsrω gene locus, providing a dynamic sink for proteins that need to be transiently withdrawn from active nuclear compartments under stress conditions [14]. As already noted above, ISWI mutant condition causes the omega speckles to form nucleoplasmic “trails” in unstressed cells (Figure S9A, S9B). Although heat shock caused clustering of the omega speckles or “trails” in wild type and ISWI mutant cells, respectively, the numbers of clusters in the latter cells were much less (Figure S9C, S9D), suggesting that speckle dynamics under heat shock is also compromised because of ISWI mutant background. Finally, the “trail”-like organization of hsrω ncRNA and its associated proteins in ISWI mutants is not limited to Malpighian tubule or salivary gland polytene cells (Figure 2A, 2B and Figure S10A, S10B), but is also observed in ISWI mutant diploid cells (), indicating that disorganization of omega speckles is a general consequence of loss of ISWI function.
Chromatin-Associated ISWI Interacts In Vivo and In Vitro with Nucleoplasm-Associated Omega Speckles
Unlike the association of ISWI with different bands and interbands on polytene chromosomes [4], [11], the hsrω-n ncRNA localizes in the nucleoplasm in proximity or at the edges of chromosome spreads, without any apparent overlap with the chromatin associated ISWI (Figure 4A, 4B). However, examination of confocal images of intact nuclei revealed some chromosome-nucleoplasm sites where ISWI and the hsrω-n ncRNA are adjacent and seem to form connecting bridges between nucleoplasm and chromatin (Figure 4C, 4D). Barring a few exceptions, Squid and other omega speckles associated hnRNPs also showed no overlap with ISWI on polytene chromosome spreads (Figure 4E, 4F and Figure S11A, S11B). Significantly, like the hsrω ncRNA they too were found to partially overlap with ISWI in several nucleoplasmic foci in intact nuclei (see Figure 4G, 4H and Figure S11C, S11D), suggesting that ISWI may indeed partially interact directly or indirectly, at least transiently, with omega speckles in the three-dimensional nuclear space.
Fig. 4. ISWI interacts in vivo with omega speckles. 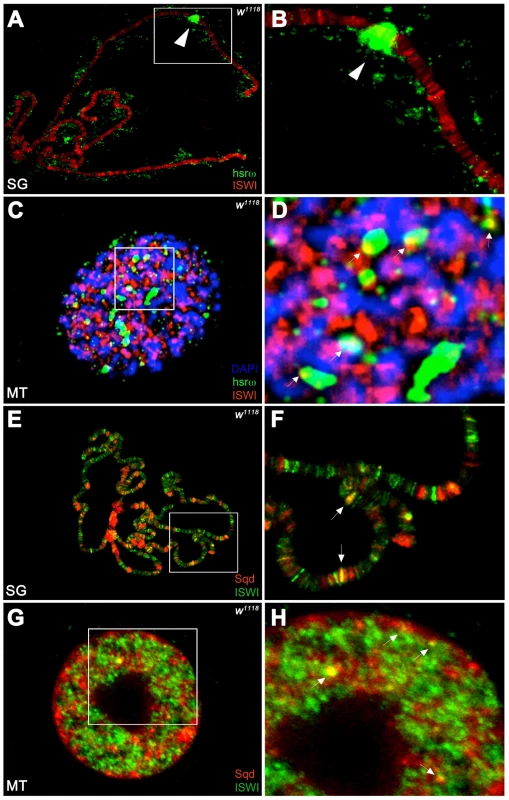
(A) Immunostaining for the ISWI protein (red) combined with FRISH for hsrω-n RNA (green) on wild type (w1118) salivary gland (SG) squashed nuclei shows omega speckles decorating polytene chromosome edges without any significant overlap with chromosome-bound ISWI. Arrowhead indicates the 93D cytologenetic location. (B) A magnified image of ISWI and hsrω-n signals corresponding to the white boxed area in A. (C) Immuno-FRISH confocal section of wild type (w1118) Malpighian tubule (MT) whole nucleus shows that ISWI (red) and hsrω-n (green) partially overlap (yellow areas) at several sites. DAPI stained DNA is in blue. (D) A magnified view of DAPI (blue), ISWI (red) and hsrω-n RNA (green) signals corresponding to the white boxed area in C shows typical examples of partial overlap (yellow, white arrowheads) between ISWI and hsrω-n RNA. (E) Double immunostaining for the ISWI (green) and Squid (red) proteins on w1118 salivary gland (SG) squashed nucleus, shows that with the exception of a few sites, there is little overlap between the two proteins on polytene chromosomes. DAPI stained DNA is in blue. (F) A magnified image of Squid and ISWI signals corresponding to the white boxed area in E shows the few co-immunostained (yellow) regions (white arrows). (G) Double immunostained confocal section to show partial overlap between ISWI (green) and Squid (red) proteins in the nucleoplasm in w1118 Malpighian tubule (MT) whole nucleus. The black empty area corresponds to the nucleolus. (H) A magnified image of Squid and ISWI signals corresponding to the white boxed area in G shows representative sites where ISWI and Squid proteins partially overlap (white arrows). In order to directly investigate whether the chromatin remodeling factor ISWI physically interacts with omega speckles, we used an affinity purified ISWI antibody [4] to conduct classic RNA immunoprecipitation. Our semi-quantitative RT-PCR analysis revealed the presence of hsrω-n ncRNA in larval nuclear extracts immunoprecipitated with ISWI antibody (Figure 5A, 5B). To rule out a non-specific association of ncRNAs with ISWI, we used the same immunoprecipitate to detect U4 and Rox1 [22] ncRNAs by RT-PCR. Significantly, neither of these two otherwise abundant ncRNAs were detectable (Figure 5A, 5B) in the mmunoprecipitate. This confirms the specificity of the physical interaction between ISWI and hsrω-n RNA in native larval extracts. Further, to exclude the possibility that the physical association observed between ISWI and hsrω was due to fortuitous interactions occurring during nuclear extract preparation, we conducted the CLIP assay (Cross-Linking & Immuno Precipitation) using the affinity purified anti-ISWI antibody [4] on fixed larval nuclear extracts. The CLIP data confirmed a highly specific interaction between ISWI and the hsrω ncRNA in the nucleus (Figure 5C), as observed with the native extracts (Figure 5B). Moreover, as shown in Figure 5D, RNA pull down assay confirmed that ISWI is also specifically pulled down by immobilized hsrω-n ncRNA along with the other known omega speckles associated hnRNPs [13] while a control generic RNA does not pull down ISWI or the other hnRNPs (Figure 5D).
Fig. 5. ISWI physically and functionally interacts with the hsrω-n ncRNA. 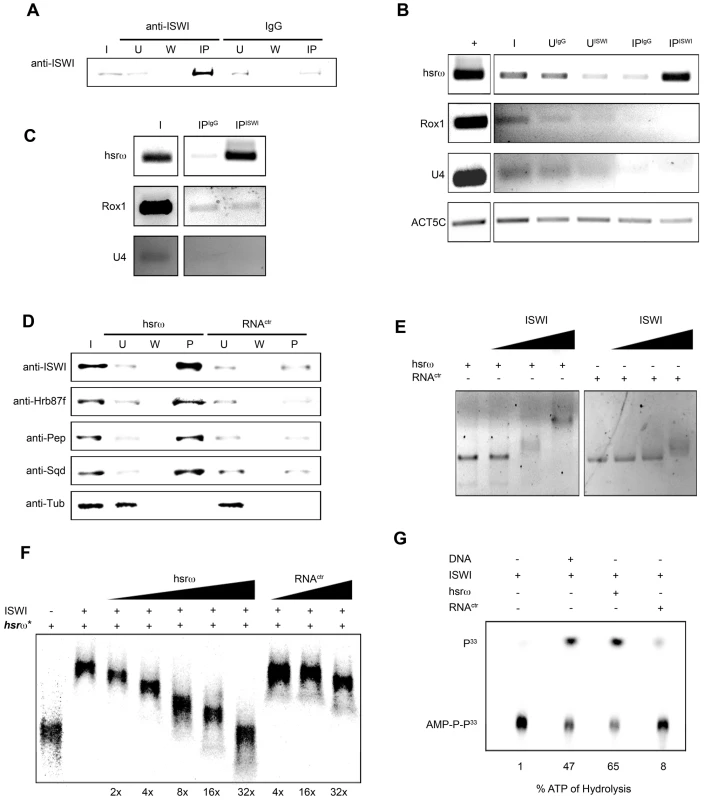
(A) RNA-Immunoprecipitation (RIP) was conducted on larval nuclear extracts [35] using an affinity purified ISWI antibody [4]. The immunoprecipitated material was split in two parts. The first aliquot was analyzed by Western blotting using the ISWI antibody. Generic rabbit IgG was used as control. I = Input, U = Unbound, W = Wash, IP = Immunoprecipitated material. (B) Total RNA was extracted from the second immunoprecipitated aliquot for RT-PCR using primers that specifically amplify the 280b tandem repeat characteristic of the nuclear hsrω–n ncRNA. RT-PCR amplification of the U4 and Rox1 ncRNAs was also carried out using specific primers for each to assess the specificity of amplification of hsrω–n repeat unit in the ISWI immunoprecipitate. RT-PCR amplicon generated with primers for Act5C RNA was used to normalize the RT-PCR signals. + = RT-PCR positive control amplification, I = Input, UISWI = Unbound from anti-ISWI RIP, UIgG = Unbound from IgG RIP, IPISWI = Immunoprecipitated material from anti-ISWI RIP, IPIgG = Immunoprecipitated material from IgG RIP. (C) To validate the physical interaction between ISWI and hsrω observed by RIP on native larval nuclear extracts (see B), CLIP (Cross-Linking & Immuno Precipitation) was carried out with affinity purified anti-ISWI antibody [4] on fixed larval nuclear extracts. I = Input, IPISWI = Immunoprecipitated material from anti-ISWI CLIP, IPIgG = Immunoprecipitated material from IgG CLIP. The immunoprecipitate was analyzed by RT-PCR using primers for the 280b tandem repeat unit of the nuclear hsrω–n ncRNA; primers amplifying the U4 and Rox1 ncRNAs were used as specificity controls. (D) Immobilized hsrω-n ncRNA and a generic control RNA were used as baits to pull down protein complexes from native larval nuclear extracts. Pulled down material was detected by Western blotting using antibodies against ISWI, Hrb87F, Pep and Sqd; Tubulin (Tub) was used as the loading control. (E) Gel mobility assay of the in vitro transcribed 280b tandem repeat unit of the hsrω-n ncRNA was carried out in the presence of increasing amounts of recombinant full length ISWI, with the ISWI/RNA molar ratios 0∶1, 1∶1, 5∶1 and 10∶1 nmoles, respectively. A generic RNA (RNActr) of approximately 300 bp was used as control. (F) Gel mobility assay was carried out with P33 radiolabeled in vitro transcribed hsrω-n ncRNA (hsrω*: 280b tandem repeat unit) in the presence of a fixed amount of recombinant full length ISWI (molar ratio ISWI/hsrω* of 8∶1). Increasing amounts of cold hsrω 280b ncRNA (2×, 4×, 8×, 16×, 32× fold excess with respect to hsrω*) specifically compete the ISWI binding with radiolabeld hsrω RNA by changing its mobility. The same fold excess of the control generic RNA (RNActr) very poorly changes the mobility of the retarded ISWI/hsrω* complex. It is important to note that when an excess of cold hrsω ncRNA is added in the ISWI/hsrω* binding reaction, the retarded complex changes its mobility instead of going away to form free probe at the bottom of the gel. Our data suggest that more than one ISWI protein binds one hrsω ncRNA molecule, so that as the amount of cold hsrω ncRNA increases, fewer ISWI molecules bind with each unit and thereby retard its movement to a lesser extent. (G) Stimulation of the ATPase activity of ISWI by generic DNA, in vitro transcribed 280b repeat unit of the hsrω-n or a generic RNA (RNActr) was assayed in vitro by thin layer chromatography in the presence of radioactive ATP-γ33P. Stimulation of the ATPase activity by the distinct nucleic acids assayed is noted below each lane as percentage of the ATP hydrolysed (P33 = P33 radioactive labeled hydrolyzed gamma phosphate, AMP-P-P33 = radioactive labeled non-hydrolyzed ATP). Classic gel shift assay using in vitro transcribed hsrω-n ncRNA repeat unit (280b) and full length recombinant ISWI clearly shows that ISWI effectively retards hsrω-n ncRNA mobility, but that of a generic control RNA is retarded poorly (Figure 5E). Moreover, the mobility shift of the hsrω-n RNA by ISWI binding is specifically competed by hsrω-n but not by a generic RNA (Figure 5F). This further confirms the specific nature of ISWI/hsrω physical interaction in vitro.
A functional significance of the physical interaction between ISWI and hsrω-n ncRNA is indicated by the stimulation of ISWI ATPase activity. Remarkably, as also reported previously [23], while the generic control RNA very poorly stimulates the ISWI ATPase activity, the hsrω-n ncRNA was found to specifically stimulate the ISWI ATPase activity to levels greater than those normally seen with DNA but lower than nucleosome-stimulation (Figure 5G) [23].
The 280b hsrω-n nuclear ncRNA repeat unit used for the binding and ATPase assays is predicted to organize into a stable double stranded RNA molecule containing a few loops (Figure S12). This secondary organization is common to many RNAs, but this structure is also reminiscent of a double stranded DNA molecule. Therefore, it remained possible that the recognition of a double stranded nucleic acid (RNA or DNA) may provide a basis for the observed binding and stimulation of ATPase activity of ISWI by the hsrω-n ncRNA. When we checked the ability of the double stranded DNA sequence encoding the hsrωncRNA to elicit ISWI ATPase activity, we found that ISWI was stimulated to levels similar to those reported for other generic linear double stranded DNA molecules [23] (Figure S13A). Furthermore, co-presence of hsrω-n ncRNA and nucleosomes in a classic ATPase assay with ISWI clearly shows that both substrates compete for ISWI binding and its ATPase activity stimulation (Figure S13B).
The ISWI protein has two functional domains (Figure 6A), the N-terminal (ISWI-N) ATPase domain and the C-terminal (ISWI-C) nucleosomal DNA recognizing domain [24]. Results presented in Figure 6B and 6C, show that the hsrω-n binds with the ISWI-N fragment and stimulate its ATPase activity, suggesting that ISWI could interact with hsrω-n ncRNA through its ATPase domain. Therefore, we further checked if the presence of ATP, ATP-γ-S (a non-hydrolizable form of ATP) or ADP could affect ISWI binding or determine a conformational change in the ISWI/hsrω complexes resolved by gel shift. Our data show that all the three nucleotides have no effect on ISWI binding (Figure 6D), probably suggesting that the ATPase activity of ISWI may not be necessary for physical interaction between ISWI and hsrω RNA.
Fig. 6. The N-terminal portion of ISWI interacts with the hsrω-n ncRNA, but its ATPase activity is not necessary for binding. 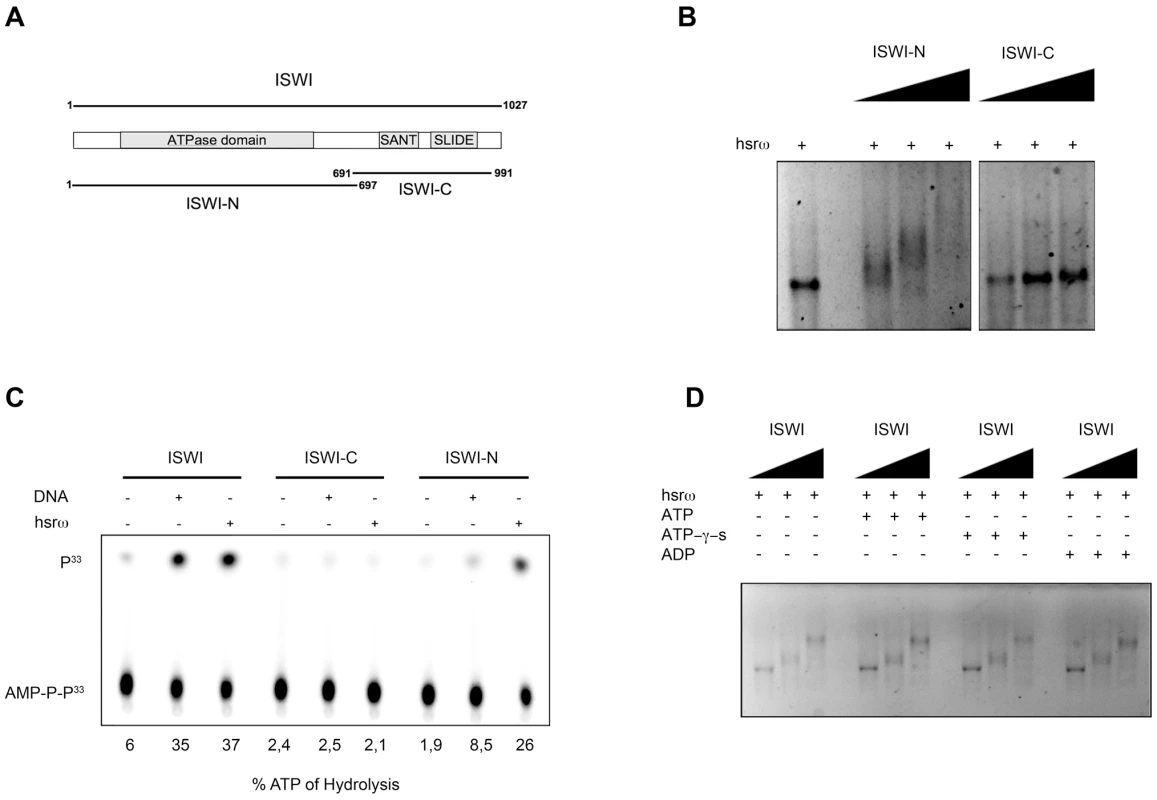
(A) Schematic representation of the two known functional domains of ISWI, the amino-terminal (ISWI-N) comprising the ATPase domain and the carboxy-terminal (ISWI-C) that specifically recognizes DNA in the context of nucleosomes through the SANT and SLIDE domains [24]. (B) Gel mobility shift assay of the 280b hsrω-n repeat unit transcript in the presence of increasing amount of ISWI-N or ISWI-C with ISWI/RNA molar ratios of 0∶1, 5∶1, 10∶1 and 20∶1 nmoles, respectively. (C) The ATPase activity of full length ISWI or its sub-domains (ISWI-N or ISWI-C) in the presence of DNA or the 280b hsrω-n repeat unit transcript was assayed in vitro by thin layer chromatography. Percentage of ATP hydrolysis, calculated as before (Figure 5G) is noted below each lane. (D) Gel mobility shift assay with hsrω-n ncRNA, in the presence of ATP, ATP-γ-S or ADP, with increasing amount of recombinant full length ISWI (ISWI/RNA molar ratios 0∶1, 5∶1 and 10∶1 nmoles, respectively). Discussion
Factors that coordinate nuclear activities occurring on chromatin and the nucleoplasmic compartments remain unidentified and uncharacterized. Therefore, an important open question in nuclear organization field is how nuclear speckles localize and organize themselves near transcriptionally active genes to cross talk with chromatin factors for processing of the nascent RNAs. Our data indicate that ISWI may provide a functional ‘bridge’ between chromatin and nuclear speckle compartments. Indeed, ISWI can directly or indirectly contact the omega speckles in intact nuclei, through hsrω-n ncRNA or some of the associated hnRNPs. Our confocal analysis suggested a functional ‘bridge’ between a chromatin factor (ISWI) and nucleoplasmic omega speckle components (hsrω ncRNA and hnRNPs). However, not all omega speckles show partial overlap with ISWI. Indeed, these molecular “bridges” between chromatin and nucleoplasm are probably transient, since time-lapse movies on live cells with fluorescently tagged chromatin and omega-speckle components clearly show very high mobility of these speckles (see Video S1), which probably may explain the absence of classic co-localization between ISWI and omega speckle components.
The observed direct physical interaction between ISWI and hsrω-n ncRNA together with the stimulation of ISWI-ATPase activity in light of the partial overlap revealed by confocal microscopy suggests that ISWI may interact with hsrω-forming speckles only transiently, probably to help the hsrω ncRNA to properly associate with or release the various omega speckle-associated hnRNPs. Loss of ISWI may impair the correct maturation, organization or localization of omega speckles resulting in the observed omega “trail” phenotype.
Our data also provide a possible explanation for the suppression of ISWI defects by hsrω-RNAi. In ISWI mutants carrying normal levels of hsrω transcripts, the limited maternally derived ISWI [3] is shared between chromatin remodelling and omega speckle organization reactions so that its sub-threshold levels in either compartments severely compromises both functions (see Video S2). However, when hsrω transcript levels are reduced by RNAi in ISWI null background, most of the maternal ISWI may become available for chromatin remodelling reactions, so that a minimal threshold level of chromosome organization can be achieved. This would permit initiation of close to normal developmental gene activity programs resulting in suppression of the ISWI eye and chromosome defects or in the postponement of the larval lethality to pupal stage. Additionally, it is known that when hsrω ncRNA is down regulated through RNAi, levels of free hnRNPs and other chromatin factors (i.e. CBP) are also elevated [25]. Therefore, we cannot formally exclude the possibility that these changes may also counteract ISWI defects by as yet unknown mechanisms.
Our work provides the first example of modulation of an ATP-dependent chromatin remodeler by a ncRNA, and to our knowledge the first in vivo and in vitro demonstration of a role of chromatin remodeler in organization of a nuclear compartment. However, the mechanism underlying stimulation of the ATPase activity of ISWI by the hsrω-n ncRNA, which may facilitate the organization of omega speckles, remains to be understood. Given the evolutionary derivation of the ISWI ATPase-domain from RNA-helicase-domains [1], a provocative hypothesis is that ISWI could “remodel” speckles by structurally helping the assembly or release of specific hnRNPs with the hsrω-n ncRNA to generate mature omega speckles. Chromatin remodelers, nuclear speckles and their associated long ncRNAs are emerging as essential components of gene regulatory networks, and their deregulation may underlie complex diseases [15], [25]–[27]. The functional homology of the human noncoding sat III transcripts with the Drosophila hsrω ncRNA [13], [27], highlights the relevance and translational significance of studies unraveling the functional connections between ncRNA-containing nuclear compartments and chromatin remodelers.
Materials and Methods
Fly Strains and Genetic Interaction
Flies were raised at 22°C on K12 medium [28]. Unless otherwise stated, strains were obtained from Bloomington, Szeged or VDRC (Vienna Drosophila RNAi Center). Genetic tests for dominant modifier (enhancement or suppression) of ISWI-EGUF and ISWIK159R phenotypes were conducted as previously described [10], [11]. The tissue specific expression of the UAS-ISWIK159R [4], the UAS-hsrωRNAi3 and the EP93D transgenic lines [15] was obtained with ey-GAL4 (for eyes and larval salivary glands) or Act5C-GAL4 driver (for larval Malpighian tubules and testis). The surface architecture of adult eyes was examined by the nail polish imprint method [26]. For the larval lethality assay, numbers of larvae of different genotypes that pupated and the numbers of pupae emerging as flies in a given cross were separately counted.
Antibodies, Plasmids, and RNA Probes
Mouse monoclonal antibodies against the following proteins were used at the indicated dilutions: Hrb87F (P11) [14] dilution 1∶5 for IF and 1∶100 for WB; Squid (1B11) [29] dilution 1∶100 for IF and 1∶2000 for WB; NonA [30] dilution 1∶50 for IF and 1∶1000 for WB; PEP [31] dilution 1∶2000 for WB. Affinity purified rabbit ISWI antibody [4] was diluted 1∶200 for IF and 1∶2000 for WB. FITC - and Rhodamine - conjugated anti-mouse and anti-rabbit secondary antibodies (Jackson Immuno Research) were diluted 1∶200 for IF and 1∶2000 for WB, respectively. The biotin-labeled anti-sense hsrω-n RNA 280b riboprobe was generated from the pDRM30 plasmid [32] and used for FRISH. For gel mobility assays the sense hsrω-n RNA riboprobe was generated from the same plasmid.
Immunofluorescence, FRISH, and ImmunoFRISH
Single and double immunofluorescence on polytene chromosome spreads were conducted as previously described [11]. Larval tissues (salivary glands, Malpighian tubules and testis) were dissected from third-instar larvae grown at 22°C. Fully or partially squashed tissue preparations were used for FRISH and Immuno-FRISH assays as previously described [14] with some modifications (Text S1).
Protein Extraction and Western Blotting (WB)
Total proteins from salivary glands and Malpighian tubules were extracted as previously described [11]. The SDS-PAGE separated proteins were transferred onto nitrocellulose membrane (Whatman Schleicher & Schuell) for Western detection using SuperSignal West Femto substrate (Pierce). Chemiluminescent signals were acquired with the ChemiDoc XRS imager (BioRad).
RNA–Immunoprecipitation
Native larval nuclear protein extracts from third instar w1118 larvae were prepared as previously described [11] and RNA-immunoprecipitations were conducted as published earlier [33] with small modifications (Text S1).
ISWI/hsrω-n Gel Mobility Assay
Recombinant full length ISWI or ISWI-N or ISWI-C proteins [23], [24] were incubated with in vitro transcribed sense 280b tandem repeat unit of the hsrω-n ncRNA or a generic RNA of the same size (RNActr, Roche) as a control, in increasing ratios of 1∶1, 5∶1,10∶1 and 20∶1 nmoles. The hsrω-n ncRNA or the RNActr were incubated with the desired protein for 30 min at 25°C in RB2 buffer (20% Glycerol, 0.2 mM EDTA, 20 mM Tris-HCl pH 7.5, 1 mM MgCl2, 150 mM NaCl, 1 mM DTT and RNAsin). After incubation, the RNA/protein complexes were resolved on 1.4% agarose gel in 0.5× TBE at 4°C for 105 minutes at 70 volts. RNA molecules were visualized by ethidium bromide staining. ATP, ATP-γ-S and ADP (Roche) were added in the gel shift assay at a final concentration of 100 µM. Excess of cold hsrω-n repeat unit or a generic RNActr transcript was used as competitor for ISWI/hsrω binding detected by gel mobility shift using P33 radiolabeled hsrω280b sense repeat unit and recombinant ISWI. RNA/protein complexes were resolved as above. After gel drying, RNA/protein complexes were visualized using the BioRad Phosphoimager system.
ATPase Assay
ATPase assay was conducted as previously described [23]. Extent of ATP hydrolysis was calculated with the following formula [P33/(P33+AMP-P−P33)]*100 (Figure 5G). The ATPase activity of 4 nmoles of full length ISWI was assayed for 1 hour; 4 nmoles of ISWI-N and ISWI-C were assayed for 30 minutes in the presence of 2 nmole of either supercoiled plasmid DNA, 280 bp hsrω-repeat unit encoding double stranded DNA, hsrω-n 280 bp tandem repeat ncRNA or a 300 bp generic RNA (RNActr; Roche) as a control.
Supporting Information
Zdroje
1. CoronaDFTamkunJW 2004 Multiple roles for ISWI in transcription, chromosome organization and DNA replication. Biochim Biophys Acta 1677 113 119
2. DirscherlSSKrebsJE 2004 Functional diversity of ISWI complexes. Biochem Cell Biol 82 482 489
3. CoronaDFSiriacoGArmstrongJASnarskayaNMcClymontSA 2007 ISWI regulates higher-order chromatin structure and histone H1 assembly in vivo. PLoS Biol 5 e232 doi:10.1371/journal.pbio.0050232
4. DeuringRFantiLArmstrongJASarteMPapoulasO 2000 The ISWI chromatin-remodeling protein is required for gene expression and the maintenance of higher order chromatin structure in vivo. Mol Cell 5 355 365
5. ParrishJZKimMDJanLYJanYN 2006 Genome-wide analyses identify transcription factors required for proper morphogenesis of Drosophila sensory neuron dendrites. Genes Dev 20 820 835
6. XiRKirillyDXieT 2005 Molecular mechanisms controlling germline and somatic stem cells: similarities and differences. Curr Opin Genet Dev 15 381 387
7. HoganCVarga-WeiszP 2007 The regulation of ATP-dependent nucleosome remodelling factors. Mutat Res 618 41 51
8. FerreiraREberharterABonaldiTChiodaMImhofA 2007 Site-specific acetylation of ISWI by GCN5. BMC Mol Biol 8 73
9. CoronaDFClapierCRBeckerPBTamkunJW 2002 Modulation of ISWI function by site-specific histone acetylation. EMBO Rep 3 242 247
10. ArancioWOnoratiMCBurgioGCollesanoMIngrassiaAM The nucleosome remodeling factor ISWI functionally interacts with an evolutionarily conserved network of cellular factors. Genetics 185 129 140
11. BurgioGLa RoccaGSalaAArancioWDi GesuD 2008 Genetic identification of a network of factors that functionally interact with the nucleosome remodeling ATPase ISWI. PLoS Genet 4 e1000089 doi:10.1371/journal.pgen.1000089
12. BendenaWGAyme-SouthgateAGarbeJCPardueML 1991 Expression of heat-shock locus hsr-omega in nonstressed cells during development in Drosophila melanogaster. Dev Biol 144 65 77
13. JollyCLakhotiaSC 2006 Human sat III and Drosophila hsr omega transcripts: a common paradigm for regulation of nuclear RNA processing in stressed cells. Nucleic Acids Res 34 5508 5514
14. PrasanthKVRajendraTKLalAKLakhotiaSC 2000 Omega speckles - a novel class of nuclear speckles containing hnRNPs associated with noncoding hsr-omega RNA in Drosophila. J Cell Sci 113 Pt 19 3485 3497
15. MallikMLakhotiaSC 2009 RNAi for the large non-coding hsromega transcripts suppresses polyglutamine pathogenesis in Drosophila models. RNA Biol 6 464 478
16. BuszczakMPaternoSLighthouseDBachmanJPlanckJ 2007 The carnegie protein trap library: a versatile tool for Drosophila developmental studies. Genetics 175 1505 1531
17. WangYZhangWJinYJohansenJJohansenKM 2001 The JIL-1 tandem kinase mediates histone H3 phosphorylation and is required for maintenance of chromatin structure in Drosophila. Cell 105 433 443
18. CarreCCiurciuAKomonyiOJacquierCFagegaltierD 2008 The Drosophila NURF remodelling and the ATAC histone acetylase complexes functionally interact and are required for global chromosome organization. EMBO Rep 9 187 192
19. GarbeJCPardueML 1986 Heat shock locus 93D of Drosophila melanogaster: a spliced RNA most strongly conserved in the intron sequence. Proc Natl Acad Sci U S A 83 1812 1816
20. MallikMLakhotiaSC 2011 Pleiotropic consequences of misexpression of developmentally active and stress-inducible non-coding hsrω gene in Drosophila. J Biosciences In Press
21. MukherjeeTLakhotiaSC 1979 3H-uridine incorporation in the puff 93D and in chromocentric heterochromatin of heat shocked salivary glands of Drosophila melanogaster. Chromosoma 74 75 82
22. OhHParkYKurodaMI 2003 Local spreading of MSL complexes from roX genes on the Drosophila X chromosome. Genes Dev 17 1334 1339
23. CoronaDFLangstGClapierCRBonteEJFerrariS 1999 ISWI is an ATP-dependent nucleosome remodeling factor. Mol Cell 3 239 245
24. GruneTBrzeskiJEberharterAClapierCRCoronaDF 2003 Crystal structure and functional analysis of a nucleosome recognition module of the remodeling factor ISWI. Mol Cell 12 449 460
25. MallikMLakhotiaSC Improved activities of CREB binding protein, heterogeneous nuclear ribonucleoproteins and proteasome following downregulation of noncoding hsromega transcripts help suppress poly(Q) pathogenesis in fly models. Genetics 184 927 945
26. MallikMLakhotiaSC 2009 The developmentally active and stress-inducible noncoding hsromega gene is a novel regulator of apoptosis in Drosophila. Genetics 183 831 852
27. PrasanthKVSpectorDL 2007 Eukaryotic regulatory RNAs: an answer to the ‘genome complexity’ conundrum. Genes Dev 21 11 42
28. GenoveseSCoronaDFV 2007 A New Medium to Grow Live Insects. European Patent MI2007A001420/8145 PTIT 2007
29. GoodrichJSClouseKNSchupbachT 2004 Hrb27C, Sqd and Otu cooperatively regulate gurken RNA localization and mediate nurse cell chromosome dispersion in Drosophila oogenesis. Development 131 1949 1958
30. BuchenauPArndt-JovinDJSaumweberH 1993 In vivo observation of the puff-specific protein no-on transient A (NONA) in nuclei of Drosophila embryos. J Cell Sci 106 Pt 1 189 199
31. AmeroSAElginSCBeyerAL 1991 A unique zinc finger protein is associated preferentially with active ecdysone-responsive loci in Drosophila. Genes Dev 5 188 200
32. LakhotiaSCSharmaA 1995 RNA metabolism in situ at the 93D heat shock locus in polytene nuclei of Drosophila melanogaster after various treatments. Chromosome Res 3 151 161
33. KeeneJDKomisarowJMFriedersdorfMB 2006 RIP-Chip: the isolation and identification of mRNAs, microRNAs and protein components of ribonucleoprotein complexes from cell extracts. Nat Protoc 1 302 307
34. StowersRSSchwarzTL 1999 A genetic method for generating Drosophila eyes composed exclusively of mitotic clones of a single genotype. Genetics 152 1631 1639
35. La RoccaGBurgioGCoronaDF 2007 A protein nuclear extract from D. melanogaster larval tissues. Fly (Austin) 1 343 345
Štítky
Genetika Reprodukční medicína
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2011 Číslo 5- Akutní intermitentní porfyrie
- Růst a vývoj dětí narozených pomocí IVF
- Vliv melatoninu a cirkadiálního rytmu na ženskou reprodukci
- Délka menstruačního cyklu jako marker ženské plodnosti
- Intrauterinní inseminace a její úspěšnost
-
Všechny články tohoto čísla
- Structural and Functional Differences in the Long Non-Coding RNA in Mouse and Human
- Identification, Replication, and Functional Fine-Mapping of Expression Quantitative Trait Loci in Primary Human Liver Tissue
- A −436C>A Polymorphism in the Human Gene Promoter Associated with Severe Childhood Malaria
- A Decline in p38 MAPK Signaling Underlies Immunosenescence in
- The Operon Balances the Requirements for Vegetative Stability and Conjugative Transfer of Plasmid R388
- Novel and Conserved Protein Macoilin Is Required for Diverse Neuronal Functions in
- Ixr1 Is Required for the Expression of the Ribonucleotide Reductase Rnr1 and Maintenance of dNTP Pools
- Genome of Strain SmR1, a Specialized Diazotrophic Endophyte of Tropical Grasses
- A Deficiency of Ceramide Biosynthesis Causes Cerebellar Purkinje Cell Neurodegeneration and Lipofuscin Accumulation
- A Latent Pro-Survival Function for the Mir-290-295 Cluster in Mouse Embryonic Stem Cells
- Association of Genetic Variants in Complement Factor H and Factor H-Related Genes with Systemic Lupus Erythematosus Susceptibility
- DNA Methylation Dynamics in Human Induced Pluripotent Stem Cells over Time
- Prion Formation and Polyglutamine Aggregation Are Controlled by Two Classes of Genes
- Integrated Genome-Scale Prediction of Detrimental Mutations in Transcription Networks
- Post-Embryonic Nerve-Associated Precursors to Adult Pigment Cells: Genetic Requirements and Dynamics of Morphogenesis and Differentiation
- A Novel Mouse Synaptonemal Complex Protein Is Essential for Loading of Central Element Proteins, Recombination, and Fertility
- STAT Is an Essential Activator of the Zygotic Genome in the Early Embryo
- A Genetic and Structural Study of Genome Rearrangements Mediated by High Copy Repeat Ty1 Elements
- A Missense Mutation in Causes a Major QTL Effect on Ear Size in Pigs
- A Flurry of Folding Problems: An Interview with Susan Lindquist
- Meiotic Recombination Intermediates Are Resolved with Minimal Crossover Formation during Return-to-Growth, an Analogue of the Mitotic Cell Cycle
- A Nervous Origin for Fish Stripes
- The ISWI Chromatin Remodeler Organizes the hsrω ncRNA–Containing Omega Speckle Nuclear Compartments
- The Telomerase Subunit Est3 Binds Telomeres in a Cell Cycle– and Est1–Dependent Manner and Interacts Directly with Est1
- Nodal-Dependent Mesendoderm Specification Requires the Combinatorial Activities of FoxH1 and Eomesodermin
- SHINE Transcription Factors Act Redundantly to Pattern the Archetypal Surface of Arabidopsis Flower Organs
- Characterizing Genetic Risk at Known Prostate Cancer Susceptibility Loci in African Americans
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Nodal-Dependent Mesendoderm Specification Requires the Combinatorial Activities of FoxH1 and Eomesodermin
- SHINE Transcription Factors Act Redundantly to Pattern the Archetypal Surface of Arabidopsis Flower Organs
- Association of Genetic Variants in Complement Factor H and Factor H-Related Genes with Systemic Lupus Erythematosus Susceptibility
- STAT Is an Essential Activator of the Zygotic Genome in the Early Embryo
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



