-
Články
- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Genome of Strain SmR1, a Specialized Diazotrophic Endophyte of Tropical Grasses
The molecular mechanisms of plant recognition, colonization, and nutrient exchange between diazotrophic endophytes and plants are scarcely known. Herbaspirillum seropedicae is an endophytic bacterium capable of colonizing intercellular spaces of grasses such as rice and sugar cane. The genome of H. seropedicae strain SmR1 was sequenced and annotated by The Paraná State Genome Programme—GENOPAR. The genome is composed of a circular chromosome of 5,513,887 bp and contains a total of 4,804 genes. The genome sequence revealed that H. seropedicae is a highly versatile microorganism with capacity to metabolize a wide range of carbon and nitrogen sources and with possession of four distinct terminal oxidases. The genome contains a multitude of protein secretion systems, including type I, type II, type III, type V, and type VI secretion systems, and type IV pili, suggesting a high potential to interact with host plants. H. seropedicae is able to synthesize indole acetic acid as reflected by the four IAA biosynthetic pathways present. A gene coding for ACC deaminase, which may be involved in modulating the associated plant ethylene-signaling pathway, is also present. Genes for hemagglutinins/hemolysins/adhesins were found and may play a role in plant cell surface adhesion. These features may endow H. seropedicae with the ability to establish an endophytic life-style in a large number of plant species.
Published in the journal: . PLoS Genet 7(5): e32767. doi:10.1371/journal.pgen.1002064
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1002064Summary
The molecular mechanisms of plant recognition, colonization, and nutrient exchange between diazotrophic endophytes and plants are scarcely known. Herbaspirillum seropedicae is an endophytic bacterium capable of colonizing intercellular spaces of grasses such as rice and sugar cane. The genome of H. seropedicae strain SmR1 was sequenced and annotated by The Paraná State Genome Programme—GENOPAR. The genome is composed of a circular chromosome of 5,513,887 bp and contains a total of 4,804 genes. The genome sequence revealed that H. seropedicae is a highly versatile microorganism with capacity to metabolize a wide range of carbon and nitrogen sources and with possession of four distinct terminal oxidases. The genome contains a multitude of protein secretion systems, including type I, type II, type III, type V, and type VI secretion systems, and type IV pili, suggesting a high potential to interact with host plants. H. seropedicae is able to synthesize indole acetic acid as reflected by the four IAA biosynthetic pathways present. A gene coding for ACC deaminase, which may be involved in modulating the associated plant ethylene-signaling pathway, is also present. Genes for hemagglutinins/hemolysins/adhesins were found and may play a role in plant cell surface adhesion. These features may endow H. seropedicae with the ability to establish an endophytic life-style in a large number of plant species.
Introduction
Soil bacteria can interact in many ways with plant partners ranging from beneficial to pathogenic. Among beneficial interactions the rhizobia play a central role, forming symbioses with legume species to produce nitrogen-fixing nodules, which supply most of the required fixed nitrogen to many agriculturally important crops such as soybean, pea, beans and clover.
A now well-characterized class of diazotrophic bacteria capable of establishing endophytic associations and promoting plant-growth of important cereal and forage grasses such as wheat, rice and maize has been investigated in recent years. Among such well-known species are Azospirillum brasilense, Gluconacetobacter diazotrophicus and H. seropedicae [1]. The colonization of plant tissues by these bacteria may involve the interplay of many as yet unidentified biochemical signals and gene products from both partners. H. seropedicae is an aerobic, prototrophic, endophytic nitrogen-fixing, plant-growth promoting bacterium, of the Betaproteobacteria found inside tissues of important crops such as corn, sugar-cane, rice, wheat and sorghum without causing disease to the plant partner [2]–[9], and has a low survival rate in plant-free soil [5]. It fixes nitrogen under conditions of ammonium and oxygen limitation [5] and can express nif genes in planta [6]–[11]. Moreover, H. seropedicae is an active plant colonizer and has been shown to promote plant growth and increase grain production [4], [9], [12]. Aluminum tolerant varieties of rice were shown by the 15N2 dilution technique to incorporate significant amount of nitrogen derived from nitrogen fixation [4], [9]. Ecological, agronomic, physiological, genetic and biochemical aspects of this organism have been reviewed [1], [12]–[14].
Results/Discussion
General features
The genome of H. seropedicae strain SmR1, a spontaneous streptomycin resistant mutant of strain Z78 [15] (ATCC 35893) was sequenced and annotated by the Paraná State Genome Programme (Genopar Consortium, www.genopar.org). Reads from the Sanger automatic sequencing (125,000) and from a full 454 FLX Titanium Roche Pyrosequencer run (1,220,352), corresponding to 100 times the coverage of the estimated genome size, were assembled to produce the genome sequence. End-sequencing of approximately 700 cosmids with an average insert of 40 kb was used to validate the final assembly.
The genome consists of a single circular chromosome of 5,513,887 base pairs with 63.4% G+C content (Table 1) and a total of 4,735 potential ORFs, encoding 3,108 proteins with assigned functions, 497 with general function prediction only and 1,130 with no known function, covering 88.3% of the genome. Coding sequences for 55 tRNA representing all 20 protein amino acids were also identified. The genome has 3 complete rRNA operons, one in the positive and two in the negative strand, all containing a pair of Ile-tRNA/Ala-tRNA genes in the intergenic region between the 16SrRNA and 23SrRNA genes (Figure 1). Genes for 19 of the 20 aminoacyl-tRNA synthetases are present with the exception of a gene coding for asparaginyl-tRNA synthetase. The biosynthesis of aspartyl-tRNAAsn or glutamyl-tRNAGln occurs via transamidation catalysed by an Asp-tRNAAsn/Glu-tRNAGln amidotransferase, an enzyme coded by the gatBAC operon as in most Bacteria [16]. These genes are widely spread among bacteria and are found in the genomes of other closely-related Betaproteobacteria such as Herminiimonas arsenicoxydans, bacteria of the Burkholderia genus, and Minibacterium massiliensis (Janthinobacterium species Marseille).
Fig. 1. The genome of Herbaspirillum seropedicae SmR1. 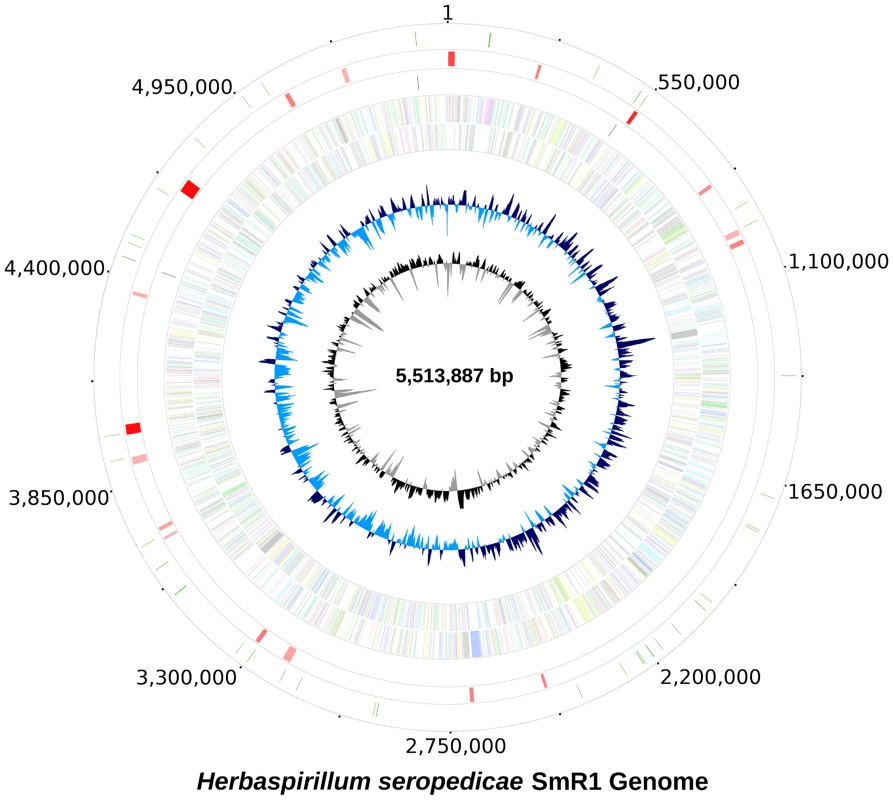
From inside to outside 1) G+C content; 2) GC skew; 3) genes color-coded according the COG functional categories; genes in the + strand and − strand are represented in the inside and outside circles respectively; 4) rRNAS operons; 5) putative horizontally transferred regions identified using IVOM: light red indicates low score and dark red indicates high score; 6) regions of H. seropedicae genome identical to castor bean (Ricinus communis) sequences (minimum of 200 bp in length and higher than 90% in identity). Tab. 1. General features of the genome of <i>Herbaspirillum seropedicae</i> SmR1. 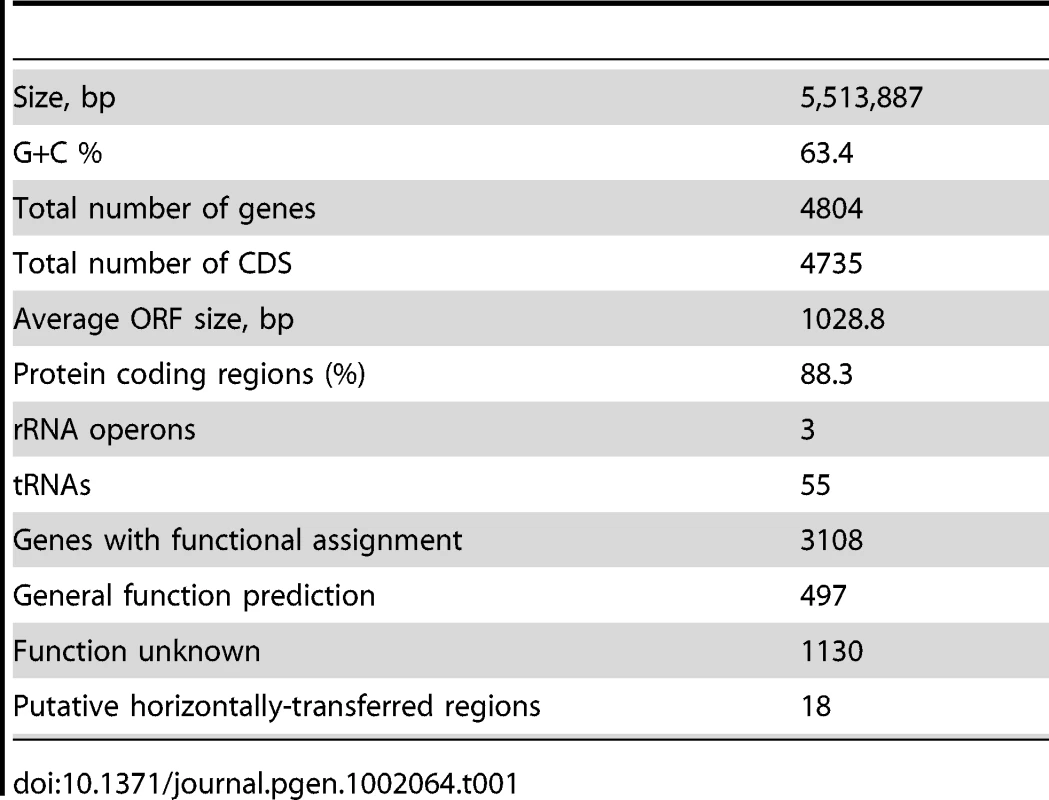
The probable origin of replication was identified based on the GC skew [17] and the positions of the genes dnaA, dnaN and gyrB. It maybe contained in the dnaA-dnaN intergenic region or upstream dnaA, where DnaA binding sequences were found. The region upstream of dnaA is unique, since instead of the rpmHrnpA operon present in most Proteobacteria it contains a probable glutamine amido transferase type II gene. Downstream from the dnaA, dnaN and gyrB genes there is a low G+C content (52%) region spanning 16.6 kbp of probable lateral transfer origin containing a reverse transcriptase gene of bacterial retrotransposons (RT_Bac_retron_I).
In the H. seropedicae genome 18 regions of probable lateral transfer origin, such as insertion sequences and phages were found. The two largest regions contain genes of bacteriophage origin. Region 1 (213,067 to 238,374) has a higher G+C content (66.4%) than the genome and contains 33 ORFs related to phage capsid assembly, regulation and phage transcription. Region 2 (967,869 to 1,006,417) has a lower G+C content (58.1%) with 52 ORFs, many related to phage P2.
One of the low G+C content (53.9%) regions contains a plasmid addiction module (operon phd/doc) coding for the PHD (prevents-host-death) and DOC (death-on-curing) proteins constituting a toxin-antitoxin (TA) module [18]. There are 3,412 PHD (http://www.ebi.ac.uk/interpro/IEntry?ac=IPR006442) and 707 DOC (http://www.ebi.ac.uk/interpro/IEntry?ac=IPR006440) protein sequences described in the Domain Bacteria, suggesting the widespread occurrence of this protection mechanism. ‘Toxin–antitoxin (TA) modules’ have been recognized as playing important roles in bacterial stress physiology and genome stabilization [18]–[20].
Genes coding for two partial and two complete transposases and 5 phage-related (three complete and two partial) and two genomic (xerC and xerD) recombinases/integrases were found in the H. seropedicae genome. The relative few number of genes related to mobile elements seems to be a common feature in all the genomes of endophytic bacteria (http://www.expasy.ch/sprot/hamap/interactions.html#Plant_endophyte?) sequenced to date. The exception is G. diazotrophicus Pal5, with 138 transposases and 223 insertion sequences [21]. The paucity in the number of putative transposable elements may suggest a low recombination/rearrangement in the genome of H. seropedicae SmR1, possibly reflecting its evolutionary adaptation to an endophytic lifestyle, and indicating a low rate of recent gene transfer that is presumably due to adaptation to a stable microenvironment, as suggested by Krause et al. [22] for the genome of Azoarcus sp.strain BH72.
General metabolism
H. seropedicae strain SmR1 is capable of growing on mono-saccharides such as D-glucose, D-fructose, D-galactose and L-arabinose, with sugar alcohols and organic acids such as L-malate or L-lactate but failed to grow on oligo - or polysaccharides [5], [15]. Accordingly, the genome of H. seropedicae contains the complete set of genes for the Entner-Doudoroff and pentose phosphate pathways. The Embden-Meyerhoff-Parnas (EMP) pathway lacks the gene coding for the classical 6-phosphofructokinase (PFK, E.C. 2.7.1.11), suggesting that H. seropedicae probably requires the involvement of the Entner-Doudoroff and the pentose phosphate pathways to metabolize D-glucose, D-fructose or D-mannose to pyruvate via the EMP pathway. Several ABC-type sugar-transport systems and one PEP/PTS transport system are present in the genome of H. seropedicae, consistent with its capacity to grow on a large number of monossaccharides [5], [15]. H. seropedicae has all the genes needed for gluconeogenesis: the EMP pathway plus those coding for fructose-1,6-biphosphatase, phosphoenolpyruvate dikinase, D-lactate and L-lactate dehydrogenases, and from two-carbon substrates such as ethanol via the glyoxalate cycle.
All genes necessary for the metabolism of D-galactose via 2-dehydro-3-deoxy-D-galactonate-6-phosphate leading to pyruvate and D-glyceraldehyde-3-phosphate are present in the genome. Subsequent conversion of pyruvate to acetyl-CoA is via the pyruvate dehydrogenase complex, while lactate dehydrogenase serves as an entry point of lactate during lactate-dependent growth. The conversion of 2-dehydro-3-deoxy-L-arabinonate to 2-keto-glutarate involves the sequential action of a dehydrase and NAD(P)-dehydrogenase. No such specific enzymes were found although several dehydrases and dehydrogenases are present in the genome of H. seropedicae SmR1.
The pathway for L-arabinose metabolism was shown to involve non-phosphorylated intermediates to produce 2-ketoglutarate [23]. This pathway probably involves the enzymes of the D-galactose breakdown pathway due to the identical configuration of C-2, C-3 and C-4 to those of L-arabinose.
The genome of H. seropedicae has all the genes for the citric acid cycle. Pathways replenishing intermediates of the cycle include the glyoxylate cycle (isocitrate lyase and malate synthase), the complete fatty acid β-oxidation pathway, the malic enzyme, phosphoenolpyruvate carboxykinase, phosphoenolpyruvate carboxylase and, from the degradation of the L-aminoacids alanine, glutamate, aspartate, asparagine and glutamine.
H. seropedicae grows in ethanol-containing media via alcohol dehydrogenase and aldehyde dehydrogenase to yield acetyl-CoA which can feed into the citric acid cycle.
H. seropedicae is an aerobic bacterium capable of fixing nitrogen under conditions of oxygen limitation. The genome of H. seropedicae has genes for four terminal oxidases: cytochrome c oxidase aa3 and the three alternative terminal oxidases bd, cbb3 and o, suggesting a branched respiratory chain. It has all the genes for the synthesis of NADH dehydrogenase, succinate dehydrogenase, cytochrome c reductase and also the complete set of genes for ATP synthase. The high affinity terminal oxidase cbb3 presumably supports ATP synthesis under the limiting oxygen conditions essential for nitrogenase synthesis and activity, as in other aerobic diazotrophs [24].
Polybetaalkanoates
H. seropedicae SmR1 synthesizes poly(3-hydroxybutyrate) under diazotrophic growth conditions and, as in other bacteria, it can reach up to 60% of the cell dry weight [25]. In silico analysis of the genome of H. seropedicae revealed 13 genes potentially involved in poly(3-hydroxybutyrate/alkanoate) synthesis and degradation. A main cluster containing phbF, phbB and phbC coding respectively for a transcription regulator, acetoacetyl-CoA reductase and poly(3-hydroxybutyrate) synthase was found between bases 3,411,979 and 3,415,628. In addition there are three phbA (acetyl-CoA acyltransferase), one phbC (poly(3-hydroxybutyrate) synthase), two phaC (poly(3-hydroxyalkanoate) synthase), one phaB (3-keto-acyl-CoA reductase), two phaP (phasin) and two poly3-hydroxyalkanoate depolymerase (phaZ) genes. The data suggests the presence of two systems for the synthesis of poly(3-hydroxyalkanoate) and one specific for poly(3-hydroxybutyrate) in H. seropedicae strain SmR1, which is consistent with the isolation of poly(3-hydroxybutyrate) and poly(3-hydroxybutyrate/valerate) co-polymer from strain Z67 [25].
Amino acid metabolism
The genome of H. seropedicae contains genes coding for the synthesis of all 20 protein amino acids. However, it has limited ability to grow on amino acids as carbon sources. It can grow on L-proline, L-tyrosine, D/L-alanine, β-alanine, L-isoleucine and L-glutamate but failed to grow on L-phenylalanine, L-histidine, L-arginine or L-lysine [5], [15], [26], [27]. In silico analysis of the genome content suggested that the pathway for the degradation of L-histidine and L-lysine is incomplete. No specific L-arginine transporter was found, supporting the observation that this molecule cannot serve a sole N-source for H. seropedicae growth [26]. On the other hand, endogenously synthesized L-arginine can be catabolised to agmatine, putrescine and to 4-aminobutanoate which could be further converted to succinate in H. seropedicae. A strain of H. seropedicae carrying a Tn5-lacZ insertion in the speB gene coding for arginase is induced under low ammonium conditions [28], suggesting the presence of a second pathway for arginine degradation under conditions of ammonium limitation.
Urea metabolism
H. seropedicae is capable of synthesizing and degrading urea. Genes coding for the complete urea cycle enzymes, the probable pathway for arginine biosynthesis in this bacterium, using proline and carbamoyl phosphate as a precursors were found. Urea is degraded by urease. The urease operon contains the structural genes ureA, ureB and ureC and the accessory genes ureD, ureE, ureF, ureG and ureJ. This operon is very similar to that of Janthinobacterium sp. Marseille, although it is lacking in the Herminiimonas arsenicoxidans genome. A complete ABC-type urea transport operon (urtABCDE) was found upstream from the ure gene cluster in both the H. seropedicae and Janthinobacterium genomes, similar to that of Corynebacterium glutamicum [29]. Analysis of a mutant strain of H. seropedicae Z78, containing a Tn5-lacZ insertion in the urtE gene and obtained by random Tn5-lacZ insertion and screening for differential expression under N-limiting conditions, led to the suggestion that both the urt and ure genes are expressed under N deprivation [28] and are probably controlled by the Ntr system since a σ54-dependent promoter is located upstream of urtA.
Nitrogen fixation
The nitrogen fixation genes (nif) of H. seropedicae, including nifA, nifB, nifZ, nifZ1, nifH, nifD, nifK, nifE, nifN, nifX, nifQ, nifW, nifV, nifU and nifS were found in a region spanning 37,547 bp interspersed with fix, mod, hes, fdx, hsc and other genes. The 46 ORFs of this cluster are organized in 7 NifA-, σ54-dependent operons. This cluster is flanked by two 348 bp fragments 93% identical (325 out of 348 bp), probably derived from a partial duplication of the gloA gene, corresponding to the region coding for the 99 aminoacid residues of the C-terminus of GloA. Just upstream the nif cluster a sequence reminiscent of a transposase gene is present, as in the nif cluster of Burkholderia vietnamiensis strain G4 chromosome 3. These are suggestive that H. seropedicae acquired the nif cluster by lateral transfer.
Two globin-like genes expressed from a putative NifA-regulated promoter are present in the nif cluster of H. seropedicae, while a single globin-like gene is found in this cluster in Burkholderia xenovorans LB400 chromosome 2 and Burkholderia vietnamiensis G4 chromosome 3. Since NifA in H. seropedicae is transcriptionally active only under limiting oxygen tensions, these globin-like proteins may support the delivery of oxygen to energy production under nitrogen-fixing conditions in both the free-living and the endophytic state. The nif cluster carries all the genes necessary for nitrogenase synthesis and activity, including molybdenum uptake, electron transport and metal cluster synthesis, and the nif operon regulatory gene, therefore a cluster capable of endowing an organism with the full capacity to fix nitrogen. No genes for alternative nitrogenases nor for both hydrogenase types were found in the genome of H. seropedicae.
Nitrate metabolism
H. seropedicae is capable of growing aerobically with nitrate as sole N source, but is unable to denitrify anaerobically [15]. In silico analysis revealed that the genome of H. seropedicae contains the genes for an assimilatory and a dissimilatory nitrate reductase. The genes for nitrate assimilation are located in two genomic regions: the first contains the genes for the ABC-type nitrate transport (nasFED) and the second contains the gene narK, a nitrate/nitrite transporter, nirBD coding for the assimilatory nitrite reductase and nasA the structural gene for the assimilatory nitrate reductase. In the same operon, upstream of nasA, a gene coding for a probable FAD-dependent pyridine nucleotide-disulphide oxidoreductase could fulfill the function of NasC in H. seropedicae. This latter operon organization is common in the Ralstonia eutropha H16, R. solanacearum and R. metallidurans genomes. The complete set of narGHJI genes coding for a respiratory nitrate reductase is present in H. seropedicae located downstream from two nitrate/nitrite transporters narK1 and narU and upstream of the regulatory pair narXnarL. No genes coding for dissimilatory nitrite reductase, nitric oxide reductase or nitrous oxide reductase are present in H. seropedicae SmR1. This is consistent with the observation by Baldani et al. [15] who found no evidence of denitrification as the release of N2O by H. seropedicae compared with that from Azospirillum lipoferum.
Presumably the nitrite formed by this respiratory nitrate reductase can only be converted to ammonium ions and not dissimilated to N2. The role of this dissimilatory nitrate reductase in H. seropedicae is not clear, however, it may be involved in NO production and in survival under hypoxia as described for Mycobacterium tuberculosis [30]. A gene coding for a nitric oxide dioxygenase was found in the genome of H. seropedicae transcribed in the opposite direction to the norR gene located immediately upstream. Nitric oxide mediates plant defense responses against pathogens and is used as a signaling molecule [31]–[34] and the role of this nitric oxide dioxygenase may be NO detoxification during the initial stages of the H. seropedicae endophytic colonization of plants.
Plant–bacterial interaction
H. seropedicae is capable of the rapid colonization of several Gramineae [11]. Monteiro et al. [35] showed H. seropedicae in cortical cell layers of maize roots 12 hours after inoculation and xylem occupation after 24 hours. Three important aspects of the H. seropedicae beneficial association with plants are its ability to invade and colonize plant hosts, to thrive on plant exudates and to benefit associated plants. Interestingly, genes coding for plant cell wall degradation enzymes such as glycosidases, cellulases and hemi-cellulases associated with bacterial penetration were not found in the H. seropedicae genome. It is likely therefore that this organism relies only on natural discontinuities of the plant root epidermis for penetration as suggested by Olivares et al. [6].
During the annotation of the H. seropedicae genome a large number of Blast returned hits with high levels of identity to ESTs and genomic sequences of Ricinus communis. A total of 686 of such sequences was found: these varied in size from 100 to 2000 bp and were distributed randomly on the genome. This result suggests that the ricinus plant used to construct the libraries had an active Herbaspirillum endophyte. Recently we showed that H. seropedicae can colonize Phaseolus vulgaris [36], and Herbaspirillum lusitanum was isolated from Phaseolus nodules [37]. Together these results indicate that Herbaspirillum species may have a broader host range than previously described.
Protein secretion
The genome of H. seropedicae has genes involved in Sec-dependent and Sec-independent protein export systems. The Sec-dependent secretion systems present are type II (T2SS), type V (auto-transporters; T5SS) and the type IV pili, while the Sec-independent secretion systems are type I (ABC transporters), type III (T3SS) and the type VI (T6SS). T3SS, type IV pili and T6SS have been implicated in delivering toxic effector proteins directly into the cytoplasm of eukaryotic cells by pathogenic bacteria. In non-pathogenic bacteria, such as H. seropedicae, the latter secretion systems may be involved in plant–bacterial recognition. In addition to the Sec translocase system, twin arginine translocase (tat genes) are also present in the H. seropedicae SmR1 genome. Genes for the type IV secretion system are absent from the H. seropedicae genome.
Effector proteins delivered by the T3SS of pathogenic bacteria can circumvent plant defense mechanisms and control host metabolism to their advantage. However, the T3SS system may also optimize beneficial host-bacteria interactions, a phenomenon first demonstrated for Rhizobium NGR234 which secrete effector proteins via the T3SS in response to flavonoids exudated by the plant host roots. The effect of the secreted effector can either enhance or diminish nodulation depending on the host legume [38]–[40]. In the H. seropedicae genome the T3SS gene region, potentially involved in plant/bacterial interactions, spans a 22 kb region of DNA which contains 7 hrp (hypersensitive response and pathogenicity), 8 hrc (hypersensitive response conserved), and 11 hypothetical ORFs (Figure 2). Two protein T3SS related genes hrpG, coding a transcription activator, and hpaB, that codes for a chaperone involved in protein secretion, are found at 10 kb downstream from the hrp/hrc cluster. The G+C content of the hrp/hrc region (66.1%) is slightly higher than the chromosomal average of 63.4%. Furthermore, no transposition elements flanking this region are present, suggesting that this region is not a recent acquisition by H. seropedicae or was laterally transferred from a closely related species. Gram negative bacteria that contain the hrp genes are divided into two main groups, according to the regulatory circuitry controlling T3SS gene expression and organization. In group I hrp genes are regulated by HrpL, a member of the ECF family of alternative sigma factors [41]–[43]. Induction of the hrpL gene requires the σ54 activator HrpS (Erwinia spp., Pantoea stewartii), or HrpS and HrpR (P. syringae). In organisms of group II the hrp genes are activated by an AraC-like activator, HrpB (R. solanacearum) or HrpX (Xanthomonas spp) [44]–[46], and the hrpX and hrpB genes are activated by the HrpG protein [46], [47]. H. seropedicae contains a gene for the ECF-like sigma factor HrpL resembling group I bacteria such as Pseudomonas syringae, Erwinia amylovora, and Pantoea stewartii. In contrast, H. seropedicae contains a gene for the HrpG protein, a transcriptional activator characteristic of group II bacteria, suggesting a hybrid regulatory system, involving regulatory elements from both groups. In addition, hrp-box motifs were found upstream of the hrp/hrc operons.
Fig. 2. The type III secretion system gene cluster of H. seropedicae SmR1 and other organisms. 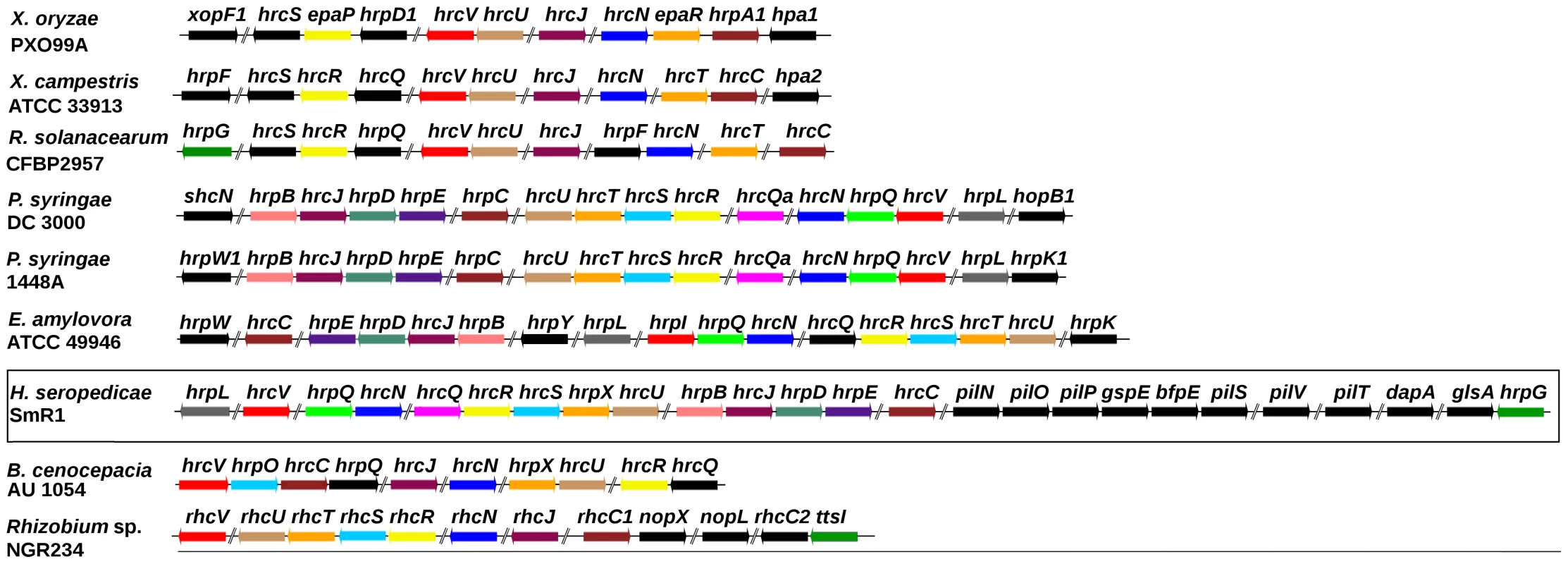
Genes of the same color in different organisms are homologous. Genes colored in black have no counterpart in the genomic regions shown. Contiguous to the hrp/hrc cluster were found the genes pilNOPgspEbfpEpilSVTdapAglsA (Figure 2). These code for proteins of the type IV pili, a system responsible for processes such as attachment to surfaces, twitching motility, biofilm formation, virulence and protein secretion [48]–[50]. In this region a gene coding for a lytic transglycosylase was also found. This protein is probably involved in partial degradation of the peptidoglycan to allow the efficient assembly and anchoring of supramolecular transport complexes such as T2SS, T3SS and type IV pili to the cell envelope. Interestingly, downstream from the genes of the type IV pili a methionyl-tRNA gene is present, suggesting that the hrp/hrc-type IV pili genes may form a genomic island.
A proteomic investigation of the secretome of H. seropedicae grown in minimal medium indicated a large number of proteins involved in cellular processes (45.4%), metabolism (36%), and hypothetical and conserved hypothetical (14.1%) proteins [51]. However, no type III proteins were detected among the secreted proteins, suggesting that specific physiological conditions may be required for expression and activity of the T3SS and synthesis of effector proteins in H. seropedicae.
Osmotic stress
A probable operon involved in the synthesis and degradation of a homopolymer of D-glucose, composed by the genes glgA (glycogen synthase), glgB (1,4-alpha-glucan branching enzyme), glgX (glycogen debranching enzyme), treZ (malto-oligosyltrehalose trehalohydrolase), malQ (4-alpha-glucanotransferase) and treY (malto-oligosyl trehalose synthase), is located in the complementary strand spanning bases 2,843,031 to 2,856,518. Neighbor gene analysis using the String server (http://string-db.org) revealed a similar gene organization in the Alphaproteobacteria Rhizobiales, in the Betaproteobacteria Burkholderia spp. and Gammaproteobacteria Xanthomonas spp. Synthesis of an amylopectin-like polysaccharide may be related to osmotic stress protection and energy storage. This may reflect the potential of these bacteria to interact with plants in an endophytic or pathogenic mode. Environmental Oxalobacteraceae such as Janthinobacterium sp. (strain Marseille) (Minibacterium massiliensis) and Heminiimonas arsenicoxidans, of the same family as H. seropedicae, lack these genes.
There are genes for two trehalose synthesis systems in the genome of H. seropedicae. One of these involves otsA coding an alpha,alpha-trehalose-phosphate synthase (UDP-forming) and otsB, coding a trehalose-6-phosphate phosphatase, and a glucoamylase gene. The other system involves an alpha-amylase (Hsero_2325), trehalose synthase (Hsero_2326), and a 1,4-alpha-glucan branching enzyme (Hsero_2327) and constitute an operon.
Furthermore, four Na+(K+)/H+ antiporter (nhaA, nhaP, arsB and Hsero_3967) genes are present in the genome of H. seropedicae which may contribute to the defense against osmotic/saline stress.
Polyphosphate
Polyphosphates are involved in the response of bacteria to extreme stress conditions of salinity, osmolarity, desiccation, N-starvation, UV radiation, barometric pressure, pH, and temperature [52]. Two genes coding for polyphosphate kinase (Hsero_0611 and ppk), the enzyme responsible for the synthesis of polyphosphate, and one coding for an exopolyphosphatase (ppx), are present in the genome of H. seropedicae. These systems may constitute adaptative defense mechanism for the endophytic life style of H. seropedicae.
Siderophores
The rhizosphere and the rhizoplan are highly competitive areas for bacterial survival and development; the capacity to acquire siderophores complexed with Fe3+ in Fe-limited soils would be advantageous in such competition. H. seropedicae has at least 27 genes involved in iron transport and metabolism. A very large gene (27,483 bp) coding for a modular peptide synthase is the only protein of H. seropedicae probably involved in siderophore synthesis (Hsero_2343). This gene is located downstream from cirA, a TonB-dependent siderophore receptor, and prfI, an ECF sigma factor. The genome has 17 TonB-dependent siderophore receptors and one ABC-type hydroxamate-type ferric siderophore uptake system. Presumably iron uptake is via active transport involving an ABC-type system and TonB/ExbB/ExbD. The rice endophyte Azoarcus also contains a plethora of TonB dependent siderophore receptors [22], [53]. This large number of iron receptors may endow organisms such as H. seropedicae and Azoarcus with a high competitiveness in iron-limited environments and may confer the ability to out-compete other bacteria. Also present in the genome is the global iron regulator gene fur.
Auxin biosynthesis
The plant growth-promoting bacteria probably owe some of their ability to the production and secretion of phytohormones [54]. There are four possible pathways in H. seropedicae for the production of indoleacetic acid (IAA) from tryptophan. The most probable route is via indolepyruvate, to indole-acetic acid catalysed by tryptophan transaminase and indolepyruvate ferredoxin oxidoreductase. Genes for the other possible metabolic routes are also present: 1) tryptophan to indoleacetate via indoleacetamide; 2) from indoleacetamide to indoleacetate via indoleacetonitrile and 3) tryptophan to indoleacetate via tryptamine and indoleacetaldehyde.
Modulation of endogenous ethylene levels by ACC deaminase
Ethylene is a known plant hormone synthesized from S-adenosylmethionine by 1-aminocyclopropane 1-carboxylate (ACC) synthase, an enzyme activated by IAA under biotic and abiotic stress conditions [55]. ACC is converted to ethylene by ACC oxidase. A gene coding ACC deaminase is present in the H. seropedicae genome and is known to compete with ACC oxidase, modulating the levels of ethylene in plants, thus decreasing the stress response promoted by ethylene and allowing plant growth under stress conditions [56]. The coordinated production of IAA and ACC deaminase by H. seropedicae is a likely mechanism for plant growth promotion by this microorganism as shown for the Herbaspirillum-related endophytic, nitrogen-fixing, plant growth-promoting Betaproteobacterium, Burkholderia phytofirmans PsJN [57].
Metabolism of aromatic compounds
H. seropedicae genome contains genes coding for degradation of benzoate, benzamide, benzonitrile, hydroxy-benzoate, and vanillate (Figure 3). In separate clusters, genes coding for a nicotinic acid degradation pathway similar to that of Pseudomonas putida [58] and a meta pathway of an as yet unknown phenolic compound were found. These pathways may be important to allow H. seropedicae to thrive on plant tissues by conferring both metabolic flexibility and defense against plant-derived toxic chemicals.
Fig. 3. Proposed pathways for aromatic compounds metabolism in <i>H. seropedicae</i> SmR1. 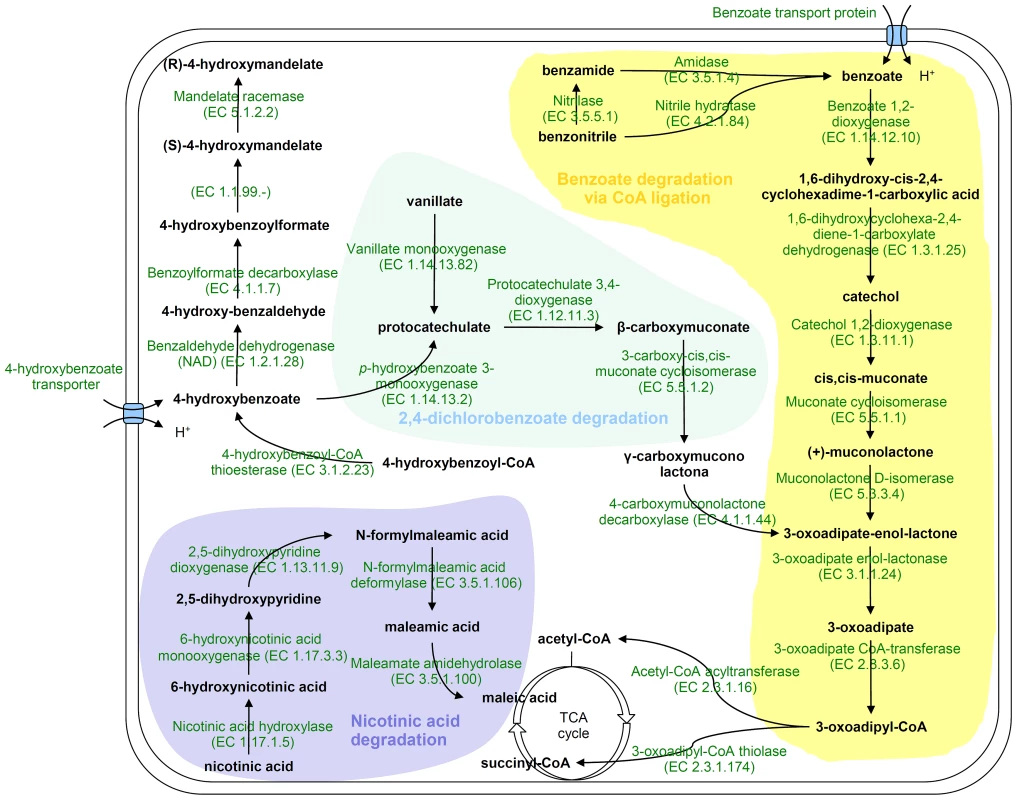
Hemagglutins/hemolysins
Hemagglutins/hemolysins are cytotoxic proteins implicated in animal pathogenesis, but a large number of genes coding for such proteins have been found in plant pathogens and plant-interacting bacteria [59]. Twenty genes related to hemagglutinin/hemolysin are present in the genome of H. seropedicae SmR1, and 9 additional genes code for hemagglutinin/hemolysin accessory proteins such as transporters/activators. Three genes code for hemagglutinins with adhesin-like domains, two of which are homologous to fhaB of Xanthomonas axonopodis pv citri [59] and are associated with genes coding for the accessory FhaC protein. The products of these genes may be required for surface attachment and biofilm formation during plant tissue colonization [60].
Concluding remarks
The genome of H. seropedicae revealed a metabolically versatile bacterium, with the ability to thrive on a range of plant metabolites from sugars to phenolic compounds (Figure 4). It is capable of synthesizing plant-growth modulators such as auxins and gibberellins, although only potential pathways for IAA synthesis were found in the genome; the cryptic genes for gibberellins and citokinins syntheses remain to be identified. It is surprising that an aggressive plant colonizer such as H. seropedicae [35] is devoid of glycohydrolases involved in plant cell wall degradation. However, H. seropedicae displays an impressive variety of protein secretion systems and hemagglutinins/hemolysins/adhesins that may facilitate plant invasion, colonization and an endophytic life, following penetration through natural epidermal wounds. Additional contributors to the plant-growth-promoting capacity of H. seropedicae may depend on the many genes involved in nitrogen fixation, NO3− and NO2− assimilation, NO oxidation, and ACC deamination (Figure 4). The presence of ACC deaminase may modulate ethylene production stimulated by IAA from bacterial origin, thus allowing plant resistance to biotic and abiotic stress conditions. These non-specific plant-interaction systems may endow H. seropedicae with the ability to establish an endophytic life-style in a large number of plant species.
Fig. 4. Molecular mechanisms probably involved in plant colonization and plant growth promotion identified in the H. seropedicae SmR1 genome. 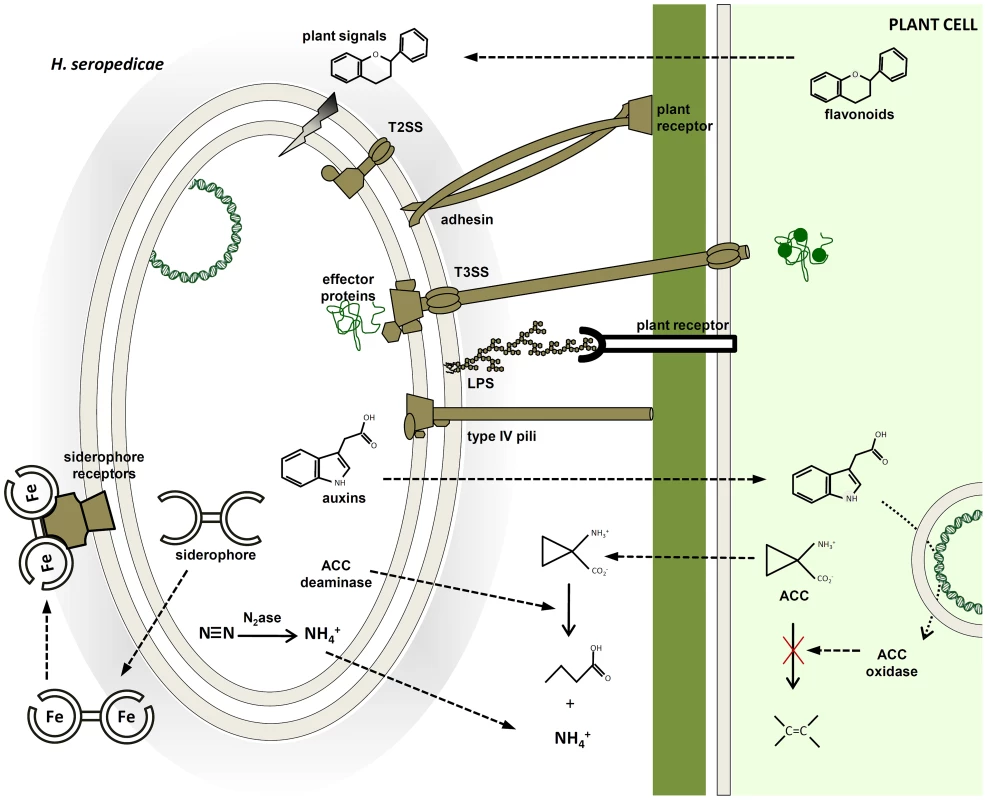
Plant signals can modulate the expression of bacterial genes coding for adhesins, type IV pili and enzymes of lipopolysaccharide (LPS) synthesis, triggering bacterial attachment to root surfaces. The molecular communication involves bacterial protein secretion and phytohormones to stimulate plant growth and modulate plant defense response. In addition, modulation of plant ethylene levels by ACC deaminase may contribute to plant growth promotion. The success of the endophytic association depends on a compatible genetic background that leads to benefits for both organisms. Materials and Methods
Organisms and DNA purification
H. seropedicae strain SmR1 was grown in liquid NFbHP medium containing 20 mM NH4Cl and 0.5% potassium malate, as described by Klassen et al. [26]. DNA was purified using phenol-chloroform extraction of cells lysed with lysozyme and SDS.
E. coli strain hosts XL1-Blue and DH10B were grown in LB or Terrific broth [61].
Genome sequencing and assembly
The genome sequence of H. seropedicae strain SmR1 total DNA was determined by the whole genome sequencing strategy [62] using short fragment (1.5–3.0 kb) libraries in pUC18 and pUC19 (Amersham Biosciences) and cosmid libraries in Supercos (Promega). DNA inserts were sequenced using the DYEnamic ET kit (GE HealthCare) and MegaBace 1000 automatic sequencers. Plasmid and cosmid DNA template preparation was performed by alkaline lysis and sequenced in 96-well plates according to standard procedures. A full DNA sequence run was performed in a Roche 454 GS-FLX Titanium by Creative Genomics, USA.
The genome was assembled using the Phred/Phrap/Consed package (www.phrap.org) and the Roche NewBler assembler. End sequences of cosmids were used to validate the genome assembly. Contig scaffolding was suggested by Autofinisher (www.phrap.org) and gaps were closed using PCR and whole insert sequencing of selected plasmid clones. The average final Phred score value was higher than 70.
Genome annotation
Potential protein coding regions (ORFs) were identified by an integrated automatic annotation platform with Glimmer 2 [63], [64], and Blast softwares [65]. Probable functions of translation products of potential orfs were inferred using the Blast package to search the public databases GenBank (L), COG [66], KEGG [67] and pFAM [68]. The output of an in-house annotation platform was reviewed by human annotators for gene assignment and proposed function. Each proposed gene sequence and annotation was validated with individual inspection by Artemis V11 [69]. tRNAs were located using tRNAscan-SE [70]. Ribosomal RNA operons were located using Blastn [65]. Putative horizontally transferred DNA regions were identified using IVOM [71].
Zdroje
1. PedrosaFOElmerichC 2007 Regulation of nitrogen fixation and ammonium assimilation in associative and endophytic nitrogen fixing bacteria. ElmerichCNewtonWE Associative and endophytic nitrogen fixing bacteria and cyanobacterial associations 47 71 Kluwer: The Netherlands
2. PimentelJPOlivaresFLPitardRMUrquiagaSAkibaF 1991 Dinitrogen fixation and infection of grass leaves by Pseudomonas rubrisubalbicans and Herbaspirillum seropedicae. Plant Soil 137 61 65
3. BaldaniVLDBaldaniJIOlivaresFLDöbereinerJ 1992 Identification and ecology of Herbaspirillum seropedicae and closely related Pseudomonas rubrisubalbicans. Symbiosis 13 65 73
4. GyaneshwarPJamesEKReddyPMLadhaJK 2002 Herbaspirillum colonization increases growth and nitrogen accumulation in aluminum-tolerant rice varieties. New Phytol 154 131 145
5. BaldaniJIPotBKirchhofGFalsenEBaldaniVLD 1996 Emended description of Herbaspirillum; inclusion of (Pseudomonas) rubrisubalbicans, a mild pathogen, as Herbaspirillum rubrisubalbicans comb. nov., and classification of a group of clinical isolates (EFgroup 1) as Herbaspirillum species 3. Int J Syst Bacteriol 46 802 810
6. OlivaresFLJamesEKBaldaniJIDöbereinerJ 1997 Infection of mottled stripe disease-susceptible and resistant sugar cane varieties by the endophytic diazotroph Herbaspirillum. New Phytol 135 723 737
7. JamesEKOlivaresFLBaldaniJIDöbereinerJ 1997 Herbaspirillum, an endophytic diazotroph colonizing vascular tissue of Sorghum bicolor L. Moench. J Exp Bot 48 785 798
8. OlivaresFLBaldaniVLDReisVMBaldaniJIDöbereinerJ 1996 Occurrence of endophytic diazotrophs Herbaspirillum spp. in roots, stems and leaves predominantly of Gramineae. Biol Fertil Soils 21 197 200
9. JamesEKGyaneshwarPMathanNBarraquioWLReddyPM 2002 Infection and colonization of rice seedlings by the plant growth-promoting bacterium Herbaspirillum seropedicae Z67. Mol Plant Microbe In 15 894 906
10. ElbeltagyANishiokaKSatoTSuzukiHYeB 2001 Endophytic colonization and in planta nitrogen fixation by a Herbaspirillum sp. isolated from wild rice species. Appl Environ Microb 67 5285 93
11. Roncato-MaccariLDBRamosHJOPedrosaFOAlquiniYChubatsuLS 2003 Endophytic Herbaspirillum seropedicae expresses nif genes in gramineous plants. FEMS Microbiol Ecol 45 39 47
12. BoddeyRMDe OliveiraOCUrquiagaSReisVMOlivaresFL 1995 Biological Nitrogen-Fixation Associated with Sugar-Cane and Rice - Contributions and Prospects for Improvement. Plant Soil 174 195 209
13. DöbereinerJPedrosaFO 1987 Nitrogen-fixing Bacteria in Nonleguminous Crop Plants Madison Science Tech 155
14. PedrosaFOBenelliEMYatesMGWassenRMonteiroRA 2001 Recent developments in the structural organization and regulation of nitrogen fixation genes in Herbaspirillum seropedicae. J Biotechnol 91 189 195
15. BaldaniJIBaldaniVLDSeldinLDöbereinerJ 1986 Characterization of Herbaspirillum seropedicae gen. nov., sp. nov., a new root-associated nitrogen-fixing bacterium. Int J Syst Bacteriol 36 86 93
16. IbbaMSöllD 2000 Aminoacyl-tRNA Synthesis. Ann Rev of Biochem 69 617 650
17. FrancinoMPOchmanH 1997 Strand asymmetries in DNA evolution. Trends Genet 13 240 245
18. LehnherrHMaguinEJafriSYarmolinskyMB 1993 Plasmid addiction genes of bacteriophage P1: doc, which causes cell death on curing of prophage, and phd, which prevents host death when prophage is retained. J Mol Biol 233 414 428
19. ButsLLahJDao-ThiMWynsLLorisR 2005 Toxin–antitoxin modules as bacterial metabolic stress managers. Trends Biochem Sci 30 672 679
20. Van MelderenLSaavedra De BastM 2009 Bacterial Toxin–Antitoxin Systems: More Than Selfish Entities? PLoS Genet 5 e1000437 doi:10.1371/journal.pgen.1000437
21. BertalanMAlbanoRPáduaVRouwsLRojasC 2009 Complete genome sequence of the sugarcane nitrogen-fixing endophyte Gluconacetobacter diazotrophicus Pal5. BMC Genomics 10 450
22. KrauseARamakumarABartelsDBattistoniFBekelT 2008 Complete genome of the mutualistic, N2-fixing grass endophyte Azoarcus sp. strain BH72. Nat Biotechnol 24 1385 1391
23. MathiasALRigoLUFunayamaSPedrosaFO 1989 L-arabinose metabolism in Herbaspirillum seropedicae. J Bacteriol 171 5206 5209
24. FischerHM 1994 Genetic regulation of nitrogen fixation in rhizobia. Microbiol Rev 58 352 386
25. CatalánALFerreiraFGillPRBatistaS 2007 Production of polyhydroxyalkanoates by Herbaspirillum seropedicae grown with different sole carbon sources and on lactose when engineered to express the lacZlacY genes. Enzyme Microb Tech 40 1352 1357
26. KlassenGPedrosaFOSouzaEMFunayamaSRigoLU 1997 Effect of nitrogen compounds on nitrogenase activity in Herbaspirillum seropedicae strain SMR1. Can J Microbiol 43 841 846
27. GussoCLSouzaEMRigoLUPedrosaFOYatesMG 2008 Effect of an ntrC mutation on amino acid or urea utilization and on nitrogenase switch-off in Herbaspirillum seropedicae. Can J Microbiol 54 235 239
28. SchwabSRamosHJSouzaEMPedrosaFOYatesMG 2007 Identification of NH4+-regulated genes of Herbaspirillum seropedicae by random insertional mutagenesis. Arch Microbiol 187 379 86
29. BeckersGBendtAKKrämerRBurkovskiA 2004 Molecular Identification of the Urea Uptake System and Transcriptional Analysis of Urea Transporter - and Urease-Encoding Genes in Corynebacterium glutamicum. J Bacteriol 186 7645 7652
30. SohaskeyDC 2008 Nitrate Enhances the Survival of Mycobacterium tuberculosis during Inhibition of Respiration. J Bacteriol 190 2981 2986
31. WojtaszekP 2000 Nitric oxide in plants: To NO or not to NO. Phytochem 54 1 4
32. HuangXKieferEvon RadUErnstDFoissnerI 2002 Nitric oxide burst and nitric oxide-dependent gene induction in plants. Plant Physiol Biochem 40 625 631
33. YamasakiH 2005 The NO world for plants: achieving balance in an open system. Plant, Cell Environ 28 78 84
34. Besson-BardACourtoisCGauthierADahanJDobrowolskaG 2008 Nitric Oxide in Plants: Production and Cross-talk with Ca2+ Signaling. Mol Plant 1 218 228
35. MonteiroRASchmidtMABauraVABalsanelliEWassemR 2008 Early colonization pattern of maize (Zea mays L. Poales, Poaceae) roots by Herbaspirillum seropedicae (Burkholderiales, Oxalobacteraceae). Genet Mol Biol 31 932 937
36. SchmidtMASouzaEMBauraVAWassemRYatesMG 2011 Evidence for the endophytic colonization of Phaseolus vulgaris (common bean) roots by the diazotroph Herbaspirillum seropedicae. Braz J Med Biol Res 44 182 185
37. ValverdeAVelázquezEGutierrezCCervantesEVentosaA 2003 Herbaspirillum lusitanum sp. nov., a novel nitrogen-fixing bacterium associated with root nodules of Phaseolus vulgaris. Int J Syst Evol Micr 53 1979 1983
38. DeakinWJBroughtonWJ 2009 Symbiotic use of pathogenic strategies: rhizobial protein secretion. Nat Rev Microbiol 7 312 320
39. MarieCDeakinWJVipreyVKopciñskaJGolinowski 2003 Characterization of Nops, nodulation outer proteins, secreted via the type III secretion system of NGR234. Mol Plant Microbe In 16 743 751
40. VipreyVDel GrecoAGolinowskiWBroughtonWJPerretX 1998 Symbiotic implications of type III protein secretion machinery in Rhizobium. Mol Microbiol 28 1381 1389
41. FrederickRDMajerczakDRCoplinDL 1993 Erwinia stewartii WtsA, a positive regulator of pathogenicity gene expression, is similar to Pseudomonas syringae pv. phaseolicola HrpS. Mol Microbiol 9 477 485
42. WeiZMBeerSV 1995 hrpL activates Erwinia amylovora hrp gene transcription and is a member of the ECF subfamily of sigma factors. J Bacteriol 177 6201 6210
43. XiaoYHutchesonSW 1994 A single promoter sequence recognized by a newly identified alternate sigma factor directs expression of pathogenicity and host range determinants in Pseudomonas syringae. J Bacteriol 176 3089 3091
44. GeninSGoughCLZischekCBoucherCA 1992 Evidence that the hrpB gene encodes a positive regulator of pathogenicity genes from Pseudomonas solanacearum. Mol Microbiol 6 3065 3076
45. KamdarHVKamounSKadoCI 1993 Restoration of pathogenicity of avirulent Xanthomonas oryzae pv. oryzae and X. campestris pathovars by reciprocal complementation with the hrpXo and hrpXc genes and identification of HrpX function by sequence analyses. J Bacteriol 175 2017 2025
46. WengelnikKBonasU 1996 HrpXv, an AraC-type regulator, activates expression of five of the six loci in the hrp cluster of Xanthomonas campestris pv. vesicatoria. J Bacteriol 178 3462 3469
47. BritoBMarendaMBarberisPBoucherCGeninS 1999 prhJ and hrpG, two new components of the plant signal-dependent regulatory cascade controlled by PrhA in Ralstonia solanacearum. Mol Microbiol 31 237 251
48. O'TooleGAKolterR 1998 Flagellar and twitching motility are necessary for Pseudomonas aeruginosa biofilm development. Mol Microbiol 30 295 304
49. HagerAJBoltonDLPelletierMRBrittnacherMJGallagherLA 2006 Type IV pili-mediated secretion modulates Francisella virulence. Mol Microbiol 62 227 237
50. HanXKennanRMParkerDDaviesJKRoodJI 2007 Type IV fimbrial biogenesis is required for protease secretion and natural transformation in Dichelobacter nodosus. J Bacteriol 189 5022 5033
51. ChavesDFSouzaEMMonteiroRAPedrosaFO 2009 A two-dimensional electrophoretic profile of the proteins secreted by Herbaspirillum seropedicae strain Z78. J Proteomics 73 50 56
52. SeufferheldMJAlvarezHMFariasME 2008 Role of Polyphosphates in Microbial Adaptation to Extreme Environments. Appl Environ Microb 74 5867 5874
53. HaunbergLSchmidtFScharfCDörrJVölkerUReinhold-HurekB 2010 Proteomic characterization of a pilR regulatory mutant of Azoarcus sp. strain BH72 with the aid of gel-based and gel-free approaches. Proteomics 10 458 469
54. BastiánFCohenAPiccoliPLunaVBaraldiR 1998 Production of indole-3-acetic acid and gibberellins A1 and A3 by Acetobacter diazotrophicus and Herbaspirillum seropedicae in chemically-defined culture media. Plant Growth Regul 24 7 11
55. WangKLiHEckerJR 2002 Ethylene Biosynthesis and Signaling Networks. Plant Cell 14 s131 151
56. GlickBRTodorovicBCzarnyJChengZDuanJMcConkeyB 2007 Promotion of plant growth by bacterial ACC deaminase. Crit Rev Plant Sci 26 227 242
57. SunYChengZGlickBR 2009 The presence of a 1-aminocyclopropane-1-carboxylate (ACC) deaminase deletion mutation alters the physiology of the endophytic plant growth-promoting bacterium Burkholderia phytofirmans PsJN. FEMS Microbiol Lett 296 131 136
58. JiménezJICanalesAJiménez-BarberoJGinalskiKRychlewskiL 2008 Deciphering the genetic determinants for aerobic nicotinic acid degradation: The nic cluster from Pseudomonas putida KT2440. Proc Natl Acad Sci 105 11329 11334
59. Van SluysMAMonteiro-VitorelloCBCamargoLEAMenckCFMda SilvaACR 2002 Comparative genomic analysis of plant-associated bacteria. Ann Rev Phytopathol 40 169 189
60. GottigNGaravagliaBSGarofaloCGOrellanoEGOttadoJ 2009 A Filamentous Hemagglutinin-Like Protein of Xanthomonas axonopodis pv.citri, the Phytopathogen Responsible for Citrus Canker, Is Involved in Bacterial Virulence. PLoS ONE 4 e4358 doi:10.1371/journal.pone.0004358
61. SambrookJFritschEFManiatisT 1989 Molecular cloning: a laboratory manual. 2ed New York Cold Spring Harbor Laboratory Press
62. HeidelbergJFEisenJANelsonWCClaytonRAGwinnML 2000 DNA sequence of both chromosomes of the cholera pathogen Vibrio cholera. Nature 406 477 484
63. SalzbergSDelcherAKasifSWhiteO 1998 Microbial gene identification using interpolated Markov models. Nucleic Acids Res 26 544 548
64. DelcherALHarmonDKasifSWhiteOSalzbergSL 1999 Improved microbial gene identification with GLIMMER. Nucleic Acids Res 27 4636 4641
65. AltschulSFMaddenTLSchäfferAAZhangJZhangZ 1997 Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25 3389 402
66. TatusovRLFedorovaNDJacksonJDJacobsARKiryutinB 2003 The COG database: an updated version includes eukaryotes. BMC Bioinformatics 11 41
67. KanehisaMGotoSHattoriMAoki-KinoshitaKFItohM 2006 From genomics to chemical genomics: new developments in KEGG. Nucleic Acids Res 34 D354 357
68. FinnRDMistryJTateJCoggillPHegerA 2009 The Pfam protein families database. Nucleic Acids Res 38 D211 D222
69. CarverTBerrimanMTiveyAPatelCBöhmeU 2008 Artemis and ACT: viewing, annotating and comparing sequences stored in a relational database. Bioinformatics 24 2672 2676
70. LoweTMEddySR 1997 tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res 25 955 964
71. VernikosGSParkhillJ 2006 Interpolated variable order motifs for identification of horizontally acquired DNA: revisiting the Salmonella pathogenicity islands. Bioinformatics 22 2196 2203
Štítky
Genetika Reprodukční medicína
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2011 Číslo 5- Růst a vývoj dětí narozených pomocí IVF
- Délka menstruačního cyklu jako marker ženské plodnosti
- Vztah užívání alkoholu a mužské fertility
- Šanci na úspěšný průběh těhotenství snižují nevhodné hladiny progesteronu vznikající při umělém oplodnění
- Vliv melatoninu a cirkadiálního rytmu na ženskou reprodukci
-
Všechny články tohoto čísla
- Structural and Functional Differences in the Long Non-Coding RNA in Mouse and Human
- Identification, Replication, and Functional Fine-Mapping of Expression Quantitative Trait Loci in Primary Human Liver Tissue
- A −436C>A Polymorphism in the Human Gene Promoter Associated with Severe Childhood Malaria
- A Decline in p38 MAPK Signaling Underlies Immunosenescence in
- The Operon Balances the Requirements for Vegetative Stability and Conjugative Transfer of Plasmid R388
- Novel and Conserved Protein Macoilin Is Required for Diverse Neuronal Functions in
- Ixr1 Is Required for the Expression of the Ribonucleotide Reductase Rnr1 and Maintenance of dNTP Pools
- Genome of Strain SmR1, a Specialized Diazotrophic Endophyte of Tropical Grasses
- A Deficiency of Ceramide Biosynthesis Causes Cerebellar Purkinje Cell Neurodegeneration and Lipofuscin Accumulation
- A Latent Pro-Survival Function for the Mir-290-295 Cluster in Mouse Embryonic Stem Cells
- Association of Genetic Variants in Complement Factor H and Factor H-Related Genes with Systemic Lupus Erythematosus Susceptibility
- DNA Methylation Dynamics in Human Induced Pluripotent Stem Cells over Time
- Prion Formation and Polyglutamine Aggregation Are Controlled by Two Classes of Genes
- Integrated Genome-Scale Prediction of Detrimental Mutations in Transcription Networks
- Post-Embryonic Nerve-Associated Precursors to Adult Pigment Cells: Genetic Requirements and Dynamics of Morphogenesis and Differentiation
- A Novel Mouse Synaptonemal Complex Protein Is Essential for Loading of Central Element Proteins, Recombination, and Fertility
- STAT Is an Essential Activator of the Zygotic Genome in the Early Embryo
- A Genetic and Structural Study of Genome Rearrangements Mediated by High Copy Repeat Ty1 Elements
- A Missense Mutation in Causes a Major QTL Effect on Ear Size in Pigs
- A Flurry of Folding Problems: An Interview with Susan Lindquist
- Meiotic Recombination Intermediates Are Resolved with Minimal Crossover Formation during Return-to-Growth, an Analogue of the Mitotic Cell Cycle
- A Nervous Origin for Fish Stripes
- The ISWI Chromatin Remodeler Organizes the hsrω ncRNA–Containing Omega Speckle Nuclear Compartments
- The Telomerase Subunit Est3 Binds Telomeres in a Cell Cycle– and Est1–Dependent Manner and Interacts Directly with Est1
- Nodal-Dependent Mesendoderm Specification Requires the Combinatorial Activities of FoxH1 and Eomesodermin
- SHINE Transcription Factors Act Redundantly to Pattern the Archetypal Surface of Arabidopsis Flower Organs
- Characterizing Genetic Risk at Known Prostate Cancer Susceptibility Loci in African Americans
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Nodal-Dependent Mesendoderm Specification Requires the Combinatorial Activities of FoxH1 and Eomesodermin
- SHINE Transcription Factors Act Redundantly to Pattern the Archetypal Surface of Arabidopsis Flower Organs
- Association of Genetic Variants in Complement Factor H and Factor H-Related Genes with Systemic Lupus Erythematosus Susceptibility
- STAT Is an Essential Activator of the Zygotic Genome in the Early Embryo
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



