-
Články
- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Programmed Ribosomal Frameshift Alters Expression of West Nile Virus Genes and Facilitates Virus Replication in Birds and Mosquitoes
Programmed ribosomal frameshift (PRF) is a strategy used by some viruses to regulate expression of viral genes and/or generate additional gene products for the benefit of the virus. Encephalitic flaviruses from Japanese encephalitis virus serogroup encode PRF motif in the beginning of nonstructural gene NS2A that results in production of an additional nonstructural protein NS1′ which for West Nile virus (WNV) consists of NS1 protein with 52 amino acid addition at the C terminus. Our previous studies showed that abolishing PFR and NS1′ production attenuated WNV virulence in mice. Here we show by using wild type and PRF-deficient WNV mutant that PRF induces overproduction of structural proteins, which facilitates virus replication in birds and mosquitoes while having no advantage for virus replication in cell lines in vitro. Presence of PRF/NS1′ allowed more efficient virus dissemination in the body of mosquitoes after taking infected blood meal and subsequent accumulation of the virus in saliva to facilitate transmission. Combined with our previous data in mice, the results obtained in this study demonstrate that while having no advantage for WNV replication in vitro, PRF provides advantage for WNV replication in vivo in mammalian, avian and mosquito hosts most likely by overproducing viral structural proteins and generating NS1′.
Published in the journal: . PLoS Pathog 10(11): e32767. doi:10.1371/journal.ppat.1004447
Category: Research Article
doi: https://doi.org/10.1371/journal.ppat.1004447Summary
Programmed ribosomal frameshift (PRF) is a strategy used by some viruses to regulate expression of viral genes and/or generate additional gene products for the benefit of the virus. Encephalitic flaviruses from Japanese encephalitis virus serogroup encode PRF motif in the beginning of nonstructural gene NS2A that results in production of an additional nonstructural protein NS1′ which for West Nile virus (WNV) consists of NS1 protein with 52 amino acid addition at the C terminus. Our previous studies showed that abolishing PFR and NS1′ production attenuated WNV virulence in mice. Here we show by using wild type and PRF-deficient WNV mutant that PRF induces overproduction of structural proteins, which facilitates virus replication in birds and mosquitoes while having no advantage for virus replication in cell lines in vitro. Presence of PRF/NS1′ allowed more efficient virus dissemination in the body of mosquitoes after taking infected blood meal and subsequent accumulation of the virus in saliva to facilitate transmission. Combined with our previous data in mice, the results obtained in this study demonstrate that while having no advantage for WNV replication in vitro, PRF provides advantage for WNV replication in vivo in mammalian, avian and mosquito hosts most likely by overproducing viral structural proteins and generating NS1′.
Introduction
West Nile virus (WNV) is a flavivirus that circulates in a bird-mosquito enzootic cycle with humans and horses as incidental hosts [1]. It belongs to the Japanese encephalitis subgroup that also includes Japanese encephalitis virus (JEV), St Louis encephalitis virus, and Murray Valley encephalitis virus [1]. The genome of WNV consists of a single-stranded, positive sense mRNA-like RNA molecule of ∼11,000 nucleotides which serves as template for a complementary negative sense RNA. Translation of the positive sense viral RNA produces a single polyprotein that is cleaved during and after translation into 3 structural proteins (C, prM/M, and E) and seven non-structural proteins (NS1, NS2A, NS2B, NS3, NS4A, NS4B, and NS5) [1]. The structural proteins are part of immature and mature virions while the C protein is the sole protein component of the nucleocapsid [1], [2]. Non-structural proteins perform many vital functions of the virus lifecycle including replication (NS1, NS2A) [3]–[7], protein processing (NS3, NS2B) [8]–[10] and virus assembly [11], [12]. Additionally, NS proteins are shown to be involved in modulation of the host cell antiviral responses including inhibition of interferon a/b (IFNα/β) induction (NS2A) [13], IFNα/β/signalling [14]–[18], TLR-3 signal transduction (NS1) [19], and complement activation (NS1) [20].
A feature unique to Flaviviruses in the Japanese encephalitis subgroup is the production of an 11th viral protein; the non-structural protein NS1′. The NS1′ was detected 25 years ago in JEV infected cells [21] but the mechanism of its synthesis was only recently discovered. Firstly, the occurrence of programmed ribosomal frameshift (PRF) in the 5′ terminus of the NS2A gene was established by computational modelling of viral RNA structures by Firth and Atkins [22]. Later, the NS1′ protein synthesis, its amino acid sequence, and RNA sequence requirements for PRF were experimentally demonstrated in mosquito cells, mammalian cells, and cell-free settings [22], [23]. PRF occurred in ∼50% of translational events and led to the production of NS1′ protein containing the entire NS1 sequence, the first 9 aa of NS2A protein, and 43 aa unique to NS1′ (Figure 1A). Translation of NS1′ protein culminated with a stop codon which impeded any further translation in the −1 open reading frame (Figure 1A).
Fig. 1. WT and A30A′ replicons show similar rates of replication in BHK cells electroporated with KUNRep-WT or KUNRep-A30A transcribed RNAs. 
(A) Schematic depiction of the translational path of non-PRF and PRF Flavivirus polyprotein. Numbers represent amino acid positions from WNVKUN genome (Accession number AY274504). The underlined sequence represents the start of the NS2A coding region and the box indicates the amino acids encoded by the frameshifted region in NS2A gene. Arrow indicates cleavage between NS1 and NS2A proteins. (B) Schematic representation of plasmid DNA encoding WNVKUN replicon KUN-rep. SP6- SP6 RNA polymerase promoter, C20aa – WNVKUN coding sequence encompassing fist 20aa of C gene, PAC- puromycin resistance gene F2A – foot and mouth virus 2A autoprotease, LacZ – β-galactosidase gene, E22 – WNVKUN coding sequence representing last 22 aa of E gene, NS1-NS5 proteins – WNVKUN sequence coding for entire non-structural region, HDVr – hepatitis delta virus ribozyme, pA- polyA signal. (C) WNVKUN nucleotide and amino acid sequence in the frameshift region encoded in the WT and A30A′ mutant replicons. The underlined sequence indicates the slippery heptanucleotide frameshift motif and in bold are the two silent nucleotide changes disrupting downstream pseudoknot interactions, introduced in KUNRep-A30A′ replicon. Numbers represent nucleotide positions in WNVKUN genomic RNA. (D) In vitro transcribed KUNRep-WT and KUNRep-A30A′ RNAs (left panel) was electroporated into BHK cells and expression levels were measured by β-gal staining (48, 72, 96 hpe) (middle panel) and by Northern hybridization (72 hpe) with WNV 3′UTR-specific, 32P-labelled DNA probe (right panel). Although the specific function(s) of NS1′ have not been determined several studies had investigated potential roles for NS1′ protein [23]–[27]. We reported that WNVKUN mutants (e.g. A30A′) carrying silent mutations abolishing PRF (and NS1′) without affecting viral accumulation showed attenuated virulence in a mouse model of encephalitis [23]. Interestingly, the JEV strain SA14-14-2 recommended for use as live attenuated vaccine by the World Health Organization contains the single mutation G66A in the NS2A gene which ablated production of NS1′ and reduced neurovirulence and neuroinvasiveness in mice [28]. G66A and G67A mutants in NS2A of another strain of JEV, JaOArS982c, abolishing NS1′ production, have also shown reduced replication in avian DF-1 cells and in embryonated chicken eggs [25]. Interestingly, NS1′ protein produced by the wt JaOArS982 virus was shown to co-localize with NS5 protein [25], indicating a role for NS1′ in RNA replication. Furthermore, Winkelmann et al. demonstrated that mutations eliminating PRF/NS1′ enhanced trans-encapsidation of WNV replicons suggesting a role for NS1′ in virion assembly [26]. Our group previously demonstrated that NS1′ co-localized with NS1 and dsRNA and rescued replication in trans of a replication-deficient WNVKUN genome containing large deletion in NS1 gene [27]. More recently studies in other laboratories also showed successful complementation of NS1-deleted WNV genome by the WNV NS1′ protein expressed from an alphavirus replicon vector [29]. These data from different laboratories strongly suggest that NS1′, like NS1, may be one of the components of viral RNA replication complex and therefore plays a role in viral RNA replication.
Despite these insights a clear function for NS1′ remains to be elucidated and some of the results seem to be contradictory. For example, our report that the NS1′-deficient silent mutant A30A′ accumulated to the same levels as WTKUN in C6/36 and mammalian cells seems to indicate that NS1′ is not required for efficient replication and/or virion formation in vitro. Additionally, titers of WNVKUN or A30A′ in the serum of infected mice (weanling and adult IRF3−/−/7−/−) and levels of CPE induced in IFNAR−/− MEFs were similar [23] and unpublished data) suggesting that NS1′ protein may not have an essential function in the viral lifecycle in these models of infection [23], [30].
Moreover, an area of investigation that has been seemingly neglected is the possibility that the frameshift itself has a function in addition to or independent from potential function(s) of the NS1′ protein itself. Since the viral polyprotein is translated as a single molecule (Figure 1A top panel) in the absence of PRF all proteins should be produced in equimolar amounts. It is logical to assume that in the presence of PRF (during WT infection) proteins located upstream of the PRF will be produced in excess to those proteins located downstream of the PRF site (Figure 1A). Since expression of all NS proteins (with the exception of NS1′) is affected by PRF, the viral polyprotein expression event when PRF occurs is essentially separated into structural and non-structural proteins (Figure 1). Conversely, if our hypothesis is correct, in infections with PRF-deficient mutants, such as A30A′ and A30P mutant viruses, all viral proteins would be produced in equimolar amounts and the ratios of structural to non-structural proteins would differ from those observed during WT/PRF-competent infections.
In this context, we decided to perform a wider characterization of the effect of infections with PRF-competent (WT) and PRF-deficient (A30A′ and A30P) viruses. We investigated potential functions of NS1′ protein/PRF such as replication and virion formation during infection of mammalian, avian, and mosquito cell lines. Our studies also included analysis of single nucleotide polymorphisms (SNPs) during infection since SNPs formation may be a manifestation of aberrant replication. We also performed a microarray analysis to determine whether the presence/absence of NS1′/PRF induces significant changes in the transcriptome profile of the infected cells. Importantly, we investigated for the first time the role of NS1′ (PRF) in virus infection, replication, and transmission following infection of Culex mosquitoes, the primary vectors of WNV [31] and virus replication following infection of house sparrows, an experimental model of WNV infection in avian hosts [32], [33].
Results
Deficiency in PRF/NS1′ does not affect WNVKUN replicon RNA replication
Previously we did not detect significant differences in accumulation of WT and A30A′ WNVKUN viruses in the supernatant of infected Vero 76 or C6/36 mosquito cells after performing multiple cycle growth curves (MSGC) [23]. However, our recent findings demonstrated that NS1′ and NS1 co-localise within infected cells and more importantly that NS1′ protein is able to complement replication of replication-deficient NS1-deletion mutants in trans [27]. These findings suggest a potential role in replication for the NS1′ protein and therefore we decided to employ replicons which allow focusing on RNA replication and exclude processes of virion formation and/or secretion.
To this end we used 2 WNVKUN replicons, SP6KUNrep3βgal (PRF-competent, wt) and SP6KUNrep3βgal/NS2A-A30A′(PRF-deficient). The latter was constructed by replacing the BsrGI 7905–9192 fragment of SP6KUNrep3βgal with the BsrGI 6327–7614 fragment from FLSDX/NS2A-A30A′. This fragment contains two silent nucleotide mutations in the NS2A gene (genome positions 3615 and 3618 C to T and G to A, respectively) resulting in retention of Alanine at position 30 and Lysine at position 31 of the NS2A protein). These mutations disrupt formation of pseudoknot structures required for PRF but do not change the amino acid sequence of the NS2A protein (Figure 1 B and C) [23]. These WNVKUN replicons were named Rep-WT and Rep-A30A′, respectively.
BHK cells were electroporated with similar amounts of in vitro transcribed RNAs (Figure 1D left panel) from Rep-WT or Rep-A30A′ replicons and X-gal staining to detect β-gal expressing cells (as means of detecting replicon RNA replication and accumulation) was performed at 48, 72, and 96 hours post-electroporation. X-gal staining of electroporated cells showed similar numbers of β-gal expressing cells at all time-points tested for both replicons (Figure 1D central panel) indicating similar levels of replication for Rep-WT and Rep-A30A′ constructs. These results were confirmed by Northern blot of total RNAs from electroporated cells using a radiolabelled 3′-UTR probe (Figure 1D right panel). Overall, these findings indicate that absence/presence of NS1′/PRF does not have a significant effect on the replication and accumulation of replicon RNAs confirming previous results with full length infectious clones [23].
Deficiency in PRF/NS1′ does not affect WNVKUN replication and/or spread in different cell lines
A recent publication by Winkelmann et al. suggested a role for NS1 and NS1′ in virion assembly [26]. Using serially passaged WNV/Dengue or WNV/JEV chimeric replicons the authors concluded that a mutation in the frameshift motif in NS2A was selected to eliminate PRF; genomes that were deficient in PRF and failed to make NS1′ (similar to A30A′ and A30P mutants in this work) were more efficiently packaged than WT genomes that were competent in PRF and producing NS1′ [26]. Considering that we did not observe differences in viral accumulation in the supernatant of infected cells or β-gal activity in cells transfected with PRF-deficient replicons, we decided to investigate whether the viral spread was affected by PRF-eliminating mutations by using IFA of cells infected at low MOI.
Vero 76 and BHK cells were infected at MOI of 0.01 with WT or A30A′ WNVKUN viruses and progress of infection was followed by IFA with E-protein specific antibodies. We used a low MOI to ensure a small number of infected cells at early time points and measured the spread of the virus over time. At 24 hpi only a few cells were infected with either virus but by 36–48 hpi most cells were infected (Figure 2 A and B). Measurement of the percentage of cells positive for E-protein at different time points indicated that both viruses spread at a similar rate in Vero 76 and BHK cells (Figure 2 C, D). Additionally, we did not observed any variation in the fluorescent signal intensity (Fig. 2A and B) indicating that both viruses accumulated to similar levels within the infected cells. These findings indicate that a role for PRF in virion formation, or secretion, or spread during viral infections seems unlikely.
Fig. 2. WNVKUN WT and WNVKUN A30A′ viruses spread to neighbouring cells at similar rates. 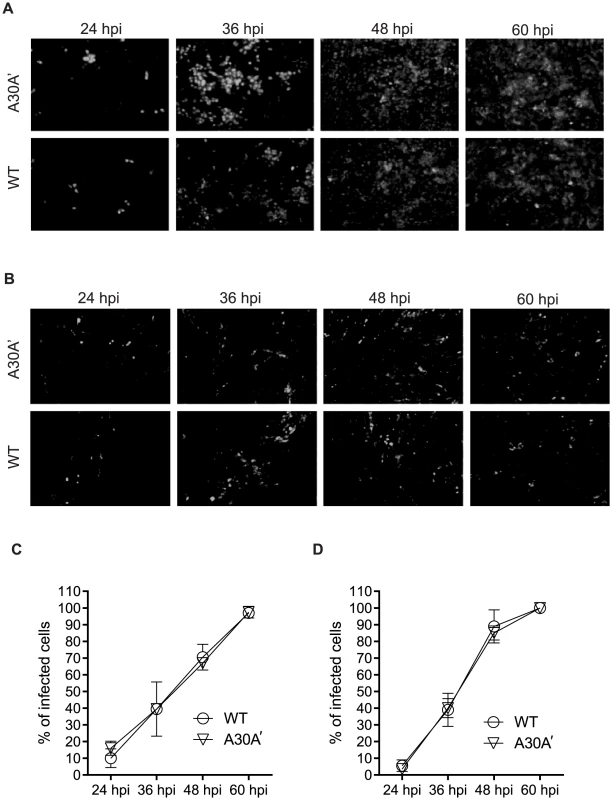
Vero (A,C) and BHK cells (B,D) were infected at MOI 0.01 with WT or A30A′ viruses. The kinetics of infection was followed by IFA with anti-E monoclonal antibodies [56] at 24, 36, 48, and 60 hpi. The percentage of infected cells was determined automatically by Incell-1000 Workstation software using the equation [(number of cells @480 nm)/(number of cells @ 360 nm)]KUNV×100−[(number of cells @480 nm)/(number of cells @ 360 nm)]mock×100. Shown are results from two independent experiments. We also analysed replication and cytopathicity (CPE) of WT and A30A′ WNVKUN viruses in another mammalian cell line LLC-MK2, derived from African green monkey kidney, as well in immortalized chicken embryo fibroblasts DF-1 to determine whether PRF/NS1′ have any effect on virus replication these experimental systems. In addition to A30A′ mutant we also included A30P virus because this mutation also eliminates PRF and production of NS1′; in addition it also changes amino acid 30 in NS2A protein from Ala to Pro. This mutant has been extensively characterized by us previously and has been shown to induce reduced CPE in various mammalian cells including mouse embryonic fibroblasts and reduced virulence in mice [13], [30], [34].
A30P WNVKUN virus showed delayed replication in DF-1 cells compared to WT and A30A′ WNVKUN viruses (Fig. 3A) and significantly reduced CPE even at lower viral loads (MOI = 1 in Figure 3B left pane; MOI = 0.1 data not shown) indicating that the attenuated phenotype of the A30P virus is a characteristic feature of this mutation in the NS2A gene. WT and A30A′ infections however, were indistinguishable and both viruses caused significant and similar severity of CPE upon infection of DF-1 cells (Figure 3A–B). This is in contrast to significant differences between wt and PRF-deficient virus replication in DF-1 cells reported for JEV [25]. It is not unreasonable to assume that PRF deficiency may affect replication of JEV and WNV differently.
Fig. 3. PRF does not impact viral accumulation in avian and mammalian cell lines. 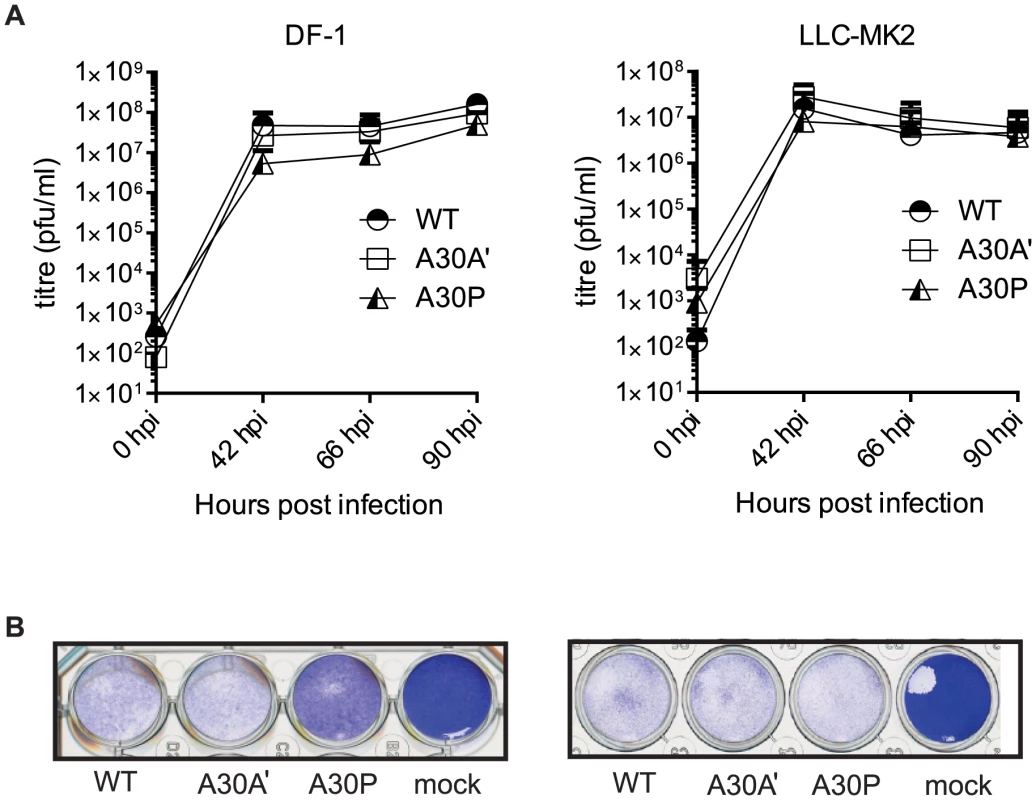
DF-1 and LLC-MK2 cell lines were infected with WNVKUN WT, WNVKUN A30A′, and WNVKUN A30P viruses at MOI of 1 and (A) Viral titres measured by plaque assay at 0, 42, 66, and 90 hpi. Error bars represent standard deviation. (B) Cytopathic effect illustrated by crystal violet staining at 5 dpi of DF-1 and LLC-MK2 cells infected with WT, A30A′, or A30P viruses at MOI of 1. Shown is a representative staining of two independent experiments. In contrast to DF-1 cells, A30P WNVKUN virus showed significant CPE in LLC-MK2 cells and was indistinguishable from WT and A30A′ WNVKUN viruses in growth kinetics and CPE (Fig. 3A–B right panels). Again, WT and A30A′ viruses were indistinguishable in growth properties and in CPE also in LLC-MK2 cells demonstrating no apparent role for PRF in virus replication and virus-induced CPE in vitro.
Global gene expression in MEFs infected with WT, A30A, and A30P WNVKUN viruses
Next we decided to investigate global expression pattern of primary mouse embryonic fibroblasts (MEFs) infected with WT and PRF-deficient WNVKUN mutants to detect potential variations in the host gene expression that may be related to PRF or mutated NS2A protein. We selected WT and IRF3−/−/7−/− knockout MEFs because of our previous studies indicating that WNVKUN mutants A30A′ and A30P were attenuated in WT weanling mice while WNVKUN mutant A30P was also attenuated in IRF3−/−/7−/− adult animals. This raised the possibility that interferon-independent pathways may be involved in this attenuation, at least for A30P virus [13], [23], [30], [34].
After infection of WT and IRF3−/−/7−/− MEFs with WT, A30A′, or A30P WNVKUN viruses at MOI 1 total RNA was extracted 48 hpi and global gene expression was measured by microarrays. The titers of viruses accumulated in the supernatant of infected cells at this time point indicated that A30P and A30A′ viruses reached lower titers than WT virus during infection of WT MEFs but titers of all viruses in IRF3−/−/7−/− MEFs were similar (Figure 4A). As expected the expression of several hundred of genes was affected by viral infection of WT MEFs with 368 genes showing upregulation and 169 genes showing down regulation when compared with non-infected cells (Figure 4B and Table 1). Similarly, after infection of IRF3−/−/7−/− MEFs with WT virus, 1639 and 1665 transcripts were upregulated and down regulated, respectively (Table 1). These results indicate that WNVKUN infection induces profound changes in the transcriptome of murine cells and these changes are reflected across numerous pathways.
Fig. 4. Infection of WT and IRF3−/−7−/− MEFs with PRF-deficient WNVKUN viruses induces differential expression of several genes. 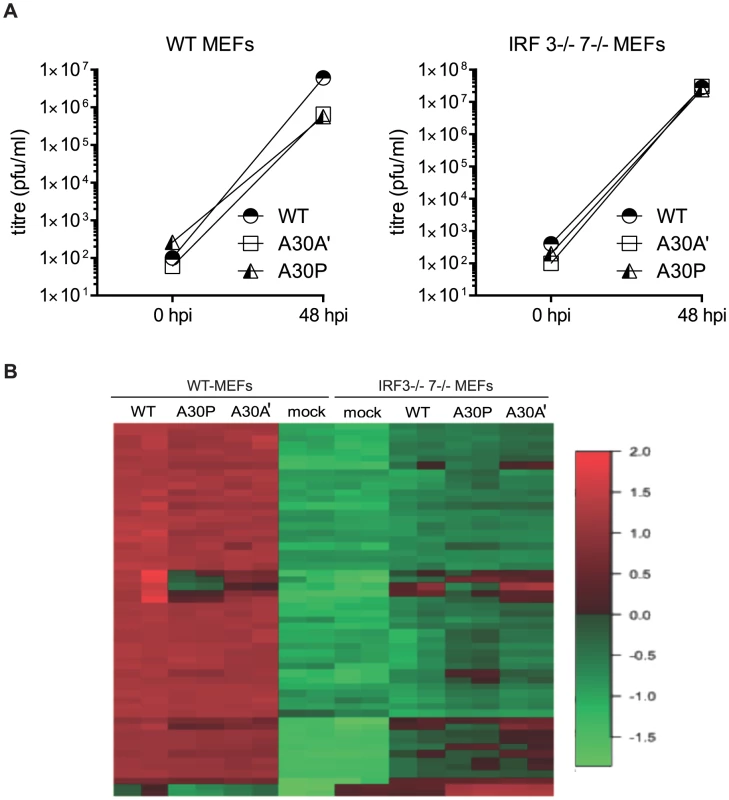
Low passage primary WT and IRF3−/−7−/− MEFs were infected at MOI of 1 with WNVKUN WT, WNVKUN A30A′, or WNVKUN A30P viruses and 48 hpi total RNA was extracted and used for analysis of global gene expression by microarrays. (A) Virus titres in the infected cell culture supernatants were assayed by plaque assay at 0 and 48 hpi. (B) Heat map showing top differentially expressed genes ranked by log fold change between different experimental conditions. Tab. 1. Summary of microarray analysis of global gene expression in MEFs infected with WT and mutant WNV<sub>KUN</sub> viruses. 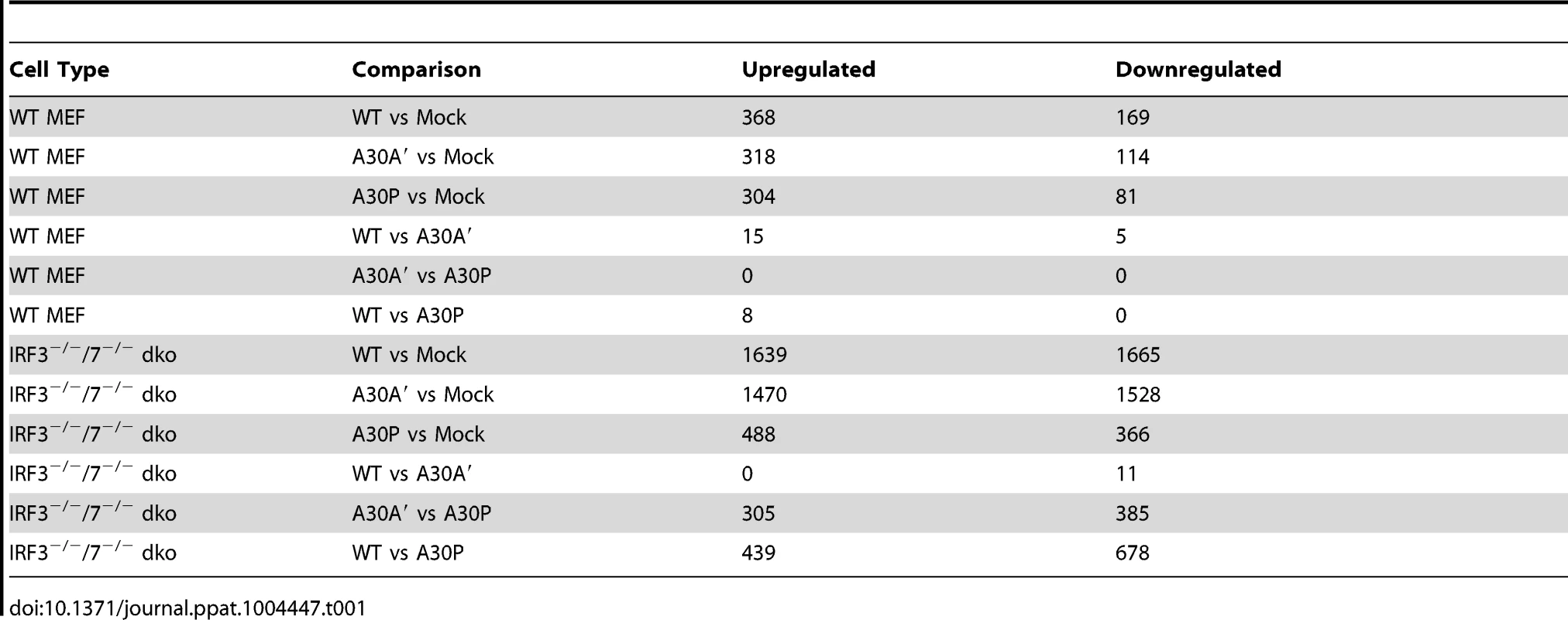
Infection with WT or A30A′ WNVKUN viruses in WT or IRF3−/−/7−/− MEFs produced similar transcriptional profiles and showed that only a small number of genes were differentially affected between these two infections (Tables 1, 2 and 3). For example, in comparison of WT vs A30A′ infection, 15 transcripts were upregulated and 5 transcripts downregulated in infected WT MEFs (Table 1) and the differences detected were relatively small (1.4–2 fold, Table 2) despite ∼10-fold difference in the viral titres (Fig. 3A). Infection of IRF3−/−/7−/− MEFs also resulted in little difference in transcript regulation between WT and A30A′ viruses (Table 1) and again the differences although significant, were small (Table 3). The data indicate that frameshift per se doesn't appear to have a significant effect on host response to WNVKUN infection in MEFs.
Tab. 2. Selected genes differentially up-regulated in WT MEFs infected with WT and A30A′ mutant WNV<sub>KUN</sub> viruses. 
Tab. 3. Selected genes differentially up-regulated in IRF3<sup>−/−</sup>/7<sup>−/−</sup> MEFs infected with WT and mutant WNV<sub>KUN</sub> viruses. 
In contrast, differentially expressed genes were abundant when comparisons included transcriptional changes induced by A30P WNVKUN virus infections but only on the background of IRF3−/−/7−/− MEFs (Table 3). The A30P mutation played a significant role in the differential regulation of hundreds of genes compared to the WT or A30A′ virus infections in IRF3−/−/7−/− MEFs (Tables 1 and 3). Surprisingly, little or no difference in the host gene transcription levels was detected between WT and A30P infections or between A30A′ and A30P infections in WT MEFs (Table 1). All 3 viruses, however, did induce upregulation and downregulation of a large number of host transcripts in both cell types when compared to Mock cells (Table 1) suggesting that MEFs mount significant antiviral response upon infection with these viruses.
Single nucleotide polymorphisms in WT and A30A′ WNVKUN genomes
To investigate whether the presence or lack of PRF may affect selection of predominant virus population during infection, we analysed WT and A30A′ viral genome pools for the presence of single nucleotide variants by sequencing using the Ion torrent sequencing platform (see Material and Methods). After quality control of the raw sequence data using fastq, trimmomatic and QFAB proprietary methods, high quality reads were mapped to the reference genomes of WT and A30A′ WNVKUN viruses. Table 4 shows a list of variants detected within the WT (15/52) and A30A′ (15/51) sequence collections and represent rare variants (1% or more) where reads were observed on both sequence strands and were observed in a least 25 independent sequence reads.
Tab. 4. Ion Torrent sequencing analysis of variants in WT and A30A′ mutant WNV<sub>KUN</sub> virus populations. 
We did not find significant differences between WT and A30A′ WNVKUN genomes related to the presence or abundance of variants (Table 4) with the exception of the two nucleotide mutations introduced in the A30A′ genome by mutagenesis which obliterate PRF formation [23] (positions 3615 and 3618) (Table 5). However, we did find that ∼11% of the WT reads across the pseudoknot region contained a variant at the critical 3615 position, similar to A30A′ mutant (underlined in Table 5), suggesting that WT genome pool does contain mutated genomes unable to elicit PRF events.
Tab. 5. Variations at nucleotide positions 3615 and 3618 in WT and A30A′ WNV<sub>KUN</sub> virus populations. 
PRF alters ratio of structural to non-structural proteins
Studies on PRF in the NS2A gene of flaviviruses have so far focused on the presence/absence of NS1′ and its possible functions on replication, virion formation/secretion, and neuroinvasiveness [23]–[26], [28], [35]. An unexplored consequence of PRF in the NS2A gene is its potential impact on altering the expression of viral proteins. Because Flavivirus proteins are produced from a large single polyprotein, all viral proteins in the absence of PRF should be produced in equimolar amounts. We assumed that occurrence of PRF at the start of the NS2A gene should change this 1∶1 ratio and should produce a surplus of structural proteins. That is, proteins located in the viral genome before the PRF site will be produced from each translatable molecule while proteins located after the PRF will be only produced in molecules not undergoing PRF (Figure 1A). In fact, PRF affects also NS1 protein synthesis (located upstream of PRF) since molecules producing NS1′ do not produce NS1 due to the deficiency in NS1/NS2A cleavage [27]. As a consequence, the occurrence of PRF should affect translation of all non-structural proteins and cause a de-facto change in the ratio of structural to non-structural proteins during translation of the Flaviviral genome (Figure 1A).
To investigate the hypothesis that PRF regulates the ratio of structural to non-structural proteins, a replicon system where the structural genes were replaced by β galactosidase (β-gal) gene was employed. Replacing secretable structural proteins with cytoplasmic β-gal eliminates likely underestimation of structural protein levels when calculating the protein expression ratio due to the secretable nature of structural proteins. Semi-quantitative Western blot was performed to measure the accumulation of β-gal protein and NS proteins (NS3 and NS5) in cells electroporated with WNVKUN Rep-WT and Rep-A30A′ RNAs. If the working hypothesis is correct, the accumulation of NS3 or NS5 proteins relative to β-gal accumulation should be increased in Rep-A30A′ transfected cells, leading to decreased β-gal/NS3 and β-gal/NS5 ratios relative to Rep-WT transfected cells.
Electroporated cells were lysed with RIPA buffer at 48, 72 and 96 hours post electroporation. Total protein lysates were separated by SDS-PAGE and Western blot analysis was performed with anti-β-gal, anti-NS3, and anti-NS5 antibodies (Figure 5 A, B). The integrated intensities of each targeted protein was determined and ratios of β-gal/NS proteins were calculated for Rep-A30A′ and Rep-WT lysates. For the clarity of presentation, ratio of β-gal/NS proteins in Rep-WT were set to 1.0 so that the β-gal/NS proteins ratios for Rep-A30A′ could be expressed relative to those determined for Rep-WT. Results in Figure 5 showed that the NS2A-A30A′ mutation present in Rep-A30A′ indeed decreased both, β-gal/NS3 and β-gal/NS5 ratio by 20%∼40% relatively to Rep-WT and this trend was observed at all time-points measured. Interestingly, these results are in line with the frequency of PRF events reported for WT WNVKUN virus [23].
Fig. 5. PRF disrupts equimolarity of viral protein synthesis in replicon-transfected and virus-infected cells. 
(A) KUNRep-WT and KUNRep-A30A′ replicons were electroporated into BHK cells and 48, 72, 96 hpe lysates were harvested and total proteins were separated by SDS-PAGE and detected by Western blot with anti-β-gal and anti-NS3 or (B) anti-β-gal and anti-NS5 antibodies. Ratios of β-gal/NS proteins were calculated and plotted as a function of KUNRep-WT ratios. (C) BHK cells were infected with WNVKUN WT or WNVKUN A30A′ virus and at 48 and 72 hpi proteins were pulse-labelled with 35S methionine/cysteine with and without brefedrin A (BFA) treatment, harvested with RIPA buffer and immuno-precipitated with anti-E and anti-NS5 antibodies. Precipitated proteins were separated by SDS-PAGE and visualised by exposure to X-ray film. Ratios of E/NS5 were calculated as above and represent 2 independent experiments. Error bars represent standard error of the mean of 2 independent experiments. We also determined the effect of PRF on viral proteins expression in WNVKUN-infected cells. However, during viral infection structural proteins assemble into viral particles that are secreted into media during viral secretion. This is likely to reduce the amount of intracellular structural proteins available in cell lysates and potentially skew intracellular structural/NS proteins ratio. To inhibit virion secretion, cells were treated with metabolic inhibitor brefeldin A (BFA) [36]. The addition of BFA late in the infection drastically reduces the amounts of secreted structural proteins (virions) without affecting viral protein synthesis [36] and hence it should reduce the effect of secretion on ratio of structural to non-structural proteins.
BHK-21 cells were infected with WNVKUN WT and A30A′ viruses and at 48 and 72 hours post infection metabolic labelling with 35S methionine-cysteine in the presence or absence of BFA was performed. After 2 hours of labelling and 4 hours of chase the cell monolayers were harvested and lysed in RIPA buffer. Radiolabelled lysates were then immuno-precipitated with anti-E and anti-NS5 monoclonal antibodies and precipitated proteins were separated by SDS-PAGE and visualized after exposure to a phosphor-imaging screen. Again, quantification of the proteins of interest and calculation of E/NS5 ratios indicated that the A30A′ mutation was associated with a decrease in the E/NS5 ratio as compared to WT WNVKUN infection (Figure 5C). This reduction was more evident in cells treated with BFA, indicating that virus secretion depleted viral structural proteins within the infected cell, as expected. These results indicate that the presence of PRF in the NS2A gene of WNV alters the equimolar ratio of viral structural and NS proteins, in the replicon or virus infection settings, demonstrating a role for PRF in translational regulation of viral protein synthesis.
PRF-deficient WNVNY99 virus is slightly attenuated in birds
Birds serve as natural reservoir vertebrate hosts for WNV. Specifically, house sparrows (HOSPs) have been used previously as a natural avian host in order to assess genetic determinants of host competence as representative WNV avian model hosts [33], [37]–[39]. We inoculated groups of ten HOSPs with 1500 PFU of WT WNVNY99 and A30A′ mutant WNVNY99 viruses and collected serum daily for 7 days after infection to analyze virus titers by plaque assay on BHK cells. WT WNVNY99 virus reached peak average titers of 4.4 log10 PFU at day 3, while A30A′ mutant WNVNY99 virus reached an average titer of 3.7 log10 PFU at day 2 after infection (Figure 6). WT WNVNY99 titers remained higher than A30A′ WNVNY99 virus titers until day 4 after infection and both viruses were then essentially cleared by 7 dpi (Figure 6). It should be noted that significant variations in susceptibility to WNV infection are common between individual wild-caught HOSPs [40]. Although the differences in virus titres between WT and A30A′ WNVNY99 infections did not reach statistical significance by two way ANOVA comparison, the A30A′ WNVNY99 virus nevertheless showed noticeable attenuation at the peak time of infection (Figure 6), suggesting a role for PRF/NS1′ in WNVNY99 virulence in avian hosts.
Fig. 6. Viremia profiles of HOSPs inoculated with WNVNY99 WT (NY99 WT) or WNVNY99 A30A′ (NY99 A30A′) viruses. 
Groups of ten HOSPs were infected with 1500PFU of each virus and bled daily for 7 days after infection. Virus titres in serum were determined by plaque assay on BHK cells and plotted using GraphPad PRISM version 6.0a software. Error bars represent standard errors of the mean. PRF-deficient WNVKUN viruses show reduced replication and spread in Culex annulirostris mosquitoes
To examine the role of PRF/NS1′ on replication and transmission in mosquitoes, Cx. annulirostris were exposed per os to infectious blood meals containing approximately 107 TCID50/mL of each of the WT, A30A′ and A30P WNVKUN viruses in two separate experiments. The ability for the virus to infect and disseminate from the midgut was assessed in both experiments, while the ability for mosquitoes to transmit the viruses was also assessed in experiment 2. In both experiments, the body infection rates were higher in the mosquitoes exposed to WT WNVKUN viruses than either of the mutant WNVKUN viruses, with the difference being significant (P<0.001) in experiment 2 (Figure 7A). Similarly, the virus disseminated at lower rates in the mosquitoes exposed to the A30A′ and A30P viruses than the WT virus, with the difference not being significant (P = 0.3915) for A30A′ in experiment 1 only. Infection and dissemination rates were lower in mosquitoes exposed to A30P than those exposed to A30A′, although the difference was not significant. Finally, both the A30A′ and A30P viruses were transmitted at significantly lower rates (P<0.005) than the WT virus. The lack of PRF/NS1′ affected the ability for the virus to replicate in the mosquito body and the quantity of virus expectorated, with the titers in the bodies and saliva expectorates being significantly lower (P<0.0001) in mosquitoes infected with A30A′ and A30P, than those in the WT viruses (Figure 7B).
Fig. 7. PRF/NS1′-deficient WNVKUN mutants show reduced replication and dissemination in Culex annulirostris mosquitoes. 
(A) Infection (bodies), dissemination (legs and wings) and transmission (saliva) rates in mosquitoes exposed to either WT virus or mutant viruses by feeding on virus infected blood meals containing approximately 107TCID50/mL of virus or intrathoracic inoculation with 220 nL of a 106 TCID50/mL virus stock. A two-tailed Fisher's exact test was used to determine significant difference between infection, dissemination and transmission rates between each of the mutant viruses and the WT virus (*P<0.05) and between the mutant viruses (†P<0.05). (B) and (C) Mean ± 95% CI (confidence interval) virus titers in bodies and saliva expectorates from mosquitoes exposed to the mutant or WT-viruses by oral feeding (B) or intrathoracic inoculation (C). Twelve days post exposure, mosquitoes were processed by removing the legs and wings, and collecting the saliva using a forced salivation technique. Homogenized and filtered mosquito components (bodies, legs and wings, and saliva expectorates) were inoculated onto confluent monolayers of C6/36 cells in a 96 well microtiter plate, and infection detected by a cell culture enzyme immunoassay (CC-EIA). Viral titers of the bodies and saliva expectorates was determined by inoculation of ten-fold dilutions of the filtered homogenates in C6/36 cells with virus detected using the CC-EIA. A one-way ANOVA was employed to determine significant differences in mean viral titers within the mosquito bodies and saliva expectorates (****P<0.0001; ***P<0.001; **P<0.005; *P<0.05; only statistically significant differences are shown). We also examined the ability for the virus to replicate in and be transmitted by Cx. annulirostris after IT inoculation, which is a mode of infection that bypasses the midgut of the mosquito. Bypassing the midgut produced higher rates of infection with the mutant viruses, with almost all mosquitoes becoming infected, irrespective of the virus inoculated (Figure 7A). Despite the high rates of infection for the 3 viruses, the titers of virus within the bodies were significantly lower in the mutant viruses than the WT virus (Figure 7C). This potentially led to the lower transmission rate observed in the mutant viruses when compared to the WT virus, with the rate being significantly lower in mosquitoes inoculated with A30P (Figure 7A).
Discussion
A novel translational mechanism for the synthesis of the NS1′ protein by encephalitic flaviviruses from the Japanese encephalitis serogroup was recently predicted by computer modelling [22] and then demonstrated experimentally for WNVKUN [23]. The uniqueness of NS1′ expression by encephalitic flaviviruses had sparked significant interest in NS1′ functions related to viral virulence. Indeed, recent reports had focused on the potential functions of the NS1′ protein in mammalian cell systems and mouse models of disease and identified several potential functions for NS1′ protein including RNA replication, virion formation, and neurovirulence [23], [24], [26], [28], [30].
Data presented in this manuscript extended our previous knowledge of PRF/NS1′ and its role(s) during infection of mammalian, avian, and mosquito cells. WT and A30A′ replicons and viruses accumulated at similar levels in different cells suggesting that the presence or absence of PRF/NS1′ does not affect the rates of viral RNA synthesis or virion accumulation and/or spread from cell to cell. This is in contradiction to the findings of Winkelmann et al. who found that abolishment of PRF and NS1′ production impacted the ability of the mutated replicon to be encapsidated into virus-like particles [26]. However, it is possible that the effect observed in those experiments was due to the use of a heterologous trans-encapsidation model where WNV replicon RNAs were packaged by dengue virus structural proteins since when WNV structural proteins were used for packaging WNV replicons, the difference in packaging efficiency between PRF-competent and PRF-deficient replicon RNAs was greatly reduced [26].
Our transcriptome analysis of WT and IRF3−/−/7−/− MEFs upon infection with WT and PRF-deficient viruses (A30A′ and A30P) provided a detailed overview of host gene expression changes after infection and a guide for future investigations. Changes in the global gene expression induced by infections with WT or A30A′ viruses were similar with only a small number of genes differentially expressed and also with relatively small differences in their levels of expression. These differentially expressed genes included transcription factors, protease inhibitors, and ER stress response elements. Importantly however, all of these differences despite being relatively small were highly significant indicating that even relatively small changes in gene expression may have biological significance. Further studies should focus on determining the role for each of these differentially expressed host genes in WNV-host interactions in vivo.
In contrast to small differences observed between WT and A30A′ viruses, significantly higher number of differentially expressed transcripts and higher fold changes for them were found between WT and A30P or A30A′ and A30P infections, suggesting that the Ala to Pro substitution in the NS2A protein has a more dramatic effect than PRF alone on the host response to infection. These differentially expressed genes in response to A30P infection include transcription factors, anti-apoptotic signals, and thermo-tolerance factors. Further studies are required to determine the role for the host genes shown the most differences in induction/repression between WT and A30P or A30A′ and A30P viruses in WNV-host interactions both in vitro and in vivo.
Variant analysis of virus populations by Ion Torrent sequencing detected a significant population of genomes in the WT swarm that contained PRF-abolishing mutation at position 3615 not present in the WT genomic sequence (proportion column in Table 5, 0.889 = ∼11%). On the other hand, the A30A′ swarm did not contain mutations to revert to WT sequence at position 3615 (proportion column, 0.999 = 0.1%) indicating that A30A′ had lost the need and/or ability to revert to WT through “error-prone” replication events. The significance of the 3615 position, and the PRF events, was underscored by our observations in position 3618. In our initial study [23], we introduced the extra G to A change at position 3618 of the A30A′ mutant with the purpose of minimizing a chance of reverse mutation(s) back to the WT sequences; this position per se does not play a role in pseudoknot formation and PRF. In contrast to the ∼11% of WT population reads containing a mutation at position 3615 less than 1% of reads showed mutations at position 3618. This suggest the intriguing possibility that during WT infection a significant proportion of the genomes produced (∼11%) are PRF-deficient, a de-facto compensation for at least a proportion of the −1 programmed ribosomal frameshifting events occurring during WT infection [23]. Further studies will be required to elucidate the true meaning of the existence of this virus sub-population, however it is likely that regulation of PRF frequency is employed by the virus to maintain the right balance between higher and lower replication efficiencies as the result of more or less efficient PRF event.
We demonstrated that PRF altered the levels of structural and non-structural proteins synthesis creating a de-facto over-expression of structural proteins over non-structural proteins; to our knowledge, this is the first time that the effect of the PRF on the expression of flavivirus proteins has been demonstrated. We hypothesize that such “excess” of structural proteins may lead to an increase in prM-E particles and/or act as an elegant modulator of viral protein expression, which may indirectly regulate host responses to viral infections. Pseudoknot formation and −1 ribosomal frameshifting in coding regions of viral genomes have been described previously [41], [42]. In retroviruses, precise ratios of frameshifting allows expression of the viral Gag-Pol polyprotein and sets an optimal Gag∶Gag-Pol ratio for virion assembly and packaging of reverse transcriptase [43]. Similarly, in SARS-CoV pp1a and pp1ab, which are expressed by frameshifting, are predicted to form a heterodimer with a stoichiometry of 8/1 as part of the replication machinery [44], [45]. In both examples maintaining precise ratios of frameshifting is crucial for virus accumulation and we would argue that similar mechanisms might be required during infection of mosquitoes and birds with WT and PRF-deficient WNVs. Indeed, we demonstrated that PRF affects virus infectivity and/or spread in mosquito vectors and in the generation of elevated viremias in an avian host species, HOSPs. PRF-deficient mutant (A30A′) showed lower titers in birds and mosquitoes and also reduced ability to be transmitted by mosquitoes as shown by lower percentage of infected saliva for these mutants. A role in replication of PRF/NS1′ in birds and mosquitoes would explain the phenotypes observed despite this and other reports where differences in replication in several cells lines infected with PRF-deficient mutants [23], [30] were not observed. The difference between in vitro and in vivo results is often seen for the bird phenotype [39]. Interestingly, in contrast to our results in avian DF-1 cells, JEV PRF mutant virus replicated significantly less efficiently than wild type JEV virus in DF-1 cells [25], suggesting that difference exists also between different albeit closely related flaviviruses in their ability to replicate in the absence of PRF in vitro.
It is important to note that upon viral infection, vertebrates establish an antiviral state mostly through type I interferon responses [46], [47] while insects respond primarily through RNA interference (RNAi) [48], [49] or the activation of immune pathways, including Toll, Imd and JAK/STAT pathways [50]. Our data suggest the existence of interactions between NS1′ protein (or an effect of PRF) and host response pathway(s)/factors that are efficient (present) in whole mosquitoes but not in cellular models of infection, including mosquito cell lines, some of which are known to be deficient in a host factor, e.g. C6/36 cells are deficient in dicer 2 [51]. In this context, the role of insect gut barriers and its interaction with the flavivirus genome is critical. Increased viral accumulation in the gut has been reported to correlate with the ability of the arboviral infection to spread throughout the mosquito and determine its capacity to transmit infection by bite [52]. Efficient infection of the midgut is likely to trigger a robust innate response by the host including, but not limited to, stimulation of enzymes involved in the oxidative stress response and re-epithelialization of damaged cells of the midgut wall [53], [54]. It is intriguing that the PRF motif is highly conserved among the JEV serocomplex viruses that utilize both Culex mosquito vector and avians as their primary vertebrate replication hosts while other flaviviruses that utilize primate and Aedes vectors such as Dengue virus 1–4 and Yellow fever viruses or rodents and ticks have not demonstrated to encode PRF. The results presented herein show the role of PRF as mechanism for modulation of the stoichiometric ratio of structural to non-structural genetic determinants that could be involved in modulating mosquito RNAi responses or potentially avian innate immune responses.
Materials and Methods
Cells and viruses
BHK, Vero76, LLC-MK2 and DF-1 and MEF cells were propagated and maintained in Dulbecco's Modified Eagle Medium (DMEM, Gibco) containing 5% fetal calf serum (FCS), 50 U/ml penicillin, 50 µg/ml streptomycin, 2 mM glutamax and 10 mM HEPES at 37°C and 5% CO2.
KUN WT, A30A′, and A30P viruses were obtained by in vitro transcription of corresponding DNA plasmids [23] and electroporation of viral RNA transcripts into BHK cells. NY99 WT and A30A′ viruses were generated from in vitro ligated two plasmid system [55]. Passage 0 viruses were harvested 1–3 days post electroporation, titrated by plaque assays, and used at noted multiplicity of infection (MOI). Mutations introduced into mutant viruses and replicons were confirmed by RT-PCR and sequencing from viral and replicon RNAs.
In vitro transcription and electroporation
Linearized DNA template from KUNV replicon plasmids was transcribed using SP6 RNA polymerase (Roche) as per manufacturer instructions and the resulting KUN replicon RNA transcripts were electroporated into BHK cells (Bio-Rad GenePulser II apparatus; 25 µF capacitance, 1.5 kV voltage, infinite resistance, two pulses with 10 second interval, with the optimal time constant between 0.7 and 0.8 ms). Electroporated cells were then resuspended in 10 ml of DMEM containing 5% FCS, seeded into culture plates, and incubated at 37°C under 5% CO2 and humid conditions. 72 h post electroporation (hpe) total cellular RNA was extracted using Trizol reagent (Invitrogen) following manufacturer instructions and separated in a denaturing 1% agarose gel. After transfer of total RNA onto nitrocellulose membrane viral genome-specific RNAs were detected by hybridization with P32-labelled KUNV 3′-UTR-specific probe.
For in-situ X-gal staining replicon-transfected cells in a 24-well plate were washed with PBS, fixed with 500 µl 4% paraformaldehyde for 20 min at room temperature and washed with 500 µl PBS 3 times. B-gal activity was detected by adding 200 µl of X-gal stain solution (50 µl K4Fe(CN)6-3H2O, 50 µl K3Fe(CN)6, 2 µl 1 M MgCl2, 50 µl X-gal stock(20 mg/ml) and 850 µl PBS) to each well, and incubating at 37°C for 1 h.
Immunofluorescence (IFA) analysis
Vero 76 and BHK cells were seeded into 24-well plates and infected at MOI 0.01. Infected cells were fixed and permeabilised (4% paraformaldehyde 20 min, 0.1% Triton X-100 for 10 min) at 24, 36, 48, 60 hours post infection and stained for 1 h at room temperature with mouse anti-E monoclonal antibodies 3.91D [56], followed by 30 min staining with the secondary Alexa Fluor 488 goat anti-mouse antibodies and DAPI (nuclear stain, Sigma-Aldrich), with intercalating 3×5 min washes with PBS between each step. After a final wash with PBS, cells were imaged using IN-cell 1000 analyser (GE Healthcare) at the required excitation and emission wavelengths and the percentage of E-positive cells (% infected) was determined using IN-Cell 1000 Workstation software where: % infected cells = [(number of cells at 480 nm)/(number of cells at 360 nm)]KUNV×100−[(number of cells at 480 nm)/(number of cells at 360 nm)]mock×100.
Growth curves and cytopathicity (CPE) assay
DF-1 and LLC-MK2 cells were seeded into 6-well plates and infected with WT, A30A, and A30P viruses at MOI of 1. Culture supernatant was harvested and viral tiers were determined by plaque assay [55] at 0, 42, 66, and 90 hpi. CPE was analysed by seeding DF-1 or LLC-MK2 cells into a 24-well plate and infecting at MOI 1. At 5 dpi cells were fixed with 4% formaldehyde for 1 h and stained with 0.2% Crystal Violet for 20 min. Images of CPE staining were obtained using Epson perfection V700 photo scanner.
Microarray analysis
Low passage primary WT and IRF3−/−/7−/− MEFs were infected with WT, A30A′, or A30P viruses at MOI of 1. At 48 hpi total RNA was extracted with Trizol and an Illumina DNA microarray was run on 16 samples (8 experimental samples with duplicates). The raw signal intensity was extracted from the encrypted binary.idat files using the IDATreader package (version 0.2.0). The raw data were processed to stabilise the variance [57] and the Quantile normalization methods within the Limma package of Bioconductor were implemented. In addition to the normalization of the experimental data, an averaging of the duplicated array probes was performed so the number of analytical features decreased to the number of uniquely identified probes. The statistical analysis of the differences in gene expression pattern were again performed by methods implemented within the Limma Bioconductor package and differential expression was defined when a gene had a log-fold-change of <−1 or >1. The expectation value for maximum accepted error corrected probability was defined as 0.05 and error correction for the p-values was performed according to methods described by Benjamini and Hochberg [58].
Variant analysis by Ion Torrent sequencing
The quality and characteristics of the Ion torrent sequences from WT and A30A viruses, in FASTQ format, was investigated using FASTQC. The distribution of the base quality scores across the reads were typical for Ion Torrent data and in both samples the median sequence length was approximately 220 nucleotides in length. The Ion Torrent FASTQ format DNA sequence reads were mapped onto the reference genomes (WT and A30A′) using the bwa software and an indexed BAM file was prepared using the Picard software. Review of the BAM alignment using Tablet revealed that the bulk of the reference sequences were covered by between 3000× to 6000× genome coverage and only a very small fraction of the reference sequence was not sampled to at least 50× coverage. A manual review of the sequence alignment was performed and identified single nucleotide deletion events occurring in both genomes but only present in the forward strand were excluded from further analysis; these deletion variants were deemed a consequence of the Ion Torrent sequencing technology.
Protein analysis
Cells were lysed with RIPA buffer and with NuPAGE LDS sample loading buffer (Invitrogen) for 20 min and heated at 95% for 3 min before loading total protein samples (∼10 µl) onto a 15-well precast Express PAGE gel (Genescript). The proteins were separated by electrophoresis at 120 V for 1 h and transferred to PDVF membrane (GE Healthcare) for 1.5 h at 45 V using a wet transfer apparatus (BioRad). Membranes were blocked for 1 h in 5% non-fat milk (BioRad) and then probed for 1 h at room temperature with gentle rocking with primary antibodies diluted in 5% non-fat milk. The membranes were washed 3 times with TBST (at least 5 min each time) and incubated for 1 h with secondary antibodies diluted in PBS. After a final wash with TBST, the blots were developed by LI-COR Odyssey Infrared Imaging System (LI-COR Biosciences) and integrated intensity of each band was used for calculating the ratios. Ratios of structural (or reporter β-gal activity) vs non-structural proteins observed during WT virus infections or WT replicon RNA replication were set to 1 and this was used as the reference for calculating corresponding ratios in PRF-deficient virus or PRF-deficient replicon replication.
Characterization of NY99 WT and A30A′ viruses in house sparrows
House sparrows (HOSPs) were collected under approved animal care and use protocols and field studies did not involve endangered or protected species. All animal studies presented herein were approved by Institutional Animal Care and Use Committees at the Division of Vector-Borne Diseases, Centers for Disease Control and Prevention (approval number 13-009). All protocols and practices for the handling and manipulation of sparrows were in accordance with the guidelines of the American Veterinary Medical Association (AVMA) for humane treatment of laboratory animals as well as the “Guidelines to the Use of Wild Birds in Research” published by the ornithological council 3rd edition (2010).
HOSPs were trapped by seed-baited ground traps in Larimer County, Colorado. Birds were bled and sera assayed for neutralizing antibodies against WNV and SLEVs as previously described [32]. Ten WNV/SLEV seronegative HOSPs were inoculated subcutaneously in the breast region by 28-gauge needle with 1500 PFU of the WT and A30A′ viruses. Viral inocula were diluted in PBS (0.1 mL final volume). Inoculated HOSPs were bled once daily by jugular venipuncture through 7 dpi. 0.1 mL of whole blood was drawn and expelled into a cryovial containing 0.45 mL of BA-1 media. Samples were allowed to clot at room temperature for >20 min and spun at 3500-g for 10 min. The 1∶10 sera sample dilutions were frozen at −80°C until titrated by plaque assay on BHK clone 15 cells for infectious titer determination as previously described [32]. The virus titres were plotted using GraphPad PRISM version 6.0a software and statistical analyses was performed using two-way ANOVA comparison using the same software package.
Characterization of wild-type and mutant KUN viruses in mosquitoes
We exposed Culex annulirostris mosquitoes to WT, A30A, and A30P viruses via infectious blood meal or intrathoracic (IT) inoculation. These mosquitoes were colonized from material collected from the Boondall Wetlands, Brisbane, Australia in 1998 and have been maintained in a colony by the Australian Army Malaria Institute for >100 generations. Previous experiments had demonstrated high infection and transmission rates with a wild type WNVKUN strain in this colony of Cx. annulirostris [59].
For virus exposure by blood feeding, 5–10 days old mosquitoes were allowed to feed on a blood meal consisting of WT or mutant viruses diluted in commercially available defibrinated sheep blood using the pledget method of Wells [60]. We also explored accumulation and spread of the WT and mutant viruses following IT inoculation. IT inoculation bypasses the midgut and associated barriers while also allowing for standard amounts of virus to be delivered directly producing a more accurate determination of the replication abilities of the virus within the mosquito [61]. For IT inoculation, 5–10 day old mosquitoes were inoculated with approximately 220 nl of virus suspension.
Following feeding or inoculation, mosquitoes were maintained on 10% sucrose at 28°C, high relative humidity and a 12L∶12D lighting regime in an environmental growth chamber. After 12 days incubation, mosquitoes were processed to determine infection, dissemination and transmission rates. Mosquitoes were anaesthetized with CO2 gas and the legs and wings removed. Detection of the virus in the legs and wings indicates that the virus has escaped from the midgut and has disseminated through the hemocoel [52]. To assess transmission potential, saliva was collected by inserting the proboscis of the mosquito into a capillary tube containing 25 µL of growth media, supplemented with 20% FCS. Following transmission attempts, the saliva expectorate were placed in 2 ml vials containing 0.6 ml of growth media with 3% FCS, and the bodies, and legs and wings were placed in separate 2 ml vials containing 1 ml of growth media with 3% FCS and three sterile 5 mm glass beads. All samples were stored at −80°C prior to analysis.
The bodies, and legs and wings were homogenized using a SPEX 8000 mixer/mill (Spex Industries, Edison, NJ). All samples were filtered through a 0.2 µm Supor membrane filter (Pall Corporation, Ann Arbor, MI) to reduce bacterial and fungal contamination. The filtrates were inoculated into the wells of a 96-well microtiter plate seeded with confluent C6/36 (Aedes albopictus) cell monolayers. Plates were incubated at 28°C in the absence of CO2. To determine the virus titers, the bodies and saliva expectorates were titrated as ten-fold dilutions, whilst the legs and wings were assessed for infection only. After 7 days, plates were fixed with PBS/acetone and virus infection detected by a cell culture enzyme immunoassay [62] with the monoclonal anti-E antibody 3.91D [56].
Statistical analysis of mosquito data
For the characterization of the wild type and mutant viruses in Cx. annulirostris, body infection rate was defined as the number percentage of mosquitoes containing virus in their bodies (number positive/number tested). The disseminated infection rate was defined as the percentage of mosquitoes containing virus in their legs and wings (number positive/number tested). The transmission rate was defined as the percentage of mosquito expectorates in which virus was detected (number of positive expectorates/number tested). Infection, dissemination and transmission rates were analysed using Chi-square and Fisher's exact test. Differences in viral titers within mosquito bodies and saliva expectorates were analysed using multiple factor ANOVA test and homogeneity of variance was performed using Bartlett's test. P values<0.05 were considered statistically significant. All analyses were performed using R and/or GraphPad statistical packages.
Zdroje
1. Diamond MS (2009) West Nile Encephalitis Virus Infection; I.W F, editor. New York: Springer. 485 p.
2. ZhangY, KaufmannB, ChipmanPR, KuhnRJ, RossmannMG (2007) Structure of immature West Nile virus. J Virol 81 : 6141–6145.
3. MackenzieJM, JonesMK, YoungPR (1996) Immunolocalization of the dengue virus nonstructural glycoprotein NS1 suggests a role in viral RNA replication. Virology 220 : 232–240.
4. MackenzieJM, KhromykhAA, JonesMK, WestawayEG (1998) Subcellular localization and some biochemical properties of the flavivirus Kunjin nonstructural proteins NS2A and NS4A. Virology 245 : 203–215.
5. KhromykhAA, SedlakPL, WestawayEG (2000) cis - and trans-acting elements in flavivirus RNA replication. J Virol 74 : 3253–3263.
6. YounS, LiT, McCuneBT, EdelingMA, FremontDH, et al. (2012) Evidence for a genetic and physical interaction between nonstructural proteins NS1 and NS4B that modulates replication of West Nile virus. J Virol 86 : 7360–7371.
7. RossiSL, FayzulinR, DewsburyN, BourneN, MasonPW (2007) Mutations in West Nile virus nonstructural proteins that facilitate replicon persistence in vitro attenuate virus replication in vitro and in vivo. Virology 364 : 184–195.
8. YamshchikovVF, CompansRW (1994) Processing of the intracellular form of the west Nile virus capsid protein by the viral NS2B-NS3 protease: an in vitro study. J Virol 68 : 5765–5771.
9. RoosendaalJ, WestawayEG, KhromykhA, MackenzieJM (2006) Regulated cleavages at the West Nile virus NS4A-2K-NS4B junctions play a major role in rearranging cytoplasmic membranes and Golgi trafficking of the NS4A protein. J Virol 80 : 4623–4632.
10. ChernovAV, ShiryaevSA, AleshinAE, RatnikovBI, SmithJW, et al. (2008) The two-component NS2B-NS3 proteinase represses DNA unwinding activity of the West Nile virus NS3 helicase. J Biol Chem 283 : 17270–17278.
11. LeungJY, PijlmanGP, KondratievaN, HydeJ, MackenzieJM, et al. (2008) Role of nonstructural protein NS2A in flavivirus assembly. J Virol 82 : 4731–4741.
12. KummererBM, RiceCM (2002) Mutations in the yellow fever virus nonstructural protein NS2A selectively block production of infectious particles. J Virol 76 : 4773–4784.
13. LiuWJ, WangXJ, ClarkDC, LobigsM, HallRA, et al. (2006) A single amino acid substitution in the West Nile virus nonstructural protein NS2A disables its ability to inhibit alpha/beta interferon induction and attenuates virus virulence in mice. J Virol 80 : 2396–2404.
14. AshourJ, Laurent-RolleM, ShiPY, Garcia-SastreA (2009) NS5 of dengue virus mediates STAT2 binding and degradation. J Virol 83 : 5408–5418.
15. Laurent-RolleM, BoerEF, LubickKJ, WolfinbargerJB, CarmodyAB, et al. (2010) The NS5 protein of the virulent West Nile virus NY99 strain is a potent antagonist of type I interferon-mediated JAK-STAT signaling. J Virol 84 : 3503–3515.
16. WermeK, WigeriusM, JohanssonM (2008) Tick-borne encephalitis virus NS5 associates with membrane protein scribble and impairs interferon-stimulated JAK-STAT signalling. Cell Microbiol 10 : 696–712.
17. LinRJ, ChangBL, YuHP, LiaoCL, LinYL (2006) Blocking of interferon-induced Jak-Stat signaling by Japanese encephalitis virus NS5 through a protein tyrosine phosphatase-mediated mechanism. J Virol 80 : 5908–5918.
18. BestSM, MorrisKL, ShannonJG, RobertsonSJ, MitzelDN, et al. (2005) Inhibition of interferon-stimulated JAK-STAT signaling by a tick-borne flavivirus and identification of NS5 as an interferon antagonist. J Virol 79 : 12828–12839.
19. WilsonJR, de SessionsPF, LeonMA, ScholleF (2008) West Nile virus nonstructural protein 1 inhibits TLR3 signal transduction. J Virol 82 : 8262–8271.
20. ChungKM, LiszewskiMK, NybakkenG, DavisAE, TownsendRR, et al. (2006) West Nile virus nonstructural protein NS1 inhibits complement activation by binding the regulatory protein factor H. Proc Natl Acad Sci U S A 103 : 19111–19116.
21. MasonPW (1989) Maturation of Japanese encephalitis virus glycoproteins produced by infected mammalian and mosquito cells. Virology 169 : 354–364.
22. FirthAE, AtkinsJF (2009) A conserved predicted pseudoknot in the NS2A-encoding sequence of West Nile and Japanese encephalitis flaviviruses suggests NS1′ may derive from ribosomal frameshifting. Virol J 6 : 14.
23. MelianEB, HinzmanE, NagasakiT, FirthAE, WillsNM, et al. (2010) NS1′ of flaviviruses in the Japanese encephalitis virus serogroup is a product of ribosomal frameshifting and plays a role in viral neuroinvasiveness. J Virol 84 : 1641–1647.
24. SunJ, YuY, DeubelV (2012) Japanese encephalitis virus NS1′ protein depends on pseudoknot secondary structure and is cleaved by caspase during virus infection and cell apoptosis. Microbes Infect 14 : 930–940.
25. TakamatsuY, OkamotoK, DinhDT, YuF, HayasakaD, et al. (2014) NS1′ protein expression facilitates production of Japanese encephalitis virus in avian cells and embryonated chicken eggs. J Gen Virol 95 : 373–383.
26. WinkelmannER, WidmanDG, SuzukiR, MasonPW (2011) Analyses of mutations selected by passaging a chimeric flavivirus identify mutations that alter infectivity and reveal an interaction between the structural proteins and the nonstructural glycoprotein NS1. Virology 421 : 96–104.
27. YoungLB, MelianEB, KhromykhAA (2013) NS1′ co-localizes with NS1 and can substitute for NS1 in West Nile virus replication. J Virol 87 : 9384–9390.
28. YeQ, LiXF, ZhaoH, LiSH, DengYQ, et al. (2012) A single nucleotide mutation in NS2A of Japanese encephalitis-live vaccine virus (SA14-14-2) ablates NS1′ formation and contributes to attenuation. J Gen Virol 93 : 1959–1964.
29. YounS, AmbroseRL, MackenzieJM, DiamondMS (2013) Non-structural protein-1 is required for West Nile virus replication complex formation and viral RNA synthesis. Virol J 10 : 339.
30. MelianEB, EdmondsJH, NagasakiTK, HinzmanE, FlodenN, et al. (2013) West Nile virus NS2A protein facilitates virus-induced apoptosis independently of interferon response. J Gen Virol 94 : 308–313.
31. TurellMJ, O'GuinnML, JonesJW, SardelisMR, DohmDJ, et al. (2005) Isolation of viruses from mosquitoes (Diptera: Culicidae) collected in the Amazon Basin region of Peru. J Med Entomol 42 : 891–898.
32. BraultAC, LangevinSA, BowenRA, PanellaNA, BiggerstaffBJ, et al. (2004) Differential virulence of West Nile strains for American crows. Emerg Infect Dis 10 : 2161–2168.
33. LangevinSA, BraultAC, PanellaNA, BowenRA, KomarN (2005) Variation in virulence of West Nile virus strains for house sparrows (Passer domesticus). Am J Trop Med Hyg 72 : 99–102.
34. LiuWJ, ChenHB, WangXJ, HuangH, KhromykhAA (2004) Analysis of adaptive mutations in Kunjin virus replicon RNA reveals a novel role for the flavivirus nonstructural protein NS2A in inhibition of beta interferon promoter-driven transcription. J Virol 78 : 12225–12235.
35. LiSH, LiXF, ZhaoH, DengYQ, YuXD, et al. (2013) Development and characterization of the replicon system of Japanese encephalitis live vaccine virus SA14-14-2. Virol J 10 : 64.
36. MackenzieJM, WestawayEG (2001) Assembly and maturation of the flavivirus Kunjin virus appear to occur in the rough endoplasmic reticulum and along the secretory pathway, respectively. J Virol 75 : 10787–10799.
37. BraultAC, LangevinSA, RameyWN, FangY, BeasleyDW, et al. (2011) Reduced avian virulence and viremia of West Nile virus isolates from Mexico and Texas. Am J Trop Med Hyg 85 : 758–767.
38. LangevinSA, BowenRA, RameyWN, SandersTA, MaharajPD, et al. (2011) Envelope and pre-membrane protein structural amino acid mutations mediate diminished avian growth and virulence of a Mexican West Nile virus isolate. J Gen Virol 92 : 2810–2820.
39. LangevinSA, BowenRA, ReisenWK, AndradeCC, RameyWN, et al. (2014) Host Competence and Helicase Activity Differences Exhibited by West Nile Viral Variants Expressing NS3-249 Amino Acid Polymorphisms. PLoS ONE 9: e100802.
40. DuggalN, Bosco-LauthA, BowenR, WheelerS, ReisenW, et al. (2014) Evidence for co-evolution of West Nile virus and house sparrows in North America. PLoS Negl Trop Dis 8: e3262.
41. BrierleyI, JennerAJ, InglisSC (1992) Mutational analysis of the “slippery-sequence” component of a coronavirus ribosomal frameshifting signal. J Mol Biol 227 : 463–479.
42. BrierleyI, PennellS, GilbertRJ (2007) Viral RNA pseudoknots: versatile motifs in gene expression and replication. Nat Rev Microbiol 5 : 598–610.
43. Shehu-XhilagaM, CroweSM, MakJ (2001) Maintenance of the Gag/Gag-Pol ratio is important for human immunodeficiency virus type 1 RNA dimerization and viral infectivity. J Virol 75 : 1834–1841.
44. ZhaiY, SunF, LiX, PangH, XuX, et al. (2005) Insights into SARS-CoV transcription and replication from the structure of the nsp7-nsp8 hexadecamer. Nat Struct Mol Biol 12 : 980–986.
45. ImbertI, GuillemotJC, BourhisJM, BussettaC, CoutardB, et al. (2006) A second, non-canonical RNA-dependent RNA polymerase in SARS coronavirus. EMBO J 25 : 4933–4942.
46. DaffisS, SutharMS, SzretterKJ, GaleMJr, DiamondMS (2009) Induction of IFN-beta and the innate antiviral response in myeloid cells occurs through an IPS-1-dependent signal that does not require IRF-3 and IRF-7. PLoS Pathog 5: e1000607.
47. PeskoKN, EbelGD (2012) West Nile virus population genetics and evolution. Infect Genet Evol 12 : 181–190.
48. DingSW (2010) RNA-based antiviral immunity. Nat Rev Immunol 10 : 632–644.
49. WuQ, WangX, DingSW (2010) Viral suppressors of RNA-based viral immunity: host targets. Cell Host Microbe 8 : 12–15.
50. ArjonaA, WangP, MontgomeryRR, FikrigE (2011) Innate immune control of West Nile virus infection. Cell Microbiol 13 : 1648–1658.
51. BrackneyDE, ScottJC, SagawaF, WoodwardJE, MillerNA, et al. (2010) C6/36 Aedes albopictus cells have a dysfunctional antiviral RNA interference response. PLoS Negl Trop Dis 4: e856.
52. TurellMJ, GarganTP2nd, BaileyCL (1984) Replication and dissemination of Rift Valley fever virus in Culex pipiens. Am J Trop Med Hyg 33 : 176–181.
53. DavisMM, EngstromY (2012) Immune response in the barrier epithelia: lessons from the fruit fly Drosophila melanogaster. J Innate Immun 4 : 273–283.
54. Tchankouo-NguetcheuS, KhunH, PincetL, RouxP, BahutM, et al. (2010) Differential protein modulation in midguts of Aedes aegypti infected with chikungunya and dengue 2 viruses. PLoS One 5: e13149.
55. AudsleyM, EdmondsJ, LiuW, MokhonovV, MokhonovaE, et al. (2011) Virulence determinants between New York 99 and Kunjin strains of West Nile virus. Virology 414 : 63–73.
56. AdamsSC, BroomAK, SammelsLM, HartnettAC, HowardMJ, et al. (1995) Glycosylation and antigenic variation among Kunjin virus isolates. Virology 206 : 49–56.
57. LinSM, DuP, HuberW, KibbeWA (2008) Model-based variance-stabilizing transformation for Illumina microarray data. Nucleic Acids Res 36: e11.
58. BenjaminiY, HochbergY (1995) Controlling the False Discovery Rate - a Practical and Powerful Approach to Multiple Testing. J R Stat Soc Series B-Methodol 57 : 289–300.
59. JohnsonPH, Hall-MendelinS, WhelanPI, FrancesSP, JansenCC, et al. (2009) Vector competence of Australian Culex gelidus Theobald (Diptera: Culicidae) for endemic and exotic arboviruses. Aust J Entomol 48 : 234–240.
60. WellsPJ, ValeTG, RussellRC, CloonanMJ (1994) Comparison of a pledget technique with other methods for bloodfeeding mosquitoes (Diptera: Culicidae) for virus studies. J Aust Entomol Soc 33 : 211–212.
61. van den HurkAF, Hall-MendelinS, PykeAT, FrentiuFD, McElroyK, et al. (2012) Impact of Wolbachia on infection with chikungunya and yellow fever viruses in the mosquito vector Aedes aegypti. PLoS Negl Trop Dis 6: e1892.
62. BroomAK, HallRA, JohansenCA, OliveiraN, HowardMA, et al. (1998) Identification of Australian arboviruses in inoculated cell cultures using monoclonal antibodies in ELISA. Pathology 30 : 286–288.
Štítky
Hygiena a epidemiologie Infekční lékařství Laboratoř
Článek Acidification Activates Motility and Egress by Enhancing Protein Secretion and Cytolytic ActivityČlánek Plasticity between MyoC- and MyoA-Glideosomes: An Example of Functional Compensation in InvasionČlánek Coronavirus Cell Entry Occurs through the Endo-/Lysosomal Pathway in a Proteolysis-Dependent MannerČlánek NK Cell Activation in Human Hantavirus Infection Explained by Virus-Induced IL-15/IL15Rα Expression
Článek vyšel v časopisePLOS Pathogens
Nejčtenější tento týden
2014 Číslo 11- Stillova choroba: vzácné a závažné systémové onemocnění
- Perorální antivirotika jako vysoce efektivní nástroj prevence hospitalizací kvůli COVID-19 − otázky a odpovědi pro praxi
- Jak souvisí postcovidový syndrom s poškozením mozku?
- Diagnostika virových hepatitid v kostce – zorientujte se (nejen) v sérologii
- Infekční komplikace virových respiračních infekcí – sekundární bakteriální a aspergilové pneumonie
-
Všechny články tohoto čísla
- Peculiarities of Prion Diseases
- Inhibitors of Peptidyl Proline Isomerases As Antivirals in Hepatitis C and Other Viruses
- War and Infectious Diseases: Challenges of the Syrian Civil War
- Microbial Contamination in Next Generation Sequencing: Implications for Sequence-Based Analysis of Clinical Samples
- Acidification Activates Motility and Egress by Enhancing Protein Secretion and Cytolytic Activity
- Co-dependence of HTLV-1 p12 and p8 Functions in Virus Persistence
- Shed GP of Ebola Virus Triggers Immune Activation and Increased Vascular Permeability
- Plasticity between MyoC- and MyoA-Glideosomes: An Example of Functional Compensation in Invasion
- The Type III Translocon Is Required for Biofilm Formation at the Epithelial Barrier
- Retromer Regulates HIV-1 Envelope Glycoprotein Trafficking and Incorporation into Virions
- IFI16 Restricts HSV-1 Replication by Accumulating on the HSV-1 Genome, Repressing HSV-1 Gene Expression, and Directly or Indirectly Modulating Histone Modifications
- Coronavirus Cell Entry Occurs through the Endo-/Lysosomal Pathway in a Proteolysis-Dependent Manner
- Silencing by H-NS Potentiated the Evolution of
- Crystal Structure of Cytomegalovirus IE1 Protein Reveals Targeting of TRIM Family Member PML via Coiled-Coil Interactions
- GAPDH-A Recruits a Plant Virus Movement Protein to Cortical Virus Replication Complexes to Facilitate Viral Cell-to-Cell Movement
- Genomic Insights into the Fungal Pathogens of the Genus : Obligate Biotrophs of Humans and Other Mammals
- Unravelling Human Trypanotolerance: IL8 is Associated with Infection Control whereas IL10 and TNFα Are Associated with Subsequent Disease Development
- The Skin Microbiome: A Focus on Pathogens and Their Association with Skin Disease
- Human Cytomegalovirus Vaccine Based on the Envelope gH/gL Pentamer Complex
- IL-37 Inhibits Inflammasome Activation and Disease Severity in Murine Aspergillosis
- Host-Specific Parvovirus Evolution in Nature Is Recapitulated by Adaptation to Different Carnivore Species
- Activation of HIV Transcription with Short-Course Vorinostat in HIV-Infected Patients on Suppressive Antiretroviral Therapy
- PUL21a-Cyclin A2 Interaction is Required to Protect Human Cytomegalovirus-Infected Cells from the Deleterious Consequences of Mitotic Entry
- Programmed Ribosomal Frameshift Alters Expression of West Nile Virus Genes and Facilitates Virus Replication in Birds and Mosquitoes
- Aminoterminal Amphipathic α-Helix AH1 of Hepatitis C Virus Nonstructural Protein 4B Possesses a Dual Role in RNA Replication and Virus Production
- NK Cell Activation in Human Hantavirus Infection Explained by Virus-Induced IL-15/IL15Rα Expression
- Structure and Specificity of the Bacterial Cysteine Methyltransferase Effector NleE Suggests a Novel Substrate in Human DNA Repair Pathway
- Genetics, Receptor Binding Property, and Transmissibility in Mammals of Naturally Isolated H9N2 Avian Influenza Viruses
- A Gatekeeper Chaperone Complex Directs Translocator Secretion during Type Three Secretion
- A Conserved Peptide Pattern from a Widespread Microbial Virulence Factor Triggers Pattern-Induced Immunity in
- Succinate Dehydrogenase is the Regulator of Respiration in
- The Plasmodesmal Protein PDLP1 Localises to Haustoria-Associated Membranes during Downy Mildew Infection and Regulates Callose Deposition
- Dysregulated B Cell Expression of RANKL and OPG Correlates with Loss of Bone Mineral Density in HIV Infection
- Restriction of Genetic Diversity during Infection of the Vector Midgut
- The Epithelial αvβ3-Integrin Boosts the MYD88-Dependent TLR2 Signaling in Response to Viral and Bacterial Components
- The Relationship between Host Lifespan and Pathogen Reservoir Potential: An Analysis in the System
- Multiple Roles of the Cytoskeleton in Bacterial Autophagy
- The Evolution and Genetics of Virus Host Shifts
- ChIP-seq and In Vivo Transcriptome Analyses of the SREBP SrbA Reveals a New Regulator of the Fungal Hypoxia Response and Virulence
- PLOS Pathogens
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Coronavirus Cell Entry Occurs through the Endo-/Lysosomal Pathway in a Proteolysis-Dependent Manner
- Peculiarities of Prion Diseases
- Host-Specific Parvovirus Evolution in Nature Is Recapitulated by Adaptation to Different Carnivore Species
- War and Infectious Diseases: Challenges of the Syrian Civil War
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



