-
Články
- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Multiple Means to the Same End: The Genetic Basis of Acquired Stress Resistance in Yeast
In nature, stressful environments often occur in combination or close succession, and thus the ability to prepare for impending stress likely provides a significant fitness advantage. Organisms exposed to a mild dose of stress can become tolerant to what would otherwise be a lethal dose of subsequent stress; however, the mechanism of this acquired stress tolerance is poorly understood. To explore this, we exposed the yeast gene-deletion libraries, which interrogate all essential and non-essential genes, to successive stress treatments and identified genes necessary for acquiring subsequent stress resistance. Cells were exposed to one of three different mild stress pretreatments (salt, DTT, or heat shock) and then challenged with a severe dose of hydrogen peroxide (H2O2). Surprisingly, there was little overlap in the genes required for acquisition of H2O2 tolerance after different mild-stress pretreatments, revealing distinct mechanisms of surviving H2O2 in each case. Integrative network analysis of these results with respect to protein–protein interactions, synthetic–genetic interactions, and functional annotations identified many processes not previously linked to H2O2 tolerance. We tested and present several models that explain the lack of overlap in genes required for H2O2 tolerance after each of the three pretreatments. Together, this work shows that acquired tolerance to the same severe stress occurs by different mechanisms depending on prior cellular experiences, underscoring the context-dependent nature of stress tolerance.
Published in the journal: . PLoS Genet 7(11): e32767. doi:10.1371/journal.pgen.1002353
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1002353Summary
In nature, stressful environments often occur in combination or close succession, and thus the ability to prepare for impending stress likely provides a significant fitness advantage. Organisms exposed to a mild dose of stress can become tolerant to what would otherwise be a lethal dose of subsequent stress; however, the mechanism of this acquired stress tolerance is poorly understood. To explore this, we exposed the yeast gene-deletion libraries, which interrogate all essential and non-essential genes, to successive stress treatments and identified genes necessary for acquiring subsequent stress resistance. Cells were exposed to one of three different mild stress pretreatments (salt, DTT, or heat shock) and then challenged with a severe dose of hydrogen peroxide (H2O2). Surprisingly, there was little overlap in the genes required for acquisition of H2O2 tolerance after different mild-stress pretreatments, revealing distinct mechanisms of surviving H2O2 in each case. Integrative network analysis of these results with respect to protein–protein interactions, synthetic–genetic interactions, and functional annotations identified many processes not previously linked to H2O2 tolerance. We tested and present several models that explain the lack of overlap in genes required for H2O2 tolerance after each of the three pretreatments. Together, this work shows that acquired tolerance to the same severe stress occurs by different mechanisms depending on prior cellular experiences, underscoring the context-dependent nature of stress tolerance.
Introduction
All organisms must respond to stressful stimuli that result from external environmental changes or internal defects caused by mutation and disease. Decades of research have characterized the mechanisms for surviving individual stresses, by mapping downstream protection systems as well as upstream signaling pathways that mediate these responses [1]–[7]. However, much less is known about the effects of combinatorial stress treatments and how cells defend against compound stresses. For example, stressful environmental changes in nature likely occur together, either simultaneously or in close succession, especially for microbes living in natural conditions. How the mechanisms of stress defense differ when cells experience successive stresses rather than a single insult is poorly understood.
Successive stress treatments can cause cells to acquire resistance to a severe (‘secondary’) stress after experiencing an initial mild (‘primary’) dose of stress. Acquired stress resistance can occur if the mild and severe treatments represent the same stressor but also across different mild and severe stresses (known as ‘cross-stress’ protection). Acquired stress resistance has been observed in diverse organisms, including yeast, bacteria, archaea, plants, flies, and mammals including mice and humans [8]–[20]. A better understanding of how cells are able to increase their resistance to further insults has potential medical application for decreasing cell death and improving human recovery from stressful events such as chemotherapy treatments and ischemia following heart attack or stroke [21]–[23].
In yeast, it had been suggested that acquired stress resistance in general, and cross-stress protection specifically, may be due to activation of the Environmental Stress Response (ESR) [24]–[30]. The ESR is a gene expression response commonly activated by a wide variety of stressful conditions [24], [25]. It includes induced expression of ∼300 genes involved in stress defense, and reduced expression of ∼600 genes broadly involved in protein synthesis and growth. However, we previously showed that ESR activation alone is insufficient to explain cross-stress protection [31]. Moreover, the ‘general-stress’ transcription factors MSN2 and MSN4 are conditionally required for acquired stress resistance, depending on the precise combination of mild and severe stress treatments [31]. These results revealed that the mechanism of acquired stress resistance is more complex than previously suspected and suggested that the response occurs through different mechanisms depending on the mild stress pretreatment.
Many studies have identified genes required to survive a single dose of oxidative stress, and several studies characterized increased tolerance after preconditioning (reviewed in [5], [6], [32]). The majority of these studies used single-gene approaches, though several used the yeast deletion collection to interrogate the entire genome [33]–[36]. Kelley et al. (2009) identified genes required to survive an acute dose of H2O2 and genes necessary to acquire H2O2 resistance following a mild H2O2 pretreatment. They found that the genes required for acquisition of H2O2 tolerance only partially overlapped the genes required to survive the acute dose alone, indicating that the mechanism of acquired H2O2 tolerance is distinct from the mechanism of basal H2O2 resistance [34]. The mechanism of cross-stress protection, in which the mild pretreatment is a different stressor than the subsequent severe stress, is largely unexplored.
Here, we leveraged the power of yeast genetics and high-throughput analysis to identify genes and processes important for acquired resistance to severe H2O2 stress after each of three mild pretreatments (mild NaCl, heat shock, or DTT treatment). We used the pooled yeast deletion collection [37], [38], including ∼4,800 homozygous diploid nonessential genes (homozygous profiling), ∼1,300 heterozygous diploid essential genes (haploinsufficiency profiling), and 1,140 strains harboring DAmP alleles of the essential genes (in which the transcript is destabilized due to insertion of a drug marker into the 3' UTR [39]) to query the vast majority of the yeast genome in a single experiment. We found that, although each pretreatment provided similar levels of subsequent H2O2 resistance, different genes and processes were required depending on the mild stress used. Functional analysis of the genes required during each pretreatment provided new insights into the relationships between regulators and processes. Acquired stress resistance thus serves as a unique phenotype through which to uncover new insights into stress biology.
Results
Methods summary
We exposed the pooled yeast deletion libraries [37]–[39] to severe doses of H2O2 after pretreatment with one of three mild stresses (Figure 1). These mild stresses were chosen because they each produce increased H2O2 tolerance in wild type but present different initial challenges to the cell. The pooled library was exposed to either 60 min of 0.7 M NaCl, 60 min at 40C after a 30°C–40°C heat shock, or 2 h exposure to 2.5 mM DTT - each treatment produces roughly equivalent levels of subsequent H2O2 tolerance in wild-type cells. After the pretreatment, cells from the culture were washed and then exposed for 2 hours to either 1.0 mM or 1.2 mM H2O2. Exposure to these H2O2 doses kills >85% of untreated wild-type cells but results in >80% viability in cells previously exposed to mild stress (data not shown). To identify mutant strains with defects in acquired H2O2 tolerance, an aliquot of the pooled library was removed from stress at each sample point (Figure 1) and outgrown for precisely 10 generations to dilute dead cells from the population. Relative strain abundances were then measured by quantifying the unique ‘barcode’ sequences (identified by microarray and/or deep sequencing analysis). A defect in acquired H2O2 tolerance was identified based on the log2 change in strain abundance before and after treatments (see Figure 1 and Materials and Methods for details). We also identified 202 strains that were sensitive to a low dose of 0.4 mM H2O2 in the absence of any pretreatment (e.g. Sample 2 versus Sample 1, false discovery rate (FDR)<0.05, Table S1); these included many genes and regulators known to be important for the H2O2 response [33]–[35]. Because we were interested only in genes important for the acquisition of stress tolerance, we removed from consideration strains with equal fitness defects at both the low and ‘secondary’ doses of H2O2 and strains sensitive to the mild stress treatment alone (identified by comparing Sample 3 versus Sample 1). Strains that met all of these criteria in replicate experiments were defined as having a specific defect in acquiring resistance to H2O2.
Fig. 1. Experimental overview. 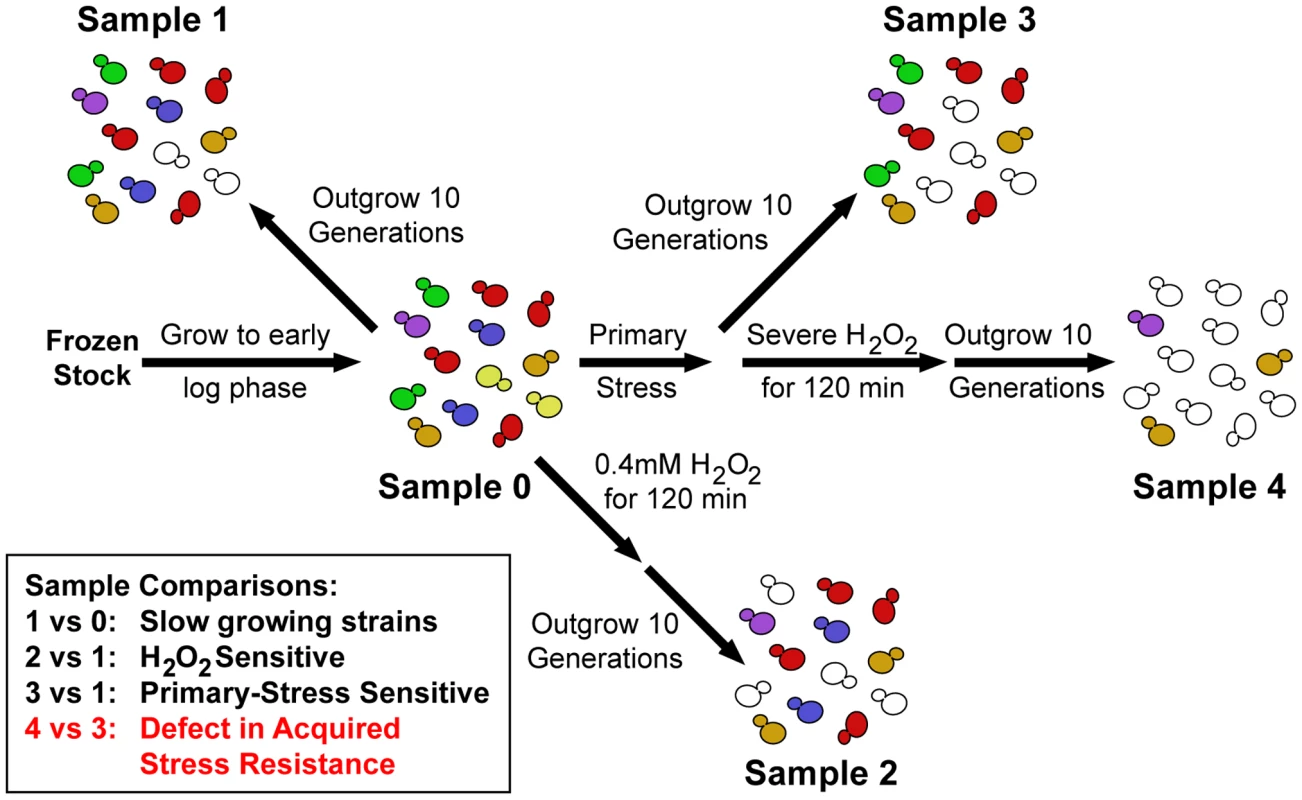
Pooled mutant libraries were grown >7 generations in log phase (Sample 0) before being exposed to one of three mild (‘Primary’) stress pretreatments. Cells were then either outgrown 10 generations (Sample 3) or washed and exposed to severe H2O2 for 2 hours followed by 10 generations outgrowth (Sample 4). Strains sensitive to a mild dose of H2O2 were identified in a separate control experiment (Sample 2). Strains of interest were determined through the sample comparisons listed (see text for details). Little overlap in genes required for acquisition of stress resistance after each pretreatment
A substantial fraction of the yeast genome was required for acquisition of normal H2O2 resistance after at least one of the three pretreatments. In all, 841 strains (∼13% of measured genes) displayed a defect in acquiring H2O2 tolerance, with 225 strains identified following mild NaCl treatment, 308 after heat shock, and 497 after DTT treatment (Table S2). Validation experiments were performed for 48 strains, the majority of which were predicted to have a defect after one or more pretreatments and three that were predicted to have no defect after any pretreatment. We measured mutant phenotypes in response to all three mild stresses, allowing us to quantify false positive and false negative rates, by competing each identified strain or the isogenic wild type against a GFP-marked strain (see Materials and Methods for details). This defined an upper limit of ∼25% false positives and ∼25% false negatives; however, these values are almost certainly inflated, because our validation assay does not precisely mirror the selection experiments and was performed using the haploid deletion library. Nonetheless, the results validate that the majority of our strain identifications are accurate.
There was surprisingly little overlap between the genes necessary for acquired H2O2 resistance following each mild stress (Figure 2A) – only 28 strains had defects following all three primary-stress conditions (Table 1). This observation cannot be explained by the nominally high false-negative rate: of the 48 strains validated, 34 were predicted to have conditional defects - only two of these 34 (6%) proved to have a universal defect in the validation experiments. There was also low overlap between the genes necessary following these primary stresses compared to genes required after mild H2O2 pretreatment [34]. There is little functionality in common to the 28 shared genes, with a few exceptions. There were several genes involved DNA damage repair and vacuolar processes, along with negative regulators of Ras (IRA1) and TOR (NPR2 and NPR3) signaling, which themselves suppress the stress response (reviewed in [4], [40]). However, even among the 28 strains with universal defects, the magnitude of their fitness defects varied dramatically depending on the initial mild stress used (Figure 2B). Thus, even the genes necessary in all three cases were not equally important following each mild-stress pretreatment.
Fig. 2. Genes important for acquired H2O2 resistance after each pretreatment. 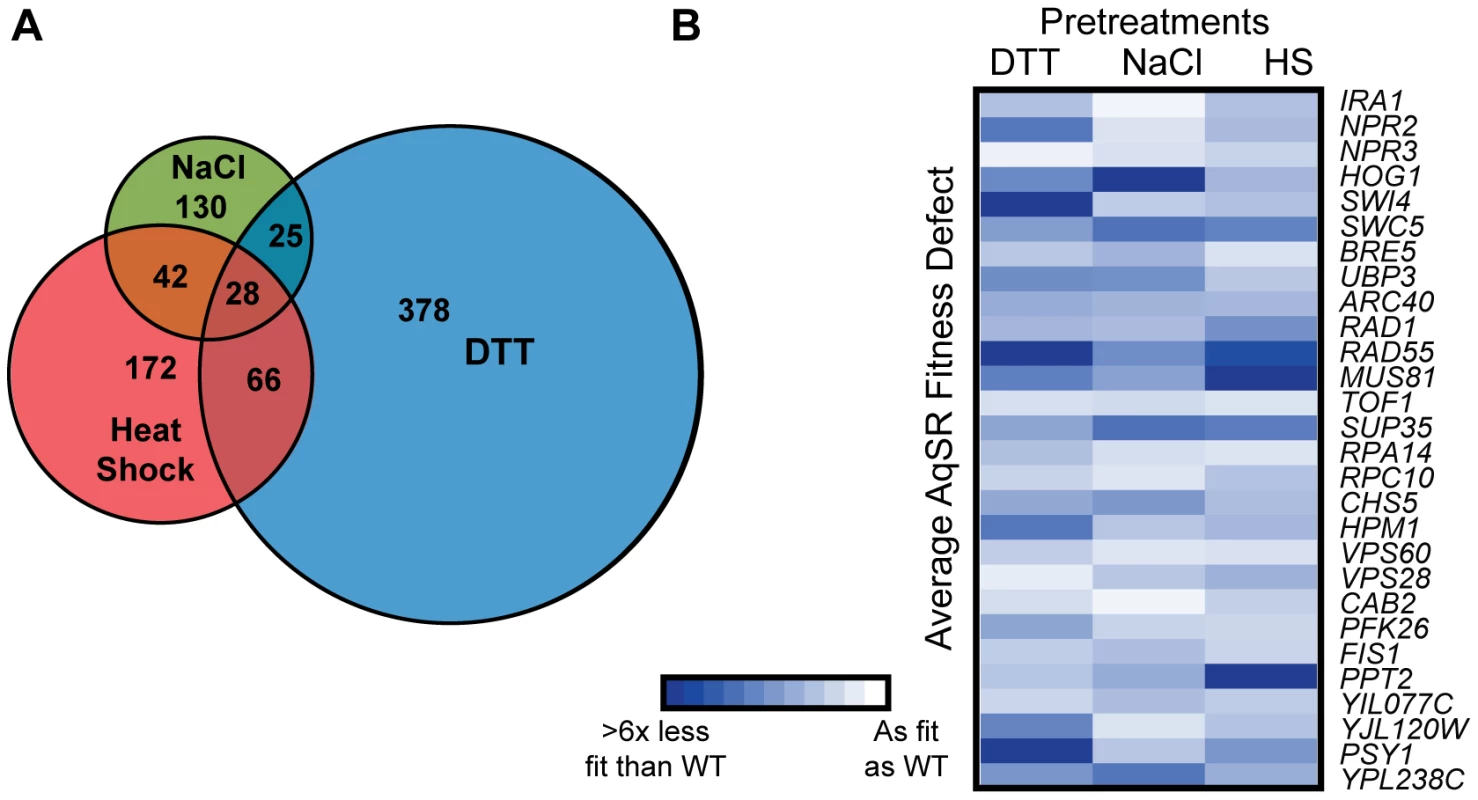
(A) The number of genes necessary for acquiring H2O2 resistance after NaCl, heat shock, or DTT treatment are shown in the Venn diagram, with shared genes identified in the overlap. (B) The average fitness defect following exposure to 1.0 mM H2O2 is shown for 28 strains that had a defect after all three pretreatments. Each row represents an individual strain, and each column represents a single pretreatment. Average fitness defects in acquired stress resistance are represented by increased color intensity, according to the key. Data shown are for deletion strains, with the exception of three genes (ARC40, CAB2, and YPL238C) for which fitness scores were taken from strains expressing the DAmP alleles. Gene annotations are found in Table 1. Tab. 1. Genes required following all 3 pretreatments. 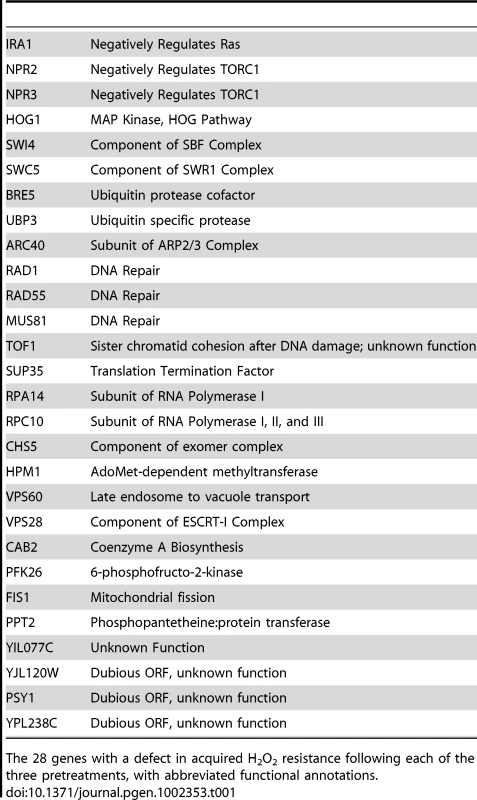
The 28 genes with a defect in acquired H2O2 resistance following each of the three pretreatments, with abbreviated functional annotations. We also found limited overlap in functional processes enriched in each group of required genes (Table 2). Genes necessary for acquired H2O2 tolerance after NaCl pretreatment were involved in proteolysis as well as HOG signaling, a pathway well known to respond to NaCl. By contrast, genes important following heat shock were enriched for DNA damage repair, protein transport, and late endosome to vacuole transport. Functions enriched in the group of the DTT-required genes included ubiquitin-dependent and -independent protein catabolism, ribosomal proteins, and regulation of translation. Other processes were shared for two of the three mild stressors (Table 2). Considering this and the above results, we conclude that genes and processes necessary to acquire H2O2 resistance are largely distinct and determined by each pretreatment.
Tab. 2. Functional enrichments in genes necessary to acquire H2O2 tolerance. 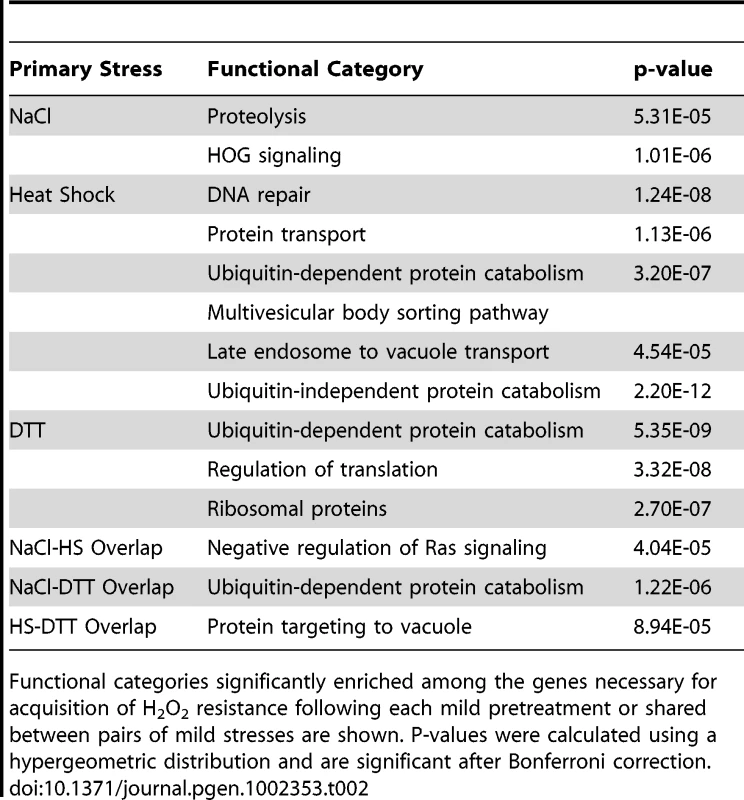
Functional categories significantly enriched among the genes necessary for acquisition of H2O2 resistance following each mild pretreatment or shared between pairs of mild stresses are shown. P-values were calculated using a hypergeometric distribution and are significant after Bonferroni correction. Low correlation between fitness contribution and gene expression
Previous studies showed little correlation between a gene's expression change during stress and its requirement to survive prolonged treatment with that stressor [37], [41]–[46]. However, we and others showed that gene expression changes are not required to survive the initial stress treatment, but rather are critical for acquired resistance to the secondary stress [31], [47]–[49]. We therefore wondered if gene expression changes were more correlated with genes' involvement in acquired, rather than basal, stress tolerance. However, we too found low correlation between a gene's fitness effect and its expression change during the mild-stress treatment. Roughly 24% of genes necessary for acquired H2O2 tolerance after mild NaCl or heat shock were induced in expression during pretreatment (a slight enrichment above that expected by chance, p = 0.048). In fact, genes necessary for acquired H2O2 tolerance after DTT treatment were actually enriched for DTT-repressed genes (p = 0.0003). Conversely, the majority of genes whose expression increased during each mild stress treatment played no role in subsequent H2O2 tolerance. Thus, gene induction is a poor predictor of gene requirement for both basal [37] and acquired stress tolerance (see Discussion).
Initiation of the yeast ESR was originally proposed to give rise to cross-stress protection [24]–[30]; however, we showed that initiation of the ESR cannot explain acquired stress resistance [31]. Consistent with this notion, we observed little enrichment of ESR genes in any of the gene lists identified above (with the exception of repressed-ESR genes among those required after DTT pretreatment). While individual ESR genes can contribute substantially to the acquisition of stress tolerance (see below), the ESR as a whole seems not to be the sole determinant of the resistance acquired.
The results above indicate that acquired H2O2 tolerance occurs through distinct modes, rather than a common mechanism, for each mild-stress pretreatment. We were interested in exploring the possible reasons for the low overlap in required genes. Below we present example cases of three models that explain the low overlap in required genes.
Condition-specific regulators are only required during specific pretreatments
One possibility is that different upstream signaling pathways mediate the cellular response, even if the downstream effectors of acquired H2O2 tolerance may be the same across pretreatments. Indeed, an example of condition-specific signaling is seen if NaCl is the pretreatment. Several transcriptional regulators and signaling molecules were important for acquired H2O2 resistance after NaCl stress, including the stress-activated transcription factor MSN2 [25]–[27], [50] and the majority of HOG signaling components (including HOG1, PBS2, SSK2, SSK1, STE50, and CDC42) (Figure S1). Notably, none of the corresponding deletion strains was sensitive to a 1 h exposure to 0.7 M NaCl (data not shown), but all had major defects in acquired H2O2 tolerance. The Hog1 pathway regulates expression of stress-responsive genes specifically during osmotic shock and related stresses but not other conditions (J. Clarke and APG, unpublished data). Consistently, none of the HOG mutant strains validated with an acquired-stress defect after other mild stresses (although the hog1Δ strain had a general recovery defect, perhaps due to its separate role in cell-cycle progression [51]–[55] (Table 1)). Thus, Hog1 components are required for acquisition of H2O2 tolerance if NaCl is the mild treatment but not after pretreatments that do not activate the pathway.
This explanation also holds for other pretreatments. The transcription factor Hsf1p, a critical regulator of the heat-shock response that plays an overlapping role with Msn2p [56], [57], was required for full acquisition of H2O2 tolerance following heat, but not NaCl or DTT, pretreatments. Interestingly, several regulators not previously known to respond to DTT exposure were required after this pretreatment. These included RTG transcriptional regulators (RTG1, RTG2, RTG3) and members of the Snf1p signaling system (GAL83, STD1, and SNF3) that respond to mitochondria-to-nucleus retrograde signaling and nutrient availability, respectively [58], [59]. This suggests that additional, novel regulators of the primary responses are likely being uncovered.
Alternative lines of H2O2 defense are mobilized after each pretreatment
Although different upstream regulators were involved in each mild-stress response, we wondered if the same downstream effectors might be universally required for subsequent H2O2 tolerance. We focused on the cytosolic catalase Ctt1p, which reduces H2O2 to water and oxygen, as an obvious mechanism for detoxifying H2O2. CTT1 was the most important gene for acquiring H2O2 resistance after mild NaCl treatment (Figure S1). Importantly, cells lacking CTT1 had no observable sensitivity to H2O2 in the absence of pretreatment, consistent with the low basal expression of this gene ([60] and S. Haroon and APG, data not shown).
Somewhat surprisingly, CTT1 was not universally required for acquisition of H2O2 tolerance: although the gene was critical if NaCl was the mild stressor, CTT1 was completely dispensable after heat shock or DTT pretreatments (Figure S2). Instead, both heat shock and mild DTT treatments required the glutathione system for acquisition of H2O2 tolerance. Glutathione peroxidases provide an independent mode of H2O2 reduction that is coupled to glutathione oxidation [5]. Deletion of either of the glutathione peroxidases GPX1 or GPX2 did not result in an acquired stress defect (likely due to their known functional redundancy [61]). However, deletion of genes involved in glutathione metabolism, including GSH1 that encodes the first step of glutathione synthesis and the glutathione reductase Glr1p that recycles the oxidized peptide, produced a defect after heat shock or DTT pretreatments but not NaCl. Thus, cells appear to rely on different modes of H2O2 detoxification after NaCl versus heat or DTT pretreatments.
We wondered why cells would utilize different detoxification mechanisms for different pretreatments. At least part of the answer lies in the gene-expression response. Although CTT1 transcript was induced by all three mild stresses (albeit to different levels), Ctt1 protein accumulated to significant levels only after NaCl treatment (Figure S3). Neither Gsh1p nor Glr1p increased in abundance after any treatment (data not shown). However, glutathione peroxidases did increase under different conditions: Gpx2p was induced nearly 2.5-fold in response to DTT but only marginally (1.3-fold) after heat or NaCl exposure (Figure S3C). We were unable to measure Gpx1p levels by Western, although GPX1 transcript increased after heat and NaCl treatments (data not shown and Figure S3A). These results show that the differential requirement for CTT1 and genes involved in glutathione metabolism correlates with the conditional induction of Ctt1p or Gpx2p. Consistent with this result, we found that a double mutant lacking CTT1 and GSH1 had no additional defect in acquired H2O2 tolerance compared to the single mutants (data not shown).
Unique challenges are presented depending on the mild-severe stress combination
A third model for condition-specific mechanisms of acquired H2O2 tolerance is that the mode of resistance depends on the unique cellular conditions after each pretreatment. This model implies that the cell may experience H2O2 differently depending on its internal status immediately before treatment. As an example, we focused on the stress-specific poly-ubiquitin Ubi4p, which was necessary for acquired H2O2 tolerance after heat and DTT treatments but dispensable following NaCl. Ubi4p plays an important role in protein degradation and turnover in response to heat shock (reviewed in [2] and [62]). Consistent with previous observations [63], cells lacking UBI4 were not sensitive to mild heat shock, based on viability (data not shown) or growth rate (Figure 3A). Interestingly, the ubi4Δ strain was able to acquire H2O2 resistance after heat shock, since it had wild-type viability after secondary-stress treatment (Figure S4). However, the mutant had a significant growth defect upon recovery from H2O2 stress that persisted until ∼8 h after removal from H2O2 (Figure 3B and 3C). The temporary recovery defect recapitulated the ubi4Δ fitness defect observed in the selection experiments.
Fig. 3. The ubi4? strain has a specific defect in growth recovery after H2O2 treatment. 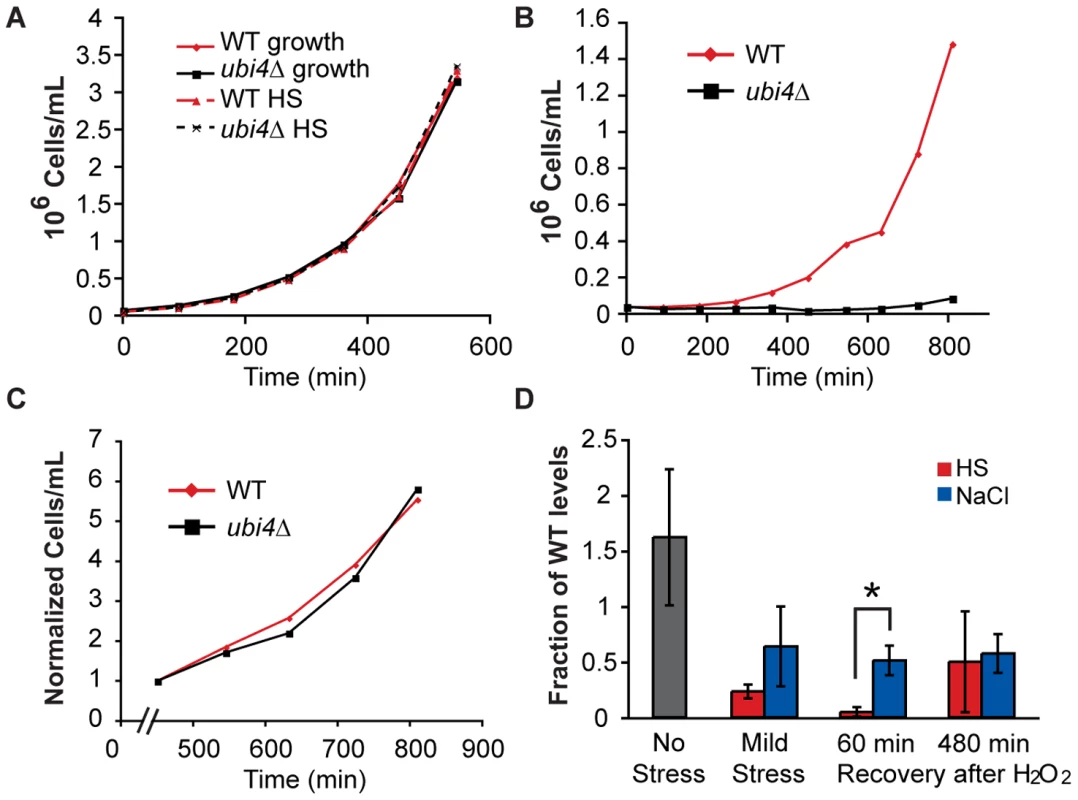
Wild-type (red) and ubi4Δ (black) cells were grown separately to log phase, exposed to a either a mock treatment (A, solid lines), 30–40°C heat shock (HS) for 60 min (A, dashed lines), or heat shock followed by 2 h treatment with 1.0 mM H2O2 (B). Cells were then removed from stress and monitored for growth by cell counting on a flow cytometer. (C) The growth rate normalized to cell density at 450 min after the removal from H2O2 stress is shown for wild-type (red) and ubi4Δ (black) cells. These growth curves are representative examples of several replicates. (D) Free ubiquitin was measured by Western analysis in wild type and ubi4Δ cells exposed to 1 h mild heat shock (HS, red) or NaCl (green) alone (‘Mild Stress’) and when cells were exposed to 1.0 mM H2O2 for 1 h immediately after mild stress and allowed to recover 60 or 480 min in stress-free media. Levels of free ubiquitin (normalized to an actin loading control) are shown relative to the paired wild-type sample from each of three biological replicates. A significant difference between heat- and NaCl-pretreated cells was seen 60 min after H2O2 recovery (asterisk, p = 0.018). To assess why Ubi4p was required after heat shock but not NaCl treatment, we measured free ubiquitin levels before, during, and after stress treatments (Figure 3D). Mono-ubiquitin was diminished but measurable in the ubi4Δ strain exposed to mild NaCl or heat shock alone. In contrast, free ubiquitin was virtually undetectable in cells treated with heat shock followed by H2O2 (Figure 3D). Mono-ubiquitin levels were again observable in the ubi4Δ strain 8 h after removal from H2O2, when the growth rate recovered. In contrast to the case of heat pretreatment, mono-ubiquitin was not depleted in the ubi4Δ strain treated with successive NaCl and H2O2. Thus, the combined effects of heat followed by H2O2 treatment require ubiquitin synthesis from the UBI4 gene to supplant the consumed ubiquitin.
Another possible example of context-dependent stress defense was seen when H2O2 stress followed mild DTT treatment, which invoked a large number of unique genes. To examine the connections between these genes, we constructed a network based on their genetic or physical interactions (Figure 4). The resulting network was heavily connected and pointed to a few key processes. Ribosomal proteins and proteins involved ubiquitin metabolism showed a large number of physical interactions, both within and between processes, while proteins involved in chromatin biology and actin cytoskeleton/cell wall showed the most genetic connections. This highly interconnected network demonstrates that the long list of genes important after DTT treatment can be collapsed into a smaller subset of processes.
Fig. 4. Interaction network of genes necessary for acquired H2O2 resistance following DTT pretreatment. 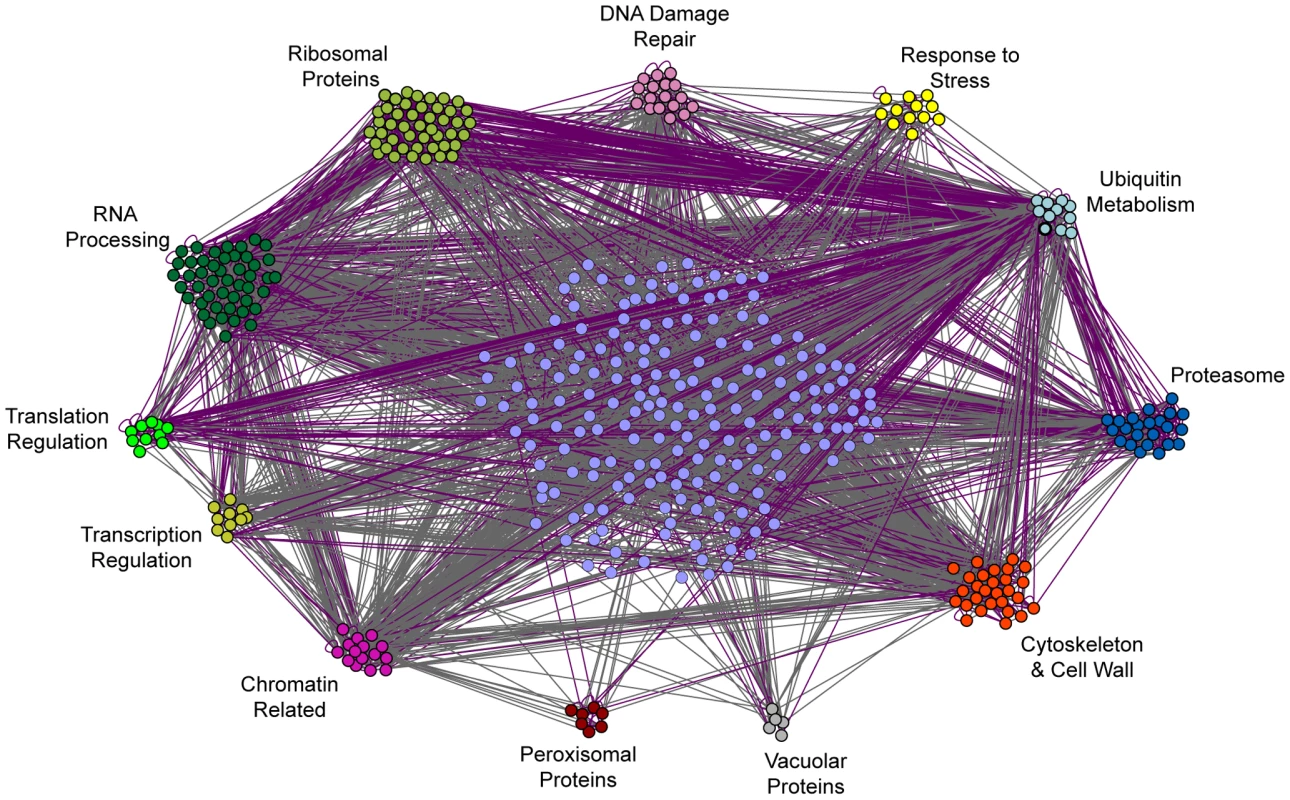
The 497 genes necessary during DTT pretreatment were organized according to genetic (grey) and physical (purple) interactions and enriched GO-slim categories using the program GOlorize v2.4 and Cytoscape v2.4.1. The resulting network was then manually adjusted to subdivide GO-slim categories and further distinguish functional groups. Genes that could not be placed into one of the listed GO-slim categories are shown in the center of the network and were organized by GOlorize according to their interactions only. To delineate whether the roles of these processes were related to DTT's reducing potential or to specific effects on ER function through the unfolded protein response (UPR), we repeated the selection using tunicamycin as a primary stress, to induce the UPR by blocking N-glycosylation in the ER [64]. We found that some, but not all, of the genes important after DTT treatment were also required after tunicamycin pretreatment (Table S2). Most notably, ribosomal proteins were important after both pretreatments (p = 6×10−7). Other genes required after DTT pretreatment were in fact necessary for tunicamycin survival; these were enriched for vacuolar/lysosomal transport (p = 8×10−6) and protein deubiquitination (p = 6×10−6), and included several genes linked to RNA processing. Why genes related to ribosome synthesis and protein and RNA metabolism are necessary for acquired H2O2 resistance after DTT, and to some extent tunicamycin, treatment remains unclear. However, hints from the literature suggest a connection between ER function and RNA catabolism [65]–[70]. These processes may be particularly susceptible to H2O2 attack if ER function is already disrupted (see Discussion).
Discussion
Our results show that the genes and processes necessary to acquire resistance to the same severe stress (H2O2 in this case) are distinctly different depending on the mild stress to which cells are previously exposed. Although there were some shared processes required for pairs of pretreatments, there were surprisingly few genes required for acquisition of H2O2 tolerance after all three mild-stress treatments. Even among these shared genes, their contributions varied dramatically depending on the pretreatment. Thus, the vast majority of genes function in a condition-specific manner to produce the same end result - increased H2O2 tolerance.
We have presented three different models explaining the low degree of mechanistic overlap, including 1) condition-specific signaling, 2) use of different downstream effectors that enact the same roles, and 3) application of entirely different defense strategies based on each pretreatment. Furthermore, we note that the genes and processes involved in acquired stress resistance could function in two fundamentally different ways. Induced production and/or function of some gene products may be sufficient to boost H2O2 resistance. For example, an exogenous pulse of CTT1 expression in the absence of stress is sufficient to increase H2O2 tolerance (S. Haroon and APG, unpublished). Alternatively, some genes and processes may be necessary, but not sufficient on their own, for acquired H2O2 resistance. Their action may instead be important to combat compounded stress, which may render some cellular processes more susceptible to oxidative attack. This model may explain the requirement for fundamental cellular processes, including RNA metabolism, ribosome biogenesis, and actin cytoskeleton, when DTT is the pretreatment. These processes are unlikely to produce H2O2 tolerance, but may instead become sensitive to H2O2 attack after DTT. Indeed, a prior study showed that ribosomal proteins are particularly prone to DTT-induced aggregation when the thioredoxin defense system is abolished [71]. Furthermore, the recent links between genes involved in RNA catabolism, P-body formation, and normal ER function [65]–[70], [72]–[74] may explain why mild stresses that trigger the UPR uniquely require these genes for subsequent stress survival.
Previous studies showed that <1% of genes required for long-term NaCl treatment showed increased expression in response to that condition [37]. Here we found that up to 24% of genes necessary for subsequent H2O2 tolerance are induced during the NaCl pretreatment. This enrichment was not true for all pretreatments, particularly DTT exposure during which most important genes showed reduced expression. Nonetheless, it suggests that gene expression is more closely correlated with, but still a relatively poor predictor of, a gene's requirement in acquired stress tolerance. The low correlation could reflect pervasive post-transcriptional regulation during mild-stress treatment. Alternatively, many genes necessary but not sufficient for acquired H2O2 tolerance may not be actively regulated in response to stress, but rather are already present at a required basal activity. Many other genes are induced during pretreatment but unnecessary for survival of either the mild stress or severe H2O2 treatment ([37], [75] and this study). It is likely that subsets of these genes (including many in the ESR) are important for acquiring resistance to other secondary stresses [31].
Beyond the mechanisms that underlie acquired stress resistance, a remaining question is its purpose. Cross-stress protection may simply be a byproduct of the overlapping effects of two stresses. For example, very high doses of NaCl can produce oxidative damage [76]; it is possible that oxidative defense mechanisms are induced during mild NaCl treatment to prepare for severe NaCl treatment rather than H2O2. Alternatively, cells may have evolved to prepare for impending stress if successive stressful environments are frequently encountered in nature, or if surviving infrequent compound stresses provides a sufficient selective advantage [31], [77], [78]. In E. coli, stresses that occur sequentially as bacteria travel through the gastrointestinal tract can provide cross-stress protection, and this acquired resistance is lost if cells evolve in the absence of sequential exposure [77], [78]. The role of acquired stress resistance in nature will become clearer as more is learned about the natural ecology of yeast. In the meantime, acquired stress resistance serves as an important phenotype to provide new insights into stress resistance and the complex relationship between phenotype and environment.
Materials and Methods
Strains and growth conditions
Strains used are shown in Table S3. We used normalized pools of the diploid homozygous non-essential yeast knockout (YKO) collection (BY4743 MATa/α his3Δ1/his3Δ1 leu2 Δ0/leu2Δ0 lys2Δ0/LYS2 MET15/met15Δ0 ura3Δ0/ura3Δ0 background), diploid heterozygous essential YKO collection (BY4743), and DAmP yeast library (derived from BY4741/Y6683 strains (MATa/α his3Δ1/his3Δ1 leu2Δ0/leu2Δ0 ura3Δ0/ura3Δ0 met15Δ0/met15Δ0 CYH2+/cyh2) [39]. GFP-marked strain AGY0231 (MATa ura3Δ0 lys2Δ0 dORF-SWH1::Ptdh3-yEGFP-Tcyc1) used for competition experiments was graciously provided by Barry Williams. Unless otherwise noted, cells were grown in batch culture in YPD (1% yeast extract, 2% peptone, 2% glucose) at 30°C.
Acquired stress resistance selection experiments
Pools of the three deletion collections were grown separately for ∼7.5 generations in YPD to an optical density (OD600) of 0.3. The cultures were then mixed such that each strain was roughly equally represented in the resulting pools of barcodes, and an aliquot was removed as the unstressed, time 0 control sample (Sample 0). One fraction of the culture was outgrown in YPD for 10 generations (with washing and dilution in fresh YPD medium after 2, 5, and 10 generations to maintain log-phase growth). The resulting outgrown culture was collected as Sample 1 and compared to Sample 0 to identify slow-growing strains (Figure 1). A second fraction of the original culture was exposed to 0.4 mM H2O2 for two hours, centrifuged, washed, returned to fresh YPD, and outgrown 10 generations before collection as Sample 2. The remainder of the culture was exposed to one of three primary stresses, including 1 h exposure to 0.7 M NaCl, 2 h exposure to 2.5 mM DTT, or 1 h growth at 40°C after a 30°C culture was collected and resuspended in fresh 40°C medium. Following primary-stress exposure cells were centrifuged, washed, and a fraction of the culture was outgrown for 10 generations and collected as each respective Sample 3. The remaining culture was exposed to secondary stress (1.0 mM or 1.2 mM H2O2) for 2 hours, then centrifuged, washed and returned to fresh YPD medium for 10 generations outgrowth before collection as Sample 4 (1.0 mM H2O2) or Sample 4A (1.2 mM H2O2). Comparing Sample 4 to Sample 3 identified strains with a defect in acquiring resistance to severe H2O2 after mild-stress pretreatment. Most experiments were performed in at least duplicate from start to finish, with the exception of essential-gene mutants done once for NaCl and heat shock pretreatments (see Table S4). All samples were characterized by microarray analysis, and two of each experiment were also interrogated by deep sequencing (see below). Selections were also performed as above using 20 µM Tunicamycin (Sigma) for four hours as a primary stress, in biological duplicate. Fitness scores for all experiments are listed in Table S5.
Barcode microarrays
The barcode microarrays were performed as in Pierce et al. 2007. Briefly, ‘up’ and ‘down’ barcodes were separately amplified from genomic DNA using common primers, and resulting PCR products were hybridized to 16K TAG4 barcode microarrays (Affymetrix part no. 511331) as previously described [79]. Each ‘up’ and ‘down’ barcode tag is represented five times on the array, for a total of 10 measurements per deletion strain. For each array, signal intensities of ‘up’ and for ‘down’ tags were averaged separately, excluding clear outliers. Quantile normalization was performed across all arrays and done separately for averaged ‘up’ and ‘down’ tag signal intensities. Following normalization, a correction factor was applied to correct for feature saturation [79], and the relative abundance of each barcoded deletion strain was then determined. Negative log2 ratios of strain abundance signify decreased strain fitness. Strains with positive log2 values, which may represent a fitness advantage, were generally not confirmed in validation assays and are not discussed further (data not shown).
Barcode sequencing
The sequencing protocol was adapted from [80]. Barcodes were amplified from genomic DNA using primers that included the common YKO barcode amplification sequences, the Illumina anchor sequences, and multiplex indexes for sample multiplexing (sequences available in Table S6), using Herculase II Fusion DNA polymerase (Agilent). PCR products of ∼150 bp were purified using the e-Gel gel purification system and SybrGreen (Invitrogen). Two biological replicates of Samples 1, 2, 3, and 4 were sequenced for each selection.
Finished libraries were sent to the University of Wisconsin Sequencing Facility for Illumina sequencing. Briefly, quality and quantity of the finished libraries were assessed using an Agilent DNA 1000 series chip assay and QuantIT PicoGreen dsDNA Kit (Invitrogen), respectively. Each library was standardized to 10 µM, then 12 uniquely indexed upstream barcode libraries and 12 uniquely indexed downstream barcode libraries were pooled in each lane (representing ‘up’ and ‘down’ tags from 12 different Samples above). Cluster generation was performed using a standard Cluster Kit (v4) and the Illumina Cluster Station, or a standard cBot Kit (v4) and the Illumina cBot. Single-end 50 bp or 75 bp reads were collected using standard SBS kits (v4) and SCS 2.5 software, on an Illumina Genome Analyzer IIx. Images were analyzed using the standard Illumina Pipeline, version 1.5, and the sequence reads were mapped back to YKO barcode sequences using custom scripts, allowing one mismatch per 6-bp multiplex sequence and two mismatches per 20 bp ‘up’ or ‘down’ tag (discarding mismatches that did not map uniquely).
Validation
Survival experiments were performed as in [31], except viability was scored using an EasyCyte flow cytometer (Millipore) and LIVE/DEAD Fungalite Yeast Viability Kit (Invitrogen). Briefly, cells were exposed to mild stress or mock treatment in flasks, collected by centrifugation, and resuspended in YPD. Cells were then exposed to 12 doses of H2O2 (0–5 mM) in 96-well plates for 2 hours, and incubated with dye for 30–60 min before fluorescence was scored at each H2O2 dose. Survival scores shown in the figures were based on the fraction of pretreated cells that survived each dose, minus the fraction of mock treated cells that survived that dose – a single score was then computed as the sum of those values across all doses of secondary stress [31]. CTT1 results were also validated in an independent ctt1Δ::URA3 strain that was then complimented with CTT1 on a plasmid (data not shown).
GFP competition experiments were performed by competing a GFP-marked strain against either wild-type BY4741 or single-gene deletion strains from the haploid yeast deletion library (Open Biosystems). Cells were grown separately overnight to early log phase (OD600 0.3) and mixed at a 1∶5 ratio of GFP-marked: unmarked cells. Mixed cultures were exposed to no stress, primary stress alone, 0.4 mM H2O2 alone, or primary stress followed by 1.0 mM H2O2 for 2 hours; cells were then washed with YPD and grown 10 generations in YPD in the absence of stress. Relative strain abundance was inferred based on the proportion of GFP-expressing cells assayed using the EasyCyte flow cytometer (Millipore) before and after outgrowth [81]. The proportion of GFP-expressing cells when mixed with a given deletion strain was compared to proportion of GFP-expressing cells mixed with wild-type BY4741; an increase in the number of GFP-marked: mutant cells, relative to the wild-type control, indicated a competition defect in the deletion strain of interest.
Western blots
BY4741 and ubi4Δ cells were grown at least 7 generations to early log phase and a sample of each culture was collected for an unstressed control. The culture was exposed to 30–40°C heat shock or 0.7 M NaCl for one hour, then washed and exposed to 1.0 mM H2O2 for 2 hours, and washed and outgrown in YPD. Cell samples were collected before and after pretreatment and at 1 h and 8 h during YPD outgrowth.
Whole-cell lysate was assayed by Western analysis with the following primary antibodies: polyclonal rabbit anti-ubiquitin (kindly provided by R. Vierstra), monoclonal mouse anti-FLAG (F3165, Sigma), polyclonal rabbit anti-TAP (CAB1001, Open Biosystems), or monoclonal mouse anti-actin (MAB1501; Millipore,Billerica, MA). Secondary antibodies included LiCor (Lincoln, NE) IRDye 680LT goat anti-rabbit (926–68021) or goat anti-mouse (926–32210) fluorescent antibodies. Blots were visualized and analyzed using an Odyssey Infrared Imaging System v3.0.21. Free ubiquitin, FLAG-Ctt1p, or Gpx2-TAPp were normalized to actin in each lane.
Quantitative PCR
Quantitative PCR was done as previously described [82] using iQSYBR Green Supermix (Bio-Rad, Hercules, CA) on a MyiQ2 Bio-Rad Cycler. Primers spanned a 3' 100–200 bp region of each ORF. Cycle numbers were normalized ERV25 mRNA as an internal control unaffected by stress.
Data analysis
For each strain, fitness after a particular treatment was taken as the log2 change in strain abundance between each Sample and its corresponding control (see Figure 1). Strains were identified as defective in acquired stress resistance if they met the following criteria in the microarray and/or sequencing experiments: 1) Strains displayed a fitness defect in response to 1.0 mM H2O2 following primary treatments (e.g. Sample 4 compared to Sample 3) that was at least 1 standard deviation from the mean of all strains. 2) The fitness defect following primary-stress treatment alone (Sample 3 versus Sample 1) was <1 standard deviation from the mean of all strains. 3) The fitness defect in response to 0.4 mM H2O2 (Sample 2 versus Sample 1) was less than the defect in 1.0 mM (or 1.2 mM) H2O2. 4) These criteria were true in at least two replicates. These stringent lists were expanded by manually adding strains whose fitness phenotypes were highly correlated with identified mutants. For libraries with only one replicate (for example, the heterozygous deletion collection used in the NaCl selection), identified strains were required to meet the stringent criteria for both 1.0 mM and 1.2 mM H2O2 doses or in corresponding mutants from multiple libraries (e.g. a significant defect in both the heterozygous-gene deletion strain and DAmP strain).
Clustering was done in Cluster 3.0 (http://bonsai.hgc.jp/~mdehoon/software/cluster/software.htm) using hierarchical clustering and uncentered Pearson correlation as the metric [83]. Enrichment of gene functional categories was performed using the hypergeometric distribution in Excel or the program Funspec [84] with Bonferroni-corrected p-values <0.01 taken as significant. Network graphs were constructed using Cytoscape 2.8 [85]. Genetic and physical interactions were downloaded from BioGRID release 3.0.66 [86]. Enrichment of genetic or physical interactions, compared to random chance, was determined for 1000 randomly sampled networks with the same number of genes and assessing the number of trials with equal or greater number of total pairwise connections to the observed networks. Genes with defects in acquired stress resistance were defined as induced or repressed during pretreatments if the average (n> = 3) expression change was greater than 1.5X higher or lower than unstressed cells 45 min after 0.7 M NaCl or 15 min after a 30–37°C heat shock [31], or 90 min after 2.5 mM DTT (S. Topper and APG, unpublished).
Supporting Information
Zdroje
1. GaschAP 2007 Comparative genomics of the environmental stress response in ascomycete fungi. Yeast
2. ParsellDALindquistS 1993 The function of heat-shock proteins in stress tolerance: degradation and reactivation of damaged proteins. Annu Rev Genet 27 437 496
3. CraigEAGambillBDNelsonRJ 1993 Heat shock proteins: molecular chaperones of protein biogenesis. Microbiol Rev 57 402 414
4. EstruchF 2000 Stress-controlled transcription factors, stress-induced genes and stress tolerance in budding yeast. FEMS Microbiol Rev 24 469 486
5. JamiesonDJ 1998 Oxidative stress responses of the yeast Saccharomyces cerevisiae. Yeast 14 1511 1527
6. HohmannSMagerPWillemH 2003 Yeast Stress Responses. Heidelberg Springer-Verlag
7. MagerWHFerreiraPM 1993 Stress response of yeast. Biochem J 290 (Pt1) 1 13
8. HahnGMNingSCElizagaMKappDSAndersonRL 1989 A comparison of thermal responses of human and rodent cells. Int J Radiat Biol 56 817 825
9. LouYYousefAE 1997 Adaptation to sublethal environmental stresses protects Listeria monocytogenes against lethal preservation factors. Appl Environ Microbiol 63 1252 1255
10. MaleckKLevineAEulgemTMorganASchmidJ 2000 The transcriptome of Arabidopsis thaliana during systemic acquired resistance. Nat Genet 26 403 410
11. SchenkPMKazanKWilsonIAndersonJPRichmondT 2000 Coordinated plant defense responses in Arabidopsis revealed by microarray analysis. Proc Natl Acad Sci U S A 97 11655 11660
12. ChinnusamyVSchumakerKZhuJK 2004 Molecular genetic perspectives on cross-talk and specificity in abiotic stress signalling in plants. J Exp Bot 55 225 236
13. DurrantWEDongX 2004 Systemic acquired resistance. Annu Rev Phytopathol 42 185 209
14. CharngYYLiuHCLiuNYHsuFCKoSS 2006 Arabidopsis Hsa32, a novel heat shock protein, is essential for acquired thermotolerance during long recovery after acclimation. Plant Physiol 140 1297 1305
15. HeckerMPane-FarreJVolkerU 2006 SigB-Dependent General Stress Response in Bacillus subtilis and Related Gram-Positive Bacteria. Annu Rev Microbiol
16. JeongWSJunMKongAN 2006 Nrf2: a potential molecular target for cancer chemoprevention by natural compounds. Antioxid Redox Signal 8 99 106
17. KenslerTWWakabayashiNBiswalS 2007 Cell survival responses to environmental stresses via the Keap1-Nrf2-ARE pathway. Annu Rev Pharmacol Toxicol 47 89 116
18. MatsumotoHHamadaNTakahashiAKobayashiYOhnishiT 2007 Vanguards of paradigm shift in radiation biology: radiation-induced adaptive and bystander responses. J Radiat Res (Tokyo) 48 97 106
19. ScholzHFranzMHeberleinU 2005 The hangover gene defines a stress pathway required for ethanol tolerance development. Nature 436 845 847
20. LuDMaulikNMoraruIIKreutzerDLDasDK 1993 Molecular adaptation of vascular endothelial cells to oxidative stress. Am J Physiol 264 C715 722
21. RaffaghelloLLeeCSafdieFMWeiMMadiaF 2008 Starvation-dependent differential stress resistance protects normal but not cancer cells against high-dose chemotherapy. Proc Natl Acad Sci U S A 105 8215 8220
22. ZhaoHSapolskyRMSteinbergGK 2006 Interrupting reperfusion as a stroke therapy: ischemic postconditioning reduces infarct size after focal ischemia in rats. J Cereb Blood Flow Metab 26 1114 1121
23. ZhaoH 2009 Ischemic postconditioning as a novel avenue to protect against brain injury after stroke. J Cereb Blood Flow Metab 29 873 885
24. CaustonHCRenBKohSSHarbisonCTKaninE 2001 Remodeling of yeast genome expression in response to environmental changes. Mol Biol Cell 12 323 337
25. GaschAPSpellmanPTKaoCMCarmel-HarelOEisenMB 2000 Genomic expression programs in the response of yeast cells to environmental changes. Mol Biol Cell 11 4241 4257
26. Martinez-PastorMTMarchlerGSchullerCMarchler-BauerARuisH 1996 The Saccharomyces cerevisiae zinc finger proteins Msn2p and Msn4p are required for transcriptional induction through the stress response element (STRE). Embo J 15 2227 2235
27. SchmittAPMcEnteeK 1996 Msn2p, a zinc finger DNA-binding protein, is the transcriptional activator of the multistress response in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A 93 5777 5782
28. GornerWDurchschlagEMartinez-PastorMTEstruchFAmmererG 1998 Nuclear localization of the C2H2 zinc finger protein Msn2p is regulated by stress and protein kinase A activity. Genes Dev 12 586 597
29. AmorosMEstruchF 2001 Hsf1p and Msn2/4p cooperate in the expression of Saccharomyces cerevisiae genes HSP26 and HSP104 in a gene - and stress type-dependent manner. Mol Microbiol 39 1523 1532
30. EstruchFCarlsonM 1993 Two homologous zinc finger genes identified by multicopy suppression in a SNF1 protein kinase mutant of Saccharomyces cerevisiae. Mol Cell Biol 13 3872 3881
31. BerryDBGaschAP 2008 Stress-activated genomic expression changes serve a preparative role for impending stress in yeast. Mol Biol Cell 19 4580 4587
32. TempleMDPerroneGGDawesIW 2005 Complex cellular responses to reactive oxygen species. Trends Cell Biol 15 319 326
33. ThorpeGWFongCSAlicNHigginsVJDawesIW 2004 Cells have distinct mechanisms to maintain protection against different reactive oxygen species: oxidative-stress-response genes. Proc Natl Acad Sci U S A 101 6564 6569
34. KelleyRIdekerT 2009 Genome-wide fitness and expression profiling implicate Mga2 in adaptation to hydrogen peroxide. PLoS Genet 5 e1000488 doi:10.1371/journal.pgen.1000488
35. TuckerCLFieldsS 2004 Quantitative genome-wide analysis of yeast deletion strain sensitivities to oxidative and chemical stress. Comp Funct Genomics 5 216 224
36. LeeWSt OngeRPProctorMFlahertyPJordanMI 2005 Genome-wide requirements for resistance to functionally distinct DNA-damaging agents. PLoS Genet 1 e24 doi:10.1371/journal.pgen.0010024
37. GiaeverGChuAMNiLConnellyCRilesL 2002 Functional profiling of the Saccharomyces cerevisiae genome. Nature 418 387 391
38. WinzelerEAShoemakerDDAstromoffALiangHAndersonK 1999 Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science 285 901 906
39. YanZCostanzoMHeislerLEPawJKaperF 2008 Yeast Barcoders: a chemogenomic application of a universal donor-strain collection carrying bar-code identifiers. Nat Methods 5 719 725
40. de la Torre-RuizMAMozo-VillariasAPujolNPetkovaMI 2010 How budding yeast sense and transduce the oxidative stress signal and the impact in cell growth and morphogenesis. Curr Protein Pept Sci 11 669 679
41. Yeger-LotemERivaLSuLJGitlerADCashikarAG 2009 Bridging high-throughput genetic and transcriptional data reveals cellular responses to alpha-synuclein toxicity. Nat Genet 41 316 323
42. JinYHDunlapPEMcBrideSJAl-RefaiHBushelPR 2008 Global transcriptome and deletome profiles of yeast exposed to transition metals. PLoS Genet 4 e1000053 doi:10.1371/journal.pgen.1000053
43. TaiSLSnoekILuttikMAAlmeringMJWalshMC 2007 Correlation between transcript profiles and fitness of deletion mutants in anaerobic chemostat cultures of Saccharomyces cerevisiae. Microbiology 153 877 886
44. WarringerJEricsonEFernandezLNermanOBlombergA 2003 High-resolution yeast phenomics resolves different physiological features in the saline response. Proc Natl Acad Sci U S A 100 15724 15729
45. ChangMBellaouiMBooneCBrownGW 2002 A genome-wide screen for methyl methanesulfonate-sensitive mutants reveals genes required for S phase progression in the presence of DNA damage. Proc Natl Acad Sci U S A 99 16934 16939
46. BirrellGWBrownJAWuHIGiaeverGChuAM 2002 Transcriptional response of Saccharomyces cerevisiae to DNA-damaging agents does not identify the genes that protect against these agents. Proc Natl Acad Sci U S A 99 8778 8783
47. LewisJAElkonIMMcGeeMAHigbeeAJGaschAP 2010 Exploiting natural variation in Saccharomyces cerevisiae to identify genes for increased ethanol resistance. Genetics 186 1197 1205
48. Flattery-O'BrienJCollinsonLPDawesIW 1993 Saccharomyces cerevisiae has an inducible response to menadione which differs from that to hydrogen peroxide. J Gen Microbiol 139 501 507
49. WestfallPJPattersonJCChenREThornerJ 2008 Stress resistance and signal fidelity independent of nuclear MAPK function. Proc Natl Acad Sci U S A 105 12212 12217
50. SchullerCBrewsterJLAlexanderMRGustinMCRuisH 1994 The HOG pathway controls osmotic regulation of transcription via the stress response element (STRE) of the Saccharomyces cerevisiae CTT1 gene. Embo J 13 4382 4389
51. ClotetJEscoteXAdroverMAYaakovGGariE 2006 Phosphorylation of Hsl1 by Hog1 leads to a G2 arrest essential for cell survival at high osmolarity. Embo J 25 2338 2346
52. AlexanderMRTyersMPerretMCraigBMFangKS 2001 Regulation of cell cycle progression by Swe1p and Hog1p following hypertonic stress. Mol Biol Cell 12 53 62
53. BelliGGariEAldeaMHerreroE 2001 Osmotic stress causes a G1 cell cycle delay and downregulation of Cln3/Cdc28 activity in Saccharomyces cerevisiae. Mol Microbiol 39 1022 1035
54. EscoteXZapaterMClotetJPosasF 2004 Hog1 mediates cell-cycle arrest in G1 phase by the dual targeting of Sic1. Nat Cell Biol 6 997 1002
55. EscoteXMirandaMRodriguez-PorrataBMasACorderoR 2011 The stress-activated protein kinase Hog1 develops a critical role after resting state. Mol Microbiol
56. WuC 1995 Heat shock transcription factors: structure and regulation. Annu Rev Cell Dev Biol 11 441 469
57. GaschAP 2002 The Environmental Stress Response: a common yeast response to environmental stresses. HohmannSMagerP Yeast Stress Responses Heidelberg Springer-Verlag 11 70
58. HedbackerKCarlsonM 2008 SNF1/AMPK pathways in yeast. Front Biosci 13 2408 2420
59. LiaoXButowRA 1993 RTG1 and RTG2: two yeast genes required for a novel path of communication from mitochondria to the nucleus. Cell 72 61 71
60. LipsonDRazTKieuAJonesDRGiladiE 2009 Quantification of the yeast transcriptome by single-molecule sequencing. Nat Biotechnol 27 652 658
61. AveryAMAverySV 2001 Saccharomyces cerevisiae expresses three phospholipid hydroperoxide glutathione peroxidases. J Biol Chem 276 33730 33735
62. HochstrasserM 1996 Ubiquitin-dependent protein degradation. Annu Rev Genet 30 405 439
63. FinleyDOzkaynakEVarshavskyA 1987 The yeast polyubiquitin gene is essential for resistance to high temperatures, starvation, and other stresses. Cell 48 1035 1046
64. TkaczJSLampenO 1975 Tunicamycin inhibition of polyisoprenyl N-acetylglucosaminyl pyrophosphate formation in calf-liver microsomes. Biochem Biophys Res Commun 65 248 257
65. GalaoRPChariAAlves-RodriguesILobaoDMasA 2010 LSm1-7 complexes bind to specific sites in viral RNA genomes and regulate their translation and replication. Rna 16 817 827
66. NoueiryAODiezJFalkSPChenJAhlquistP 2003 Yeast Lsm1p-7p/Pat1p deadenylation-dependent mRNA-decapping factors are required for brome mosaic virus genomic RNA translation. Mol Cell Biol 23 4094 4106
67. Restrepo-HartwigMAhlquistP 1999 Brome mosaic virus RNA replication proteins 1a and 2a colocalize and 1a independently localizes on the yeast endoplasmic reticulum. J Virol 73 10303 10309
68. HollienJWeissmanJS 2006 Decay of endoplasmic reticulum-localized mRNAs during the unfolded protein response. Science 313 104 107
69. HollienJLinJHLiHStevensNWalterP 2009 Regulated Ire1-dependent decay of messenger RNAs in mammalian cells. J Cell Biol 186 323 331
70. DeckerCJParkerR 2006 CAR-1 and trailer hitch: driving mRNP granule function at the ER? J Cell Biol 173 159 163
71. RandJDGrantCM 2006 The thioredoxin system protects ribosomes against stress-induced aggregation. Mol Biol Cell 17 387 401
72. PrinzAHartmannEKaliesKU 2000 Sec61p is the main ribosome receptor in the endoplasmic reticulum of Saccharomyces cerevisiae. Biol Chem 381 1025 1029
73. JonikasMCCollinsSRDenicVOhEQuanEM 2009 Comprehensive characterization of genes required for protein folding in the endoplasmic reticulum. Science 323 1693 1697
74. WiedmannBPrehnS 1999 The nascent polypeptide-associated complex (NAC) of yeast functions in the targeting process of ribosomes to the ER membrane. FEBS Lett 458 51 54
75. ChenYFeldmanDEDengCBrownJADe GiacomoAF 2005 Identification of mitogen-activated protein kinase signaling pathways that confer resistance to endoplasmic reticulum stress in Saccharomyces cerevisiae. Mol Cancer Res 3 669 677
76. PastorMMProftMPascual-AhuirA 2009 Mitochondrial function is an inducible determinant of osmotic stress adaptation in yeast. J Biol Chem 284 30307 30317
77. TagkopoulosILiuYCTavazoieS 2008 Predictive behavior within microbial genetic networks. Science 320 1313 1317
78. MitchellARomanoGHGroismanBYonaADekelE 2009 Adaptive prediction of environmental changes by microorganisms. Nature 460 220 224
79. PierceSEDavisRWNislowCGiaeverG 2007 Genome-wide analysis of barcoded Saccharomyces cerevisiae gene-deletion mutants in pooled cultures. Nat Protoc 2 2958 2974
80. SmithAMHeislerLEMellorJKaperFThompsonMJ 2009 Quantitative phenotyping via deep barcode sequencing. Genome Res 19 1836 1842
81. BreslowDKCameronDMCollinsSRSchuldinerMStewart-OrnsteinJ 2008 A comprehensive strategy enabling high-resolution functional analysis of the yeast genome. Nat Methods 5 711 718
82. LeeMVTopperSEHublerSLHoseJWengerCD 2011 A dynamic model of proteome changes reveals new roles for transcript alteration in yeast. Mol Syst Biol 7 514
83. EisenMBSpellmanPTBrownPOBotsteinD 1998 Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci U S A 95 14863 14868
84. RobinsonMDGrigullJMohammadNHughesTR 2002 FunSpec: a web-based cluster interpreter for yeast. B M C Bioinformatics 3 35
85. ShannonPMarkielAOzierOBaligaNSWangJT 2003 Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13 2498 2504
86. StarkCBreitkreutzBJRegulyTBoucherLBreitkreutzA 2006 BioGRID: a general repository for interaction datasets. Nucleic Acids Res 34 D535 539
Štítky
Genetika Reprodukční medicína
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2011 Číslo 11- Akutní intermitentní porfyrie
- Souvislost haplotypu M2 genu pro annexin A5 s opakovanými reprodukčními ztrátami
- Příjem alkoholu a menstruační cyklus
- Doporučení pro diagnostiku a léčbu akutních jaterních porfyrií
- Doc. Miloš Kubánek: Nemocní se srdeční amyloidózou jsou často skryti a sledováni pod jinými diagnózami
-
Všechny články tohoto čísla
- De Novo Origins of Human Genes
- Duplication Hotspots Are Associated with Late-Replicating Regions of the Genome
- De Novo Origin of Human Protein-Coding Genes
- Cyclin D/CDK4 and Cyclin E/CDK2 Induce Distinct Cell Cycle Re-Entry Programs in Differentiated Muscle Cells
- Short Day–Mediated Cessation of Growth Requires the Downregulation of AINTEGUMENTALIKE1 Transcription Factor in Hybrid Aspen
- Physiological IRE-1-XBP-1 and PEK-1 Signaling in Larval Development and Immunity
- Role of Pirh2 in Mediating the Regulation of p53 and c-Myc
- Signatures of Environmental Genetic Adaptation Pinpoint Pathogens as the Main Selective Pressure through Human Evolution
- FOXO Regulates Organ-Specific Phenotypic Plasticity In
- Heritable Epigenetic Variation among Maize Inbreds
- Foxn1 Regulates Lineage Progression in Cortical and Medullary Thymic Epithelial Cells But Is Dispensable for Medullary Sublineage Divergence
- Attenuation of the Sensing Capabilities of PhoQ in Transition to Obligate Insect–Bacterial Association
- A Novel Protein LZTFL1 Regulates Ciliary Trafficking of the BBSome and Smoothened
- Activation of Bmp2-Smad1 Signal and Its Regulation by Coordinated Alteration of H3K27 Trimethylation in -Induced Senescence
- Histone H3K56 Acetylation, CAF1, and Rtt106 Coordinate Nucleosome Assembly and Stability of Advancing Replication Forks
- The SUN Protein Mps3 Is Required for Spindle Pole Body Insertion into the Nuclear Membrane and Nuclear Envelope Homeostasis
- Evidence-Based Annotation of Gene Function in MR-1 Using Genome-Wide Fitness Profiling across 121 Conditions
- Effect of Host Species on the Distribution of Mutational Fitness Effects for an RNA Virus
- Pch2 Acts through Xrs2 and Tel1/ATM to Modulate Interhomolog Bias and Checkpoint Function during Meiosis
- SOX9 Governs Differentiation Stage-Specific Gene Expression in Growth Plate Chondrocytes via Direct Concomitant Transactivation and Repression
- from the Aphid : A Missing Link from Facultative to Obligate Insect Endosymbiont
- Recessive Antimorphic Alleles Overcome Functionally Redundant Loci to Reveal Function in Flowers and Meristems
- Over-Expression of DSCAM and COL6A2 Cooperatively Generates Congenital Heart Defects
- Consequences of Eukaryotic Enhancer Architecture for Gene Expression Dynamics, Development, and Fitness
- Distinct Genetic Architectures for Male and Female Inflorescence Traits of Maize
- Capture of MicroRNA–Bound mRNAs Identifies the Tumor Suppressor miR-34a as a Regulator of Growth Factor Signaling
- For Male , Sperm Activation Is a “Just-in-Time” Event
- PcG Complexes Set the Stage for Epigenetic Inheritance of Gene Silencing in Early S Phase before Replication
- The Gene Contains Hotspots for L1 Endonuclease-Dependent Insertion
- Relative Burden of Large CNVs on a Range of Neurodevelopmental Phenotypes
- Multiple Means to the Same End: The Genetic Basis of Acquired Stress Resistance in Yeast
- Genome-Wide Crossover Distribution in Meiosis Reveals Sex-Specific Patterns along Chromosomes
- TRY-5 Is a Sperm-Activating Protease in Seminal Fluid
- Homologs of Retinoblastoma-Associated Protein 46/48 Associate with a Histone Deacetylase to Act Redundantly in Chromatin Silencing
- Genetic Interaction Maps in Reveal Functional Crosstalk among Cell Envelope Biogenesis Pathways
- The ERI-6/7 Helicase Acts at the First Stage of an siRNA Amplification Pathway That Targets Recent Gene Duplications
- PBX1 Genomic Pioneer Function Drives ERα Signaling Underlying Progression in Breast Cancer
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Evidence-Based Annotation of Gene Function in MR-1 Using Genome-Wide Fitness Profiling across 121 Conditions
- De Novo Origins of Human Genes
- Capture of MicroRNA–Bound mRNAs Identifies the Tumor Suppressor miR-34a as a Regulator of Growth Factor Signaling
- TRY-5 Is a Sperm-Activating Protease in Seminal Fluid
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



