-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaActin-Related Protein Arp6 Influences H2A.Z-Dependent and -Independent Gene Expression and Links Ribosomal Protein Genes to Nuclear Pores
Actin-related proteins are ubiquitous components of chromatin remodelers and are conserved from yeast to man. We have examined the role of the budding yeast actin-related protein Arp6 in gene expression, both as a component of the SWR1 complex (SWR-C) and in its absence. We mapped Arp6 binding sites along four yeast chromosomes using chromatin immunoprecipitation from wild-type and swr1 deleted (swr1Δ) cells. We find that a majority of Arp6 binding sites coincide with binding sites of Swr1, the catalytic subunit of SWR-C, and with the histone H2A variant Htz1 (H2A.Z) deposited by SWR-C. However, Arp6 binding detected at centromeres, the promoters of ribosomal protein (RP) genes, and some telomeres is independent of Swr1 and Htz1 deposition. Given that RP genes and telomeres both show association with the nuclear periphery, we monitored the ability of Arp6 to mediate the localization of chromatin to nuclear pores. Arp6 binding is sufficient to shift a randomly positioned locus to nuclear periphery, even in a swr1Δ strain. Arp6 is also necessary for the pore association of its targeted RP promoters possibly through cell cycle-dependent factors. Loss of Arp6, but not Htz1, leads to an up-regulation of these RP genes. In contrast, the pore-association of GAL1 correlates with Htz1 deposition, and loss of Arp6 reduces both GAL1 activation and peripheral localization. We conclude that Arp6 functions both together with the nucleosome remodeler Swr1 and also without it, to mediate Htz1-dependent and Htz1-independent binding of chromatin domains to nuclear pores. This association is shown to have modulating effects on gene expression.
Published in the journal: . PLoS Genet 6(4): e32767. doi:10.1371/journal.pgen.1000910
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1000910Summary
Actin-related proteins are ubiquitous components of chromatin remodelers and are conserved from yeast to man. We have examined the role of the budding yeast actin-related protein Arp6 in gene expression, both as a component of the SWR1 complex (SWR-C) and in its absence. We mapped Arp6 binding sites along four yeast chromosomes using chromatin immunoprecipitation from wild-type and swr1 deleted (swr1Δ) cells. We find that a majority of Arp6 binding sites coincide with binding sites of Swr1, the catalytic subunit of SWR-C, and with the histone H2A variant Htz1 (H2A.Z) deposited by SWR-C. However, Arp6 binding detected at centromeres, the promoters of ribosomal protein (RP) genes, and some telomeres is independent of Swr1 and Htz1 deposition. Given that RP genes and telomeres both show association with the nuclear periphery, we monitored the ability of Arp6 to mediate the localization of chromatin to nuclear pores. Arp6 binding is sufficient to shift a randomly positioned locus to nuclear periphery, even in a swr1Δ strain. Arp6 is also necessary for the pore association of its targeted RP promoters possibly through cell cycle-dependent factors. Loss of Arp6, but not Htz1, leads to an up-regulation of these RP genes. In contrast, the pore-association of GAL1 correlates with Htz1 deposition, and loss of Arp6 reduces both GAL1 activation and peripheral localization. We conclude that Arp6 functions both together with the nucleosome remodeler Swr1 and also without it, to mediate Htz1-dependent and Htz1-independent binding of chromatin domains to nuclear pores. This association is shown to have modulating effects on gene expression.
Introduction
Genomic DNA is complexed with histones and non-histone proteins to form chromatin, which is organized into active and inactive domains within the interphase nucleus [1]–[3]. Histone tail modifications, chromatin compaction, and the subnuclear positioning of chromatin domains contribute epigenetic information that helps to determine gene expression patterns. While the enzymology of histone modification has been well characterized, little is known about the mechanisms that determine the spatial organization of chromatin in interphase nuclei.
In both vertebrates and yeast, transcriptionally inactive heterochromatin is enriched around the nucleolus or at the nuclear envelope (NE). In vertebrates, perinuclear anchoring appears to require the nuclear lamina, while in yeast integral proteins of the inner nuclear membrane tether repressed chromatin domains peripherally [reviewed in 4]. Recent work has shown that in addition to silent heterochromatic loci, some euchromatic yeast genes are found at the NE as well. Indeed, inducible budding yeast genes such as INO1, GAL1, and HXK1 form a stable association with the nuclear pore complex (NPC) upon activation. In some cases, this interaction ensures maximal expression and fine-tuning of induction rates [5]–[7]. The up-regulated X chromosome in male flies may also be associated with nuclear pores [8], as are the highly transcribed ribosomal protein (RP) genes of yeast [9].
Besides nuclear pore proteins, little is known about the components that position active chromatin domains within the nucleus. Nuclear actin and myosin, as well as myosin-like and actin-related proteins have been proposed as candidates that could contribute to the organization of transcription in the interphase nucleus [8], [10]–[17]. Indeed, actin itself is not only found as part of the filamentous cytoskeleton, but in various large chromatin modifying complexes, which are exclusively nuclear.
In all organisms from yeast to man, the actin family includes a number of proteins that are structurally similar to actin, called actin-related proteins or ARPs. The yeast S. cerevisiae alone harbors ten ARP genes, numbered from 1 to 10, with ascending degrees of dissimilarity to actin [18]–[19]. Arp1-3 and Arp10 are cytoplasmic and help regulate cytoskeletal structures, while the other six (Arp4 to Arp9) are nuclear proteins [20]–[23]. Nuclear ARPs, like nuclear actin, are often found in ATP-dependent chromatin modifying complexes that shift or displace nucleosomes, or in complexes that acetylate histone tails (e.g. NuA4 complex) [reviewed in 23]. Exactly how ARPs contribute to nucleosome modification, however, is unknown.
Arp6 is an evolutionarily conserved nuclear ARP [24]–[26]. The budding yeast Arp6, along with two other actin-family members, Act1 and Arp4, are part of the 14-component SWR1 chromatin remodeling complex (SWR-C), which is called SRCAP or Snf2-Related CREB-binding Activator Protein in mammals [27]. In addition to the ATPase subunit, Swr1, SWR-C includes Swc1/Fun36, Swc2/Vps72, Swc3, Swc4/God1, Swc5/Aor1, Swc6/Vps71, Swc7, Yaf9, Bdf1, Rvb1, and Rvb2 [28]. The SWR-C holocomplex can exchange H2A with its variant H2A.Z (Htz1 in budding yeast) in assembled nucleosomes [29], [30]. Arp6 appears to form a subcomplex with Swc2, Swc3, and Swc6, and helps bridge this subcomplex with a second one containing Swr1, to form the functional SWR-C [28]. Since Swc2 component is responsible for binding Htz1, the Arp6-mediated bridging is necessary for Htz1 deposition [28], [29].
Nucleosomes containing acetylated H2A.Z are specifically enriched at promoters in higher eukaryotes, and consistently, Htz1 is found in nucleosomes flanking the nucleosome-free region located at transcription start sites in yeast [31]–[36]. Additionally, Htz1 prevents the spreading of heterochromatin proteins in subtelomeric domains [35], [37], [38]. In vitro analyses have indicated that Arp6 contributes to transcriptional regulation by mediating Htz1 deposition [28], yet empirical evidence probing Arp6 function in vivo is very limited.
In addition to a role in chromatin modulating complexes, some nuclear ARPs are thought to have functions beyond that of the remodeling complex to which they belong. In other cases, such as Arp4 (BAF53 in mammals), ARP proteins are involved in multiple complexes, so that the phenotypes associated with ARP gene deletion are more extensive than those provoked by loss of a complex-specific ATPase subunit. Indeed, ARP4 is essential for viability in yeast, and the protein is a component of both INO80 - and SWR-C remodeling complexes and the NuA4 histone acetyltransferase, which carry out distinct nuclear functions [22], [39], [40]. Intriguingly, biochemical fractionation suggests that even these three complexes do not account for the entire nuclear complement of Arp4 [22], [39]. Other support for independent functions for ARPs comes from genome-wide screens for synthetic lethality. The 125 gene deletions that are lethal for cells lacking Arp6, for instance, are not necessarily lethal for cells lacking the Swr1 ATPase subunit [41]. Finally, the human Arp8 (hArp8) was implicated in mitotic chromosome phenotypes that could not be attributed to the hINO80 chromatin remodeling complex to which it belongs [42].
Here we have localized Arp6 along budding yeast chromosomes, both in the absence and presence of Swr1. We find that most Arp6 co-localizes with Swr1, being enriched in the promoters of divergently transcribed genes. This correlates with the deposition of the histone H2A variant H2A.Z/Htz1, and is consistent with the proposal that Arp6, as part of SWR-C, contributes to transcriptional regulation by exchanging H2A for Htz1 [33], [35]. Intriguingly, however, Arp6 binds some promoters in a Swr1-independent manner, including promoters of ribosomal protein genes. Indeed, transcript measurements show that Arp6 alters RP gene regulation independently of Swr1-mediated Htz1 deposition. We find that Arp6 can relocate chromatin to the NE independently of Swr1, and that arp6 deletion reduces the association of RP genes with the NPC. This leads to a slight elevation in RP gene expression. We argue that Arp6 not only modulates local chromatin organization by facilitating Htz1 deposition, but also contributes to long-range chromatin organization that can fine-tune expression levels independently of SWR-C.
Results
Co-localization of Arp6 and Swr1 at yeast promoter regions
We determined the localization of Arp6 and Swr1 along budding yeast chromosomes by chromatin immunoprecipitation using high-density microarray chips (ChIP-chip assay). A 3FLAG epitope tag was introduced at the 3′ end of the genomic copy of either ARP6 or SWR1, and the functionality of the fusion constructs was confirmed by monitoring growth rates and sensitivity to DNA damaging agents (Figure S1). Genomic DNA fragments (mean size ≅ 100 bp) were recovered, together with either Arp6 or Swr1 by anti-FLAG immunoprecipitation from formaldehyde-fixed and sonicated cells grown asynchronously in glucose-containing media (YPD). The fragments isolated by ChIP, as well as those in the total extract, were labeled with fluorescent dyes and hybridized to a high-density oligonucleotide array covering chromosomes 3, 4, 5, and 6R at either 100-bp or 300-bp resolution. With eleven 25-nucleotide probes for each 300-bp locus, the reliability of signal strength could be evaluated robustly by calculating the p-value for each locus (p<0.025) [43]. Reliability of the log2 ratio of the ChIP fraction recovery to the supernatant fraction was scored, and allowed discrimination of significant positive (yellow bars) and negative signals (open bars) for binding with overall resolution of ∼300 bp [43] (data available at www.ncbi.nlm.nih.gov/geo under the access number GSE9213).
Arp6 binding sites were found widely dispersed along chromosomes 3, 4, 5, and 6R, occasionally spreading over regions of several kb (Figures S2, S3, S5, and Figure 1, respectively). Among 8441 detectable chromosomal loci, 1498 loci were evaluated as positive for Arp6 binding (Table 1). Among the Arp6-binding sites, 71% also tested positive for Swr1, suggesting that Arp6 co-localizes with Swr1 at the majority of its chromosomal binding sites. The frequency of co-localization does not vary significantly among the chromosomes tested (Table 1). In Figure 1A, vertical arrows indicate the top ten peaks (signal log ratio >0.8) for Arp6 binding on Chr 6. Interestingly, one of the Arp6 peaks encompasses the centromere (arrow I), and a second is subtelomeric, containing unique sequences adjacent to the TG-rich repeats (arrow X). Arp6 binding sites generally coincided well with those of Swr1 (Figure 1A and 1B), except in subtelomeric zones where Arp6 binds alone (arrow X). Co-localization patterns were similar on all chromosome arms analysed (Figures S2, S3, S5). A closer examination of Arp6 promoter binding on Chr 6, indicates that Arp6 and Swr1 bind preferentially at the promoters of divergently transcribed genes (Figure S6, gray shade). These sites also contain Htz1 [35], and the coincidence of all three signals suggests that SWR-C stays bound after depositing Htz1 (H2A.Z), presumably to modulate promoter accessibility.
Fig. 1. Localization of Arp6 and Swr1 on chromosome 6R. 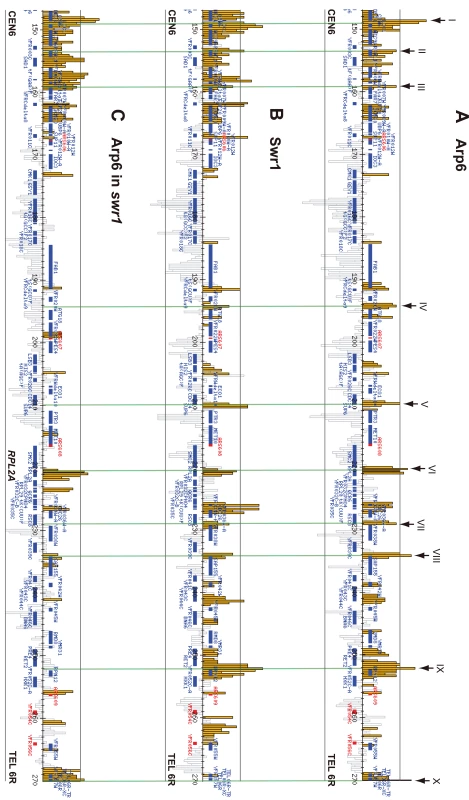
Vertical bars represent the binding ratio of proteins at each locus. Filled bars with yellow and black were determined to be significantly positive, for loci covering 300-bp or 100-bp regions, respectively. Open bars were not significantly positive. The scale of the vertical axis is log2 and the upper and lower horizontal lines represent 1.0 and -1.0, respectively. The central horizontal axis shows kilobase units. (A) Localization of Arp6-FLAG in SWR1 wild-type cells. (B) Localization of Swr1-FLAG. (C) Localization of Arp6-FLAG in swr1 cells. Vertical arrows (I to X) in A and vertical green lines represent the position of the highest ten clusters consisting of at least two continuous binding loci in both Arp6-and Swr1-FLAG ChIP. Tab. 1. Correlation of localization of Arp6 and Swr1 on budding yeast chromosomes. 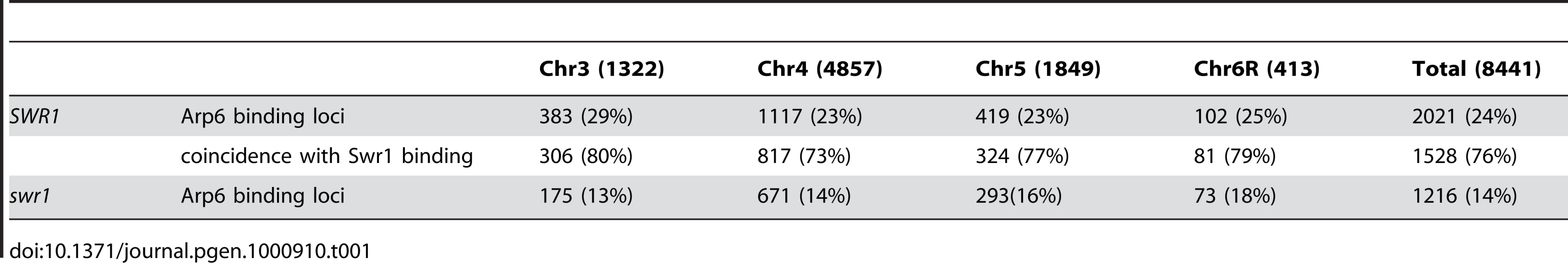
In conclusion, our ChIP-chip analysis suggests that Arp6 binds chromatin as a component of SWR-C at most euchromatic sites, and in particular in the promoters of divergently transcribed genes. However, at centromeres, some telomeres and select promoters, Arp6 appears to bind independently of Swr1 (Figure 1C, Table S1, and below).
Comparison of Arp6 - and Swr1-containing complexes
The fact that Arp6 ChIP recovered the sites where Swr1 does not bind suggests that Arp6 may associate with other nuclear proteins or complexes, although to date it has only been reported to be a component of SWR-C. To test this possibility, we analyzed the native molecular masses of Swr1 - and Arp6-containing complexes using gel filtration chromatography (Figure 2). Swr1 is recovered almost exclusively in fractions of ∼1–2 MDa (Figure 2A, second panel, lanes 5 and 6), and fractionates similarly to the catalytic ATPase of the SWI/SNF remodeler Snf2 (Figure 2A, first panel). In the presence of Swr1, Arp6 was distributed in both the 1–2 MDa Swr1-containing fractions, and in fractions that correspond to a molecular weight of 100–250 kDa (Figure 2A, third panel). In this latter fraction the complexes are still likely to be larger than the 57-kDa Arp6 monomer. Upon deletion of Swr1, the presence of Arp6 in the high MW fractions was significantly reduced (Figure 2A, fourth panel, and Figure 2B), and its abundance in the lower MW fractions increased. Our observations are consistent with the observation that Swr1 serves as platform for the assembly of SWR-C subunits, and that in the absence of Swr1, Arp6 only retains association with Swc6 [28], [44]. Nonetheless, a small amount of Arp6 was still recovered in a high MW complex in the swr1Δ strain (>1 MDa, Figure 2A, fourth panel). This may reflect participation of Arp6 in another large complex, albeit one of lower abundance. Quantitation of Arp6 recovery in both wild-type and swr1Δ cells, suggests that 30% of Arp6 is part of SWR-C or another large complex (Figure 2B), while the majority of Arp6 self-dimerizes or forms a complex with other small proteins. The nature of these is unknown, but the only reported partners of Arp6 in a swr1Δ strain are Swc6 and nucleosomes [28].
Fig. 2. Sizing column Superose6 analysis of Arp6-complexes. 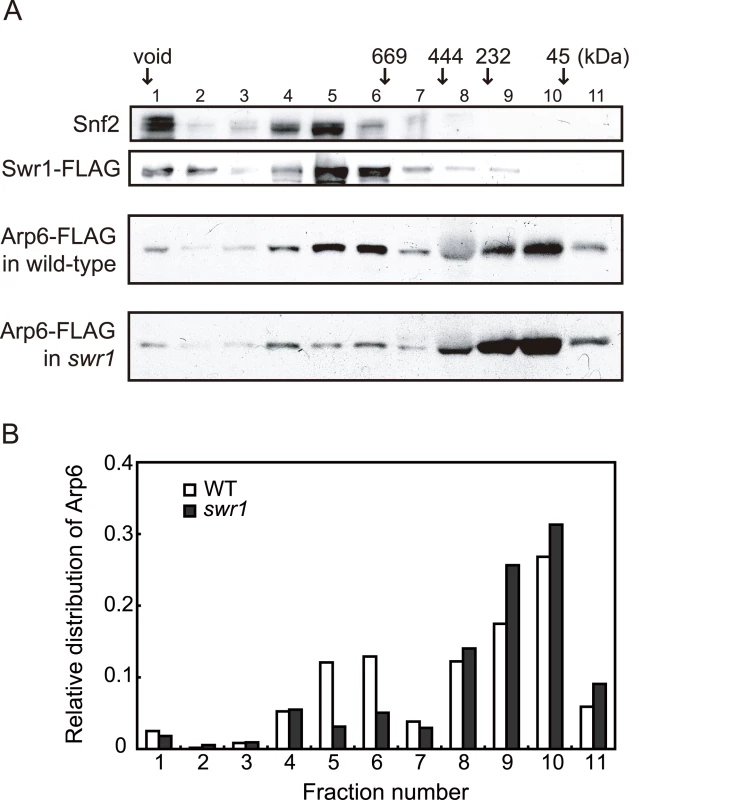
(A) Extracts from cells expressing Arp6-FLAG under wild-type and swr1 background were fractionated on a Superose6 column, and Arp6-FLAG in the fractions was detected on a Western blot with an anti-FLAG antibody (two bottom panels). The extract from cells expressing Swr1-FLAG was fractionated as well, and endogenous Snf2 and Swr1-FLAG were detected with an anti-Snf2 antibody and an anti-FLAG antibody, respectively (two top panels). The numbers of fractions and the positions of molecular mass standards are shown over the panels. (B) The intensity of Arp6-FLAG in the fractions were quantified, and relative distributions of Arp6 in the fractions of wild-type (WT) and swr1 cells were shown. SWR-C independent chromatin association of Arp6
To examine the ability of Arp6 to bind chromatin with or without Swr1, we fractionated yeast cells into a chromatin and a soluble protein fraction, using the well-established TritonX-100 lysis procedure [45]. This analysis confirmed that the majority of Arp6 is associated with chromatin in wild-type cells (Figure 3B). This is also true for tightly bound chromatin proteins like Ino80, topoisomerase II and ORC (Figure 3B and data not shown). Consistent with the finding that most Arp6-binding sites coincide with those of Swr1, we found that the association of Swr1 with chromatin required the presence of Arp6 (Figure 3B, arp6, lane Chr). This is unlikely to reflect an indirect effect on chromatin, since the association of Ino80 or topoisomerase II with chromatin was unaffected by arp6 deletion (Figure 3B). In contrast, a large fraction of Arp6 (39% compared to wild-type) remained associated with chromatin even in the absence of Swr1 (Figure 3B, swr1, lane Chr). This result suggests that the physical association of Arp6 with chromatin is at least in part independent of SWR-C, and is consistent with the ChIP-chip data which show partially non-overlapping distributions of Arp6 and Swr1 (Figure 1).
Fig. 3. Arp6 partitions between soluble and insoluble chromatin fractions. 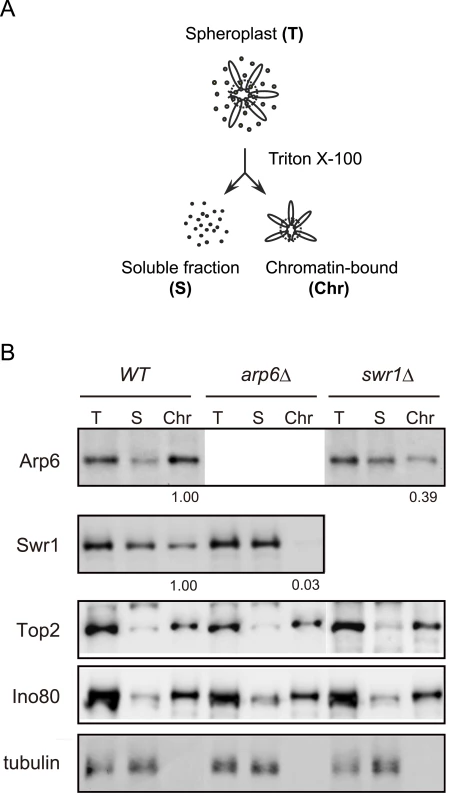
(A) The fractionation protocol is shown. Yeast spheroplasts from appropriate strains were lysed with Triton-X100. A gentle centrifugation step separates a supernatant containing the bulk of cellular proteins from a chromatin pellet. (B) Wild-type, swr1, and arp6 cells were subjected to the fractionation protocol described in (A), and the spheroplast (T), soluble fraction (S), and chromatin-bound (Chr) samples were probed using Western blot for Arp6-FLAG, Swr1-FLAG, topoisomerase II (Top2), the enzymatic component of the INO80 chromatin remodeling complexes (Ino80-MYC), and the soluble non-chromatin protein, tubulin. Numbers under panels show the ratios of chromatin-bound Arp6 and Swr1 in the mutants relative to WT. Their intensities were normalized with chromatin-preparation efficiencies obtained by quantification of the Western blot for Ino80. Detection of the Arp6 binding loci in the absence of Swr1
To elucidate the SWR-C-dependent and -independent functions of Arp6, we performed ChIP-chip analysis for Arp6 in a strain lacking SWR1 entirely (swr1Δ). Consistent with the reduced level of chromatin-bound Arp6 (Figure 3B), fewer Arp6-binding sites were found in swr1Δ cells (Table 1). The major binding sites lost were those where both Arp6 and Swr1 colocalize in 5′ promoter regions (Figure 1, Figure S6, and Table 1), including the intergenic region 5′ of the SWR1 gene itself (Figure S4).
To quantify the effect of swr1 deletion on Arp6 binding patterns, we compared the log ratio of each Arp6-binding site in wild-type and swr1Δ cells (Figure S7A and S7B) with the log ratio of Swr1 binding at that locus. Generally, at the sites where both Arp6 and Swr1 were bound in wild-type cells, Arp6 binding was lost in swr1Δ cells (Figure 1 and Figures S2, S3, S5, S6). However, the overall distribution of Arp6 in the swr1Δ strain changes; notably, values increased at sites where Swr1 was not bound in the wild-type background (Figure S7B). This argues that in addition to an overall reduction in Arp6 binding, preferred Arp6 binding positions were altered in the absence of Swr1. This change in Arp6 binding suggests that SWR-C either competes for a limiting pool of Arp6 or alters chromatin such that some Arp6 binding sites are inaccessible, possibly reflecting indirect effects of Htz1 deposition.
Importantly, a subfraction of Arp6 binding sites persist in both wild-type and swr1Δ cells (see Figure 1). This argues, consistent with the fractionation data (Figure 3B), that Arp6 binds a subset of chromosomal loci independently of SWR-C, even in wild-type cells. Examples of this are RP gene promoters (e.g. RPL2A, Figure 1, arrow VI) and the Tel6R subtelomeric zone, which contains the inducible gene HXK1 (Figure 1, arrow X). Indeed, Swr1-independent binding of Arp6 was enriched generally in a number of subtelomeric regions (Table S1). Despite the difficulty of analyzing subtelomeric domains on microarrays due to the presence of repetitive sequences, persistent Arp6 binding could be confirmed at Tel6R, Tel3L, Tel3R, and Tel4R in the absence of Swr1 (Figure 1, Figures S2, S3).
Involvement of Arp6 in the expression of RP genes independently of H2A.Z-deposition
To examine the independent contributions of Arp6 and Swr1 to gene expression, we performed a yeast whole-genome microarray with wild-type, arp6Δ, and swr1Δ cells (Table S2) (details available at www.ncbi.nlm.nih.gov/geo under the access number GSE17780). This expression microarray analysis was repeated at least three times for each strain, and the statistical differences were determined by t-test. Changes with p<0.05 were considered significant. We found a larger number of genes to be differentially regulated in both arp6Δ and swr1Δ cells (Figure 4A). When we compared the misregulated genes between the two mutants, we found that 87 out of 506 genes repressed in swr1Δ (see down-regulated in swr1, Figure 4A) are also down-regulated in arp6Δ cells (17% overlap), and 56 out of 375 genes induced in swr1Δ (see up-regulated in swr1, Figure 4A) are also up-regulated in arp6Δ cells (15% overlap; Figure 4A and Table S2). This overlap of down - or up-regulated genes between swr1Δ and arp6Δ strains was less than that reported for down - or up-regulated genes between swr1 and htz1 mutants (44% and 38%, respectively) [30]. Our expression data are consistent with our biochemical and ChIP analyses, which suggest that a majority of Arp6 is not recovered with SWR-C by sedimentation analysis (Figure 2B). The divergence of misregulated genes in the two mutants suggests that Arp6 can influence gene expression independently of SWR-C.
Fig. 4. Transcript analysis in swr1 and arp6 mutant cells. 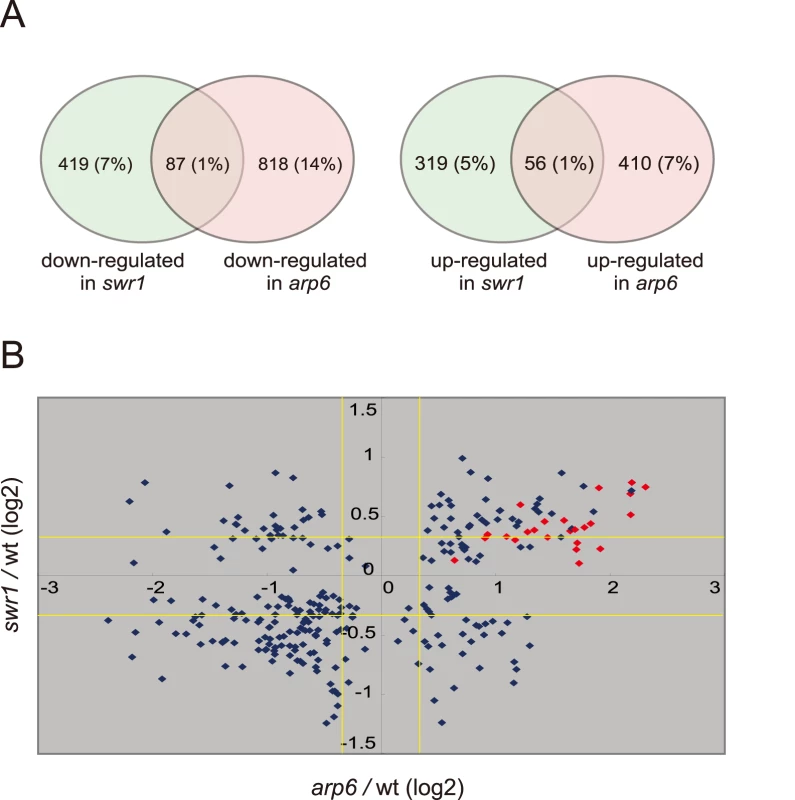
Microarray analysis was repeated at least three times for swr1Δ and arp6Δ strain, and the statistical differences were determined using a t-test. p<0.05 was considered significant. (A) The Venn diagrams illustrate the degree of overlap between genes whose RNA levels were changed by 1.25-fold in arp6 and swr1 mutants. Numbers correspond to misregulated genes in the total genes. Relative numbers of the misregulated genes (%/total number of yeast gene) are indicated in parentheses. (B) Genes whose transcriptional changes were statistically significant (p<0.05) both in arp6 and swr1 cells were plotted according to their log2 ratios. The yellow lines show the 1.25-fold changes. The red diamonds represents RP genes. To extract more information on the differential effects of Arp6 and Swr1 in gene expression, we compared the degree of change in transcript level for each gene whose misregulation was significant (p<0.05) in both arp6Δ and swr1Δ strains (Figure 4B). Intriguingly, the degree of transcriptional change as a consequence of arp6 deletion was greater than that provoked by swr1 deletion. On the other hand, the deposition of Htz1 to promoters were similarly impaired in arp6Δ and swr1Δ cells, as previously reported [28], [29] (Figure S8). This analysis further indicates that Arp6 contributes to gene expression not only through Htz1 deposition, but also through a Swr1-independent mechanism.
Strikingly, among the 40 most up-regulated genes in arp6Δ cells we found 21 ribosomal protein genes (Table 2). Some of these RP genes were also modestly up-regulated in swr1Δ cells. Consistently, the ChIP-chip analysis in chromosomes 3, 4, 5, and 6R (Figures S2, S3, S5, and Figure 1, respectively) revealed that Arp6 and Swr1 bind to 25, and respectively 24, of the 27 RP genes on these chromosomes (Table S3). Importantly, and in contrast to most other euchromatic loci, Arp6 remained bound to all of these RP genes (including RPS16B, RPL13A, RPP1A, RPL31A, and RPL2A), even after deletion of SWR1 (Figure 1, Figure 5, Figures S2, S3, S5, and Table S3). When we compared the transcription of RP genes between arp6Δ and swr1Δ cells, we find these genes more significantly up-regulated by loss of Arp6 than by loss of Swr1 (Figure 4B, red diamonds, and Table S4). From this we conclude that an Arp6-dependent, but Swr1 - and Htz1-independent, mechanism modulates RP gene expression.
Fig. 5. Swr1-independent binding of Arp6 to RP genes. 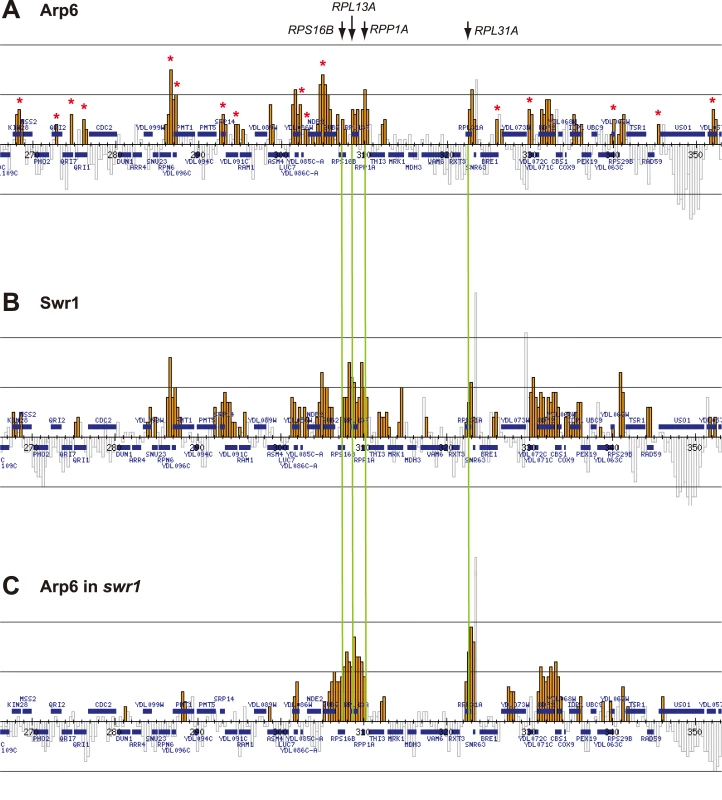
Vertical bars represent the binding ratio of proteins in each locus. The binding of Arp6-FLAG (top), Swr1-FLAG (middle), and Arp6-FLAG in swr1 cells (bottom) in the region 266K-353K of Chr 4L were compared. The positions of the RP genes (RPS16B, RPL13A, RPP1A, and RPL31A) in the region are shown with arrows and green lines. Red asterisks indicate those Arp6-gene promoter bindings which disappeared in the absence of Swr1. Tab. 2. Genes markedly up-regulated in arp6 cells. 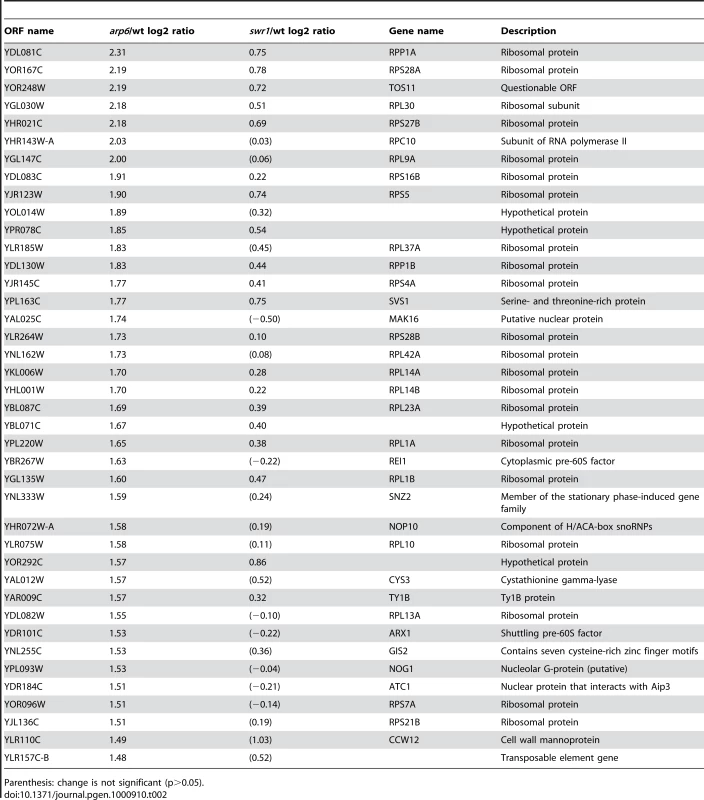
Parenthesis: change is not significant (p>0.05). Consistent with our analysis, it was reported earlier that Htz1 is excluded from RP genes [34], [35]. Moreover, microarray analyses have shown that the absence of Htz1 does not have any significant effects on RP gene expression [30], [37], [46]. To confirm this, we examined the expression of the relevant RP genes by quantitative rtPCR. We could confirm that transcript levels were not significantly altered by loss of Htz1, yet were increased in arp6Δ (Figure 6A). In contrast, other Arp6-bound promoters that are known to be regulated by SWR-C mediated deposition of Htz1 (e.g. GAL1 [31], [47]) showed a reduction or delay in induction by galactose that was similar in both htz1Δ and arp6Δ cells (Figure 6B). This could be extended to several non-inducible genes, to which Arp6 binds in Swr1-dependent manner such as RDS1 (YCR106W) and UBX3 (YDL091C) (Figure S9). These genes, like the inducible GAL1, showed a similar decrease in expression in both htz1Δ and arp6Δ cells (Figure S9, filled and gray bars, respectively). Our observations argue that Arp6 is involved in gene expression in both the Htz1-dependent and Htz1-independent pathways. Moreover, Arp6 binding can both increase and lower transcript levels: at GAL1, where Htz1 is deposited in an Arp6 - and Swr1-dependent manner, expression is less efficient in the absence of Arp6 or Htz1 deposition, while at RP genes, where Arp6 binding is independent of Swr1 and Htz1, its absence increases expression levels.
Fig. 6. Quantitative analysis of transcripts in cells lacking Arp6 (arp6) and H2A.Z (htz1) using RT–PCR. 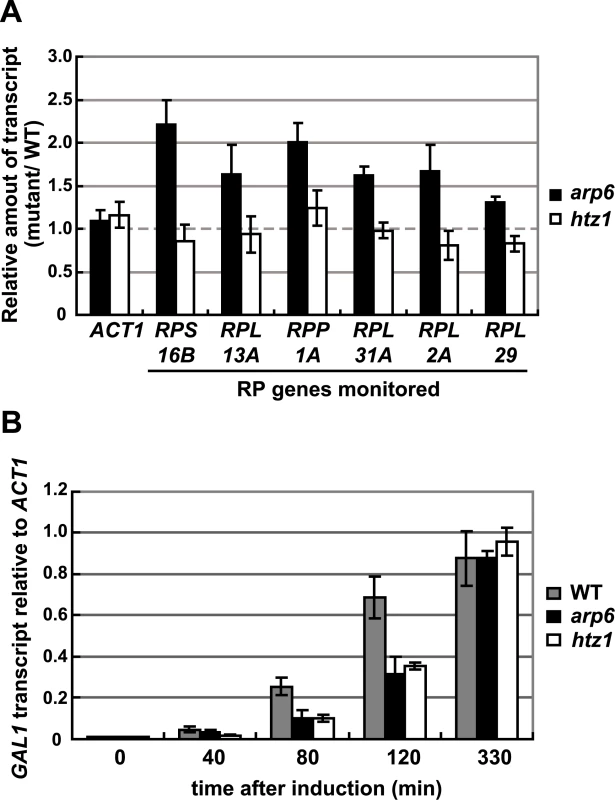
(A) The same amount of total RNA from wild-type, arp6, and htz1 cells was analyzed using quantitative RT–PCR by using primer sets specific for each of the RP genes. The ACT1 gene was analyzed as a control. The relative amount of the transcript of the genes in arp6 and htz1 to wild-type is shown by filled and open bars, respectively. The data points represent the mean ± SD for at least three experiments. (B) Total RNA was prepared from wild-type, arp6, and htz1 cells (gray, filled, and open bars, respectively) at the indicated times after induction of GAL1 expression in galactose. The amounts of GAL1 transcript in these strains were analyzed using quantitative RT–PCR, and shown as relative amounts compared to ACT1 transcript. The data points represent the mean ± SD for at least three experiments. Arp6 mediates relocalization of chromatin in a Swr1-independent manner
Recent studies have suggested that not only local changes in chromatin organization, but also long-range chromatin organization can influence gene expression. Genome-wide ChIP-chip analysis for nuclear pore components has shown that RP genes associate with components of the NPC [9]. Given that Arp6 associates with most RP genes in the absence of Swr1, we wondered whether the Arp6-specific effect on transcription might be mediated through an interaction of the target gene with nuclear pores. To examine this possibility, we made use of an assay that scores for the ability of a protein fused to LexA to shift a randomly positioned chromosomal locus to the nuclear periphery [2]. This assay has been used to identify protein domains that are sufficient for interaction with structural components of the NE. The locus chosen is a constitutively expressed gene near ARS607 (PES4) at which we have inserted 4 LexA binding sites and a lac operator array that allows visualization with GFP-LacI (Figure 7A) [2]. Proteins that are to be tested for perinuclear relocalization activity are expressed as LexA fusions in a strain expressing GFP-Nup49 to tag the NE. Unlike the expression of LexA alone (Figure 7B, LexA), the LexA-Arp6 fusion protein led to an enrichment of the PES4 locus in the outermost nuclear zone (zone 1) in both G1 - and S-phase cells (Figure 7B, LexA-Arp6 in WT). Importantly, the relocalization activity of LexA-Arp6 was independent of Swr1 (Figure 7B, LexA-Arp6 in swr1). Expression of LexA alone does not shift the position of the tagged locus, allowing us to conclude that LexA targeted Arp6 is sufficient to favor the association of a chromatin locus with the NE. This is independent of Htz1 deposition, since it requires the catalytic activity of Swr1. We note that there is a low level of endogenous Arp6 detected near the PES4 locus in swr1 mutant cells (Figure 1), yet this is insufficient to tether a significant fraction of the sites to the NE (see LexA alone).
Fig. 7. Perinuclear anchoring activity of Arp6. 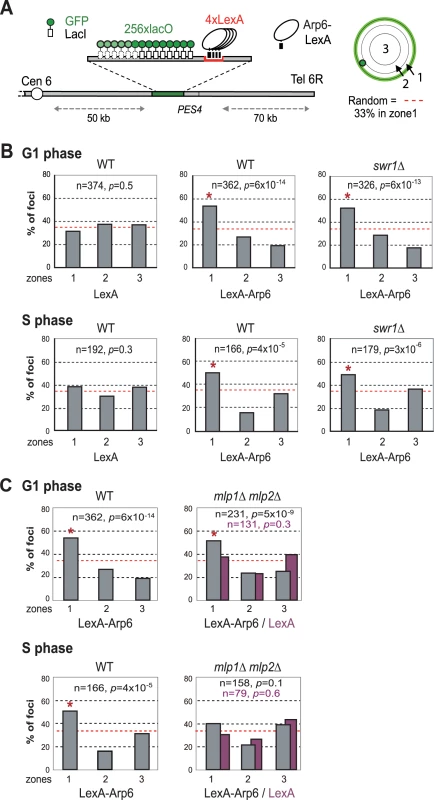
(A) The ability of a LexA fusion to relocate the lacO-tagged PES4 locus bearing LexA binding sites was tested using a strain GA-1461. PES4 is located 70 kb and 50 kb from Tel6R and Cen6, respectively. The lacO array was visualized by binding a GFP-LacI fusion, and the nuclear envelope is visualized through a Nup49-GFP fusion. The focal plane in which the GFP spot was brightest was used to monitor distances, which were reported as a ratio to nuclear diameter. These values were binned into one of three concentric zones of equal surface and are presented as percentage of total spots scored. The position of PES4 was mapped in wild-type (WT) and swr1 cells expressing LexA alone or a LexA-Arp6 fusion. (B) Arp6 can relocalize an internal chromatin locus (PES4) to the nuclear periphery. Cells were classified as G1 (unbudded) or S phase (budded with spherical nucleus). (C) Mlp1/Mlp2 is required for the perinuclear anchoring by Arp6 in S-phase cells. The position of the lacO arrays was scored on PES4::lacO tagged cells expressing LexA (purple) or LexA-Arp6 (grey). The bar graphs show the percentage of spots (y-axis) per zone (x-axis) for G1-phase (upper panels) and S-phase (lower panels) cells. The number of cells analyzed (n) and the confidence values (p) for the χ2 analysis between random and test distributions are indicated. The red dashed horizontal line at 33% indicates a random distribution, and zone 1 distributions that are significantly different from random are indicated by an asterisk (p<0.05). Arp6 is required for the association of chromatin with the NPC
To confirm that the Arp6-bound locus associates with nuclear pores, as opposed to other perinuclear sites, we performed the relocalization assay in a strain that expresses a nuclear pore protein Nup133 that lacks its N-terminal domain (nup133ΔN) [48]. In this mutant, functional NPCs cluster on one side of the nucleus allowing us to monitor whether a LexA-Arp6 targeted locus moves to pores or to other sites on the nuclear envelope (Figure 8A). Compared to cells expressing LexA alone, the LYS2 locus bound by LexA-Arp6 not only accumulated in the nuclear peripheral zone like the PES4 locus (49% in zone 1 vs 34% for LexA alone in G1-phase cells, data not shown) but also colocalized significantly with clustered NPC (Figure 8A; 22.4%, n = 322, p<0.01). Previous studies have shown that a randomly distributed tagged locus would coincide with a pore cluster in 9% of the cells, while a locus that has a predisposition to be perinuclear (i.e. 60% occupation of zone 1) would coincide with a pore cluster in 10% of cell scored [49]. The 22% scored for Arp6 relocation versus the 8% scored for the control LexA is thus highly significant. It is comparable to the ∼two-fold increase in colocalization achieved by targeting LexA-Nup84 vs LexA alone [49]. This rate of colocalization suggests that a component of the NPC is able to bind Arp6.
Fig. 8. Involvement of Arp6 in intranuclear organization through the NPC. 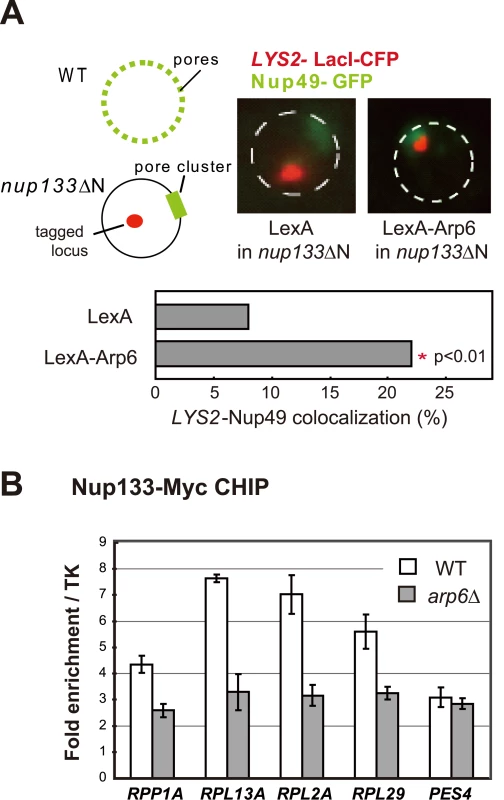
(A) The positions of lacO-tagged LYS2 (red) and of CFP-Nup49 (green) were observed in a nup133ΔN background, in which nuclear pores cluster on one side of the nucleus. Bar graphs represent the percentage of complete red-green signal overlap counted in cells expressing LexA alone or a LexA-Arp6 fusion. The confidence values (p) for the χ2 analysis between them is indicated. The predicted colocalization for a randomly positioned locus is 9% [49]. (B) Requirement of Arp6 for the interaction of RP genes with the NPC. The association of Nup133-Myc with RP genes, RPP1A, RPL13A, RPL2A, and RPL29, was quantified using ChIP analysis combined with quantitative PCR in wild-type (WT) and arp6 cells, and is plotted over a background control of the TK gene [80]. The PES4 locus was analyzed as a control. The data points represent the mean ± SD for at least three experiments. We next used quantitative ChIP analysis to test whether the loss of Arp6 influences the association of endogenous RP genes with the NPC. Immunoprecipitation of Nup133-Myc confirmed that the RP genes tested previously [9] are associated with pores and that deletion of arp6 reduces the recovery of these genes with Nup133 (Figure 8B). PES4, a randomly positioned locus with no natural affinity for nuclear pores, did not precipitate significantly with Nup133 and was unaffected by arp6 deletion (Figure 8B). From this we conclude that Arp6 is required for the RP gene-NPC interaction (Figure 8B).
We asked whether Nup133 was the only site of interaction for these genes with the nuclear envelope. In other words, we checked by lacO-tagging and scoring of subnuclear position, whether RP or GAL1 genes would lose all perinuclear localization in absence of Arp6. We found that the galactose-induced relocalization of GAL10 (which shares the GAL1 promoter) to the NE was indeed lost in S-phase arp6Δ cells (Figure 9A), as was the constitutive association of the RP gene RPL9A (Figure 9B). Inexplicably, however, the loss of association provoked by arp6 deletion was cell-cycle stage specific, arguing that an alternative, possibly redundant mechanism allowed loci to remain peripheral, although probably not associated with Nup133, in G1-phase cells. The effect was also at least partially locus - or context-specific, since a second tagged RP gene cluster at RPP1A was enriched at the nuclear periphery in both wild-type and arp6Δ strains (data not shown). Taken together our data argue that Arp6, while being sufficient to relocate loci to the NE (Figure 7), is not the only pathway that tethers active genes at nuclear pores. This was already suggested from the results from the Rosbash, Silver, Stutz, Hurt, Nehrbass, Brickner and Proudfoot laboratories, who have identified both SAGA-dependent and SAGA-independent pathways for locating active loci at nuclear pores [4]. The fact that GAL10 and RPL9A association was ablated in S-phase by arp6 deletion, suggests that a redundant pathway of anchoring functions primarily in G1 phase. Although it is unclear why transcription-regulated association with the NE, should be cell-cycle controlled, this is highly reminiscent of the distinct G1 - and S-phase specific tethering mechanisms that mediate anchoring of telomeres [2], [50] and DNA damage [49].
Fig. 9. Arp6 is required for the peripheral association of galactose-induced GAL10 and constitutively expressed RP gene RPL9A. 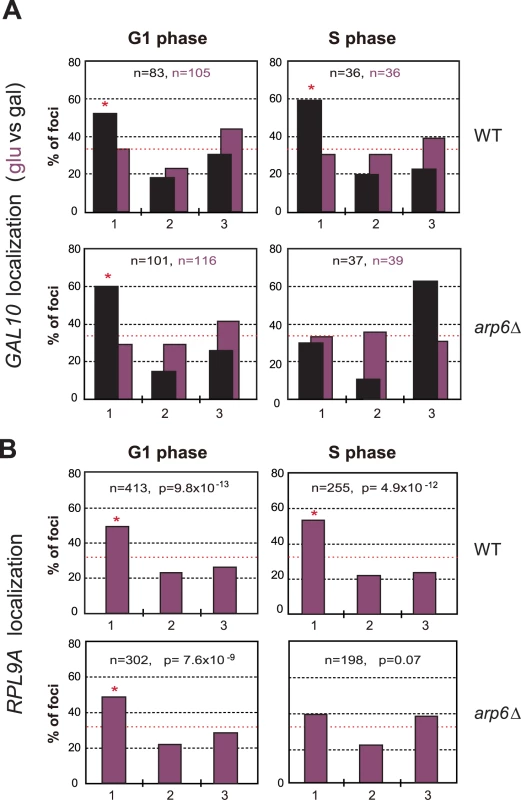
(A) The GAL1-GAL10 locus was tagged by inserting 256 lac operators in a haploid wild-type or arp6 deletion strain bearing GFP-lacI and Nup49-GFP fusions (wild-type; GA-4098, arp6; GA-6024) [81]. The position of the lacO arrays relative to the nuclear envelope was scored on images take of living cells growth either on glucose (purple) or after 2 hours of gene induction on 2% galactose (black). Three zone scoring was carried out as in Figure 7. The number of cells analyzed for each stage of the cell cycle are indicated, and the confidence values (p) for the χ2 analysis between random and test distributions on galactose are: wild-type (G1, p = 4×10−4; S, p = 6×10−4) and arp6 (G1, p = 2.7×10−8; S, p = 0.44) none of the values on glucose are significantly different from random (p>0.05). The G1-S differences on galactose are not significant in WT, but are in the arp6 mutant (p = 0.024). Note that in this analysis we omitted the rare, very small budded cells. (B) The RPL9A locus was tagged by inserting 256 lac operators in a haploid wild-type or arp6 deletion strain bearing GFP-lacI and Nup49-GFP fusions (wild-type; GA-3635, arp6; GA-5132) [81]. The position of the lacO arrays relative to the nuclear envelope was scored as in Figure 7 on image stacks taken on living cells grown on SC. Symbols and quantitation are as in A. RPL9A locus p values for test vs random distributions are: wild-type (G1, p = 9.8×10−13; S, p = 4.9×10−12) and arp6 (G1, p = 7.6×10−9; S, p = 0.07). The G1 vs S distributions in wild-type are not significantly different (p = 0.33) while in arp6 cells the difference is significant (p = 0.028). Whereas values for Zone 1 in wild-type vs arp6 cells in G1 are not significantly different (p = 0.82), the difference in mid-to-late S phase cells is (p = 0.0018). An asterisk indicates that values that have a nonrandom distribution (p<0.05). The number of cells analyzed (n) and the confidence values (p) for the χ2 analysis between random and test distributions are indicated. We next examined whether other pore-associated proteins, namely, Mlp1 and Mlp2, myosin-like proteins associated with the inner nuclear basket, are involved in either the Arp6-mediated pathway of gene anchoring. They were likely candidates due to their implication in the association of GAL10 and HSP104 with nuclear pores, through Mex67 and Yra1 [51]–[53]. To test this, the position of the tagged PES4 locus bound by LexA-Arp6 or by LexA alone was determined in strains carrying mlp1Δ mlp2Δ deletions (Figure 7C, mlp1Δ mlp2Δ). Intriguingly, we again see that LexA-Arp6 anchoring activity was dependent on Mlp1 and Mlp2 exclusively in S-phase cells. We conclude that Arp6 is able to mediate association with nuclear pores in an Mlp1/Mlp2-dependent manner, yet again our data indicate that a second pathway for Arp6 binding is functional in G1 phase. Arp6 may interact with the coiled-coil proteins Mlp1 and Mlp2 directly, although it is more likely to bind through Yra1 (see Discussion). Overall, our results support the notion that Arp6 has a role both in local chromatin modulation through H2A.Z-deposition, and in long-range chromatin organization through its ability to bind proteins associated with the NPC; an interaction which depends at least partially on the myosin-like proteins 1 and 2.
Discussion
SWR-C–dependent and–independent binding of Arp6 to chromatin
Our high resolution ChIP-chip assay has shown that Arp6 co-localizes with Swr1 at most of its euchromatic sites, presumably as a component of SWR-C [28], [30]. No conserved sequence motif for the binding of Arp6 or Swr1 could be identified (data not shown), partly because the targeted regions are relatively large and transcription factor binding sites are often degenerate. Moreover, it is possible that Arp6 is targeted by recognition of a specifically modified nucleosome and not a DNA binding factor [28]. Interestingly, the highest Arp6 occupancy was detected within a 300-bp fragment containing the start ATG codon of the SWR1 gene. This coincides with a peak of Swr1 and argues for an auto-regulatory loop for SWR1 expression (Figure S4) [35].
More generally, the ChIP-chip assay showed a coordinated enrichment for both Arp6 and Swr1 in intergenic regions particularly near the 5′ ends of divergently transcribed genes (Figure 1A and 1B). Of the top ten loci for Arp6 binding on Chr 6 (arrows, Figure 1A), five contain the start ATG codon of genes, two are located within 200 bp of an ATG codon, and two others are within 400 bp of an ATG codon. Only at telomeres are Arp6 sites more than 1 kb from the nearest ATG. The localization of Htz1 on yeast chromosomes has been previously examined in detail, and was shown to be present at the 5′ ends of most genes [33]–[35]. This suggests that Arp6 and Swr1 remain chromatin-bound at sites where they incorporate Htz1.
Unexpectedly, we also detected numerous loci that were positive for Arp6 but negative for Swr1 interaction (Figure 1A and 1B, Table 1). A large fraction of Arp6 was shown not be integrated in the SWR-C complex by gel filtration analysis. Moreover, although Swr1 association with chromatin was dependent on Arp6, about 40% of total Arp6 remained chromatin-bound in the absence of Swr1 (Figure 2 and Figure 3). Swr1 contributes not only to the catalytic activity of SWR-C, but also provides critical protein-protein contacts that maintain the integrity of the holocomplex [28]. Importantly, in a swr1 deletion strain Swc2, the Htz1-binding module of SWR-C, dissociates from Arp6 and only Swc6 remains Arp6 bound [28]. Consistently, sites that bind Arp6 in the absence of Swr1 are anti-correlated with the presence of Htz1.
ChIP-chip analysis showed that the SWR-C-independent binding of Arp6 is observed at RP genes and in some subtelomeric zones, which are both depleted for Htz1 [37], [38], [54]. Intriguingly, we have also found that the perinuclear tethering of Tel6R is impaired in arp6, but not in swr1 mutants (K.S., A.T. and S.M.G. unpublished data). The role of Arp6 at telomeres is therefore not restricted to the deposition of Htz1.
Binding of Arp6 to RP genes
The RP genes are among the most important genes for cell metabolism, and the fine-tuning of RP gene transcription responds to a variety of environmental effects, ultimately coordinated by the TORC1 complex (Target of Rapamycin) [reviewed in 55]. However, exactly how these crucial genes are regulated at a transcriptional level is unclear. Several transcription factors, including Rap1, Fhl1, and the high mobility group protein, Hmo1, have been shown to play roles in the expression of RP genes. Previous studies suggested that Rap1 may recruit Fhl1 and Hmo1 to RP promoters [56], [57]. As shown here, Arp6 binds to most of the RP genes present on the chromosomes we analyzed. Given that Rap1 has significant genetic interactions with Arp6 [58], we speculate that Arp6 may cooperate with Rap1 to regulate the association of other factors to RP gene promoters. A particular constellation of factors may also contribute to the association of these genes with nuclear pores.
Kasahara et al. [59] compared the binding of Hmo1 with those of Fhl1 and Rap1 to RP genes using ChIP-chip analysis, and divided RP genes into classes that have either Hmo1-dependent or Hmo1-independent binding of Fhl1 and Rap1. We find no correlation of either class with the presence of Arp6 (data not shown). Moreover, Arp6 is bound at the promoters of RPP1A, RPL4B, and RPP2B, which belong to a subgroup that binds neither Hmo1, Fhl1, nor Rap1 [59]. Thus, while it is possible that Arp6 influences the binding of these factors, the converse is not true. We also note that, unlike loss of Hmo1, Fhl1 or Rap1, the absence of Arp6 leads to an increase in the expression level of genes such as RPP1A (Table 2). This argues that the binding of Arp6 reduces rather than enhances RP gene expression. It is important to note that RP genes are highly expressed, and therefore even a 50% drop in expression means that the gene is still actively transcribed. Thus the localization of RP genes to pores by Arp6 binding reduces but does not eliminate expression. This is not the first report of pore association leading to reduced expression: a gene in the heat-shock family, HSP104, which is associated with the NPC by an mRNA - and Mlp1/Mlp2-dependent pathway, also had higher expression levels when its association with the NPC was impaired [51].
Arp6 is required for gene expression in a H2A.Z-dependent and -independent mechanism
We show here that the loss of Arp6 increases expression of RP genes by 1.5 - to 2-fold, in a manner independent of Htz1 and SWR-C (Figure 4, Figure 6A, and Table 2). Arp6 binds many of these RP gene promoters and is required for their tight association with the nuclear pore protein Nup133 (Figure 8B). Given that chromatin-bound Arp6 can relocate genes to pores, we can conclude that Arp6 either directly or indirectly mediates the association of RP genes with pores. In general, the RP and non-RP genes that are most activated by arp6 deletion (Table 2, e.g. YOR248W) are among those associated with NPCs [9]. This establishes for the first time a strong correlation between association with the NPC and down-regulation for a class of coordinately regulated genes.
There have been several reports showing that genes induced by non-glucose carbon sources, inositol starvation or heat shock associate with the NPC for optimal induction [5]–[7], [9], [60]. We confirm here that the association of GAL1 with the NPC is Htz1 - and Arp6 - dependent [61] (Figures S8, S10), and that in the absence of either factor, induction occurs less rapidly, although the final mRNA level is unchanged (Figure 6B). Since Arp6 is required for H2A.Z-deposition, this result is not surprising. Still, it is important to contrast this result with that observed for RP genes: at GAL1, Arp6 contributes to gene activation in an Htz1 - and Swr1-dependent manner, while at RP genes it contributes to down-regulation in an Htz1 - and Swr1-independent manner. Both Arp6-mediated activities correlate with localization to the nuclear pore.
Our data extend and support previous studies that show that final mRNA levels from galactose - and stress-induced genes decrease if association with pores is impaired [6], [7], [61]. We find that among the 40 loci most significantly down-regulated by loss of Arp6, ten are heat-shock or stress-induced genes (Table S5). We confirm by ChIP-chip that Arp6 also binds these genes, albeit less avidly than it binds RP promoters (Figures S2, S3, S5). Since our analyses were not done under conditions of stress, we propose that basal level expression is also affected by Arp6-mediated Htz1 deposition. The induction of these genes often requires the SAGA histone acetyltransferase complex which further contributes to NPC tethering through Sus1 [17], complementing the Htz1 contact [61], [62]. Thus our results confirm that Arp6-pore association acts on two pathways relevant to a number of genes: binding through SWR-C and Htz1 deposition facilitates expression of stress-induced genes, and binding independently of Htz1 at RP promoters leads to a 1.5 to 2-fold down-regulation. This allows us to propose that nuclear pores are platforms for fine-tuning gene expression and not necessarily for enhancing initiation. The control steps may involve RNA processing, export or even RNA Pol II elongation.
Two essential mRNA export proteins, Mex67 and Yra1, have been implicated in NPC-gene association and gene expression [4], [51], [52]. Mex67 and Yra1 physically interact with Mlp1/Mlp2 proteins [52], [53], [63], [64], which we show here to be required for Arp6-mediated relocation to the NE in S-phase cells (Figure 7C). Interestingly, proteome analysis has identified Yra1 as a binding partner of Arp6, but not of Swr1 or Htz1 [65]. This raises the possibility that the Mlp-Yra1-Arp6 interaction allows for the perinuclear tethering of Arp6-bound RP genes (Figure 7). A redundant mechanism in G1 phase cells may account for the fact that Arp6 continues to anchor in this phase of the cell cycle in the mlp1 mlp2 mutant.
Pore association may fine-tune RP gene expression through feed-back mechanisms that are driven by ribosomal protein levels [66]–[69]. If the loss of association of RP genes with the NPC in arp6 cells initially reduces mRNA processing and export [70], then ensuing reduction in levels of ribosomal proteins themselves may feed-back to counteract repression, enhancing RP gene expression [66]. Consistently, the RPS14B/CRY2 transcript levels are increased in cells defective in mRNA transport [68].
Arp6 and long-range chromatin organization
In addition to facilitating mRNA processing and export, the association of euchromatic domains with the NPC may facilitate the formation of nuclear subcompartments by creating boundaries [6] or by recruiting proteins required for genetic function or epigenetic control [reviewed in 4]. We note that a large fraction of Arp6 is chromatin-bound even in the absence of Swr1 (Figure 3), and that 25% of total Arp6 can be recovered in a nuclease-resistant nuclear scaffold fraction (data not shown). The association of Arp6 with an insoluble fraction of the nucleus, together with its ability to influence the localization of genes, argue that Arp6 can contribute to long-range organization of chromatin in the interphase nucleus. The ability of Arp6 to relocate chromatin to pores is not characteristic of all Arp proteins; the targeting of Arp5, a component of the INO80 chromatin remodeling complex with related molecular properties, does not change the random distribution of the tagged PES4 locus (H. van Attikum and S.M.G., personal communication). The perinuclear binding activity may thus reflect a unique domain of Arp6 or a binding partner with affinity for the NPC.
The positioning of chromatin in the interphase nucleus not only influences transcription, mRNA processing and export, but genome stability as well. Several laboratories have reported that critically short telomeres, irreparable DNA double-strand breaks and collapsed replication forks shift to the NPC for a repair pathway controlled by SUMO-dependent ubiquitin ligase [71]–[74]. Since arp6 mutants show hypersensitivity to various DNA damaging agents, Arp6 may also contribute to repair through its perinuclear relocalization activity.
We have recently analyzed chicken DT40 cells carrying a conditional knockout for Arp6, and found that the radial distribution of chromosome territories was altered in the absence of Arp6 (Ohfuchi et al., submitted). We therefore entertain the hypothesis that the contribution of Arp6 to long-range chromatin organization is evolutionarily conserved. In vertebrates there is as yet no compelling data implicating the NPC in gene expression or DNA repair, although other intranuclear structures such as PML bodies or transcription factories may replace pores in this function. We note that the reduction of human Arp4 by siRNA, unlike the loss of BRG-1, BRM, or Tip49, causes an expansion of the nuclear volume occupied by individual chromosomes (chromosome territories, [75]). While the mechanism remains obscure, this is consistent with the proposal that ARPs have roles in the long-range organization of chromatin that are independent of chromatin remodeling activities. The challenge remains to understand how cells regulate the interaction of chromatin with ARPs, nuclear actin, myosin and known structural proteins like lamins and nuclear pores.
Materials and Methods
Plasmids, strains, and yeast imaging methods
The LexA-Arp6 fusion was constructed as in Taddei et al. [7]. Yeast transformations were done using the lithium acetate procedure, and PCR-based gene deletions and tagging were performed as described [76]. The genotypes of all strains used in this study are listed in Table S6. Standard culture conditions at 30°C were used unless otherwise indicated. A 6His-3FLAG tag was fused to the C-terminus of Arp6 or Swr1 using the cassette amplified from pU6H3FLAG (gift from Dr. De Antoni) [43]. Live fluorescence microscopy and quantification was performed according to Hediger et al. [77] and Taddei et al. [2].
Chromatin immunoprecipitation (ChIP)–chip analysis
A chromosome III, IV, V, and VI right-arm high-density oligonucleotide chip was produced by Affymetrix Custom Express Service (SC3456a520015F, P/N, Affymetrix). Sequence and position of oligonucleotides on the microarrays are available from Affymetrix. ChIP was carried out as previously described [43] with a few modifications. Yeast cells were grown in 200 ml YPD medium for 12 hr at 30°C, cross-linked, and disrupted using a multi-beads shocker (MB400C, Yasui Kikai), which was able to keep cells precisely at lower than 6°C during disruption by Zr beads. The anti-FLAG monoclonal antibody M2 (Sigma-Aldrich) was used for ChIP. ChIP DNA was purified and amplified by random priming as previously described [78]. The total of amplified DNA was digested with DNaseI to a mean size of 100 bp, purified, and the fragments were end-labeled with biotin-N6-ddATP. Hybridization, washing, staining, and scanning were performed according to the manufacturer's instructions (Affymetrix). Data analyses were carried out as described previously [43].
Microarray analysis
For microarray analysis, total RNAs were prepared from cultures grown at 30°C in YPD medium to OD600 = 1.0 using TRIzol (Invitrogen). Microarray detection was performed as previously described [79], and carried out on at least three independent cultures.
Gel filtration analysis
The native molecular mass of complexes was monitored by gel filtration analysis according to Harata et al. [22] with modifications. Yeast extract from 100-ml culture of log-phase cells were applied to a Superose 6 column, and proteins were eluted at a flow rate of 0.2 ml/min. 1-ml fractions were collected and subjected to Western blot with the anti-FLAG M2 antibody to detect Arp6-Flag and Swr1-Flag. Snf2 was detected by using an anti-Snf2 antibody (Upstate).
Chromatin fractionation assay
The chromatin fractionation assay was performed as previously described [45] with the following modification. After spheroplasting, cells ware washed twice in 50 mM Hepes-KOH pH 7.5, 20 mM KCl, 2 mM EDTA-KOH, 0.05 mM spermine, 0.125 mM spermidine, 1 M sorbitol, 1% Trasylol, and 1 mM PMSF. The pellet of spheroplasts (∼4×108cells) was then re-suspended in 1 ml 50 mM Hepes-KOH pH 7.5, 2.5 mM MgCl2, 10 mM glycerol 2-phosphate, 0.1 mM Na3VO4, 0.25% Triton X-100, 300 µg/ml benzamidine, 1 µg/ml pepstatin A, 2 µg/ml antipain, 0.5 µg/ml leupeptin, 100 µg/ml TPCK, 50 µg/ml TLCK.
Quantitative PCR and ChIP analysis
For quantitative RT-PCR analysis, total RNAs were prepared from cultures grown at 30°C in YPD or YPG medium to OD600 = 1.0 by using RNeasy Mini Kit (Qiagen). Total RNAs were reverse-transcribed using the High Capacity cDNA Reverse Transcription kit (ABI), and subjected to quantitative real-time PCR with a SYBR Green Master Mix (ABI Prism 7000 Sequence Detector System and Software). ChIP was performed as for ChIP-chip analysis, but purified ChIP DNA was subjected to quantitative real-time PCR rather than microarray hybridization. For the primer sets, see Text S1. Real-time PCR monitors the threshold cycle at which the exponential curve of the accumulated product passes a threshold. PCR reactions were performed at least three times. TK normalization was performed as described in Shimada et al. [80].
Supporting Information
Zdroje
1. SpectorDL
2003 The dynamics of chromosome organization and gene regulation. Annu Rev Biochem 72 573 608
2. TaddeiA
HedigerF
NeumannFR
BauerC
GasserSM
2004 Separation of silencing from perinuclear anchoring functions in yeast Ku80, Sir4 and Esc1 proteins. EMBO J 23 1301 1312
3. SchneiderR
GrosschedlR
2007 Dynamics and interplay of nuclear architecture, genome organization, and gene expression. Genes Dev 21 3027 3043
4. AkhtarA
GasserS
2007 The nuclear envelope and transcriptional control. Nature Rev 8 507 517
5. BricknerJH
WalterP
2004 Gene recruitment of the activated INO1 locus to the nuclear membrane. PLoS Biol 2 e342 doi:10.1371/journal.pbio.0020342
6. SchmidM
AribG
LaemmliC
NishikawaJ
DurusselT
2006 Nup-PI: the nucleopore-promoter interaction of genes in yeast. Mol Cell 21 379 391
7. TaddeiA
Van HouweG
HedigerF
KalckV
CubizollesF
2006 Nuclear pore association confers optimal expression levels for an inducible yeast gene. Nature 441 774 778
8. MendjanS
TaipaleM
KindJ
HolzH
GebhardtP
2006 Nuclear pore components are involved in the transcriptional regulation of dosage compensation in Drosophila. Mol Cell 21 811 823
9. CasolariJM
BrownCR
KomiliS
WestJ
HieronymusH
SilverPA
2004 Genome-wide localization of the nuclear transport machinery couples transcriptional status and nuclear organization. Cell 117 427 439
10. PedersonT
AebiU
2005 Nuclear actin extends, with no contraction in sight. Mol Biol Cell 16 5055 5060
11. ChuangCH
CarpenterAE
FuchsovaB
JohnsonT
de LanerolleP
2006 Long-range directional movement of an interphase chromosome site. Curr Biol 16 825 831
12. HofmannWA
de LanerolleP
2006 Nuclear actin: to polymerize or not to polymerize. J Cell Biol 172 495 496
13. HofmannWA
JohnsonT
KlapczynskiM
FanJL
de LanerolleP
2006 From transcription to transport: emerging roles for nuclear myosin I. Biochem Cell Biol 84 418 426
14. DundrM
OspinaJK
SungMH
JohnS
UpenderM
2007 Actin-dependent intranuclear repositioning of an active gene locus in vivo. J Cell Biol 179 1095 1103
15. YooY
WuX
GuanJL
2007 A novel role of the actin-nucleating Arp2/3 complex in the regulation of RNA polymerase II-dependent transcription. J Biol Chem 282 7616 7623
16. HuQ
KwonYS
NunezE
CardamoneMD
HuttKR
OhgiKA
2008 Enhancing nuclear receptor-induced transcription requires nuclear motor and LSD1-dependent gene networking in interchromatin granules. Proc Natl Acad Sci U S A 105 19199 19204
17. LuthraR
KerrSC
HarremanMT
ApponiLH
FaskenMB
2007 Actively transcribed GAL genes can be physically linked to the nuclear pore by the SAGA chromatin modifying complex. J Biol Chem 282 3042 3049
18. PochO
WinsorB
1997 Who's who among the Saccharomyces cerevisiae actin-related proteins? A classification and nomenclature proposal for a large family. Yeast 13 1053 1058
19. MullerJ
OmaY
VallarL
FriederichE
PochO
2005 Sequence and Comparative Genomic Analysis of Actin-related Proteins. Mol Biol Cell 16 5736 5748
20. WeberV
HarataM
HauserH
WintersbergerU
1995 The actin-related protein Act3p of Saccharomyces cerevisiae is located in the nucleus. Mol Biol Cell 6 1263 1270
21. GravaS
DumoulinP
MadaniaA
TarassovI
WinsorB
2000 Functional analysis of six genes from chromosomes XIV and XV of Saccharomyces cerevisiae reveals YOR145c as an essential gene and YNL059c/ARP5 as a strain-dependent essential gene encoding nuclear proteins. Yeast 16 1025 1033
22. HarataM
OmaY
TabuchiT
ZhangY
StillmanDJ
2000 Multiple actin-related proteins of Saccharomyces cerevisiae are present in the nucleus. J Biochem (Tokyo) 128 665 671
23. ChenM
ShenX
2007 Nuclear actin and actin-related proteins in chromatin dynamics. Curr Opin Cell Biol 19 326 330
24. KatoM
SasakiM
MizunoS
HarataM
2001 Novel actin-related proteins in vertebrates: similarities of structure and expression pattern to Arp6 localized on Drosophila heterochromatin. Gene 268 133 140
25. JinJ
CaiY
LiB
ConawayRC
WorkmanJL
2005 In and out: histone variant exchange in chromatin. Trends Biochem Sci 30 680 687
26. OhfuchiE
KatoM
SasakiM
SugimotoK
OmaY
2006 Vertebrate Arp6, a novel nuclear actin-related protein, interacts with heterochromatin protein 1. Eur J Cell Biol 85 411 421
27. CaiY
JinJ
FlorensL
SwansonSK
KuschT
2005 The mammalian YL1 protein is a shared subunit of the TRRAP/TIP60 histone acetyltransferase and SRCAP complexes. J Biol Chem 280 13665 13670
28. WuWH
AlamiS
LukE
WuCH
SenS
2005 Swc2 is a widely conserved H2AZ-binding module essential for ATP-dependent histone exchange. Nat Struct Mol Biol 12 1064 1071
29. KroganNJ
KeoghMC
DattaN
SawaC
RyanOW
2003 A Snf2 family ATPase complex required for recruitment of the histone H2A variant Htz1. Mol Cell 12 1565 1576
30. MizuguchiG
ShenX
LandryJ
WuWH
SenS
2004 ATP-driven exchange of histone H2AZ variant catalyzed by SWR1 chromatin remodeling complex. Science 303 343 348
31. AdamM
RobertF
LarochelleM
GaudreauL
2001 H2A.Z is required for global chromatin integrity and for recruitment of RNA polymerase II under specific conditions. Mol Cell Biol 21 6270 6279
32. GuillemetteB
BatailleAR
GevryN
AdamM
BlanchetteM
2005 Variant histone H2A.Z is globally localized to the promoters of inactive yeast genes and regulates nucleosome positioning. PLoS Biol 3 e384 doi:10.1371/journal.pbio.0030384
33. LiB
PattendenSG
LeeD
GutierrezJ
ChenJ
2005 Preferential occupancy of histone variant H2AZ at inactive promoters influences local histone modifications and chromatin remodeling. Proc Natl Acad Sci U S A 102 18385 18390
34. RaisnerRM
HartleyPD
MeneghiniMD
BaoMZ
LiuCL
2005 Histone variant H2A.Z marks the 5′ ends of both active and inactive genes in euchromatin. Cell 123 233 248
35. ZhangH
RobertsDN
CairnsBR
2005 Genome-wide dynamics of Htz1, a histone H2A variant that poises repressed/basal promoters for activation through histone loss. Cell 123 219 231
36. MillarCB
XuF
ZhangK
GrunsteinM
2006 Acetylation of H2AZ Lys 14 is associated with genome-wide gene activity in yeast. Genes Dev 20 711 722
37. MeneghiniMD
WuM
MadhaniHD
2003 Conserved histone variant H2A.Z protects euchromatin from the ectopic spread of silent heterochromatin. Cell 112 725 736
38. BabiarzJE
HalleyJE
RineJ
2006 Telomeric heterochromatin boundaries require NuA4 - dependent acetylation of histone variant H2A.Z in Saccharomyces cerevisiae. Genes Dev 20 700 710
39. GalarneauL
NouraniA
BoudreaultAA
ZhangY
HeliotL
2000 Multiple links between the NuA4 histone acetyltransferase complex and epigenetic control of transcription. Mol Cell 5 927 937
40. SunadaR
GorzerI
OmaY
YoshidaT
SukaN
2005 The nuclear actin-related protein Act3p/Arp4p is involved in the dynamics of chromatin-modulating complexes. Yeast 22 753 768
41. KroganNJ
BaetzK
KeoghMC
DattaN
SawaC
2004 Regulation of chromosome stability by the histone H2A variant Htz1, the Swr1 chromatin remodeling complex, and the histone acetyltransferase NuA4. Proc Natl Acad Sci U S A 101 13513 13518
42. AoyamaN
OkaA
KitayamaK
KurumizakaH
HarataM
2008 The actin-related protein hArp8 accumulates on the mitotic chromosomes and functions in chromosome alignment. Exp Cell Res 314 859 868
43. KatouY
KanohY
BandoM
NoguchiH
TanakaH
2003 S-phase checkpoint proteins Tof1 and Mrc1 form a stable replication-pausing complex. Nature 424 1078 1083
44. WuWH
WuCH
LadurnerA
MizuguchiG
WeiD
2009 N terminus of Swr1 binds to histone H2AZ and provides a platform for subunit assembly in the chromatin remodeling complex. J Biol Chem 284 6200 6207
45. PaseroP
DunckerBP
SchwobE
GasserSM
1999 A role for the Cdc7 kinase regulatory subunit Dbf4p in the formation of initiation-competent origins of replication. Genes Dev 13 2159 2176
46. KoborMS
VenkatasubrahmanyamS
MeneghiniMD
GinJW
JenningsJL
2004 A protein complex containing the conserved Swi2/Snf2-related ATPase Swr1p deposits histone variant H2A.Z into euchromatin. PLoS Biol 2 e131 doi:10.1371/journal.pbio.0020131
47. GligorisT
ThireosG
TzamariasD
2007 The Tup1 corepressor directs Htz1 deposition at a specific promoter nucleosome marking the GAL1 gene for rapid activation. Mol Cell Biol 27 4198 4205
48. DoyeV
WepfR
HurtEC
1994 A novel nuclear pore protein Nup133p with distinct roles in poly(A)+ RNA transport and nuclear pore distribution. EMBO J 13 6062 6075
49. SchoberH
FerreiraH
KalckV
GehlenLR
GasserSM
2009 Yeast telomerase and the SUN domain protein Mps3 anchor telomeres and repress subtelomeric recombination. Genes Dev 23 928 938
50. HedigerF
NeumannFR
Van HouweG
DubranaK
GasserSM
2002 Live imaging of telomeres: yKu and Sir proteins define redundant telomere-anchoring pathways in yeast. Curr Biol 12 2076 2089
51. DieppoisG
IglesiasN
StutzF
2006 Cotranscriptional recruitment to the mRNA export receptor Mex67p contributes to nuclear pore anchoring of activated genes. Mol Cell Biol 26 7858 7870
52. VinciguerraP
IglesiasN
CamblongJ
ZenklusenD
StutzF
2005 Perinuclear Mlp proteins downregulate gene expression in response to a defect in mRNA export. EMBO J 24 813 823
53. OeffingerM
WeiKE
RogersR
DeGrasseJA
ChaitBT
2007 Comprehensive analysis of diverse ribonucleoprotein complexes. Nat Methods 4 951 956
54. ShiaWJ
LiB
WorkmanJL
2006 SAS-mediated acetylation of histone H4 Lys 16 is required for H2A.Z incorporation at subtelomeric regions in Saccharomyces cerevisiae. Genes Dev 20 2507 2512
55. MartinDE
PowersT
HallMN
2006 Regulation of ribosome biogenesis: where is TOR? Cell Metab 4 259 260
56. HallDB
WadeJT
StruhlK
2006 An HMG protein, Hmo1, associates with promoters of many ribosomal protein genes and throughout the rRNA gene locus in Saccharomyces cerevisiae. Mol Cell Biol 26 3672 3679
57. ZhaoY
McIntoshKB
RudraD
SchawalderS
ShoreD
2006 Fine-structure analysis of ribosomal protein gene transcription. Mol Cell Biol 26 4853 4862
58. CollinsSR
MillerKM
MaasNL
RoguevA
FillinghamJ
2007 Functional dissection of protein complexes involved in yeast chromosome biology using a genetic interaction map. Nature 446 806 810
59. KasaharaK
OhtsukiK
KiS
AoyamaK
TakahashiH
2007 Assembly of regulatory factors on rRNA and ribosomal protein genes in Saccharomyces cerevisiae. Mol Cell Biol 27 6686 6705
60. CabalGG
GenovesioA
Rodriguez-NavarroS
ZimmerC
GadalO
2006 SAGA interacting factors confine sub-diffusion of transcribed genes to the nuclear envelope. Nature 441 770 773
61. BricknerDG
CajigasI
Fondufe-MittendorfY
AhmedS
LeePC
2007 H2A.Z-mediated localization of genes at the nuclear periphery confers epigenetic memory of previous transcriptional state. PLoS Biol 5 e81 doi:10.1371/journal.pbio.0050081
62. DilworthDJ
TackettAJ
RogersRS
YiEC
ChristmasRH
2005 The mobile nucleoporin Nup2p and chromatin-bound Prp20p function in endogenous NPC-mediated transcriptional control. J Cell Biol 171 955 965
63. FischerT
StrasserK
RaczA
Rodriguez-NavarroS
OppizziM
2002 The mRNA export machinery requires the novel Sac3p-Thp1p complex to dock at the nucleoplasmic entrance of the nuclear pores. EMBO J 21 5843 5852
64. HieronymusH
SilverPA
2003 Genome-wide analysis of RNA-protein interactions illustrates specificity of the mRNA export machinery. Nat Genet 33 155 161
65. KroganNJ
CagneyG
YuH
ZhongG
GuoX
2006 Global landscape of protein complexes in the yeast Saccharomyces cerevisiae. Nature 440 637 643
66. ZhaoY
SohnJH
WarnerJR
2003 Autoregulation in the biosynthesis of ribosomes. Mol Cell Biol 23 699 707
67. PresuttiC
CiafréSA
BozzoniI
1991 The ribosomal protein L2 in S. cerevisiae controls the level of accumulation of its own mRNA. EMBO J 10 2215 2221
68. LiZ
PaulovichAG
WoolfordJLJr
1995 Feedback inhibition of the yeast ribosomal protein gene CRY2 is mediated by the nucleotide sequence and secondary structure of CRY2 pre - mRNA. Mol Cell Biol 15 6454 6464
69. TashevaES
RoufaDJ
1995 Regulation of human RPS14 transcription by intronic antisense RNAs and ribosomal protein S14. Genes Dev 9 304 316
70. RougemailleM
DieppoisG
Kisseleva-RomanovaE
GudipatiRK
LemoineS
2008 THO/Sub2p functions to coordinate 3′-end processing with gene-nuclear pore association. Cell 135 308 321
71. NagaiS
DubranaK
Tsai-PflugfelderM
DavidsonMB
RobertsTM
2008 Functional targeting of DNA damage to a nuclear pore-associated SUMO-dependent ubiquitin ligase. Science 322 597 602
72. AbdallahP
LucianoP
RungeKW
LisbyM
GéliV
2009 A two-step model for senescence triggered by a single critically short telomere. Nat Cell Biol 11 988 993
73. KalocsayM
HillerNJ
JentschS
2009 Chromosome-wide Rad51 spreading and SUMO-H2A.Z-dependent chromosome fixation in response to a persistent DNA double-strand break. Mol Cell 33 335 343
74. OzaP
JaspersenSL
MieleA
DekkerJ
PetersonCL
2009 Mechanisms that regulate localization of a DNA double-strand break to the nuclear periphery. Genes Dev 23 912 927
75. LeeK
KangMJ
KwonSJ
KwonYK
KimKW
2007 Expansion of chromosome territories with chromatin decompaction in BAF53-depleted interphase cells. Mol Biol Cell 18 4013 4023
76. LongtineMS
McKenzieA3rd
DemariniDJ
ShahNG
WachA
1998 Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14 953 961
77. HedigerF
DubranaK
GasserSM
2002 Myosin-like proteins 1 and 2 are not required for silencing or telomere anchoring, but act in the Tel1 pathway of telomere length control. J Struct Biol 140 79 91
78. IyerVR
HorakCE
ScafeCS
BotsteinD
SnyderM
2001 Genomic binding sites of the yeast cell-cycle transcription factors SBF and MBF. Nature 409 533 538
79. IwahashiH
KitagawaE
SuzukiY
UedaY
IshizawaYH
2007 Evaluation of toxicity of the mycotoxin citrinin using yeast ORF DNA microarray and Oligo DNA microarray. BMC genomics 8 95
80. ShimadaK
OmaY
SchlekerT
KugouK
OhtaK
2008 Ino80 chromatin remodeling complex promotes recovery of stalled replication forks. Curr Biol 18 566 575
81. HedigerF
TaddeiA
NeumannFR
GasserSM
2004 Methods for visualizing chromatin dynamics in living yeast. Methods Enzymol 375 345 365
Štítky
Genetika Reprodukční medicína
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2010 Číslo 4- Srdeční frekvence embrya může být faktorem užitečným v předpovídání výsledku IVF
- Délka menstruačního cyklu jako marker ženské plodnosti
- Vztah užívání alkoholu a mužské fertility
- Šanci na úspěšný průběh těhotenství snižují nevhodné hladiny progesteronu vznikající při umělém oplodnění
- Růst a vývoj dětí narozených pomocí IVF
-
Všechny články tohoto čísla
- Whole-Genome SNP Association in the Horse: Identification of a Deletion in Myosin Va Responsible for Lavender Foal Syndrome
- Human Telomeres Are Hypersensitive to UV-Induced DNA Damage and Refractory to Repair
- Fragilities Caused by Dosage Imbalance in Regulation of the Budding Yeast Cell Cycle
- Admixture Mapping Scans Identify a Locus Affecting Retinal Vascular Caliber in Hypertensive African Americans: the Atherosclerosis Risk in Communities (ARIC) Study
- Activation of Estrogen-Responsive Genes Does Not Require Their Nuclear Co-Localization
- Genetic Tests for Ecological and Allopatric Speciation in Anoles on an Island Archipelago
- A Pax3/Dmrt2/Myf5 Regulatory Cascade Functions at the Onset of Myogenesis
- Two New Loci for Body-Weight Regulation Identified in a Joint Analysis of Genome-Wide Association Studies for Early-Onset Extreme Obesity in French and German Study Groups
- Hypomethylation of a LINE-1 Promoter Activates an Alternate Transcript of the MET Oncogene in Bladders with Cancer
- Candidate Causal Regulatory Effects by Integration of Expression QTLs with Complex Trait Genetic Associations
- Combined Inactivation of pRB and Hippo Pathways Induces Dedifferentiation in the Retina
- Allele-Specific Virulence Attenuation of the HopZ1a Type III Effector via the ZAR1 Resistance Protein
- Down-Regulation of Honey Bee Gene Biases Behavior toward Food Rich in Protein
- A Microarray-Based Genetic Screen for Yeast Chronological Aging Factors
- Actin-Related Protein Arp6 Influences H2A.Z-Dependent and -Independent Gene Expression and Links Ribosomal Protein Genes to Nuclear Pores
- Phosphorylation of the Conserved Transcription Factor ATF-7 by PMK-1 p38 MAPK Regulates Innate Immunity in
- A -Regulatory Signature for Chordate Anterior Neuroectodermal Genes
- Genetic Analysis of Fin Development in Zebrafish Identifies Furin and Hemicentin1 as Potential Novel Fraser Syndrome Disease Genes
- Assembly of a 40 Mb Eukaryotic Genome from Short Sequence Reads: , a Model Organism for Fungal Morphogenesis
- Trait-Associated SNPs Are More Likely to Be eQTLs: Annotation to Enhance Discovery from GWAS
- Absence of Evidence for MHC–Dependent Mate Selection within HapMap Populations
- The TALE Class Homeobox Gene Defines the Anterior Compartment for Head Regeneration
- Cyclic Expression of Lhx2 Regulates Hair Formation
- Genetic Evidence for Hybrid Trait Speciation in Butterflies
- Epigenetic Regulation of a Murine Retrotransposon by a Dual Histone Modification Mark
- Chromosome 9p21 SNPs Associated with Multiple Disease Phenotypes Correlate with Expression
- S Phase Progression in Human Cells Is Dictated by the Genetic Continuity of DNA Foci
- The Next Generation Becomes the Now Generation
- Acts as a Tumor Suppressor in a Murine Retinoblastoma Model by Facilitating Tumor Cell Death
- Genome-Wide Association Study of Lp-PLA Activity and Mass in the Framingham Heart Study
- The Five Zinc Transporters Undergo Different Evolutionary Fates towards Adaptive Evolution to Zinc Tolerance in
- MicroRNA–Directed siRNA Biogenesis in
- Deletion of the WD40 Domain of LRRK2 in Zebrafish Causes Parkinsonism-Like Loss of Neurons and Locomotive Defect
- Incipient Balancing Selection through Adaptive Loss of Aquaporins in Natural Populations
- GTPase Activity Plays a Key Role in the Pathobiology of LRRK2
- Natural Single-Nucleosome Epi-Polymorphisms in Yeast
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Whole-Genome SNP Association in the Horse: Identification of a Deletion in Myosin Va Responsible for Lavender Foal Syndrome
- Admixture Mapping Scans Identify a Locus Affecting Retinal Vascular Caliber in Hypertensive African Americans: the Atherosclerosis Risk in Communities (ARIC) Study
- Genetic Tests for Ecological and Allopatric Speciation in Anoles on an Island Archipelago
- Human Telomeres Are Hypersensitive to UV-Induced DNA Damage and Refractory to Repair
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



