-
Články
- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
RNA Biology in Fungal Phytopathogens
article has not abstract
Published in the journal: . PLoS Pathog 9(10): e32767. doi:10.1371/journal.ppat.1003617
Category: Pearls
doi: https://doi.org/10.1371/journal.ppat.1003617Summary
article has not abstract
RNA-dependent processes are essential to determine when, where, and how much of a protein is synthesized. In eukaryotes, these processes start with transcription in the nucleus and end with mRNA translation at distinct cytoplasmic sites followed by mRNA degradation [1]. In between, a number of defined steps occur such as 5′ capping, splicing, polyadenylation, nuclear export, and cytoplasmic transport. Most of the steps are tightly regulated to achieve highly precise spatiotemporal expression. RNA-binding proteins serve as key factors that are essential at each level of posttranscriptional control [1]. Small RNAs are additional factors that can inhibit translation or are able to promote degradation of mRNAs.
Due to the central importance of RNA biology in regulating protein synthesis, it is not surprising that fungal pathogens intensely rely on RNA-dependent processes to control infection. Here, we focus on RNA biology in fungal plant pathogenesis, which is best studied in the corn smut Ustilago maydis [2], [3].
RNA Biology Orchestrates Fungal Infection
In U. maydis, a major pathogenicity determinant is the homeodomain transcription factor bW/bE that triggers a cascade of transcriptional responses essential for full virulence [4], [5]. Progression toward infection is regulated by several bE/bW-induced factors, including Clp1 and Cib1. Importantly, their functions are precisely regulated at the posttranscriptional level. clp1 is a direct target gene of bW/bE, which is induced early during formation of infectious filaments. However, the Clp1 protein is only synthesized later at the specific stage of plant penetration when it acts as an antagonist of bW/bE [6]. This is indicative for a tight translational regulation of clp1 mRNA by a currently unknown mechanism (Figure 1, #1). Cib1 constitutes a direct interaction partner of Clp1. The corresponding cib1 gene is constitutively expressed but regulated at the level of mRNA splicing. Splicing is induced during infection, resulting in the expression of a Cib1 variant containing a functionally important Clp1 interaction domain [7]. Thus, a complex network of transcriptional and posttranscriptional controls is essential to fine-tune infection.
Fig. 1. The role of fungal RNA biology during plant infection. 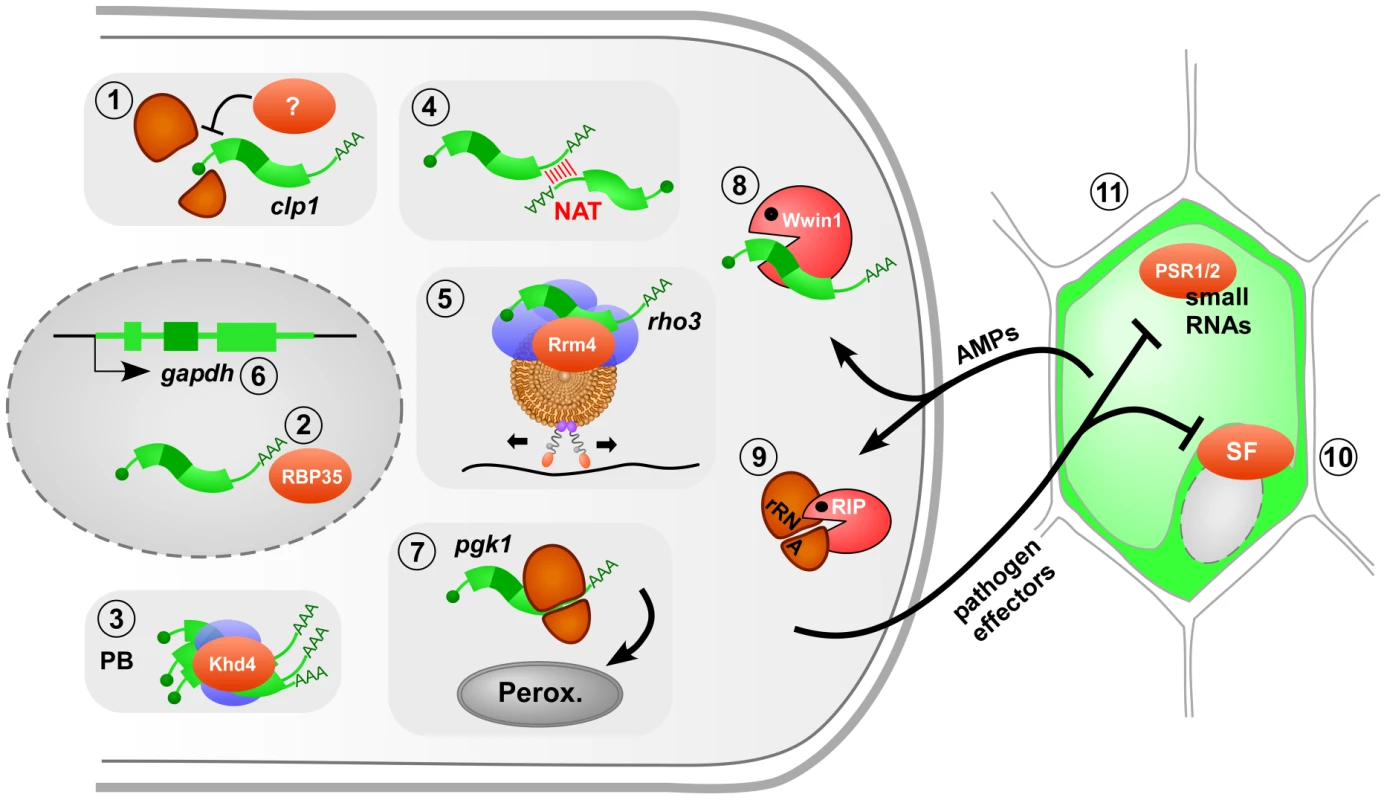
Model of a filamentous pathogen (left) interacting with its plant host (right). RNA-dependent processes crucial for infection are numbered as follows: 1 translational regulation of clp1 mRNA (ribosomal subunits in orange, mRNA in green); 2 3′ end processing by the RNA-binding protein RBP35 in the nucleus (gray oval); 3 modulation of mRNA stability in processing bodies (PB, blue: accessory factors); 4 natural antisense transcripts (NAT) regulate mRNA stability; 5 endosomal mRNA transport along microtubule (black line, brown: endosome with motor proteins attached); 6 alternative splicing of gapdh mRNA to generate an enzyme with a peroxisomal targeting sequence; 7 translational read-through adding a peroxisomal targeting sequence; 8 antimicrobial peptide (AMP) Wheatwin1(Wwin1) with RNase activity; 9 ribosome-inactivating protein (RIP) alters rRNA; 10 pathogen effectors target host splice factors (SF) as shown for the bacterial effector HopU1 inactivating AtGRP7; 11 pathogen effectors interfere with the generation of small RNAs in the host as shown for oomycetes effectors. Further details are given in the text. RNA-binding proteins are key factors in RNA biology. In the rice blast fungus Magnaporthe grisea, loss of the RRM (RNA recognition motif) protein RBP35 causes defects in virulence and development. Protein interaction studies revealed that RBP35 functions as a novel auxiliary protein of the polyadenylation machinery of plant pathogens (Figure 1, #2) [8]. A systematic approach in U. maydis deleting genes encoding various RNA-binding proteins revealed that mutants lacking Khd4 exhibit defects in morphology and pathogenicity [9]. Khd4 is a multi-KH-domain protein with homologs in other pathogens such as Cryptococcus neoformans and Candida albicans. It specifically binds the sequence AUACCC and is most likely involved in regulation of mRNA stability [10]. This is supported by the observation that Khd4 localizes to processing bodies (unpublished observation), which are known centers for mRNA degradation (Figure 1, #3). The importance of regulating mRNA stability was recently further underlined by showing that natural antisense transcripts might also be important for the regulation of mRNA stability and pathogenicity (Figure 1, #4) [11].
In addition to regulation through natural antisense transcripts, RNA interference (RNAi) is an important mechanism to control endogenous and foreign RNA in most organisms. Noteworthily, genome comparison with Ustilago hordei revealed that the RNAi machinery has been specifically lost in U. maydis during evolution, indicating that this mode of RNA regulation is clearly dispensable for a successful plant pathogen [12]. It has been rationalized that RNAi deficiency enables hosting of double-stranded RNA viruses. These can carry the genetic information to produce toxins to kill sensitive neighbors, which might account for an evolutionary advantage [13]. Taken together, posttranscriptional control at the level of splicing, polyadenylation, translation, and stability is an important regulatory tool of fungal pathogens to coordinate infection.
mRNA Transport Is Crucial during Early Infection
In higher eukaryotes, active microtubule-dependent transport of mRNAs by molecular motors functions in spatial protein expression during developmental and neuronal processes [14]. Similarly, in U. maydis the ELAV (embryonic lethal abnormal vision)-type protein Rrm4 functions in long-distance mRNA transport, a process that is specifically important during formation of infectious unipolar filaments [15]–[17]. Studying the mechanism of transport revealed that shuttling of Rrm4 along microtubules is connected to membrane trafficking [17], [18]. Rrm4 colocalizes almost exclusively with motile Rab5a-positive endosomes. Similarly to endosome shuttling [19], trafficking of Rrm4 is mediated by the concerted action of the minus end–directed motor dynein Dyn1/2 and the plus end–directed kinesin Kin3 [17]. Since colocalization of Rrm4 with endosomes occurs even in the absence of Kin3, endosomal hitchhiking was proposed as mode of transport (Figure 1, #5) [17]. Importantly, Rrm4 is dispensable for endosomal shuttling, but the RNA-binding capacity of Rrm4 is needed for unipolar growth [15], [17].
Rrm4 transports specific mRNAs such as ubi1 and rho3, encoding ubiquitin fused to ribosomal protein Rpl40 and the small GTPase Rho3, respectively [16]. Rho3 accumulates at retraction septa of infectious filaments and rrm4 deletion strains are disturbed in septum formation. Hence, microtubule-dependent transport of rho3 mRNA might act in correct septal localization of Rho3, which could promote septum insertion in infectious filaments [16].
In summary, endosomal mRNA transport along microtubules is a novel aspect of RNA biology and this posttranscriptional process is particularly important during early infection. Since the RNA-binding protein She3 functions in actin-dependent mRNA transport during infection of C. albicans [20], a molecular link of mRNA trafficking and infection might be widespread in fungal pathogenicity.
RNA Biology Determines the Precise Subcellular Distribution of Proteins during Infection
Correct targeting of proteins to subcellular compartments is an essential process and defects can affect virulence. Generally proteins are directed to their subcellular location by signal sequences, whose presence can be regulated by alternative splicing of pre-mRNA. A novel example is the glycolytic enzyme glyceraldehyde-3-phosphate dehydrogenase (GAPDH) of U. maydis, which is usually localized in the cytoplasm. However, 10% of its mRNA is alternatively spliced, generating an enzyme with a C-terminal peroxisomal targeting sequence (PTS1 type). This results in peroxisomal localization of GAPDH, a process termed cryptic peroxisomal targeting (Figure 1, #6) [21]. Similarly, the 3-phosphoglycerate kinase (PGK) resides to a certain extent in peroxisomes. Here, ribosomal read-through of the termination codon creates an isoform with a functional C-terminal PTS1 (Figure 1, #7). The apparently “sloppy” translation accounts for the necessary fraction of peroxisomal enzyme [21]. In peroxisomes, GAPDH and PGK might function together with other NADH-dependent dehydrogenases in redox homeostasis. This regulatory process appears to be particularly important during pathogenic development, since mutants specifically lacking the peroxisomal isoforms are viable but less virulent [21]. Thus, RNA biology in form of alternative splicing and translational read-through determines the correct subcellular targeting of the encoded enzymes and this contributes to virulence.
RNA Biology in Plant-Microbe Interactions
The importance of RNA biology in fungal infection is further underlined by the fact that posttranscriptional regulation is targeted during host defense. Plants react to infection by producing antimicrobial peptides (AMPs) that are proposed to degrade the fungal cell wall or to permeabilize the membrane. Some AMPs also enter the cell to exert their function [22]. An important example for the latter is Wheatwin1, a pathogenesis-related protein (PR4) with antifungal activity [23]. Wheatwin1 is targeted to the cytoplasm of intruding fungi where it functions as RNase and inhibits pathogenic growth by hydrolyzing fungal RNA (Figure 1, #8) [23]. Furthermore, plants express ribosome-inactivating proteins with antifungal activity. These attack ribosomes by exhibiting N-glycosidase activity, removing a single adenine from the rRNA, and thereby translation is inhibited (Figure 1, #9) [24]. Thus, plants interfere with fungal RNA biology for defense.
Intriguingly, the opposite is true as well: pathogens interfere with posttranscriptional control of the host to promote infection. Although this has so far only been shown for bacterial and oomycete phytopathogens [25], we predict that similar mechanisms will be discovered for fungal phytopathogens. For example, it has been known for many years that U. maydis secretes ribonucleases that might function as virulence factors in degrading host RNAs [26]. A prominent case for bacterial effectors is HopU1 from Pseudomonas syringae that is essential for virulence on Arabidopsis thaliana [27]. HopU1 functions as mono-ADP-ribosyltransferase and modifies the RNA-binding domain of AtGRP7, a splice factor that regulates expression of the immune receptor FLS2 (Figure 1, #10). Thus, bacterial effectors appear to inactivate host RNA-binding proteins by posttranslational modification to overcome plant defense.
Moreover, microbial effectors interfere with plant microRNA activity. miR393, for instance, targets the F-box auxin receptor TIR1 and thereby represses auxin signaling, a process involved in plant immunity [28]. The bacterial effector AvrPto interferes with microRNA function in the plant host both at the transcriptional and posttranscriptional level [29]. This seems to be a broad concept also present in oomycete pathogens. The effector PSR1 inhibits DICER-dependent functions and thereby it suppresses biogenesis of a broad range of mi - and siRNAs involved in regulating plant immunity. The effector PSR2 specifically targets a subset of trans-acting siRNAs, resulting in altered expression of host resistance proteins (Figure 1, #11) [30]. In summary, plant defense targets fungal RNA biology and vice versa pathogen effectors suppress host RNA silencing during plant protection.
Conclusions and Future Directions
The different levels of RNA biology such as alternative splicing, 3′ end processing, endosomal mRNA transport, and translational regulation are essential to orchestrate plant infection of fungal pathogens. RNA-binding proteins are key components in executing these functions. After improving transcriptome-wide approaches such as iCLIP [31], we are now able to investigate the complete RNA-binding landscape of RNA-binding proteins involved in fungal infection [32]. Moreover, it has become apparent that RNA biology is also particularly important during plant/microbe communication. Mutually exchanged effectors act on RNA components of the defence/virulence pathway of the partner. Thus, RNA biology emerges as an important research field with the potential to identify new Achilles' heels of fungal pathogens. Taking plants as an inspiring model, drugs interfering with RNA-dependent processes of pathogens should be considered for host protection in the future.
Zdroje
1. MooreMJ (2005) From birth to death: the complex lives of eukaryotic mRNAs. Science 309 : 1514–1518.
2. FeldbrüggeM, ZarnackK, VollmeisterE, BaumannS, KoepkeJ, et al. (2008) The posttranscriptional machinery of Ustilago maydis. Fungal Genet Biol 45: S40–S46.
3. VollmeisterE, FeldbrüggeM (2010) Posttranscriptional control of growth and development in Ustilago maydis. Curr Opin Microbiol 13 : 693–699.
4. BrefortT, DoehlemannG, Mendoza-MendozaA, ReissmannS, DjameiA, et al. (2009) Ustilago maydis as a pathogen. Annu Rev Phytopathol 47 : 423–445.
5. VollmeisterE, SchipperK, BaumannS, HaagC, PohlmannT, et al. (2012) Fungal development of the plant pathogen Ustilago maydis. FEMS Microbiol Rev 36 : 59–77.
6. SchererM, HeimelK, StarkeV, KämperJ (2006) The Clp1 protein is required for clamp formation and pathogenic development of Ustilago maydis. Plant Cell 18 : 2388–2401.
7. HeimelK, SchererM, SchulerD, KämperJ (2010) The Ustilago maydis Clp1 protein orchestrates pheromone and b-dependent signaling pathways to coordinate the cell cycle and pathogenic development. Plant Cell 22 : 2908–2922.
8. FranceschettiM, BuenoE, WilsonRA, TuckerSL, Gómez-MenaC, et al. (2011) Fungal virulence and development is regulated by alternative pre-mRNA 3′end processing in Magnaporthe oryzae. PLoS Pathog 7: e1002441 doi:10.1371/journal.ppat.1002441
9. BechtP, VollmeisterE, FeldbrüggeM (2005) Role for RNA-binding proteins implicated in pathogenic development of Ustilago maydis. Eukaryot Cell 4 : 121–133.
10. VollmeisterE, HaagC, ZarnackK, BaumannS, KönigJ, et al. (2009) Tandem KH domains of Khd4 recognize AUACCC and are essential for regulation of morphology as well as pathogenicity in Ustilago maydis. RNA 15 : 2206–2218.
11. DonaldsonME, SavilleBJ (2013) Ustilago maydis natural antisense transcript expression alters mRNA stability and pathogenesis. Mol Microbiol 89 : 29–51.
12. LaurieJD, AliS, LinningR, MannhauptG, WongP, et al. (2012) Genome comparison of barley and maize smut fungi reveals targeted loss of RNA silencing components and species-specific presence of transposable elements. Plant Cell 24 : 1733–1745.
13. DrinnenbergIA, FinkGR, BartelDP (2011) Compatibility with killer explains the rise of RNAi-deficient fungi. Science 333 : 1592.
14. PrattCA, MowryKL (2013) Taking a cellular road-trip: mRNA transport and anchoring. Curr Opin Cell Biol 25 : 99–106.
15. BechtP, KönigJ, FeldbrüggeM (2006) The RNA-binding protein Rrm4 is essential for polarity in Ustilago maydis and shuttles along microtubules. J Cell Sci 119 : 4964–4973.
16. KönigJ, BaumannS, KoepkeJ, PohlmannT, ZarnackK, et al. (2009) The fungal RNA-binding protein Rrm4 mediates long-distance transport of ubi1 and rho3 mRNAs. EMBO J 28 : 1855–1866.
17. BaumannS, PohlmannT, JungbluthM, BrachmannA, FeldbrüggeM (2012) Kinesin-3 and dynein mediate microtubule-dependent co-transport of mRNPs and endosomes. J Cell Sci 125 : 2740–2752.
18. GöhreV, VollmeisterE, BölkerM, FeldbrüggeM (2012) Microtubule-dependent membrane dynamics of Ustilago maydis: trafficking and function of Rab5a-positive endosomes. Commun Integr Biol 5 : 482–487.
19. SteinbergG (2012) The transport machinery for motility of fungal endosomes. Fungal Genet Biol 49 : 675–676.
20. ElsonSL, NobleSM, SolisNV, FillerSG, JohnsonAD (2009) An RNA transport system in Candida albicans regulates hyphal morphology and invasive growth. PLoS Genet 5: e1000664 doi:10.1371/journal.pgen.1000664
21. FreitagJ, AstJ, BölkerM (2012) Cryptic peroxisomal targeting via alternative splicing and stop codon read-through in fungi. Nature 485 : 522–525.
22. van der WeerdenNL, BleackleyMR, AndersonMA (2013) Properties and mechanisms of action of naturally occurring antifungal peptides. Cell Mol Life Sci 70 : 3545–3570.
23. BertiniL, ProiettiS, AleandriMP, MondelloF, SandiniS, et al. (2012) Modular structure of HEL protein from Arabidopsis reveals new potential functions for PR-4 proteins. Biol Chem 393 : 1533–1546.
24. StirpeF (2013) Ribosome-inactivating proteins: from toxins to useful proteins. Toxicon 67 : 12–16.
25. StaigerD, KorneliC, LummerM, NavarroL (2013) Emerging role for RNA-based regulation in plant immunity. New Phytol 197 : 394–404.
26. BlankA, DekkerCA (1975) Differential activity staining: its use in characterization of guanylyl-specific ribonuclease in the genus Ustilago. Proc Natl Acad Sci U S A 72 : 4914–4917.
27. NicaiseV, JoeA, JeongBR, KorneliC, BoutrotF, et al. (2013) Pseudomonas HopU1 modulates plant immune receptor levels by blocking the interaction of their mRNAs with GRP7. EMBO J 32 : 701–712.
28. NavarroL, DunoyerP, JayF, ArnoldB, DharmasiriN, et al. (2006) A plant miRNA contributes to antibacterial resistance by repressing auxin signaling. Science 312 : 436–439.
29. NavarroL, JayF, NomuraK, HeSY, VoinnetO (2008) Suppression of the microRNA pathway by bacterial effector proteins. Science 321 : 964–967.
30. QiaoY, LiuL, XiongQ, FloresC, WongJ, et al. (2013) Oomycete pathogens encode RNA silencing suppressors. Nat Genet 45 : 330–333.
31. KönigJ, ZarnackK, RotG, CurkT, KayikciM, et al. (2010) iCLIP reveals the function of hnRNP particles in splicing at individual nucleotide resolution. Nat Struct Mol Biol 17 : 909–915.
32. CastelloA, FischerB, EichelbaumK, HorosR, BeckmannBM, et al. (2012) Insights into RNA biology from an atlas of mammalian mRNA-binding proteins. Cell 149 : 1393–1406.
Štítky
Hygiena a epidemiologie Infekční lékařství Laboratoř
Článek vyšel v časopisePLOS Pathogens
Nejčtenější tento týden
2013 Číslo 10- Stillova choroba: vzácné a závažné systémové onemocnění
- Diagnostický algoritmus při podezření na syndrom periodické horečky
- Diagnostika virových hepatitid v kostce – zorientujte se (nejen) v sérologii
- Perorální antivirotika jako vysoce efektivní nástroj prevence hospitalizací kvůli COVID-19 − otázky a odpovědi pro praxi
- První vakcína proti klíšťové encefalitidě: vakcína FSME-IMMUN
-
Všechny články tohoto čísla
- Are We There Yet? Recent Progress in the Molecular Diagnosis and Novel Antifungal Targeting of and Invasive Aspergillosis
- Fungal Iron Availability during Deep Seated Candidiasis Is Defined by a Complex Interplay Involving Systemic and Local Events
- Emergence of Azole-Resistant Strains due to Agricultural Azole Use Creates an Increasing Threat to Human Health
- Fungal Adenylyl Cyclase Acts As a Signal Sensor and Integrator and Plays a Central Role in Interaction with Bacteria
- Sensing of the Microbial Neighborhood by
- Antivirulence Therapy for Animal Production: Filling an Arsenal with Novel Weapons for Sustainable Disease Control
- The Cell Biology of : How to Teach Using Animations
- A Structure-Guided Mutation in the Major Capsid Protein Retargets BK Polyomavirus
- RNA Biology in Fungal Phytopathogens
- , , and the Human Mouth: A Sticky Situation
- The Gene Is Essential for Resistance to Human Serum in
- Unisexual Reproduction Drives Evolution of Eukaryotic Microbial Pathogens
- Bacterial Pathogens Activate a Common Inflammatory Pathway through IFNλ Regulation of PDCD4
- Bats and Viruses: Friend or Foe?
- Protein Trafficking through the Endosomal System Prepares Intracellular Parasites for a Home Invasion
- IL-22 Mediates Goblet Cell Hyperplasia and Worm Expulsion in Intestinal Helminth Infection
- B Cells Enhance Antigen-Specific CD4 T Cell Priming and Prevent Bacteria Dissemination following Genital Tract Infection
- Alternative Roles for CRISPR/Cas Systems in Bacterial Pathogenesis
- Chemicals, Climate, and Control: Increasing the Effectiveness of Malaria Vector Control Tools by Considering Relevant Temperatures
- Dengue Vaccines: Strongly Sought but Not a Reality Just Yet
- Feeding Uninvited Guests: mTOR and AMPK Set the Table for Intracellular Pathogens
- Driven Enforced Viral Replication in Dendritic Cells Contributes to Break of Immunological Tolerance in Autoimmune Diabetes
- IL-4Rα-Associated Antigen Processing by B Cells Promotes Immunity in Infection
- A Gammaherpesvirus Uses Alternative Splicing to Regulate Its Tropism and Its Sensitivity to Neutralization
- MicroRNA-155 Promotes Autophagy to Eliminate Intracellular Mycobacteria by Targeting Rheb
- Epigenetic Dominance of Prion Conformers
- MAIT Cells Detect and Efficiently Lyse Bacterially-Infected Epithelial Cells
- The Role of TcdB and TccC Subunits in Secretion of the Tcd Toxin Complex
- A Mechanism for the Inhibition of DNA-PK-Mediated DNA Sensing by a Virus
- PLOS Pathogens
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Dengue Vaccines: Strongly Sought but Not a Reality Just Yet
- MicroRNA-155 Promotes Autophagy to Eliminate Intracellular Mycobacteria by Targeting Rheb
- Alternative Roles for CRISPR/Cas Systems in Bacterial Pathogenesis
- Feeding Uninvited Guests: mTOR and AMPK Set the Table for Intracellular Pathogens
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání


