-
Články
- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Molecular profiles of high-grade and low-grade pseudomyxoma peritonei
Abstract:
Pseudomyxoma peritonei (PMP) is a rare disease exhibiting a distinct clinical feature caused by cancerous cells that produce mucinous fluid in the abdominal cavity. PMPs originate most frequently from the appendix and less frequently from the ovary. This disease can range from benign to malignant, and histologically, PMP is classified into two types: disseminated peritoneal adenomucinosis (DPAM) representing the milder phenotype, and peritoneal mucinous adenocarcinomas (PMCA) representing the aggressive phenotype. Although histological classification is clinically useful, the pathogenesis of PMP remains largely unknown. To elucidate the molecular mechanisms underlying PMP, we analyzed 18 PMP tumors comprising 10 DPAMs and 8 PMCAs. DNA was extracted from tumor and matched non-tumorous tissues, and was sequenced using Ion AmpliSeq Cancer Panel containing 50 cancer-related genes. Analysis of the data identified a total of 35 somatic mutations in 10 genes, and all mutations were judged as pathological mutations. Mutations were frequently identified in KRAS (14/18) andGNAS (8/18). Interestingly, TP53 mutations were found in three of the eight PMCAs, but not in the DPAMs. PIK3CA and AKT1 mutations were also identified in two PMCAs, but not in the DPAMs. These results suggested that KRAS and/or GNAS mutations are common genetic features of PMP, and that mutations in TP53 and/or genes related to the PI3K-AKT pathway may render malignant properties to PMP. These findings may be useful for the understanding of tumor characteristics, and facilitate the development of therapeutic strategies.Keywords:
DPAM ; mutation; PMCA ; pseudomyxoma peritonei; TP53
Authors: Rei Noguchi 1; Hideaki Yano 2; Yoshimasa Gohda 2; Ryuichiro Suda 2; Toru Igari 3; Yasunori Ohta 4; Naohide Yamashita 5; Kiyoshi Yamaguchi 1; Yumi Terakado 1; Tsuneo Ikenoue 1; Yoichi Furukawa 1
Authors place of work: Division of Clinical Genome Research, The Institute of Medical Science, The University of Tokyo, Tokyo, Japan 1; Department of Surgery, National Center for Global Health and Medicine, Tokyo, Japan 2; Pathology Division of Clinical Laboratory, National Center for Global Health and Medicine, Tokyo, Japan 3; Department of Pathology, Research Hospital, The Institute of Medical Science, The University of Tokyo, Tokyo, Japan 4; Department of Advanced Medical Science, Research Hospital, The Institute of Medical Science, The University of Tokyo, Tokyo, Japan 5
Published in the journal: Cancer Medicine 2015; Early View(Early View)
Category: Original Research
doi: https://doi.org/10.1002/cam4.542© 2015 The Authors. Cancer Medicine published by John Wiley & Sons Ltd.
This is an open access article under the terms of the Creative Commons Attribution License, which permits use, distribution and reproduction in any medium, provided the original work is properly cited.
© 2015 The Authors. Cancer Medicine published by John Wiley & Sons Ltd.Summary
Abstract:
Pseudomyxoma peritonei (PMP) is a rare disease exhibiting a distinct clinical feature caused by cancerous cells that produce mucinous fluid in the abdominal cavity. PMPs originate most frequently from the appendix and less frequently from the ovary. This disease can range from benign to malignant, and histologically, PMP is classified into two types: disseminated peritoneal adenomucinosis (DPAM) representing the milder phenotype, and peritoneal mucinous adenocarcinomas (PMCA) representing the aggressive phenotype. Although histological classification is clinically useful, the pathogenesis of PMP remains largely unknown. To elucidate the molecular mechanisms underlying PMP, we analyzed 18 PMP tumors comprising 10 DPAMs and 8 PMCAs. DNA was extracted from tumor and matched non-tumorous tissues, and was sequenced using Ion AmpliSeq Cancer Panel containing 50 cancer-related genes. Analysis of the data identified a total of 35 somatic mutations in 10 genes, and all mutations were judged as pathological mutations. Mutations were frequently identified in KRAS (14/18) andGNAS (8/18). Interestingly, TP53 mutations were found in three of the eight PMCAs, but not in the DPAMs. PIK3CA and AKT1 mutations were also identified in two PMCAs, but not in the DPAMs. These results suggested that KRAS and/or GNAS mutations are common genetic features of PMP, and that mutations in TP53 and/or genes related to the PI3K-AKT pathway may render malignant properties to PMP. These findings may be useful for the understanding of tumor characteristics, and facilitate the development of therapeutic strategies.Keywords:
DPAM ; mutation; PMCA ; pseudomyxoma peritonei; TP53Introduction
Pseudomyxoma peritonei (PMP) is a disease exhibiting clinical manifestations such as extended abdomen, abdominal discomfort, obstruction in digestive tract, and nutritional compromise due to accumulated gelatinous ascites and/or disseminated lesions of neoplastic mucin-secreting cells in the peritoneal cavity. Primary tumor cells of PMP develop most frequently in the appendix and occasionally in other organs including the ovary, colorectum, gallbladder, stomach, pancreas, fallopian tube, urachus, lung, and breast [1]. It is a rare condition with an estimated incidence of 1–2 per million per year [2]. Based on their architectural complexity and degree of cytologic atypia, the tumors are histologically divided into two groups: [3] disseminated peritoneal adenomucinosis tumors (DPAM) showing low-grade cytologic atypia (nucleomegaly, nuclear stratification, rate mitotic figures, and single cell necrosis) and minimal architectural complexity (villiform, flat epithelial proliferation, and small papillary excrescences), and peritoneal mucinous adenocarcinomas tumors (PMCA) demonstrating any of the following: destructive invasion of the organ's wall, high-grade cytologic atypia (extensive full-thickness nuclear stratification, vesicular nuclei, marked nuclear membrane irregularities, prominent nucleoi, and brisk mitotic activity), or complex epithelial proliferation (complex papillary fronds and cribriform glandular spaces). The histological classification is reportedly associated with prognosis of the patients with PMP; a median 5-year survival of 62.5% in DPAMs and 37.7% in PMCAs [4].
Several molecular studies have disclosed profiles of PMP including frequent mutations in KRAS andGNAS, decreased expression of E-cadherin, and amplification of MCL1 and JUN. However, the precise molecular mechanism(s) underlying the development and progression of PMP and genetic factors associated with the response of their treatment remain to be resolved.
In this study, we have analyzed a total of 18 Japanese PMPs comprising 10 DPAMs and 8 PMCAs using multiplex-PCR-based sequence, and identified 35 somatic mutations. Activating mutations inRAS family members and GNAS were observed in both PMCAs and DPAMs, and deregulation of TP53 or PI3K-AKT pathway was found in PMCAs but not DPAMs. These data may be useful for the understanding of the development and progression of PMP.
Materials and Methods
Patients and clinical tissues
This study was approved by the institutional review boards of The Institute of Medical Science, The University of Tokyo (IMSUT-IRB #26-67), and National Center for Global Health and Medicine. All patients were given written informed consent for the study including genetic analysis. All samples were obtained from National Center for Global Health and Medicine (2012–2014). Tumor tissues and corresponding non-cancerous tissues were obtained from surgical specimens of 18 patients with PMP, and frozen in deep freezer until analysis. In 12 of the 18 cases, primary tumors were available, but in the remaining six cases, metastatic lesions were used for the analysis. Clinicopathological information, treatment, and prognosis of the patients are summarized in Table 1. Eighteen patients underwent surgical operations including cytoreductive surgery (CRS) for 12 patients, debulking operation for 5, and exploratory laparotomy for one. Intraoperative hyperthermic intraperitoneal chemotherapy (HIPEC) with 10 mg/m2 mitomycin C at the temperature between 41 and 42°C was administered to the 12 CRS and 2 debulking operations. To seven patients treated with CRS and HIPEC, early postoperative intraperitoneal chemotherapy (EPIC) comprising 15 mg/kg fluorouracil was added intraperitoneally on days 1–4 after surgery. Residual disease was scored according to the completeness of cytoreduction (CCR) score [5]. CCR0 indicates no macroscopic residual tumor; CCR1, no nodule larger than 2.5 mm in diameter remained; CCR2, nodules between 2.5 mm and 2.5 cm in diameter remained; and CCR3, nodules larger than 2.5 cm in diameter remained. All 18 tumors were histologically diagnosed as PMP and classified into 10 DPAMs and 8 PMCAs according to the definitions proposed by Misdraji et al. [3]. Histological examination did not find component of signet ring cells in the 18 samples.
Tab. 1. Clinicopathological features of the 18 PMP patients. 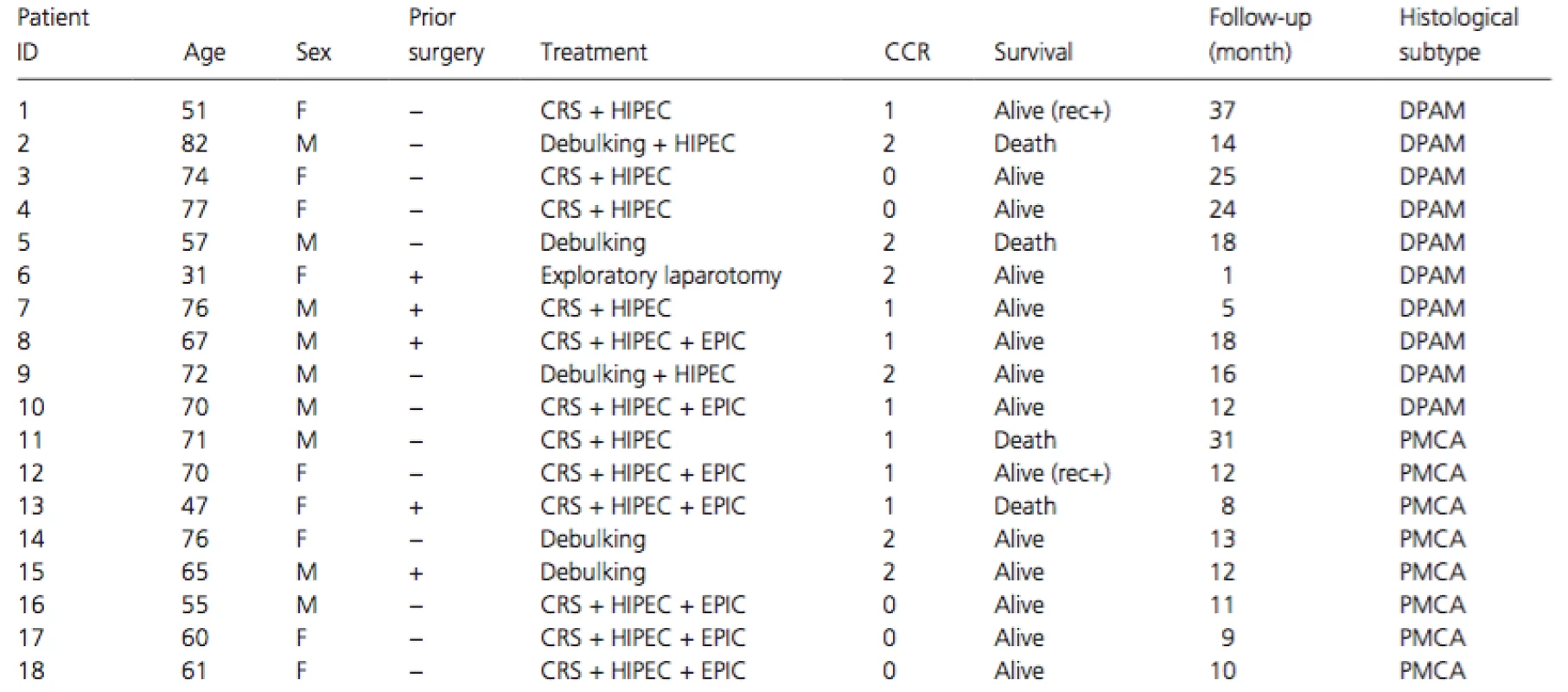
CRS, cytoreductive surgery; HIPEC, hyperthermic intraperitoneal chemotherapy; EPIC, early postoperative intraperitoneal chemotherapy; CCR, completeness of cytoreduction; DPAM, disseminated peritoneal adenomucinosis; PMCA, peritoneal mucinous carcinomatosis; rec+, recurrence+. Extraction and quantification of DNA
Frozen sections (10 μm in thickness) of tumor and nontumorous tissues were fixed in ice-cold 4% formalin for 10 min, washed by water for 5 min, and subsequently stained by hematoxylin. Tumor and non-tumorous cells were collected from the sections by laser microdissection (LMD) using LMD 7000 (Leica Microsystems, Inc., Bensheim, Germany). Genomic DNA was extracted from the collected cells using a QIAamp DNA formalin-fixed, paraffin-embedded tissue kit (Qiagen, Valencia, CA) according to the manufacturer's instructions. Concentration of DNA was assessed by e-SPECT (Malcom, Tokyo, Japan) and Qubit2 fluorometer (Invitrogen, CA). All genomic DNA was stored at −20°C until use.
Multiplex PCR and DNA sequencing
Ten nanograms of DNA were used for multiplex PCR amplification of a panel covering 207 areas in 50 cancer-related genes including ABL1, AKT1, ALK, APC, ATM, BRAF, CDH1, CDKN2A, CBF1R,CTNNB1, EGFR, ERBB2, ERBB4, EZH2, FBXW7, FGFR1, FGFR2, FGFR3, FLT3, GNA11, GNAQ, GNAS,HNF1A, HRAS, IDH1, IDH2, JAK2, JAK3, KDR, KIT, KRAS, MET, MLH1, MPL, NOTCH1, NPM1, NRAS,PDGFRA, PIK3CA, PTEN, PTPN11, RB1, RET, SMAD4, SMARCB1, SMO, SRC, STK11, TP53, and VHL(Ion AmpliSeq Cancer Panel v2; Life Technologies, Carlsbad, CA). The library construction and subsequent enrichment of the paired samples were performed using the OneTouch system according to the manufacturer's protocol. Sequencing was performed on their 316 chip with a capacity of 300-500 megabases using the Ion PGM™ System (Life Technologies). Sequencing reads were mapped to the University of California, Santa Cruz (UCSC) human genome (GRCh37/hg19) using Torrent Suite™ software (Life Technologies).
Variant calling and classification of somatic mutations
Sequence data were analyzed using Variant Caller™ and Ion Reporter™ (Life Technologies). Calls of single nucleotide variants (SNV) less than 2% and calls of insertions and deletions (indels) less than 5% in tumor tissues were excluded from further analysis. Calls at positions with sequence coverage greater than 50 reads in both tumor and non-tumorous cells were analyzed, and those present in the tumor sample but not in the matched normal were regarded as somatic mutations. Fisher's exact test was carried out for the variants present in both tumor and corresponding normal tissue, and those detected at a significantly higher frequency in tumor tissues than the matched normal controls with p-values less than 0.01 were classified as somatic mutations. All somatic mutations were reviewed by Integrative Genomic Viewer (IGV).
The somatic mutations were classified into three groups, namely intronic, splice site, and exonic mutations by their location, and the exonic mutations were further divided into four types, namely exonic indels, nonsense, synonymous, and non-synonymous mutations. Pathological significance of the mutations was evaluated using several databases: ClinVar, COSMIC, ONCOMINE, Human Gene Mutation Database (HGMD), Leiden Open Variation Database (LOVD) in International Society for Gastrointestinal Hereditary Tumors (InSiGHT), TP53 Database in International Agency for Research on Cancer (IARC), dbSNP, and Human Genetic Variation Database (HGVD) in Kyoto University. Mutations reported to play a role in tumorigenesis, and those regarded deleterious in ClinVar, COSMIC, ONCOMINE, HGMD, LOVD, and TP53 Database were judged as pathological mutations. In addition, nonsense mutations, splice site mutations, and exonic indels were included as pathological mutations. Among the remaining mutations, we evaluated missense mutations using three prediction tools: SIFT, PolyPhen, and PANTHER. In this study, we considered those predicted to be damaged/deleterious/pathological by all three methods as pathological mutations, and other missense mutations as variants of uncertain significance (VUS). The remaining synonymous mutations were regarded as non-pathological alterations.
Histological analysis
Histological diagnosis was performed with all the tissue samples obtained by surgical resection and finally classified into PMCA and DPAM. Presence of signet ring cells was judged positive if the cells were observed at any percentages in the tumorous components but not in isolated mucin pools. Tissue sections of the 18 tumors were further analyzed by immunohistochemical staining with anti-TP53 antibody (sc-126, Santa Cruz Biotechnology, Inc., Delaware, CA), Histofine SAB-PO(R) Kit (Nichirei, Tokyo, Japan), and ImmPACT DAB systems (VECTOR LABORATORIES, Burlingame, CA). Expression of TP53 was scored into three classes, namely normal, negative, and excessive, according to previous reports [6, 7]. Normal staining referred to a weak focal nuclear staining, resembling a staining pattern for normal epithelium in less than 50% of tumor nuclei. A negative score indicated a complete lack of stained tumor nuclei (even though stromal cell nuclei showed normal pattern). The staining was classified as excessive if a strong nuclear staining was present in the majority of tumor cells (>50%).
Statistical analysis
Statistical differences were analyzed using the Fisher's exact test. Differences were considered statistically significant when the p-value was below 0.01.
Results
Treatments and clinicopathological features of the 18 PMP patients
The 18 PMPs originated from the appendix and were histologically classified into 10 DPAMs and 8 PMCAs. Components of signet ring cells were not detected in the 18 PMPs. A typical histological manifestation is shown in Figure 1. The gender and average age of the patients were not significantly different between the 10 DPAMs and the 8 PMCAs (Table 1). Patients underwent CRS (12 cases), debulking operation (5 cases), or exploratory laparotomy (1 case), and HIPEC was administered to the operations in 14 patients. EPIC was added to CRS-HIPEC in seven cases. The follow-up periods ranged from 1 to 37 months (median: 13 months). The periods are too short to compare the prognosis between the patients with DPAMs and those with PMCAs.
Figure 1. Hematoxylin and eosin (HE) staining (A–C) and immunohistochemical staining (D–F) of TP53. Typical histological appearance of DPAM (A) and PMCAs (B, C). Normal staining (D, F) and excessive staining (E). Magnification: ×200 (A–F) 
Genetic analysis
Genetic analysis of the 18 tumors and the matched normal tissues was performed by amplicon sequencing with multiplex PCR using Cancer Panel v2. The average sequence throughput per sample was approximately 22 Mb, and the average number of reads per amplicon was 951. Among the 207 regions analyzed, 203 (98.3%) were covered by more than 100 reads on average, and all regions (207/207) were covered by at least 20 reads on average. Subsequent mutation analysis with Valiant Caller™ and Ion Reporter™ detected a total of 62 variant calls that were candidates for somatic mutations. However, 24 of the 62 calls were filtered out for further analysis because they were observed in matched normal tissues and the frequency was not statistically different between the tumors and non-cancerous tissues. Among the remaining 38 alterations, three were located within homopolymers and they turned out to be miscalls by reviewing with IGV. We finally identified a total of 35 somatic mutations in the 18 tumors. All mutations are located in exons and comprised 33 non-synonymous mutations, 1 deletion, and 1 insertion.
The most frequently mutated gene was KRAS (14/18) and the second was GNAS (8/18), followed byTP53 (4/18), SMAD4 (3/18), AKT1 (1/18), NRAS (1/18), PIK3CA (1/18), PDGFRA (1/18), RET (1/18), andVHL (1/18) (Table 2). All 14 KRAS mutations were located at codons 12 or 13, the two major hot spots, and were missense mutations (p.G12D, p.G12V, p.G12S, and p.G13D). The eight GNASmutations were located at codon 201 (p.R201H and p.R201C). Since these mutants are well-known oncogenic forms, the mutations of KRAS and GNAS should play an active role in the tumorigenesis of PMP. All four TP53 mutations (p.R175H, p.P250L, p.G266R, and p.R273C) were located in the DNA-binding domain region and were reported to be pathological. Three SMAD4 mutations included two indels and one missense mutation (p.C499R). This missense mutation was located in the MAD homology 2-domain where many pathological mutations have been reported. We considered this mutation as a pathological mutation since all three prediction tools, PolyPhen SIFT, and PANTHER, judged it as a deleterious or pathological mutation. Thus, the three SMAD4 mutations should result in loss of SMAD4 function and play a vital role in the tumorigenesis.
Tab. 2. List of pathological mutations 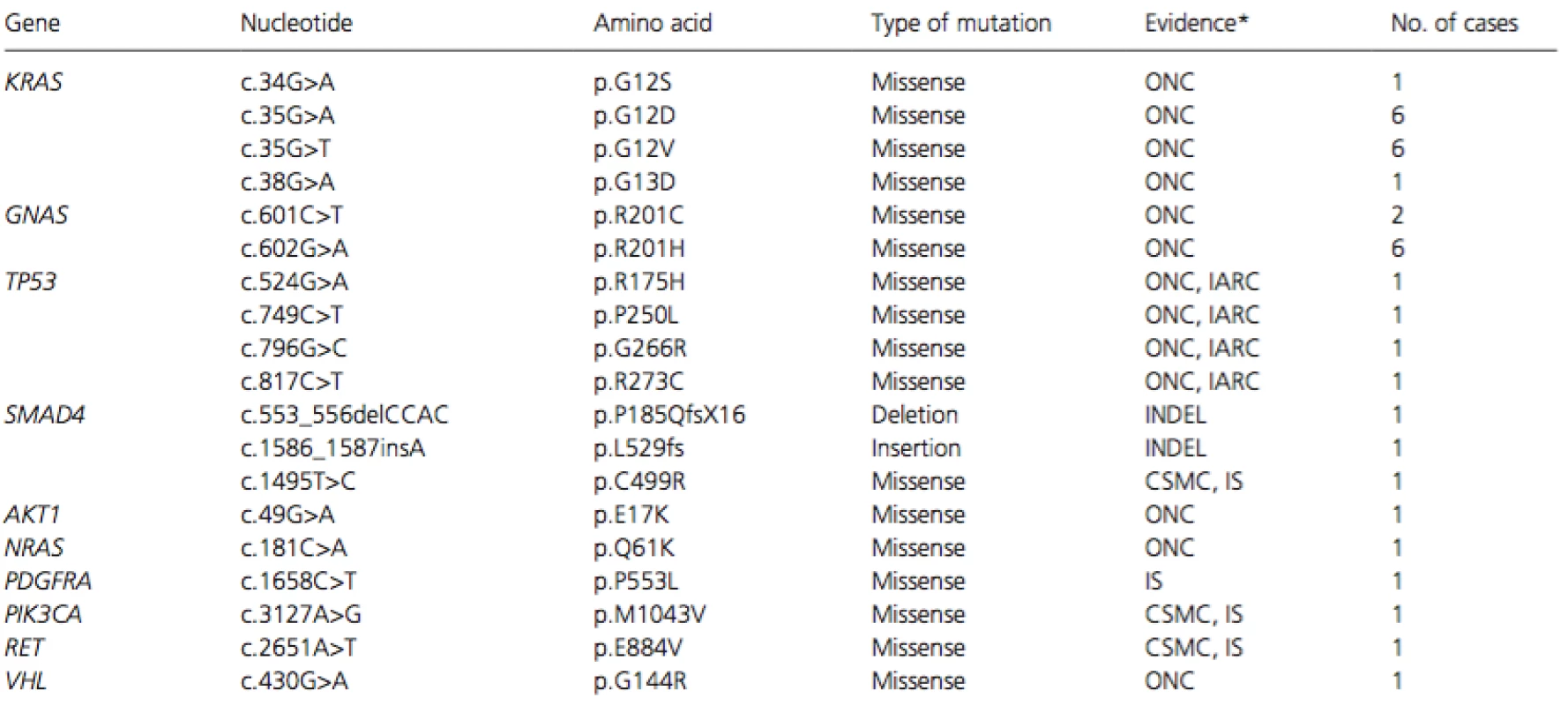
*Evidence of pathological significance was judged from the type of mutation (INDEL), databases including ONCOMINE (ONC), TP53 database in International Agency for Research on Cancer (IARC), and COSMIC (CSMC), or in silico (IS) analyses. We identified in this study a missense PDGFRA mutation of c.1658C>T (p.P553L) that has not been reported in literatures or public databases. In silico analysis of the mutation using the three algorithms predicted it as a pathological mutation; probably damaging (score:1) by PolyPhen, deleterious (score:0) by SIFT, and deleterious (subPSEC: −6.28, Pdeterious: 0.96) by PANTHER. Taken together, all 35 somatic mutations were classified as pathological mutations.
Mutation profiles of DPAMs and PMCAs
We further compared mutation profiles between the 10 DPAMs and the 8 PMCAs. It is of note thatKRAS mutations were observed in both types of tumors (eight of the ten DPAMs and six of the eight PMCAs), and that a mutation of NRAS was found in a PMCA without KRAS mutation. These data suggest that the activation of RAS family members is commonly involved in both DPAMs and PMCAs.GNAS mutations were detected in five DPAMs and three PMCAs, implying that activation of GNAS may also play a key role in the development of both types of tumors. On the other hand, TP53mutations were found in three PMCAs but not in the 10 DPAMs, suggesting that the inactivation of TP53 may render malignant properties to PMCAs. Similarly, mutations in PIK3CA, AKT1, or PDGFRAwere found in the PMCAs, but not in the DPAMs. It is worthwhile to note that mutations of PIK3CA orAKT1 were found in two PMCAs without TP53 mutations but not in the DPAMs, and that the mutations were mutually exclusive. Therefore, activation of the PI3K-AKT pathway may also play an important role in the malignant transformation of PMP.
Accumulation of TP53 in PMCAs
To corroborate the deregulation of TP53 in PMCAs, we performed immunohistochemical staining of TP53 using paraffin-embedded tissues (Fig. 1). Consistent with the mutation data, strong accumulation of TP53 was not observed in the DPAMs, but three of the eight PMCAs revealed excessive TP53 staining. As expected, we found the accumulated TP53 protein in the three tumors carrying TP53 mutation (Table 3).
Tab. 3. Mutation status and IHC of TP53 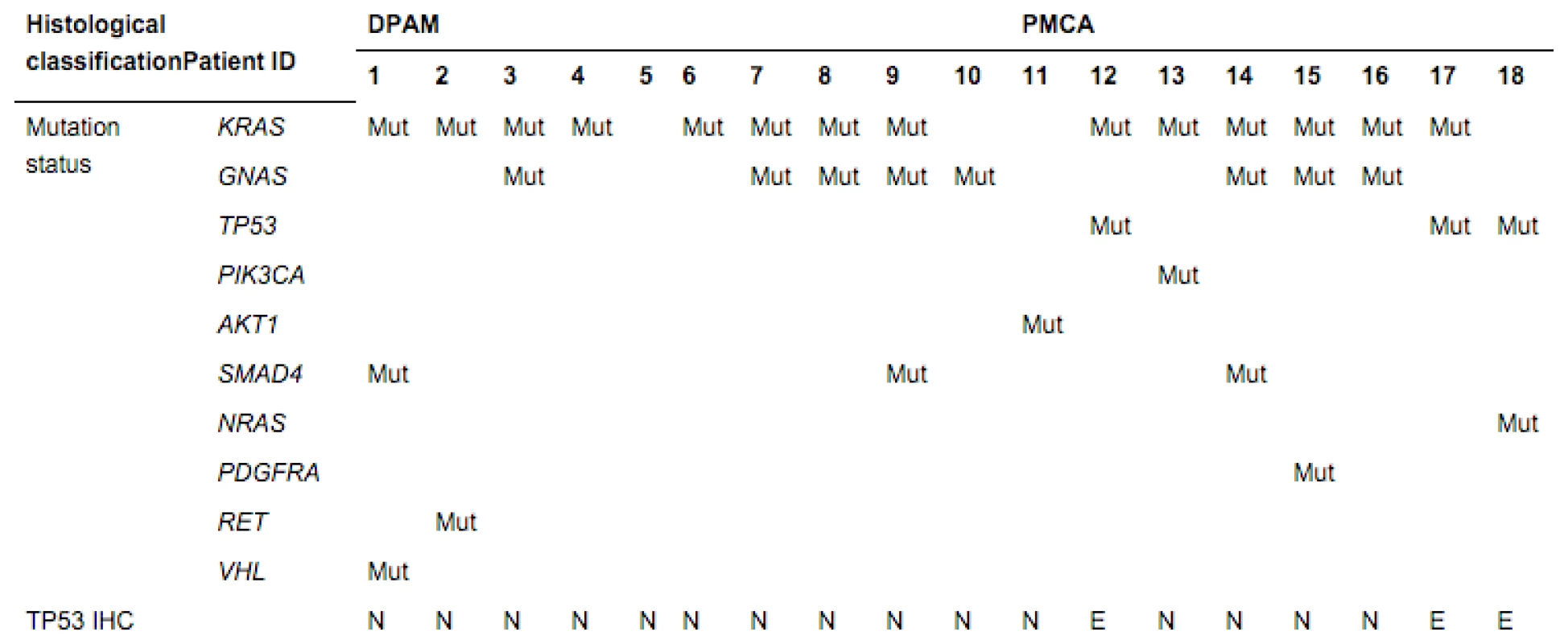
Mut, a pathological mutation (somatic) was identified; blank, no somatic mutation was identified; N, normal staining; E, excessive staining. Discussion
In this study, we performed genetic analysis of 18 Japanese PMPs and unveiled their mutation profiles. Analysis of the data disclosed that both DPAMs and PMCAs carry frequent mutations in RASfamily members and GNAS, and that mutations in genes associated with TP53 and/or PI3K-AKT pathways may play a crucial role in the malignant transformation of PMP.
In accordance with our data, several groups have already reported frequent KRAS mutations in PMP at a frequency between 58% and 94% (37 [57.8%] of 64 PMP, Shetty et al. [8]; 33 [94.3%] of 35 PMP, Nishikawa et al. [9]; 70%, Sio et al. [10]; and 75%, Liu et al. [11]). Since the frequency of KRASmutation in colorectal cancer is approximately 40%, KRAS mutation in PMP is markedly high. In regard to the difference of KRAS mutation between PMCAs and DPAMs, two groups showed more frequent mutations in PMCAs than DPAMs, but other groups did not observe significant difference between the two types of tumors [8, 9]. Our data support the latter view and revealed frequent mutations in DPAMs as well as PMCAs. Although we did not find KRAS mutations in four tumors, one of the four had an NRAS mutation, suggesting that the remaining three tumors may carry mutations in other regions of KRAS or NRAS, or in other genes associated with the RAS-MAPK pathway. These data imply that the activation of RAS-MAPK pathway plays a crucial role in both types of PMP, and that anticancer drugs targeting this pathway should be a rational strategy to treat PMP.
In good agreement with previous reports [9-11], we identified GNAS mutations at a similar frequency in PMPs (44.4%). Although Nishikawa et al. [9] identified GNAS mutations in DPAMs but not in PMCAs, we found GNAS mutations in both PMCAs (3/8, 37.5%) and DPAMs (5/10, 50%). This result suggested that GNAS mutation may also play an important role in the development of both types of PMP. In this study, GNAS mutations were not associated with any clinicopathological factors including gender, age, and histological stages (data not shown). The GNAS gene encodes the α-subunit of stimulatory G protein, and the R201C and R201H mutants at codon 201, the mutation hot spot, enhance the activity of adenylyl cyclase. Consequently, mutated GNAS leads to the activation of the protein kinase A (PKA) pathway through the induction of cyclin AMP (cAMP). Recently, a group reported that GNAS regulates expression of MUC2 and MUC5AC in colon cancer cells, and that its expression does not associate with cell proliferation both in vitro and in vivo [9]. GNAS mutations are also frequently identified in intraductal papillary mucinous neoplasms (IPMN) of the pancreas [12]. Since it is reported that the secreted mucin from PMP contains MUC2, MUC5AC, and MUC5B, activated GNAS may induce the production of these mucin in both PMP and IPMN.
In this study, TP53 mutations were shown in three of the eight PMCAs, but not in any of the ten DPAMs. Consistently, immunohistochemical staining showed excessive accumulation of TP53 protein in the three PMCAs, but not in other PMCAs with wild-type TP53 or the 10 DPAMs, corroborating that impaired TP53 pathway is associated with PMCA. Two groups have reported low frequencies of TP53mutations in PMP so far; one of ten high-grade PMPs [7], and two of eight low-grade/well-differentiated mucinous adenocarcinoma with PMP [11]. On the other hand, several groups found accumulation of TP53 at a higher frequency in PMCA than DPAM by immunohistochemical staining [7, 8]. Although the frequency of TP53 accumulation is different among the studies, possibly because of different antibodies and/or condition of the tissues analyzed, these studies have suggested that TP53 deregulation is more frequently associated with PMCA than DPAM. Since TP53mutation is involved in the step of malignant transformation, impaired TP53 pathway may render malignant properties to PMCA. In addition, immunohistochemical staining of TP53 may be useful as a biomarker of PMCA.
Although the number of mutations is small, we identified mutations in PIK3CA and AKT1 in PMCAs, but not in DPAMs. PI3K-AKT pathway plays a crucial role in regulating a number of cellular processes, such as apoptosis, migration, angiogenesis, cell proliferation, and glucose metabolism. It is of note that 14–32% of colorectal cancer harbored somatic mutations in PIK3CA and that the mutations are rare in premalignant lesions of the colon [13, 14]. Thus, deregulation of PI3K-AKT pathway may also play a crucial role in the progression of PMP. Since the oncogenic mutations in PIK3CA induce downstream signaling through the phosphorylation of AKT and subsequent activation of mTOR complex 1 (mTORC1), inhibition of PI3K, AKT, and/or mTORC1 may be an effective therapeutic strategy to treat the tumors with an oncogenic PIK3CA mutation. Additionally, inhibitors of AKT and/or mTORC1 may benefit the patients with an oncogenic AKT1 mutation.
In this study, we identified a novel mutation in PDGFRA of c.1658C>T (p.P553L). Although the most frequent mutation of PDGFRA (p.D842V) is located in the tyrosine kinase 2 domain, the juxtamembrane domain (exon12) also harbors frequent mutations such as p.D561V. Taking the location together with the in silico data using the three tools into our consideration, we judged that p.P553L mutation was a pathological mutation. PDGFRA mutations are reported in human cancers including colorectal cancer (6.0%) and stomach cancer (2.6%) in the public data of COSMIC v71. It is of note that frequent PDGFRA mutations have been reported in gastrointestinal stromal tumors (GIST) (10%) [15]. Most of the mutations are considered to activate its kinase activity and autonomously induce its downstream molecules. Importantly, tumors carrying PDGFRA mutations were shown to be sensitive to the in vitro treatment with Imatinib, an inhibitor of kinase activity of bcr-abl, cKit, and PDGFRA [16, 17]. Therefore, patients with the c.1658C>T (p.P553L) mutation may benefit from Imatinib treatment.
Recently, PMPs with signet ring cells are reported to be associated with aggressive characteristics and poor outcomes by Davison et al. and Shetty et al. [18, 19]. Although we did not find presence of signet ring cells in the 18 PMPs, further studies are necessary for the better classification of PMP and the elucidation of biological properties and genetic profiles of PMP with signet cell component.
In conclusion, we have found in the present study that KRAS and/or GNAS mutations are frequently involved in the development of PMPs, and that disruption of TP53 or PI3K-AKT pathway may play a crucial role in their progression. In addition, we have shown here that immunohistological staining of TP53 may be useful for the classification of PMPs. Importantly, analysis of mutation profiles may be useful not only for the comprehensive understanding of their tumorigenesis, but also the personalized therapy for patients with PMP.
Acknowledgments
The authors thank Hatakeyama, Yamaguchi, Yusa, and Onoda for their excellent technical assistance. This work was supported in part by the Grant-in-Aid for Young Scientists to K. Yamaguchi (#26860206) from the Japan Society for the Promotion of Science.
Conflict of Interest
None declared.
Funding Information
This work was supported in part by the Grant-in-Aid for Young Scientists to
K. Yamaguchi (#26860206) from the Japan Society for the Promotion of Science.Received: 8 May 2015;
Revised: 18 August 2015;
Accepted: 23 August 2015
* Correspondence
Yoichi Furukawa, Division of Clinical Genome Research, Advanced Clinical Research Center, Institute of Medical Science, The University of Tokyo, 4-6-1 Shirokanedai, Minato-ku, Tokyo 108-8639, Japan. Tel: +81 3 6409 2100; Fax: +81 3 6409 2101; E-mail: furukawa@ims.u-tokyo.ac.jp
Zdroje
1. Prayson, R. A., W. R. Hart, and R. E. Petras. 1994. Pseudomyxoma peritonei. A clinicopathologic study of 19 cases with emphasis on site of origin and nature of associated ovarian tumors. Am. J. Surg. Pathol.18 : 591–603.
2. Smeenk, R. M., M. L. van Velthuysen, V. J. Verwaal, and F. A. Zoetmulder. 2008. Appendiceal neoplasms and pseudomyxoma peritonei: a population based study. Eur. J. Surg. Oncol. 34 : 196–201.
3. Misdraji, J., R. K. Yantiss, F. M. Graeme-Cook, U. J. Balis, and R. H. Young. 2003. Appendiceal mucinous neoplasms: a clinicopathologic analysis of 107 cases. Am. J. Surg. Pathol. 27 : 1089–1103.
4. Bradley, R. F., J. Ht. Stewart, G. B. Russell, E. A. Levine, and K. R. Geisinger. 2006. Pseudomyxoma peritonei of appendiceal origin: a clinicopathologic analysis of 101 patients uniformly treated at a single institution, with literature review. Am. J. Surg. Pathol. 30 : 551–559.
5. Jacquet, P., and P. H. Sugarbaker. 1996. Current methodologies for clinical assessment of patients with peritoneal carcinomatosis. J. Exp. Clin. Canc. Res. 15 : 49–58.
6. Vartiainen, J., H. Lassus, P. Lehtovirta, P. Finne, H. Alfthan, R. Butzow, et al. 2008. Combination of serum hCG beta and p53 tissue expression defines distinct subgroups of serous ovarian carcinoma. Int. J. Cancer122 : 2125–2129.
7. Nummela, P., L. Saarinen, A. Thiel, P. Jarvinen, R. Lehtonen, A. Lepisto, et al. 2015. Genomic profile of pseudomyxoma peritonei analyzed using next-generation sequencing and immunohistochemistry. Int. J. Cancer 136:E282–E289.
8. Shetty, S., P. Thomas, B. Ramanan, P. Sharma, V. Govindarajan, and B. Loggie. 2013. Kras mutations and p53 overexpression in pseudomyxoma peritonei: association with phenotype and prognosis. J. Surg. Res.180 : 97–103.
9. Nishikawa, G., S. Sekine, R. Ogawa, A. Matsubara, T. Mori, H. Taniguchi, et al. 2013. Frequent GNAS mutations in low-grade appendiceal mucinous neoplasms. Br. J. Cancer 108 : 951–958.
10. Sio, T. T., A. S. Mansfield, T. E. Grotz, R. P. Graham, J. R. Molina, F. G. Que, et al. 2014. Concurrent MCL1 and JUN amplification in pseudomyxoma peritonei: a comprehensive genetic profiling and survival analysis. J. Hum. Genet. 59 : 124–128.
11. Liu, X., K. Mody, F. B. de Abreu, J. M. Pipas, J. D. Peterson, T. L. Gallagher, et al. 2014. Molecular profiling of appendiceal epithelial tumors using massively parallel sequencing to identify somatic mutations. Clin. Chem.60 : 1004–1011.
12. Furukawa, T., Y. Kuboki, E. Tanji, S. Yoshida, T. Hatori, M. Yamamoto, et al. 2011. Whole-exome sequencing uncovers frequent GNAS mutations in intraductal papillary mucinous neoplasms of the pancreas. Sci. Rep.1 : 161.
13. Samuels, Y., Z. Wang, A. Bardelli, N. Silliman, J. Ptak, S. Szabo, et al. 2004. High frequency of mutations of the PIK3CA gene in human cancers. Science 304 : 554.
14. Velho, S., C. Oliveira, A. Ferreira, A. C. Ferreira, G. Suriano, S. Schwartz Jr, et al. 2005. The prevalence of PIK3CA mutations in gastric and colon cancer. Eur. J. Cancer 41 : 1649–1654.
15. Corless, C. L. 2014. Gastrointestinal stromal tumors: what do we know now? Mod. Pathol.27(Suppl 1):S1–S16.
16. Corless, C. L., A. Schroeder, D. Griffith, A. Town, L. McGreevey, P. Harrell, et al. 2005. PDGFRA mutations in gastrointestinal stromal tumors: frequency, spectrum and in vitro sensitivity to imatinib. J. Clin. Oncol.23 : 5357–5364.
17. Hirota, S., A. Ohashi, T. Nishida, K. Isozaki, K. Kinoshita, Y. Shinomura, et al. 2003. Gain-of-function mutations of platelet-derived growth factor receptor α gene in gastrointestinal stromal tumors.Gastroenterology 125 : 660–667.
18. Shetty, S., B. Natarajan, P. Thomas, V. Govindarajan, P. Sharma, and B. Loggie. 2013. Proposed classification of pseudomyxoma peritonei: influence of signet ring cells on survival. Am. Surg. 79 : 1171–1176.
19. Davison, J. M., H. A. Choudry, J. F. Pingpank, S. A. Ahrendt, M. P. Holtzman, A. H. Zureikat, et al. 2014.Clinicopathologic and molecular analysis of disseminated appendiceal mucinous neoplasms: identification of factors predicting survival and proposed criteria for a three-tiered assessment of tumor grade. Mod. Pathol. 27 : 1521–1539.
Štítky
Onkologie
Článek vyšel v časopiseCancer Medicine
Nejčtenější tento týden
2015 Číslo Early View- Nejlepší kůže je zdravá kůže: 3 úrovně ochrany v moderní péči o stomii
- Metamizol v léčbě různých bolestivých stavů – kazuistiky
- Metamizol v terapii bolesti v ambulanci praktického lékaře i pediatra
- Nejasný stín na plicích – kazuistika
- Zatím roční přežití bez progrese ve 2. linii léčby metastatického karcinomu pankreatu – kazuistika
-
Všechny články tohoto čísla
- Pretreatment biopsy analysis of DAB2IP identifies subpopulation of high-risk prostate cancer patients with worse survival following radiation therapy
- Sorafenib for the treatment of advanced hepatocellular carcinoma with extrahepatic metastasis: a prospective multicenter cohort study
- Molecular profiles of high-grade and low-grade pseudomyxoma peritonei
- Association between epithelial-mesenchymal transition and cancer stemness and their effect on the prognosis of lung adenocarcinoma
- National treatment patterns in patients presenting with Stage IVC head and neck cancer: analysis of the National Cancer Database
- Cancer Medicine
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Molecular profiles of high-grade and low-grade pseudomyxoma peritonei
- National treatment patterns in patients presenting with Stage IVC head and neck cancer: analysis of the National Cancer Database
- Association between epithelial-mesenchymal transition and cancer stemness and their effect on the prognosis of lung adenocarcinoma
- Pretreatment biopsy analysis of DAB2IP identifies subpopulation of high-risk prostate cancer patients with worse survival following radiation therapy
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



