-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaExpression Profiling during Arabidopsis/Downy Mildew Interaction Reveals a Highly-Expressed Effector That Attenuates Responses to Salicylic Acid
A comprehensive understanding of host-pathogen interactions requires knowledge of the dynamics of gene expression changes in both the host and the pathogen during a time course of infection. However, expression profiling has often focused on either the host or the pathogen due to limitations of methods that involve microarrays. We report here gene expression changes in both Arabidopsis and its parasite Hyaloperonospora arabidopsidis (Hpa) simultaneously during infection using a high-throughput RNA sequencing method. By resequencing Hpa isolate Waco9, we found it evades Arabidopsis resistance gene RPP1 through deletion of cognate recognized effector ATR1. We also found that Hpa suppresses responsiveness to salicylic acid (SA) in haustoriated cells into which host-translocated effectors are delivered. An Hpa effector HaRxL62, previously shown to enhance host susceptibility, was highly expressed in this assay, and we found it suppresses responsiveness to SA. Expression profiling of both pathogen effector genes and host genes involved in immunity allows us to suggest distinct mechanisms of effector-mediated susceptibility and reveals interesting Hpa effectors for detailed mechanistic investigation in future experiments.
Published in the journal: . PLoS Pathog 10(10): e32767. doi:10.1371/journal.ppat.1004443
Category: Research Article
doi: https://doi.org/10.1371/journal.ppat.1004443Summary
A comprehensive understanding of host-pathogen interactions requires knowledge of the dynamics of gene expression changes in both the host and the pathogen during a time course of infection. However, expression profiling has often focused on either the host or the pathogen due to limitations of methods that involve microarrays. We report here gene expression changes in both Arabidopsis and its parasite Hyaloperonospora arabidopsidis (Hpa) simultaneously during infection using a high-throughput RNA sequencing method. By resequencing Hpa isolate Waco9, we found it evades Arabidopsis resistance gene RPP1 through deletion of cognate recognized effector ATR1. We also found that Hpa suppresses responsiveness to salicylic acid (SA) in haustoriated cells into which host-translocated effectors are delivered. An Hpa effector HaRxL62, previously shown to enhance host susceptibility, was highly expressed in this assay, and we found it suppresses responsiveness to SA. Expression profiling of both pathogen effector genes and host genes involved in immunity allows us to suggest distinct mechanisms of effector-mediated susceptibility and reveals interesting Hpa effectors for detailed mechanistic investigation in future experiments.
Introduction
During co-evolution with pathogens, plants have evolved multiple immune signaling mechanisms that successful pathogens have evolved to evade or suppress. The first layer is based on recognition of broadly conserved pathogen molecules (pathogen/microbe-associated molecular patterns, PAMP/MAMPs) by plant cell surface pattern-recognition receptors (PRRs), resulting in PAMP - (or pattern)-triggered immunity (PTI) [1]. However, PTI can be suppressed by pathogen proteins, termed effectors, that are delivered into the apoplast or plant cell cytoplasm, resulting in effector-triggered susceptibility. Plants also carry a second layer of defense, so-called effector triggered immunity (ETI), in which cytoplasmic disease resistance (R) proteins recognize directly or indirectly the presence of pathogen effectors. Recognized effectors are often known as avirulence (AVR) proteins [2], [3]. A hallmark of ETI is the hypersensitive response (HR), which involves programmed cell death at pathogen infection sites and helps resist biotrophic pathogens.
In many oomycetes, such as Phytophthora spp. and downy mildews, the most common host-translocated effectors are the RxLR-type proteins that contain an N-terminal signal peptide and a RxLR (or RxLR-EER) motif involved in secretion and host uptake, and a C-terminal domain carrying the effector activity [3]–[5]. Hyaloperonospora arabidopsidis (Hpa; formerly Peronospora parasitica or Hyaloperonospora parasitica) is an obligate biotrophic oomycete that causes downy mildew in Arabidopsis thaliana. The Arabidopsis-Hpa pathosystem has been extensively used to study host/pathogen co-evolution, and has enabled identification of cognate host R and pathogen AVR genes, termed RPP (recognition of Peronospora parasitica) and ATR (Arabidopsis thaliana recognized), respectively [6]. Genome analysis of Hpa isolate Emoy2 identified 134 high-confidence effector candidates (HaRxL genes) [7]. Comprehensive screening of HaRxL effectors revealed that the majority of HaRxLs contribute positively to pathogen fitness [8], [9]. In addition, HaRxLs can be located in different subcellular compartments in planta [10]. Some have been shown by yeast two hybrid screens to interact with various plant proteins [11]. However, the mechanisms by which most Hpa effectors promote virulence remain to be elucidated.
Salicylic acid (SA) is a phytohormone essential for the immune response against biotrophic pathogens [12]. SA biosynthesis is triggered during both PTI and ETI [13]. Signaling downstream of SA is largely controlled by the regulatory protein NON-EXPRESSOR OF PR GENES1 (NPR1), which upon activation by SA acts as a transcriptional coactivator of a large set of defense-related genes, such as PATHOGENESIS-RELATED GENE 1 (PR1) [14]. Another phytohormone, jasmonic acid (JA), is synthesized upon pathogen and herbivore attack, and is essential for the immune response against necrotrophic pathogens and herbivores [15]. Multiple studies revealed a mutually antagonistic interaction between SA - and JA-dependent signaling [16], [17]. Some pathogens and herbivores appear to induce SA-JA crosstalk [18]–[23]. For example, Pseudomonas syringae produces coronatine, a toxin that mimics the bioactive jasmonate JA-isoleucine [24] and promotes stomatal reopening and bacterial propagation in both local and systemic tissues by inhibiting SA signaling and accumulation [20], [23]. In addition to SA and JA, recent studies have revealed involvement of other phytohormones, such as ethylene (ET), abscisic acid (ABA), gibberellin and auxin, in biotic interactions [25]. Remarkably, several pathogens produce phytohormones and phytohormone mimics like coronatine in P. syringae.
To dissect the Arabidopsis-Hpa interaction, changes in expression of Arabidopsis or Hpa genes during infection were previously investigated by microarray analysis for Arabidopsis genes [26]–[29] and by cDNA-amplified fragment length polymorphism and expressed sequence tag analysis for Hpa genes [30]–[32]. In Hpa, however, these approaches were not sensitive enough to enable genome-wide quantification of changes in gene expression during infection. Expression profiling in Arabidopsis or Hpa was carried out with different Arabidopsis accessions, Hpa isolates, plant ages and infection time courses, hindering comparison of these data. Recently, we established a high-throughput mRNA expression-profiling method (Expression Profiling through Random Sheared cDNA tag Sequencing [EXPRSS]) enabling the detection of differential expression of more genes, with higher sensitivity, than microarray and traditional RNA sequencing methods [33]. Briefly, EXPRSS is a restriction enzyme-independent tag-sequencing method and generates one tag per transcript at a relatively defined position from the 3′ end of a gene, ensuring no length-based data transformation and enabling expression data to be obtained at a ∼10× greater read depth than standard Illumina RNA sequencing. This is helpful when we investigate low-level transcripts, such as pathogen transcripts in host-pathogen interactions. Using EXPRSS, we monitored mRNA levels for both Arabidopsis and Hpa genes during infection. Here, we report the expression patterns of Hpa predicted effectors and Arabidopsis genes on the basis of transcriptome data in Arabidopsis Col-0 inoculated with the avirulent Hpa isolate Emoy2 (recognized by RPP4 [34]) or the virulent isolate Waco9. From this analysis, we found that ATR1 (recognized by RPP1 [35]) is not expressed in Hpa Waco9, and after resequencing the Waco9 genome, we found the ATR1 region is deleted. An Hpa effector HaRxL62, previously shown to enhance host susceptibility [8], [9], was highly expressed in this assay, and was shown here to suppress responsiveness to SA.
Results
Expression profiling of host and pathogen during Arabidopsis-Hpa interaction
Arabidopsis Col-0 was inoculated with either the avirulent isolate Emoy2 (incompatible interaction) or the virulent isolate Waco9 (compatible interaction) of Hpa, and infected plants were harvested at 1, 3 and 5 days post-inoculation (dpi) prior to Illumina sequencing using EXPRSS [33]. Hpa haustoria are formed in both compatible and incompatible interactions till 1 dpi, and HR cell death is observed only in incompatible interactions [36]. HR was observed in Hpa Emoy2-inoculated leaves of Col-0 from 3 dpi, whereas no visible HR was observed at 1 dpi (Figure 1A). After Hpa Waco9 inoculation, extensive growth of intercellular mycelium was evident on leaves from 3 dpi, and then sporulation (conidiophores bearing conidiospores) was observed at 5 dpi (Figure 1A). In addition to the infectious stages, samples were taken from intact plants (0 dpi) and water-sprayed (mock-treated) plants as control samples for transcriptome analysis in Arabidopsis. Further, to evaluate the expression pattern of Hpa genes, samples were taken from conidiospores before inoculation. The experiment was carried out with three independent biological replicates.
Fig. 1. Hpa development and scheme for aligning Illumina sequence reads. 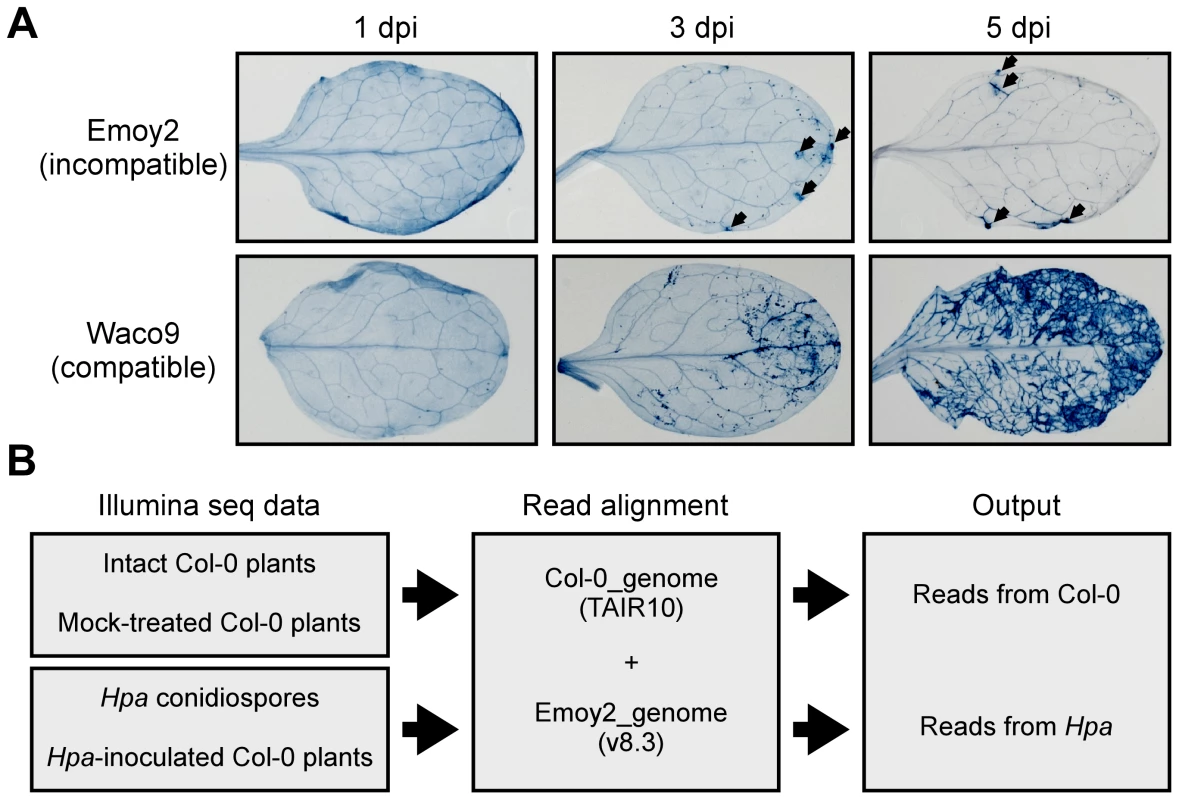
(A) Trypan blue staining in three-week-old Arabidopsis Col-0 plants at 1, 3 and 5 dpi with Hpa Emoy2 and Waco9. Black arrows indicate the parts in which HR cell death was observed. (B) Work-flow scheme to separate Illumina sequencing reads from Arabidopsis and Hpa. Total RNA was prepared from infected plants, and libraries for EXPRSS were prepared. Although 36 bp sequencing reads are sufficient to identify Arabidopsis genes distinctly using EXPRSS [33], longer sequencing reads (80 bp) were used in this study to avoid cross-mapping to the Arabidopsis and Hpa genomes. The Illumina sequencing reads were mapped to the combined genome of Arabidopsis TAIR10 and Hpa Emoy2 v8.3 [7] (Figure 1B). Mapped-reads to Arabidopsis and Hpa genomes were counted separately and the distribution of mean expression of each gene was represented as TPM (tags per million) of total reads mapped to Arabidopsis or Hpa genomes. To provide sufficient depth for expression analysis of Hpa genes in infected plants, Illumina sequencing was carried out twice for the incompatible interaction (Hpa Emoy2-inoculated plants) and for the early time point (at 1 dpi) of the compatible interaction (Hpa Waco9-inoculated plants). In this study, we did the analyses using uniquely mapped or up to 10 matching reads (Table S1 and Datasets S1, S2 and S3; see Materials and Methods). Using only uniquely mapped reads would give a minimum estimate of high confidence in gene expression, but we might even discard the information for homologous genes. Although we cannot rule out the presence of some false positives and false negatives in the data using up to 10 matching reads, the data would contain more information including homologous genes. For these reasons, the data with up to 10 matching reads were used in the following analyses. Most reads in intact and mock-treated plants were mapped to the Arabidopsis genome (i.e. % Hpa reads <0.005), whereas most reads from Hpa conidiospores were mapped to the Hpa genome (i.e. % Hpa reads >91.7) (Table 1 and Figure S1). The reads mapped to the Arabidopsis genome in samples from Hpa conidiospores are likely to be due to Arabidopsis contamination in the spore inoculum, as Hpa was propagated on susceptible Arabidopsis accessions and its conidiospores were collected from infected Arabidopsis leaf tissues. The results suggest high gene-identification accuracy between Arabidopsis and Hpa in this study.
Tab. 1. Summary of transcriptome data in Arabidopsis inoculated with Hpa. 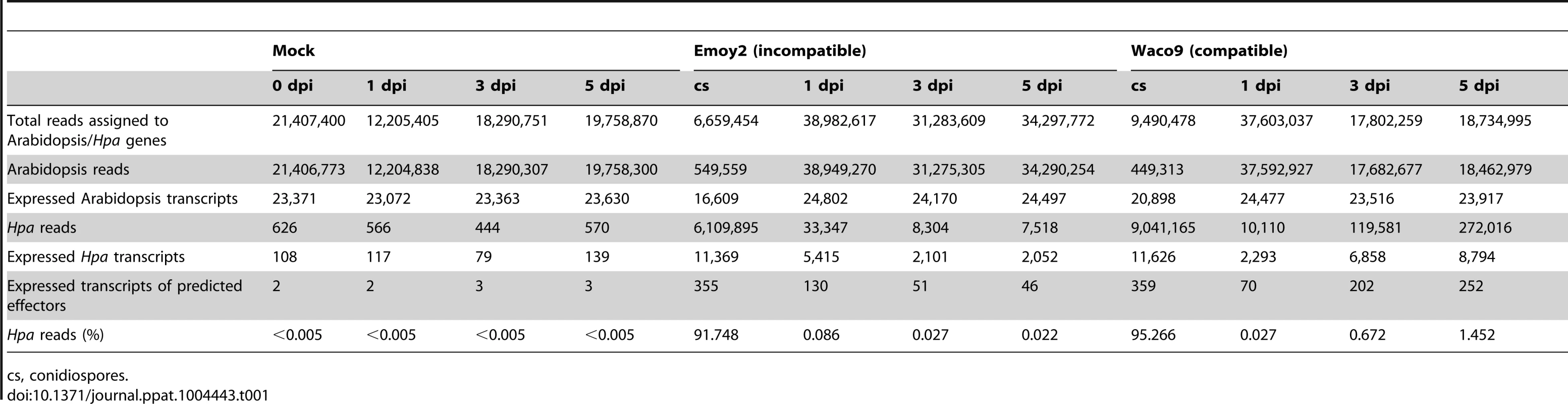
cs, conidiospores. In the incompatible interaction, the number of Hpa reads clearly decreased from 1 dpi, whereas the population of Hpa reads increased in the compatible interaction (Figure S1 and Table 1). This indicates that Hpa Emoy2 dies upon recognition after 1 dpi, corresponding to visible HR from 3 dpi with Emoy2 (Figure 1A). Hence, the data at 3 and 5 dpi with Emoy2 were omitted from the Hpa transcriptome data. The analysis of the overall transcriptome data revealed that out of 27,416 protein coding genes in Arabidopsis TAIR10 and 14,489 genes in Hpa v8.3, 24,559 (89.6%) for Arabidopsis and 11,394 (78.6%) and 11,690 (80.7%) for Hpa Emoy2 and Waco9, respectively, were expressed in at least one of the samples (Table 2 and Datasets S1, S2 and S3).
Tab. 2. The number of genes detected in this study. 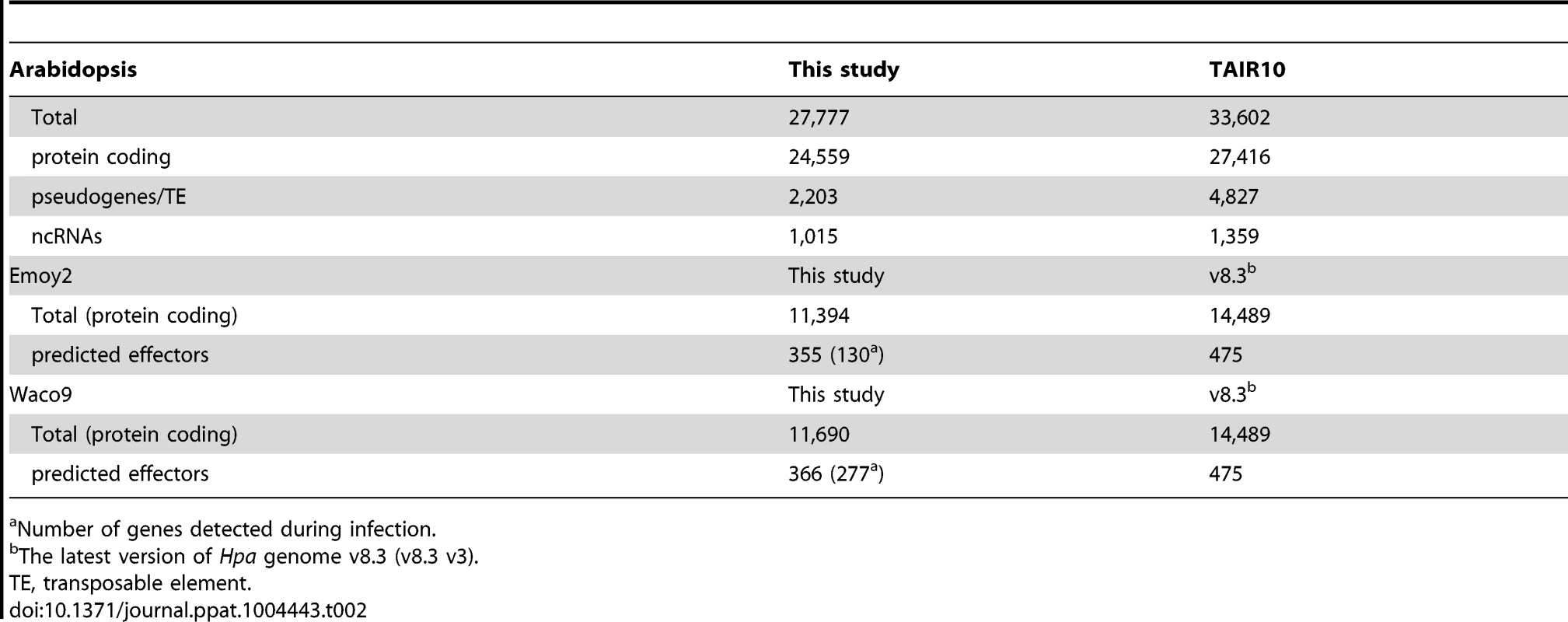
Number of genes detected during infection. Expression pattern of Hpa predicted effectors during infection
The Hpa Emoy2 genome analysis revealed 134 high-confidence effector candidates (HaRxLs) with a signal peptide and canonical RxLR (or RxLR-EER) motif [7]. These include effector candidates HaRxL17, HaRxL44 and HaRxL96 [10], [18], [37] and avirulent effectors ATR1, ATR13 and ATR39 [35], [38], [39]. ATR5 containing a signal peptide and canonical EER motif, but not a canonical RxLR motif, was identified as an avirulence gene recognized by RPP5 [40]. This report suggests the existence of effector candidates without canonical RxLR motif. In our study, we defined a total of 475 genes as predicted effectors (Table S2). The selection criteria for predicted effectors were the following: (1) high-confidence effector candidates (HaRxLs), (2) RxLR-like genes with at least one non-canonical feature, as for ATR5 (HaRxLLs), (3) putative Crinkler-homologous genes with RxLR motif (HaRxLCRNs) [4], (4) homologous genes based on amino acid sequence similarity over the 5′ region including a signal peptide and RxLR motif (e.g. HaRxL1b).
Transcriptome analysis of the compatible interaction revealed that 277 predicted effectors were expressed in at least one infection time point (Table 2). By quantifying the expression level, we found predicted effectors expressed highly during infection, e.g. HaRxL76 and HaRxL62 (about 0.2% and 0.1% of total Hpa mRNA at 3 dpi, respectively). In addition, most of the highly-expressed predicted effectors were upregulated more than two fold at 3 dpi compared to the expression level in conidiospores (Figure 2A). These findings suggested specific regulation of expression of some predicted effector genes upregulated at 3 dpi. To predict potential cis-regulatory elements in the upstream regions of Hpa genes, we categorized genes into five groups as follows; 87 predicted effectors which were induced more than two fold at 3 dpi (induced effectors), 115 predicted effectors which were detected at 3 dpi but were not induced more than two fold at 3 dpi (non-induced effectors), 1,880 genes excluding predicted effectors which were induced more than two fold at 3 dpi (induced genes exc effectors), 4,776 genes excluding predicted effectors which were detected at 3 dpi but were not induced more than two fold (non-induced genes exc effectors), and 14,489 genes predicted in Hpa v8.3 (all genes) (Table S3). The expression pattern of “induced effectors” and “non-induced effectors” was similar to “induced genes exc effectors” and “non-induced genes exc effectors”, respectively (Figure 2B). The sets of promoters of “induced effectors” and “non-induced effectors” were searched separately for conserved motifs using MEME [41], and then the motifs found were evaluated for over-representation in other groups using FIMO [42]. The INR-FPR motif, known as a core promoter element in oomycete genes [43], [44], was over-represented within 200 nt upstream of the start codon of “induced effectors” (E-value = 9.3e-068) (Figure 2C and D). The motif was also significantly over-represented in “non-induced effectors” and “induced genes exc effectors” (Figure 2D and Table S4), suggesting that INR-FPR motif is enriched in promoters of predicted effectors and genes induced during infection in Hpa. We also found two novel motifs (Motif I and II) within 500 nt upstream of the start codon that do not show any significant similarity to known motifs as determined by a TOMTOM search against the JASPAR database [45]. Interestingly, Motif I was overrepresented in only “induced effectors” (E-value = 8.0e-003), whereas Motif II was overrepresented in only “non-induced effectors” (E-value = 1.1e-003) (Figure 2F, G, I and J). The results suggest that Motif I and II might play a role in the regulation of the expression of predicted effector genes in Hpa.
Fig. 2. Expression pattern of Hpa predicted effectors and potential cis-regulatory elements in Hpa. 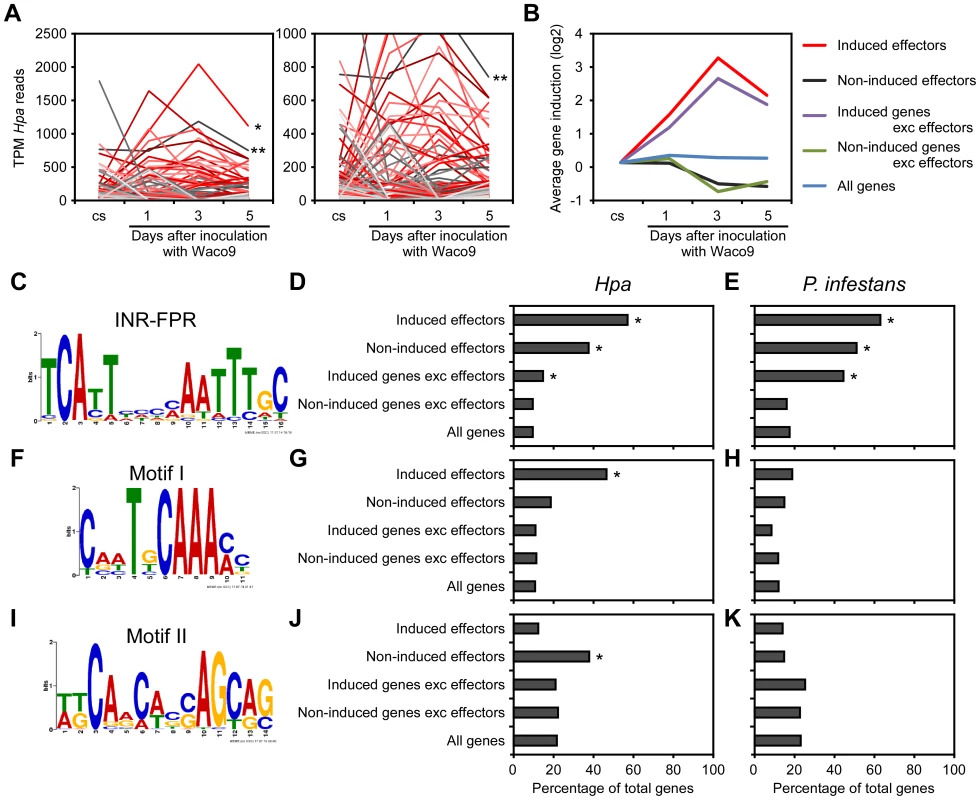
(A) Expression pattern of predicted effectors expressed in at least one of three infections (1, 3, and 5 dpi) with Hpa Waco9. Expression levels were represented as TPM (tags per million) of total reads mapped to Hpa genome. Red lines indicate predicted effectors induced more than two fold at 3 dpi compared to the expression level in conidiospores (cs). Single and double asterisks indicate expression pattern of HaRxL76 and HaRxL62, respectively. A right line chart is magnification of left one. (B) Average expression pattern of genes in the indicated groups during the infection with Hpa Waco9. The induction levels compared to the level in cs were indicated by value of log2. (C to K) Distribution of motifs in coexpressed genes of Hpa and P. infestans. Nucleotide conservation of (C) the INR-FPR motif in “induced effectors”, (F) Motif I in “induced effectors” and (I) Motif II in “non-induced effectors” is displayed as sequence logos, based on hits within 200 nt (INR-FPR) and 500 nt (Motif I and II) upstream of the start codon. Bar charts indicate the percent of promoters within each group that contain (D, E) the INR-FPR motif within 200 nt and (G, H) Motif I and (J, K) Motif II within 500 nt upstream of the start codon. The analysis was done in promoters from (D, G, J) Hpa and (E, H, K) P. infestans. Asterisks indicate statistically significant over-representation of the motifs compared to population in “all genes” (p<1e-4), which is shown in Table S4. To evaluate whether these motifs are conserved in other oomycetes, we checked the presence of these motifs in promoters of Phytophthora infestans genes co-expressed during infection according to microarray data [46]. As reported previously [43], [44], INR-FPR was over-represented in P. infestans RxLR effectors and genes induced during infection as observed for Hpa (Figure 2E and Table S4). Motif I and Motif II were not significantly over-represented in promoters of P. infestans genes (Figure 2H and K), suggesting that these novel motifs might be Hpa-specific cis-regulatory elements.
Hpa Waco9 overcomes RPP1-mediated resistance through deletion of ATR1
Transcriptome analysis revealed that 355 and 366 predicted effectors were expressed in conidiospores and/or infections with Hpa Emoy2 and Waco9, respectively (Table 2). Of these, 339 predicted effectors were expressed in both Hpa Emoy2 and Waco9, whereas 16 and 27 predicted effectors were expressed in only Hpa Emoy2 and Waco9, respectively (Figure 3A and Table S5). ATR5, an effector recognized by RPP5 [40], was found among the 339 predicted effectors expressed in both Hpa Emoy2 and Waco9 (Figure 3B and Table S5). The Waco9 allele of ATR5 is identical to the Emoy2 allele. Surprisingly, while ATR1 was expressed in Hpa Emoy2, no tag corresponding to ATR1 in Hpa Waco9 was detected (Figure 3B and Table S5). We resequenced Hpa Waco9 genome using an Illumina Genome Analyzer II, and found that the genomic region that includes ATR1 is deleted in Waco9 (Figure 3C). These results suggest that Hpa Waco9 can infect plants containing functional RPP1, but not plants containing functional RPP5. To evaluate this possibility, several Arabidopsis accessions were inoculated with Hpa Emoy2 and Waco9. ATR1 from Hpa Emoy2 is recognized by RPP1-Nd from Arabidopsis Nd-1 accession and RPP1-WsA and RPP1-WsB from Arabidopsis Ws-2 accession (the accession previously reported as Ws-0 in our laboratory is in fact Ws-2) [35]. As expected, Arabidopsis Nd-1 and Ws-2 are resistant to Hpa Emoy2, but susceptible to Hpa Waco9 (Figure S2). We also checked the phenotype on an Arabidopsis RIL 3860 (3860), a recombinant inbred line from a cross between Col-5 and Nd-1 that lacks RPP1-Nd, and a transgenic 3860 line containing the functional RPP1-Nd gene (3860:RPP1Nd) [35]. Like Arabidopsis Nd-1 and Ws-2, 3860:RPP1Nd is resistant to Hpa Emoy2, but susceptible to Hpa Waco9, whereas Arabidopsis 3860 is susceptible to both Hpa Emoy2 and Waco9 (Figure 3D). On the other hand, no Hpa sporulation was observed on Arabidopsis Ler-0 accession containing functional RPP5, RPP5-Ler, inoculated with Hpa Emoy2 and Waco9 (Figure S2). To confirm if Hpa Emoy2 and Waco9 are recognized by RPP5-Ler, Arabidopsis CW84, a broadly Hpa-susceptible recombinant inbred line generated from a cross between Col-0 and Ws-2 [47], and CW84 transformants containing RPP5-Ler (CW84:RPP5Ler) [40] were inoculated with Hpa Emoy2 and Waco9. Like Arabidopsis Ler-0, CW84:RPP5Ler is resistant to both Hpa Emoy2 and Waco9, whereas Arabidopsis CW84 is susceptible to both Hpa isolates (Figure 3D). These results indicate that Hpa Waco9 overcomes recognition by RPP1, but not RPP5, through the deletion of ATR1 from its genome.
Fig. 3. Hpa Waco9 overcomes recognition by RPP4, but not RPP5. 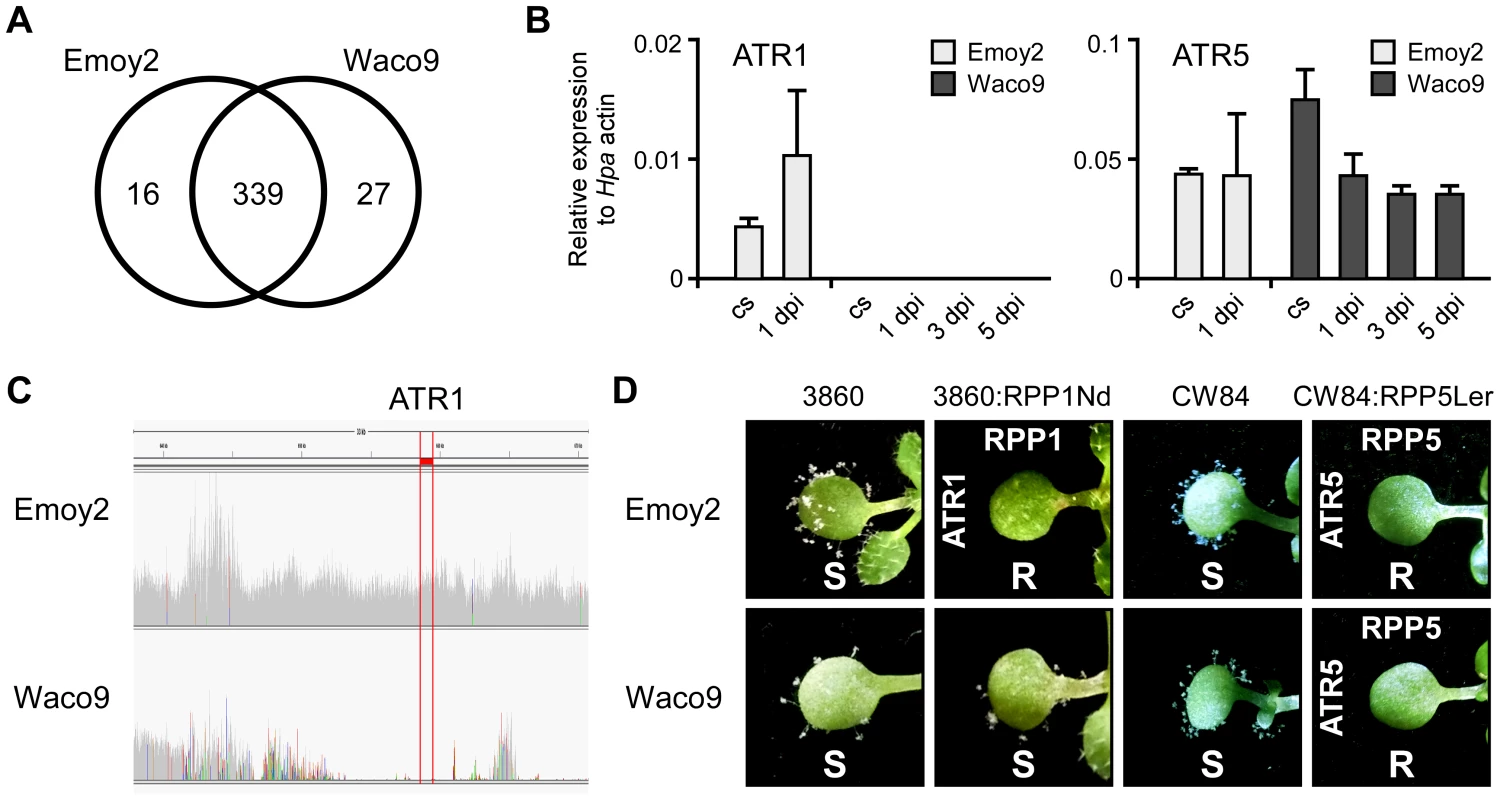
(A) The number of predicted effectors expressed in Hpa Emoy2 and/or Waco9. (B) Expression of ATR1 and ATR5 in Hpa Emoy2 and Waco9 conidiospores (cs) and the infections in Arabidopsis Col-0. The expression level was determined by qRT-PCR using specific primers for ATR1 and ATR5. Expression of Hpa actin was used to normalize the expression value in each sample. Data are means ± SDs from three biological replicates. (C) Illumina sequencing reads coverage in genomic region including ATR1. Region indicated in red is of ATR1. (D) Resistance (R) and susceptibility (S) to Hpa Emoy2 and Waco9 in seven-day-old 3860, RPP1-Nd-transformed 3860 (3860:RPP1Nd), CW84 and RPP5-Ler_transformed CW84 (CW84:RPP5Ler) plants. The plants inoculated with Hpa Emoy2 and Waco9 were photographed at 6 dpi. Expression pattern of Arabidopsis genes in compatible and incompatible interactions with Hpa
We investigated Arabidopsis gene expression during infection with Hpa Emoy2 and Waco9. The expression of 24,559 Arabidopsis protein-coding genes (89.6% of the 27,416 protein-coding genes predicted in Arabidopsis TAIR10) was detected in at least one time point (Tables 2 and Dataset S1). Of these, 1,048 Arabidopsis genes showed significant changes in gene expression (FDR = 0.001) after inoculation with Hpa Emoy2 or Waco9 (Table S6). To reveal compatible - or incompatible-interaction-specific changes in gene expression, we determined the level of overlap of differentially expressed Arabidopsis genes between infections with Hpa Emoy2 and Waco9 (Figure 4A). We found that many genes were specifically upregulated at 1 dpi with Hpa Emoy2 (80 genes) and at 3 and 5 dpi with Hpa Waco9 (335 and 863 genes, respectively) (Figure 4A). The Arabidopsis genes upregulated at 1 dpi with Hpa Emoy2, but not Waco9, might be induced upon recognition by RPP4 (i.e. ETI), while the genes upregulated in the interaction with Hpa Waco9, but not Emoy2, might be genes targeted by Hpa to enhance susceptibility. Therefore, we focused on upregulated Arabidopsis genes at 1 dpi with Hpa Emoy2 and at 3 and 5 dpi with Hpa Waco9, and categorized them into four groups: Group I, 81 upregulated Arabidopsis genes at 1 dpi with Hpa Emoy2; Group II, 98 upregulated Arabidopsis genes at only 3 dpi with Hpa Waco9; Group III, 297 upregulated Arabidopsis genes at both 3 and 5 dpi with Hpa Waco9; Group IV, 516 upregulated Arabidopsis genes at only 5 dpi with Hpa Waco9 (Figure 4B, C and Table S7). Interestingly, 86.4% of Arabidopsis genes in Group I (70 genes) were also upregulated at 3 and/or 5 dpi with Hpa Waco9. Gene Ontology (GO) term enrichment analysis showed that responses involved in disease resistance (e.g. defense response, GO:0006952; response to salicylic acid stimulus, GO:0009751) were significantly enriched in all Groups (Figure 4D left). These findings suggest that defense-related Arabidopsis genes upregulated at early time points in the incompatible interaction are similarly regulated at late time points in the compatible interaction. This is consistent with previous reports on expression profiling in Arabidopsis and Hpa interactions [26]–[29]. On the other hand, genes responsive to ET (GO:0009723) and hormones (GO:0009725), such as ABA (GO:0009737) and auxin (GO:0009733), were overrepresented in Group II, III and/or IV but absent in Group I, highlighting genes induced specifically during a compatible interaction (Figure 4D right). In these Groups, we also found overrepresentation of genes related to nitrate transport (GO:0015706), water deprivation (GO:0009414) and starvation (GO:0042594) (Figure 4D right).
Fig. 4. Arabidopsis genes differentially expressed after inoculation with Hpa Emoy2 and Waco9. 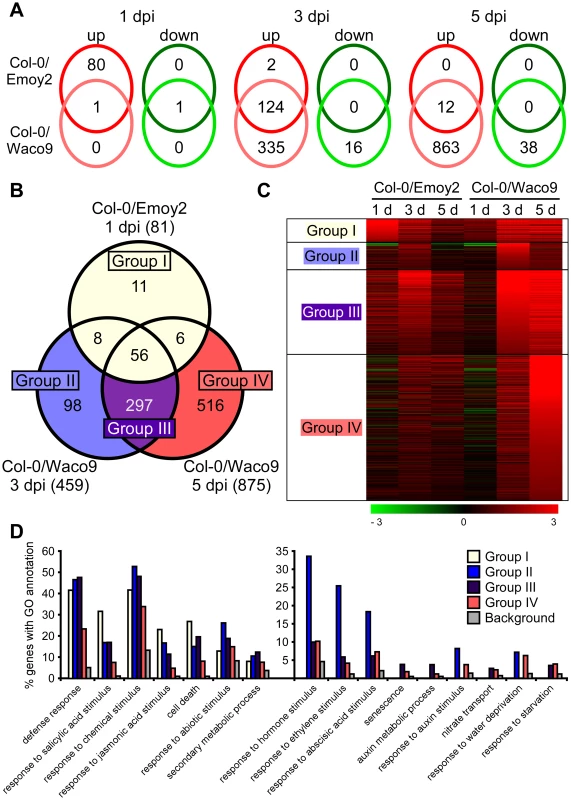
(A) The number of Arabidopsis genes significantly upregulated or downregulated at 1, 3 and 5 dpi with Hpa Emoy2 and Waco9. (B) Assessment of overlap of genes significantly upregulated at 1 dpi with Hpa Emoy2 and at 3 and 5 dpi with Hpa Waco9, and classification into Group I (yellow), II (blue), III (purple) and IV (red). (C) Expression pattern of genes categorized into Group I, II, III and IV. The relative expression (in log2 ratios) is colored red for induction and green for repression as illustrated in the fold change color bars. (D) Percentage of genes with significantly enriched gene ontology (GO) terms in Group I (yellow), II (blue), III (purple) and IV (red), compared to the background (grey). Y-axis: percentage of genes that fall within each given GO annotation class. Hpa infection suppresses SA-inducible PR1 expression in Arabidopsis
Defense-related Arabidopsis genes including SA-responsive genes were found to be upregulated not only at 1 dpi with Hpa Emoy2 but also at 3 and 5 dpi with Hpa Waco9 (Figure 4). Indeed, there was a positive correlation between these genes and genes upregulated by treatment with benzothiadiazole S-methylester (BTH; a functional analog of SA) [48] (Figure 5A and Table S8). At 1 dpi, BTH-inducible genes, such as PR1, were upregulated by inoculation with Hpa Emoy2, but not Hpa Waco9, whereas these genes were upregulated at 3 and 5 dpi with Hpa Waco9 (Figure 5A and B).
Fig. 5. Hpa suppresses PR1 expression induced by SA in infected cells. 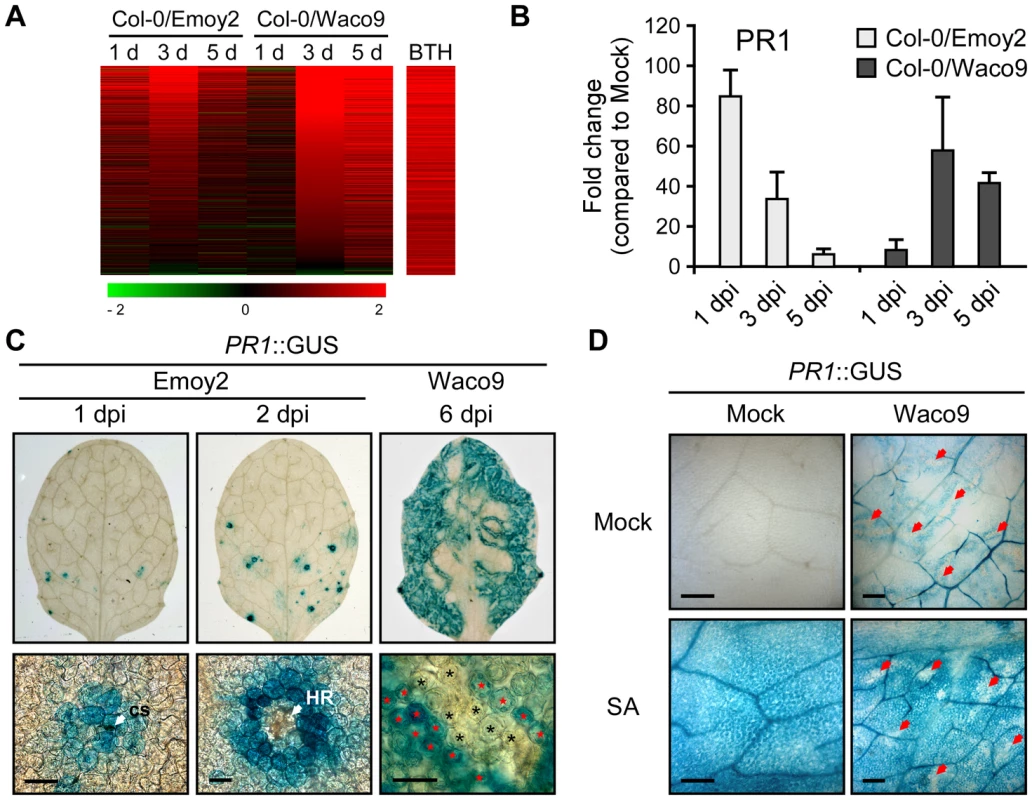
(A) Expression pattern of 871 BTH-inducible genes reported by Wang et al. (2006) [48] after inoculation with Hpa Emoy2 and Waco9. The relative expression (in log2 ratios) is colored red for induction and green for repression as illustrated in the fold change color bars. (B) Expression of PR1 in Arabidopsis at 1, 3, and 5 dpi with Hpa Emoy2 and Waco9. The expression level was determined by qRT-PCR using specific primers for PR1 and indicated as relative fold induction compared to water-treated samples (mock). Expression of EF-1α was used to normalize the expression value in each sample. Data are means ± SDs from three biological replicates. (C) GUS staining in three-week-old Arabidopsis leaves containing PR1 promoter fused GUS (PR1::GUS) at 1 and 2 dpi with Hpa Emoy2 and at 6 dpi with Hpa Waco9. Lower images are magnified upper images. Black and red asterisks indicate Hpa-haustoriated and non-haustoriated mesophyll cells, respectively. cs, conidiospore. Scale bars = 40 µm. (D) GUS staining in Hpa-infected PR1::GUS lines 8 hours after treatment with SA (200 µM). The leaves at 4 dpi with Hpa Waco9 or spraying water (mock) were infiltrated with SA or water (mock). Red arrows indicate Hpa-haustoriated cells. Scale bars = 100 µm. Recently, we reported the cell-specific expression pattern of PR1 in a compatible interaction by infecting PR1::GUS lines with Hpa Waco9 [18]. PR1::GUS expression is suppressed in haustoriated cells, but not in non-haustoriated adjacent cells (Figure 5C) [18], but this could arise either via suppression of SA biosynthesis or SA responsiveness in these cells. To distinguish these possibilities, we investigated the effect of Hpa infection on SA - and BTH-inducible PR1::GUS expression. PR1::GUS lines at 4 dpi with Hpa Waco9 or mock infected were treated with SA, BTH or water. As expected, we observed GUS staining in non-infected PR1::GUS lines after treatment with SA and BTH (Figures 5D and S3). In Hpa-infected PR1::GUS lines, although GUS staining was observed in non-haustoriated cells after SA and BTH treatment, Hpa-haustoriated cells were not stained (Figures 5D and S3). These results suggest that Hpa suppresses the expression of PR1 induced by treatment with SA and BTH. Thus, Hpa suppresses SA responsiveness by interfering with signaling, but not by promoting SA degradation.
We also investigated the cell-specific expression pattern of PR1::GUS in the incompatible interaction. GUS staining was observed in cells that Hpa Emoy2 had infected and the surrounding cells at 1 dpi, and observed in the cell layer surrounding cells in which HR cell death had occurred at 2 dpi (Figure 5C). These results are consistent with expression profiling data derived from whole Hpa-infected tissues (Figure 5A and B).
A highly expressed Hpa effector, HaRxL62, suppresses responsiveness to SA
Histochemical GUS analysis in Hpa-infected PR1::GUS lines showed that Hpa suppresses SA-inducible PR1 expression specifically in the haustoriated cells into which RxLR effectors are delivered (Figure 5D). To identify Hpa effectors which participate in the suppression, the level of PR1 expression after treatment with SA was checked in transgenic lines expressing Hpa predicted effectors and the SA-insensitive npr1 mutants [49], as a positive control. Nine Hpa effector-expressing lines showed more susceptibility to Hpa compared to wild type (WT) Col-0 plants [8], [10] (Figure S4 and Table S9). HaRxL62-expressing lines showed a five-fold reduction in expression level of PR1 compared to WT after SA treatment, whereas no significant reduction was observed in eight other Hpa effector-expressing lines, including HaRxLL464-expressing lines (Figure 6A). To evaluate the effect of HaRxL62 on Hpa growth after treatment with SA, WT plants, npr1 mutants and HaRxL62 - and HaRxLL464-expressing lines were treated with SA or water as a mock treatment and, 24 hours later, inoculated with Hpa Waco9 (Figure 6B). Although water-treated WT plants were susceptible to Hpa Waco9, no Hpa growth was observed in SA-treated WT plants. As expected, SA did not trigger resistance to Hpa in npr1 mutants. In HaRxLL464-expressing plants treated with SA, essentially no Hpa spores were observed as observed for WT plants, whereas there were countable Hpa spores in HaRxL62-expressing plants treated with SA (Figure 6B), consistent with reduction in expression level of PR1 after treatment with SA (Figure 6A). As shown in Figure 2A, HaRxL62 was the second-highest expressed Hpa effector at 3 dpi. These results suggest that HaRxL62, a highly-expressed effector during infection, reduces responsiveness to SA.
Fig. 6. HaRxL62 reduces responsiveness to SA. 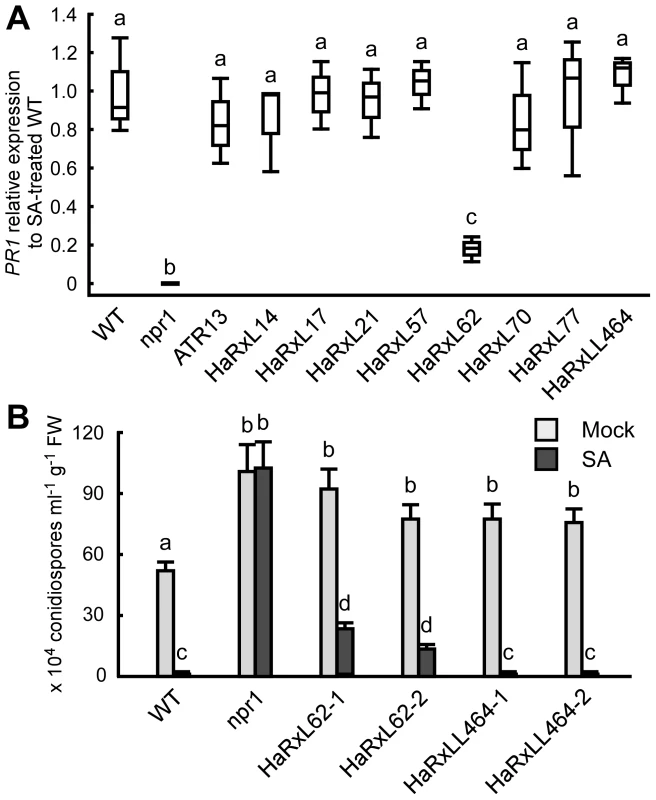
(A) Expression level of PR1 8 hours after treatment with SA (100 µM) in ten-day-old Col-0 plants (WT), npr1 mutants and transgenic lines expressing the indicated Hpa predicted effectors. The expression level was determined by qRT-PCR using specific primers for PR1 and indicated as relative fold induction compared to the expression level in WT after SA treatment. Expression of EF-1α was used to normalize the expression value in each sample. Data are means from three biological replicates showing quantiles. Data analysis was carried using one-way ANOVA followed by Tukey's HSD (honestly significant difference). Genotypes showing significant differences (p<0.01) are marked with different alphabets (B) Hpa growth on three-week-old Col-0 plants (WT), npr1 mutants and two independent transgenic lines expressing HaRxL62 (HaRxL62-1 and HaRxL62-2) and HaRxLL464 (HaRxLL464-1 and HaRxLL464-2) pretreated with SA (10 µM) or water (mock). The plants 24 hours after spray treatment with SA or water were inoculated with Hpa Waco9. Conidiospores were harvested and counted at 6 dpi. Different letters indicate significantly different values at p<0.05 (one-way ANOVA, Tukey's HSD). Discussion
A comprehensive understanding of host-pathogen interactions requires knowledge of the associated gene expression changes in both the host and the pathogen. However, in most cases, expression profiling has focused on either the host or the pathogen due to limitations and obstacles of older methods that involve microarrays [50]. In this study, using a high-throughput expression profiling method, EXPRSS [33], the transcriptomes of both Arabidopsis and Hpa in compatible and incompatible interactions were analyzed in parallel. With comparative genomics, we revealed that Hpa Waco9 evades RPP1-mediated resistance through deletion of cognate AVR gene ATR1. Histochemical analysis showed that Hpa suppresses SA-inducible PR1 expression specifically in infected cells. Finally, we found a highly-expressed Hpa effector candidate involved in suppression of responsiveness to SA.
SA has been implicated as an important signal in plant immune signaling [51], [52]. For example, Arabidopsis eds5/sid1 and ics1/sid2 mutants in which SA levels are reduced [53], [54] are more susceptible to both virulent and avirulent forms of P. syringae and Hpa [51]. Expression profiling in Arabidopsis showed that SA-responsive genes including PR1 are activated not only at early time points in the incompatible interaction but also at late time points in the compatible interaction (Figure 5A and B), consistent with previous reports [26]–[29]. Most recently, we reported that Hpa suppresses expression of PR1::GUS specifically in cells containing haustoria, into which host-translocated effectors are delivered, but not in non-haustoriated adjacent cells, which show high expression levels of PR1::GUS [18]. Here, we showed less PR1::GUS expression in Hpa-haustoriated cells after treatment with SA and BTH, indicating that Hpa interferes with the recognition of SA and/or downstream signaling after the recognition (Figure 5D). HaRxL62-expressing plants showed significant reduction in SA-induced expression of PR1 and compromised resistance to Hpa after treatment with SA (Figure 6). HaRxL62 may make an important contribution to the virulence of Hpa because of its high expression levels during infection (Figure 2A). However, the suppression of SA-inducible resistance to Hpa in HaRxL62-expressing plants was moderate even though HaRxL62-expressing plants and npr1 mutant plants showed comparable susceptibility to Hpa (Figure 6B). These findings suggest that HaRxL62 also targets other defense pathway(s) than the SA pathway and other Hpa effectors must also participate in suppression of responsiveness to SA. Anderson et al. (2012) [37] showed that HaRxL96 suppresses PR1 expression, but not SA biosynthesis, induced by inoculation with an avirulent isolate of Hpa. HaRxL44 attenuates SA-dependent transcription through interfering with Mediator function by degrading MED19a, a transcriptional component involved in SA/JA crosstalk [18].
Our cell biology analysis also reveals a shortcoming of transcriptome analysis using whole tissues. We show that during Hpa infection, PR1 is expressed in non-haustoriated adjacent cells, but not in haustoriated cells. We presume that recognition of diffusible PAMPs from Hpa leads to PTI, resulting in SA biosynthesis and PR1 expression, and Hpa suppresses the responses in colonized cells by delivering effectors. Better methods are required for cell-type specific expression profiling specifically in haustoriated cells.
In addition to SA and JA, other phytohormones, such as ET, ABA and auxin, are also implicated in plant immunity [25]. ETHYLENE INSENSITIVE3 (EIN3) and ETHYLENE INSENSITIVE3-LIKE1 (EIL1), two closely related Arabidopsis transcription factors known to regulate the ET pathway, repress biosynthesis of SA by binding directly to the promoter of the SA biosynthetic gene ICS1/SID2 [55]. Consistent with this, plants mutated in EIN3/EIL1 and the key ET-signaling protein EIN2 exhibit enhanced resistance to P. syringae [55] in spite of suppressed signaling of FLS2 which recognizes the bacterial PAMP flagellin [56]. Increased susceptibility to P. syringae and Hpa is observed in plants treated with ABA and in ABA over-accumulating plants, and vice versa in ABA-deficient mutants [57]–[59]. Similarly, elevated auxin signaling correlates with increase in susceptibility to P. syringae and Hpa [60]–[63]. Collectively, these findings suggest that ET, ABA and auxin behave as negative regulators of defense responses. Some bacterial effectors appear to target these signaling systems. Conditional expression of P. syringae effector AvrPtoB increases in planta ABA levels and enhances bacterial growth [64]. AvrBs3, a type three effector from Xanthomonas campestris pv. vesicatoria, induces auxin responsive genes, resulting in cell hypertrophy [65]. Our expression profiling in Hpa-infected Arabidopsis revealed overrepresentation of genes related to responses to ET (GO:0009723), ABA (GO:0009737) and auxin (GO:0009733) in Group II, III and/or IV, genes upregulated at 3 and/or 5 dpi with Hpa Waco9, but not at 1 dpi with Hpa Emoy2 (Figure 4). Consistent with this finding, previous expression profiling using microarrays in Arabidopsis Ler-0 inoculated with compatible (Cala2) and incompatible (Waco9, recognized by RPP5) Hpa isolates revealed that many compatible-specific genes are ABA responsive [28]. Interestingly, we also found that genes involved in nitrate transport (GO:0015706) were overrepresented in Group III and IV (Figure 4D). Hpa lacks genes for nitrate and nitrite reductases and a nitrate transporter [7], which is also true for another obligate biotrophic powdery mildew fungi [66]. Expression profiling in Hpa revealed 202 and 252 predicted effectors expressed at 3 and 5 dpi with Hpa Waco9, respectively (Table 1). Conceivably, some of these effectors target these phytohormone signaling and host nitrate transporter systems.
This study also showed expression patterns and levels of Hpa predicted effectors, which may help select bona fide virulence effectors. Indeed, the second-highest expressed Hpa effector at 3 dpi, HaRxL62, appears to enhance susceptibility at least in part by suppressing responsiveness to SA. In a previous screening of Hpa predicted effectors that enhance the virulence and/or that suppress PTI, HaRxL62 was selected as the most effective Hpa effector [8], [9]. HaRxL76, the highest-expressed Hpa effector at 3 dpi, was not in the list for our previous screenings. HaRxL76 and other highly-expressed Hpa predicted effectors will be investigated in future studies.
To evade recognition by cognate R genes, the majority of RxLR effector genes are subject to diversifying selection, resulting in a diverse set of effector alleles in the pathogen population [4], [5]. ATR1 and ATR13 have a high level of sequence polymorphism in the C-terminal regions that confer effector activity and are recognized by RPP1 and RPP13, respectively [35], [38]. In this study, we revealed that ATR1 is deleted in Hpa Waco9 genome, resulting in loss of recognition by RPP1 (Figure 3). Qutob et al. (2009) and (2013) [67], [68] reported that virulent strains of Phytophthora sojae escape detection by R gene Rps3a through silencing a cognate AVR effector Avr3a. In virulent pathogens, the effectors recognized by cognate R genes would be deleted and polymorphic like ATR1 and ATR13, or not expressed like Avr3a. These possibilities can be evaluated by comparative genomics and transcriptomics.
In this study, we found overrepresentation of oomycete core element INR-FPR and two novel motifs, Motif I and II, in the promoter of Hpa predicted effectors (Figure 2). The INR-FPR motif is associated with higher levels of transcripts and pathogenesis-related genes including RxLR effectors in P. infestans [43]. Consistent with this, the genes with the INR-FPR motif were highly enriched for both Hpa predicted effectors and P. infestans RxLR effectors, especially effectors induced during infection referred to as “induced effectors”. On the other hand, we found association of Motif I and II with “induced effectors” and “non-induced effectors”, respectively, in Hpa, but not in P. infestans. While Hpa and P. infestans may have a common pre-initiation complex for transcription, there might be distinct regulatory mechanisms for specific gene expression, perhaps resulting from different lifestyles. Although the findings may be useful for predicting potential effectors in related oomycetes, it will be difficult to investigate functions of these motifs in Hpa because transformation of biotrophic oomycete pathogens is difficult.
Here, we explored gene expression changes in both Arabidopsis and Hpa simultaneously during infection using a high-throughput RNA sequencing method, EXPRSS [33]. Although we cannot rule out the possibility that differences in effector sets between Hpa Emoy2 and Waco9 confer distinct transcriptional changes in Arabidopsis genes during infection, expression profiling of both pathogen effector genes and host genes involved in immunity allows us to suggest distinct mechanisms of effector-mediated susceptibility. When stably expressed in planta, some Hpa effectors cause diverse developmental phenotypes, highlighting that the effectors might interfere with fundamental plant regulatory mechanisms [69]. Further comparative investigations of transcriptional changes in Arabidopsis genes between Hpa infections and effector(s)-expressing plants would be interesting. Recently, using a custom-designed combined pathogen and host whole-genome microarray, Jupe et al. (2013) [70] reported a simultaneous overview of gene expression changes in both Phytophthora capsici and its host tomato during the infection. In comparison to their approach using a custom microarray, our approach using EXPRSS can be more easily applied to host-pathogen interactions for which both host and pathogen genome sequences are available. This work opens the door towards transcriptome studies in infection biology that should help unravel pathogen infection strategies and the mechanisms by which host defense responses are overcome.
Materials and Methods
Plant material and growth
Arabidopsis accessions used in this study were obtained from the Nottingham Arabidopsis Stock Centre. Arabidopsis RIL 3860 and 3860:RPP1Nd were kindly provided by Jim L. Beynon, University of Warwick, UK [35], and Arabidopsis CW84 and CW84:RPP5Ler were from Bailey et al. (2011) [40]. PR1::GUS lines were from Caillaud et al. (2013) [18], and plants expressing Hpa predicted effectors other than HaRxL62 were from Fabro et al. (2011) [8] and Caillaud et al. (2012) [10] (Table S9). A construct for expressing HaRxL62 in planta was generated by recombining the corresponding ORF from the signal peptide cleavage site cloned in pENTR/SD/D-TOPO (Invitrogen) into the Gateway destination binary vector pENS-StrepII-3×HA-GW under the control of Cauliflower mosaic virus 35S promoter [71]. The construct was transferred to Agrobacterium tumefaciens strain GV3101 (pMP90 RK) [72] and transformed into Arabidopsis accession Col-0 by the floral dipping method [73]. Primary transformants (T1) were selected on soil containing BASTA (Bayer CropScience, Wolfenbüttel, Germany) and checked for expression of HaRxL62 by Western blot analysis as described by Asai et al. (2008) [74]. The progeny of the T2 generation was observed and 3∶1 (BASTA-resistant/BASTA-susceptible) segregating lines were taken further. Homozygous lines were selected by examining the BASTA resistance of T3 seedlings. Two independent transgenic lines were analyzed.
For Hpa-inoculation assay, Arabidopsis plants were grown at 22°C and 60% humidity under a 10-h photoperiod and a 14-h dark period in environmentally controlled growth cabinets. For SA-induced PR1 expression analysis, Arabidopsis plants were grown on 0.7% agar plates of MS medium at 22°C under a 16-h photoperiod and an 8-h dark period in environmentally controlled growth cabinets.
Pathogen assays
For Hpa infection, Arabidopsis plants were spray-inoculated to saturation with a spore suspension of 5×104 conidiospores/ml. Plants were covered with a transparent lid to maintain high humidity (90–100%) conditions in a growth cabinet at 16°C under a 10-h photoperiod until the day for sampling.
To evaluate hyphae growth and HR cell death, leaves inoculated with Hpa Emoy2 or Waco9 were stained with trypan blue as described by Asai and Yoshioka (2009) [75].
To evaluate conidiospore production, 5 pools of 3 plants for each Arabidopsis line were harvested in 1 ml of water. After vortexing, the amount of conidiospores released was determined using a haemocytometer.
RNA extraction, cDNA synthesis and qRT-PCR
Total RNAs were extracted using TRI reagent (Sigma) and 1-bromo-3-chloropropane (Sigma) according to the procedure of the manufacturer. RNAs were precipitated with half volume of isopropanol and half volume of high salt precipitation buffer (0.8 M sodium citrate and 1.2 M sodium chloride). RNA samples were treated with DNaseI (Roche) and purified by RNeasy Mini Kit (Qiagen) according to the procedure of the manufacturers.
Total RNAs (3 µg) were used for generating cDNAs in a 20 µl volume reaction according to Invitrogen Superscript II Reverse Transcriptase protocol. The obtained cDNAs were diluted five times, and 1 µl were used for 10 µl qPCR reaction.
qPCR was performed in 10 µl final volume using 5 µl SYBR Green mix (Sigma), 1 µl diluted cDNAs, and primers. qPCR was run on the CFX96 Real-Time System C1000 thermal cycler (Biorad) using the following program: (1) 95°C, 3 min; (2) [95°C, 30 sec, then 60°C, 30 sec, then 72°C, 30 sec]×45, 72°C, 10 min followed by a temperature gradient from 55°C to 95°C. The relative expression values were determined using EF-1α (At5g60390) as a reference gene and the comparative cycle threshold method (2−ΔΔCt). Primers used for qPCR are listed in Table S10.
Hpa Waco9 genome sequencing
Genomic DNA was extracted from Hpa Waco9 conidiospores using a Nucleon PhytoPure DNA extraction kit (GE Healthcare) according to the procedure of the manufacturer. A paired-end 400 bp insert size library was prepared and sequenced on Illumina Genome Analyzer II. The sequence reads were aligned in a paired end fashion to the Hpa Emoy2 v8.3 [7] using BWA [76]. Trailing nucleotides with a quality score of less than 10 were trimmed using the -q option. In order to maximize the number of aligned reads, unaligned reads were aligned using a more sensitive aligner, Stampy [77]. SAMtools [78] was used to generate a BAM file that enables visualization of the alignment with the Integrative Genomics Viewer [79], as seen in Figure 3C.
For correction of Hpa genome by Waco9 SNVs, genetic variations between Hpa Emoy2 and Waco9 were predicted using SAMtools [78]. Hpa Emoy2 v8.3 genome sequence [7] was corrected by substituting Hpa Waco9 SNVs, using a custom Perl script. Insertion and deletion variations were ignored. The sequence data have been deposited in NCBI's Short Read Archive (SRA) and are accessible through SRA accession number SRX493773.
RNA sequencing
RNA sequencing was performed as described previously [33]. Purified double stranded cDNAs were subjected to Covaris shearing (parameters: intensity, 5; duty cycle, 20%; cycles/burst, 200; duration, 60 sec). The libraries were sequenced on Illumina Genome Analyzer II. The sequence data have been deposited in NCBI's Gene Expression Omnibus (GEO) and are accessible through GEO Series accession number GSE53641. Sequence reads to gene associations were carried out using the considerations described previously [33]. Quality-filtered libraries were aligned to the combined genome of Arabidopsis TAIR10 and Hpa Emoy2 v8.3 [7] using Bowtie version 0.12.8 [80]. Unaligned reads from previous step were aligned to the combined genome reference using Novoalign v2.08.03 (http://www.novocraft.com/). Remaining reads were aligned to transcript sequences of Arabidopsis Col-0 (ftp://ftp.Arabidopsis.org/home/tair/Sequences/blast_datasets/TAIR10_blastsets/TAIR10_cdna_20101214_updated) using Bowtie version 0.12.8 [80]. The reads with up to 10 reportable alignments or uniquely aligned reads were selected for downstream analysis. Differential expression analysis was performed using the R statistical language version 2.11.1 with the Bioconductor [81] package, edgeR version 1.6.15 [82] with the exact negative binomial test using tagwise dispersions.
Identification of DNA motifs
For identifying cis-regulatory elements, 200 and 500 nt upstream of the start codon of coexpressed Hpa genes categorized into five groups as shown in Figure 2B and Table S3 were extracted from Waco9-SNVs-corrected v8.3 genome sequence using a custom Perl script. The sets of sequences extracted from genes categorized into “induced effectors” and “non-induced effectors” were searched separately using MEME version 4.9.1 (http://meme.nbcr.net/meme/cgi-bin/meme.cgi) [41]. MEME was run with minimum width of 6 and maximum width of up to 20 and zero or one per sequence was allowed.
The abundance of each motif found by MEME analysis in other groups was evaluated per individual motif using FIMO (http://meme.nbcr.net/meme/cgi-bin/fimo.cgi) [42] with a q-value cutoff 1e-4. Similarity to known motifs was assessed using TOMTOM (http://meme.nbcr.net/meme/cgi-bin/tomtom.cgi) [45] against the JASPAR database.
In P. infestans isolate T30-4, genes were categorized into five groups according to whether genes were significantly upregulated at 2 and 3 dpi in microarray data of Cooke et al. (2012) [46]. As described above, 200 and 500 nt upstream of the start codon of coexpressed P. infestans genes were extracted, and then the abundance of each motif was evaluated using FIMO [42].
GO enrichment analysis
To investigate enrichment of specific gene ontologies in Arabidopsis genes categorized into four groups (Group I to IV) as shown in Figure 4D and Table S7, the Singular Enrichment Analysis was done with FDR = 0.05 using AgriGO (http://bioinfo.cau.edu.cn/agriGO/analysis.php).
GUS staining
GUS activity was assayed histochemically with 5-bromo-4-chloro-3-indolyl-β-D-glucuronic acid (1 mg/ml) in a buffer containing 100 mM sodium phosphate pH 7.0, 0.5 mM potassium ferrocyanide, 0.5 mM potassium ferricyanide, 10 mM EDTA, 0.1% Triton. Arabidopsis leaves were vacuum-infiltrated with staining solution and then incubated overnight at 37°C in the dark. Destaining was performed in 100% ethanol followed by incubation in chloral hydrate solution. Stained leaves were observed using a Zeiss Axioplan 2 microscope (Jena, Germany).
SA-induced PR1 expression analysis
For SA-induced PR1 expression analysis as shown in Figure 6A, ten-day-old plants grown on MS medium plates were used. The plants were equilibrated in water overnight, and water was changed for 100 µM SA (Sigma) solution in the morning. After 8 h of incubation with SA, the plants were quickly dried and flash-frozen in liquid nitrogen. Five plants per condition were used for RNA extraction.
Accession numbers
Sequence data of 475 Hpa predicted effectors can be found in NCBI's GenBank data library under accession numbers described in Table S2.
Supporting Information
Zdroje
1. SegonzacC, ZipfelC (2011) Activation of plant pattern-recognition receptors by bacteria. Curr Opin Microbiol 14 : 54–61.
2. JonesJD, DanglJL (2006) The plant immune system. Nature 444 : 323–329.
3. HeinI, GilroyEM, ArmstrongMR, BirchPR (2009) The zig-zag-zig in oomycete-plant interactions. Mol Plant Pathol 10 : 547–562.
4. WinJ, MorganW, BosJ, KrasilevaKV, CanoLM, et al. (2007) Adaptive evolution has targeted the C-terminal domain of the RXLR effectors of plant pathogenic oomycetes. Plant Cell 19 : 2349–2369.
5. VleeshouwersVG, RaffaeleS, VossenJH, ChampouretN, OlivaR, et al. (2011) Understanding and exploiting late blight resistance in the age of effectors. Annu Rev Phytopathol 49 : 507–531.
6. CoatesME, BeynonJL (2010) Hyaloperonospora Arabidopsidis as a pathogen model. Annu Rev Phytopathol 48 : 329–345.
7. BaxterL, TripathyS, IshaqueN, BootN, CabralA, et al. (2010) Signatures of adaptation to obligate biotrophy in the Hyaloperonospora arabidopsidis genome. Science 330 : 1549–1551.
8. FabroG, SteinbrennerJ, CoatesM, IshaqueN, BaxterL, et al. (2011) Multiple candidate effectors from the oomycete pathogen Hyaloperonospora arabidopsidis suppress host plant immunity. PLoS Pathog 7: e1002348.
9. BadelJL, PiquerezSJ, GreenshieldsD, RallapalliG, FabroG, et al. (2013) In planta effector competition assays detect Hyaloperonospora arabidopsidis effectors that contribute to virulence and localize to different plant subcellular compartments. Mol Plant Microbe Interact 26 : 745–757.
10. CaillaudMC, PiquerezSJM, FabroG, SteinbrennerJ, IshaqueN, et al. (2012) Subcellular localization of the Hpa RxLR effector repertoire identifies a tonoplast-associated protein HaRxL17 that confers enhanced plant susceptibility. Plant J 69 : 252–265.
11. MukhtarMS, CarvunisAR, DrezeM, EppleP, SteinbrennerJ, et al. (2011) Independently evolved virulence effectors converge onto hubs in a plant immune system network. Science 333 : 596–601.
12. VlotAC, DempseyDA, KlessigDF (2009) Salicylic Acid, a multifaceted hormone to combat disease. Annu Rev Phytopathol 47 : 177–206.
13. MishinaTE, ZeierJ (2007) Pathogen-associated molecular pattern recognition rather than development of tissue necrosis contributes to bacterial induction of systemic acquired resistance in Arabidopsis. Plant J 50 : 500–513.
14. DongX (2004) NPR1, all things considered. Curr Opin Plant Biol 7 : 547–552.
15. BallareCL (2011) Jasmonate-induced defenses: a tale of intelligence, collaborators and rascals. Trends Plant Sci 16 : 249–257.
16. GlazebrookJ (2005) Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annu Rev Phytopathol 43 : 205–227.
17. Gimenez-IbanezS, SolanoR (2013) Nuclear jasmonate and salicylate signaling and crosstalk in defense against pathogens. Front Plant Sci 4 : 72.
18. CaillaudMC, AsaiS, RallapalliG, PiquerezSJM, FabroG, et al. (2013) A downy mildew effector attenuates salicylic acid-triggered immunity in Arabidopsis by interacting with the host mediator complex. PLoS Biol 11: e1001732.
19. JiangS, YaoJ, MaKW, ZhouH, SongJ, et al. (2013) Bacterial effector activates jasmonate signaling by directly targeting JAZ transcriptional repressors. PLoS Pathog 9: e1003715.
20. ZhengXY, SpiveyNW, ZengW, LiuPP, FuZQ, et al. (2012) Coronatine promotes Pseudomonas syringae virulence in plants by activating a signaling cascade that inhibits salicylic acid accumulation. Cell Host Microbe 11 : 587–596.
21. El OirdiM, El RahmanTA, RiganoL, El HadramiA, RodriguezMC, et al. (2011) Botrytis cinerea manipulates the antagonistic effects between immune pathways to promote disease development in tomato. Plant Cell 23 : 2405–2421.
22. DiezelC, von DahlCC, GaquerelE, BaldwinIT (2009) Different lepidopteran elicitors account for cross-talk in herbivory-induced phytohormone signaling. Plant Physiol 150 : 1576–1586.
23. UppalapatiSR, IshigaY, WangdiT, KunkelBN, AnandA, et al. (2007) The phytotoxin coronatine contributes to pathogen fitness and is required for suppression of salicylic acid accumulation in tomato inoculated with Pseudomonas syringae pv. tomato DC3000. Mol Plant Microbe Interact 20 : 955–965.
24. FonsecaS, ChiniA, HambergM, AdieB, PorzelA, et al. (2009) (+)-7-iso-Jasmonoyl-L-isoleucine is the endogenous bioactive jasmonate. Nat Chem Biol 5 : 344–350.
25. Robert-SeilaniantzA, GrantM, JonesJD (2011) Hormone crosstalk in plant disease and defense: more than just jasmonate-salicylate antagonism. Annu Rev Phytopathol 49 : 317–343.
26. MaleckK, LevineA, EulgemT, MorganA, SchmidJ, et al. (2000) The transcriptome of Arabidopsis thaliana during systemic acquired resistance. Nat Genet 26 : 403–410.
27. EulgemT, WeigmanVJ, ChangHS, McDowellJM, HolubEB, et al. (2004) Gene expression signatures from three genetically separable resistance gene signaling pathways for downy mildew resistance. Plant Physiol 135 : 1129–1144.
28. HuibersRP, de JongM, DekterRW, Van den AckervekenG (2009) Disease-specific expression of host genes during downy mildew infection of Arabidopsis. Mol Plant Microbe Interact 22 : 1104–1115.
29. WangW, BarnabyJY, TadaY, LiH, TorM, et al. (2011) Timing of plant immune responses by a central circadian regulator. Nature 470 : 110–114.
30. van der BiezenEA, JuwanaH, ParkerJE, JonesJD (2000) cDNA-AFLP display for the isolation of Peronospora parasitica genes expressed during infection in Arabidopsis thaliana. Mol Plant Microbe Interact 13 : 895–898.
31. Bittner-EddyPD, AllenRL, RehmanyAP, BirchP, BeynonJL (2003) Use of suppression subtractive hybridization to identify downy mildew genes expressed during infection of Arabidopsis thaliana. Mol Plant Pathol 4 : 501–507.
32. CabralA, StassenJH, SeidlMF, BautorJ, ParkerJE, et al. (2011) Identification of Hyaloperonospora arabidopsidis transcript sequences expressed during infection reveals isolate-specific effectors. PLoS One 6: e19328.
33. RallapalliG, KemenEM, Robert-SeilaniantzA, SegonzacC, EtheringtonGJ, et al. (2014) EXPRSS: an Illumina based high-throughput expression-profiling method to reveal transcriptional dynamics. BMC Genomics 15 : 341.
34. van der BiezenEA, FreddieCT, KahnK, ParkerJE, JonesJD (2002) Arabidopsis RPP4 is a member of the RPP5 multigene family of TIR-NB-LRR genes and confers downy mildew resistance through multiple signalling components. Plant J 29 : 439–451.
35. RehmanyAP, GordonA, RoseLE, AllenRL, ArmstrongMR, et al. (2005) Differential recognition of highly divergent downy mildew avirulence gene alleles by RPP1 resistance genes from two Arabidopsis lines. Plant Cell 17 : 1839–1850.
36. KochE, SlusarenkoA (1990) Arabidopsis is susceptible to infection by a downy mildew fungus. Plant Cell 2 : 437–445.
37. AndersonRG, CasadyMS, FeeRA, VaughanMM, DebD, et al. (2012) Homologous RXLR effectors from Hyaloperonospora arabidopsidis and Phytophthora sojae suppress immunity in distantly related plants. Plant J doi: []10.1111/j.1365-313X.2012.05079.x [epub ahead of print]
38. AllenRL, Bittner-EddyPD, Grenvitte-BriggsLJ, MeitzJC, RehmanyAP, et al. (2004) Host-parasite coevolutionary conflict between Arabidopsis and downy mildew. Science 306 : 1957–1960.
39. GoritschnigS, KrasilevaKV, DahlbeckD, StaskawiczBJ (2012) Computational prediction and molecular characterization of an oomycete effector and the cognate Arabidopsis resistance gene. PLoS Genet 8: e1002502.
40. BaileyK, CevikV, HoltonN, Byrne-RichardsonJ, SohnKH, et al. (2011) Molecular cloning of ATR5Emoy2 from Hyaloperonospora arabidopsidis, an avirulence determinant that triggers RPP5-mediated defense in Arabidopsis. Mol Plant Microbe Interact 24 : 827–838.
41. BaileyTL, BodenM, BuskeFA, FrithM, GrantCE, et al. (2009) MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res 37: W202–208.
42. GrantCE, BaileyTL, NobleWS (2011) FIMO: scanning for occurrences of a given motif. Bioinformatics 27 : 1017–1018.
43. RoyS, PoidevinL, JiangT, JudelsonHS (2013) Novel core promoter elements in the oomycete pathogen Phytophthora infestans and their influence on expression detected by genome-wide analysis. BMC Genomics 14 : 106.
44. SeidlMF, WangRP, Van den AckervekenG, GoversF, SnelB (2012) Bioinformatic inference of specific and general transcription factor binding sites in the plant pathogen Phytophthora infestans. PLoS One 7: e51295.
45. GuptaS, StamatoyannopoulosJA, BaileyTL, NobleWS (2007) Quantifying similarity between motifs. Genome Biol 8: R24.
46. CookeDE, CanoLM, RaffaeleS, BainRA, CookeLR, et al. (2012) Genome analyses of an aggressive and invasive lineage of the Irish potato famine pathogen. PLoS Pathog 8: e1002940.
47. BotellaMA, ParkerJE, FrostLN, Bittner-EddyPD, BeynonJL, et al. (1998) Three genes of the Arabidopsis RPP1 complex resistance locus recognize distinct Peronospora parasitica avirulence determinants. Plant Cell 10 : 1847–1860.
48. WangD, AmornsiripanitchN, DongXN (2006) A genomic approach to identify regulatory nodes in the transcriptional network of systemic acquired resistance in plants. PLoS Pathog 2: e123.
49. CaoH, GlazebrookJ, ClarkeJD, VolkoS, DongXN (1997) The Arabidopsis NPR1 gene that controls systemic acquired resistance encodes a novel protein containing ankyrin repeats. Cell 88 : 57–63.
50. WestermannAJ, GorskiSA, VogelJ (2012) Dual RNA-seq of pathogen and host. Nat Rev Microbiol 10 : 618–630.
51. NawrathC, MetrauxJP (1999) Salicylic acid induction-deficient mutants of Arabidopsis express PR-2 and PR-5 and accumulate high levels of camalexin after pathogen inoculation. Plant Cell 11 : 1393–1404.
52. DebRoyS, ThilmonyR, KwackYB, NomuraK, HeSY (2004) A family of conserved bacterial effectors inhibits salicylic acid-mediated basal immunity and promotes disease necrosis in plants. Proc Natl Acad Sci U S A 101 : 9927–9932.
53. SerranoM, WangBJ, AryalB, GarcionC, Abou-MansourE, et al. (2013) Export of Salicylic Acid from the Chloroplast Requires the Multidrug and Toxin Extrusion-Like Transporter EDS5. Plant Physiol 162 : 1815–1821.
54. WildermuthMC, DewdneyJ, WuG, AusubelFM (2001) Isochorismate synthase is required to synthesize salicylic acid for plant defence. Nature 414 : 562–565.
55. ChenHM, XueL, ChintamananiS, GermainH, LinHQ, et al. (2009) ETHYLENE INSENSITIVE3 and ETHYLENE INSENSITIVE3-LIKE1 repress SALICYLIC ACID INDUCTION DEFICIENT2 expression to negatively regulate plant innate immunity in Arabidopsis. Plant Cell 21 : 2527–2540.
56. BoutrotF, SegonzacC, ChangKN, QiaoH, EckerJR, et al. (2010) Direct transcriptional control of the Arabidopsis immune receptor FLS2 by the ethylene-dependent transcription factors EIN3 and EIL1. Proc Natl Acad Sci U S A 107 : 14502–14507.
57. MohrPG, CahillDM (2003) Abscisic acid influences the susceptibility of Arabidopsis thaliana to Pseudomonas syringae pv. tomato and Peronospora parasitica. Funct Plant Biol 30 : 461–469.
58. de Torres-ZabalaM, BennettMH, TrumanWH, GrantMR (2009) Antagonism between salicylic and abscisic acid reflects early host-pathogen conflict and moulds plant defence responses. Plant J 59 : 375–386.
59. FanJ, HillL, CrooksC, DoernerP, LambC (2009) Abscisic acid has a key role in modulating diverse plant-pathogen interactions. Plant Physiol 150 : 1750–1761.
60. NavarroL, DunoyerP, JayF, ArnoldB, DharmasiriN, et al. (2006) A plant miRNA contributes to antibacterial resistance by repressing auxin signaling. Science 312 : 436–439.
61. ParkJE, ParkJY, KimYS, StaswickPE, JeonJ, et al. (2007) GH3-mediated auxin homeostasis links growth regulation with stress adaptation response in Arabidopsis. J Biol Chem 282 : 10036–10046.
62. Robert-SeilaniantzA, MacLeanD, JikumaruY, HillL, YamaguchiS, et al. (2011) The microRNA miR393 re-directs secondary metabolite biosynthesis away from camalexin and towards glucosinolates. Plant J 67 : 218–231.
63. MutkaAM, FawleyS, TsaoT, KunkelBN (2013) Auxin promotes susceptibility to Pseudomonas syringae via a mechanism independent of suppression of salicylic acid-mediated defenses. Plant J 74 : 746–754.
64. de Torres-ZabalaM, TrumanW, BennettMH, LafforgueG, MansfieldJW, et al. (2007) Pseudomonas syringae pv. tomato hijacks the Arabidopsis abscisic acid signalling pathway to cause disease. EMBO J 26 : 1434–1443.
65. MaroisE, Van den AckervekenG, BonasU (2002) The Xanthomonas type III effector protein AvrBs3 modulates plant gene expression and induces cell hypertrophy in the susceptible host. Mol Plant Microbe Interact 15 : 637–646.
66. SpanuPD, AbbottJC, AmselemJ, BurgisTA, SoanesDM, et al. (2010) Genome expansion and gene loss in powdery mildew fungi reveal tradeoffs in extreme parasitism. Science 330 : 1543–1546.
67. QutobD, Tedman-JonesJ, DongSM, KufluK, PhamH, et al. (2009) Copy number variation and transcriptional polymorphisms of Phytophthora sojae RXLR effector genes Avr1a and Avr3a. PLoS One 4: e5066.
68. QutobD, ChapmanBP, GijzenM (2013) Transgenerational gene silencing causes gain of virulence in a plant pathogen. Nat Commun 4 : 1349..
69. CaillaudMC, WirthmuellerL, FabroG, PiquerezSJ, AsaiS, et al. (2012) Mechanisms of nuclear suppression of host immunity by effectors from the Arabidopsis downy mildew pathogen Hyaloperonospora arabidopsidis (Hpa). Cold Spring Harb Symp Quant Biol 77 : 285–293.
70. JupeJ, StamR, HowdenAJ, MorrisJA, ZhangR, et al. (2013) Phytophthora capsici-tomato interaction features dramatic shifts in gene expression associated with a hemi-biotrophic lifestyle. Genome Biol 14: R63.
71. GarciaAV, Blanvillain-BaufumeS, HuibersRP, WiermerM, LiG, et al. (2010) Balanced nuclear and cytoplasmic activities of EDS1 are required for a complete plant innate immune response. PLoS Pathog 6: e1000970.
72. KonczC, SchellJ (1986) The promoter of TL-DNA gene 5 controls the tissue-specific expression of chimeric genes carried by a novel type of Agrobacterium binary vector. Mol Gen Genet 204 : 383–396.
73. CloughSJ, BentAF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16 : 735–743.
74. AsaiS, OhtaK, YoshiokaH (2008) MAPK signaling regulates nitric oxide and NADPH oxidase-dependent oxidative bursts in Nicotiana benthamiana. Plant Cell 20 : 1390–1406.
75. AsaiS, YoshiokaH (2009) Nitric oxide as a partner of reactive oxygen species participates in disease resistance to necrotrophic pathogen Botrytis cinerea in Nicotiana benthamiana. Mol Plant Microbe Interact 22 : 619–629.
76. LiH, DurbinR (2009) Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25 : 1754–1760.
77. LunterG, GoodsonM (2011) Stampy: a statistical algorithm for sensitive and fast mapping of Illumina sequence reads. Genome Res 21 : 936–939.
78. LiH, HandsakerB, WysokerA, FennellT, RuanJ, et al. (2009) The Sequence Alignment/Map format and SAMtools. Bioinformatics 25 : 2078–2079.
79. ThorvaldsdottirH, RobinsonJT, MesirovJP (2013) Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinform 14 : 178–192.
80. LangmeadB, TrapnellC, PopM, SalzbergSL (2009) Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10: R25.
81. GentlemanRC, CareyVJ, BatesDM, BolstadB, DettlingM, et al. (2004) Bioconductor: open software development for computational biology and bioinformatics. Genome Biol 5: R80.
82. RobinsonMD, McCarthyDJ, SmythGK (2010) edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26 : 139–140.
Štítky
Hygiena a epidemiologie Infekční lékařství Laboratoř
Článek Identification of the Microsporidian as a New Target of the IFNγ-Inducible IRG Resistance SystemČlánek Human Cytomegalovirus Drives Epigenetic Imprinting of the Locus in NKG2C Natural Killer CellsČlánek APOBEC3D and APOBEC3F Potently Promote HIV-1 Diversification and Evolution in Humanized Mouse ModelČlánek Role of Non-conventional T Lymphocytes in Respiratory Infections: The Case of the Pneumococcus
Článek vyšel v časopisePLOS Pathogens
Nejčtenější tento týden
2014 Číslo 10- Stillova choroba: vzácné a závažné systémové onemocnění
- Diagnostika virových hepatitid v kostce – zorientujte se (nejen) v sérologii
- Diagnostický algoritmus při podezření na syndrom periodické horečky
- Perorální antivirotika jako vysoce efektivní nástroj prevence hospitalizací kvůli COVID-19 − otázky a odpovědi pro praxi
- Infekční komplikace virových respiračních infekcí – sekundární bakteriální a aspergilové pneumonie
-
Všechny články tohoto čísla
- Theory and Empiricism in Virulence Evolution
- -Related Fungi and Reptiles: A Fatal Attraction
- Adaptive Prediction As a Strategy in Microbial Infections
- Antimicrobials, Stress and Mutagenesis
- A Novel Function of Human Pumilio Proteins in Cytoplasmic Sensing of Viral Infection
- Social Motility of African Trypanosomes Is a Property of a Distinct Life-Cycle Stage That Occurs Early in Tsetse Fly Transmission
- Autophagy Controls BCG-Induced Trained Immunity and the Response to Intravesical BCG Therapy for Bladder Cancer
- Identification of the Microsporidian as a New Target of the IFNγ-Inducible IRG Resistance System
- mRNA Structural Constraints on EBNA1 Synthesis Impact on Antigen Presentation and Early Priming of CD8 T Cells
- Infection Causes Distinct Epigenetic DNA Methylation Changes in Host Macrophages
- Neutrophil Crawling in Capillaries; A Novel Immune Response to
- Live Attenuated Vaccine Protects against Pulmonary Challenge in Rats and Non-human Primates
- The ESAT-6 Protein of Interacts with Beta-2-Microglobulin (β2M) Affecting Antigen Presentation Function of Macrophage
- Characterization of Uncultivable Bat Influenza Virus Using a Replicative Synthetic Virus
- HIV Acquisition Is Associated with Increased Antimicrobial Peptides and Reduced HIV Neutralizing IgA in the Foreskin Prepuce of Uncircumcised Men
- Uses a Unique Ligand-Binding Mode for Trapping Opines and Acquiring A Competitive Advantage in the Niche Construction on Plant Host
- Involvement of a 1-Cys Peroxiredoxin in Bacterial Virulence
- Ethanol Stimulates WspR-Controlled Biofilm Formation as Part of a Cyclic Relationship Involving Phenazines
- Densovirus Is a Mutualistic Symbiont of a Global Crop Pest () and Protects against a Baculovirus and Bt Biopesticide
- Insights into Intestinal Colonization from Monitoring Fluorescently Labeled Bacteria
- Mycobacterial Antigen Driven Activation of CD14CD16 Monocytes Is a Predictor of Tuberculosis-Associated Immune Reconstitution Inflammatory Syndrome
- Lipoprotein LprG Binds Lipoarabinomannan and Determines Its Cell Envelope Localization to Control Phagolysosomal Fusion
- Dampens the DNA Damage Response
- MicroRNAs Suppress NB Domain Genes in Tomato That Confer Resistance to
- Novel Cyclic di-GMP Effectors of the YajQ Protein Family Control Bacterial Virulence
- Vaginal Challenge with an SIV-Based Dual Reporter System Reveals That Infection Can Occur throughout the Upper and Lower Female Reproductive Tract
- Detecting Differential Transmissibilities That Affect the Size of Self-Limited Outbreaks
- One Small Step for a Yeast - Microevolution within Macrophages Renders Hypervirulent Due to a Single Point Mutation
- Expression Profiling during Arabidopsis/Downy Mildew Interaction Reveals a Highly-Expressed Effector That Attenuates Responses to Salicylic Acid
- Human Cytomegalovirus Drives Epigenetic Imprinting of the Locus in NKG2C Natural Killer Cells
- Interaction with Tsg101 Is Necessary for the Efficient Transport and Release of Nucleocapsids in Marburg Virus-Infected Cells
- The N-Terminus of Murine Leukaemia Virus p12 Protein Is Required for Mature Core Stability
- Sterol Biosynthesis Is Required for Heat Resistance but Not Extracellular Survival in
- Allele-Specific Induction of IL-1β Expression by C/EBPβ and PU.1 Contributes to Increased Tuberculosis Susceptibility
- Host Cofactors and Pharmacologic Ligands Share an Essential Interface in HIV-1 Capsid That Is Lost upon Disassembly
- APOBEC3D and APOBEC3F Potently Promote HIV-1 Diversification and Evolution in Humanized Mouse Model
- Structural Basis for the Recognition of Human Cytomegalovirus Glycoprotein B by a Neutralizing Human Antibody
- Systematic Analysis of ZnCys Transcription Factors Required for Development and Pathogenicity by High-Throughput Gene Knockout in the Rice Blast Fungus
- Epstein-Barr Virus Nuclear Antigen 3A Promotes Cellular Proliferation by Repression of the Cyclin-Dependent Kinase Inhibitor p21WAF1/CIP1
- The Host Protein Calprotectin Modulates the Type IV Secretion System via Zinc Sequestration
- Cyclophilin A Associates with Enterovirus-71 Virus Capsid and Plays an Essential Role in Viral Infection as an Uncoating Regulator
- A Novel Alpha Kinase EhAK1 Phosphorylates Actin and Regulates Phagocytosis in
- The pH-Responsive PacC Transcription Factor of Governs Epithelial Entry and Tissue Invasion during Pulmonary Aspergillosis
- Sensing of Immature Particles Produced by Dengue Virus Infected Cells Induces an Antiviral Response by Plasmacytoid Dendritic Cells
- Co-opted Oxysterol-Binding ORP and VAP Proteins Channel Sterols to RNA Virus Replication Sites via Membrane Contact Sites
- Characteristics of Memory B Cells Elicited by a Highly Efficacious HPV Vaccine in Subjects with No Pre-existing Immunity
- HPV16-E7 Expression in Squamous Epithelium Creates a Local Immune Suppressive Environment via CCL2- and CCL5- Mediated Recruitment of Mast Cells
- Dengue Viruses Are Enhanced by Distinct Populations of Serotype Cross-Reactive Antibodies in Human Immune Sera
- CD4 Depletion in SIV-Infected Macaques Results in Macrophage and Microglia Infection with Rapid Turnover of Infected Cells
- A Sialic Acid Binding Site in a Human Picornavirus
- Contact Heterogeneity, Rather Than Transmission Efficiency, Limits the Emergence and Spread of Canine Influenza Virus
- Myosins VIII and XI Play Distinct Roles in Reproduction and Transport of
- HTLV-1 Tax Stabilizes MCL-1 via TRAF6-Dependent K63-Linked Polyubiquitination to Promote Cell Survival and Transformation
- Species Complex: Ecology, Phylogeny, Sexual Reproduction, and Virulence
- A Critical Role for IL-17RB Signaling in HTLV-1 Tax-Induced NF-κB Activation and T-Cell Transformation
- Exosomes from Hepatitis C Infected Patients Transmit HCV Infection and Contain Replication Competent Viral RNA in Complex with Ago2-miR122-HSP90
- Role of Non-conventional T Lymphocytes in Respiratory Infections: The Case of the Pneumococcus
- Kaposi's Sarcoma-Associated Herpesvirus Induces Nrf2 during Infection of Endothelial Cells to Create a Microenvironment Conducive to Infection
- A Relay Network of Extracellular Heme-Binding Proteins Drives Iron Acquisition from Hemoglobin
- Glutamate Secretion and Metabotropic Glutamate Receptor 1 Expression during Kaposi's Sarcoma-Associated Herpesvirus Infection Promotes Cell Proliferation
- Reduces Malaria and Dengue Infection in Vector Mosquitoes and Has Entomopathogenic and Anti-pathogen Activities
- PLOS Pathogens
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Novel Cyclic di-GMP Effectors of the YajQ Protein Family Control Bacterial Virulence
- MicroRNAs Suppress NB Domain Genes in Tomato That Confer Resistance to
- The ESAT-6 Protein of Interacts with Beta-2-Microglobulin (β2M) Affecting Antigen Presentation Function of Macrophage
- Characterization of Uncultivable Bat Influenza Virus Using a Replicative Synthetic Virus
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



