-
Články
- Vzdělávání
- Časopisy
Top články
- Témata
- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Relacin, a Novel Antibacterial Agent Targeting the Stringent Response
Finding bacterial cellular targets for developing novel antibiotics has become a major challenge in fighting resistant pathogenic bacteria. We present a novel compound, Relacin, designed to inhibit (p)ppGpp production by the ubiquitous bacterial enzyme RelA that triggers the Stringent Response. Relacin inhibits RelA in vitro and reduces (p)ppGpp production in vivo. Moreover, Relacin affects entry into stationary phase in Gram positive bacteria, leading to a dramatic reduction in cell viability. When Relacin is added to sporulating Bacillus subtilis cells, it strongly perturbs spore formation regardless of the time of addition. Spore formation is also impeded in the pathogenic bacterium Bacillus anthracis that causes the acute anthrax disease. Finally, the formation of multicellular biofilms is markedly disrupted by Relacin. Thus, we establish that Relacin, a novel ppGpp analogue, interferes with bacterial long term survival strategies, placing it as an attractive new antibacterial agent.
Published in the journal: . PLoS Pathog 8(9): e32767. doi:10.1371/journal.ppat.1002925
Category: Research Article
doi: https://doi.org/10.1371/journal.ppat.1002925Summary
Finding bacterial cellular targets for developing novel antibiotics has become a major challenge in fighting resistant pathogenic bacteria. We present a novel compound, Relacin, designed to inhibit (p)ppGpp production by the ubiquitous bacterial enzyme RelA that triggers the Stringent Response. Relacin inhibits RelA in vitro and reduces (p)ppGpp production in vivo. Moreover, Relacin affects entry into stationary phase in Gram positive bacteria, leading to a dramatic reduction in cell viability. When Relacin is added to sporulating Bacillus subtilis cells, it strongly perturbs spore formation regardless of the time of addition. Spore formation is also impeded in the pathogenic bacterium Bacillus anthracis that causes the acute anthrax disease. Finally, the formation of multicellular biofilms is markedly disrupted by Relacin. Thus, we establish that Relacin, a novel ppGpp analogue, interferes with bacterial long term survival strategies, placing it as an attractive new antibacterial agent.
Introduction
The emergence of multi drug resistant bacteria dictates the need to develop novel antibiotics that target key components of essential bacterial processes. The pleiotropic response to starvation, known as the Stringent Response, provides a potential target, as it is crucial for activation of survival strategies such as stationary phase, sporulation and biofilm formation [1]–[4]. Further, the Stringent Response has been recently shown to mediate antibiotic tolerance in nutrient-limited bacteria [5]. The Stringent Response is induced by the accumulation of the bacterial signaling molecules 5′-triphosphate-3′-diphosphate and 5′-3′-bis-diphosphate, collectively called (p)ppGpp [6]. Synthesis of (p)ppGpp has been characterized as a ribosome-dependent pyrophosphate transfer of the β and γ phosphates from an ATP donor to the 3′ hydroxyl group of GTP or GDP [7].
In Gram negative bacteria (p)ppGpp is mostly synthesized by RelA and hydrolyzed by SpoT, while in Gram positive bacteria a bifunctional enzyme, Rel/Spo, both synthesizes and hydrolyses (p)ppGpp [8], [9]. Upon nutrient deprivation, Rel proteins bind to ribosomes blocked by uncharged tRNA and catalyze the synthesis of (p)ppGpp [10]. It has been proposed that Rel proteins hop between stalled ribosomes in order to achieve the (p)ppGpp concentration required to induce the Stringent Response [10]. A recent report, however, proposes that RelA actually synthesizes ppGpp only after it is dissociated from the ribosome [11]. The Rel proteins comprise two major domains: a catalytic domain located in the N-terminus and a regulatory domain in the C-terminus [12]. Crystal structure analysis of the N-terminal domain of Rel/Spo from Streptococcus equisimilis (S. equisimilis) revealed two conformations with opposing hydrolase and synthetase states [13]. Further, the N-terminal domain was found to harbor two catalytic subdomains: N-terminal which hydrolyses (p)ppGpp and C-terminal that catalyzes its synthesis [12].
When ppGpp accumulates within the bacterial cell it affects transcription and a plethora of physiological activities [14]–[16]. Indeed, the activation of many stress-induced genes is partially or totally dependent on ppGpp [17], [18]. The importance of (p)ppGpp as a regulator of bacterial survival prompted us to develop a series of non-hydrolyzable ppGpp analogues [19] potentially targeting Rel proteins. Here we present Relacin, a potent inhibitor of Rel proteins. We demonstrate that Relacin inhibits RelA and Rel/Spo in vitro and impairs growth, sporulation and biofilm formation in Gram positive bacteria.
Results
Relacin inhibits (p)ppGpp production by Rel proteins
Based on the Rel/Spo crystal structure [13], we designed Relacin (Figure 1A), a 2′-deoxyguanosine-based analogue of ppGpp, in which the original pyrophosphate moieties at positions 5′ and 3′ were replaced by glycyl-glycine dipeptides linked to the sugar ring by a carbamate bridge (see Text S1; Figure S1A and S1B). Modeling the binding of Relacin to the Rel/Spo synthetase site shows that it occupies a considerable volume of the binding pocket and forms a range of hydrogen bonds and hydrophobic interactions (Figure S1C), providing a structural basis for the inhibitory effect of Relacin.
Fig. 1. Relacin inhibits the activity of Rel proteins. 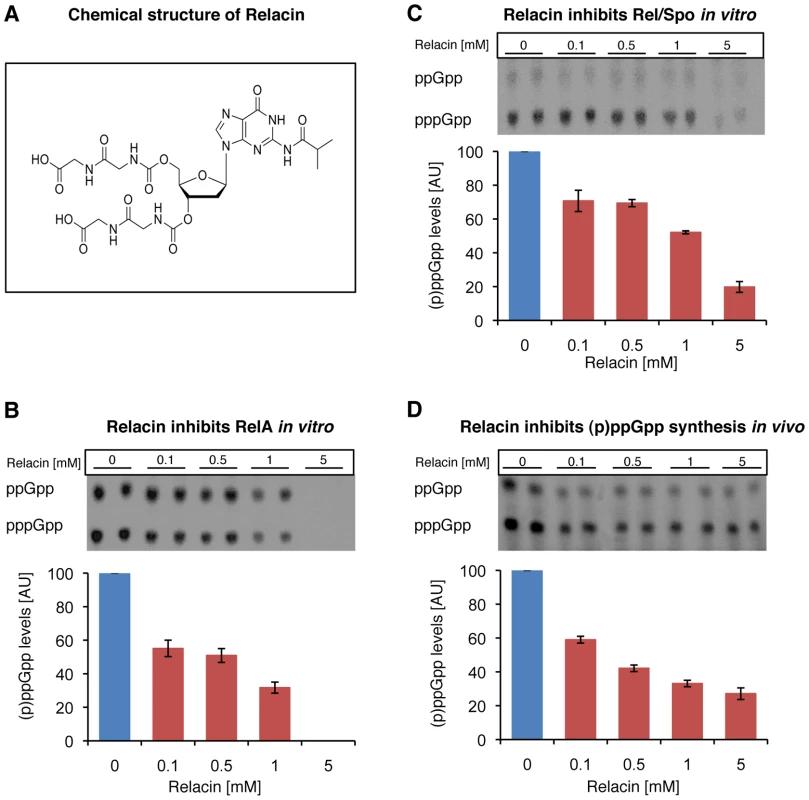
(A) Chemical structure of Relacin. (B–C) Relacin inhibits (p)ppGpp synthesis in vitro. Representative autoradiograms of PEI thin-layer chromatography showing a decrease in labeled (p)ppGpp synthesized from α-32P-GTP precursor by purified RelA (E. coli) (B) or Rel/Spo (D. radiodurans) (C) with increasing concentrations of Relacin, as indicated (see Materials and Methods). Shown is the average of duplicates of a representative experiment. Error bars represent the range. (D) Relacin inhibits (p)ppGpp synthesis in living B. subtilis (PY79) cells. The accumulation of (p)ppGpp in response to amino acid starvation, induced by the addition of SHX, was monitored in the absence or presence of increasing concentrations of Relacin, as indicated. The (p)ppGpp level was determined using PEI thin-layer chromatography as in (B–C) (see Materials and Methods). Shown is the average of duplicates of a representative experiment. Error bars represent the range. To investigate the biological activity of Relacin, we first evaluated its inhibitory potential on the (p)ppGpp synthetase activity of RelA and Rel/Spo purified from Escherichia coli (E. coli) and Deinococcus radiodurans (D. radiodurans), respectively. Relacin was shown to inhibit both Rel proteins in a dose-dependent manner. Remarkably, at the highest Relacin concentration, the Rel enzymes from Gram negative and positive bacteria were inhibited by approximately 100% and 80%, respectively (Figure 1B and 1C). Notably, the synthesis of ppGpp and pppGpp by both Rel proteins was similarly inhibited (Figure S2A and S2B).
Next, we examined the effect of Relacin on the interaction between Rel/Spo and stalled ribosomes. Ribosomes purified from D. radiodurans were incubated with Rel/Spo in the presence of increasing concentrations of Relacin, and the relative amount of Rel/Spo molecules associated with 70S complexes was examined. Western blot analysis revealed that Relacin increases the levels of Rel/Spo locked on the ribosomes (Figure 2A), suggesting that the presence of Relacin reduces the pool of protein molecules available for (p)ppGpp re-synthesis [10]. To further investigate whether ribosomes are actually required for Relacin activity, we took advantage of a RelA mutant protein (RelAC638F), which exerts its activity in a ribosome-independent manner. Relacin was equally able to inhibit the mutant protein (Figure 2B), indicating a direct Relacin-RelA interaction.
Fig. 2. The effect of Relacin on Rel-ribosomes interaction. 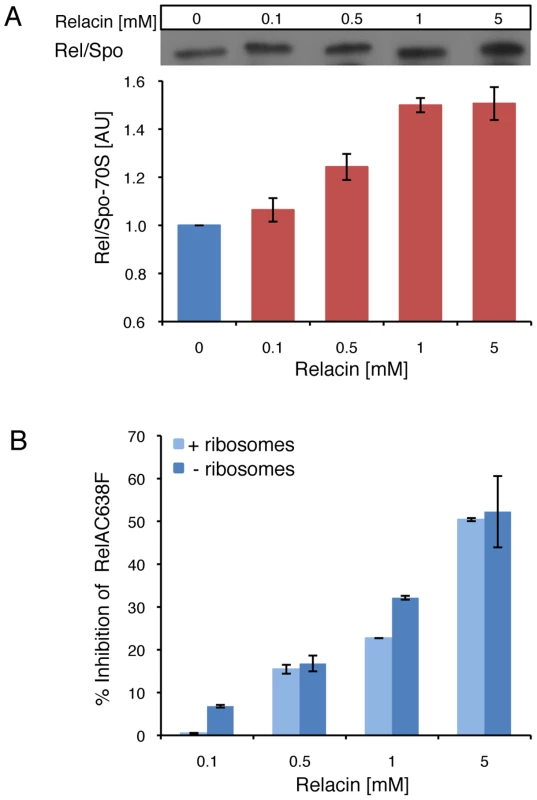
(A) Relacin inhibits dissociation of Rel/Spo from the ribosome. The relative amount of Rel/Spo (D. radiodurans) bound to purified ribosomes was quantified following the addition of increasing levels of Relacin. Rel/Spo molecules associated with 70S complexes were detected by Western blot analysis (see Materials and Methods). Histogram indicates the average of two independent biological repeats. Error bars represent the range. (B) Ribosome independent inhibition of (p)ppGpp synthesis. The constitutively active, ribosome-independent RelAC638F (E. coli) protein was treated with increasing concentrations of Relacin, as indicated (see Materials and Methods) in the presence or absence of isolated ribosomes. Shown is the average of duplicates of a representative experiment. Error bars represent the range. We then examined the influence of Relacin on (p)ppGpp production in living cells upon induction of the Stringent Response. To this end, cells of the Gram positive spore forming bacterium Bacillus subtilis (B. subtilis) were incubated with Relacin and treated with serine hydroxamate (SHX) to simulate amino acid starvation [20], [21]. Subsequently, the accumulated levels of (p)ppGpp were monitored from cell extracts. In line with the inhibitory activity observed in vitro, Relacin markedly reduced (p)ppGpp production in vivo (Figure 1D). Interestingly, although Relacin was found to completely inhibit the activity of purified RelA from the Gram negative bacterium E. coli (Figure 1B), no obvious effect of the compound on bacterial (p)ppGpp synthesis was observed (Figure S2C). This is most likely due to the inability of Relacin to penetrate the E. coli cell and reach its target.
Relacin reduces survival of Gram positive bacteria
Having ascertained that Relacin affects the production of (p)ppGpp in vivo by B. subtilis cells and given the vital role of the Stringent Response in bacterial survival, we investigated the impact of Relacin on cell growth and viability. Interestingly, in the presence of Relacin, cells exhibited an extended logarithmic phase as indicated by the increase in OD600 values, implying that they failed to properly enter into stationary phase (Figure 3A). Of note, a similar phenomenon was observed for spoT null mutant of Helicobacter pylori [22]. This failure led to substantial dose-dependent cell death after 24 hours, with an estimated IC50 of 200 µM, as monitored by the reduction in colony forming units (Figures 3B and S3). Moreover, after 48 hours the deleterious effect of Relacin persisted, reducing the number of colonies by approximately five orders of magnitude relative to untreated cultures (Figure 3C). A similar viability pattern was observed in untreated B. subtilis cells lacking Rel/Spo (Figure 3C), suggesting that this enzyme is indeed the main target for Relacin action. Consistent with this observation, the survival of the mutant strain was not affected by the addition of Relacin (Figure 3C). Notably, the effect of Relacin on survival is not likely to be dependent on spore formation, as only few spores, if any, were present in untreated cultures. On the other hand, the appearance of dead cells as well as disintegrated cells was largely increased within the treated population over time (Figure S4). Consistent with the inability of Relacin to perturb (p)ppGpp production in E. coli (Figure S2C), no effect on growth and viability was detected in these cells.
Fig. 3. Relacin affects bacterial growth and survival. 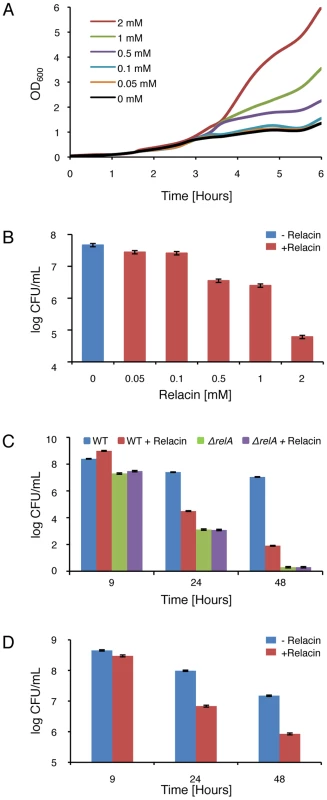
(A) Relacin influences entry into stationary phase. Shown are growth curves of wild type B. subtilis (PY79) cells grown in CH medium at 37°C in the absence or presence of increasing concentrations of Relacin added at OD600 0.2. (B) Relacin exerts a toxic effect. The viability of B. subtilis (PY79) cells was evaluated by counting colony forming units (CFU) after 24 hours of incubation in CH medium at 37°C in the absence or presence of increasing concentrations of Relacin added at OD600 0.2. Shown is a representative experiment, in which SD was calculated from at least three repeats for each concentration. (C) Long term effect of Relacin treatment. The effect of Relacin (2 mM) on the viability of wild type B. subtilis (PY79) cells or ΔrelA (ME215) cells was measured. Cells were incubated in CH medium at 37°C, and viability was determined by counting colony forming units (CFU). Relacin was added at OD600 0.2. Shown is a representative experiment, in which SD was calculated from at least three repeats for each point. (D) The toxic effect of Relacin on GAS. The effect of Relacin (2 mM) on the viability of wild type GAS (JRS4) cells, incubated in THY medium at 37°C, was evaluated as in (C). The biological activity of Relacin was further explored in non-spore-forming Gram positive bacteria. Treating the Group A streptococcus (GAS) with Relacin revealed that, although growth rate was only slightly affected, cell viability was largely reduced after 24 hours (Figure S5A and S5B). This toxic effect was enhanced after 48 hours (Figure 3D) and was associated with the formation of very small colonies. Additionally, as observed for B. subtilis, entering stationary phase was perturbed by Relacin in the extremely slow growing bacterium D. radiodurans (Figure S5C). Furthermore, the addition of Relacin to D. radiodurans cells diminished bacterial viability, as indicated by plating efficiency assay carried out after 56 and 72 hours of incubation (Figure S5D). Thus, we established that Relacin functions as an antibacterial agent that impairs entry into stationary phase and causes bacterial death.
Relacin perturbs long term survival strategies
In addition to switching into stationary phase some bacteria, such as Bacilli, respond to nutrient limitation by producing highly resilient dormant spores as a strategy for long term survival [23]–[25]. Entry into sporulation is triggered by a decrease in the intracellular GTP pools, in part due to conversion of GTP into (p)ppGpp by RelA [26]. At the onset of sporulation, an asymmetric septum is generated, dividing the cell into a nurturing mother cell and a smaller forespore compartment that develops into a spore. Subsequently, the forespore is engulfed by the mother cell to form a fully mature spore. Remarkably, when nutrients become available the spore can rapidly convert into an actively growing cell [23]–[25]. To explore whether Relacin affects sporulation, B. subtilis cells were induced to sporulate in the presence or absence of Relacin and sporulation progression was monitored by observing polar septa formation. Indeed, sporulation was largely inhibited, with asymmetric septa exhibited by only 8% and 0.5% of the cells treated with 200 µM and 1 mM of Relacin, respectively. In comparison, 47% of untreated cells displayed polar septa at the same time point (Figure 4A). In line with these observations, Relacin lowered the number of cells expressing early (SpoIIE), middle (SpoIIQ) and late (SspE) sporulation-specific proteins along the process [25] (Figures 4B and S6). Subsequently, a fivefold drop in the formation of mature heat resistant spores was measured at the highest Relacin concentration (Figure 4A and 4C). Remarkably, adding Relacin to sporulating cells at different time points, up to six hours after the induction of sporulation, strongly inhibited spore formation regardless of the time of addition (Figure 4E). These findings indicate that the ppGpp signal is crucial throughout the entire pathway of sporulation, and demonstrate the potency of Relacin to impede this process. Importantly, spore formation in the pathogenic bacterium Bacillus anthracis, the causative agent of anthrax disease, was inhibited by Relacin in a similar fashion (Figure 4D), establishing the compound as a general inhibitor of the Bacilli sporulation process.
Fig. 4. Relacin influences the sporulation process in Bacilli. 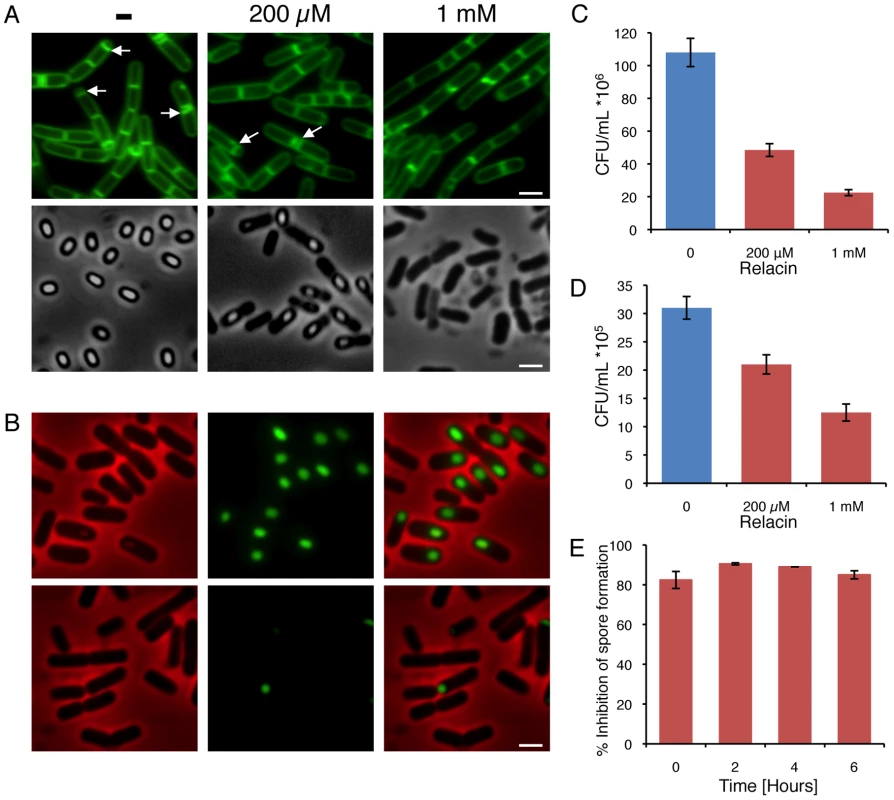
(A) Relacin inhibits sporulation. Microscopy images of sporulating wild type B. subtilis (PY79) cells in the absence or presence of Relacin, added at time 0 of sporulation at the indicated concentrations. Upper panels: cells at t = 2 hr of sporulation stained with the fluorescent membrane dye FM1–43. Arrows indicate position of polar septa. Lower panels: phase contrast images of cells at t = 24 hr of sporulation. Scale bars correspond to 1 µm. (B) Relacin inhibits expression of the mid-sporulation protein SpoIIQ. Fluorescence microscopy images of B. subtilis (PE128) cells harboring spoIIQ-gfp at t = 4 hr of sporulation, in the absence (upper panels) or presence (lower panels) of Relacin (1 mM), added at time 0 of sporulation. Shown are phase contrast (red), GFP fluorescence (green) and overlay images. Scale bar corresponds to 1 µm. (C–D) Relacin inhibits Bacilli spore formation. The formation of heat resistant B. subtilis (PY79) (C) and B. anthracis (Sterne) (D) spores was monitored in the absence or presence of Relacin, added at the indicated concentrations at time 0 of sporulation (see Text S1). Shown are representative experiments, in which SD was calculated from at least three repeats for each concentration. (E) Relacin added at different time points during sporulation inhibits spore formation. Inhibition of spore formation by wild type B. subtilis (PY79) cells was evaluated after addition of Relacin (1 mM) at the indicated time points of sporulation. Inhibition was determined using a heat resistance assay (see Text S1) and is expressed relative to untreated cultures. Shown is a representative experiment, in which SD was calculated from at least three repeats for each time point. Since it has been reported that relA mutant cells fail to properly form multicellular biofilm structures [2], the effect of Relacin on the ability of B. subtilis cells to produce biofilms was evaluated. Indeed, a disrupted pellicle was visualized at the air/liquid interface of standing cell cultures grown in the presence of the compound (Figure 5A). Importantly, the effect on biofilm formation was found to be dose-dependent (Figure 5A). Consistent with this observation, Relacin also inhibited the development of biofilm on solid medium, leading to the formation of colonies with altered morphology that were smaller in size than the untreated ones (Figure 5B). To visualize cell assembly within the biofilm pellicle in higher resolution upon Relacin treatment, we took advantage of a strain harboring the rrnE promoter fused to gfp. This promoter was found to be constitutively active [27], and therefore reports cell viability and localization. Observing biofilm pellicles by confocal laser scanning microscopy revealed that the untreated cells formed homogeneous biofilm layers, while the treated cell pellicles contained large gaps, indicating their disintegrated state (Figure 5C). Moreover, staining the biofilm with propidium iodide (PI), indicative of unviable cells, showed the signal to be higher within the treated biofilm (Figure 5E). Finally, quantifying GFP fluorescence from recovered pellicles revealed a clear reduction in the viable biomass upon Relacin treatment, as the measured fluorescence level was significantly reduced (Figure 5D). Taken together, we conclude that Relacin interferes with biofilm formation, an alternative bacterial developmental pathway.
Fig. 5. Relacin affects biofilm formation in Bacillus subtilis. 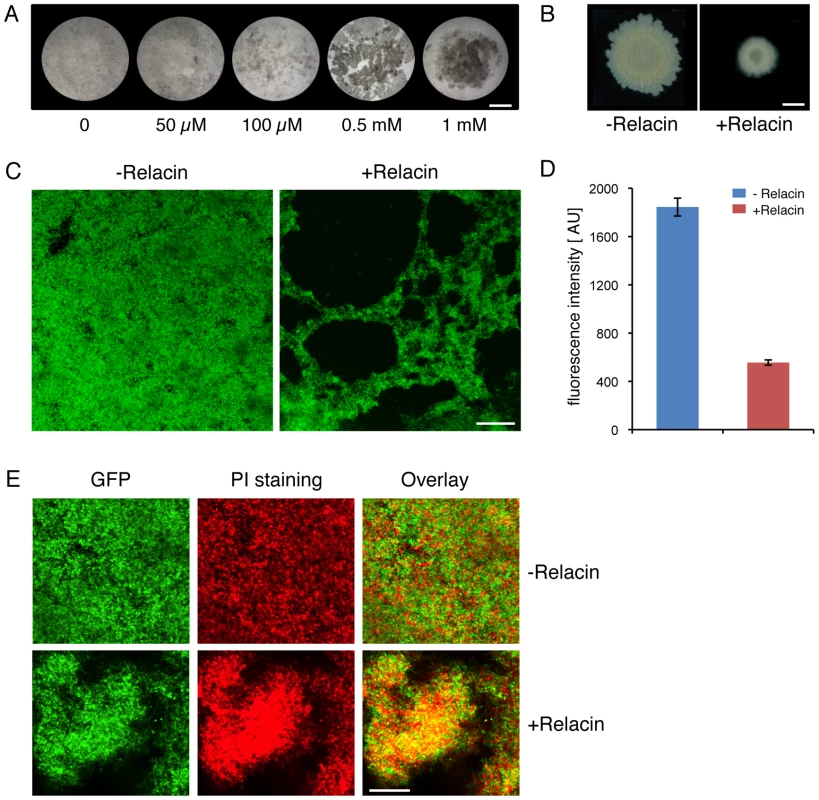
(A) Relacin inhibits pellicle biofilm formation. Wild type B. subtilis (3610) cells were induced to form biofilms in liquid standing cultures in the absence or presence of Relacin at the indicated concentrations (see Text S1). Cultures were photographed after 3 days. Scale bar corresponds to 5 mm. (B) Relacin inhibits biofilm colony formation. Wild type B. subtilis (3610) cells were induced to form biofilms on solid medium, in the absence or presence of Relacin (1 mM) (see Text S1). Colonies were photographed after 24 hours. Scale bar corresponds to 3 mm. (C) Relacin causes biofilm disintegration. B. subtilis (YA224) cells harboring PrrnE-gfp fusion were induced to form biofilm in liquid standing cultures in the absence or presence of Relacin (1 mM) as indicated (see Text S1). Biofilms were visualized after 3 days using confocal microscopy (see Materials and Methods). Green signal corresponds to GFP produced from PrrnE. Scale bar corresponds to 50 µm. (D) Relacin reduces biofilm biomass. B. subtilis (YA224) cells harboring PrrnE-gfp fusion were induced to form biofilm in liquid standing cultures in the absence or presence of Relacin (1 mM) as indicated. Biofilm pellicles were disintegrated and cell biomass was evaluated by GFP fluorescence measurements and is displayed in arbitrary units [AU] (see Text S1). Shown is the average of two independent biological repeats. Error bars represent the range. (E) Relacin leads to cell death within the biofilm. B. subtilis (YA224) cells harboring PrrnE-gfp fusion were induced to form biofilm in liquid standing cultures in the absence or presence of Relacin (1 mM) as indicated (see Text S1). After 3 days, biofilms were stained with PI to indicate cell death and observed by confocal microscopy (see Materials and Methods). Shown are GFP fluorescence produced from PrrnE (green), PI staining (red), and overlay images. Scale bar corresponds to 50 µm. Discussion
In this report, we established Relacin as a novel antibacterial agent. By specifically interfering with the activation of the Stringent Response, Relacin perturbs the switch into stationary phase in several tested Gram positive bacteria and leads to bacterial death. Although Relacin did not affect growth and survival of the Gram negative E. coli, it was found to effectively inhibit the E. coli RelA in vitro, implying that improving the delivery of Relacin to Gram negative bacteria may lead to an effective outcome. Relacin was found to block every tested stage of B. subtilis sporulation, proving the essentiality of the Stringent Response throughout this process. Finally, we demonstrate that Relacin affects the production of multicellular biofilm communities, formed in response to challenging conditions. Taken together, we present evidence that Relacin impedes bacterial long term survival pathways, placing the compound as a new promising antibacterial agent.
By utilizing the crystal structure of Rel/Spo from the S. equisimilis, we were able to model the interaction of Relacin with amino acid residues located within the Rel/Spo synthetase site. This analysis yielded the identification of a putative binding mode of Relacin, presumably adopting the conformation shown in Figure S1C. In this conformation, Relacin forms a net of hydrogen bonds and hydrophobic interactions that are most likely to provide a more efficient binding in comparison to previously identified inhibitors exhibiting lower activity [19].
Relacin appears to specifically target Rel proteins, as the effect of the compound was nearly undetectable when tested on Rel/Spo mutant cells. Consistently, Relacin activity in vivo resulted in a sharp decrease in (p)ppGpp synthesis. Since ppGpp inhibits the enzyme inosine monophosphate dehydrogenase, it causes the cellular GTP levels to decrease [28]. The intracellular levels of GDP/GTP are known to determine the initiation of several developmental pathways such as sporulation and biofilm formation [26], [29], [30] that were indeed shown to be influenced by Relacin. Interestingly, we also observed that Relacin treatment resulted in a large decrease in Rel/Spo ability to dissociate from ribosomes in vitro. This deficiency could be explained by the model proposed by Wendrich et al., [10] in which the rapid accumulation of ppGpp during amino acid starvation is attributed to the ability of RelA to ‘hop’ between ribosomes. This potential hopping is probably a consequence of the synthesis of (p)ppGpp that releases RelA from the ribosome, liberating it for another synthesis cycle.
The emergence of bacterial resistance to the current array of antimicrobial agents demands the development of novel strategies to eradicate pathogenic bacteria. The traditional cellular antibiotic targets include ribosomes, cell wall constituents and components of nucleic acids synthesis [31]. These cellular targets are mainly active during the bacterial vegetative phase, making the available antibiotics effective mostly during growth. However, the ability of bacteria to reside in nature within biofilm communities or as durable spores, as well as to become persistent to antibiotic treatment [32], sets the need to tackle these alternative modes. In this regard, Relacin affects specifically the Stringent Response, a pathway crucial for the activation of bacterial survival strategies. Since Relacin can persist for a relatively long period of time, and exert its effect even a few days post addition, it might become a valuable antagonist of these long term survival approaches. Taken together, Relacin may be combined with antibiotics currently in use, to eradicate non-homogenous bacterial populations with cells residing in diverse life cycles.
Cellular components, which are conserved throughout the bacterial kingdom and crucial for cellular survival, provide attractive antimicrobial targets as long as they lack eukaryotic counterparts. One of such targets is the highly conserved bacterial tubulin-like cell division protein FtsZ, which provides the basis for the assembly of the division machinery [33]. Indeed, a promising inhibitor of FtsZ with potent and selective activity against Staphylococci has been described [34]. In a similar fashion, the ubiquity of Rel enzymes among bacteria, combined with the absence of known (p)ppGpp synthetases in mammalian cells [35], [36], strengthen the potential of Relacin to turn into a therapeutic antibiotic. The profound influence of Relacin on long term bacterial survival makes it an attractive compound to serve as a scaffold for generating an array of new antibacterial agents.
Materials and Methods
Synthesis and modeling of Relacin
Synthesis of Relacin and a structural model for its interaction with Rel/Spo (p)ppGpp synthetase binding pocket are described in details Text S1.
Bacterial growth conditions
Bacterial strains used in this study are described in Table S1. Plasmid construction is described in Text S1. All general methods for B. subtilis were carried out as described previously [37]. B. subtilis cells were grown in hydrolyzed casein (CH) at 37°C [37], unless indicated differently. GAS strain was grown at 37°C without shaking in Todd-Hewitt medium supplemented with 0.2% yeast extract (THY) [38]. D. radiodurans R1 cells were grown in TYG which contains: 0.5% tryptone, 0.3% yeast extract and 0.1% glucose at 30°C with shaking. E. coli cells were grown at 37°C in LB medium. Cultures were inoculated to an OD600 of 0.05 using an overnight culture grown in the same medium, unless indicated differently. Sporulation conditions and biofilm colony and pellicle formation are described in Text S1.
Purification of Rel proteins and crude ribosomes
Purification of RelA or RelA-C638F from E. coli (CF9467) harboring ΔrelA and over-expressing pQE30-RelA or pQE30-RelA-C638F respectively, was carried out as described previously [19]. Purification of Rel/Spo from D. Radiodurans R1 was performed under identical conditions; however, the protein was expressed in E. coli BL21 CodonPlus (Stratagene) cells. Of note, Rel/Spo from D. Radiodurans R1, is the only known full length active protein purified from Gram positive bacteria. Isolation of crude ribosomes (containing 70S, mRNA, tRNA) from E. coli (CF9467) was carried out as described previously [19]. Isolation of crude ribosomes from D. Radiodurans was carried out in a similar fashion with the following modifications: D. radiodurans R1 cells were grown in LB(+) over night at 30°C, cells were diluted 1∶100 in LB(+) medium and incubated at 30°C for additional 48 hours.
Measuring (p)ppGpp synthesis in vitro
For measuring (p)ppGpp synthesis by RelA, RelA-C638F or Rel/Spo proteins in vitro: 1 µg of purified Rel protein, 20 µg of isolated ribosomes and 10 µCi of α-32P labeled GTP, were mixed in a buffer [0.5 mM GTP, 4 mM ATP, 50 mM Tris-HCl (pH 7.4), 1 mM DTT, 10 mM MgCl2, 10 mM KCl and 27 mM (NH4)2SO4] to a final volume of 20 µL without or with increasing amounts of Relacin as indicated. Reactions were stopped by the addition of 5 µL formic acid. Each reaction was loaded (5 µL) and separated on Cellulose PEI TLC plates (Merck) using 1.5 M KH2PO4 as mobile phase. Plates were autoradiographed using the Fijix Bas100 PhosphorImager (Japan). (p)ppGpp signal was measured using TINA 2.0 software (Raytest, Strauben-Hardt). The total amount of (p)ppGpp was the sum of signals from ppGpp and pppGpp.
Measuring (p)ppGpp synthesis in vivo
B. subtilis (PY79) or E. coli (W3110) cells were grown in MOPS glucose minimal medium [39] supplemented with all amino acids except glutamine and glutamate. At OD600 0.1, cells were supplemented with H332PO4 and incubated for 45 minutes, after which Relacin was added at the indicated concentrations. Cells were incubated for additional 15 minutes. Next, amino acid starvation was induced by adding serine-hydroxamate (SHX, Sigma) 1 mg/mL [20]. Samples were withdrawn ten minutes after addition of SHX and analyzed for their (p)ppGpp content as described above (Measuring (p)ppGpp synthesis in vitro).
Measuring Rel/Spo - 70S association
The reaction was carried out as described above for measuring (p)ppGpp synthesis in vitro, without the addition of radiolabeled GTP, with or without increasing amounts of Relacin as indicated. Reactions were centrifuged for 4 hours at 35,000 g (4°C), ribosomal fractions were separated by 12% SDS-polyacrylamide gel electrophoresis, transferred to PVDF membrane (Millipore Bedford) and processed for immunoreaction using mouse anti-His antibody (1∶10,000; Amersham). Immunoreactive proteins were detected using a chemiluminescence kit (Biological Industries) according to the manufacturer's protocol.
Fluorescence microscopy
Fluorescence microscopy was carried out as previously described [40]. Samples (0.5 mL) of a given culture were removed, centrifuged briefly, and resuspended in 10 µL of PBS×1 (Phosphate-Buffered Saline) supplemented with 1 µg/mL membrane stain FM1–43 or FM4–64 (Molecular Probes, Invitrogen). Cells were visualized and photographed using an Axioplan2 microscope (Zeiss) equipped with CoolSnap HQ camera (Photometrics, Roper Scientific) or an Axioobserver Z1 microscope (Zeiss) equipped with a CoolSnap HQII camera (Photometrics, Roper Scientific). System control and image processing were performed using MetaMorph 7.2r4 software (Molecular Devices).
Confocal laser scanning microscopy
For observing biofilm pellicles, the medium of 3 day-old pellicles grown in microplates was gently removed and minimal volume of PBS ×1 with or without 10 µg/ml PI (Molecular Probes, Invitrogen) was added. Cells were visualized and photographed using a confocal laser scanning fluorescence microscope LSM700 (Zeiss). System control and image processing were performed using Zen 2009 (Zeiss) and MetaMorph 7.2r4 (Molecular Devices) softwares.
Supporting Information
Zdroje
1. JainV, KumarM, ChatterjiD (2006) ppGpp: Stringent response and survival. J Microbiol 44 : 1–10.
2. LemosJAC, BrownTA, BurneRA (2004) Effects of RelA on key virulence properties of planktonic and biofilm populations of Streptococcus mutans. Infect Immun 72 : 1431–1440.
3. OchiK, KandalaJC, FreeseE (1981) Initiation of Bacillus subtilis sporulation by the stringent response to partial amino-acid deprivation. J Biol Chem 256 : 6866–6875.
4. PotrykusK, CashelM (2008) (p)ppGpp: Still Magical? Annu Rev Microbiol 35–51.
5. NguyenD, Joshi-DatarA, LepineF, BauerleE, OlakanmiO, et al. (2011) Active starvation responses mediate antibiotic tolerance in biofilms and nutrient-limited bacteria. Science 334 : 982–986.
6. CashelM, GallantJ (1969) Two compounds implicated in the function of the RC gene of Escherichia coli. Nature 221 : 838–841.
7. Cashel M GD, Hernandez VH, Vinella D (1996) The stringent response. In: Neidhardt, FC et al., editors. Escherichia coli and Salmonella typhimurium: Cellular and Molecular Biology (2nd edn). ASM Press. pp. 1458–1496.
8. MetzgerS, SarubbiE, GlaserG, CashelM (1989) Protein sequences encoded by the relA and the spoT genes of Escherichia coli are interrelated. J Biol Chem 264 : 9122–9125.
9. WendrichTM, MarahielMA (1997) Cloning and characterization of a relA/spoT homologue from Bacillus subtilis. Mol Microbiol 26 : 65–79.
10. WendrichTM, BlahaG, WilsonDN, MarahielMA, NierhausKH (2002) Dissection of the mechanism for the stringent factor RelA. Mol Cell 10 : 779–788.
11. EnglishBP, HauryliukV, SanamradA, TankovS, DekkerNH, et al. (2011) Single-molecule investigations of the stringent response machinery in living bacterial cells. Proc Natl Acad Sci U S A 108: E365–373.
12. MecholdU, MurphyH, BrownL, CashelM (2002) Intramolecular regulation of the opposing (p)ppGpp catalytic activities of Rel(Seq), the Rel/Spo enzyme from Streptococcus equisimilis. J Bacteriol 184 : 2878–2888.
13. HoggT, MecholdU, MalkeH, CashelM, HilgenfeldR (2004) Conformational antagonism between opposing active sites in a bifunctional RelA/SpoT homolog modulates (p)ppGpp metabolism during the stringent response. Cell 117 : 57–68.
14. BarkerMM, GaalT, JosaitisCA, GourseRL (2001) Mechanism of regulation of transcription initiation by ppGpp. I. Effects of ppGpp on transcription initiation in vivo and in vitro. J Mol Biol 305 : 673–688.
15. KrasnyL, GourseRL (2004) An alternative strategy for bacterial ribosome synthesis: Bacillus subtilis rRNA transcription regulation. EMBO J 23 : 4473–4483.
16. VrentasCE, GaalT, BerkmenMB, RutherfordST, HaugenSP, et al. (2008) Still looking for the magic spot: The crystallographically defined binding site for ppGpp on RNA polymerase is unlikely to be responsible for rRNA transcription regulation (vol 377, pg 551, 2008). J Mol Biol 379 : 1130–1130.
17. Gruber TM, Gross CA (2003) Multiple sigma subunits and the partitioning of bacterial transcription space. In: Ornston LN, editor. Annu Rev Microbiol Volume 57. pp. 441–466.
18. MagnussonLU, FarewellA, NystromT (2005) ppGpp: a global regulator in Escherichia coli. Trends Microbiol 13 : 236–242.
19. WexselblattE, KatzhendlerJ, Saleem-BatchaR, HansenG, HilgenfeldR, et al. (2010) ppGpp analogues inhibit synthetase activity of Rel proteins from Gram negative and Gram positive bacteria. Bioorg Med Chem 18 : 4485–4497.
20. GroppM, StrauszY, GrossM, GlaserG (2001) Regulation of Escherichia coli RelA requires oligomerization of the C-terminal domain. J Bacteriol 183 : 570–579.
21. TosaT, PizerLI (1971) Effect of serine hydroxamate on the growth of Escherichia coli. J Bacteriol 106 : 966–971.
22. ZhouYN, ColemanWG, YangZX, YangY, HodgsonN, et al. (2008) Regulation of Cell Growth during Serum Starvation and Bacterial Survival in Macrophages by the Bifunctional Enzyme SpoT in Helicobacter pylori. J Bacteriol 190 : 8025–8032.
23. ErringtonJ (2003) Regulation of endospore formation in Bacillus subtilis. Nat Rev Microbiol 1 : 117–126.
24. PiggotPJ, HilbertDW (2004) Sporulation of Bacillus subtilis. Curr Opin Microbiol 7 : 579–586.
25. StragierP, LosickR (1996) Molecular genetics of sporulation in Bacillus subtilis. Annu Rev Genet 30 : 297–341.
26. LopezJM, MarksCL, FreeseE (1979) Decrease of guanine-nucleotides initiates sporulation of Bacillus subtilis. Biochim Biophys Acta 587 : 238–252.
27. RosenbergA, SinaiL, SmithY, Ben-YehudaS (2012) Dynamic Expression of the Translational Machinery during Bacillus subtilis Life Cycle at a Single Cell Level. PLoS One 7: e41921.
28. OchiK, KandalaJ, FreeseE (1982) Evidence that Bacillus subtilis sporulation induced by the stringent response is caused by the decrease in GTP or GDP. J Bacteriol 151 : 1062–1065.
29. HsuehYH, SomersEB, WongAC (2008) Characterization of the codY gene and its influence on biofilm formation in Bacillus cereus. Arch Microbiol 189 : 557–568.
30. Ratnayake-LecamwasamM, SerrorP, WongKW, SonensheinAL (2001) Bacillus subtilis CodY represses early-stationary-phase genes by sensing GTP levels. Genes Dev 15 : 1093–1103.
31. KohanskiMA, DwyerDJ, CollinsJJ (2010) How antibiotics kill bacteria: from targets to networks. Nat Rev Microbiol 8 : 423–435.
32. CostertonJW, StewartPS, GreenbergEP (1999) Bacterial biofilms: a common cause of persistent infections. Science 284 : 1318–1322.
33. GoehringNW, BeckwithJ (2005) Diverse paths to midcell: assembly of the bacterial cell division machinery. Curr Biol 15: R514–526.
34. HaydonDJ, StokesNR, UreR, GalbraithG, BennettJM, et al. (2008) An inhibitor of FtsZ with potent and selective anti-staphylococcal activity. Science 321 : 1673–1675.
35. MittenhuberG (2001) Comparative genomics and evolution of genes encoding bacterial (p)ppGpp synthetases/hydrolases (the Rel, RelA and SpoT proteins). J Mol Microbiol Biotech 3 : 585–600.
36. SunD, LeeG, LeeJH, KimH-Y, RheeH-W, et al. (2010) A metazoan ortholog of SpoT hydrolyzes ppGpp and functions in starvation responses. Nat Struct Mol Biol 17 : 1188–1194.
37. Harwood CR, Cutting SM (1990) Modern microbiological methods molecular biological methods for Bacillus. John Wiley & Sons Press. 618 p.
38. VanderijnI, KesslerRE (1980) Growth-characteristics of Group-A streptococci in a new chemically defined medium. Infect Immun 27 : 444–448.
39. NeidhardF, BlochPL, SmithDF (1974) Culture medium for enterobacteria. J Bacteriol 119 : 736–747.
40. Bejerano-SagieM, Oppenheimer-ShaananY, BerlatzkyI, RouvinskiA, MeyerovichM, et al. (2006) A checkpoint protein that scans the chromosome for damage at the start of sporulation in Bacillus subtilis. Cell 125 : 679–690.
Štítky
Hygiena a epidemiologie Infekční lékařství Laboratoř
Článek vyšel v časopisePLOS Pathogens
Nejčtenější tento týden
2012 Číslo 9- Stillova choroba: vzácné a závažné systémové onemocnění
- Choroby jater v ordinaci praktického lékaře – význam jaterních testů
- Diagnostický algoritmus při podezření na syndrom periodické horečky
- Perorální antivirotika jako vysoce efektivní nástroj prevence hospitalizací kvůli COVID-19 − otázky a odpovědi pro praxi
- Diagnostika virových hepatitid v kostce – zorientujte se (nejen) v sérologii
-
Všechny články tohoto čísla
- A Population Genomics Perspective on the Emergence and Adaptation of New Plant Pathogens in Agro-Ecosystems
- A Novel Rhabdovirus Associated with Acute Hemorrhagic Fever in Central Africa
- Ifitm3 Limits the Severity of Acute Influenza in Mice
- Copper at the Front Line of the Host-Pathogen Battle
- The Human Cytomegalovirus Assembly Compartment: A Masterpiece of Viral Manipulation of Cellular Processes That Facilitates Assembly and Egress
- Insights from Genomics into Bacterial Pathogen Populations
- Slc15a4, a Gene Required for pDC Sensing of TLR Ligands, Is Required to Control Persistent Viral Infection
- Relacin, a Novel Antibacterial Agent Targeting the Stringent Response
- Global Assessment of Genomic Regions Required for Growth in
- The Battle over mTOR: An Emerging Theatre in Host–Pathogen Immunity
- Very Long O-antigen Chains Enhance Fitness during -induced Colitis by Increasing Bile Resistance
- and Beyond—Inositol Utilization and Its Implications for the Emergence of Fungal Virulence
- Antifungal Drug Discovery: Something Old and Something New
- Deep Sequencing of Antiviral T-Cell Responses to HCMV and EBV in Humans Reveals a Stable Repertoire That Is Maintained for Many Years
- PLOS Pathogens
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Insights from Genomics into Bacterial Pathogen Populations
- Relacin, a Novel Antibacterial Agent Targeting the Stringent Response
- Ifitm3 Limits the Severity of Acute Influenza in Mice
- Antifungal Drug Discovery: Something Old and Something New
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



