-
Články
- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Comparison of independent screens on differentially vulnerable motor neurons reveals alpha-synuclein as a common modifier in motor neuron diseases
The term “motor neuron disease” refers to a group of disorders, causing progressive paralysis of affected patients due to the degeneration of motor neurons cells which control voluntary movements. Importantly, not all motor neurons appear to be affected in the same way, with those that control the face being affected less that those that control the abdomen. The reason why some motor neurons are more vulnerable is unknown; however, understanding this may provide new targets for therapeutics to slow motor neuron degeneration either as stand-alone therapeutics or in combination with SMN-inducing compounds. In this study, we analysed gene expression in different groups of motor neurons and compared this to previously published expression data to identify commonalities. One of the common transcripts was alpha-synuclein (SNCA), which was consistently expressed at lower levels in vulnerable motor neurons. Importantly, when SNCA levels were increased in a mouse model of motor neuron disease, the disease phenotype was significantly reduced, including an extension in survival and reduction in motor neuron pathology. Collectively, these results demonstrate that this approach can identify disease modifiers that can reduce disease severity in models of motor neuron disease and potentially identify new therapeutic targets.
Published in the journal: . PLoS Genet 13(3): e32767. doi:10.1371/journal.pgen.1006680
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1006680Summary
The term “motor neuron disease” refers to a group of disorders, causing progressive paralysis of affected patients due to the degeneration of motor neurons cells which control voluntary movements. Importantly, not all motor neurons appear to be affected in the same way, with those that control the face being affected less that those that control the abdomen. The reason why some motor neurons are more vulnerable is unknown; however, understanding this may provide new targets for therapeutics to slow motor neuron degeneration either as stand-alone therapeutics or in combination with SMN-inducing compounds. In this study, we analysed gene expression in different groups of motor neurons and compared this to previously published expression data to identify commonalities. One of the common transcripts was alpha-synuclein (SNCA), which was consistently expressed at lower levels in vulnerable motor neurons. Importantly, when SNCA levels were increased in a mouse model of motor neuron disease, the disease phenotype was significantly reduced, including an extension in survival and reduction in motor neuron pathology. Collectively, these results demonstrate that this approach can identify disease modifiers that can reduce disease severity in models of motor neuron disease and potentially identify new therapeutic targets.
Introduction
The term “motor neuron disease” refers to a group of disorders in which motor neurons are a prominent pathological target. Such disorders are generally severely disabling and frequently fatal within months to years of diagnosis. Effective treatments for many motor neuron diseases are currently lacking. Motor neuron diseases can be categorized into various types. For example, Amytrophic Lateral Sclerosis (ALS) affects upper and lower motor neurons and disease onset is typically in adulthood between the ages of 30 and 50. While approxitamely 10% of ALS cases are familial, the majority of new cases are sporadic[1]. Spinal Muscular Atrophy (SMA) refers to a type of motor neuron disease that is caused by homozygous loss of the SMN1 gene[2, 3], resulting in the loss of lower motor neurons. Due to the presence of an additional partially functional copy of SMN, termed SMN2, which can exist in a range of copy numbers, SMA severity can vary widely[4]. However, the most common form of this disease has an onset of less than 6 months of age and a life expectancy of under 2 years without significant respiratory support. Spinal and Bulbar Muscular Atrophy (SBMA) is an X-linked motor neuron disease caused by an expansion of a trinucleotide repeat in the androgen receptor gene[5]. SBMA also appears to result in the degeneration of lower motor neurons with onset between 30 and 50 years of age. SBMA disease progression is typically slower than other types of motor neuron diseases and patients typically have normal life expectancies.
Distinct motor neuron diseases with their own specific cause, onset and prognosis are united by the common vulnerability and loss of motor neurons. Importantly, however, in each disease motor neurons are not uniformly vulnerable. In both patients and animal models, some motor neuron populations are lost very early in the disease, whilst others remain remarkably intact, even at late stages of disease. For example, in SMA the pattern of motor neuron pathology is highly predictable. This has been extensively characterised in mouse models of the disease[6–9]. The pattern of selective vulnerability in patients is less-well documented but it has been described as highly stereotyped, even within muscles groups[10]. One of the last groups of motor neurons to be affected are those supplying the muscles of the face, in particular those supplying the extra-ocular muscles[11]. The location of disease onset in patients with ALS is more variable; however, there appears to be a sparing of the motor neurons which supply the extra-ocular muscles[12]. This finding has been corroborated in mouse models of ALS in which there appears to be a marked differential vulnerability of specific cranial nerve nuclei[13, 14]. Therefore, despite subtleties in the different patterns of selective vulnerability between different motor neuron diseases, there are also important pathologenic similarities. This point was highlight in a study by Comley et al., demonstrating a shared pattern of selective vulnerability in mouse models of ALS and SMA [6]. Recent work has also shown significant overlap in the molecular mechanisms which govern distinct subtypes of motor neuron diseases [15]. Identifying the common mechanisms giving rise to selective protection or vulnerability of motor neurons will provide important biological insight into motor neuron development, but can also lead to the identification of novel therapeutic targets for motor neuron diseases.
This observed selective vulnerability of motor neurons creates a valuable opportunity to investigate the mechanism of motor neuron vulnerability in motor neuron diseases. Indeed, we have recently utilised this observation to investigate the transcriptional differences occurring pre-symptomatically in a mouse model of SMA[16]. In this study, motor neuron vulnerability was defined by the level of pathology observed at the neuromuscular junction (NMJ). Pathology at the NMJ was defined as denervation, pre-synaptic swelling and decrease in pre - and post-synaptic complexity. We identified an increase in vulnerability in the NMJs from the abdominal muscles compared to those in the cranial muscles. The motor neuron cell bodies which corresponded to these differentially vulnerable NMJs were isolated and RNAseq was performed to generate transcriptional profiles for abdominal and cranial motor neurons from SMA and WT mice. The purpose of this study was to identify transcriptional changes which correlate with a decrease in Smn levels, and those which correlate with an increase in motor neuron pathology. However, an additional benefit of this screen was to profile the transcriptomes of vulnerable and resistant motor neurons from wild-type mice. We suggest that genes which are differentially regulated between these two populations of healthy motor neurons have the potential to be important modifiers of disease. Indeed, identifying the modifiers in selectively resistant motor neuron pools which are responsible for their decreased their vulnerability could provide key insight for the development of strategies to protect more vulnerable motor neurons.
This idea has previously been exploited by a number of independent groups who have observed predictable patterns of selective vulnerability in different motor neuron diseases, and aimed to identify transcriptional changes between vulnerable and resistant motor neuron pools[17–19]. Each of these studies identified motor neurons which were predictably vulnerable or resistant in SMA, SBMA or ALS patients or animal models, and used laser capture microdissection to isolate these equivalent motor neurons from neurologically healthy humans, wild-type rats or mice. These 3 screens have identified a large number of transcriptional changes between differentially vulnerable motor neurons in healthy individuals.
The transcriptional profiles from these screens, therefore, represent a valuable set of data, detailing expression changes occurring between vulnerable and resistant motor neurons from a range of species and ages, all from healthy individuals. Such changes cannot, therefore, be due to any pathology, and are rather reflective of instrinsic differences between motor neuron pool which may alter their vulnerability to pathological situations. In our search for transcriptional modifiers of motor neurons, we suggest that common features between these transcriptional changes have a high chance of being modifiers of motor neuron pathology. Features which are common across transcriptional screens are also likely to be modifiers across multiple motor neuron diseases, rather than just one MND subtype. Gaining knowledge of these modifiers will give insight into shared mechanism of disease, and therefore potential shared therapeutic options.
In this study, we compared the transcriptome of vulnerable (innervating abdominal muscles) and resistant (innervating cranial muscles) motor neurons from P10 wild-type mice which are differentially vulnerable in mouse models of SMA. In order to refine this data set, we reanalyzed the raw data from 3 published independent microarray screens on healthy but differentially vulnerable neurons and compared it to our RNAseq data. We identified 6 transcripts that share common directional changes in all 4 screens: CELF5, Col5a2, PGEMN1, SNCA, STMN1 and HOXa5. Functional clustering of the transcripts that were changed in 2 or more of the 4 screens revealed an enrichment for synaptic and axonal transcripts. Introduction of the differentially expressed genes into a Drosophila model of ALS8 rescued hallmarks of the neurodegenerative phenotype, demonstrating that the differentially expressed genes can function in disease-relevant pathways. Due to the lack of Drosophila homologue for SNCA, and because of evidence from the literature implying SNCA may have neuroprotective qualities, we investigated whether increasing levels of SNCA could amleriorate the phenotype in a mouse model of motor neuron disease. ScAAV9-SNCA was delivered to a mouse model of SMA, resulting in a significant decrease in disease severity, including an extension in survival and increased weight gain. Importantly, NMJ pathology in scAAV9-SNCA treated mice was significantly improved, providing evidential support for the notion that differentially expressed genes from susceptible motor neurons can serve as disease modifiers.
Results
Differential levels of neuromuscular junction pathology correlate with differential expression levels of a large number of transcripts
Differentially vulnerable motor neurons have been reported in patients and in mouse models of SMA [6–11]. In the Smn2B/- SMA mouse model, selective vulnerability can be observed at the neuromuscular junction (NMJ). Analysis of NMJs in the abdominal muscles revealed a high level of denervation, alongside other markers of pathology such as neurofilament accumulation, shrinkage of endplates and a decrease in endplate complexity (Fig 1)[8, 16]. This represents a “vulnerable” population. Analysis of a group of cranial muscles, which are innervated by motor neurons residing in the facial nucleus of the brainstem, show no evidence of denervation and minimal evidence of other markers of NMJ pathology and therefore represent a “resistant” population (Fig 1) [8, 16].
Fig. 1. Intermuscular variability in the levels of neuromuscular junction pathology in the Smn2B/- mouse model of SMA. 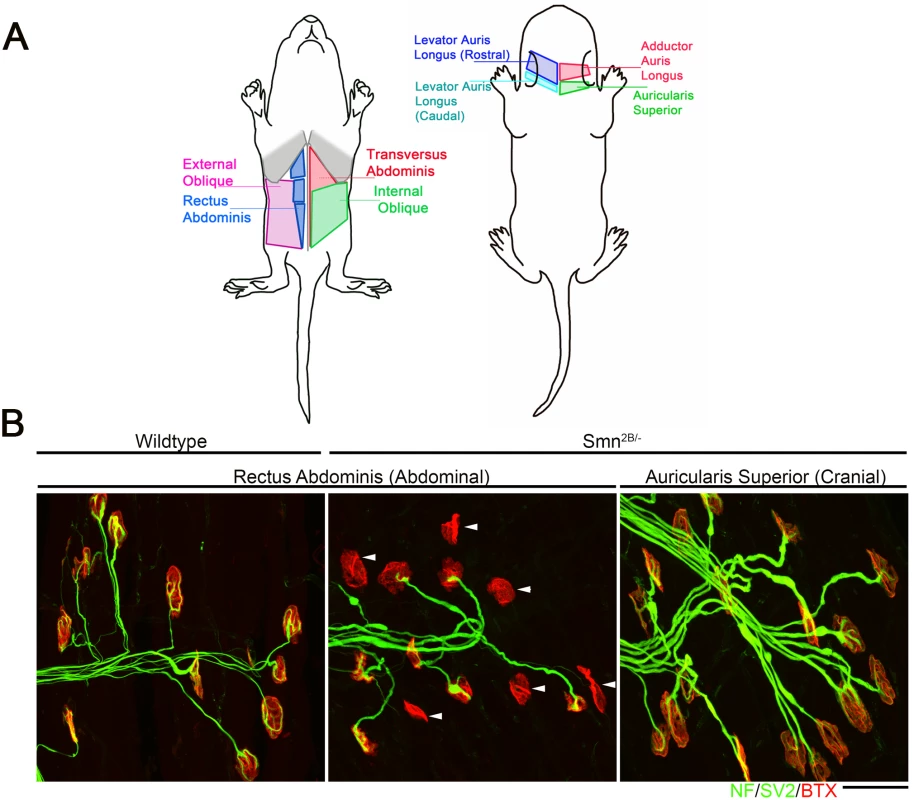
A) Schematic diagram showing location of muscles which were innervated by either vulnerable or resistant motor neurons in a mouse model of SMA. Vulnerable muscles, as defined by increased neuromuscular junction (NMJ) pathology, include: external and internal oblique; transversus abdominis; and rectus abdominis. Resistant muscles, as defined by a low level of NMJ pathology, include: levator auris longus; auricularis superior; and adductor auris longus. B) Confocal micrographs showing NMJs with the pre-synaptic terminal labeled with antibodies against neurofilament (NF; Green) and synaptic vesicle protein 2 (green) and the muscle endplate labeled with alpha-bungarotoxin (red) from rectus abdominis and auricularis superior muscles. Note that in the wild-type abdominal and Smn2B/- cranial muscle, all endplates appear fully innervated where each endplate is covered by the pre-synaptic terminal labeled with SV2 and NF. In the rectus abdominis from the Smn2B/-, mouse there is evidence of significant NMJ pathology, as evidenced by endplates lacking a pre-synaptic terminal (white arrow heads). Scale bar = 40μm. In previous work, we used intramuscular injection of dextran molecules to trace the motor neurons’ cell bodies which correspond to these differentially vulnerable groups of NMJs[16]. Cell bodies were isolated by laser capture microdissection and RNAseq was performed on extracted RNA. Parallel experiments were performed on wild-type and Smn2B/- SMA mice. This study[16] focused on the differences between SMA and WT mice at a pre-symptomatic time point (P10). However, this work also produced a transcriptional profile of thoracic (vulnerable) and cranial (resistant) motor neurons from wild-type mice. In the current study, we address the hypothesis that the transcriptional changes occurring between vulnerable and resistant motor neurons in wildtype mice reflect intrinsic differences which contribute to the differential vulnerability observed in the mouse model of disease. Comparison of the transcriptional data between resistant and vulnerable motor neurons from wild-type mice resulted in 910 significantly altered transcripts with a fold change of >1.5 fold, with 218 up-regulated and 692 down regulated in vulnerable versus resistant motor neurons (Table 1, S1 Table). Functional clustering of these transcriptional changes using DAVID bioinformatics resources version 6.8 revealed an enrichment for extracellular matrix and glycoproteins (Table 2).
Tab. 1. Top 20 transcriptional changes identified between vulnerable (abdominal) and resistant (cranial) motor neurons from P10 wild-type mice. 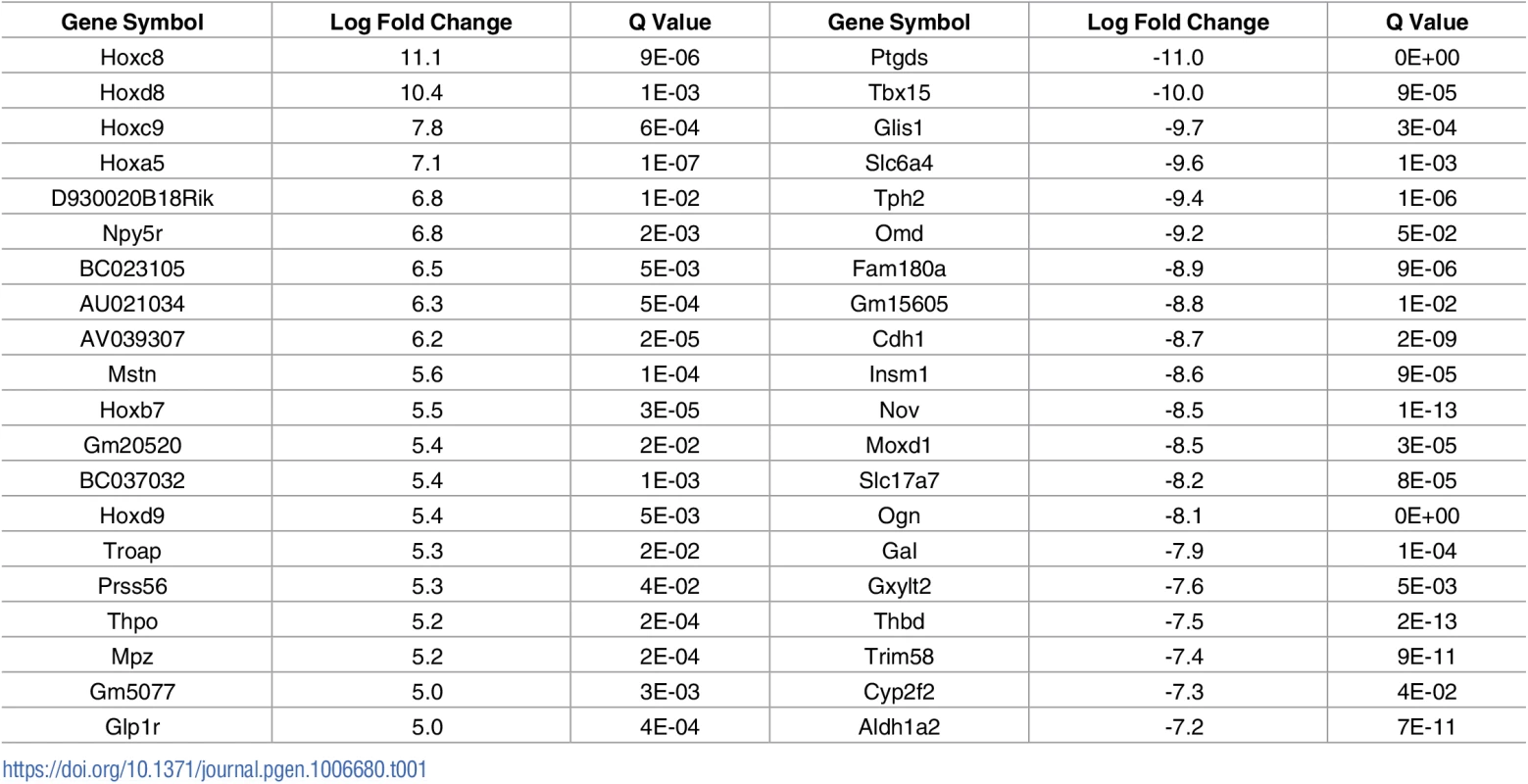
Tab. 2. Top 5 functional clustering of transcriptional changes identified between vulnerable and resistant motor neurons. 
Comparison of RNAseq data with published screens refines list of potential modifiers
This screen has identified a large number of transcriptional changes. However, it is difficult to differentiate those which merely correlate with differential vulnerability from those which actually contribute to differential vulnerability. In order to identify those changes which had the highest probability of being modifiers and perhaps play also a causative role in motor neuron pathology, we compared the results of our screen with the results of other screens on different populations of differentially vulnerable motor neurons which have been previously published (Table 3).
Tab. 3. Summary of independent screens on differentially vulnerable motor neurons. 
Raw microarray data were re-analysed and a list of differentially expressed genes for each screen was generated. As screens were performed in different species, genetic homologues were identified, and all genes were listed corresponding to the mouse official gene symbol. Comparison of the results of the 3 microarray studies with our own RNAseq data revealed a large number of common changes, with the majority occurring the same direction of change (Fig 2). Transcriptional changes from these 4 transcriptional screens were sorted based on direction of change. This resulted in the identification of 595 transcripts which were altered in a common direction in 2 screens (S2 Table), 62 transcripts which were common in 3 screens (S3 Table) and 6 transcripts which were common in all 4 screens (Table 4). Functional clustering of the transcriptional changes occurring in 2 or more screens revealed an enrichment for axonal and synaptic proteins (Table 5).
Tab. 4. Transcriptional changes which are common across all 4 screens on differentially vulnerable motor neurons. 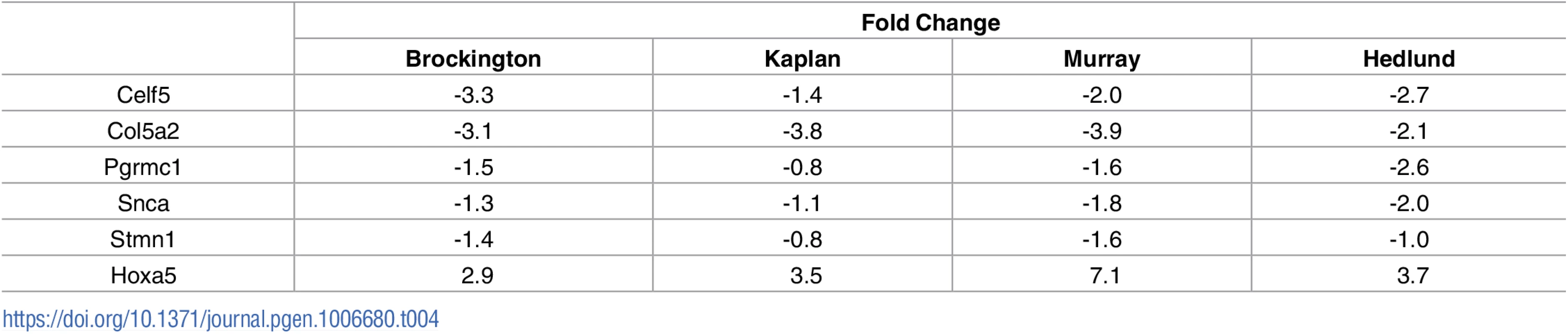
Tab. 5. Functional clustering of transcriptional changes which are common in 2 or more of the screens on differentially vulnerable motor neurons. 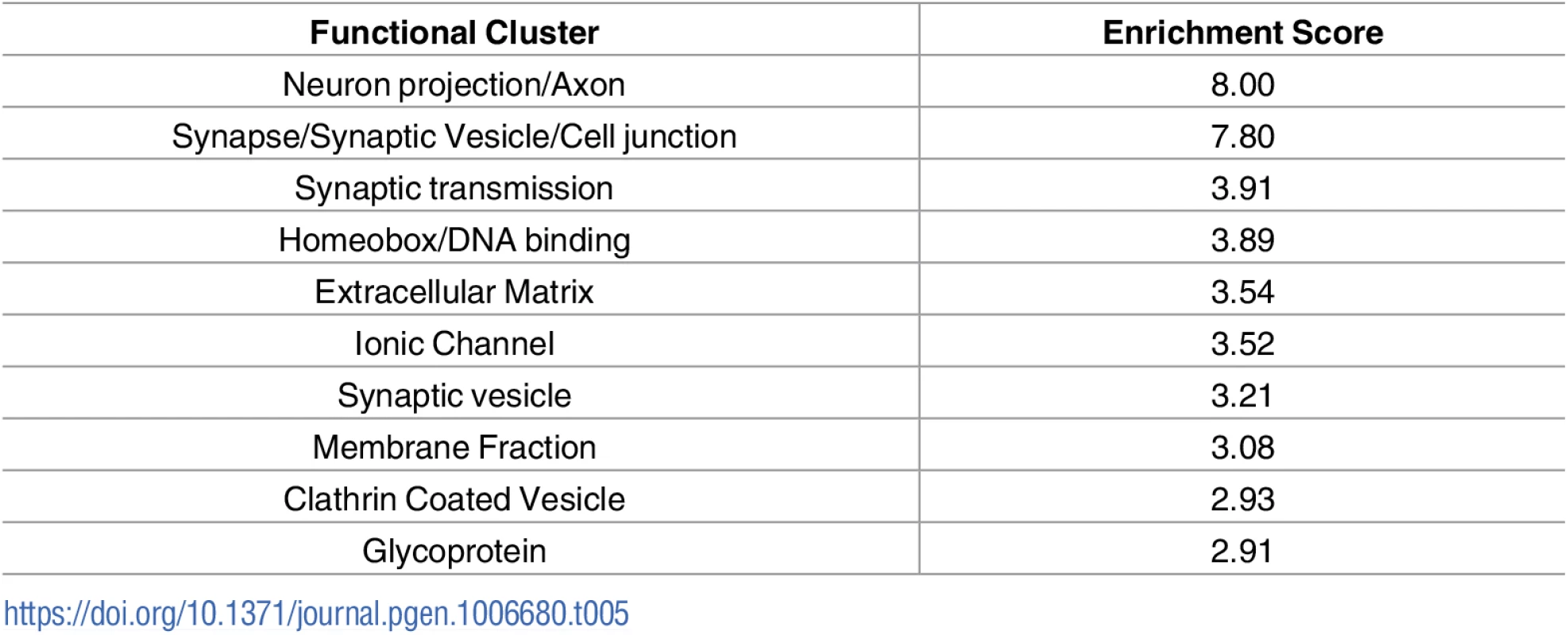
Fig. 2. Comparison of 4 independent screens on differentially vulnerable motor neurons reveals a large number of common transcriptional changes. 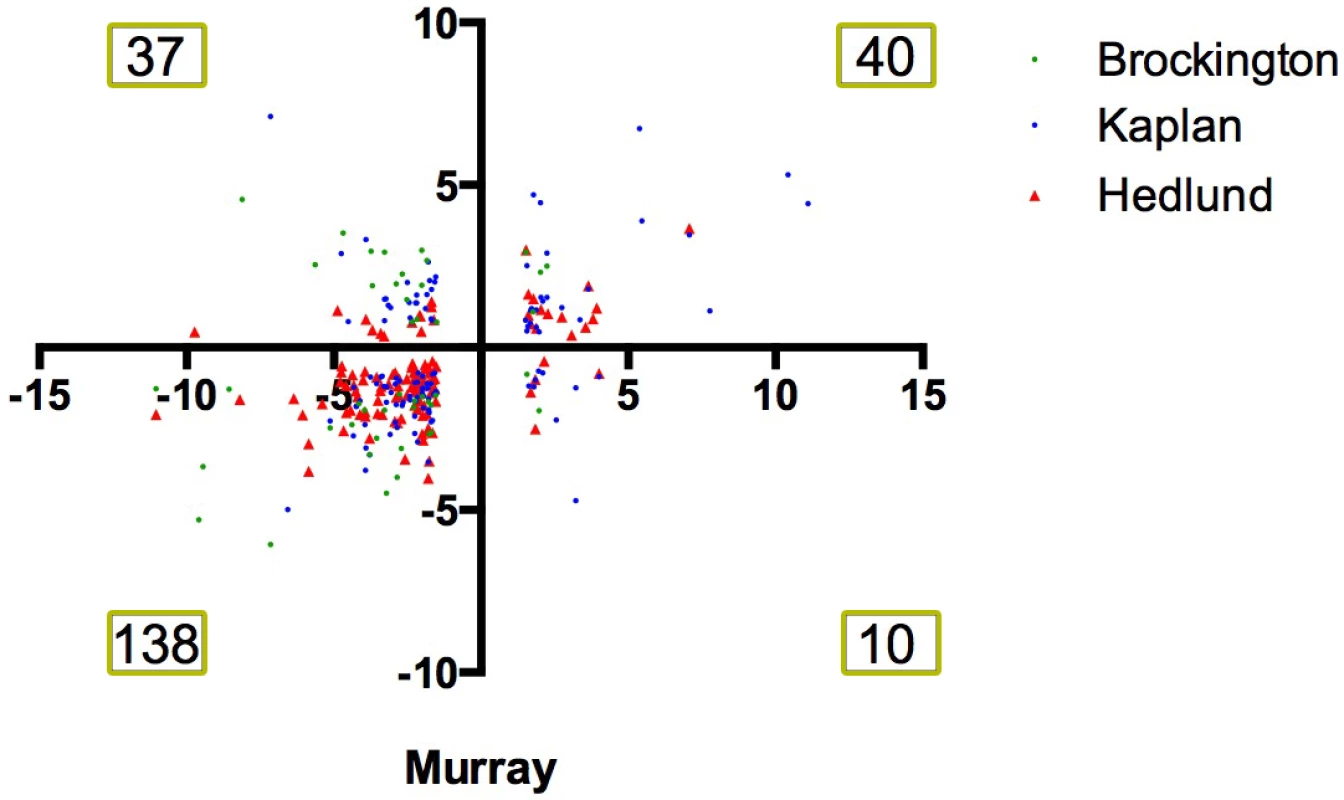
Scatter plot showing the fold change of transcripts which were differentially expressed in differentially vulnerable motor neurons in the RNAseq performed by Murray et al., 2015 [16] and in the microarray study on differentially vulnerable motor neuron performed by Brockington et al., 2013 [17] (green), Kaplan et al., 2014 [19] (blue) and Hedlund et al., 2010 [18](red). Numbers denote number of number of common transcriptional screens within each quadrant of the plot. Note that the majority of changes occur with a common directional change i.e. fall within the bottom left or top right quadrant of the graph. Differentially expressed transcripts modify the degenerative eye phenotype in a Drosophila model of ALS8
To determine whether the identified transcripts can modify neurodegenerative pathways, a Drosophila model of ALS was used to functionally validate the candidate genes by driving expression of the candidate genes or by transiently knocking-down expression. The purpose of this screen was to determine whether a significant number of transcripts identified here were capable of modifying the phenotype in an independent model of neurodegeneration induced by an ALS causing mutation. Transcripts were selected that either changed in 3 or more of the transcriptional screens (Table 4 and S2 Table) or were featured in one of the two top functional clusters (axonal or synaptic transcripts) (Table 5). In this model, a Drosophila line expresses the P58S mutation in the VAMP associated protein B gene (VAPB). This is equivalent to the P56S mutation in human VAMP which is a causative mutation of human motor neuron disease, including ALS8 [20]. This mutation has been shown to affect a range of cellular processes which have been implicated in MND, including the unfolded protein response, endocytosis, vesicular trafficking, mitochondrial defects and autophagy [21–26]. The Drosophila homologue of VAPB is termed VAP-33-1, or DVAP. In previous work, DVAP-P38S expression was driven in the eye of Drosophila using the UAS/GAL4 system[27], with an eyeless-GAL4 (ey-GAL4) driver. This resulted in a roughness of the adult Drosophila eye and a significant reduction its size, which could be attributed to a decrease cell survival[28]. This model and the eye phenotype readout has previously been used in a large-scale enhancer and suppressor screen for genetic modifiers of ALS8 pathology [23].
As outlined above, for this in vivo validation, we chose to include all transcripts which were changed in 3 or more screens, as well as those pertaining to the top two functional clusters, of axonal or synaptic transcripts. This resulted in a list of 160 transcripts. Drosophila homologues were predicted for each transcript using the DRSC Integrative Ortholog Prediction tool (http://www.flyrnai.org/cgi-bin/DRSC_orthologs.pl). Results were filtered to return only the best match where more than one homologue was found and restricted to those with a DIOPT score of greater than 2. Where more than one homologue was found with an equivalent weighted score, all potential homologues were included. From the 160 transcripts listed, we identified an homologue for 116, which includes 39 transcripts up-regulated and 77 down regulated transripts in vulnerable motor neurons.
For transcripts which were up-regulated in vulnerable motor neurons, we identified publically available lines carrying RNAi constructs to knock down transcripts of interest. For the majority of lines (38/46), we selected those with zero off target effect predicted. For a small number of lines this was not possible. In this case, lines with the minimum number of off target effects were used. For transcripts which were down-regulated in vulnerable motor neurons, we identified publically available lines carrying a P-element insertion which, based on the position and orientation, would be predicted to result in an over expression of the transcripts of interest.
From this, 66 publically available lines were available to decrease or increase the expression of these transcripts respectively (Table 6). Lines designed to decrease or increase expression our candidate modifiers were crossed with DVAP-P58S flies. Those lines which increase the size of the eye compared to the DVAP-58S flies were categorised as suppressors, and those which decreased the size of the eye were categorised as enhancers of the neurodegenerative phenotype. Overall, 11 transcripts modified the neurodegenerative eye phenotype observed in DVAP-P58S flies, with 7 suppressors and 4 enhancers (Fig 3). Overall, 17% of transcripts displayed an ability of modify the phenotype. Whilst it is difficult to draw direct parallels to other screens, previous screen using Drosophila models of motor neuron disease or disease causing mutations, and performing unbiased screen to identifiy modiers have resulted in 0.4 to 4% of genes being identified as enhancers or suppressors. Our increased hit rate compared to these unbiased or enriched suggests that this bioinformatics approach has led to a list which is enriched for disease modifying genes. This suggest that this approach is identifying relevant transcripts which are capable of modifying neurodegenerative pathways associated with motor neuron disease.
Tab. 6. Mouse genes and their Drosophila homologue which were tested in the DVAPP-P58S Drosophila screen. 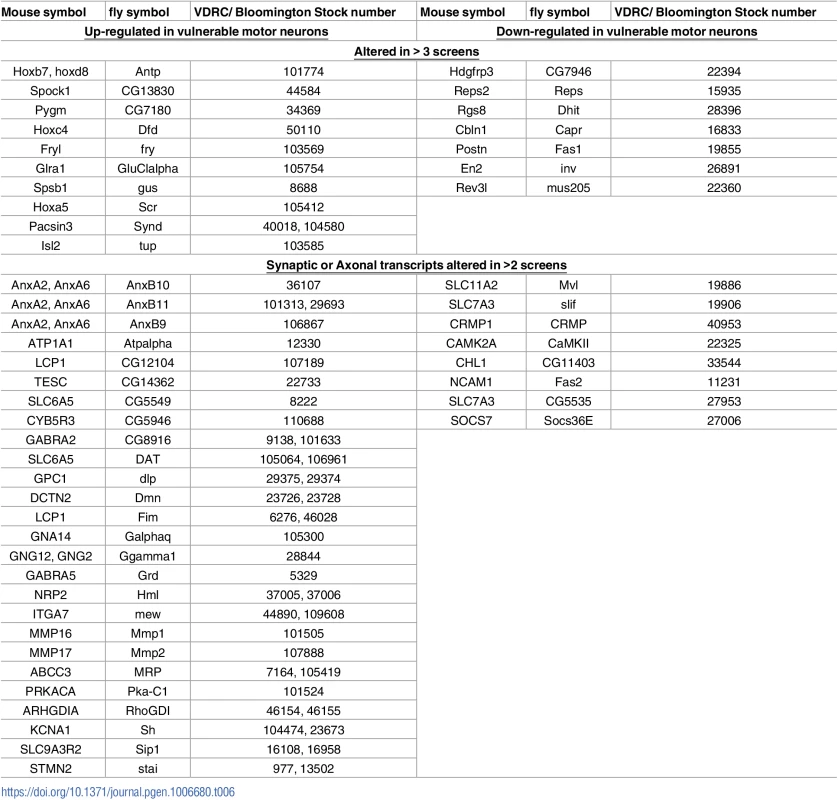
Fig. 3. In vivo validation of transcripts in a Drosophila model of ALS8. 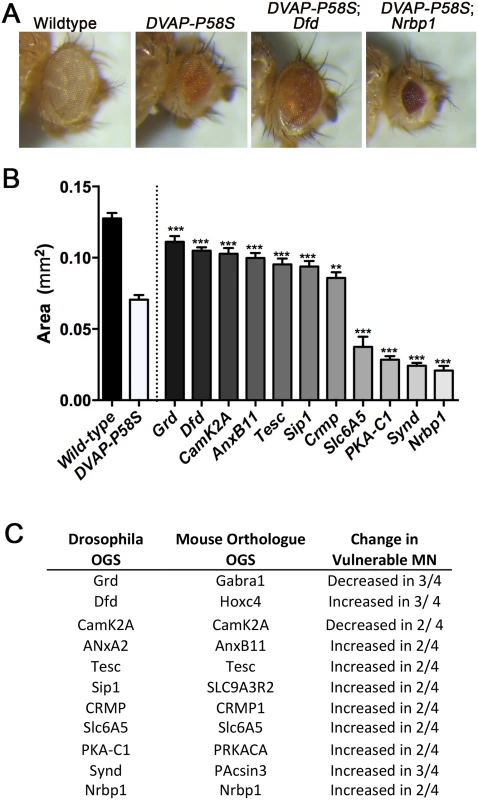
A) Images show eyes from wild-type, DVAP-P58S (carrying ALS8 patient mutation), DVAP-P58S Dfd (AL8 patient mutation with decreased expression of Dfd, the Drosophila homologue of Hoxc4) and DVAP-P58S Dfd (AL8 patient mutation with decreased expression of Nrbp1) Drosophila. Note the decrease in eye size observed in DVAP-P58S flies. This phenotype was supressed by decreased expression of dfd, and enhanced by decreased expression of Nrbp1. B) Bar chart (Mean ± SEM) showing the area (mm2) of the eye in Drosophila lines which over or under express specified transcripts in DVAP-P58S flies. *** P<0.001, **P<0.01 by ANOVA with Holm-Sidak’s post hoc test. N = approx. 12 flies per group with each value reflecting average of 2 eyes per fly. C) Table details the transcripts which were identified as modifiers of the eye phenotype in DVAP-P58S flies, denoting the Drosphila official gene symbol, the official gene symbol of the mouse homologue and the directional change observed in vulnerable motor neurons in the independent transcriptional screens [16–19]. For those transcripts which were decreased in vulnerable motor neurons, their expression was increased in DVAP-P58S flies, and for those transcripts which were increased in vulnerable motor neurons, their expression was decreased in DVAP-P58S flies. Over expression of alpha-synuclein modifies phenotype in a mouse model of SMA
Alpha-synuclein (SNCA) was consistently decreased in vulnerable motor neurons across all four screens. This was of particular interest as a decrease in SNCA levels have been reported in SMA patient spinal cord, patient fibroblasts and NSC-34 motor neuron-like cells [29]. There are also a number of studies indicating that, in certain scenarios, over expression of SNCA can be neuroprotective[30–33]. As SNCA is a strong candidate to modify neuronal pathology, we sought to further investigate the effects of SNCA over expression in models of motor neuron disease. Unfortunately, there is no homologue for SNCA, which makes the effect of over expression of SNCA in DVAP-P58S flies difficult to interpret. For this reason we turned to a mammalian model, and sought to determine the impact of SNCA transient expression in the Smn2B/- mouse model of SMA. To provide widespread expression of the SNCA transgene, an scAAV9-SNCA vector was developed. AAV9 has a broad tropism for many tissues within the periphery and the central nervous system, including astrocytes and neuronal lineages [34]. At postnatal day 1, a single injection of 1e11 or 3e11 viral particles of scAAV9-SNCA was delivered via an intracerebroventricular injection into the Smn2B/- mouse model of SMA. The lower dose was selected based upon the amount of vector that provides a robust phenotypic rescue using scAAV9-SMN[35]. Injection of 1e11 viral particles scAAV9-SNCA has no discernable effect on life span or weight gain, however, the higher dose of 3e11 viral particles resulted in an ~88% (23 day) increase in median life span and a significant increase in average body weight from approximately P20 onwards (Fig 4A and 4B). Since the initial transcriptomic screen was predicated upon the differential pathology observed at the NMJ, we next examined whether scAAV9-SNCA treatment improved the NMJ phenotype in SMA mice. Importantly, analysis of NMJs from P18 scAAV9-SNCA injected mice revealed a significant increase in the percentage of fully occupied endplates compared to untreated controls, indicative of a decrease in denervation and motor neuron pathology (Fig 4C and 4D).
Fig. 4. Overexpression of SNCA ameliorate phenotype and neuromuscular junction pathology in the Smn2B/- mouse model of SMA. 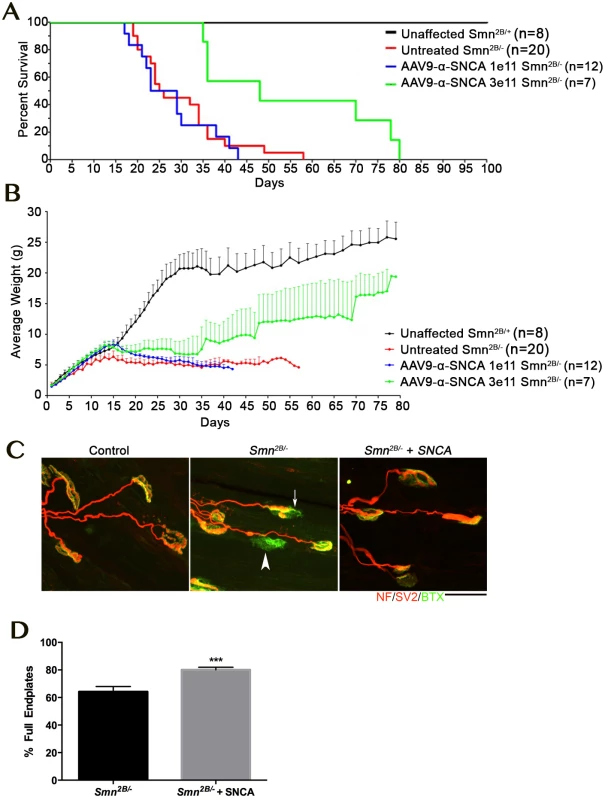
A, B) Kaplan Maier plot (A) and weight curve (B) showing profile of control (Smn2B/+; black) and untreated Smn2B/- (red) compared to mice treated with a low dose (1e11; blue) or high dose (3e11; green) of AAV9 expressing SNCA. Note that while the low dose only increases weight gain (Student’s t-test p < 0.0001, the high dose of AAV9-SNCA significantly increased weight (Student’s t-test p<0.0001) and lifespan in the Smn2B/- mouse model (Mantel-Cox Survival Curve Comparison Test p = 00027). C) Confocal micrographs showing NMJs with the pre-synaptic terminal labeled with antibodies against neurofilament (NF; red) and synaptic vesicle protein 2 (red) and the muscle endplate labeled with alpha-bungarotoxin (green) from the transversus abdominis muscle from P18 mice. Note the presence of fully (arrowhead) and partially (arrow) denervated endplates in the untreated Smn2B/- mouse which were less commonly observed in the Smn2B/- mouse treated with high dose AAV9-SNCA. Scale bar = 20μm. D) Bar chart ± SEM showing the increase in the percentage of fully occupied endplates in untreated Smn2B/- mice (black bars) compared to Smn2B/- treated with high dose AAV9-SNCA. *** P <0.001 by Mann-Whitney U test where n = 4/8 mice/muscles per group. Together this work demonstrates that in a mouse model of SMA, over expression of SNCA can impact upon the neurodegenerative pathways, and has the capacity to extend lifespan and ameliorate the phenotype. This result is a clear proof of principle that this approach can identify relevant phenotypic modifiers that have the capacity to impact disease development in an important model of neurodegeneration.
Discussion
In this study we have compared multiple screens performed across 3 species to generate a list of common differentially expressed transcripts. We have used a Drosophila model to demonstrate that a high proportion of these can modify the neurodegenerative phenotype caused by a human ALS8 mutation. Importantly we have also shown that over expression of SNCA can extend life span and decrease NMJ pathology in a mouse models of SMA. This demonstrates that this approach can identify relevant phenotypic modifiers. It also suggests that SNCA is an exciting candidate which deserves further attention, and that the remaining candidate list is likely to have some exciting transcripts within it.
The use of transcriptional screen methodologies to dissect the mechanism of disease is central in the study of most pathologies. Screening techniques have been instrumental in identifying affected pathways and perturbed cellular functions. However, when the intended resolution of a transcriptional screen is to identify individual differentially expressed transcripts, it can be difficult to separate those which are changed due to the process of disease, and those which are changed to compensate for a loss of function or due to the altered activity of the organism, organ or cell. Even at pre-symptomatic stages of disease, there are likely to be transcriptional changes occurring as a secondary response to the original pathology. A significant advantage of the current work is to compare the transcriptomes of distinct differentially vulnerable motor neuron populations from normal healthy individuals. This approach allows us to identify those changes which are present in a normal situation, but could potentially impact upon the disease process. Clearly dissecting apart those which are changed by coincidence from those which actually have the potential to modify disease is challenging. This is before the added challenge of identifying those which are actually relevant in humans. However, by comparing across 4 screens, across 3 or more different MNDs, and across 3 species, including humans, we feel we have created an experimental design which has a high chance of identifying clinically relevant modifiers of motor neuron disease.
Common regulators of vulnerability in motor neuron disease
A number of studies have implicated SMN1 and SMN2 copy number in the incidence of sporadic ALS[36]. The observation that ALS causing mutations in FUS and TDP-43 can alter the localisation and associations of Smn has also led to some suggestion of shared mechanism between diseases[36]. Furthermore, although there are certainly some important distinctions in the patterns of selective vulnerability between distinct motor neuron diseases, there are some common themes, in that motor neurons originating in brainstem motor nuclei appear consistently comparatively spared, particularly those supplying the extra-ocular muscles [6, 11–14]. There is therefore good reason to suppose that the mechanisms mediated selective vulnerability in motor neuron disease can, to at least some extent, be shared. In an extension of this, resultant neuroprotective therapies should have the potential to benefit a range of conditions.
The work presented in this study has generated some exciting candidates to be cross-disease modifiers. Aside from SNCA (discussed below) there are a number of transcripts which warrant further investigation. CUGBP, elav—like family member 5 (CELF5) belongs to a family of developmentally expressed RNA binding proteins, with a proposed role in pre-mRNA splicing [37]. As this is a function shared by many MND causing mutations, it is easy to generate hypothesis about how differential CELF5 levels may modify pathology. Progesterone receptor membrane component 1 (Pgrmc1) is best characterized due to its role in cancer. However, its oncogenic actions are due in part to its ability to promote cell survival and inhibit apoptosis. PGRMC1 is thought to mediate the protective effects of progesterone on rats modeling Alzheimer’s disease via inhibition of the mitochondrial apoptotic mechanism[38]. It has also shown to be an important mediator in the neuro-protective effect of a synthetic progesterone in the degenerative eye disease retinitis pigmentosa[39]. The observation that Pgrmc1 is decreased in all 4 screens in vulnerable motor neurons could be associated with the increase in cell death of this subpopulation of cells. Stathmin (Stmn1) is a well characterized microtubule binding protein and as such, has important roles in cellular functions dependent upon microtubules, including in mitosis, motility, process formation and intracellular transport[40]. Stathmin has been shown to be dysregulated in a mouse model of ALS, and knockout of stathmin produces a mouse displaying peripheral and central axon degeneration [41, 42]. Interestingly, decreased stathmin levels have previously been shown to increase body weight, motor performance and NMJ maturation is a mouse model of SMA[43]. Therefore amongst our top differentially expressed transcripts we have some very exciting candidates to be modifiers of motor neuron diseases.
How does SNCA function as a disease modifier
SNCA performs a number of cellular roles, but has been implicated as a causative factor of Parkinsons disease [44]. Mutations in SNCA are strongly associated with aggregate formation, leading to the degeneration of nigrostriatal neurons, causing the well characterised and common disorder of the basal ganglia. Although the pathogenic mechanisms of SNCA aggregates in Parkinson’s is relatively well characterised, the normal function of SNCA is less well defined. It is known to be a small protein of about 140 amino acids localising to the pre-synaptic terminal [45]. It is thought to have an important role in neurotransmitter release. Indeed, over expression of SNCA in primary hippocampal neuron cultures and hippocampal slice culture has been shown to inhibit synaptic vesicle exocytosis, potentially by slowing the recycling of synaptic vesicles and decreasing the number of vesicles in the readily releasable pool[46]. Furthermore, alpha-synuclein knockout mice display an increased rate of vesicle filling under repetitive stimulation[47]. How then might this role of SNCA be a protective modifier in motor neuron diseases? The idea of SNCA possessing neuroprotective qualities is not novel. Whilst some have suggested that over expression of wild-type SNCA could increase vulnerability to certain insults such as oxidopamine (a toxin specific to dopiminergic neurons)[33], other work has shown that increased SNCA can decrease toxicity caused by the pesticide paraquat [32], the apoptotic ages staurosporin and etoposide [30] and oxidative stress induced by hydrogen peroxide [31]. The resistance to oxidative stress observed was proposed to be due to a down regulation of the cell stress induced c-Jun N-terminal kinase pathway which promotes apoptosis[31]. The mechanism by which SNCA could be a protective modifier in the context of this study is currently unclear. However given the multiple scenarios in which SNCA has been shown to be neuroprotective, further work is justified to explore this mechanism.
Why are transcripts differentially expressed in motor neurons?
As the screens detailed in this report have been performed in exclusively healthy motor neurons, which happen to be differentially vulnerable in disease, the transcriptional changes which we are reporting likely occur for reasons unrelated to motor neuron pathology. It is therefore important to consider why the transcriptional changes exist. These transcriptional changes may reflect differences in the development, function, physiology or anatomy of the individual motor neurons. For example, we might suggest that cranial motor neurons generally have a shorter axonal length that those located elsewhere in the body. It is also possible that they have have other structural differences such as a more elaborate dendritic tree, or a different proportion of axodendritic or axosomatic synpases. The potential differences in form and function between motor neuron pools are seemingly endless. We can also only currently hypothesize about how changes in development, form or function could result in a selective sparing in a pathological situation. For this reason, rather than dismissing transcriptional changes occurring between differentially vulnerable motor neuron as, most likely due to a difference in the development, location or function of a given pool of motor neurons, it may be useful to use the list of transcripts identified to generate ideas about how this can impact upon the anatomy and physiology of the motor neuron. Indeed, the observation that a large number of HOX genes were differentially expressed between differentially vulnerable motor units may be attributed to the different location of the different motor neuron pools. However, it may be that the actual difference in location, and the associated differences in anatomy and physiology actually contribute to their differentially vulnerabililty. Determining the reasons and consequences of the differential transcriptional expression may lead to to a broader understanding of what fundamental differences make a motor neuron more or less vulnerable during disease. We therefore suggest that future efforts, to understand the impact of differential expression of specific transcripts or alterations in specific cellular pathways relate to the development, physiology and anatomy of specific motor neuron pools may be fruitful in our search to understand the phenomenom of selectively motor unit vulnerability of motor neuron disease.
Conclusions
In this work we have employed a novel approach to identify transcripts that are functionally significant in motor neuron disease-relevant pathways. We have demonstrated that at least one of these candidates can modify the phenotype in a mouse model of SMA, and believe that the remaining list contains additional candidates that warrant further examination. Based upon the design of the experiments, these modifiers may functionally interact in more than one disease context and therefore have the ability to provide protection to motor neurons in a variety of neurodegenerative conditions. Future efforts to identify potent modifiers and their mechanisms of action will provide insight into the mechanism of disease, and aid in the development of therapeutic agents which can slow the degeneration of motor neurons in MND.
Materials and methods
Data acquisition and analysis
RNAseq data, profiling the transcriptome of cranial and abdominal motor neurons from P10 wildtype mice was obtained as detailed in Murray et. al.[16]. Further analysis of this data was allowed by generous agreement with Dr Rashmi Kothary. The raw microarray files detailing transcriptional data published in Kaplan et al., 2014 and Brockington et al., 2013 were downloaded from the gene expression omnibus using the reference numbers GSE52118 and GSE40438 respectively[17, 19]. Raw microarray files from the study by Hedlund et al. were generously provided by Dr Eva Hedlund[18].
Following acquisition of raw microarray data sets, data was normalised using a quantile method, and genes which were differentially expressed within each screen were identified. All genes with an adjusted P value of >0.05 were eliminated from the study. For RNAseq data, transcripts had been identified by alignment to the mouse mm9 genome assemble in Murray et. al.[16], and relative transcript levels were compared using CuffDiff software v1.3 using the UCSC transcript model. Significance was considered with an adjusted P value of <0.05 and a greater that 1.5 fold change in expression level. HomoloGene was use to identify the genetic homologue between species. Data was sorted in excel to reveal changes which occurred in a common direction in 2 or more screens.
Screening methods and Drosophila husbandry
Genetic schemes and Drosophila husbandry were performed as detailed in Sanhueza et al., 2015[23]. Briefly, the tester line carrying both the ey-Gal4 driver and the UAS-DVAP-P58S transgene on the second chromosome was crossed individually with RNAi and EP lines with the potential of overexpressing the gene of interest. The F1 progeny was analyzed for the suppression or the enhancement of the DVAP-P58S induced small and rough eye phenotype. In particular, 8–10 males of either the EP or RNAi line were mated to 10–15 females of the ey-Gal4, DVAP-P58S/CyO ALS8 fly stock. After two days, adults were transferred to a new vial to have a duplicate cross. Embryos from both vials were raised at 29°C in a water bath to maximize the effect of the Gal4. Both enhancing and suppressing effects of the DVAP-P58S-induced eye neurodegenerative phenotype were assessed in these conditions. RNAi lines were acquired from the Vienna Drosophila RNAi line Center while EP and EPgy lines were obtained from the Drosophila Bloomington Stock Center.
Mouse maintenance
For analysis of differentially vulnerable muscles, Smn2B/- mice and wildtype controls on a C57Bl6 background were maintained in the animal facilities at the University of Edinburgh. Mice were sacrificed by overdose of inhalation anaethetic (isofluorane) and cervical dislocation. All experiments were performed in accordance with the regulations set out by the UK Home Office. For experiments requiring the administration of AAV9, all mice were housed and handled in accordance with the Animal Care and Use Committees of the University of Missouri. Smn2B/2B mice were a kind gift from Dr. R. Kothary (Ottawa, Canada). FVB Smn+/- mice were purchased from the Jackson Laboratory. Mice were housed under a 12 hours light/dark cycle and the colonies were maintained as heterozygote breeding pairs under specific pathogen-free conditions.
Vector construction
293T HEK cells (ATCC CRL 3216, American Type Culture Collection, Manassas, VA, USA) cultured in 4 10-floor cell factories until ~85% confluent. Cells were triple transfected with Rep2Cap9 (Serotype 2 Rep proteins, Serotype 9 capsid proteins), pHelper (Adenovirus helper constructs), and scAAV-CBA-SNCA using 25 kDa Polyethyleneimine (PEI) at a molar ratio of 1 : 1:1. Media was changed 24 hours after transfection, and cells were harvested at 48 hours after transfection. Cells were suspended in 10 mmol Tris, pH = 8.0, lysed by 5 freeze-thaw-cycles in liquid nitrogen, DNAse treated, and protease treated. CsCl crystals were added to the lysate (0.631 g of CsCl per ml of the lysate) to generate a solution with a density of ∼1.4 mg/ml. After incubation at 37°C for 45 min, the solution was centrifuged at 4000 rpm in an Eppendorf 5810 R at 4°C. Virus was purified from lysate by 3 rounds of density gradient centrifugation at an average RCF of 158,000. High titer fractions were detected after each round of centrifugation using quantitative real-time PCR. The final fractions were dialyzed exhaustively against phosphate buffered saline and stored at 4°C until use.
Administration of AAV9 vectors
Viral delivery was performed by intracerebroventricular (ICV) injection using methods described previously [48, 49]. Briefly, ICV injections were performed using sterilized glass micropipettes. The needles were inserted perpendicular to the skull at the injection site approximately 0.25 mm lateral to the sagittal suture and 0.5 mm rostral to the coronary suture.
Immunofluorescent staining and quantification
For NMJ labelling, muscles were immediately dissected from recently sacrificed mice and fixed in 4% PFA (Electron Microscopy Science) in PBS for 15 min. Post-synaptic AChRs were labelled with α-bungarotoxin (BTX) for 30 min. Muscles were permeabilised in 2% Triton X-100 in PBS for 30 min, then blocked in 4% bovine serum albumin (BSA)/1% Triton X-100 in PBS for 30 min before incubation overnight in primary antibodies [Neurofilament (NF; 2H3)—Developmental Studies Hybridoma Bank; synaptic vesicle protein 2 (SV2)—Developmental Studies Hybridoma Bank; S100 –Dako; all 1 : 250] and visualised with Cy3-conjugated secondary antibodies [Cy3 goat anti-mouse; 1 : 250, Jackson]. Muscles were then whole-mounted in Dako Fluorescent mounting media. Confocal microscopy was performed using a Nikon A1R+ Resonant Scanning System (Nikon) (10x and 40x objectives; 0.3 and 1.3 oil NA; Nikon A1R+ microscope; simultaneous image acquisition). 488 and 543 nm laser lines were used for excitation. The resultant confocal Z-series produced in NIS Elements 2D Analysis software were exported and merged using Fiji ImageJ software.
The percentage of fully occupied endplates was determined by classifying each endplate in a given field of view either fully occupied (pre-synaptic terminal (SV2 and NF) completely overlies endplate (BTX)), partially occupied (pre-synaptic terminal only partially covers endplate (BTX)), or vacant (no pre-synaptic label overlies endplate). At least 4 fields of view were analysed per muscle totalling >100 endplates per muscle.
All data was assembled and analysed using Microsoft Excel and GraphPad Prism.
Supporting Information
Zdroje
1. Dion PA, Daoud H, Rouleau GA. Genetics of motor neuron disorders: new insights into pathogenic mechanisms. Nat Rev Genet. 2009;10(11):769–82. doi: 10.1038/nrg2680 19823194
2. Lefebvre S, Burglen L, Reboullet S, Clermont O, Burlet P, Viollet L, et al. Identification and characterization of a spinal muscular atrophy-determining gene. Cell. 1995;80(1):155–65. 7813012
3. Rodrigues NR, Owen N, Talbot K, Ignatius J, Dubowitz V, Davies KE. Deletions in the survival motor neuron gene on 5q13 in autosomal recessive spinal muscular atrophy. Hum Mol Genet. 1995;4(4):631–4. 7633412
4. Gavrilov DK, Shi X, Das K, Gilliam TC, Wang CH. Differential SMN2 expression associated with SMA severity. Nat Genet. 1998;20(3):230–1. doi: 10.1038/3030 9806538
5. La Spada A. Spinal and Bulbar Muscular Atrophy. In: Pagon RA, Adam MP, Ardinger HH, Wallace SE, Amemiya A, Bean LJH, et al., editors. GeneReviews(R). Seattle (WA)1993.
6. Comley LH, Nijssen J, Frost-Nylen J, Hedlund E. Cross-disease comparison of amyotrophic lateral sclerosis and spinal muscular atrophy reveals conservation of selective vulnerability but differential neuromuscular junction pathology. J Comp Neurol. 2016;524(7):1424–42. doi: 10.1002/cne.23917 26502195
7. Ling KK, Gibbs RM, Feng Z, Ko CP. Severe neuromuscular denervation of clinically relevant muscles in a mouse model of spinal muscular atrophy. Hum Mol Genet. 2012;21(1):185–95. doi: 10.1093/hmg/ddr453 21968514
8. Murray LM, Beauvais A, Bhanot K, Kothary R. Defects in neuromuscular junction remodelling in the Smn(2B/-) mouse model of spinal muscular atrophy. Neurobiol Dis. 2013;49 : 57–67. doi: 10.1016/j.nbd.2012.08.019 22960106
9. Murray LM, Comley LH, Thomson D, Parkinson N, Talbot K, Gillingwater TH. Selective vulnerability of motor neurons and dissociation of pre - and post-synaptic pathology at the neuromuscular junction in mouse models of spinal muscular atrophy. Hum Mol Genet. 2008;17(7):949–62. doi: 10.1093/hmg/ddm367 18065780
10. Deymeer F, Serdaroglu P, Parman Y, Poda M. Natural history of SMA IIIb: muscle strength decreases in a predictable sequence and magnitude. Neurology. 2008;71(9):644–9. doi: 10.1212/01.wnl.0000324623.89105.c4 18725590
11. Kubota M, Sakakihara Y, Uchiyama Y, Nara A, Nagata T, Nitta H, et al. New ocular movement detector system as a communication tool in ventilator-assisted Werdnig-Hoffmann disease. Dev Med Child Neurol. 2000;42(1):61–4. 10665977
12. Gizzi M, DiRocco A, Sivak M, Cohen B. Ocular motor function in motor neuron disease. Neurology. 1992;42(5):1037–46. 1579227
13. Haenggeli C, Kato AC. Differential vulnerability of cranial motoneurons in mouse models with motor neuron degeneration. Neurosci Lett. 2002;335(1):39–43. 12457737
14. Nimchinsky EA, Young WG, Yeung G, Shah RA, Gordon JW, Bloom FE, et al. Differential vulnerability of oculomotor, facial, and hypoglossal nuclei in G86R superoxide dismutase transgenic mice. J Comp Neurol. 2000;416(1):112–25. 10578106
15. Achsel T, Barabino S, Cozzolino M, Carri MT. The intriguing case of motor neuron disease: ALS and SMA come closer. Biochem Soc Trans. 2013;41(6):1593–7. doi: 10.1042/BST20130142 24256260
16. Murray LM, Beauvais A, Gibeault S, Courtney NL, Kothary R. Transcriptional profiling of differentially vulnerable motor neurons at pre-symptomatic stage in the Smn (2b/-) mouse model of spinal muscular atrophy. Acta Neuropathol Commun. 2015;3 : 55. doi: 10.1186/s40478-015-0231-1 26374403
17. Brockington A, Ning K, Heath PR, Wood E, Kirby J, Fusi N, et al. Unravelling the enigma of selective vulnerability in neurodegeneration: motor neurons resistant to degeneration in ALS show distinct gene expression characteristics and decreased susceptibility to excitotoxicity. Acta Neuropathol. 2013;125(1):95–109. doi: 10.1007/s00401-012-1058-5 23143228
18. Hedlund E, Karlsson M, Osborn T, Ludwig W, Isacson O. Global gene expression profiling of somatic motor neuron populations with different vulnerability identify molecules and pathways of degeneration and protection. Brain. 2010;133(Pt 8):2313–30. doi: 10.1093/brain/awq167 20826431
19. Kaplan A, Spiller KJ, Towne C, Kanning KC, Choe GT, Geber A, et al. Neuronal matrix metalloproteinase-9 is a determinant of selective neurodegeneration. Neuron. 2014;81(2):333–48. doi: 10.1016/j.neuron.2013.12.009 24462097
20. Nishimura AL, Mitne-Neto M, Silva HC, Richieri-Costa A, Middleton S, Cascio D, et al. A mutation in the vesicle-trafficking protein VAPB causes late-onset spinal muscular atrophy and amyotrophic lateral sclerosis. Am J Hum Genet. 2004;75(5):822–31. doi: 10.1086/425287 15372378
21. Chen HJ, Anagnostou G, Chai A, Withers J, Morris A, Adhikaree J, et al. Characterization of the properties of a novel mutation in VAPB in familial amyotrophic lateral sclerosis. J Biol Chem. 2010;285(51):40266–81. doi: 10.1074/jbc.M110.161398 20940299
22. Larroquette F, Seto L, Gaub PL, Kamal B, Wallis D, Lariviere R, et al. Vapb/Amyotrophic lateral sclerosis 8 knock-in mice display slowly progressive motor behavior defects accompanying ER stress and autophagic response. Hum Mol Genet. 2015;24(22):6515–29. doi: 10.1093/hmg/ddv360 26362257
23. Sanhueza M, Chai A, Smith C, McCray BA, Simpson TI, Taylor JP, et al. Network analyses reveal novel aspects of ALS pathogenesis. PLoS Genet. 2015;11(3):e1005107. doi: 10.1371/journal.pgen.1005107 25826266
24. Peretti D, Dahan N, Shimoni E, Hirschberg K, Lev S. Coordinated lipid transfer between the endoplasmic reticulum and the Golgi complex requires the VAP proteins and is essential for Golgi-mediated transport. Mol Biol Cell. 2008;19(9):3871–84. doi: 10.1091/mbc.E08-05-0498 18614794
25. Gomez-Suaga P, Paillusson S, Stoica R, Noble W, Hanger DP, Miller CC. The ER-Mitochondria Tethering Complex VAPB-PTPIP51 Regulates Autophagy. Curr Biol. 2017;27(3):371–85. doi: 10.1016/j.cub.2016.12.038 28132811
26. Han SM, El Oussini H, Scekic-Zahirovic J, Vibbert J, Cottee P, Prasain JK, et al. VAPB/ALS8 MSP ligands regulate striated muscle energy metabolism critical for adult survival in caenorhabditis elegans. PLoS Genet. 2013;9(9):e1003738. doi: 10.1371/journal.pgen.1003738 24039594
27. Perrimon N, Noll E, Mccall K, Brand A. Generating Lineage-Specific Markers to Study Drosophila Development. Dev Genet. 1991;12(3):238–52. doi: 10.1002/dvg.1020120309 1651183
28. Forrest S, Chai A, Sanhueza M, Marescotti M, Parry K, Georgiev A, et al. Increased levels of phosphoinositides cause neurodegeneration in a Drosophila model of amyotrophic lateral sclerosis. Hum Mol Genet. 2013;22(13):2689–704. doi: 10.1093/hmg/ddt118 23492670
29. Acsadi G, Li X, Murphy KJ, Swoboda KJ, Parker GC. Alpha-synuclein loss in spinal muscular atrophy. J Mol Neurosci. 2011;43(3):275–83. doi: 10.1007/s12031-010-9422-1 20640532
30. da Costa CA, Ancolio K, Checler F. Wild-type but not Parkinson's disease-related ala-53 —> Thr mutant alpha -synuclein protects neuronal cells from apoptotic stimuli. J Biol Chem. 2000;275(31):24065–9. doi: 10.1074/jbc.M002413200 10818098
31. Hashimoto M, Hsu LJ, Rockenstein E, Takenouchi T, Mallory M, Masliah E. alpha-Synuclein protects against oxidative stress via inactivation of the c-Jun N-terminal kinase stress-signaling pathway in neuronal cells. J Biol Chem. 2002;277(13):11465–72. doi: 10.1074/jbc.M111428200 11790792
32. Manning-Bog AB, McCormack AL, Purisai MG, Bolin LM, Di Monte DA. Alpha-synuclein overexpression protects against paraquat-induced neurodegeneration. J Neurosci. 2003;23(8):3095–9. 12716914
33. Zhou W, Hurlbert MS, Schaack J, Prasad KN, Freed CR. Overexpression of human alpha-synuclein causes dopamine neuron death in rat primary culture and immortalized mesencephalon-derived cells. Brain Res. 2000;866(1–2):33–43. 10825478
34. Foust KD, Nurre E, Montgomery CL, Hernandez A, Chan CM, Kaspar BK. Intravascular AAV9 preferentially targets neonatal neurons and adult astrocytes. Nat Biotechnol. 2009;27(1):59–65. doi: 10.1038/nbt.1515 19098898
35. Robbins KL, Glascock JJ, Osman EY, Miller MR, Lorson CL. Defining the therapeutic window in a severe animal model of spinal muscular atrophy. Hum Mol Genet. 2014;23(17):4559–68. doi: 10.1093/hmg/ddu169 24722206
36. Butchbach ME. Copy Number Variations in the Survival Motor Neuron Genes: Implications for Spinal Muscular Atrophy and Other Neurodegenerative Diseases. Front Mol Biosci. 2016;3 : 7. doi: 10.3389/fmolb.2016.00007 27014701
37. Ladd AN, Charlet N, Cooper TA. The CELF family of RNA binding proteins is implicated in cell-specific and developmentally regulated alternative splicing. Mol Cell Biol. 2001;21(4):1285–96. doi: 10.1128/MCB.21.4.1285-1296.2001 11158314
38. Qin Y, Chen Z, Han X, Wu H, Yu Y, Wu J, et al. Progesterone attenuates Abeta(25–35)-induced neuronal toxicity via JNK inactivation and progesterone receptor membrane component 1-dependent inhibition of mitochondrial apoptotic pathway. J Steroid Biochem Mol Biol. 2015;154 : 302–11. doi: 10.1016/j.jsbmb.2015.01.002 25576906
39. Jackson AC, Roche SL, Byrne AM, Ruiz-Lopez AM, Cotter TG. Progesterone receptor signalling in retinal photoreceptor neuroprotection. J Neurochem. 2016;136(1):63–77. doi: 10.1111/jnc.13388 26447367
40. Rubin CI, Atweh GF. The role of stathmin in the regulation of the cell cycle. J Cell Biochem. 2004;93(2):242–50. doi: 10.1002/jcb.20187 15368352
41. Liedtke W, Leman EE, Fyffe RE, Raine CS, Schubart UK. Stathmin-deficient mice develop an age-dependent axonopathy of the central and peripheral nervous systems. Am J Pathol. 2002;160(2):469–80. doi: 10.1016/S0002-9440(10)64866-3 11839567
42. Strey CW, Spellman D, Stieber A, Gonatas JO, Wang X, Lambris JD, et al. Dysregulation of stathmin, a microtubule-destabilizing protein, and up-regulation of Hsp25, Hsp27, and the antioxidant peroxiredoxin 6 in a mouse model of familial amyotrophic lateral sclerosis. Am J Pathol. 2004;165(5):1701–18. doi: 10.1016/S0002-9440(10)63426-8 15509539
43. Wen HL, Ting CH, Liu HC, Li H, Lin-Chao S. Decreased stathmin expression ameliorates neuromuscular defects but fails to prolong survival in a mouse model of spinal muscular atrophy. Neurobiol Dis. 2013;52 : 94–103. doi: 10.1016/j.nbd.2012.11.015 23268200
44. Polymeropoulos MH, Lavedan C, Leroy E, Ide SE, Dehejia A, Dutra A, et al. Mutation in the alpha-synuclein gene identified in families with Parkinson's disease. Science. 1997;276(5321):2045–7. 9197268
45. Maroteaux L, Campanelli JT, Scheller RH. Synuclein: a neuron-specific protein localized to the nucleus and presynaptic nerve terminal. J Neurosci. 1988;8(8):2804–15. 3411354
46. Nemani VM, Lu W, Berge V, Nakamura K, Onoa B, Lee MK, et al. Increased expression of alpha-synuclein reduces neurotransmitter release by inhibiting synaptic vesicle reclustering after endocytosis. Neuron. 2010;65(1):66–79. doi: 10.1016/j.neuron.2009.12.023 20152114
47. Yavich L, Tanila H, Vepsalainen S, Jakala P. Role of alpha-synuclein in presynaptic dopamine recruitment. J Neurosci. 2004;24(49):11165–70. doi: 10.1523/JNEUROSCI.2559-04.2004 15590933
48. Glascock JJ, Shababi M, Wetz MJ, Krogman MM, Lorson CL. Direct central nervous system delivery provides enhanced protection following vector mediated gene replacement in a severe model of spinal muscular atrophy. Biochem Biophys Res Commun. 2012;417(1):376–81. doi: 10.1016/j.bbrc.2011.11.121 22172949
49. Shababi M, Glascock J, Lorson CL. Combination of SMN trans-splicing and a neurotrophic factor increases the life span and body mass in a severe model of spinal muscular atrophy. Hum Gene Ther. 2011;22(2):135–44. doi: 10.1089/hum.2010.114 20804424
Štítky
Genetika Reprodukční medicína
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2017 Číslo 3- Růst a vývoj dětí narozených pomocí IVF
- Délka menstruačního cyklu jako marker ženské plodnosti
- Vztah užívání alkoholu a mužské fertility
- Akutní intermitentní porfyrie
- Intrauterinní inseminace a její úspěšnost
-
Všechny články tohoto čísla
- 2016 Reviewer and Editorial Board Thank You
- The social genome: Current findings and implications for the study of human genetics
- A rare loss-of-function mutation reduces blood eosinophil counts and protects from asthma
- A variant in the gene in a dog with ichthyosis
- Excess of genomic defects in a woolly mammoth on Wrangel island
- CUZD1 is a critical mediator of the JAK/STAT5 signaling pathway that controls mammary gland development during pregnancy
- Fishing for adaptive epistasis using mitonuclear interactions
- Comparison of independent screens on differentially vulnerable motor neurons reveals alpha-synuclein as a common modifier in motor neuron diseases
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- CUZD1 is a critical mediator of the JAK/STAT5 signaling pathway that controls mammary gland development during pregnancy
- A rare loss-of-function mutation reduces blood eosinophil counts and protects from asthma
- A variant in the gene in a dog with ichthyosis
- Fishing for adaptive epistasis using mitonuclear interactions
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



