-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaBuffers Inherent Left/Right Asymmetry Ensuring Symmetric Forelimb Formation
Externally, the human form appears bilaterally symmetric. For example, each of our pairs of arms and legs are the same length. This external symmetry masks many asymmetries found in internal organs. In most people the heart is found on the left side of the chest. The stomach, liver and spleen are also positioned asymmetrically. The authors of this study demonstrate, using a mouse model, that bilateral symmetry of the arms is not a default, passive state but that mechanisms are in place that ensure symmetrical formation of the left and right limbs. Bilateral symmetry of the arms is achieved by the action of a gene Tbx5 that masks the effects of signals that acted earlier during embryogenesis, many days before limb formation, and imposed asymmetries on the forming internal organs. Maintaining bilateral symmetry of the arms is important for them to carry out their normal functions but this process can go wrong. Holt-Oram syndrome patients have upper limb defects, including shortened arms. Consistently the defects are more severe in their left arm than right. This birth defect is caused by disruption of the TBX5 gene. By linking the action of Tbx5 to symmetrical limb formation, the authors provide an explanation for why Holt-Oram syndrome patients have more severe defects in the left arms than right.
Published in the journal: . PLoS Genet 12(12): e32767. doi:10.1371/journal.pgen.1006521
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1006521Summary
Externally, the human form appears bilaterally symmetric. For example, each of our pairs of arms and legs are the same length. This external symmetry masks many asymmetries found in internal organs. In most people the heart is found on the left side of the chest. The stomach, liver and spleen are also positioned asymmetrically. The authors of this study demonstrate, using a mouse model, that bilateral symmetry of the arms is not a default, passive state but that mechanisms are in place that ensure symmetrical formation of the left and right limbs. Bilateral symmetry of the arms is achieved by the action of a gene Tbx5 that masks the effects of signals that acted earlier during embryogenesis, many days before limb formation, and imposed asymmetries on the forming internal organs. Maintaining bilateral symmetry of the arms is important for them to carry out their normal functions but this process can go wrong. Holt-Oram syndrome patients have upper limb defects, including shortened arms. Consistently the defects are more severe in their left arm than right. This birth defect is caused by disruption of the TBX5 gene. By linking the action of Tbx5 to symmetrical limb formation, the authors provide an explanation for why Holt-Oram syndrome patients have more severe defects in the left arms than right.
Introduction
The external body plan of most metazoans is bilaterally symmetric. How this symmetry is achieved has fascinated biologists for centuries and is exemplified by Leonardo Da Vinci’s description of the “Vitruvian Man” that describes some of the uniform proportions of the human body. In vertebrates, external bilateral symmetry masks significant internal asymmetries such as the position of the heart and liver and number of lobes of the lungs. While there has been progress in identifying how asymmetry of internal organs is generated [1], we know very little about how symmetry in bilateral structures, such as the limbs is established. A classic study that compared the lengths of the skeletal elements of left and right embryonic chick wings found there was very little difference in their size [2] demonstrating the exquisite fidelity in the genetic programmes regulating limb development, a finding all the more intriguing by the lack of evidence for molecular crosstalk between the developing left and right limb buds.
Holt Oram Syndrome (HOS) [OMIM 142900] is caused by mutations in TBX5, a T-box transcription factor expressed in the forelimb and heart. The clinical features of HOS include heart defects and a spectrum of upper/fore limb defects ranging in severity from total aplasia of the radial elements to relatively mild defects such as a tri-phalangeal thumb [3]. A characteristic feature of HOS patients is that although the severity of limb defects can vary between patients, the left limb is more severely affected than the right in most patients [3]. HOS is a dominant disorder and heterozygous affected individuals carry mutations in TBX5 predicted to result in truncated proteins that fail to fold or are rapidly degraded [4] and are most likely loss-of-function alleles. HOS defects are therefore thought to arise as a result of TBX5 haploinsufficiency and indicate that both copies of the gene are required for normal function.
Attempts to recapitulate the upper limb defects associated with HOS in the mouse have previously been unsuccessful. Conditional heterozygous deletion of Tbx5 using the limb-restricted Prx1Cre (Tbx5lox/+;Prx1Cre) does not produce any obvious forelimb phenotype, while homozygous conditional deletion of Tbx5 (Tbx5lox/lox;Prx1Cre) leads to a total loss of forelimb bud initiation and subsequent forelimb formation [5]. We have developed two alternative strategies to model the left-biased, asymmetrical limb defects of HOS patients in the mouse. One strategy utilizes a gene deletion-gene replacement strategy [6] in which Tbx5 is conditionally deleted using the Prx1Cre line and hypomorphic levels of a Prx1-Tbx transgene are expressed in the forelimb regions of the same embryo. The alternative strategy uses a mosaic cre line, Prx1Cre(98), to conditionally delete Tbx5 from cells in the right and left forelimb-forming LPM. Both approaches successfully recapitulate the types of limb abnormalities observed in HOS patients and significantly, the left bias in the severity of the limb defects. We also demonstrate that the disruption of INV, a gene crucial for the establishment of the left-right pathway [7] in the presence of hypomorphic levels of Prx1-Tbx lead to the formation of right sided forelimb defects. Additionally, optimal levels of Fgf10 expression in the presence of hypomorphic Prx1-Tbx levels are not sufficient to rescue symmetrical forelimb bud initiation and formation. These data demonstrate that although the left and right limb ultimately develop to be bilaterally symmetric structures they arise from regions of the left and right embryo flank that have inherent asymmetries. We further show that threshold levels of Tbx5 are required to overcome these asymmetries to ensure bilaterally symmetric forelimb formation.
Results
Hypomorphic Tbx levels recapitulate HOS limb defects
To generate a model mouse of HOS, we used a gene deletion-gene replacement strategy (S1A Fig) [6] so that hypomorphic levels of a Tbx transgene are delivered to the forelimb-forming region. Previously, we have shown that homozygous deletion of Tbx5 using the limb-restricted Cre-deleter line, Prx1Cre, produces a forelimb-less phenotype [5]. This defect can be completely rescued by misexpression of either Tbx5 or Tbx4 transgenes driven by the same Prx1 regulatory element [8]. One chimeric construct, Prx1-5N5T4C#1, a fusion of the N-terminal and T-Box DNA binding domains of Tbx5 and the C-terminal domain of Tbx4, is also able to rescue forelimb bud formation [8]. These results indicate that Tbx5 and Tbx4 have common functions in initiation of fore and hindlimb outgrowth and Tbx5, Tbx4 and Tbx5/Tbx4 chimera proteins can act equivalently. Another transgenic line, Prx1-5N5T4C#2 (hereinafter referred to as Prx1-Tbx), harbours the same transgene as Prx1-5N5T4C#1 but in a different locus and expresses hypomorphic levels of the chimera protein compared to endogenous Tbx5 (S1B–S1E Fig). This hypomorphic misexpression of the transgene only partially rescues forelimb bud initiation defects caused by the conditional deletion of Tbx5 (Tbx5lox/lox;Prx1Cre;Prx1-Tbx) and recapitulates many features of the upper limb defects seen in HOS patients, for example an anterior bias to the structures affected (including triphalangeal thumb, absent thumb), defects in the scapula and in more severe examples, phocomelia (Fig 1A–1C). Another clinical feature of HOS is the broad range in the severity of limb defects, even within a family carrying the same TBX5 mutation, ranging from almost complete absence of the upper limb to triphalangeal thumb. This is also reproduced in our mouse model (Fig 1C). Despite the range in the severity of the limb defects between different embryos, in individual Tbx5lox/lox;Prx1Cre;Prx1-Tbx mutant embryos the forelimb defects are consistently more severe in the left forelimb than the right. In our mouse model this was observed in 100% of the embryos studied (n = 11) (S2 Table). A transverse section of Z/AP/+;Prx1Cre embryo shows the bilaterally symmetrical Cre activity in the left and right forelimb buds [9] (Fig 1D). Furthermore, we compared Cre mRNA expression levels between the left and right forelimb buds and did not detect statistically significant difference (p > 0.05 with two-tailed Student’s t-test) (Fig 1E). This result is consistent with the fact that the Prx1Cre driver has been used in combination with numerous other conditional mutant alleles and all limb defects reported to date are bilaterally symmetric. In addition, equivalent, bilateral chimera transcript levels are present in Prx1-Tbx transgenic embryos (p > 0.05 with two-tailed Student’s t-test) (Fig 1E), indicating Tbx activity that partially and asymmetrically rescues limb outgrowth is bilaterally symmetric in Tbx5lox/lox;Prx1Cre;Prx1-Tbx mutant embryos.
Fig. 1. A mouse Tbx hypomorph mutant produces left-biased asymmetric forelimb defects. 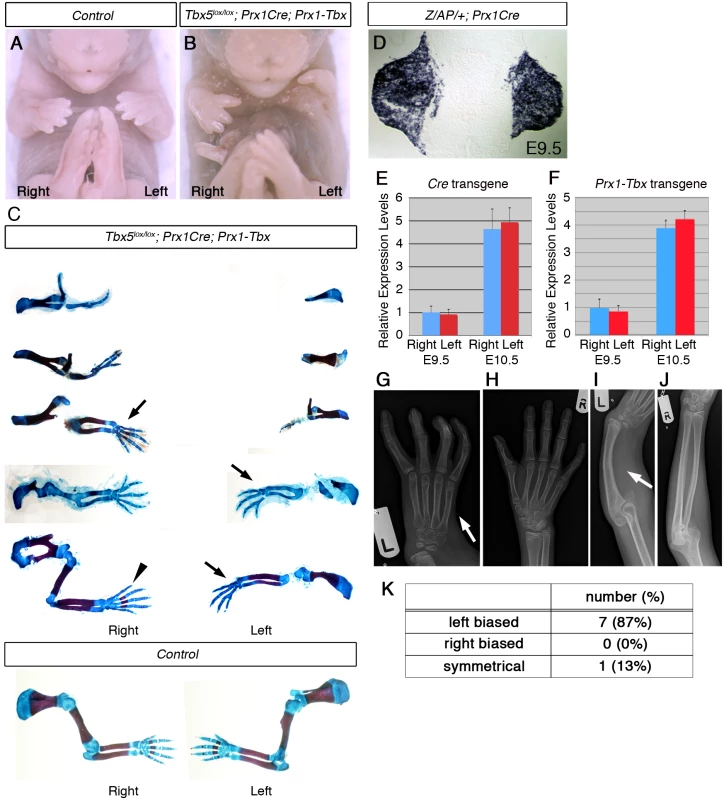
A-B, E17.5 control (A) and Tbx5lox/lox;Prx1Cre;Prx1-Tbx mutant (B). C, Tbx5lox/lox;Prx1Cre;Prx1-Tbx mutant forelimb skeletal preparations shown in descending order of severity (top to bottom) and E17.5 control forelimbs. The defects are consistently more severe in the left forelimb than the right (n = 11). Absent digit 1 (thumb) (arrows), triphalangeal digit 1 (arrowhead). D, A transverse section at the level of the forelimb bud of a E9.5 Z/AP/+;Prx1Cre embryo. E, qPCR analysis of left and right forelimb buds of Prx1Cre embryos. F, qPCR analysis of the left and right forelimb buds of Prx1-Tbx transgenic embryos. G-J. X-ray radiographs of a HOS patient. The left forelimb is more affected than the right one. The thumb is absent on the left hand (G, arrow) while it is present on the right (H). The radius is missing on the left side (I, arrow), while it is formed on the right (J). K. Numbers of patients showing left-biased, right-biased and symmetrical forelimb defects. In a previous study it was calculated that 70% of HOS individuals display asymmetrical upper limb defects, and in 91% of these, the left side is more severely affected than the right [3]. In contrast, 100% of the Tbx hypomorphic mutant mice we obtained showed left-biased defects. A possible explanation for this discrepancy in penetrance is that the original analysis of HOS patients was carried out before genetic lesions in TBX5 were identified as responsible for HOS and some of the original patients may not have had pathogenic TBX5 mutations. We searched the electronic patient record at Great Ormond St Hospital and identified 32 patients with a clinical suspicion of Holt-Oram syndrome. In 11 cases TBX5 mutations had been identified, 8 of which were pathogenic mutations (S1 Table). 7 cases show left-biased forelimb defects and one case is symmetrical (Fig 1G–1K) [10–13]. We did not observe right-biased defects in any patients with confirmed pathogenic TBX5 mutations. Therefore in our patient series when pathogenic mutations in TBX5 have been confirmed the clinical presentation includes left-biased severity with almost 100% penetrance consistent with what we observed in our mouse model.
During forelimb bud initiation, Tbx5 induces the expression of Fgf10 in the limb mesenchyme and Fgf10 subsequently induces Fgf8 expression in the overlying ectoderm. Sall4 is also proposed as a target of Tbx5 [14,15]. We examined whether the expression patterns of these genes are different between the right and left forelimb buds of Tbx5lox/lox;Prx1Cre;Prx1-Tbx mutant embryos. At E10.5 mutant embryos can be divided into two groups depending on the severity of phenotypes, which correlate with the variation in forelimb defects observed later at E17.5. In the first group, there is no obvious left limb bud and small right limb buds are formed (Fig 2B, 2E and 2H). Fgf10, Fgf8 and Sall4 are not detected in the left lateral plate mesoderm (Fig 2B, 2E and 2H arrowheads). In contrast, low levels of Fgf10 and Sall4 are detectable in the right forelimb buds and Fgf8 is present in the right AER. In this group of mutants the left limb bud lacks the first emergence of the bud, rather than a subsequent regression. In the second group, they form small left limb buds (Fig 2C, 2F and 2I, arrowheads). The right limb buds are larger than left ones but still smaller than control limb buds. Fgf10 expression in the left forelimb bud is lower than the right (Fig 2C), whereas there is no detectable difference in Sall4 expression between the left and right forelimb buds (Fig 2I). Fgf8 expression in the left AER is disrupted in contrast to the right AER where it is expressed throughout (Fig 2F).
Fig. 2. Marker gene expression in left and right forelimb buds of Tbx hypomorphs. 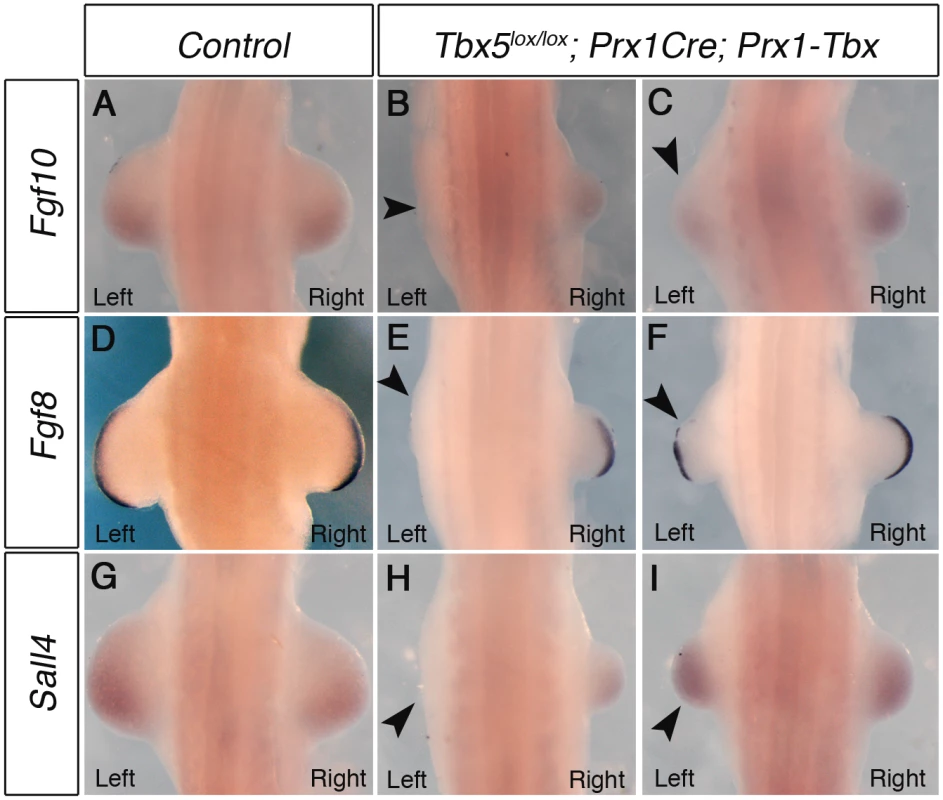
WISH analysis of E10.5 embryos. A, Fgf10 expression in a control embryo B, Left forelimb bud is not formed and Fgf10 expression is lost on the left side (arrowhead) in a severely affected embryo C, Fgf10 is weaker in left forelimb of a mildly affected embryo (arrowhead). D, Fgf8 expression in control E, Fgf8 is absent in left AER of a severely affected embryo (arrowhead). F, Fgf8 is disrupted in left AER of a mildly affected embryo (arrowhead). G, Sall4 expression in control. H, Sall4 is absent in left LPM of a severely affected embryo (arrowhead). I, Sall4 is expressed at similar levels bilaterally in a mildly affected embryo. A minimum of 4 mutants for each phenotype were analysed with each probe. Together, these results demonstrate that hypomorphic levels of a Prx1-Tbx transgene cause the limb outgrowth defect by failing to establish the correct positive feedback loop of Fgf10 and Fgf8 and this defect is consistently more severe in the left forelimbs than the right.
Mosaic Tbx5 deletion causes left-biased forelimb defects
We tested if mosaic deletion of Tbx5 in the limb mesenchyme induces similar defects as hypomorphic Tbx mutants. We hypothesized that following mosaic deletion of Tbx5, only a subset of cells would express Fgf10 so that the total amount of secreted Fgf10 available in the forelimb LPM is reduced to hypomorphic levels. For this purpose, we used the Prx1Cre(98) transgenic line [16]. In this mouse, the same Prx1Cre transgene is integrated in a different locus, resulting in a mosaic and delayed Cre recombinase activity in the early forelimb bud (Figs 3A–3F and S2). Cre activity is not detected at 16 somites stage in Prx1Cre (98) (Fig 3B), when Cre from Prx1Cre is already active (Fig 3A), but its activity is detectable in the forelimb region at 22 somites stage and at E10.5 in a mosaic manner (Figs 3D, 3F and S2). Cre activity appears to be at equivalent levels in the left and right forelimbs when analysed with a cre reporter and stained histologically (S2 Fig). Furthermore, we confirmed the symmetrical mRNA expression levels between the left and right forelimb buds (p > 0.05 with two-tailed Student’s t-test) (Fig 3G). This mosaic and delayed deletion of Tbx5 by Prx1Cre(98) is sufficient to cause forelimb defects (Fig 3H and 3I).
Fig. 3. Comparison of Cre activity in Prx1Cre and Prx1Cre(98) transgenic deleter lines. 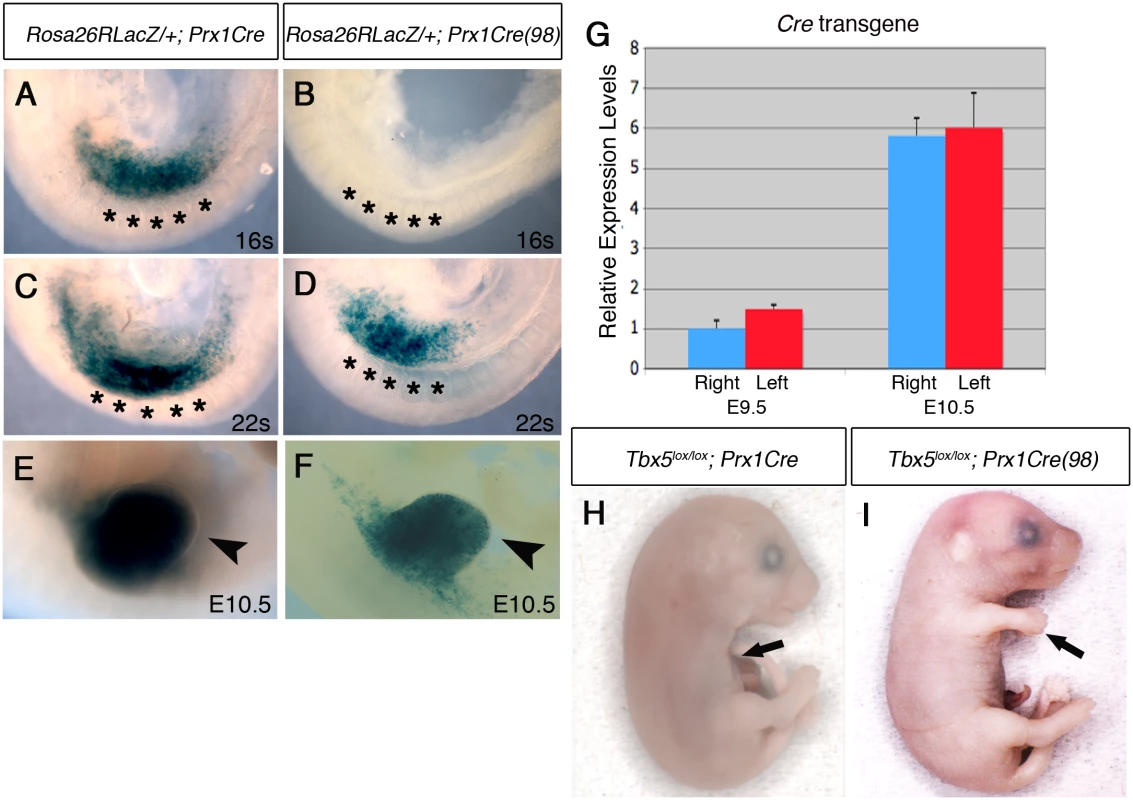
A-F, Lateral views of right side of embryos are shown. LacZ staining of Rosa26RLacZ/+;Prx1Cre embryos (A, C and E) and Rosa26RLacZ/+;Prx1Cre(98) embryos (B, D, and F). In the Prx1Cre line, Cre is active throughout the forelimb-forming region (marked by black asterisks in the adjacent somites) by 16 somites stage (A) while cre activity is not detected in Rosa26RLacZ/+;Prx1Cre(98) at this stage (B). At 22 somites stage, staining is seen throughout the nascent forelimb bud as well as the LPM rostral and caudal to the forelimb-forming region in Rosa26RLacZ/+;Prx1Cre embryos (C). At this stage cre is active in the nascent forelimb bud of Prx1Cre(98) embryo in a ‘salt and pepper’ mosaic manner (D). At E10.5 strong staining is observed throughout the forelimb buds of Rosa26RLacZ/+; Prx1Cre (E, arrowhead) and in a mosaic manner in the forelimb buds of Rosa26RLacZ/+;Prx1Cre(98) embryo (F, arrowhead). G, qPCR analysis of left and right forelimb buds of Prx1Cre(98) embryos. Cre mRNA is expressed at a similar level on both sides. H, All the elements of the forelimb have failed to form in E17.5 Tbx5lox/lox;Prx1Cre embryo (arrow). I, Abnormal forelimbs are formed in E17.5 Tbx5lox/lox;Prx1Cre(98) embryo (arrow). These Tbx5lox/lox;Prx1Cre (98) embryos display similar, although milder, defects as Tbx5lox/lox;Prx1Cre;Prx1-Tbx mutants including triphalangeal thumb, absent thumb and defects in the scapula and humerus (Fig 4A and 4B). Again, although a range in the severity of defects is observed in different embryos, consistently defects are observed with greater severity and higher frequency on the left side than the right (n = 18) (Fig 4B and S2 Table). An exception in these milder phenotypes was triphalangeal thumb that is observed with similar frequency on both sides. In mutant embryos at E10.5, forelimb buds are smaller than those of control embryos (n = 12) (Fig 4C–4H). Fgf8 expression is disrupted on the left forelimb (Fig 4F). As the defects in this mutant are milder than those of Tbx5lox/lox;Prx1Cre;Prx1-Tbx embryos, clear reduction of Fgf10 and Sall4 expression patterns were not observed (Fig 4D and 4H). These results demonstrate that mosaic and delayed deletion of Tbx5 from the left and right forelimb LPM leads to left-biased asymmetric forelimb defects.
Fig. 4. The mosaic and delayed deletion of Tbx5 results in the formation of left-biased forelimb defects. 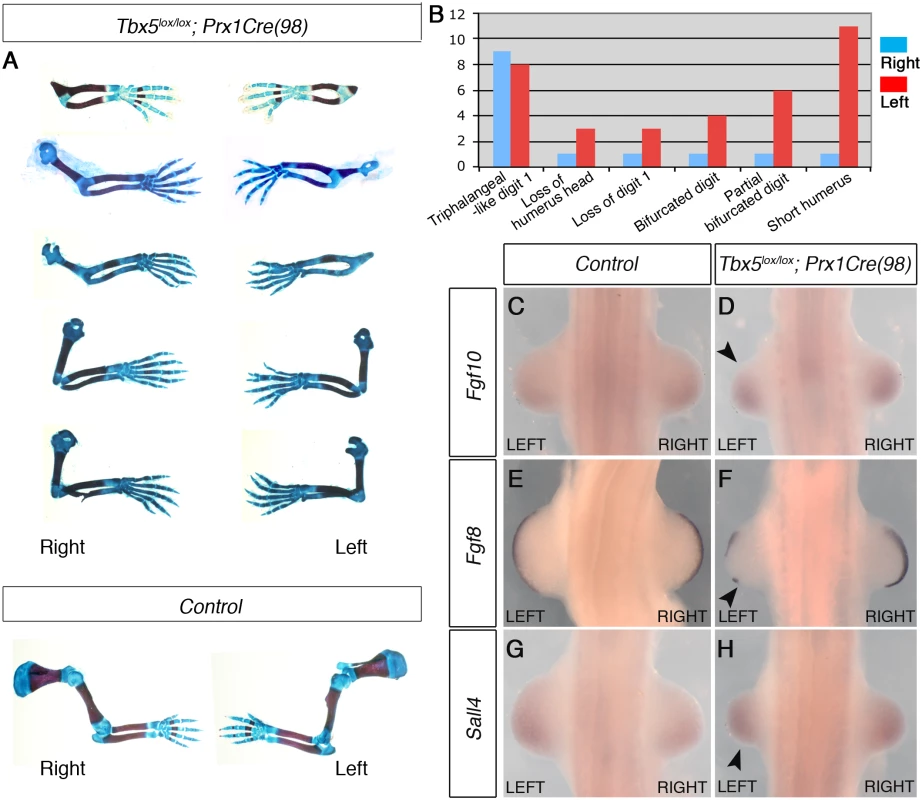
A, a selection of 5 pairs of the left and right forelimbs of E17.5 Tbx5lox/lox;Prx1Cre(98) mutants and control embryos illustrating the range in forelimb defect severity. Mutant forelimbs are shown in descending order of severity from top to bottom. B, Tabulation of forelimb defects observed in 18 mutant embryos. Defects are observed more frequently in the left than the right forelimbs. C-H, WISH analysis of E10.5 embryos. Left forelimbs are smaller than right forelimbs (n = 12) (arrowheads). Fgf8 expression is disrupted in left AER (F) while an obvious reduction of Fgf10 (D) and Sall4 (H) is not observed. Together, using two different genetic strategies, we demonstrate that bilateral hypomorphic levels of Tbx activity cause left-biased forelimb defects. These results reveal an inherent asymmetric difference in the left and right forelimb LPM and that an optimal level of Tbx activity is required to buffer the asymmetric difference to ensure bilateral, symmetric limb outgrowth.
Limb-forming LPM asymmetry is linked to the left-right pathway
We tested if the inherent asymmetric difference in the left and right limb-forming LPM is downstream of the axial left-right pathway. The establishment of the left-right axis is critical for the asymmetric patterning of the visceral organs and the disruption of the left-right pathway can cause situs inversus, a reversal of internal organ asymmetry [17–19]. Previous studies have demonstrated that the biased asymmetry of genetic and drug-induced forelimb defects is subject to mirror reversal in situs inversus embryos [20,21]. Therefore, we tested whether the left bias of forelimb defects in Tbx5lox/lox;Prx1Cre;Prx1-Tbx mutants is reversed in situs inversus embryos. We used homozygous mutants of INV as the loss of this gene induces situs inversus rather than randomising situs as reported in the IV mutant [7,22,23]. We generated 30 INV/INV embryos, 21 of which show situs solitus and the other 9 embryos show situs inversus, indicating that in our hands and in common with other reports the penetrance of situs inversus in INV/INV embryos is lower than originally reported [7,24].
Situs solitus, or normal organ asymmetry, in E14.5 Tbx5lox/lox;Prx1Cre;Prx1-Tbx mutants is shown by the left-sided position of the heart (black asterisk, Fig 5A). In this example, the left forelimb is more severely affected (3 digits) than the right forelimb (4 digits) (red arrow, Fig 5B). Because of the low penetrance of situs inversus in INV homozygous mutants we observed, we were only able to obtain 3 Tbx5lox/lox;Prx1Cre;Prx1-Tbx;INV/INV mutants that show situs inversus, indicated by right-sided heart position (black asterisk, Fig 5C). In all embryos, the right limb is more severely affected (Figs 5D, S3 and S2 Table). In the embryo shown in Fig 5D the right forelimb has 3 digits with the most anterior bifurcated (red arrow) whereas the left forelimb has 4 digits.
Fig. 5. Tbx5lox/lox;Prx1Cre;Prx1-Tbx;INV/INV mutants with situs inversus have right biased asymmetric forelimb defects. 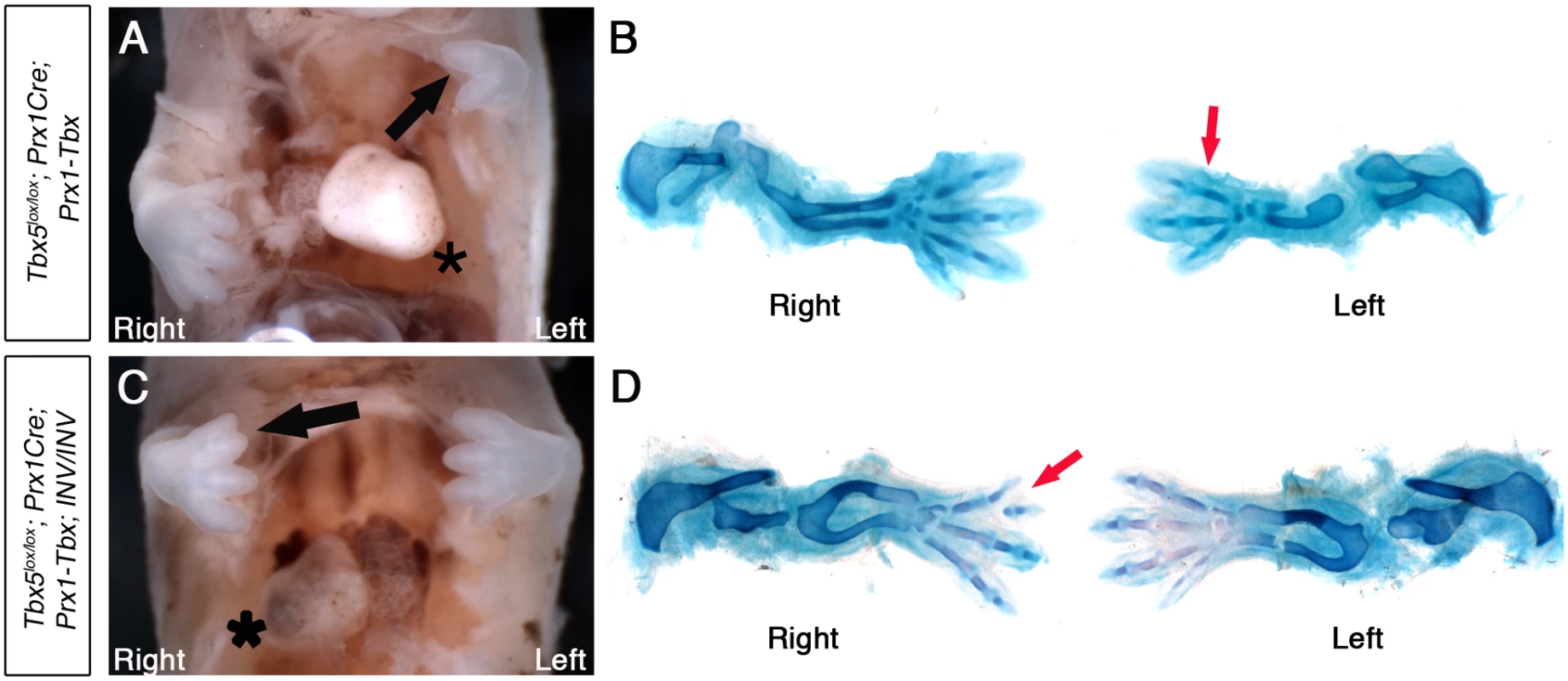
A, Tbx5lox/lox;Prx1Cre;Prx-Tbx mutant embryo at E14.5. The heart is on left (asterisk). B, Skeletal preparation. Right forelimb has 4 digits, while left forelimb has 3 digits (red arrow). C, Tbx5lox/lox;Prx1Cre;Prx1-Tbx;INV/INV mutant embryo at E14.5. The heart is on the right (asterisk), indicating situs inversus. The right forelimb is more severely affected than the left (black arrow). D, Skeletal preparation. The left forelimb has 4 digits. In contrast, the right forelimb is more severely affected having 3 digits with the most anterior bifurcated (red arrow). This indicates that the left-bias of the forelimb defects caused by hypomorphic levels of the Prx1-Tbx transgene are reversed in embryos with situs inversus and embryos now display a right-sided bias in forelimb defects.
Tbx5 ensures symmetrical forelimb formation independent of Fgf10
Since Fgf10 is a direct target of Tbx5 during forelimb initiation, we tested whether this factor is mediating the mechanism to buffer asymmetry between the left and right forelimb-forming regions. If so, optimal levels of Fgf10 in the Tbx5lox/lox;Prx1Cre; Prx1-Tbx mutant background would be sufficient to rescue bilaterally symmetric limb formation.
To carry out this assay, we used a Cre-inducible Fgf10-expressing line, Z/EGFgf10 (see Materials and Methods). By crossing with the Prx1Cre transgenic, transgene-derived Fgf10 can be expressed throughout the forelimb-forming region. First we tested the ability of the Z/EGFgf10 transgenic to rescue Fgf10 mutant phenotypes. Fgf10-/- null mutants lack all forelimb skeletal elements as the establishment of the positive feedback loop of Fgf10 and Fgf8 is required for limb bud formation and subsequent outgrowth [25] (S4D–S4F Fig). Fgf10 expression from the Z/EGFgf10 transgene, is able to fully rescue the forelimb defects in Fgf10-/- null mutants (n = 2/2) (S4G–S4I Fig and S2 Table), indicating the levels of Fgf10 produced by Z/EGFgf10 are sufficient to support normal limb formation.
We used the Z/EGFgf10 to test if Fgf10 expressed from this transgene is able to rescue the asymmetric defects observed in Tbx5lox/lox;Prx1Cre;Prx1-Tbx mutants. Tbx5lox/lox;Prx1Cre;Prx1-Tbx;Z/EGFgf10 mutants have milder forelimb outgrowth defects compared to Tbx5lox/lox;Prx1Cre;Prx1-Tbx (Fig 6A and 6B), consistent with previous studies showing that Fgf10 acts downstream of Tbx5 during forelimb initiation. Skeletal analysis of four mutants, however, demonstrates that some defects, including absent thumb, bifurcated digit, hypoplastic scapula and short humerus are still observed (Fig 6C–6E). Significantly, these defects are more severe and more frequently observed on the left side (n = 4/4) (S2 Table), indicating left-biased asymmetric differences are still present. These results demonstrate that in the presence of bilaterally symmetric, hypomorphic Tbx expression levels, raising levels of Fgf10 ligand (to a level sufficient to fully rescue limb formation in the Fgf10-/- null mutants) cannot rescue symmetrical forelimb outgrowth.
Fig. 6. Fgf10 expression at an optimal level in the forelimb LPM cannot rescue asymmetric defects of Tbx5lox/lox; Prx1Cre; Prx1-Tbx mutants. 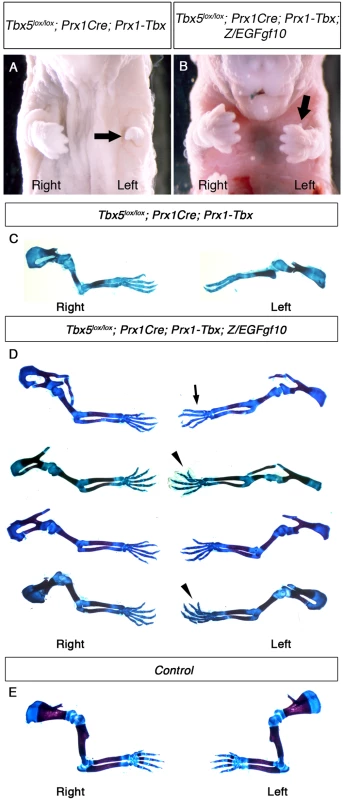
A-B, Ventral views of mutant embryos at E17.5. The left forelimb is severely truncated compared to the right in Tbx5lox/lox;Prx1Cre;Prx1-Tbx mutant (arrow) (A). Tbx5lox/lox;Prx1Cre;Prx1-Tbx;Z/EGFgf10 mutant lacks digit one in the left forelimb (arrow) (B). C-E, Skeletal preparation of Tbx5lox/lox;Prx1Cre;Prx1-Tbx (C), Tbx5lox/lox;Prx1Cre;Prx1-Tbx;Z/EGFgf10 (D) and control (E) forelimbs. Left forelimbs are more severely affected than right in both examples. Absent digit 1 (thumb) (arrow), bifurcated digit (arrowheads). Discussion
Here, we demonstrate that inherent asymmetry between the left and right LPM is buffered by threshold levels of Tbx5 that ensure bilaterally symmetric forelimb formation (Fig 7).
Fig. 7. Schematic representation of Tbx5 buffering the inherent LPM L/R asymmetry. 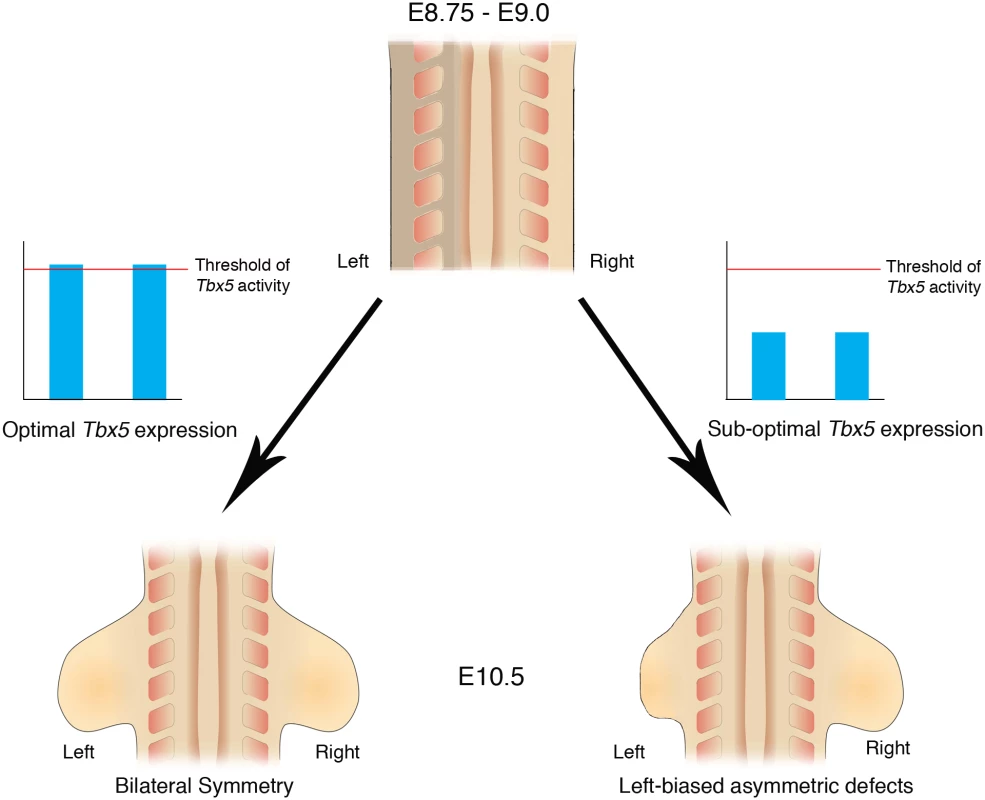
Bilateral optimal Tbx5 expression reaches above a threshold level that buffers the inherent asymmetry in the left and right LPM, which includes the future forelimb-forming regions. Bilateral, suboptimal Tbx5 expression levels that fail to reach threshold levels leads to abnormalities in limb formation and cannot buffer the inherent asymmetry in the LPM and as a result the left limb is more severely affected by lowered Tbx5 activity than the right limb. Bilaterally symmetric limb development is not the ground state
The simplest explanation for how bilateral symmetry in limb formation is achieved is by default and that the gene programmes controlling limb formation can operate with equal fidelity on the left and right sides of the embryo. Over the last 20 years however, it has become clear that the pathway that breaks L/R symmetry in the very early embryo and ultimately establishes the asymmetry in visceral organs has an impact on the LPM. Components of the L/R pathway, Nodal, Lefty and Pitx2 are expressed in the left LPM at stages prior to limb formation, however, the effects of these genes on bilateral symmetry of the limb have not been appreciated, previously. Our results indicate that bilaterally symmetric limb development is not the ground state but instead the ‘memory’ of L/R pathway genes expressed in the left LPM can interfere with the ability of LPM cells to establish Fgf10-Fgf8 positive feedback loop essential for limb outgrowth, and that threshold levels of Tbx5 are required to buffer this developmental history. Furthermore, we demonstrate that one direct target of Tbx5, Fgf10, does not have the ability to mask the bilateral asymmetry, suggesting that Tbx5 carries out this buffering role upstream of Fgf10.
Other bilaterally symmetric structures in the embryo, such as the somites, also need to override the influence of early asymmetric gene expression associated with the L/R pathway. RA signalling is essential to synchronize somite formation between left and right sides by antagonizing Fgf8 to ensure symmetric FGF signalling activity on the both sides of embryos [26–28]. Together with our study, these results suggest that buffering the effects of the left-right pathways is a general strategy employed during the formation of bilaterally symmetric structures in the developing embryo. Significantly, however, the mechanisms by which bilateral symmetry is achieved are different in different structures, such as the limbs and somites. During somitogenesis, the L/R asymmetric signals are counter-balanced by asymmetric expression of Nr2f2, a nuclear receptor that promotes the transcriptional activity of the RA signaling pathway [29]. In contrast, in the limb, bilaterally symmetric Tbx5 expression at levels above a threshold are able to buffer the effects of the L/R pathway. A buffering mechanism that operates through bilaterally symmetric gene expression may be more robust than one that relies on establishing asymmetric gene expression to counterbalance a ‘historic’ gene expression asymmetry.
Molecular mechanisms buffering bilateral limb asymmetry
While Tbx hypomorph mutants and HOS patients are unique in that they display left-biased forelimb defects, a few other mutant mice with oriented asymmetric forelimb outgrowth defects have been reported [21,30–32]. Retinaldehyde dehydrogenase 2 (Raldh2) mutants rescued by maternal administration of RA display more severely affected left forelimbs than right [32], similar to Tbx hypomorph mutants. Furthermore, the defects in these rescued forelimbs include triphalangeal thumbs, lack of thumbs and loss of humerus as seen in Tbx hypomorph mutants. Since RA signalling directly regulates Tbx5 transcription [33], Tbx5 expression may be at hypomorphic levels in these rescued embryos, which would explain the left-biased forelimb defects.
Two transgene insertion mutant mice, legless and footless, display right-biased forelimb defects [21,30,31]. The Legless mutation causes hypomorphic expression of Sp8, a zinc finger transcription factor expressed in the AER [34], and the footless mutation causes hypomorphic expression of R-spondin 2, a secreted protein also expressed in the AER [35]. Since these genes are both co-expressed in AER and Sp8 is required for R-spondin2 expression, hypomorphic levels of Sp8 in the legless mutant is likely the cause of hyopmorphic levels of R-spondin2. Although how hypomorphic levels of R-spondin2 causes the forelimb asymmetry is not understood, this mutant demonstrates that bilateral disruption of the Wnt pathway can produce right-biased asymmetric forelimb defects. Further studies will reveal how input from different signaling pathways is buffered to ensure that their ultimate output, in the form of limb structures is bilaterally symmetric.
Asymmetrical limb defects can be induced by teratogens such as cadmium, acetazolamide, MNNG and acetoxymethyl-methylnitrosamine [36–41]. The mechanisms causing these asymmetrical defects are not understood however. The affected mice do not show anterior biased limb defects or other clinical features of HOS as seen in Tbx5 hypomorph mutants suggesting these teratogens are not acting by disrupting the Tbx5 pathway or act on only one component of what could be several different pathways regulated by Tbx5.
Left-bias severity of HOS limb defects
The consensus is that HOS is caused by haploinsufficiency rather than pathogenic mutations producing dominant-negative acting forms of the protein. HOS pathogenic mutations have been identified throughout the coding sequence and are not exclusively localized to either the DNA binding T domain or regions thought to interact with cofactors. Functional studies of mutations that lie within the T domain show these forms of the protein have reduced DNA binding ability and diminished binding with interaction partners consistent with these mutations leading to reduced transcriptional activation of target genes leading to functional haploinsufficiency [42].
HOS is unique among congenital abnormalities affecting the limb in that a consistent feature of the clinical presentation is the left-biased severity in the defects [3]. Our results show that the asymmetry in the severity of limb phenotype results as a consequence of an effect on the very earliest events of limb bud formation when the precursors of limb structures are recruited. These events are sensitive to the inherent asymmetry in the limb-forming LPM when sub-threshold, hypomorphic levels of Tbx proteins are present. Our analysis of the penetrance in genetically defined HOS patients and our observations in our HOS mouse model indicate left-biased defects present with almost 100% penetrance. Our results provide an explanation for origins of the left-biased severity of HOS defects and confirm it as key a diagnostic criterion to indicate HOS.
Shoulder/girdle involvement is a cardinal feature of HOS and the humerus can also be affected and this is reflected in our mouse mutants. Overall, we observed more severe shoulder and humerus defects in our mouse mutant series than have been described in HOS patients and this may reflect the generally more severe severity range in limb defects we observe in our mouse mutant compared to HOS. In clinical descriptions of HOS, greater emphasis has been placed on description of more distal structures such as radius and hand that are expected to have greater impact on patient limb function. Shoulder girdle and humerus abnormalities are often poorly described making it difficult to compare to the defects we see in the mouse.
Materials and Methods
Ethics statement
The electronic patient record at Great Ormond Street was reviewed and approved by the hospital clinical audit committee (ref 1337). Data were extracted into a linked anonymised database. Animal work was carried out under an appropriate Home Office licence and approved by the local ethics panel (AWERB).
Embryos and mouse lines
Mice were staged according to Kaufman 2001 [43]. Noon on the day a vaginal plug was observed was taken to be 0.5 embryonic days (E) of development. Prx1Cre [9], Prx1Cre(98) [16], conditional Tbx5 [44], INV [7], Rosa26RLacZ [45] and the chimeric transgenic lines [8] have all been described previously. The Z/EGFgf10 line was produced using the Z/EG backbone [46] provided by C. Lobe. Briefly a mouse Fgf10 cDNA was inserted into the Z/EG construct upstream of the IRESeGFP cassette. The full construct was transfected into ES cells. Cells that had successfully integrated the construct were selected with G418, screened for single integration events by Southern Blott and surviving clones were used to generate chimeras from which founder animals were derived.
Whole mount in situ hybridisation
Whole mount in situ hybridisation protocols were carried out as previously described [47]. A minimum of 4 mutant embryos were analysed for each genotype at each stage. mFgf10 [48] and mFgf8 [49] probes were reported previously. We used a full-length mouse Sall4 clone as a probe template.
Skeletal preparations
E14.5 and E17.5 embryo skeletons were stained using alizarin red (for bone) and alcian blue (for cartilage) [50].
Quantitative PCR
RNA was extracted from the 10 forelimb buds of embryos according to the manufacturer’s instructions using the RNeasy mini kit (Qiagen) and cDNA was subsequently prepared by using SuperScript III Reverse Transcriptase (Invitrogen).
Primers were generated using the PrimerBlast application available online (http://www.ncbi.nlm.nih.gov/tools/primer-blast). The following primer pairs were used: Cre recombinase Fwd 5’ - GAACGAAAACGCTGGTTAGC -3’ Rev 5’ - CCCGGC AAAACAGGTAGTTA -3’, Prx1-Tbx Fwd 5’ - GAGACAGCTTTTATCGCTGTG -3’ Rev 5’ - CATCGCTGCCCCGGAATCCCT -3’, GAPDH Fwd 5’ - TGTCAGCAATGCATCCTGCA -3’ Rev 5’ - CCGTTCAGCTCTGGGATG AC -3’. The two-tailed Student’s t-test was used for statistical analysis.
Western blot analysis
Wild type and mutant forelimb buds (E10.5) were homogenized in RIPA buffer. A rabbit polyclonal Tbx5 antibody raised against a peptide spanning an N-terminal region of the protein was used (details are available on request). Densitometry was performed by scanning the original films and then analyzing the bands with ImageJ (NIH).
Analysis of HOS patients
The electronic patient record at Great Ormond Street was searched for the terms ‘Tbx5’ and ‘Holt-Oram’. Data on the cardiac, genetic and upper limb changes of affected patients were extracted by G.M. into a linked anonymised database. Patients without recorded Tbx5 mutation analysis were cross-checked with the national reference laboratory in Nottingham, and missing results retrieved. This review was approved by the hospital clinical audit committee (ref 1337).
Supporting Information
Zdroje
1. Hamada H, Tam PP (2014) Mechanisms of left-right asymmetry and patterning: driver, mediator and responder. F1000Prime Rep 6 : 110. doi: 10.12703/P6-110 25580264
2. Summerbell D, Wolpert L (1973) Precision of development in chick limb morphogenesis. Nature 244 : 228–230. 4583096
3. Newbury-Ecob RA, Leanage R, Raeburn JA, Young ID (1996) Holt-Oram syndrome: a clinical genetic study. J Med Genet 33 : 300–307. 8730285
4. Barnett P, Postma A (2014) Molecular Genetics of Holt–Oram Syndrome. eLS.
5. Rallis C, Bruneau BG, Del Buono J, Seidman CE, Seidman JG, et al. (2003) Tbx5 is required for forelimb bud formation and continued outgrowth. Development 130 : 2741–2751. 12736217
6. Minguillon C, Del Buono J, Logan MP (2005) Tbx5 and Tbx4 are not sufficient to determine limb-specific morphologies but have common roles in initiating limb outgrowth. Dev Cell 8 : 75–84. doi: 10.1016/j.devcel.2004.11.013 15621531
7. Yokoyama T, Copeland NG, Jenkins NA, Montgomery CA, Elder FF, et al. (1993) Reversal of left-right asymmetry: a situs inversus mutation. Science 260 : 679–682. 8480178
8. Minguillon C, Gibson-Brown JJ, Logan MP (2009) Tbx4/5 gene duplication and the origin of vertebrate paired appendages. Proc Natl Acad Sci U S A 106 : 21726–21730. doi: 10.1073/pnas.0910153106 19995988
9. Logan M, Martin JF, Nagy A, Lobe C, Olson EN, et al. (2002) Expression of Cre Recombinase in the developing mouse limb bud driven by a Prxl enhancer. Genesis 33 : 77–80. doi: 10.1002/gene.10092 12112875
10. Bayne LG, Klug MS (1987) Long-term review of the surgical treatment of radial deficiencies. J Hand Surg Am 12 : 169–179. 3559066
11. Blauth W (1967) [The hypoplastic thumb]. Arch Orthop Unfallchir 62 : 225–246. 4874597
12. Manske PR, McCarroll HR Jr., James M (1995) Type III-A hypoplastic thumb. J Hand Surg Am 20 : 246–253. doi: 10.1016/S0363-5023(05)80018-8 7775762
13. Smith P (2002) Lister’s The Hand: Diagnosis and Indications (4th Ed). London: Churchill-Livingstone.
14. Harvey SA, Logan MP (2006) sall4 acts downstream of tbx5 and is required for pectoral fin outgrowth. Development 133 : 1165–1173. doi: 10.1242/dev.02259 16501170
15. Koshiba-Takeuchi K, Takeuchi JK, Arruda EP, Kathiriya IS, Mo R, et al. (2006) Cooperative and antagonistic interactions between Sall4 and Tbx5 pattern the mouse limb and heart. Nat Genet 38 : 175–183. doi: 10.1038/ng1707 16380715
16. Hasson P, DeLaurier A, Bennett M, Grigorieva E, Naiche LA, et al. (2010) Tbx4 and tbx5 acting in connective tissue are required for limb muscle and tendon patterning. Dev Cell 18 : 148–156. doi: 10.1016/j.devcel.2009.11.013 20152185
17. Boettger T, Wittler L, Kessel M (1999) FGF8 functions in the specification of the right body side of the chick. Curr Biol 9 : 277–280. 10074453
18. Norris DP, Robertson EJ (1999) Asymmetric and node-specific nodal expression patterns are controlled by two distinct cis-acting regulatory elements. Genes Dev 13 : 1575–1588. 10385626
19. Varlet I, Collignon J, Norris DP, Robertson EJ (1997) Nodal signaling and axis formation in the mouse. Cold Spring Harb Symp Quant Biol 62 : 105–113. 9598342
20. Kocher-Becker U VE, Kocher W. (1991) Correlation of asymmetries of mutant polydactyly and mutant situs. Teratology 44.
21. Schreiner CM, Scott WJ Jr., Supp DM, Potter SS (1993) Correlation of forelimb malformation asymmetries with visceral organ situs in the transgenic mouse insertional mutation, legless. Dev Biol 158 : 560–562. doi: 10.1006/dbio.1993.1214 8344471
22. Layton WM Jr. (1976) Random determination of a developmental process: reversal of normal visceral asymmetry in the mouse. J Hered 67 : 336–338. 1021593
23. Supp DM, Brueckner M, Kuehn MR, Witte DP, Lowe LA, et al. (1999) Targeted deletion of the ATP binding domain of left-right dynein confirms its role in specifying development of left-right asymmetries. Development 126 : 5495–5504. 10556073
24. Okada Y, Nonaka S, Tanaka Y, Saijoh Y, Hamada H, et al. (1999) Abnormal nodal flow precedes situs inversus in iv and inv mice. Mol Cell 4 : 459–468. 10549278
25. Sekine K, Ohuchi H, Fujiwara M, Yamasaki M, Yoshizawa T, et al. (1999) Fgf10 is essential for limb and lung formation. Nat Genet 21 : 138–141. doi: 10.1038/5096 9916808
26. Kawakami Y, Raya A, Raya RM, Rodriguez-Esteban C, Izpisua Belmonte JC (2005) Retinoic acid signalling links left-right asymmetric patterning and bilaterally symmetric somitogenesis in the zebrafish embryo. Nature 435 : 165–171. doi: 10.1038/nature03512 15889082
27. Sirbu IO, Duester G (2006) Retinoic-acid signalling in node ectoderm and posterior neural plate directs left-right patterning of somitic mesoderm. Nat Cell Biol 8 : 271–277. doi: 10.1038/ncb1374 16489341
28. Vermot J, Gallego Llamas J, Fraulob V, Niederreither K, Chambon P, et al. (2005) Retinoic acid controls the bilateral symmetry of somite formation in the mouse embryo. Science 308 : 563–566. doi: 10.1126/science.1108363 15731404
29. Vilhais-Neto GC, Maruhashi M, Smith KT, Vasseur-Cognet M, Peterson AS, et al. (2010) Rere controls retinoic acid signalling and somite bilateral symmetry. Nature 463 : 953–957. doi: 10.1038/nature08763 20164929
30. Bell SM, Schreiner CM, Hess KA, Anderson KP, Scott WJ (2003) Asymmetric limb malformations in a new transgene insertional mutant, footless. Mech Dev 120 : 597–605. 12782276
31. McNeish JD, Scott WJ, Potter SS (1988) Legless, a novel mutation found in PHT1-1 transgenic mice. Science 241 : 837–839. 3406741
32. Niederreither K, Vermot J, Schuhbaur B, Chambon P, Dolle P (2002) Embryonic retinoic acid synthesis is required for forelimb growth and anteroposterior patterning in the mouse. Development 129 : 3563–3574. 12117807
33. Nishimoto S, Wilde SM, Wood S, Logan MP (2015) RA Acts in a Coherent Feed-Forward Mechanism with Tbx5 to Control Limb Bud Induction and Initiation. Cell Rep 12 : 879–891. doi: 10.1016/j.celrep.2015.06.068 26212321
34. Bell SM, Schreiner CM, Waclaw RR, Campbell K, Potter SS, et al. (2003) Sp8 is crucial for limb outgrowth and neuropore closure. Proc Natl Acad Sci U S A 100 : 12195–12200. doi: 10.1073/pnas.2134310100 14526104
35. Bell SM, Schreiner CM, Wert SE, Mucenski ML, Scott WJ, et al. (2008) R-spondin 2 is required for normal laryngeal-tracheal, lung and limb morphogenesis. Development 135 : 1049–1058. doi: 10.1242/dev.013359 18256198
36. Barr M Jr. (1973) The teratogenicity of cadmium chloride in two stocks of Wistar rats. Teratology 7 : 237–242. doi: 10.1002/tera.1420070304 4268328
37. Bochert G, Platzek T, Blankenburg G, Wiessler M, Neubert D (1985) Embryotoxicity induced by alkylating agents: left-sided preponderance of paw malformations induced by acetoxymethyl-methylnitrosamine in mice. Arch Toxicol 56 : 139–150. 3977594
38. Inouye M, Murakami U (1978) Teratogenic effect of N-methyl-N'-nitro-N-nitrosoguanidine in mice. Teratology 18 : 263–267. doi: 10.1002/tera.1420180213 715730
39. Layton WM Jr. Hallesy DW (1965) Deformity of Forelimb in Rats: Association with High Doses of Acetazolamide. Science 149 : 306–308. 14300527
40. Layton WM Jr.Layton MW (1979) Cadmium induced limb defects in mice: strain associated differences in sensitivity. Teratology 19 : 229–235. doi: 10.1002/tera.1420190213 473073
41. Levin M (1997) Left-right asymmetry in vertebrate embryogenesis. Bioessays 19 : 287–296. doi: 10.1002/bies.950190406 9136626
42. Boogerd CJ, Dooijes D, Ilgun A, Mathijssen IB, Hordijk R, et al. (2010) Functional analysis of novel TBX5 T-box mutations associated with Holt-Oram syndrome. Cardiovasc Res 88 : 130–139. doi: 10.1093/cvr/cvq178 20519243
43. Kaufman MH (2001) The Atlas of Mouse Development, 2nd edn. Cambridge, UK: Academic Press.
44. Bruneau BG, Nemer G, Schmitt JP, Charron F, Robitaille L, et al. (2001) A murine model of Holt-Oram syndrome defines roles of the T-box transcription factor Tbx5 in cardiogenesis and disease. Cell 106 : 709–721. 11572777
45. Soriano P (1999) Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat Genet 21 : 70–71. doi: 10.1038/5007 9916792
46. Novak A, Guo C, Yang W, Nagy A, Lobe CG (2000) Z/EG, a double reporter mouse line that expresses enhanced green fluorescent protein upon Cre-mediated excision. Genesis 28 : 147–155. 11105057
47. Riddle RD, Johnson RL, Laufer E, Tabin C (1993) Sonic hedgehog mediates the polarizing activity of the ZPA. Cell 75 : 1401–1416. 8269518
48. Sala FG, Curtis JL, Veltmaat JM, Del Moral PM, Le LT, et al. (2006) Fibroblast growth factor 10 is required for survival and proliferation but not differentiation of intestinal epithelial progenitor cells during murine colon development. Dev Biol 299 : 373–385. doi: 10.1016/j.ydbio.2006.08.001 16956603
49. Mahmood R, Bresnick J, Hornbruch A, Mahony C, Morton N, et al. (1995) A role for FGF-8 in the initiation and maintenance of vertebrate limb bud outgrowth. Curr Biol 5 : 797–806. 7583127
50. McLeod MJ (1980) Differential staining of cartilage and bone in whole mouse fetuses by alcian blue and alizarin red S. Teratology 22 : 299–301. doi: 10.1002/tera.1420220306 6165088
Štítky
Genetika Reprodukční medicína
Článek Epistasis and Entropy
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2016 Číslo 12- Akutní intermitentní porfyrie
- Prenatální expozice ftalátům a anogenitální vzdálenost u novorozenců
- Je „freeze-all“ pro všechny? Odborníci na fertilitu diskutovali na virtuálním summitu
- Srdeční frekvence embrya může být faktorem užitečným v předpovídání výsledku IVF
- Kryokonzervace lidských embryí – preferovaná metoda asistované reprodukce
-
Všechny články tohoto čísla
- Bringing Editors to Preprint Servers
- Epistasis and Entropy
- Middle Age Has Its Advantages
- Craniofacial Ciliopathies and the Interpretation of Hedgehog Signal Transduction
- Comment on " Is Dispensable for Mouse Development"
- Shared Information between Residues Is Sufficient to Detect Pairwise Epistasis in a Protein
- A Hox-Embedded Long Noncoding RNA: Is It All Hot Air?
- Deep Reads: Favorites from a Few Different Shelves
- Buffers Inherent Left/Right Asymmetry Ensuring Symmetric Forelimb Formation
- Intracellular Zn(II) Intoxication Leads to Dysregulation of the PerR Regulon Resulting in Heme Toxicity in
- Mutations in the Heme Exporter Cause Sensory Neurodegeneration with Loss of Pain Perception
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Intracellular Zn(II) Intoxication Leads to Dysregulation of the PerR Regulon Resulting in Heme Toxicity in
- Craniofacial Ciliopathies and the Interpretation of Hedgehog Signal Transduction
- Mutations in the Heme Exporter Cause Sensory Neurodegeneration with Loss of Pain Perception
- Shared Information between Residues Is Sufficient to Detect Pairwise Epistasis in a Protein
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



