-
Články
- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Genetic compensation: A phenomenon in search of mechanisms
Several recent studies in a number of model systems including zebrafish, Arabidopsis, and mouse have revealed phenotypic differences between knockouts (i.e., mutants) and knockdowns (e.g., antisense-treated animals). These differences have been attributed to a number of reasons including off-target effects of the antisense reagents. An alternative explanation was recently proposed based on a zebrafish study reporting that genetic compensation was observed in egfl7 mutant but not knockdown animals. Dosage compensation was first reported in Drosophila in 1932, and genetic compensation in response to a gene knockout was first reported in yeast in 1969. Since then, genetic compensation has been documented many times in a number of model organisms; however, our understanding of the underlying molecular mechanisms remains limited. In this review, we revisit studies reporting genetic compensation in higher eukaryotes and outline possible molecular mechanisms, which may include both transcriptional and posttranscriptional processes.
Published in the journal: . PLoS Genet 13(7): e32767. doi:10.1371/journal.pgen.1006780
Category: Review
doi: https://doi.org/10.1371/journal.pgen.1006780Summary
Several recent studies in a number of model systems including zebrafish, Arabidopsis, and mouse have revealed phenotypic differences between knockouts (i.e., mutants) and knockdowns (e.g., antisense-treated animals). These differences have been attributed to a number of reasons including off-target effects of the antisense reagents. An alternative explanation was recently proposed based on a zebrafish study reporting that genetic compensation was observed in egfl7 mutant but not knockdown animals. Dosage compensation was first reported in Drosophila in 1932, and genetic compensation in response to a gene knockout was first reported in yeast in 1969. Since then, genetic compensation has been documented many times in a number of model organisms; however, our understanding of the underlying molecular mechanisms remains limited. In this review, we revisit studies reporting genetic compensation in higher eukaryotes and outline possible molecular mechanisms, which may include both transcriptional and posttranscriptional processes.
Introduction
Genetic robustness is the ability of a living organism to maintain its viability and fitness despite genetic variations, including perturbations. Genetic perturbations play an important role in evolution; however, organisms require buffering systems to ensure similar developmental outcomes despite minor differences in genetic makeup or environmental conditions, a process known as robustness or canalization [1, 2]. In 1932, dosage compensation was reported as the first example of genetic robustness. Male fruit flies were reported to have a twofold increase in transcription from their single X chromosome, resulting in the same gene expression levels as females with two active X chromosomes [3, 4]. In contrast, in mammals, females undergo inactivation of one of their X chromosomes through heterochromatization, allowing for similar developmental outcomes in both sexes [5–7]. The concept of genetic robustness was further supported by several recent studies: for example, only 20% of the protein-coding genes in yeast were reported to be essential for growth in laboratory conditions [8], and a lack of phenotype was reported for several mouse [9], zebrafish [10], and Arabidopsis [11] mutants.
Genetic robustness may arise from redundant genes, whereby the loss of one gene may be compensated by another with overlapping functions and expression pattern, as reported for several mutants in a range of model organisms [12–19] (reviewed in [20]). Another form of robustness arises from tightly regulated cellular networks including metabolic, signaling, and transcriptional networks. Perturbation of a particular gene’s function in a network may alter the expression of other genes within the same network, thereby maintaining cellular wellness [21, 22]. Additionally, in response to a gene knockout, organisms such as yeast may accumulate mutations in one or more genes modulating the affected pathway, thereby partially or fully rescuing the final outcome [23, 24].
While the above-mentioned modes of genetic robustness may occur as a result of the loss of function of a specific protein, a number of studies suggest a different form of genetic robustness, one that is triggered upstream of protein function (hereafter referred to as genetic compensation or transcriptional adaptation [Table 1]) [25–27]. The increasing use of recent advances in reverse genetic tools have revealed phenotypic differences between knockouts (i.e., mutants) and knockdowns (e.g., antisense-, including morpholino [MO]-, treated animals) in a number of model systems including Arabidopsis [28–30], mouse [31–34], Drosophila [35], zebrafish [10, 36], and human cell lines [37–39]. While some studies attributed these phenotypic differences to toxicity or off-target effects of the knockdown reagents [40–43] (reviewed in [44]), a recent study in zebrafish proposed gene expression changes and consequent compensation in mutant but not knockdown animals as the reason for the observed differences [25]. While knockdown of egfl7, an endothelial extracellular-matrix (ECM) gene, leads to severe vascular defects, most egfl7 mutants exhibit no obvious defects. This discrepancy was attributed at least partly to the upregulation of other ECM proteins, specifically Emilins, in egfl7 mutants but not antisense-injected embryos. Moreover, the authors observed minor or no vascular defects upon egfl7 MO injections into egfl7 mutants, indicating that the phenotypic differences are not due to MO toxicity. In addition, this study reported upregulation of vegfab mRNA levels in vegfaa mutant but not knockdown animals. While the mechanisms triggering the transcriptional adaptation response in vegfaa mutant animals remain unknown, the authors propose that it lies upstream of protein function, as overexpression of dominant-negative Vegfaa, which causes a vegfaa mutant-like phenotype, did not lead to an increase in vegfab mRNA levels. In this review, we focus on studies reporting transcriptional adaptation and/or genetic compensation in higher eukaryotes and outline possible underlying molecular mechanisms.
Genetic compensation in response to gene knockout is a widespread phenomenon
Upregulation of related genes following a gene knockout may be a direct consequence of the loss of protein function. For example, mice lacking the ribosomal gene Rpl22 show no defects in translation owing to the upregulation of its paralogue, Rpl22l1, the expression of which is normally inhibited by RPL22 [46]. Upregulation of related genes due to the loss of a negative feedback loop may be the first hypothesis to test when a mutant fails to show a phenotype, and a knockdown approach may help test it. For example, human RBL2 mutant T lymphocytes proliferate normally and exhibit normal immune function due to RBL1 upregulation, an upregulation also detected upon RBL2 knockdown in human breast cancer cell lines [47, 48], suggesting a negative feedback loop. Similarly, both knockouts and knockdowns of HDAC-1 lead to the upregulation of HDAC-2 in several human and mouse cell lines and tissues, and vice versa [49–51].
In contrast, lack of a compensatory response in knockdown animals compared to their corresponding mutants indicates that a trigger upstream of protein function is at play, perhaps the genomic lesion itself or the mutant mRNA (Table 2). For example, small interfering RNA (siRNA)-mediated depletion of TET1, an enzyme that converts 5-methylcytosine (5mC) to 5-hydroxymethylcytosine (5hmC), in mouse embryonic stem cells (mESCs) leads to a significant reduction in 5hmC levels and a loss of undifferentiated morphology; in contrast, Tet1 mutant mESCs exhibit only a slight decrease in 5hmC levels and maintain an undifferentiated morphology [52], suggesting possible compensation by the closely related enzyme, TET2, in mutant but not knockdown mESCs [53]. In addition, while knockdown of any of the three cyclin D family members was reported to inhibit proliferation in several cell lines [54–56], mice lacking a single isoform develop minimal defects, suggesting compensation by one of the other genes [57–59]. Indeed, knockout of two Cyclin D genes in mouse leads to the upregulation of the third Cyclin D gene. Accordingly, double knockout mice show minor phenotypes only in tissues that fail to upregulate the third Cyclin D gene [60]. In addition, mouse Cyclin D2 mutant B lymphocytes exhibit no obvious proliferative phenotype due to the upregulation of Cyclin D3 [61]. Furthermore, short hairpin RNA (shRNA)-mediated knockdown of Importinα5 was reported to inhibit neural differentiation of mESCs cells [62]; however, Importinα5 mutant mice display normal brain development, possibly due to the upregulation of IMPORTINα4 expression [63]. siRNA-mediated knockdown of Kindlin-2, which encodes an integrin coactivator, in mouse embryonic fibroblasts (MEFs) was reported to decrease INTEGRIN β1 activation and prevent INTERLEUKIN 1β–mediated increase in focal adhesion number [64]. However, Kindlin-2 mutant cells were able to form focal adhesions due to the upregulation of KINDLIN-1 [65].
Tab. 2. Examples of discrepancies between mutant and knockdown phenotypes. 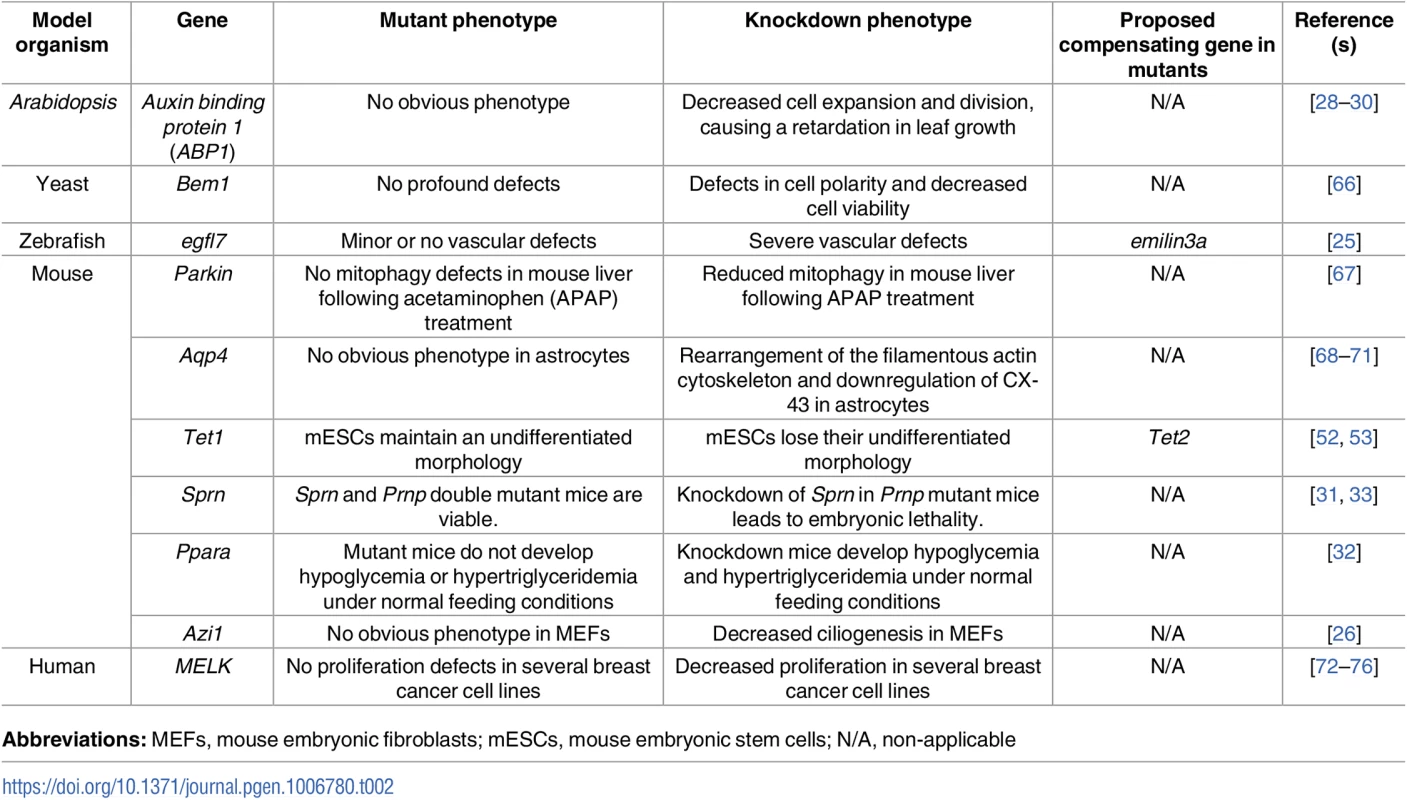
Abbreviations: MEFs, mouse embryonic fibroblasts; mESCs, mouse embryonic stem cells; N/A, non-applicable In another example, antisense-mediated knockdown of Tau was reported to inhibit axonal elongation in cultured neuronal cells [77, 78]. However, axonal elongation was not affected in cultured neurons from Tau mutants, possibly due to the upregulation of microtubule-associated protein 1A (MAP1A) [79]. Interestingly, such upregulation was not detected upon Tau knockdown in mouse oligodendrocytes [80]. Dystrophin mutant mice have been reported not to develop a severe muscular dystrophy phenotype due to the upregulation of a number of genes including that encoding the dystrophin-related protein UTROPHIN [81, 82]. Interestingly, UTROPHIN upregulation was not detected in Dystrophin knockdown mice [83].
Furthermore, β-Actin mutant mice were reported to display transcriptional upregulation of several other Actin genes, including γ-Actin and α-Actin [27, 84, 85]. Interestingly, restoration of β-Actin expression in β-Actin mutant MEFs did not lead to a reduction in the γ-Actin transcriptional upregulation response, implying that this transcriptional adaptation response is triggered upstream of β-ACTIN function [27]. In addition, γ-Actin knockout, but not knockdown, in MEFs leads to αsm-ACTIN upregulation [85]. Moreover, while siRNA-mediated depletion of the centrosomal protein AZI1 in MEFs leads to a significant decrease in ciliogenesis, MEFs derived from Azi1 mutant mice display no defects in ciliogenesis [26]. The authors also reported that Azi1 mutant MEFs were resistant to Azi1 siRNA, ruling out off-target effects of the siRNA and leading them to hypothesize the existence of a compensatory response in the mutant MEFs. Interestingly, this potential compensation is not observed during sperm flagella formation. This approach of testing the antisense reagent in mutant cells was subsequently used by Rossi et al. in zebrafish [25] and can be a powerful tool to identify cases of compensation in mutants versus nonspecific effects of the knockdown reagents.
Global versus conditional loss-of-function studies
Reduction or absence of a phenotype in several germline mutants compared to their conditional counterparts has been reported in a number of studies in mouse. For example, germline mutants for Pkm2 are viable and fertile [86]; however, conditional deletion of Pkm2 in MEFs limits nucleotide synthesis, leading to cell-cycle arrest [87]. Similarly, Sirt1 mutant mice have no obvious liver defects, while hepatocyte-specific Sirt1 mutant mice develop fatty liver [88]. Mice with conditional Fgfr3 deletion in chondrocytes exhibit more severe (and a higher incidence of) chondrona-like lesions compared to global mutant mice [89]. Moreover, conditional loss of the RETINOBLASTOMA (RB1) tumor suppressor enables cell-cycle reentry of quiescent primary MEFs, while quiescent MEFs derived from global Rb1 mutant animals are unable to reenter the cell cycle, due at least in part to the compensatory upregulation of p107 [90]. In addition, while Cd44 global mutant mice display only mild phenotypes [91, 92], keratinocyte-specific mutant mice display reduced epidermal stiffness and delayed wound healing, as well as reduced keratinocyte proliferation in response to 12-O-tetradecanoylphorbol-13-acetate [93]. While cell nonautonomous effects may underlie some of these discrepancies, an alternative hypothesis is that a compensatory network becomes established during germline maturation or embryonic development, allowing the organism to adapt to the mutation. Recent data in zebrafish, however, suggest that a mutation does not need to go through the germ line to induce a compensatory response [25], indicating that multiple mechanisms may underlie this process.
Mechanisms underlying the transcriptional adaptation response
Based on the observations reported thus far, one can identify at least two possible triggers of the transcriptional adaptation response: (1) the DNA lesion and (2) the mutant mRNA. We will first speculate about how each of these potential triggers might lead to transcriptional adaptation and then briefly review other potential triggers including some that might induce posttranscriptional adaptation.
DNA lesion as the trigger for the transcriptional adaptation response
This section will focus on the DNA lesion being the trigger for the transcriptional adaptation response and will mostly explore the potential role of epigenetic changes following DNA damage.
Following DNA damage, global chromatin reorganization and decondensation are detected [94, 95], actions mediated by several chromatin remodelers and histone-modifying enzymes (reviewed in [96]). One possibility is that in response to a mutation, global chromatin reorganization may positively affect chromatin accessibility around the compensating gene(s), thereby leading to increased expression levels (Fig 1A). Part of such a model is consistent with the process of dosage compensation in Drosophila where the male-specific lethal (MSL) proteins, together with other proteins, form a complex on the male X chromosome leading to H4K16 acetylation and subsequent induction of an open chromatin configuration, which is more accessible for transcription [97]. Along these lines, a Caenorhabditis elegans study attributed the incomplete penetrance of intestinal phenotypes in skn-1 mutants [98] to the high variability in expression of the compensating gene end-1 [99]. Interestingly, this variability in end-1 expression was attributed to differences in chromatin remodeling at loci controlling end-1 expression. It will thus be interesting to compare chromatin accessibility at the upregulated genes’ regulatory regions in wild-type, mutant, and knockdown samples.
Fig. 1. Proposed models of transcriptional adaptation. 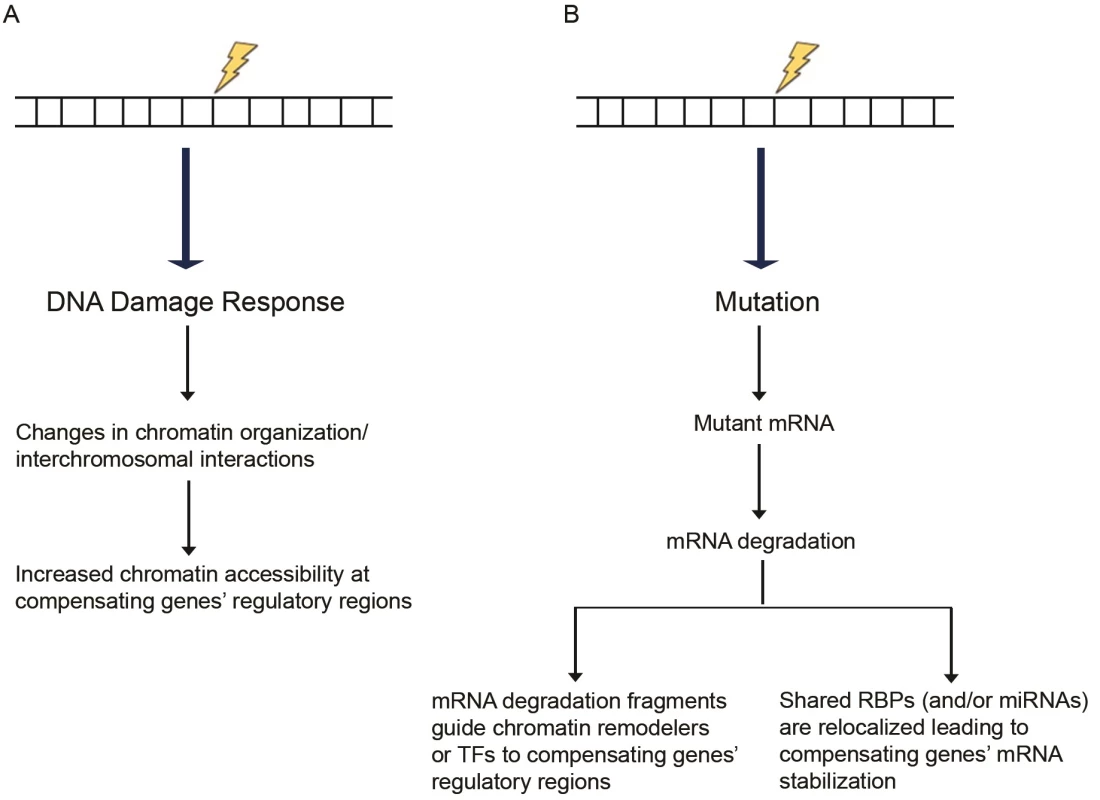
(A) DNA damage response can induce chromatin reorganization, increasing chromatin accessibility at the compensatory genes’ regulatory regions. (B) Mutations can lead to transcripts that are targeted for degradation through mRNA surveillance pathways. The resulting RNA fragments may trigger the compensatory response. As a secondary effect of the mutated gene’s mRNA degradation, RBPs or miRNAs normally acting on the mutated as well as the compensating genes’ mRNAs become more available to exert their stabilizing effects on the compensating genes’ mRNAs. Abbreviations: miRNAs, microRNAs; RBPs, RNA-binding proteins; TFs, transcription factors. Chromatin reorganization may be accompanied by changes in DNA looping and nuclear organization [100], which may also affect gene expression. Interchromosomal interactions are well documented (reviewed in [101]), and different kinds of stress, such as temperature, have been shown to increase interchromosomal interactions in Drosophila [102]. DNA damage-induced stress could similarly lead to modifications in interchromosomal interactions, including those between the mutated gene and certain other loci leading to specific gene upregulation. Chromosome-capture studies in wild-type and mutant samples to identify changes in interchromosomal interactions may help test this model.
Leading to a different potential model, a number of studies have reported the generation of small non-coding RNAs (ncRNAs) from regions spanning a double-stranded break (DSB), termed DSB-induced RNAs (diRNAs) [103, 104] (reviewed in [105]). The authors proposed that such diRNAs are essential for DNA-damage repair (DDR), possibly by acting as guides for chromatin remodelers or proteins important for DDR. Thus, diRNAs might also guide specific transcription factors (TFs) or chromatin remodelers to regulatory regions of compensating genes through homology-based interactions, leading to increased transcription. Such a model of small ncRNAs guiding specific transcription factors or chromatin remodelers to modulate gene expression is consistent with publications describing that roX1 and roX2 RNAs are essential for the dosage-compensation response in Drosophila males by guiding the assembly of the MSL protein complex on the X chromosome and subsequent histone modifications [106–108] (reviewed in [97]).
One question for these models that involve chromatin remodeling concerns the transmission of the transcriptional adaptation response to the next generation. Genomic imprinting via histone modification [109–111] (reviewed in [112]) is a possible mechanism.
In addition, induction of GADD45A expression following DNA damage has been reported to induce global DNA demethylation in HEK293T cells, leading to increased activation of methylation-silenced promoters [113]. Thus, one should also assess the methylation status of regulatory regions of the upregulated genes. However, to our knowledge, no links have been established to date between DNA lesions and changes in DNA methylation patterns at specific (i.e., compensating) loci.
Since all these models are based on DNA lesions, it will be important to assess transcriptional adaptation after inducing different types of mutations. One would expect the upregulation of the same genes following all types of mutations, including non-deleterious ones.
Mutant mRNA as the trigger for the transcriptional adaptation response
This section will focus on the mutant mRNA being the trigger for the transcriptional adaptation response. After reviewing a few examples, we will focus specifically on how RNA fragmentation by different mRNA surveillance pathways could trigger such a response.
Mutations often lead to mRNAs with a premature termination codon (PTC), secondary structures that stall ribosomal translocation, or, less frequently, mRNAs that lack a stop codon. The presence of such mRNAs triggers the nonsense-mediated decay (NMD), no-go decay, or no-stop decay pathways, respectively, which results in mRNA degradation (reviewed in [114–116]). A recent study in zebrafish reported that two different mutations in the same exon of mt2 cause different degrees of phenotypic severity. Surprisingly, the mutant allele with the milder phenotype exhibited a higher degree of NMD. Antisense-mediated knockdown of the NMD pathway and consequent decrease in mutant mRNA degradation led to a more severe phenotype, consistent with the possibility that NMD triggers a compensatory response that decreases the severity of the mutant phenotype [117]. One hypothesis is that the RNA fragments resulting from the mRNA surveillance pathways function to regulate gene expression. While the current understanding in the field is that the mRNA surveillance pathways lead to processive mRNA degradation [114, 118], it is possible that short-lived and relatively rare degradation intermediates are present.
If the fragments are long enough, one can hypothesize that they act in a fashion similar to long noncoding RNAs (reviewed in [119]) and, for example, guide specific transcription factors or chromatin remodelers to the regulatory regions of compensating genes through homology-mediated base pairing (Fig 1B). Other studies have reported that injection of short (20–22 nt) RNA fragments from a specific mRNA leads to increased transcription of the corresponding locus [120, 121]. Mechanistically, the authors report that the injected sense RNA fragments can form double-stranded RNA (dsRNA) duplexes with short antisense transcripts normally produced from the locus. The resulting dsRNAs may then be utilized by the RNA interference (RNAi) machinery in an ARGONAUTE-dependent manner to induce chromatin modifications at the locus and increase euchromatin histone marks or decrease heterochromatin histone marks. Although the exact machinery underlying such dsRNA-induced epigenetic changes remains unknown, this model is consistent with several other studies reporting transcriptional activation through histone modification following targeting of dsRNA to the promoter region of various genes [122–125]. Previous analyses of the mouse and human transcriptome have identified several antisense transcripts that can participate in forming sense/antisense pairs [126–130]. Thus, one could hypothesize that RNA fragments act in a similar fashion and form dsRNA duplexes with antisense transcripts from the compensating loci, leading to transcriptional upregulation.
RNA-binding proteins (RBPs) can also regulate gene expression in a number of ways (reviewed in [131]), one of which is by increasing gene expression through stabilizing mRNAs [132]. The highly dynamic binding of RBPs is regulated by cellular conditions; therefore, regulating RBP interactions following genotoxic stress may be a mechanism for the cell to compensate for a lost gene. Along these lines, mRNAs that encode functionally related proteins tend to be coregulated by specific RBPs, forming what is known as RNA operons or RNA regulons [133–135] (reviewed in [136]). Thus, the mutant and compensating genes might be regulated by the same RBPs, and if the mutant mRNA is subjected to degradation or if its secondary structure is affected by the mutation (thereby affecting RBP binding), RBPs would become available to stabilize the compensatory genes’ mRNAs (Fig 1B).
Besides their well-known function in silencing gene expression [137], micro-RNAs (miRNAs) can enhance gene expression through several mechanisms. Although miRNAs normally target mRNAs, miRNA-373 was reported to bind promoter regions of CDH1 and CSDC2 in PC3 (a human prostate cancer cell line) cells and induce their expression through an unknown mechanism [138]. miRNAs can also increase the translation of certain mRNAs; for example, under amino acid starvation conditions, miRNA10a was reported to bind the 5′UTR of ribosomal protein mRNAs and enhance their translation [139]. miRNAs have multiple target mRNAs [140, 141], and, thus, if a mutation leads to mRNA degradation, the miRNAs targeting the affected gene will become available to modulate other targets (Fig 1B).
Since these models rely on the generation and potential degradation of mRNAs from the mutated locus, one would not expect upregulation of potentially compensating genes in the absence of active transcription of the mutant mRNA. It will thus be important to assess transcriptional adaptation in alleles where an mRNA is not produced.
Other potential mechanisms for the compensatory response
This brief section will focus on increased translational response following the mutational loss of specific genes and will evoke processes such as mRNA modifications and upstream open reading frames.
In response to stress (such as heat shock), pseudouridylation or N6-methylation of adenosines (m6A) was reported to be enriched on certain mRNAs, thereby increasing their stability or promoting their translation [142, 143] (reviewed in [144]). However, as is the case for DNA methylation, there has been no report thus far about mRNAs from selective loci being modified in this manner.
Upstream open reading frames (uORFs) are regulatory elements present in the 5’UTRs of around 50% of vertebrate mRNAs [145, 146]. They may act as translational repressors, as the translation of the uORFs can occur at the expense of that of the mRNA’s coding sequence [147, 148]. Under cellular stress conditions, there is a tendency to inhibit global translation by phosphorylating eIF2α, which then acts as a competitive inhibitor of the translation initiation factor eIF2B, thereby reducing translation reinitiation rates [149]. This mechanism may allow the increased translation of certain mRNAs under cellular stress. For example, the yeast transcription factor gene GCN4 has 4 uORFs and under normal conditions, the 4 uORFs are translated with less reinitiation at the main ORF. Under nutritional stress, the first uORF is translated efficiently; however, due to eIF2α phosphorylation, the remaining uORFs are poorly translated, and reinitiation only occurs at the main ORF, thereby increasing GCN4 production [150]. It is thus possible that certain gene mutations induce cellular stress, allowing for uORF skipping and increased translation of compensating genes. However, as is the case for the DNA and RNA methylation modifications mentioned above, it is not clear how specificity, in terms of selective proteins being upregulated, would arise.
Conclusion
Despite its role in maintaining an organism’s robustness, the molecular mechanisms underlying genetic compensation remain poorly understood. Here, we reviewed studies reporting genetic compensation in several higher eukaryotes, outlined potential underlying mechanisms, and proposed experiments that should help test these potential mechanisms. Studying epigenetic changes following DNA damage, a major difference between mutants and knockdowns, should allow a better understanding of why a compensatory response is triggered by knockout but not knockdown approaches. Moreover, we also proposed mRNA surveillance pathways, ncRNAs, uORFs, RBPs, and miRNAs as potential players in the compensatory response.
Recently, a study of more than 500,000 human genomes identified 13 individuals harboring disease-causing mutations in 8 different genes, with no reported clinical manifestation of the disease [151]. Other studies on Icelandic and British people identified complete gene knockouts in several apparently healthy individuals [152, 153]. While functional characterization of the identified alleles still remains to be completed, it is likely that genetic compensation underlies the lack of phenotype in individuals with severe mutant alleles. Moreover, several factors have been proposed to explain the concept of incomplete penetrance, including environmental factors, different genetic backgrounds, and different expression levels of modifier genes [154, 155]; however, this concept remains poorly understood, as a recent study reported that incomplete penetrance is even common in mice with the same genetic background [156]. We propose that incomplete penetrance may be due to compensatory responses being triggered in some individuals but not in others. Investigating the molecular mechanisms underlying genetic compensation may help us understand why some mutations cause disease while others do not. It might also lead to the development of more effective therapies that enhance an organism’s robustness to a mutation rather than correct its effect, e.g., increase the expression of the compensating gene(s) rather than correct the function of the defective gene.
Zdroje
1. Mather K. Genetical control of stability in development. Heredity. 1953;7(3):297–336.
2. Waddington CH. Canalization of Development and Genetic Assimilation of Acquired Characters. Nature. 1959;183(4676):1654–5. 13666847
3. Muller HJ, editor Further studies on the nature and causes of gene mutations. Proceedings of the 6th International Congress of Genetics; 1932.
4. Mukherjee AS, Beermann W. Synthesis of ribonucleic acid by the X-chromosomes of Drosophila melanogaster and the problem of dosage compensation. Nature. 1965;207(998):785–6. Epub 1965/08/14. 5885936.
5. Barr ML, Bertram EG. A morphological distinction between neurones of the male and female, and the behaviour of the nucleolar satellite during accelerated nucleoprotein synthesis. Nature. 1949;163(4148):676. Epub 1949/04/30. 18120749.
6. Heard E, Disteche CM. Dosage compensation in mammals: fine-tuning the expression of the X chromosome. Genes & development. 2006;20(14):1848–67. Epub 2006/07/19. doi: 10.1101/gad.1422906 16847345.
7. Lyon MF. Gene action in the X-chromosome of the mouse (Mus musculus L.). Nature. 1961;190 : 372–3. Epub 1961/04/22. 13764598.
8. Giaever G, Chu AM, Ni L, Connelly C, Riles L, Veronneau S, et al. Functional profiling of the Saccharomyces cerevisiae genome. Nature. 2002;418(6896):387–91. Epub 2002/07/26. doi: 10.1038/nature00935 12140549.
9. White JK, Gerdin AK, Karp NA, Ryder E, Buljan M, Bussell JN, et al. Genome-wide generation and systematic phenotyping of knockout mice reveals new roles for many genes. Cell. 2013;154(2):452–64. Epub 2013/07/23. doi: 10.1016/j.cell.2013.06.022 23870131; PubMed Central PMCID: PMCPmc3717207.
10. Kok FO, Shin M, Ni CW, Gupta A, Grosse AS, van Impel A, et al. Reverse genetic screening reveals poor correlation between morpholino-induced and mutant phenotypes in zebrafish. Developmental cell. 2015;32(1):97–108. Epub 2014/12/24. doi: 10.1016/j.devcel.2014.11.018 25533206; PubMed Central PMCID: PMCPmc4487878.
11. Bouche N, Bouchez D. Arabidopsis gene knockout: phenotypes wanted. Current opinion in plant biology. 2001;4(2):111–7. Epub 2001/03/03. 11228432.
12. Wang Y, Schnegelsberg PN, Dausman J, Jaenisch R. Functional redundancy of the muscle-specific transcription factors Myf5 and myogenin. Nature. 1996;379(6568):823–5. Epub 1996/02/29. doi: 10.1038/379823a0 8587605.
13. von Koch CS, Zheng H, Chen H, Trumbauer M, Thinakaran G, van der Ploeg LH, et al. Generation of APLP2 KO mice and early postnatal lethality in APLP2/APP double KO mice. Neurobiology of aging. 1997;18(6):661–9. Epub 1998/02/14. 9461064.
14. Santamaria D, Barriere C, Cerqueira A, Hunt S, Tardy C, Newton K, et al. Cdk1 is sufficient to drive the mammalian cell cycle. Nature. 2007;448(7155):811–5. Epub 2007/08/19. doi: 10.1038/nature06046 17700700.
15. Cadigan KM, Grossniklaus U, Gehring WJ. Functional redundancy: the respective roles of the two sloppy paired genes in Drosophila segmentation. Proceedings of the National Academy of Sciences of the United States of America. 1994;91(14):6324–8. Epub 1994/07/05. 8022780; PubMed Central PMCID: PMCPmc44194.
16. González-Gaitán M, Rothe M, Wimmer EA, Taubert H, Jäckle H. Redundant functions of the genes knirps and knirps-related for the establishment of anterior Drosophila head structures. Proceedings of the National Academy of Sciences of the United States of America. 1994;91(18):8567–71. PMC44647. 8078924
17. Hoffmann FM. Drosophila abl and genetic redundancy in signal transduction. Trends in genetics: TIG. 1991;7(11–12):351–5. Epub 1991/11/01. 1820686.
18. Cohen R, Yokoi T, Holland JP, Pepper AE, Holland MJ. Transcription of the constitutively expressed yeast enolase gene ENO1 is mediated by positive and negative cis-acting regulatory sequences. Molecular and cellular biology. 1987;7(8):2753–61. Epub 1987/08/01. 3313003; PubMed Central PMCID: PMCPmc367892.
19. Nedvetzki S, Gonen E, Assayag N, Reich R, Williams RO, Thurmond RL, et al. RHAMM, a receptor for hyaluronan-mediated motility, compensates for CD44 in inflamed CD44-knockout mice: a different interpretation of redundancy. Proceedings of the National Academy of Sciences of the United States of America. 2004;101(52):18081–6. Epub 2004/12/15. doi: 10.1073/pnas.0407378102 15596723; PubMed Central PMCID: PMCPMC539795.
20. Tautz D. Redundancies, development and the flow of information. BioEssays: news and reviews in molecular, cellular and developmental biology. 1992;14(4):263–6. Epub 1992/04/01. doi: 10.1002/bies.950140410 1596275.
21. Barabasi AL, Oltvai ZN. Network biology: understanding the cell's functional organization. Nat Rev Genet. 2004;5(2):101–13. Epub 2004/01/22. doi: 10.1038/nrg1272 14735121.
22. Davidson E, Levin M. Gene regulatory networks. Proceedings of the National Academy of Sciences of the United States of America. 2005;102(14):4935. doi: 10.1073/pnas.0502024102 15809445
23. Teng X, Dayhoff-Brannigan M, Cheng WC, Gilbert CE, Sing CN, Diny NL, et al. Genome-wide consequences of deleting any single gene. Molecular cell. 2013;52(4):485–94. Epub 2013/11/12. doi: 10.1016/j.molcel.2013.09.026 24211263; PubMed Central PMCID: PMCPmc3975072.
24. Chen P, Wang D, Chen H, Zhou Z, He X. The non-essentiality of essential genes in yeast provides therapeutic insights into a human disease. Genome research. 2016. Epub 2016/07/22. doi: 10.1101/gr.205955.116 27440870.
25. Rossi A, Kontarakis Z, Gerri C, Nolte H, Holper S, Kruger M, et al. Genetic compensation induced by deleterious mutations but not gene knockdowns. Nature. 2015;524(7564):230–3. Epub 2015/07/15. doi: 10.1038/nature14580 26168398.
26. Hall EA, Keighren M, Ford MJ, Davey T, Jarman AP, Smith LB, et al. Acute versus chronic loss of mammalian Azi1/Cep131 results in distinct ciliary phenotypes. PLoS Genet. 2013;9(12):e1003928. Epub 2014/01/15. doi: 10.1371/journal.pgen.1003928 24415959; PubMed Central PMCID: PMCPMC3887133.
27. Tondeleir D, Lambrechts A, Muller M, Jonckheere V, Doll T, Vandamme D, et al. Cells lacking beta-actin are genetically reprogrammed and maintain conditional migratory capacity. Molecular & cellular proteomics: MCP. 2012;11(8):255–71. Epub 2012/03/27. 10.1074/mcp.M111.015099. 22448045; PubMed Central PMCID: PMCPMC3412960.
28. Gao Y, Zhang Y, Zhang D, Dai X, Estelle M, Zhao Y. Auxin binding protein 1 (ABP1) is not required for either auxin signaling or Arabidopsis development. Proceedings of the National Academy of Sciences. 2015;112(7):2275–80. doi: 10.1073/pnas.1500365112 25646447
29. Chen X, Grandont L, Li H, Hauschild R, Paque S, Abuzeineh A, et al. Inhibition of cell expansion by rapid ABP1-mediated auxin effect on microtubules. Nature. 2014;516(7529):90–3. Epub 2014/11/20. doi: 10.1038/nature13889 25409144; PubMed Central PMCID: PMCPmc4257754.
30. Braun N, Wyrzykowska J, Muller P, David K, Couch D, Perrot-Rechenmann C, et al. Conditional repression of AUXIN BINDING PROTEIN1 reveals that it coordinates cell division and cell expansion during postembryonic shoot development in Arabidopsis and tobacco. The Plant cell. 2008;20(10):2746–62. Epub 2008/10/28. doi: 10.1105/tpc.108.059048 18952781; PubMed Central PMCID: PMCPmc2590743.
31. Young R, Passet B, Vilotte M, Cribiu EP, Beringue V, Le Provost F, et al. The prion or the related Shadoo protein is required for early mouse embryogenesis. FEBS letters. 2009;583(19):3296–300. Epub 2009/09/22. doi: 10.1016/j.febslet.2009.09.027 19766638.
32. De Souza AT, Dai X, Spencer AG, Reppen T, Menzie A, Roesch PL, et al. Transcriptional and phenotypic comparisons of Ppara knockout and siRNA knockdown mice. Nucleic acids research. 2006;34(16):4486–94. Epub 2006/09/02. doi: 10.1093/nar/gkl609 16945951; PubMed Central PMCID: PMCPmc1636368.
33. Daude N, Wohlgemuth S, Brown R, Pitstick R, Gapeshina H, Yang J, et al. Knockout of the prion protein (PrP)-like Sprn gene does not produce embryonic lethality in combination with PrP(C)-deficiency. Proceedings of the National Academy of Sciences of the United States of America. 2012;109(23):9035–40. Epub 2012/05/24. doi: 10.1073/pnas.1202130109 22619325; PubMed Central PMCID: PMCPmc3384183.
34. McJunkin K, Mazurek A, Premsrirut PK, Zuber J, Dow LE, Simon J, et al. Reversible suppression of an essential gene in adult mice using transgenic RNA interference. Proceedings of the National Academy of Sciences of the United States of America. 2011;108(17):7113–8. Epub 2011/04/13. doi: 10.1073/pnas.1104097108 21482754; PubMed Central PMCID: PMCPmc3084121.
35. Yamamoto S, Jaiswal M, Charng WL, Gambin T, Karaca E, Mirzaa G, et al. A drosophila genetic resource of mutants to study mechanisms underlying human genetic diseases. Cell. 2014;159(1):200–14. Epub 2014/09/27. doi: 10.1016/j.cell.2014.09.002 25259927; PubMed Central PMCID: PMCPmc4298142.
36. Law SH, Sargent TD. The serine-threonine protein kinase PAK4 is dispensable in zebrafish: identification of a morpholino-generated pseudophenotype. PLoS ONE. 2014;9(6):e100268. Epub 2014/06/20. doi: 10.1371/journal.pone.0100268 24945275; PubMed Central PMCID: PMCPMC4063752.
37. Evers B, Jastrzebski K, Heijmans JP, Grernrum W, Beijersbergen RL, Bernards R. CRISPR knockout screening outperforms shRNA and CRISPRi in identifying essential genes. Nature biotechnology. 2016;34(6):631–3. Epub 2016/04/26. doi: 10.1038/nbt.3536 27111720.
38. Karakas B, Weeraratna AT, Abukhdeir AM, Konishi H, Gustin JP, Vitolo MI, et al. P21 gene knock down does not identify genetic effectors seen with gene knock out. Cancer biology & therapy. 2007;6(7):1025–30. Epub 2007/07/06. 17611398; PubMed Central PMCID: PMCPmc2667557.
39. Morgens DW, Deans RM, Li A, Bassik MC. Systematic comparison of CRISPR/Cas9 and RNAi screens for essential genes. Nature biotechnology. 2016;34(6):634–6. Epub 2016/05/10. doi: 10.1038/nbt.3567 27159373; PubMed Central PMCID: PMCPmc4900911.
40. Robu ME, Larson JD, Nasevicius A, Beiraghi S, Brenner C, Farber SA, et al. p53 activation by knockdown technologies. PLoS Genet. 2007;3(5):e78. Epub 2007/05/29. doi: 10.1371/journal.pgen.0030078 17530925; PubMed Central PMCID: PMCPmc1877875.
41. Baek ST, Kerjan G, Bielas SL, Lee JE, Fenstermaker AG, Novarino G, et al. Off-target effect of doublecortin family shRNA on neuronal migration associated with endogenous microRNA dysregulation. Neuron. 2014;82(6):1255–62. Epub 2014/06/20. doi: 10.1016/j.neuron.2014.04.036 24945770; PubMed Central PMCID: PMCPmc4086250.
42. Olejniczak M, Galka P, Krzyzosiak WJ. Sequence-non-specific effects of RNA interference triggers and microRNA regulators. Nucleic acids research. 2010;38(1):1–16. Epub 2009/10/22. doi: 10.1093/nar/gkp829 19843612; PubMed Central PMCID: PMCPmc2800214.
43. Olejniczak M, Urbanek MO, Jaworska E, Witucki L, Szczesniak MW, Makalowska I, et al. Sequence-non-specific effects generated by various types of RNA interference triggers. Biochimica et biophysica acta. 2016;1859(2):306–14. Epub 2015/11/28. doi: 10.1016/j.bbagrm.2015.11.005 26612823.
44. Jackson AL, Linsley PS. Recognizing and avoiding siRNA off-target effects for target identification and therapeutic application. Nature reviews Drug discovery. 2010;9(1):57–67. Epub 2010/01/01. doi: 10.1038/nrd3010 20043028.
45. Housden BE, Muhar M, Gemberling M, Gersbach CA, Stainier DY, Seydoux G, et al. Loss-of-function genetic tools for animal models: cross-species and cross-platform differences. Nat Rev Genet. 2017;18(1):24–40. Epub 2016/11/01. doi: 10.1038/nrg.2016.118 27795562; PubMed Central PMCID: PMCPMC5206767.
46. O'Leary MN, Schreiber KH, Zhang Y, Duc AC, Rao S, Hale JS, et al. The ribosomal protein Rpl22 controls ribosome composition by directly repressing expression of its own paralog, Rpl22l1. PLoS Genet. 2013;9(8):e1003708. Epub 2013/08/31. doi: 10.1371/journal.pgen.1003708 23990801; PubMed Central PMCID: PMCPmc3750023.
47. Mulligan GJ, Wong J, Jacks T. p130 is dispensable in peripheral T lymphocytes: evidence for functional compensation by p107 and pRB. Molecular and cellular biology. 1998;18(1):206–20. Epub 1998/01/07. 9418868; PubMed Central PMCID: PMCPmc121478.
48. Jackson JG, Pereira-Smith OM. Primary and compensatory roles for RB family members at cell cycle gene promoters that are deacetylated and downregulated in doxorubicin-induced senescence of breast cancer cells. Molecular and cellular biology. 2006;26(7):2501–10. Epub 2006/03/16. doi: 10.1128/MCB.26.7.2501-2510.2006 16537896; PubMed Central PMCID: PMCPmc1430319.
49. Jurkin J, Zupkovitz G, Lagger S, Grausenburger R, Hagelkruys A, Kenner L, et al. Distinct and redundant functions of histone deacetylases HDAC1 and HDAC2 in proliferation and tumorigenesis. Cell Cycle. 2011;10(3):406–12. Epub 2011/01/29. doi: 10.4161/cc.10.3.14712 21270520; PubMed Central PMCID: PMCPMC3115015.
50. Hagelkruys A, Lagger S, Krahmer J, Leopoldi A, Artaker M, Pusch O, et al. A single allele of Hdac2 but not Hdac1 is sufficient for normal mouse brain development in the absence of its paralog. Development (Cambridge, England). 2014;141(3):604–16. Epub 2014/01/23. doi: 10.1242/dev.100487 24449838; PubMed Central PMCID: PMCPmc4773893.
51. Lagger G, O'Carroll D, Rembold M, Khier H, Tischler J, Weitzer G, et al. Essential function of histone deacetylase 1 in proliferation control and CDK inhibitor repression. The EMBO journal. 2002;21(11):2672–81. Epub 2002/05/29. doi: 10.1093/emboj/21.11.2672 12032080; PubMed Central PMCID: PMCPMC126040.
52. Dawlaty MM, Ganz K, Powell BE, Hu YC, Markoulaki S, Cheng AW, et al. Tet1 is dispensable for maintaining pluripotency and its loss is compatible with embryonic and postnatal development. Cell stem cell. 2011;9(2):166–75. Epub 2011/08/06. doi: 10.1016/j.stem.2011.07.010 21816367; PubMed Central PMCID: PMCPMC3154739.
53. Freudenberg JM, Ghosh S, Lackford BL, Yellaboina S, Zheng X, Li R, et al. Acute depletion of Tet1-dependent 5-hydroxymethylcytosine levels impairs LIF/Stat3 signaling and results in loss of embryonic stem cell identity. Nucleic acids research. 2012;40(8):3364–77. Epub 2012/01/03. doi: 10.1093/nar/gkr1253 22210859; PubMed Central PMCID: PMCPMC3333871.
54. Wang J, Wang Q, Cui Y, Liu ZY, Zhao W, Wang CL, et al. Knockdown of cyclin D1 inhibits proliferation, induces apoptosis, and attenuates the invasive capacity of human glioblastoma cells. Journal of neuro-oncology. 2012;106(3):473–84. Epub 2011/09/14. doi: 10.1007/s11060-011-0692-4 21912938.
55. Radulovich N, Pham N - A, Strumpf D, Leung L, Xie W, Jurisica I, et al. Differential roles of cyclin D1 and D3 in pancreatic ductal adenocarcinoma. Molecular Cancer. 2010;9(1):1–15. doi: 10.1186/1476-4598-9-24 20113529
56. Becker KA, Ghule PN, Lian JB, Stein JL, van Wijnen AJ, Stein GS. Cyclin D2 and the CDK substrate p220(NPAT) are required for self-renewal of human embryonic stem cells. Journal of cellular physiology. 2010;222(2):456–64. Epub 2009/11/06. doi: 10.1002/jcp.21967 19890848; PubMed Central PMCID: PMCPmc3059841.
57. Sicinska E, Aifantis I, Le Cam L, Swat W, Borowski C, Yu Q, et al. Requirement for cyclin D3 in lymphocyte development and T cell leukemias. Cancer cell. 2003;4(6):451–61. Epub 2004/01/07. 14706337.
58. Sicinski P, Weinberg RA. A specific role for cyclin D1 in mammary gland development. Journal of mammary gland biology and neoplasia. 1997;2(4):335–42. Epub 2000/08/10. 10935021.
59. Huard JM, Forster CC, Carter ML, Sicinski P, Ross ME. Cerebellar histogenesis is disturbed in mice lacking cyclin D2. Development (Cambridge, England). 1999;126(9):1927–35. Epub 1999/04/02. 10101126.
60. Ciemerych MA, Kenney AM, Sicinska E, Kalaszczynska I, Bronson RT, Rowitch DH, et al. Development of mice expressing a single D-type cyclin. Genes & development. 2002;16(24):3277–89. Epub 2002/12/28. doi: 10.1101/gad.1023602 12502747; PubMed Central PMCID: PMCPmc187507.
61. Lam EW, Glassford J, Banerji L, Thomas NS, Sicinski P, Klaus GG. Cyclin D3 compensates for loss of cyclin D2 in mouse B-lymphocytes activated via the antigen receptor and CD40. The Journal of biological chemistry. 2000;275(5):3479–84. Epub 2000/02/01. 10652342.
62. Yasuhara N, Shibazaki N, Tanaka S, Nagai M, Kamikawa Y, Oe S, et al. Triggering neural differentiation of ES cells by subtype switching of importin-alpha. Nature cell biology. 2007;9(1):72–9. Epub 2006/12/13. doi: 10.1038/ncb1521 17159997.
63. Shmidt T, Hampich F, Ridders M, Schultrich S, Hans VH, Tenner K, et al. Normal brain development in importin-alpha5 deficient-mice. Nature cell biology. 2007;9(12):1337–8; author reply 9. Epub 2007/12/07. doi: 10.1038/ncb1207-1337 18059353.
64. Rajshankar D, Downey GP, McCulloch CA. IL-1β enhances cell adhesion to degraded fibronectin. The FASEB Journal. 2012;26(11):4429–44. doi: 10.1096/fj.12-207381. 22829527
65. Theodosiou M, Widmaier M, Bottcher RT, Rognoni E, Veelders M, Bharadwaj M, et al. Kindlin-2 cooperates with talin to activate integrins and induces cell spreading by directly binding paxillin. eLife. 2016;5:e10130. Epub 2016/01/29. doi: 10.7554/eLife.10130 26821125; PubMed Central PMCID: PMCPmc4749545.
66. Jost AP, Weiner OD. Probing Yeast Polarity with Acute, Reversible, Optogenetic Inhibition of Protein Function. ACS synthetic biology. 2015;4(10):1077–85. Epub 2015/06/03. doi: 10.1021/acssynbio.5b00053 26035630; PubMed Central PMCID: PMCPmc4609243.
67. Williams JA, Ni HM, Haynes A, Manley S, Li Y, Jaeschke H, et al. Chronic Deletion and Acute Knockdown of Parkin Have Differential Responses to Acetaminophen-induced Mitophagy and Liver Injury in Mice. The Journal of biological chemistry. 2015;290(17):10934–46. Epub 2015/03/11. doi: 10.1074/jbc.M114.602284 25752611; PubMed Central PMCID: PMCPMC4409255.
68. Nicchia GP, Srinivas M, Li W, Brosnan CF, Frigeri A, Spray DC. New possible roles for aquaporin-4 in astrocytes: cell cytoskeleton and functional relationship with connexin43. FASEB journal: official publication of the Federation of American Societies for Experimental Biology. 2005;19(12):1674–6. Epub 2005/08/17. doi: 10.1096/fj.04-3281fje 16103109.
69. Ma T, Yang B, Gillespie A, Carlson EJ, Epstein CJ, Verkman AS. Generation and phenotype of a transgenic knockout mouse lacking the mercurial-insensitive water channel aquaporin-4. J Clin Invest. 1997;100(5):957–62. Epub 1997/09/01. doi: 10.1172/JCI231 9276712; PubMed Central PMCID: PMCPmc508270.
70. Manley GT, Fujimura M, Ma T, Noshita N, Filiz F, Bollen AW, et al. Aquaporin-4 deletion in mice reduces brain edema after acute water intoxication and ischemic stroke. Nature medicine. 2000;6(2):159–63. Epub 2000/02/02. doi: 10.1038/72256 10655103.
71. Papadopoulos MC, Manley GT, Krishna S, Verkman AS. Aquaporin-4 facilitates reabsorption of excess fluid in vasogenic brain edema. FASEB journal: official publication of the Federation of American Societies for Experimental Biology. 2004;18(11):1291–3. Epub 2004/06/23. doi: 10.1096/fj.04-1723fje 15208268.
72. Speers C, Zhao SG, Kothari V, Santola A, Liu M, Wilder-Romans K, et al. Maternal Embryonic Leucine Zipper Kinase (MELK) as a Novel Mediator and Biomarker of Radioresistance in Human Breast Cancer. Clin Cancer Res. 2016;22(23):5864–75. Epub 2016/11/01. doi: 10.1158/1078-0432.CCR-15-2711 27225691.
73. Lin ML, Park JH, Nishidate T, Nakamura Y, Katagiri T. Involvement of maternal embryonic leucine zipper kinase (MELK) in mammary carcinogenesis through interaction with Bcl-G, a pro-apoptotic member of the Bcl-2 family. Breast cancer research: BCR. 2007;9(1):R17. Epub 2007/02/07. doi: 10.1186/bcr1650 17280616; PubMed Central PMCID: PMCPMC1851384.
74. Hebbard LW, Maurer J, Miller A, Lesperance J, Hassell J, Oshima RG, et al. Maternal embryonic leucine zipper kinase is upregulated and required in mammary tumor-initiating cells in vivo. Cancer Res. 2010;70(21):8863–73. Epub 2010/09/24. doi: 10.1158/0008-5472.CAN-10-1295 20861186; PubMed Central PMCID: PMCPMC3990264.
75. Wang Y, Lee YM, Baitsch L, Huang A, Xiang Y, Tong H, et al. MELK is an oncogenic kinase essential for mitotic progression in basal-like breast cancer cells. eLife. 2014;3:e01763. Epub 2014/05/23. doi: 10.7554/eLife.01763 24844244; PubMed Central PMCID: PMCPMC4059381.
76. Lin A, Giuliano CJ, Sayles NM, Sheltzer JM. CRISPR/Cas9 mutagenesis invalidates a putative cancer dependency targeted in on-going clinical trials. eLife. 2017;6. Epub 2017/03/25. doi: 10.7554/eLife.24179 28337968.
77. Caceres A, Kosik KS. Inhibition of neurite polarity by tau antisense oligonucleotides in primary cerebellar neurons. Nature. 1990;343(6257):461–3. Epub 1990/02/01. doi: 10.1038/343461a0 2105469.
78. Caceres A, Potrebic S, Kosik KS. The effect of tau antisense oligonucleotides on neurite formation of cultured cerebellar macroneurons. The Journal of neuroscience: the official journal of the Society for Neuroscience. 1991;11(6):1515–23. Epub 1991/06/01. 1904479.
79. Harada A, Oguchi K, Okabe S, Kuno J, Terada S, Ohshima T, et al. Altered microtubule organization in small-calibre axons of mice lacking tau protein. Nature. 1994;369(6480):488–91. Epub 1994/06/09. doi: 10.1038/369488a0 8202139.
80. Seiberlich V, Bauer NG, Schwarz L, Ffrench-Constant C, Goldbaum O, Richter-Landsberg C. Downregulation of the microtubule associated protein tau impairs process outgrowth and myelin basic protein mRNA transport in oligodendrocytes. Glia. 2015;63(9):1621–35. Epub 2015/04/08. doi: 10.1002/glia.22832 25847153.
81. Deconinck AE, Rafael JA, Skinner JA, Brown SC, Potter AC, Metzinger L, et al. Utrophin-dystrophin-deficient mice as a model for Duchenne muscular dystrophy. Cell. 1997;90(4):717–27. Epub 1997/08/22. 9288751.
82. Law DJ, Allen DL, Tidball JG. Talin, vinculin and DRP (utrophin) concentrations are increased at mdx myotendinous junctions following onset of necrosis. Journal of cell science. 1994;107 (Pt 6):1477–83. Epub 1994/06/01. 7962191.
83. Ghahramani Seno MM, Graham IR, Athanasopoulos T, Trollet C, Pohlschmidt M, Crompton MR, et al. RNAi-mediated knockdown of dystrophin expression in adult mice does not lead to overt muscular dystrophy pathology. Human molecular genetics. 2008;17(17):2622–32. Epub 2008/05/31. doi: 10.1093/hmg/ddn162 18511456.
84. Bunnell TM, Burbach BJ, Shimizu Y, Ervasti JM. beta-Actin specifically controls cell growth, migration, and the G-actin pool. Molecular biology of the cell. 2011;22(21):4047–58. Epub 2011/09/09. doi: 10.1091/mbc.E11-06-0582 21900491; PubMed Central PMCID: PMCPMC3204067.
85. Patrinostro X, O'Rourke AR, Chamberlain CM, Moriarity BS, Perrin BJ, Ervasti JM. Relative importance of betacyto - and gammacyto-actin in primary mouse embryonic fibroblasts. Molecular biology of the cell. 2017;28(6):771–82. Epub 2017/01/13. doi: 10.1091/mbc.E16-07-0503 28077619.
86. Dayton TL, Gocheva V, Miller KM, Israelsen WJ, Bhutkar A, Clish CB, et al. Germline loss of PKM2 promotes metabolic distress and hepatocellular carcinoma. Genes & development. 2016;30(9):1020–33. Epub 2016/04/30. doi: 10.1101/gad.278549.116 27125672; PubMed Central PMCID: PMCPmc4863734.
87. Lunt SY, Muralidhar V, Hosios AM, Israelsen WJ, Gui DY, Newhouse L, et al. Pyruvate kinase isoform expression alters nucleotide synthesis to impact cell proliferation. Molecular cell. 2015;57(1):95–107. Epub 2014/12/09. doi: 10.1016/j.molcel.2014.10.027 25482511; PubMed Central PMCID: PMCPmc4289430.
88. Wang RH, Li C, Deng CX. Liver steatosis and increased ChREBP expression in mice carrying a liver specific SIRT1 null mutation under a normal feeding condition. International journal of biological sciences. 2010;6(7):682–90. Epub 2010/11/26. 21103071; PubMed Central PMCID: PMCPmc2990071.
89. Zhou S, Xie Y, Tang J, Huang J, Huang Q, Xu W, et al. FGFR3 Deficiency Causes Multiple Chondroma-like Lesions by Upregulating Hedgehog Signaling. PLoS Genet. 2015;11(6):e1005214. Epub 2015/06/20. doi: 10.1371/journal.pgen.1005214 26091072; PubMed Central PMCID: PMCPmc4474636.
90. Sage J, Miller AL, Perez-Mancera PA, Wysocki JM, Jacks T. Acute mutation of retinoblastoma gene function is sufficient for cell cycle re-entry. Nature. 2003;424(6945):223–8. Epub 2003/07/11. doi: 10.1038/nature01764 12853964.
91. Protin U, Schweighoffer T, Jochum W, Hilberg F. CD44-deficient mice develop normally with changes in subpopulations and recirculation of lymphocyte subsets. Journal of immunology (Baltimore, Md: 1950). 1999;163(9):4917–23. Epub 1999/10/21. 10528194.
92. Schmits R, Filmus J, Gerwin N, Senaldi G, Kiefer F, Kundig T, et al. CD44 regulates hematopoietic progenitor distribution, granuloma formation, and tumorigenicity. Blood. 1997;90(6):2217–33. Epub 1997/10/06. 9310473.
93. Shatirishvili M, Burk AS, Franz CM, Pace G, Kastilan T, Breuhahn K, et al. Epidermal-specific deletion of CD44 reveals a function in keratinocytes in response to mechanical stress. Cell death & disease. 2016;7(11):e2461. Epub 2016/11/11. doi: 10.1038/cddis.2016.342 27831556; PubMed Central PMCID: PMCPMC5260879.
94. Takahashi K, Kaneko I. Changes in nuclease sensitivity of mammalian cells after irradiation with 60Co gamma-rays. International journal of radiation biology and related studies in physics, chemistry, and medicine. 1985;48(3):389–95. Epub 1985/09/01. 3875579.
95. Ziv Y, Bielopolski D, Galanty Y, Lukas C, Taya Y, Schultz DC, et al. Chromatin relaxation in response to DNA double-strand breaks is modulated by a novel ATM - and KAP-1 dependent pathway. Nature cell biology. 2006;8(8):870–6. Epub 2006/07/25. doi: 10.1038/ncb1446 16862143.
96. Downs JA, Nussenzweig MC, Nussenzweig A. Chromatin dynamics and the preservation of genetic information. Nature. 2007;447(7147):951–8. Epub 2007/06/22. doi: 10.1038/nature05980 17581578.
97. Stuckenholz C, Kageyama Y, Kuroda MI. Guilt by association: non-coding RNAs, chromosome-specific proteins and dosage compensation in Drosophila. Trends in genetics: TIG. 1999;15(11):454–8. Epub 1999/10/26. 10529808.
98. Bowerman B, Eaton BA, Priess JR. skn-1, a maternally expressed gene required to specify the fate of ventral blastomeres in the early C. elegans embryo. Cell. 1992;68(6):1061–75. Epub 1992/03/20. 1547503.
99. Raj A, Rifkin SA, Andersen E, van Oudenaarden A. Variability in gene expression underlies incomplete penetrance. Nature. 2010;463(7283):913–8. Epub 2010/02/19. doi: 10.1038/nature08781 20164922; PubMed Central PMCID: PMCPmc2836165.
100. Chambeyron S, Bickmore WA. Chromatin decondensation and nuclear reorganization of the HoxB locus upon induction of transcription. Genes & development. 2004;18(10):1119–30. Epub 2004/05/25. doi: 10.1101/gad.292104 15155579; PubMed Central PMCID: PMCPmc415637.
101. Wei Z, Huang D, Gao F, Chang WH, An W, Coetzee GA, et al. Biological implications and regulatory mechanisms of long-range chromosomal interactions. The Journal of biological chemistry. 2013;288(31):22369–77. Epub 2013/06/20. doi: 10.1074/jbc.R113.485292 23779110; PubMed Central PMCID: PMCPMC3829327.
102. Li L, Lyu X, Hou C, Takenaka N, Nguyen HQ, Ong CT, et al. Widespread rearrangement of 3D chromatin organization underlies polycomb-mediated stress-induced silencing. Molecular cell. 2015;58(2):216–31. Epub 2015/03/31. doi: 10.1016/j.molcel.2015.02.023 25818644; PubMed Central PMCID: PMCPMC4402144.
103. Wei W, Ba Z, Gao M, Wu Y, Ma Y, Amiard S, et al. A role for small RNAs in DNA double-strand break repair. Cell. 2012;149(1):101–12. Epub 2012/03/27. doi: 10.1016/j.cell.2012.03.002 22445173.
104. Francia S, Michelini F, Saxena A, Tang D, de Hoon M, Anelli V, et al. Site-specific DICER and DROSHA RNA products control the DNA-damage response. Nature. 2012;488(7410):231–5. Epub 2012/06/23. doi: 10.1038/nature11179 22722852; PubMed Central PMCID: PMCPMC3442236.
105. d'Adda di Fagagna F. A direct role for small non-coding RNAs in DNA damage response. Trends in cell biology. 2014;24(3):171–8. Epub 2013/10/26. doi: 10.1016/j.tcb.2013.09.008 24156824.
106. Franke A, Baker BS. The rox1 and rox2 RNAs are essential components of the compensasome, which mediates dosage compensation in Drosophila. Molecular cell. 1999;4(1):117–22. Epub 1999/08/13. 10445033.
107. Meller VH, Wu KH, Roman G, Kuroda MI, Davis RL. roX1 RNA paints the X chromosome of male Drosophila and is regulated by the dosage compensation system. Cell. 1997;88(4):445–57. Epub 1997/02/21. 9038336.
108. Amrein H, Axel R. Genes expressed in neurons of adult male Drosophila. Cell. 1997;88(4):459–69. Epub 1997/02/21. 9038337.
109. Yang Y, Li T, Vu TH, Ulaner GA, Hu JF, Hoffman AR. The histone code regulating expression of the imprinted mouse Igf2r gene. Endocrinology. 2003;144(12):5658–70. Epub 2003/09/17. doi: 10.1210/en.2003-0798 12975326.
110. Fournier C, Goto Y, Ballestar E, Delaval K, Hever AM, Esteller M, et al. Allele-specific histone lysine methylation marks regulatory regions at imprinted mouse genes. The EMBO journal. 2002;21(23):6560–70. Epub 2002/11/29. 12456662; PubMed Central PMCID: PMCPMC136958. doi: 10.1093/emboj/cdf655
111. Carr MS, Yevtodiyenko A, Schmidt CL, Schmidt JV. Allele-specific histone modifications regulate expression of the Dlk1-Gtl2 imprinted domain. Genomics. 2007;89(2):280–90. Epub 2006/11/28. doi: 10.1016/j.ygeno.2006.10.005 17126526; PubMed Central PMCID: PMCPMC1802099.
112. McEwen KR, Ferguson-Smith AC. Genomic Imprinting–A Model for Roles of HistoneModifications in Epigenetic Control. In: Ferguson-Smith AC, Greally JM, Martienssen RA, editors. Epigenomics. Dordrecht: Springer Netherlands; 2009. p. 235–58.
113. Barreto G, Schafer A, Marhold J, Stach D, Swaminathan SK, Handa V, et al. Gadd45a promotes epigenetic gene activation by repair-mediated DNA demethylation. Nature. 2007;445(7128):671–5. Epub 2007/02/03. doi: 10.1038/nature05515 17268471.
114. Lykke-Andersen S, Jensen TH. Nonsense-mediated mRNA decay: an intricate machinery that shapes transcriptomes. Nat Rev Mol Cell Biol. 2015;16(11):665–77. Epub 2015/09/24. doi: 10.1038/nrm4063 26397022.
115. Akimitsu N. Messenger RNA Surveillance Systems Monitoring Proper Translation Termination. The Journal of Biochemistry. 2008;143(1):1–8. doi: 10.1093/jb/mvm204 17981821
116. Harigaya Y, Parker R. No-go decay: a quality control mechanism for RNA in translation. Wiley interdisciplinary reviews RNA. 2010;1(1):132–41. Epub 2010/07/01. doi: 10.1002/wrna.17 21956910.
117. Schuermann A, Helker CS, Herzog W. Metallothionein 2 regulates endothelial cell migration through transcriptional regulation of vegfc expression. Angiogenesis. 2015;18(4):463–75. Epub 2015/07/23. doi: 10.1007/s10456-015-9473-6 26198291; PubMed Central PMCID: PMCPmc4596909.
118. Mitchell P, Tollervey D. An NMD pathway in yeast involving accelerated deadenylation and exosome-mediated 3'—>5' degradation. Molecular cell. 2003;11(5):1405–13. Epub 2003/05/29. 12769863.
119. Vance KW, Ponting CP. Transcriptional regulatory functions of nuclear long noncoding RNAs. Trends in genetics: TIG. 2014;30(8):348–55. Epub 2014/06/30. doi: 10.1016/j.tig.2014.06.001 24974018; PubMed Central PMCID: PMCPMC4115187.
120. Ghanbarian H, Wagner N, Michiels JF, Cuzin F, Wagner KD, Rassoulzadegan M. Small RNA-directed epigenetic programming of embryonic stem cell cardiac differentiation. Scientific reports. 2017;7 : 41799. Epub 2017/02/07. doi: 10.1038/srep41799 28165496; PubMed Central PMCID: PMCPMC5292948.
121. Wagner KD, Wagner N, Ghanbarian H, Grandjean V, Gounon P, Cuzin F, et al. RNA induction and inheritance of epigenetic cardiac hypertrophy in the mouse. Developmental cell. 2008;14(6):962–9. Epub 2008/06/10. doi: 10.1016/j.devcel.2008.03.009 18539123.
122. Li LC, Okino ST, Zhao H, Pookot D, Place RF, Urakami S, et al. Small dsRNAs induce transcriptional activation in human cells. Proceedings of the National Academy of Sciences of the United States of America. 2006;103(46):17337–42. Epub 2006/11/07. doi: 10.1073/pnas.0607015103 17085592; PubMed Central PMCID: PMCPMC1859931.
123. Janowski BA, Younger ST, Hardy DB, Ram R, Huffman KE, Corey DR. Activating gene expression in mammalian cells with promoter-targeted duplex RNAs. Nature chemical biology. 2007;3(3):166–73. Epub 2007/01/30. doi: 10.1038/nchembio860 17259978.
124. Schwartz JC, Younger ST, Nguyen NB, Hardy DB, Monia BP, Corey DR, et al. Antisense transcripts are targets for activating small RNAs. Nature structural & molecular biology. 2008;15(8):842–8. Epub 2008/07/08. doi: 10.1038/nsmb.1444 18604220; PubMed Central PMCID: PMCPMC2574822.
125. Zhang X, Li H, Burnett JC, Rossi JJ. The role of antisense long noncoding RNA in small RNA-triggered gene activation. RNA. 2014;20(12):1916–28. doi: 10.1261/rna.043968.113. 25344398
126. Katayama S, Tomaru Y, Kasukawa T, Waki K, Nakanishi M, Nakamura M, et al. Antisense transcription in the mammalian transcriptome. Science (New York, NY). 2005;309(5740):1564–6. Epub 2005/09/06. doi: 10.1126/science.1112009 16141073.
127. Kiyosawa H, Yamanaka I, Osato N, Kondo S, Riken Ger Group Laboratory for Genome Exploration Research Group RGSCRYIS-cT-kYKJ, Gslmembers Genome Science Laboratory RHWSJ, et al. Antisense Transcripts With FANTOM2 Clone Set and Their Implications for Gene Regulation. Genome research. 2003;13(6b):1324–34. doi: 10.1101/gr.982903. 12819130
128. Yelin R, Dahary D, Sorek R, Levanon EY, Goldstein O, Shoshan A, et al. Widespread occurrence of antisense transcription in the human genome. Nature biotechnology. 2003;21(4):379–86. Epub 2003/03/18. doi: 10.1038/nbt808 12640466.
129. Carninci P, Kasukawa T, Katayama S, Gough J, Frith MC, Maeda N, et al. The transcriptional landscape of the mammalian genome. Science (New York, NY). 2005;309(5740):1559–63. Epub 2005/09/06. doi: 10.1126/science.1112014 16141072.
130. Chen J, Sun M, Kent WJ, Huang X, Xie H, Wang W, et al. Over 20% of human transcripts might form sense-antisense pairs. Nucleic acids research. 2004;32(16):4812–20. Epub 2004/09/10. doi: 10.1093/nar/gkh818 15356298; PubMed Central PMCID: PMCPMC519112.
131. Glisovic T, Bachorik JL, Yong J, Dreyfuss G. RNA-binding proteins and post-transcriptional gene regulation. FEBS letters. 2008;582(14):1977–86. Epub 2008/03/18. doi: 10.1016/j.febslet.2008.03.004 18342629; PubMed Central PMCID: PMCPmc2858862.
132. Kuwano Y, Rabinovic A, Srikantan S, Gorospe M, Demple B. Analysis of nitric oxide-stabilized mRNAs in human fibroblasts reveals HuR-dependent heme oxygenase 1 upregulation. Molecular and cellular biology. 2009;29(10):2622–35. Epub 2009/03/18. doi: 10.1128/MCB.01495-08 19289500; PubMed Central PMCID: PMCPmc2682029.
133. Gerber AP, Herschlag D, Brown PO. Extensive association of functionally and cytotopically related mRNAs with Puf family RNA-binding proteins in yeast. PLoS Biol. 2004;2(3):E79. Epub 2004/03/17. doi: 10.1371/journal.pbio.0020079 15024427; PubMed Central PMCID: PMCPMC368173.
134. Keene JD, Tenenbaum SA. Eukaryotic mRNPs may represent posttranscriptional operons. Molecular cell. 2002;9(6):1161–7. Epub 2002/06/28. 12086614.
135. Keene JD, Lager PJ. Post-transcriptional operons and regulons co-ordinating gene expression. Chromosome research: an international journal on the molecular, supramolecular and evolutionary aspects of chromosome biology. 2005;13(3):327–37. Epub 2005/05/04. doi: 10.1007/s10577-005-0848-1 15868425.
136. Keene JD. RNA regulons: coordination of post-transcriptional events. Nat Rev Genet. 2007;8(7):533–43. Epub 2007/06/19. doi: 10.1038/nrg2111 17572691.
137. Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet. 2008;9(2):102–14. Epub 2008/01/17. doi: 10.1038/nrg2290 18197166.
138. Place RF, Li LC, Pookot D, Noonan EJ, Dahiya R. MicroRNA-373 induces expression of genes with complementary promoter sequences. Proceedings of the National Academy of Sciences of the United States of America. 2008;105(5):1608–13. Epub 2008/01/30. doi: 10.1073/pnas.0707594105 18227514; PubMed Central PMCID: PMCPmc2234192.
139. Orom UA, Nielsen FC, Lund AH. MicroRNA-10a binds the 5'UTR of ribosomal protein mRNAs and enhances their translation. Molecular cell. 2008;30(4):460–71. Epub 2008/05/24. doi: 10.1016/j.molcel.2008.05.001 18498749.
140. Pasquinelli AE. MicroRNAs and their targets: recognition, regulation and an emerging reciprocal relationship. Nat Rev Genet. 2012;13(4):271–82. Epub 2012/03/14. doi: 10.1038/nrg3162 22411466.
141. Jacobsen A, Silber J, Harinath G, Huse JT, Schultz N, Sander C. Analysis of microRNA-target interactions across diverse cancer types. Nature structural & molecular biology. 2013;20(11):1325–32. Epub 2013/10/08. doi: 10.1038/nsmb.2678 24096364; PubMed Central PMCID: PMCPmc3982325.
142. Schwartz S, Bernstein DA, Mumbach MR, Jovanovic M, Herbst RH, Leon-Ricardo BX, et al. Transcriptome-wide mapping reveals widespread dynamic-regulated pseudouridylation of ncRNA and mRNA. Cell. 2014;159(1):148–62. Epub 2014/09/16. doi: 10.1016/j.cell.2014.08.028 25219674; PubMed Central PMCID: PMCPMC4180118.
143. Zhou J, Wan J, Gao X, Zhang X, Jaffrey SR, Qian SB. Dynamic m(6)A mRNA methylation directs translational control of heat shock response. Nature. 2015;526(7574):591–4. Epub 2015/10/13. doi: 10.1038/nature15377 26458103; PubMed Central PMCID: PMCPMC4851248.
144. Licht K, Jantsch MF. Rapid and dynamic transcriptome regulation by RNA editing and RNA modifications. J Cell Biol. 2016;213(1):15–22. Epub 2016/04/06. doi: 10.1083/jcb.201511041 27044895; PubMed Central PMCID: PMCPMC4828693.
145. Iacono M, Mignone F, Pesole G. uAUG and uORFs in human and rodent 5'untranslated mRNAs. Gene. 2005;349 : 97–105. Epub 2005/03/22. doi: 10.1016/j.gene.2004.11.041 15777708.
146. Chew GL, Pauli A, Rinn JL, Regev A, Schier AF, Valen E. Ribosome profiling reveals resemblance between long non-coding RNAs and 5' leaders of coding RNAs. Development (Cambridge, England). 2013;140(13):2828–34. Epub 2013/05/24. doi: 10.1242/dev.098343 23698349; PubMed Central PMCID: PMCPmc3678345.
147. Morris DR, Geballe AP. Upstream open reading frames as regulators of mRNA translation. Molecular and cellular biology. 2000;20(23):8635–42. Epub 2000/11/14. 11073965; PubMed Central PMCID: PMCPmc86464.
148. Calvo SE, Pagliarini DJ, Mootha VK. Upstream open reading frames cause widespread reduction of protein expression and are polymorphic among humans. Proceedings of the National Academy of Sciences of the United States of America. 2009;106(18):7507–12. Epub 2009/04/18. doi: 10.1073/pnas.0810916106 19372376; PubMed Central PMCID: PMCPmc2669787.
149. Hinnebusch AG, Dever TE, Asano K. 9 Mechanism of Translation Initiation in the Yeast Saccharomyces cerevisiae2007.
150. Mueller PP, Hinnebusch AG. Multiple upstream AUG codons mediate translational control of GCN4. Cell. 1986;45(2):201–7. Epub 1986/04/25. 3516411.
151. Chen R, Shi L, Hakenberg J, Naughton B, Sklar P, Zhang J, et al. Analysis of 589,306 genomes identifies individuals resilient to severe Mendelian childhood diseases. Nature biotechnology. 2016;34(5):531–8. Epub 2016/04/12. doi: 10.1038/nbt.3514 27065010.
152. Narasimhan VM, Hunt KA, Mason D, Baker CL, Karczewski KJ, Barnes MR, et al. Health and population effects of rare gene knockouts in adult humans with related parents. Science (New York, NY). 2016;352(6284):474–7. Epub 2016/03/05. doi: 10.1126/science.aac8624 26940866; PubMed Central PMCID: PMCPmc4985238.
153. Sulem P, Helgason H, Oddson A, Stefansson H, Gudjonsson SA, Zink F, et al. Identification of a large set of rare complete human knockouts. Nature genetics. 2015;47(5):448–52. Epub 2015/03/26. doi: 10.1038/ng.3243 25807282.
154. Nadeau JH. Modifier genes in mice and humans. Nat Rev Genet. 2001;2(3):165–74. Epub 2001/03/21. doi: 10.1038/35056009 11256068.
155. Raj A, van Oudenaarden A. Nature, nurture, or chance: stochastic gene expression and its consequences. Cell. 2008;135(2):216–26. Epub 2008/10/30. doi: 10.1016/j.cell.2008.09.050 18957198; PubMed Central PMCID: PMCPmc3118044.
156. Dickinson ME, Flenniken AM, Ji X, Teboul L, Wong MD, White JK, et al. High-throughput discovery of novel developmental phenotypes. Nature. 2016;537(7621):508–14. Epub 2016/09/15. doi: 10.1038/nature19356 27626380.
Štítky
Genetika Reprodukční medicína
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2017 Číslo 7- Akutní intermitentní porfyrie
- Růst a vývoj dětí narozených pomocí IVF
- Farmakogenetické testování pomáhá předcházet nežádoucím efektům léčiv
- Pilotní studie: stres a úzkost v průběhu IVF cyklu
- IVF a rakovina prsu – zvyšují hormony riziko vzniku rakoviny?
Nejčtenější v tomto čísle- Genetic compensation: A phenomenon in search of mechanisms
- The gene drive bubble: New realities
- Statistical correction of the Winner’s Curse explains replication variability in quantitative trait genome-wide association studies
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání