-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaMAML1 Enhances the Transcriptional Activity of Runx2 and Plays a Role in Bone Development
Mastermind-like 1 (MAML1) is a transcriptional co-activator in the Notch signaling pathway. Recently, however, several reports revealed novel and unique roles for MAML1 that are independent of the Notch signaling pathway. We found that MAML1 enhances the transcriptional activity of runt-related transcription factor 2 (Runx2), a transcription factor essential for osteoblastic differentiation and chondrocyte proliferation and maturation. MAML1 significantly enhanced the Runx2-mediated transcription of the p6OSE2-Luc reporter, in which luciferase expression was controlled by six copies of the osteoblast specific element 2 (OSE2) from the Runx2-regulated osteocalcin gene promoter. Interestingly, a deletion mutant of MAML1 lacking the N-terminal Notch-binding domain also enhanced Runx2-mediated transcription. Moreover, inhibition of Notch signaling did not affect the action of MAML1 on Runx2, suggesting that the activation of Runx2 by MAML1 may be caused in a Notch-independent manner. Overexpression of MAML1 transiently enhanced the Runx2-mediated expression of alkaline phosphatase, an early marker of osteoblast differentiation, in the murine pluripotent mesenchymal cell line C3H10T1/2. MAML1−/− embryos at embryonic day 16.5 (E16.5) had shorter bone lengths than wild-type embryos. The area of primary spongiosa of the femoral diaphysis was narrowed. At E14.5, extended zone of collagen type II alpha 1 (Col2a1) and Sox9 expression, markers of chondrocyte differentiation, and decreased zone of collagen type X alpha 1 (Col10a1) expression, a marker of hypertrophic chondrocyte, were observed. These observations suggest that chondrocyte maturation was impaired in MAML1−/− mice. MAML1 enhances the transcriptional activity of Runx2 and plays a role in bone development.
Published in the journal: . PLoS Genet 9(1): e32767. doi:10.1371/journal.pgen.1003132
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1003132Summary
Mastermind-like 1 (MAML1) is a transcriptional co-activator in the Notch signaling pathway. Recently, however, several reports revealed novel and unique roles for MAML1 that are independent of the Notch signaling pathway. We found that MAML1 enhances the transcriptional activity of runt-related transcription factor 2 (Runx2), a transcription factor essential for osteoblastic differentiation and chondrocyte proliferation and maturation. MAML1 significantly enhanced the Runx2-mediated transcription of the p6OSE2-Luc reporter, in which luciferase expression was controlled by six copies of the osteoblast specific element 2 (OSE2) from the Runx2-regulated osteocalcin gene promoter. Interestingly, a deletion mutant of MAML1 lacking the N-terminal Notch-binding domain also enhanced Runx2-mediated transcription. Moreover, inhibition of Notch signaling did not affect the action of MAML1 on Runx2, suggesting that the activation of Runx2 by MAML1 may be caused in a Notch-independent manner. Overexpression of MAML1 transiently enhanced the Runx2-mediated expression of alkaline phosphatase, an early marker of osteoblast differentiation, in the murine pluripotent mesenchymal cell line C3H10T1/2. MAML1−/− embryos at embryonic day 16.5 (E16.5) had shorter bone lengths than wild-type embryos. The area of primary spongiosa of the femoral diaphysis was narrowed. At E14.5, extended zone of collagen type II alpha 1 (Col2a1) and Sox9 expression, markers of chondrocyte differentiation, and decreased zone of collagen type X alpha 1 (Col10a1) expression, a marker of hypertrophic chondrocyte, were observed. These observations suggest that chondrocyte maturation was impaired in MAML1−/− mice. MAML1 enhances the transcriptional activity of Runx2 and plays a role in bone development.
Introduction
Runt-related transcription factor 2 (Runx2) is a transcription factor belonging to the Runx gene family, which is homologous to Drosophila runt, a pair-rule gene involved in somitogenesis [1]. Runx2 is an essential factor for bone and hypertrophic cartilage formation that is expressed very early in bone development and continues to be present through the later phase of development [2]. Runx2 promotes the differentiation of pluripotent mesenchymal progenitor cells into the osteogenic lineage, but its role in terminal differentiation to mature osteoblasts and the production of bone matrix remains unclear. To date, it has been reported that several transcription factors and cofactors, such as TAZ [3], Grg5 [4], Rb [5], and HDAC4 [6], interact with Runx2 and positively or negatively regulate its function. However, in many cases, the physiological significance of the interaction is unclear. To further elucidate the function of Runx2, we performed luciferase assay-based screening of additional factors regulating the transcriptional activity of Runx2 using a full-length cDNA library containing approximately 10,000 clones. The screening system identified the mastermind-like (MAML) family of proteins showed especially strong potential for regulating Runx2 transcriptional activity. Overexpression of MAML1 enhanced the Runx2-mediated expression of alkaline phosphatase, an early marker of osteoblast differentiation, in C3H10T1/2 cells. Furthermore, MAML1−/− embryos at E14.5 and 16.5 had shorter bone lengths than wild type embryos. The area of primary spongiosa of the femoral diaphysis was narrowed, indicated that chondrocyte maturation was impaired. These data suggest that MAML1 enhanced the transcriptional activity of Runx2 and plays a role in bone development.
Results
MAML1 enhances the transcriptional activity of Runx2
We used a full-length cDNA library containing approximately 10,000 clones (FLJ clones, established by New Energy and Industrial Technology Development Organization [NEDO], Japan) and p6OSE2-Luc reporter assay system (Figure 1A). We identified a few novel factors that enhance Runx2 transcriptional activity. Among them, AK123604 (Homo sapiens cDNA FLJ41610), which is highly similar to Mastermind-like protein 3 (MAML3), showed especially strong activity. MAML is a human homolog of Drosophila mastermind, a protein that plays a role in the Notch signaling. MAML family members consist of MAML1, MAML2 and MAML3. We found that MAML1 and MAML2 also enhanced Runx2 transcriptional activity as well (Figure 1B). Because the establishment of knockout mice of MAML1 preceded MAML2 and MAML3, we primarily analyzed MAML1.
Fig. 1. MAML1 enhances the transcriptional activity of Runx2. 
A, 293T cells were transiently transfected with p6OSE2-Luc reporter, Runx2 expression plasmid, and about 10,000 FLJ expression plasmids. B, 293T cells were transiently transfected with a p6OSE2-Luc reporter or p6OSE2-mutant-Luc together with MAML1, MAML2, MAML3, and Runx2 expression plasmids. Luciferase levels were normalized to the Renilla luciferase activity of a cotransfected phRL-TK-Luc reporter and presented as fold activation relative to the luciferase level of the p6OSE2-Luc reporter construct alone. Error bars represent the standard deviation of triplicate transfections. MAML1 enhances Runx2 activity in a Notch-independent manner in vitro
MAML1 consists of 1016 amino acids and contains a conserved basic region and two acidic regions. To investigate which region is essential for regulating Runx2, we assessed each deletion mutant by p6OSE2-Luc reporter assay (Figure 2). The N-terminal basic region, which is essential for the interaction with Notch, and the C-terminal acidic region of MAML1 were dispensable for Runx2 transcriptional activity. On the other hand, the center region (residues 343–711), whose function is not well known, was essential.
Fig. 2. MAML1 enhances Runx2 activity in a Notch-independent manner in vitro. 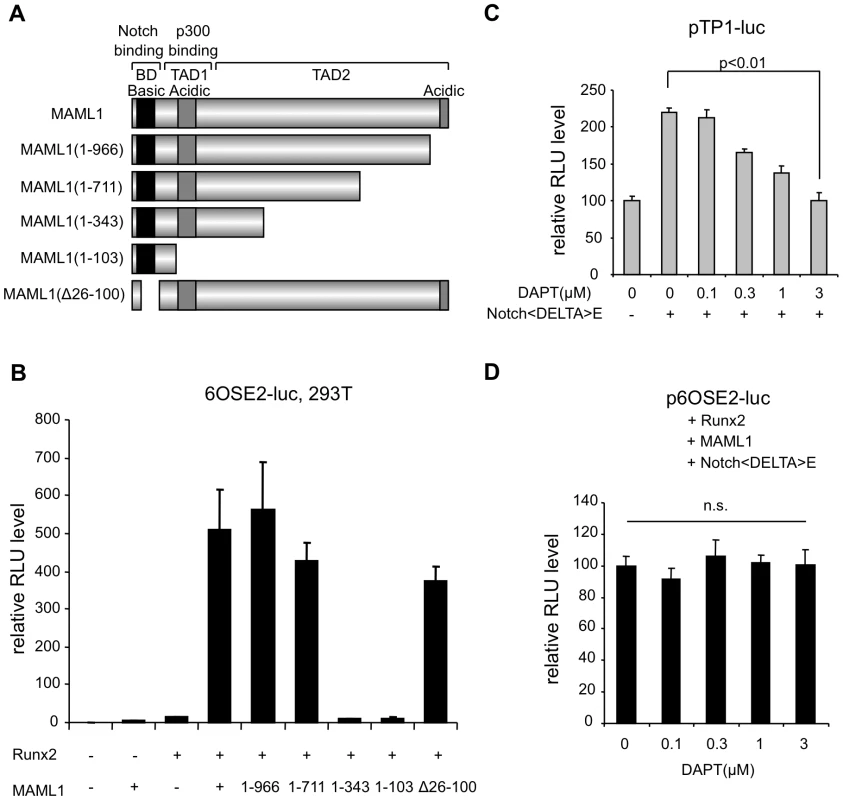
A, The structure of MAML1 and its truncated forms. B, 293T cells were transiently transfected with a p6OSE2-Luc reporter alone or together with truncated forms of MAML1 Error bars represent the standard deviation of triplicate transfections. C, 293T cells were transiently transfected with a pTP1-Luc reporter and NotchΔE expression plasmid in the presence of γ-secretase inhibitor (DAPT). Error bars represent the standard deviation of triplicate transfections. D, 293T cells were transiently transfected with a p6OSE2-Luc reporter with Runx2, MAML1 and NotchΔE expression plasmid in the presence of γ-secretase inhibitor (DAPT). Error bars represent the standard deviation of triplicate transfections. Because MAML1 is a coactivator of Notch signaling, we investigated whether or not the action of MAML1 on Runx2 was dependent on Notch. Notch1ΔE is cleaved by γ-secretase to produce the Notch intracellular domain (NICD), which translocates into the nucleus and transactivates the target gene. A Γ-secretase inhibitor DAPT inhibited the Notch1ΔE-mediated activation of pTP1-Luc, in which luciferase expression was controlled by Notch signaling (Figure 2C). On the other hand, DAPT did not affect the action of MAML1 on Runx2 in the presence of Notch1ΔE (Figure 2D). This suggests that MAML1 possibly enhance the transcriptional activity of Runx2 in a Notch-independent manner.
MAML1 promotes Runx2-mediated osteoblastic differentiation
293T cell used in the luciferase assay is derived from human embryonic kidney and does not express Runx2. Therefore, we next investigated whether MAML1 controls osteoblastic differentiation through Runx2 in the murine pluripotent mesenchymal cell line C3H10T1/2 (Figure 3). Overexpression of Runx2 promoted the expression of the ALP gene, an early osteoblast marker. Co-overexpression of MAML1 rapidly augmented the Runx2-mediated expression of ALP, whereas MAML1 alone did not induce ALP expression. However, this effect was observed only in early phase of osteoblast differentiation and later phase markers such as bone sialoprotein and osteocalcin were not changed (data not shown).
Fig. 3. MAML1 promotes Runx2-mediated osteoblastic differentiation. 
A, C3H10T1/2 cells were transiently transfected with a Runx2 and/or MAML1 expression plasmids and cultured for 2 days. B, Total RNA was isolated and reverse-transcribed and TaqMan real-time PCR was performed to investigate the expression level of alkaline phosphatase gene, a marker of osteoblast. Analysis of skeletal defects in MAML1−/− mice
We analyzed MAML1 knockout (MAML1−/−) mice [7]. Normal Mendelian ratios are observed up to E18.5, but MAML1−/− mice with C57BL/6 background die during the perinatal period. In the original paper, however, MAML1−/ − mice die within 14 days after birth [7]. The difference of lethality in the mice is thought to be due to the difference of the background. At E16.5, MAML1−/− mice were smaller than wild type mice (Figure 4A). Whole mounted embryos at E16.5 stained with Alcian Blue and Alizarin Red showed that the mineralized region in the long bones of MAML1−/− mice was relatively short compared with wild type mice (Figure 4A). Histological analysis revealed that the area of primary spongiosa of the femoral diaphysis was reduced in MAML1−/− mice compared to wild type mice (Figure 4B, 4C). At E14.5, extended zone of Col2a1 and Sox9 expression, markers of chondrocyte differentiation, and decreased zone of Col10a1 expression, a marker of hypertrophic chondrocyte, were observed (Figure 4D). These observations indicated the impairment of chondrocyte maturation in MAML1−/− mice.
Fig. 4. Analysis of skeletal defects in MAML1−/− mice. 
A, Whole mounted embryos at E16.5 were stained with Alcian Blue and Alizarin Red. B, The femoral sections at E16.5 were stained with Alcian Blue (upper panel). Arrow bar indicates the area of primary spongiosa. The expression of collagen, type 10 alpha 1, a marker of hypertrophic chondrocyte, was shown by in situ hybridization (lower panel). Green bars, primary spongiosa; red bars, hypertrophic zone; blue bars, proliferating zone. Black bars, 200 µm. C, The primary spongiosa length of MAML1 null (KO) embryos and wild type (WT) littermates. D. Femoral sections at E14.5. The expression of type 10 alpha 1 collagen, type 2 alpha 1 collagen and sox9 was shown by in situ hybridization. Bars, 100 µm. Discussion
We utilized approximately 10,000 arrayed and addressable cDNA clones, which allowed systematic, efficient, and unbiased screening of cDNAs encoding factors that could activate Runx2-mediated expression of the p6OSE2-Luc reporter construct (Figure 1). This revealed that MAML was a potential activator of Runx2-mediated luciferase expression.
MAML is a coactivator of Notch signaling. Upon ligand stimulation from neighboring cells, Notch is cleaved by γ-secretase and its intracellular domain (NICD) translocates into the nucleus [8]. NICD interacts with CSL through a RAM domain at the N-terminus that has high affinity for the β-trefoil domain of CSL. Then, the ankyrin repeats domain of NICD docks with the Rel-homology domain of CSL and creates a high-affinity binding site for MAML. MAML associates with the CSL-NICD complex through the N-terminal basic region, recruits p300, RNA polymerase II and other unknown factors, and activates the transcription of target genes such as Hes1 [9], [10]. On the other hand, Notch-independent action of MAML1 on p53 [11], beta-catenin [12], MEF2C [13] and NF-kappaB [14] has been previously reported.
Recently, two groups have published studies using genetically modified mice [15], [16]. Hilton and colleagues showed that Notch signaling inhibits osteoblast differentiation through Hes or Hey proteins, which diminish Runx2 transcriptional activity via physical interaction, and acts to maintain a pool of mesenchymal progenitors. Engin and colleagues showed that pathological gain of Notch function in established osteoblastic lineages activates expansion of the immature osteoblastic pool by increasing transcription of the genes encoding osterix, cyclin D and cyclin E and by repressing the function of Runx2 by direct interaction and inhibition of its binding. These findings suggest that Notch signaling negatively regulate the function of Runx2. We indicated that the N-terminal Notch-binding region of MAML1 is dispensable for the action of MAML1 on Runx2 and Notch signaling inhibitor does not affected the action of MAML1 on Runx2. Furthermore, knockdown of p300, a coactivator [9], [10], did not affected the activation of Runx2 transcriptional activity by MAML1 (data not shown). These data suppose that the action of MAML1 on Runx2 is Notch-independent.
To elucidate how MAML regulates Runx2-mediated transcription, we investigated the physical interaction of MAML1 with Runx2, but we could not demonstrate the interaction between Runx2 and MAML1 (data not shown), suggesting that this interaction is very weak and possibly indirect.
We showed the impairment of chondrocyte maturation in MAML1−/− mice. Because Runx2 facilitates chondrocyte maturation, the phenotype of MAML1−/− mice may be caused by the dysfunction of Runx2. On the other hand, the expression of Sox9, a transcription activator of collagen type II, was upregulated by Notch activation and this activation of Notch signaling thereby promoted differentiation of proliferative and prehypertrophic chondrocytes [17]. Therefore, from this current findings, it is not clear yet whether or not the phenotype of MAML1−/− mice is due to the dysfunction of Runx2 or Notch signaling. Other possibility to explain MAML1−/− mice bone phenotype is that other cell signaling cascades and molecules could be involved into MAML dependent gene regulation and thus bone development. For example, MEF2C, a transcription factor that regulates muscle and cardiovascular development, was reported to control bone development by activating the gene program for chondrocyte hypertrophy [18].
Taken together, our analysis revealed novel function of MAML1, Notch independent promotion of Runx2 activity and its role in bone development. Further elucidation of the precise molecular mechanisms responsible for the initiation and termination of this functional association during bone development may provide us with a new basis for understanding the molecular network in osteoblasts and potential therapeutic targets for bone diseases.
Materials and Methods
Ethics statement
All animal experiments were performed according to protocols approved by the Institutional Animal Care and Use Committee at National Institute for Child Health and Development (Protocol 2004-003).
Plasmids
The p6OSE2-Luc and p6OSE2-mut-Luc reporter construct were previously reported [19]. The pEF-BOS hMam-1 (MAML1) plasmid, its truncated forms [20], hMam-2 (MAML3), hMam-3 (MAML2) [21] and pCS2+Notch1ΔE [22] were previously reported. The pCG mRunx2 plasmid by Dr. Nakashima (Tokyo Medical and Dental University, Tokyo), and the p3xFLAG mRunx2 plasmid by Dr. Hikata (Keio University, Tokyo). The pTP1-Luc (pGa981-6) construct was provided by Dr. Ursula Strobl (Institute of Clinical Molecular Biology and Tumor Genetics, Germany).
Reporter assays
For the primary screening, we diluted approximately 10,000 FLJ clones (Full-length human cDNA sequencing project, NEDO) to 10 ng/µL in 10 mM Tris-HCl (pH 8.5), and dispensed 5 µL to each well in 384-well plates. We then added 10 ng of p6OSE2-Luc, 2 ng of pCG, 0.1 µL of Fugene6 (Roche Diagnostics), and 5 µL of Opti-MEM I Reduced-Serum Medium (Invitrogen) to each well. 293T cells were diluted to 1.25×105 cells/mL with Dulbecco's modified Eagle medium (DMEM) containing 10% heat-inactivated fetal bovine serum (FBS), 50 units/mL penicillin, and 50 µg/mL streptomycin, and seeded at 40 µL (5,000 cells) per well. After 48 hours of culture, we removed the supernatant and added 40 µL of Steady Glo Luciferase Assay Reagent (Promega) diluted 2-fold with phosphate buffered saline (PBS) to each well. After 10 minutes at room temperature, luminescence was measured using a plate reader (ARVO, Perkin Elmer). After the second screening, the assay was performed in 96-well plates. We added γ-secretase inhibitor IX, N-[N-(3, 5-difluorophenylacetyl-L-alanyl)]-S-phenylglycine t-butylester (DAPT; Calbiochem), to the medium 2 hours before transfection.
Cell culture, transfection, and differentiation assays
We purchased the C3H10T1/2 murine pluripotent mesenchymal cell line from ATCC and maintained it in DMEM containing 10% heat-inactivated FBS, 50 units/mL penicillin, and 50 µg/mL streptomycin. For differentiation assays, we seeded cells in a multi-well plate at a density of 2,000 cells/cm2 and cultured them for 3 days. The medium was then changed to the osteoblastic medium (MEM-alpha containing heat-inactivated FBS, 50 units/mL penicillin, 50 µg/mL streptomycin, 50 µM ascorbic acid 2-phosphate, 10 mM β-glycerophosphate, and 0.1 µM dexamethasone), transfected with p3xFLAG-Runx2 and/or pEF BOS-hMam1 by FugeneHD (Roche diagnostics), and cultured.
Real-time PCR
We isolated total RNA from the cultured cells using the RNeasy mini kit (QIAGEN) and reverse transcribed 2 µg of total RNA using Ready-To-Go You-Prime First-Strand Beads (GE Healthcare) and oligo-dT primer. The products were diluted 10-fold with distilled water and used as a template for real-time PCR. Real-time PCR was performed using a TaqMan Gene Expression Assay, TaqMan Universal PCR Mix and the 7900HT Fast Real-Time PCR System (Applied Biosystems).
Double staining of MAML1−/− mouse embryos
We backcrossed MAML1 null mice [7] at least 10 times onto a C57BL/6 background. We fixed mouse embryos on embryonic day 16.5 (E16.5) in ethanol overnight and then stained them overnight with Alcian blue solution (0.15 mg/mL Alcian blue 8GX in 20% acetic acid and 80% ethanol). The embryos were washed briefly with ethanol twice, treated with 2% potassium hydroxide overnight, and then stained overnight with Alizarin red solution (0.075 mg/mL Alizarin red S in 1% potassium hydroxide).
Alcian blue staining
Tissues were fixed in 4% paraformaldehyde-PBS overnight at 4°C, processed, embedded in paraffin, and sectioned. Slides were deparaffinized, washed with water, treated with 3% acetic acid, and then with 1% Alcian blue 8GX for 60 minutes. After staining, we washed the slides briefly with 3% acetic acid, then with water for 5 minutes, counterstained with Kernechtrot Stain Solution (Muto Pure Chemicals, Tokyo) for 5 minutes, washed with water for 3 minutes, and dehydrated the slides.
In situ hybridization
Tissues were fixed in 4% paraformaldehyde-PBS overnight at 4°C, processed, embedded in paraffin, and sectioned. Slides were deparaffinized, treated with proteinase K (8 µg/mL) for 10 minutes at RT, and then with 0.2% glycine in PBS for 10 minutes at RT. Slides were refixed in 4% paraformaldehyde-PBS for 10 minutes at RT, washed with PBS for 5 minutes 3 times, acetylated with 0.1 M triethanolamin-HCl (pH 8.0) for 10 minutes, washed with PBS for 30 minutes, and then prehybridized with prehybridization buffer (50% deionized formamide and 5× saline-sodium citrate (SSC)) for 60 minutes at 65°C. We hybridized the slides with DIG-labeled antisense riboprobes in hybridization buffer (50% deionized formamide, 5× SSC, 0.25 mg/mL yeast tRNA, 10% dextran sulfate, and 5× Denhardt's solution) in a humidified chamber at 65°C overnight. After hybridization, the slides were washed with 5× SSC (1× SSC: 0.15 M NaCl, 0.015 M sodium citrate) at 65°C for 20 minutes, 0.2× SSC at 65°C for 3 hours, and NT buffer (0.1 M Tris-HCl [pH 7.5], 0.15 M NaCl) for 5 minutes at RT. We incubated the slides at 4°C overnight with alkaline phosphatase (ALP)-coupled anti-DIG antibody in NT buffer containing 0.1% sheep serum. The slides were washed with NT buffer for 15 minutes 3 times and equilibrated in NTM (0.1 M NaCl, 0.1 M Tris-HCl [pH 9.5], and 0.05 M MgCl2) for 5 minutes at RT. The slides were then treated with BM Purple AP Substrate (Roche) for 3 hours at RT in a humid chamber protected from light.
Statistical analysis
The two-tailed independent Student's t-test was used to calculate all P values.
Zdroje
1. KomoriT (2002) Runx2, a multifunctional transcription factor in skeletal development. J Cell Biochem 87 : 1–8.
2. DucyP, ZhangR, GeoffroyV, RidallAL, KarsentyG (1997) Osf2/Cbfa1: a transcriptional activator of osteoblast differentiation. Cell 89 : 747–754.
3. HongJH, HwangES, McManusMT, AmsterdamA, TianY, et al. (2005) TAZ, a transcriptional modulator of mesenchymal stem cell differentiation. Science 309 : 1074–1078.
4. WangW, WangYG, ReginatoAM, GlotzerDJ, FukaiN, et al. (2004) Groucho homologue Grg5 interacts with the transcription factor Runx2-Cbfa1 and modulates its activity during postnatal growth in mice. Dev Biol 270 : 364–381.
5. BermanSD, YuanTL, MillerES, LeeEY, CaronA, et al. (2008) The retinoblastoma protein tumor suppressor is important for appropriate osteoblast differentiation and bone development. Mol Cancer Res 6 : 1440–1451.
6. VegaRB, MatsudaK, OhJ, BarbosaAC, YangX, et al. (2004) Histone deacetylase 4 controls chondrocyte hypertrophy during skeletogenesis. Cell 119 : 555–566.
7. OyamaT, HarigayaK, MuradilA, HozumiK, HabuS, et al. (2007) Mastermind-1 is required for Notch signal-dependent steps in lymphocyte development in vivo. Proc Natl Acad Sci USA 104 : 9764–9769.
8. FortiniME (2002) Gamma-secretase-mediated proteolysis in cell-surface-receptor signalling. Nat Rev Mol Cell Biol 3 : 673–684.
9. LubmanOY, KorolevSV, KopanR (2004) Anchoring notch genetics and biochemistry; structural analysis of the ankyrin domain sheds light on existing data. Mol Cell 13 : 619–626.
10. NamY, SlizP, SongL, AsterJC, BlacklowSC (2006) Structural basis for cooperativity in recruitment of MAML co-activators to Notch transcription complexes. Cell 124 : 973–983.
11. ZhaoY, KatzmanRB, DelmolinoLM, BhatI, ZhangY, et al. (2007) The notch regulator MAML1 interacts with p53 and functions as a co-activator. J Biol Chem 282 : 11969–11981.
12. Alves-GuerraMC, RonchiniC, CapobiancoAJ (2007) Mastermind-like 1 Is a specific co-activator of β-catenin transcription activation and is essential for colon carcinoma cell survival. Cancer Res 67 : 8690–8698.
13. ShenH, McElhinnyAS, CaoY, GaoP, LiuJ, et al. (2006) The Notch co-activator, MAML1, functions as a novel co-activator for MEF2C-mediated transcription and is required for normal myogenesis. Genes Dev 20 : 675–688.
14. JinB, ShenH, LinS, LiJL, ChenZ, et al. (2010) The mastermind-like 1 (MAML1) co-activator regulates constitutive NF-kappaB signaling and cell survival. J Biol Chem 285 (19)
14356–65.
15. HiltonMJ, TuX, WuX, BaiS, ZhaoH, et al. (2008) Notch signaling maintains bone marrow mesenchymal progenitors by suppressing osteoblast differentiation. Nat Med 14 : 306–314.
16. EnginF, YaoZ, YangT, ZhouG, BertinT, et al. (2008) Dimorphic effects of Notch signaling in bone homeostasis. Nat Med 14 : 299–305.
17. NakanishiK, ChanYS, ItoK (2007) Notch signaling is required for the chondrogenic specification of mouse mesencephalic neural crest cells. Mech Dev 124 : 190–203.
18. ArnoldMA, KimY, CzubrytMP, PhanD, McAnallyJ, et al. (2007) MEF2C transcription factor controls chondrocyte hypertrophy and bone development. Dev Cell 12 (3)
377–89.
19. GeoffroyV, DucyP, KarsentyG (1995) A PEBP2 alpha/AML-1-related factor increases osteocalcin promoter activity through its binding to an osteoblast-specific cis-acting element. J Biol Chem 270 (52)
30973–9.
20. KitagawaM, OyamaT, KawashimaT, YedvobnickB, KumarA, et al. (2001) A human protein with sequence similarity to Drosophila mastermind coordinates the nuclear form of notch and a CSL protein to build a transcriptional activator complex on target promoters. Mol Cell Biol 21 (13)
4337–46.
21. LinSE, OyamaT, NagaseT, HarigayaK, KitagawaM (2002) Identification of new human mastermind proteins defines a family that consists of positive regulators for notch signaling. J Biol Chem 277 (52)
50612–20.
22. JarriaultS, BrouC, LogeatF, SchroeterEH, KopanR, et al. (1995) Signalling downstream of activated mammalian Notch. Nature 377 (6547)
355–8.
Štítky
Genetika Reprodukční medicína
Článek Comparative Genome Structure, Secondary Metabolite, and Effector Coding Capacity across PathogensČlánek TATES: Efficient Multivariate Genotype-Phenotype Analysis for Genome-Wide Association StudiesČlánek Secondary Metabolism and Development Is Mediated by LlmF Control of VeA Subcellular Localization inČlánek Human Disease-Associated Genetic Variation Impacts Large Intergenic Non-Coding RNA ExpressionČlánek The Roles of Whole-Genome and Small-Scale Duplications in the Functional Specialization of GenesČlánek The Role of Autophagy in Genome Stability through Suppression of Abnormal Mitosis under Starvation
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2013 Číslo 1- Akutní intermitentní porfyrie
- Souvislost haplotypu M2 genu pro annexin A5 s opakovanými reprodukčními ztrátami
- Příjem alkoholu a menstruační cyklus
- Doporučení pro diagnostiku a léčbu akutních jaterních porfyrií
- Doc. Miloš Kubánek: Nemocní se srdeční amyloidózou jsou často skryti a sledováni pod jinými diagnózami
-
Všechny články tohoto čísla
- A Model of High Sugar Diet-Induced Cardiomyopathy
- Comparative Genome Structure, Secondary Metabolite, and Effector Coding Capacity across Pathogens
- Emerging Function of Fat Mass and Obesity-Associated Protein (Fto)
- Positional Cloning Reveals Strain-Dependent Expression of to Alter Susceptibility to Bleomycin-Induced Pulmonary Fibrosis in Mice
- Genetics of Ribosomal Proteins: “Curiouser and Curiouser”
- Transposable Elements Re-Wire and Fine-Tune the Transcriptome
- Function and Regulation of , a Gene Implicated in Autism and Human Evolution
- MAML1 Enhances the Transcriptional Activity of Runx2 and Plays a Role in Bone Development
- Predicting Mendelian Disease-Causing Non-Synonymous Single Nucleotide Variants in Exome Sequencing Studies
- A Systematic Mapping Approach of 16q12.2/ and BMI in More Than 20,000 African Americans Narrows in on the Underlying Functional Variation: Results from the Population Architecture using Genomics and Epidemiology (PAGE) Study
- Transcription of the Major microRNA–Like Small RNAs Relies on RNA Polymerase III
- Histone H3K56 Acetylation, Rad52, and Non-DNA Repair Factors Control Double-Strand Break Repair Choice with the Sister Chromatid
- Genome-Wide Association Study Identifies a Novel Susceptibility Locus at 12q23.1 for Lung Squamous Cell Carcinoma in Han Chinese
- Genetic Disruption of the Copulatory Plug in Mice Leads to Severely Reduced Fertility
- The [] Prion Exists as a Dynamic Cloud of Variants
- Adult Onset Global Loss of the Gene Alters Body Composition and Metabolism in the Mouse
- Fis Protein Insulates the Gene from Uncontrolled Transcription
- The Meiotic Nuclear Lamina Regulates Chromosome Dynamics and Promotes Efficient Homologous Recombination in the Mouse
- Genome-Wide Haplotype Analysis of Expression Quantitative Trait Loci in Monocytes
- TATES: Efficient Multivariate Genotype-Phenotype Analysis for Genome-Wide Association Studies
- Structural Basis of a Histone H3 Lysine 4 Demethylase Required for Stem Elongation in Rice
- The Ecm11-Gmc2 Complex Promotes Synaptonemal Complex Formation through Assembly of Transverse Filaments in Budding Yeast
- MCM8 Is Required for a Pathway of Meiotic Double-Strand Break Repair Independent of DMC1 in
- Comparative Genomic Analysis of the Endosymbionts of Herbivorous Insects Reveals Eco-Environmental Adaptations: Biotechnology Applications
- Integration of Nodal and BMP Signals in the Heart Requires FoxH1 to Create Left–Right Differences in Cell Migration Rates That Direct Cardiac Asymmetry
- Pharmacodynamics, Population Dynamics, and the Evolution of Persistence in
- A Hybrid Likelihood Model for Sequence-Based Disease Association Studies
- Aberration in DNA Methylation in B-Cell Lymphomas Has a Complex Origin and Increases with Disease Severity
- Multiple Opposing Constraints Govern Chromosome Interactions during Meiosis
- Transcriptional Dynamics Elicited by a Short Pulse of Notch Activation Involves Feed-Forward Regulation by Genes
- Dynamic Large-Scale Chromosomal Rearrangements Fuel Rapid Adaptation in Yeast Populations
- Heterologous Gln/Asn-Rich Proteins Impede the Propagation of Yeast Prions by Altering Chaperone Availability
- Gene Copy-Number Polymorphism Caused by Retrotransposition in Humans
- An Incompatibility between a Mitochondrial tRNA and Its Nuclear-Encoded tRNA Synthetase Compromises Development and Fitness in
- Secondary Metabolism and Development Is Mediated by LlmF Control of VeA Subcellular Localization in
- Single-Stranded Annealing Induced by Re-Initiation of Replication Origins Provides a Novel and Efficient Mechanism for Generating Copy Number Expansion via Non-Allelic Homologous Recombination
- Tbx2 Controls Lung Growth by Direct Repression of the Cell Cycle Inhibitor Genes and
- Suv4-20h Histone Methyltransferases Promote Neuroectodermal Differentiation by Silencing the Pluripotency-Associated Oct-25 Gene
- A Conserved Helicase Processivity Factor Is Needed for Conjugation and Replication of an Integrative and Conjugative Element
- Telomerase-Null Survivor Screening Identifies Novel Telomere Recombination Regulators
- Genome-Wide Analysis Reveals Selection for Important Traits in Domestic Horse Breeds
- Coordinated Degradation of Replisome Components Ensures Genome Stability upon Replication Stress in the Absence of the Replication Fork Protection Complex
- Nkx6.1 Controls a Gene Regulatory Network Required for Establishing and Maintaining Pancreatic Beta Cell Identity
- HIF- and Non-HIF-Regulated Hypoxic Responses Require the Estrogen-Related Receptor in
- Delineating a Conserved Genetic Cassette Promoting Outgrowth of Body Appendages
- The Telomere Capping Complex CST Has an Unusual Stoichiometry, Makes Multipartite Interaction with G-Tails, and Unfolds Higher-Order G-Tail Structures
- Comprehensive Methylome Characterization of and at Single-Base Resolution
- Loci Associated with -Glycosylation of Human Immunoglobulin G Show Pleiotropy with Autoimmune Diseases and Haematological Cancers
- Switchgrass Genomic Diversity, Ploidy, and Evolution: Novel Insights from a Network-Based SNP Discovery Protocol
- Centromere-Like Regions in the Budding Yeast Genome
- Sequencing of Loci from the Elephant Shark Reveals a Family of Genes in Vertebrate Genomes, Forged by Ancient Duplications and Divergences
- Mendelian and Non-Mendelian Regulation of Gene Expression in Maize
- Mutational Spectrum Drives the Rise of Mutator Bacteria
- Human Disease-Associated Genetic Variation Impacts Large Intergenic Non-Coding RNA Expression
- The Roles of Whole-Genome and Small-Scale Duplications in the Functional Specialization of Genes
- Sex-Specific Signaling in the Blood–Brain Barrier Is Required for Male Courtship in
- A Newly Uncovered Group of Distantly Related Lysine Methyltransferases Preferentially Interact with Molecular Chaperones to Regulate Their Activity
- Is Required for Leptin-Mediated Depolarization of POMC Neurons in the Hypothalamic Arcuate Nucleus in Mice
- Unlocking the Bottleneck in Forward Genetics Using Whole-Genome Sequencing and Identity by Descent to Isolate Causative Mutations
- The Role of Autophagy in Genome Stability through Suppression of Abnormal Mitosis under Starvation
- MTERF3 Regulates Mitochondrial Ribosome Biogenesis in Invertebrates and Mammals
- Downregulation and Altered Splicing by in a Mouse Model of Facioscapulohumeral Muscular Dystrophy (FSHD)
- NBR1-Mediated Selective Autophagy Targets Insoluble Ubiquitinated Protein Aggregates in Plant Stress Responses
- Retroactive Maintains Cuticle Integrity by Promoting the Trafficking of Knickkopf into the Procuticle of
- Phenome-Wide Association Study (PheWAS) for Detection of Pleiotropy within the Population Architecture using Genomics and Epidemiology (PAGE) Network
- Genetic and Functional Modularity of Activities in the Specification of Limb-Innervating Motor Neurons
- A Population Genetic Model for the Maintenance of R2 Retrotransposons in rRNA Gene Loci
- A Quartet of PIF bHLH Factors Provides a Transcriptionally Centered Signaling Hub That Regulates Seedling Morphogenesis through Differential Expression-Patterning of Shared Target Genes in
- A Genome-Wide Integrative Genomic Study Localizes Genetic Factors Influencing Antibodies against Epstein-Barr Virus Nuclear Antigen 1 (EBNA-1)
- Mutation of the Diamond-Blackfan Anemia Gene in Mouse Results in Morphological and Neuroanatomical Phenotypes
- Life, the Universe, and Everything: An Interview with David Haussler
- Alternative Oxidase Expression in the Mouse Enables Bypassing Cytochrome Oxidase Blockade and Limits Mitochondrial ROS Overproduction
- An Evolutionarily Conserved Synthetic Lethal Interaction Network Identifies FEN1 as a Broad-Spectrum Target for Anticancer Therapeutic Development
- The Flowering Repressor Underlies a Novel QTL Interacting with the Genetic Background
- Telomerase Is Required for Zebrafish Lifespan
- and Diversified Expression of the Gene Family Bolster the Floral Stem Cell Network
- Susceptibility Loci Associated with Specific and Shared Subtypes of Lymphoid Malignancies
- An Insertion in 5′ Flanking Region of Causes Blue Eggshell in the Chicken
- Increased Maternal Genome Dosage Bypasses the Requirement of the FIS Polycomb Repressive Complex 2 in Arabidopsis Seed Development
- WNK1/HSN2 Mutation in Human Peripheral Neuropathy Deregulates Expression and Posterior Lateral Line Development in Zebrafish ()
- Synergistic Interaction of Rnf8 and p53 in the Protection against Genomic Instability and Tumorigenesis
- Dot1-Dependent Histone H3K79 Methylation Promotes Activation of the Mek1 Meiotic Checkpoint Effector Kinase by Regulating the Hop1 Adaptor
- A Heterogeneous Mixture of F-Series Prostaglandins Promotes Sperm Guidance in the Reproductive Tract
- Starvation, Together with the SOS Response, Mediates High Biofilm-Specific Tolerance to the Fluoroquinolone Ofloxacin
- Directed Evolution of a Model Primordial Enzyme Provides Insights into the Development of the Genetic Code
- Genome-Wide Screens for Tinman Binding Sites Identify Cardiac Enhancers with Diverse Functional Architectures
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Function and Regulation of , a Gene Implicated in Autism and Human Evolution
- An Insertion in 5′ Flanking Region of Causes Blue Eggshell in the Chicken
- Comprehensive Methylome Characterization of and at Single-Base Resolution
- Susceptibility Loci Associated with Specific and Shared Subtypes of Lymphoid Malignancies
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



