-
Články
- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Mutations in and Cause “Splashed White” and Other White Spotting Phenotypes in Horses
During fetal development neural-crest-derived melanoblasts migrate across the entire body surface and differentiate into melanocytes, the pigment-producing cells. Alterations in this precisely regulated process can lead to white spotting patterns. White spotting patterns in horses are a complex trait with a large phenotypic variance ranging from minimal white markings up to completely white horses. The “splashed white” pattern is primarily characterized by an extremely large blaze, often accompanied by extended white markings at the distal limbs and blue eyes. Some, but not all, splashed white horses are deaf. We analyzed a Quarter Horse family segregating for the splashed white coat color. Genome-wide linkage analysis in 31 horses gave a positive LOD score of 1.6 in a region on chromosome 6 containing the PAX3 gene. However, the linkage data were not in agreement with a monogenic inheritance of a single fully penetrant mutation. We sequenced the PAX3 gene and identified a missense mutation in some, but not all, splashed white Quarter Horses. Genome-wide association analysis indicated a potential second signal near MITF. We therefore sequenced the MITF gene and found a 10 bp insertion in the melanocyte-specific promoter. The MITF promoter variant was present in some splashed white Quarter Horses from the studied family, but also in splashed white horses from other horse breeds. Finally, we identified two additional non-synonymous mutations in the MITF gene in unrelated horses with white spotting phenotypes. Thus, several independent mutations in MITF and PAX3 together with known variants in the EDNRB and KIT genes explain a large proportion of horses with the more extreme white spotting phenotypes.
Published in the journal: . PLoS Genet 8(4): e32767. doi:10.1371/journal.pgen.1002653
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1002653Summary
During fetal development neural-crest-derived melanoblasts migrate across the entire body surface and differentiate into melanocytes, the pigment-producing cells. Alterations in this precisely regulated process can lead to white spotting patterns. White spotting patterns in horses are a complex trait with a large phenotypic variance ranging from minimal white markings up to completely white horses. The “splashed white” pattern is primarily characterized by an extremely large blaze, often accompanied by extended white markings at the distal limbs and blue eyes. Some, but not all, splashed white horses are deaf. We analyzed a Quarter Horse family segregating for the splashed white coat color. Genome-wide linkage analysis in 31 horses gave a positive LOD score of 1.6 in a region on chromosome 6 containing the PAX3 gene. However, the linkage data were not in agreement with a monogenic inheritance of a single fully penetrant mutation. We sequenced the PAX3 gene and identified a missense mutation in some, but not all, splashed white Quarter Horses. Genome-wide association analysis indicated a potential second signal near MITF. We therefore sequenced the MITF gene and found a 10 bp insertion in the melanocyte-specific promoter. The MITF promoter variant was present in some splashed white Quarter Horses from the studied family, but also in splashed white horses from other horse breeds. Finally, we identified two additional non-synonymous mutations in the MITF gene in unrelated horses with white spotting phenotypes. Thus, several independent mutations in MITF and PAX3 together with known variants in the EDNRB and KIT genes explain a large proportion of horses with the more extreme white spotting phenotypes.
Introduction
Coat color is a well-studied model trait for geneticists. Coat color phenotypes are relatively easy to record, which facilitates their analysis. In mammals melanocytes cover the entire body surface and are responsible for the pigmentation of skin, hairs, and eyes. Melanocytes are formed during fetal development from melanoblasts, which originate in the neural crest and migrate across the developing fetus in order to reach their final position on the body [1]. This developmental program requires a delicate level of regulation to ensure that the correct amount of cells reaches their final destination [2]–[5]. An over-proliferation of cells that have left their surrounding tissue might have fatal consequences for the developing fetus [6]. If however too few of the migrating melanoblasts survive, this will lead to partially or completely unpigmented phenotypes [7]. Domestic animals with such unpigmented phenotypes have been highly valued due to their striking appearance and have often been actively selected in animal breeding. Consequently, our modern domestic animals provide a large repertoire of spontaneous mutants that allow the dissection of the contributions of individual genes to migration, proliferation, differentiation, and survival of melanocytes.
From a genetic point of view, white spotting is considered a complex trait [8]. Phenotypes range from tiny white spots at the extremities of the body to large unpigmented areas, in symmetrical or asymmetrical patterns, up to completely unpigmented animals [9]. Different combinations of alleles at several loci can interact and it is generally impossible to precisely predict the genotype of an animal based only on a given white spotting phenotype.
Splashed white is a distinctive but nevertheless quite variable white spotting pattern in horses, which is primarily characterized by extensive depigmentation of the head. Splashed white horses often have blue eyes and they are sometimes deaf [9]. This phenotype has not yet been characterized at the molecular level.
Mutations in two genes have been identified to cause pronounced depigmentation phenotypes in horses thus far. A total of 19 different mutant alleles at or near the KIT gene were reported to cause either completely white horses or horses with pronounced depigmentation, such as the dominant white, tobiano, and sabino-1 spotting patterns [10]–[14]. The frame overo spotting pattern, which is phenotypically overlapping with the splashed white phenotype, is caused by a missense mutation in the EDNRB gene [15]–[17]. EDNRB mutations are also found in a small fraction of human patients with Waardenburg syndrome (WS). WS is characterized by pigmentation abnormalities, such as a typical white forelock or white skin patches, strikingly blue or heterochromatic irises, and varying degrees of sensorineural deafness and skeletal dysmorphologies. Other forms of human Waardenburg syndrome are caused by mutations in EDN3, MITF, PAX3, SNAI2, and SOX10 [18]. Mouse mutants for these genes are available and show largely similar although not completely identical phenotypes [19].
In this study we report the identification of several independent mutations in horses with striking depigmentation phenotypes that show parallels to human Waardenburg syndrome.
Results
Splashed white in a Quarter Horse family
We obtained samples from a large Quarter Horse family segregating for a striking white spotting pattern termed splashed white (Figure 1). In our material this phenotype was characterized by a large blaze of variable size and shape. Many splashed white horses had an extremely large blaze extending over the eyes or even covering parts of the cheeks; such a head pattern is termed baldface. Splashed white horses also typically had high markings that extend high up on their legs and occasionally little white belly spots. Most of the splashed white horses had blue eyes or iris heterochromia. Although many splashed white horses have a very characteristic appearance, the phenotype is variable and in some splashed white horses the unpigmented areas are so small that we could not reliably distinguish these horses from horses with other subtle depigmentation phenotypes. Some, but not all of the sampled splashed white horses were deaf.
Fig. 1. Phenotypes of splashed white horses from a Quarter Horse family. 
Note that the expression of the phenotype is quite variable. Splashed white may be caused by different mutations, even within closely related horses. (A) Splashed white bay horse. The unpigmented areas are relatively small, but the horse has blue eyes. (B) Typical expression of the splashed white phenotype in a chestnut horse with blue eyes. (C) A splashed white chestnut horse with normal eye color and a relatively small blaze. (D) Splashed white coat color and brown eyes in a chestnut horse. (E, F) Two horses carrying both the PAX3C70Y and MITFprom1 alleles with typical splashed white phenotypes. Horses carrying one copy of either PAX3C70Y and/or MITFprom1 show overlapping phenotypes. None of the horses in this figure carry the EDNRBI118K (overo) allele. In this Quarter Horse family the splashed white coat color appeared to be inherited as an autosomal dominant trait. We therefore genotyped 24 cases and 7 controls from this family on the equine 50k SNP array and performed a linkage analysis (Figure S1). Parametric analysis with a fully dominant model of inheritance gave a maximum LOD score of 1.6 on ECA 6 with a corresponding maximum α of 0.78. These results suggested locus heterogeneity within the family. We also performed a case-control genome-wide association study (GWAS) with the SNP genotypes and found the strongest signal again on ECA 6 (praw = 5.6×10−7; pgenome = 0.003). The chromosomes with the next best associations were 11 and 16, each having SNPs associated at praw = 6.6×10−5, which is not genome-wide significant after permutation analysis (pgenome = 0.34).
The linked and associated region on ECA 6 contained PAX3 as a strong functional candidate gene. We sequenced the nine exons of the PAX3 gene in two cases and two controls and identified seven polymorphisms including one missense mutation, PAX3:c.209G>A (Table 1, Table S1). We then genotyped all remaining animals of the Quarter Horse family and other unrelated horses for this variant. We identified a total of 29 splashed white horses carrying the variant A-allele in heterozygous state. All these horses traced back to a female Quarter Horse born in 1987, whose genomic DNA from a hair-root sample tested homozygous wild-type. Thus, the mutation most likely arose in the germline of this animal. We did not detect the variant A-allele in 21 solid-colored Quarter Horses nor in 112 horses from 7 other breeds. We also did not find any horse with the homozygous variant A/A genotype (Table 2).
Tab. 1. Causative mutations for white spotting phenotypes. 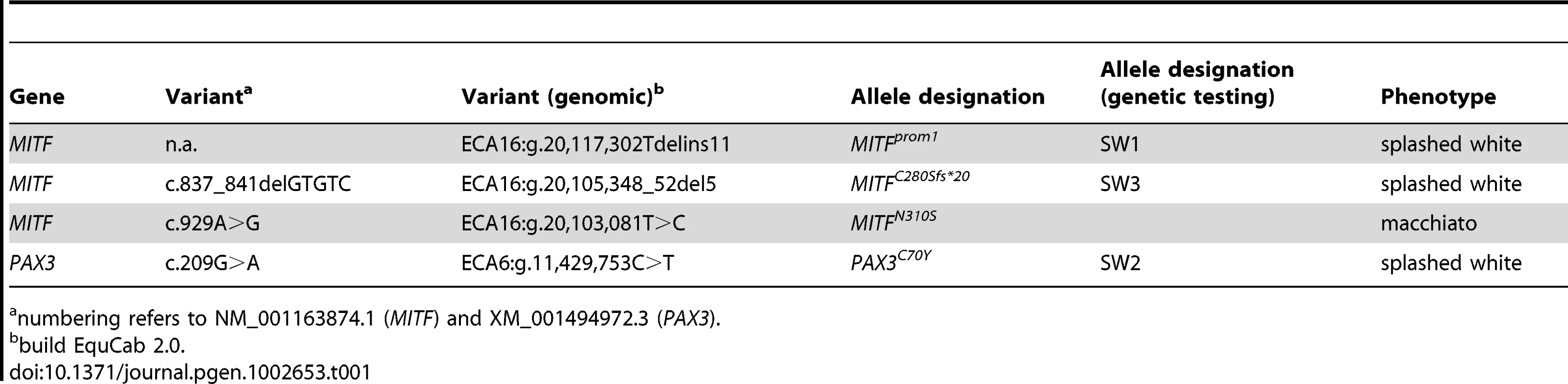
numbering refers to NM_001163874.1 (MITF) and XM_001494972.3 (PAX3). Tab. 2. Association of white spotting genotypes in different breeds. 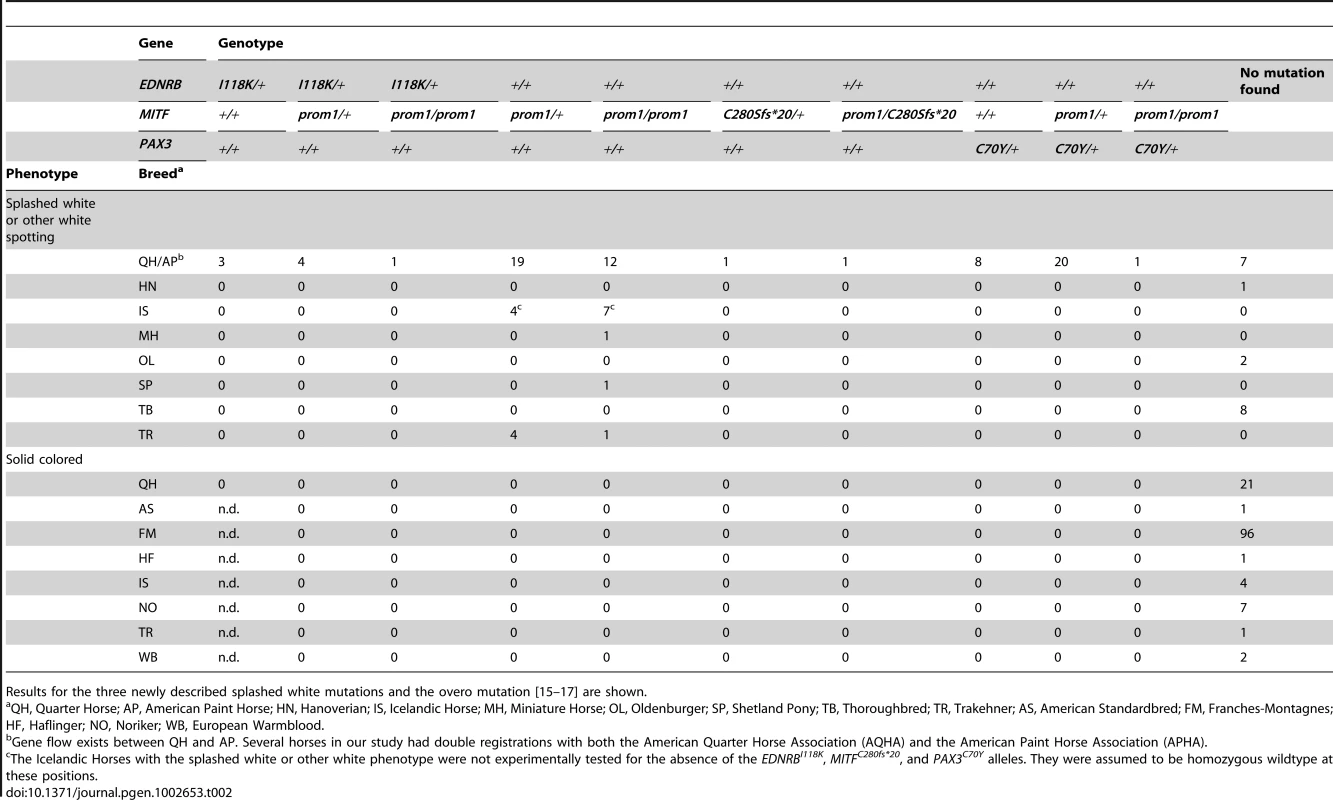
Results for the three newly described splashed white mutations and the overo mutation [15]–[17] are shown. The PAX3:c.209G>A variant is predicted to result in a non-conservative amino acid exchange p.C70Y in the so-called paired domain, which together with the homeobox domain mediates the DNA-binding of the transcription factor PAX3. The wild-type cysteine at this position is conserved across all known PAX paralogs in animals including Drosophila and Caenorhabditis elegans (Figure 2). Based on the X-ray structure of the paired domain of the human PAX6 protein, the side-chain of this cysteine is predicted to form a direct contact to the DNA backbone [20].
Fig. 2. Region around the PAX3:p.C70Y mutation. 
The cysteine at position 70 is conserved in all known PAX paralogs. The initial GWAS indicated potential secondary signals on ECA 11 and 16. We did not detect an obvious candidate gene on ECA 11. However, the MITF gene on ECA 16 represents a strong functional candidate for a white spotting phenotype. As the PAX3:p.C70Y variant did not explain the phenotype in all splashed white horses from the Quarter Horse family, we sequenced all exons and the known promoter elements from the MITF gene in two splashed white horses that did not have the PAX3C70Y allele and in two controls. We identified a total of 28 polymorphisms (Table S1). A variant in the proximal melanocyte-specific MITF promoter replacing a thymine with 11 nucleotides stood out as clear candidate for a non-coding regulatory mutation (ECA16:g.20,117,302Tdelins11; Figure 3). This variant interrupts a highly conserved binding site for PAX3 [21]–[23] and is located close to the mutation causing white spotting in dogs [24]. The variant allele was absent from 21 solid-colored Quarter Horses and from 112 horses with minimal white markings from 7 additional breeds. We found 19 splashed white horses that carried only the MITFprom1 allele and 20 splashed white horses that carried both the MITFprom1 and PAX3C70Y allele (Table 2).
Fig. 3. Proximal melanocyte-specific promoter of the MITF gene. 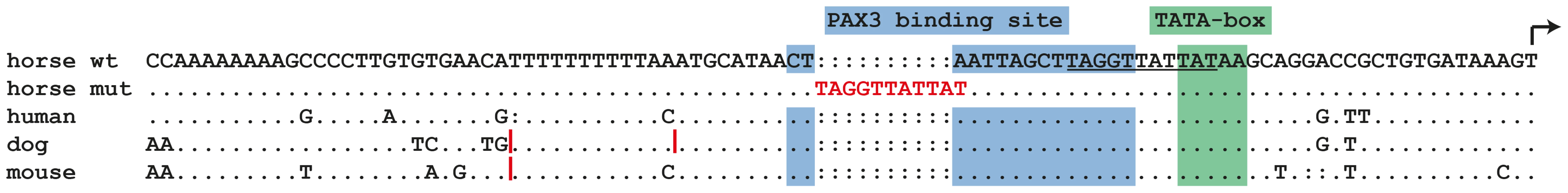
The depicted region corresponds to positions −89 to +1 with respect to the transcription start site and is in reverse complementary orientation to the genomic reference sequence (ECA16:20,117,350–20,117,261; EquCab2.0). The three vertical red lines in the dog and mouse sequences represent small insertions. In some splashed white horses, an 11 bp motif shown in red replaces an adenine, which is part of a highly conserved PAX3 transcription factor binding site in the wild-type sequence. The inserted 11 bp sequence may have arisen by duplication from an identical sequence motif a few nucleotides further downstream (underlined). We quantitatively analyzed the proportion of depigmented skin in the face area of splashed white Quarter Horses in relation to their underlying MITF and PAX3 genotypes as well as their base coat color (Figure S2, Figure S3). This analysis indicated that one copy of either the MITFprom1 or the PAX3C70Y allele has a similar effect on the face pigmentation. The presence of both the MITFprom1 and the PAX3C70Y alleles leads to a slightly more pronounced depigmentation of the face on average than either splashed white allele alone. This analysis also showed that the average white face area is more extended in splashed white chestnut horses compared to bay horses with the same splashed white genotype (Figure 4; Table S2).
Fig. 4. White face area in splashed white Quarter Horses with specific genotypes. 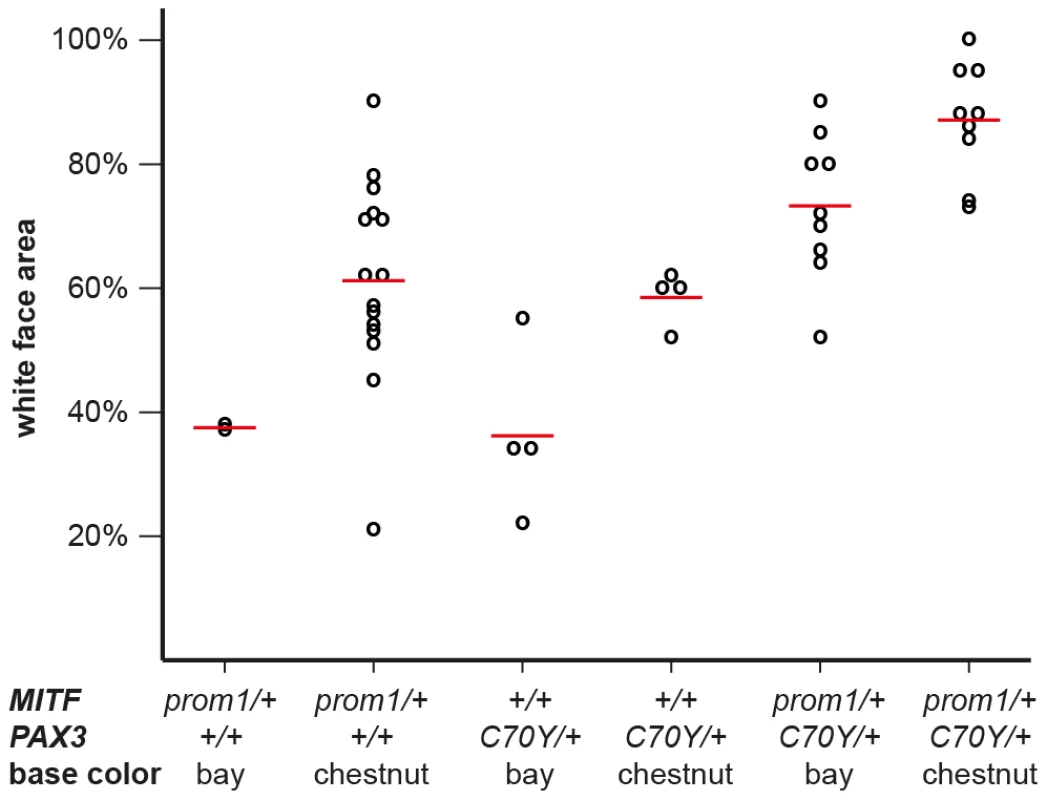
Raw phenotypes for individual horses are indicated as black circles. The average values for each genotype class are indicated by red lines. While there is considerable variation in the phenotypic expression, the average proportion of white face area is higher in chestnut horses versus bay horses and it is also higher in horses carrying both the MITFprom1 and the PAX3C70Y allele than in horses carrying only one of the two splashed white alleles (p<0.05, two-sided Welch t-test). The white face areas of all horses and the p-values between all different genotype classes are given in Table S2. Breed distribution of the MITFprom1 allele
We sequenced the coding regions and proximal promoter elements of the functional candidate genes KIT, MITF, and PAX3 in other horses with white spotting phenotypes in additional horses from various breeds. We then noticed that the MITF promoter polymorphism described above is much more widespread across breeds than we had originally hypothesized. We found the insertion allele in 58 Quarter Horses and American Paint Horses with either the splashed white or more pronounced other white spotting phenotypes (see below). We also found this allele in five Trakehner horses with white spotting phenotypes. A detailed pedigree analysis revealed that all these horses are descendants from the Thoroughbred stallion Blair Athol born in 1861. This stallion also had a very large white blaze (Figure S4). The MITFprom1 allele was also present in a Miniature Horse, a Shetland Pony, and 11 Icelandic Horses with either splashed white or more pronounced other white spotting phenotypes. Thus, the MITFprom1 allele is probably several hundred years old and arose before the foundation of the modern horse breeds.
More pronounced other white spotting phenotypes: Combinations of splashed white alleles
In our study we defined horses with ≥20% white face area and ≤10% white area on the body as “splashed white horses”. If a horse had >10% white body area, it was considered to have a more pronounced “other white spotting” phenotype. We noticed that several horses with more pronounced “other white spotting phenotypes” were offspring from two “splashed white” parents.
We identified a total of 24 horses from 5 breeds that were homozygous for the MITF promoter variant. All these MITFprom1/prom1horses showed very pronounced but still variable depigmentation phenotypes. They had at least a white belly in addition to white legs and the white head (Figure 5A–5C). One of these horses had only small pigmented areas along the dorsal midline (Figure 5A). One horse homozygous for the MITFprom1 allele and heterozygous for the PAX3C70Y allele was completely white and also deaf (Figure 5D).
Fig. 5. Phenotypes of horses with different combinations of splashed white alleles. 
(A) American Paint Horse with a very pronounced depigmentation phenotype. In addition to being homozygous for the MITFprom1 allele, it also carries a private allele at the KIT gene (p.H40Q), which may enhance the depigmentation phenotype. The functional significance of the p.H40Q variant is unclear at the moment. (B) This Quarter Horse is also homozygous for the MITFprom1 allele, but has substantially more residual pigmentation than the horse shown in panel A. (C) A Trakehner horse homozygous for the MITFprom1 allele. (D) A completely white horse with multiple splashed white alleles. (E) Quarter Horse with the rare MITFC280Sfs*20 allele. This horse has a very pronounced splashed white phenotype with blue eyes and a largely unpigmented head and belly. (F) A compound heterozygote for two different MITF mutant alleles is completely white. None of the horses in this figure carry the EDNRBI118K (overo) allele. In our mutation analysis we identified one horse with a splashed white phenotype including a white belly, which was wild-type for both the MITFprom1 and the PAX3C70Y alleles (Figure 5E). This horse carried a small deletion in exon 5 of the MITF gene (c.837_841del5). The variant leads to a frameshift and a severely truncated MITF protein (p.C280Sfs*20), which might act in a dominant negative fashion. A completely white offspring of this horse was a compound heterozygote for the MITFprom1 and the MITFC280Sfs*20 alleles (Figure 4F). The MITFC280Sfs*20 allele was absent from 21 solid Quarter Horses and 112 control horses from 7 additional breeds (Table 2).
In a few unrelated horses with large blazes we did not find any candidate causative mutations in the KIT, MITF, or PAX3 candidate genes. These horses comprised 7 Quarter Horses, 1 Hanoverian, 2 Oldenburger, and 8 Thoroughbreds (Table 2).
Macchiato in a Franches-Montagnes Horse
In 2008 a colt with a striking white-spotting phenotype was born out of two solid-colored bay Franches-Montagnes parents. The coat color resembled a combination of white-spotting and coat color dilution (Figure 6). The parentage of this colt was experimentally verified using 13 microsatellite markers. Therefore, we assumed the coat color of this colt to be the result of a spontaneous de novo mutation and subsequently termed it “macchiato”. In 2010, we performed a detailed clinical and spermatological examination, which revealed that the two year old macchiato stallion was deaf and had a low progressive sperm motility.
Fig. 6. Macchiato coat color phenotype in a Franches-Montagnes stallion. 
Note the pronounced depigmentation on the body in addition to the large white blaze on the head, the white legs, and the blue eyes. This horse has a bay basic coat color. We sequenced the coding regions of six functional candidate genes in the macchiato stallion and his solid-colored parents. The investigated candidate genes are involved in white spotting (KIT, MITF) or coat color dilution (MLPH, PMEL, SLC36A1, and SLC45A2). We identified a de novo missense mutation in exon 6 of the MITF gene in the macchiato stallion (c.929A>G). We found the mutant allele at approximately 50% intensity in DNA samples from blood, hair roots and sperm, indicating that the macchiato stallion is not a mosaic. The mutant allele was not present in DNA from blood samples from either the mother or the father. The mutation affects a highly conserved amino acid of the basic DNA binding motif of the transcription factor MITF (p.N310S). We analyzed the DNA binding properties of the mutant MITF in an electrophoretic mobility shift assay and found that DNA binding activity was reduced by about 80%, probably through changes to the kinetics of binding (Figure 7, Table S3). A mutation at the same residue in the human MITF protein leads to Tietz syndrome, which is characterized by a more generalized depigmentation and profound obligate hearing loss compared to the slightly milder Waardenburg syndrome 2A, which is caused by many other mutations in the human MITF gene [25]. The MITF:p.N310S variant was absent from 96 solid-colored Franches-Montagnes horses.
Fig. 7. Functional validation of the MITF:p.N310S mutation. 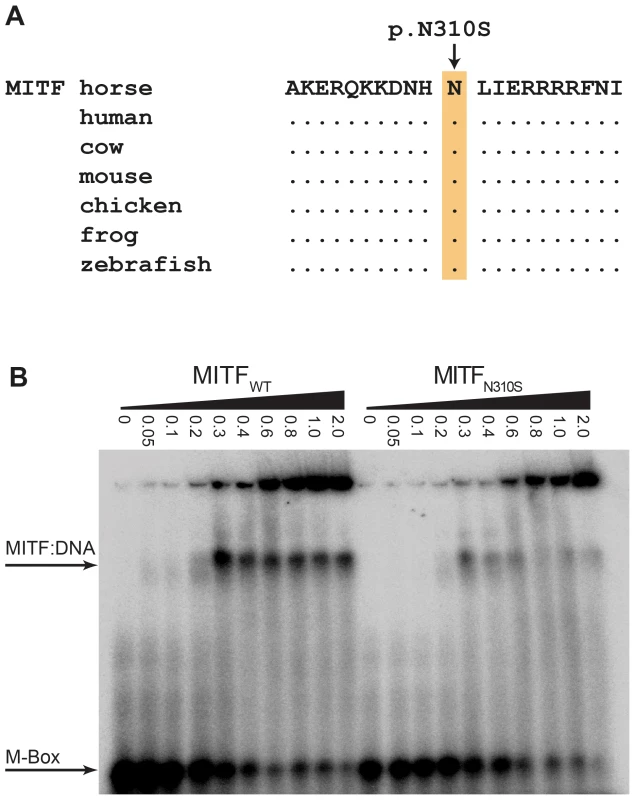
(A) The basic DNA binding domain in the vicinity of the mutation is 100% identical in vertebrates from mammals to fish. (B) Electrophoretic mobility shift assay (EMSA). Increasing concentrations (given in µM) of recombinant wild-type and mutant MITF protein from E. coli were incubated with a radioactively labeled double-stranded oligonucleotide and run on a non-denaturing polyacrylamide gel. The mutant MITF protein shows a weaker retention band than the wild-type MITF, which indicates a partially defective DNA binding activity (Table S3). Discussion
As the splashed white phenotype is rather distinctive among the many different variations of white spotting phenotypes in horses, we started this investigation with the expectation of finding only a single causative mutation. However, the linkage/GWAS data suggested locus heterogeneity even within a family of closely related Quarter Horses. Using a positional candidate gene approach we subsequently identified three causative mutations for splashed white phenotypes and one causative mutation for another similar phenotype that we termed macchiato (Table 1).
Although we do not have functional proof for all of the mutations, we obtained sufficient ancillary evidence to claim their causality. For the PAX3C70Y allele the arguments are that it (1) occurs only in horses with the splashed white phenotype, (2) the PAX3 locus was linked to this phenotype in a family, (3) mutations within the PAX3 gene cause similar phenotypes in humans and mice, and (4) the mutation affects a highly conserved cysteine residue in the paired domain, that is predicted to participate in DNA binding. We were able to determine that this variant arose in 1987 and to identify the specific founder animal.
For the three MITF mutations the support is even better than for the PAX3 mutation: (1) Multiple mutations within the same gene lead to comparable phenotypes, (2) the three variants occur exclusively in horses with characteristic depigmentation phenotypes, and (3) mutations within the MITF gene cause similar phenotypes in humans, mice, and dogs [18], [19], [24]. MITFC280Sfs*20 is the result of a frameshift mutation, which is extremely likely to affect the normal function of MITF as it truncates about half of the protein. The MITFprom1 variant affects a region of the melanocyte-specific promoter that has been shown to function as a PAX3 binding site in humans and mice [20]–[22]. For MITFN310S we demonstrated that it represents a spontaneous de novo mutation in the macchiato horse that was born out of solid-colored parents free of this allele. We also provide functional evidence that the MITFN310S protein has a reduced DNA binding capability. Taken together these data strongly argue for the causality of the four mutations in PAX3 and MITF that we report in this study.
Our findings will be of relevance to horse breeders. The MITFprom1 allele arose at least several hundred years ago, it is relatively common, and it occurs in several modern horse breeds. Horses homozygous for this allele are viable and typically have a more pronounced depigmentation phenotype than heterozygous horses. The PAX3C70Y allele is only 24 years old and occurs exclusively in Quarter and Paint Horses. We did not find a horse homozygous for this mutation, and based on data from mice it is unlikely that a homozygous PAX3C70Y/C70Y horse would be viable. As PAX3 is required for several key steps in neural development, homozygosity for this allele will most likely result in embryonic or fetal lethality [26]. Therefore, the mating of two heterozygous PAX3+/C70Y horses is not recommended in order to avoid the accidental production of an embryo homozygous for this allele. The MITFC280Sfs*20 and MITFN310S alleles are extremely rare. Data from mice again suggest that these alleles will most likely result in severe clinical phenotypes such as e.g. microphthalmia in the homozygous state [27]. Therefore, horses with such alleles should also not be mated to each other.
Depigmentation or white spotting phenotypes represent a complex trait, ranging from horses with a wild-type phenotype (no unpigmented skin areas) up to completely white horses. Previously, a number of single gene mutations in KIT and EDNRB have been shown to cause either dominant white or extreme white spotting phenotypes [10]–[17, Figure S5]. With this study we now report a series of mutations that lead to milder depigmentation phenotypes, which phenotypically overlap with the common white markings seen at the head and legs of many domestic horses. The newly reported mutations interact with other genetic factors. Chestnut horses carrying a mutation in MITF and/or PAX3 show a more pronounced depigmentation phenotype than bay horses with the same mutations. Thus, the basic coat color or more specifically the genotype at MC1R modifies the phenotypic expression of the reported mutations. The correlation between chestnut coat color and increased size of white markings has been reported before [28], [29]. The detailed analysis of depigmentation phenotypes may help us to better understand complex genetic networks on a molecular and functional level. In such networks, non-coding regulatory mutations such as MITFprom1 will probably play a major role and need to be taken into consideration.
In conclusion, we report four different mutations leading to splashed white or the new macchiato coat color phenotype in horses, which show similarities to the human Waardenburg or Tietz syndromes, respectively. This study highlights the potential of coat color genetics to gain molecular insights into complex regulatory gene networks.
Materials and Methods
Ethics statement
All animal work was conducted in accordance with the relevant local guidelines (Swiss law on animal protection and welfare - permit to the Swiss National Stud Farm no. 2227). The only animal experiment in our study was the collection of blood samples from horses by certified veterinarians.
Animals and phenotypes
We analyzed a total of 239 horses, 106 animals with white spotting-phenotypes (cases) and 133 solid-colored controls. The cases consisted of 70 horses with the splashed white phenotype and another 36 horses with more extreme white spotting phenotypes (77 Quarter or American Paint Horses, 1 Hanoverian, 11 Icelandic Horses, 1 Miniature Horse, 2 Oldenburger, 1 Shetland Pony, 8 Thoroughbreds, 5 Trakehner). The 133 solid-colored controls consisted of 21 Quarter Horses, 1 American Standardbred, 96 Franches-Montagnes horses, 1 Haflinger, 4 Icelandic Horses, 7 Noriker, 1 Trakehner, and 2 European Warmblood horses. We considered Quarter Horses and American Paint Horses to represent one joint population as several horses in our study had double registrations and as it is quite common that offspring of Quarter Horse parents with white spotting phenotypes are registered as American Paint Horses.
We considered the proportion of white face area as the primary phenotypic criterion. To quantify this proportion we drew the heads of the horses in a standard perspective from photos (Figure S2, Figure S3). We then measured the proportion of unpigmented face area in comparison to a horse that we considered to have a 100% white face. Horses with ≥20% white face area were considered to have a white spotting phenotype. If a horse had ≥20% white face area and ≤10% white area on the body, it was considered “splashed white”. If a horse had ≥20% white face area and >10% white body area, it was considered to have an “other white spotting” phenotype. Horses with ≤3% white face area and 0% white body area were considered as solid-colored controls. Horses with a white face area ranging from 3–20% were classified as unknown phenotype.
Linkage and association mapping
We isolated genomic DNA from either EDTA blood or hair root samples from all horses. We genotyped 31 horses with the illumina equine 50K SNP beadchip containing 54,602 SNPs. We used the PLINK v1.07 software for pruning of the genotype data set [30]. We removed 28,381 SNPs that did not have genotype calls in every animal and 10,753 SNPs that had minor allele frequencies below 5%. For the final analysis 31 horses and 19,319 SNPs remained. We used the Merlin software [31] and a fully dominant model of inheritance to analyze the data for parametric linkage. We used the PLINK software for genome-wide association analyses. Empirical genome-wide significance levels were determined by performing 100,000 random permutations of the assigned phenotypes.
DNA sequencing and mutation analysis
We amplified exons, flanking intron regions, and proximal promoter sequences of the KIT, MITF, and PAX3 genes as well as exon 2 of the EDNRB gene (Table S4). We subsequently sequenced the PCR products on an ABI 3730 capillary sequencer (Life Technologies). We analyzed sequencing data with the Sequencher 4.9 software for polymorphisms (Gene Codes). In the macchiato horse and its parents we sequenced all exons of the KIT, MITF, MLPH, PMEL, SLC36A1, and SLC45A2 genes.
Electrophoretic mobility shift assay (EMSA)
We expressed recombinant wild-type and N310S MITF from in E. coli Rosetta 2 cells in LB medium supplemented with 1% glucose (amino acid residues 112–207 from uniprot acc. Q95MD1). We lysed the cells by sonication in 20 mM Tris, 500 mM NaCl, 50 mM imidazole, 0.5 mM phenylmethylsulfonyl fluoride (PMSF), 0.1% (v/v) β-mercaptoethanol, pH 9. We applied the soluble fraction to Ni-NTA resin and eluted MITF with an imidazole step gradient (0.2–0.6 M) and further purified the proteins by reverse phase HPLC on a C18 column, using a gradient of 5–95% acetonitrile (0.1% TFA) over 20 ml at 1 ml/min. We lyophilised the protein-containing fractions and stored them at −20°C. The protein was refolded by resuspension in 20 mM Tris (pH 7.4), and we confirmed the correct folding by circular dichroism (CD) spectropolarimetry.
We performed the EMSA using an M-Box containing oligonucleotide (5′-GGAAAGTTAGTCATGTGCTTTTCAGAAGA-3′) as previously described [32]. The EMSA reactions contained 20 mM Tris, 50 mM NaCl, 5 mM MgCl2, 1 mM DTT, and 33 µg/ml BSA at pH 7.4. After incubation on ice for 30 min, we separated the samples on a 6% (w/v) non-denaturing polyacrylamide gel, in 0.5× TBE (45 mM Tris, 45 mM boric acid, 2.5 mM EDTA, pH 8.3) and analyzed the gels using a PhosphorImager (Molecular Dynamics). We quantified the bands from three repeated experiments and calculated average values and standard deviations.
Supporting Information
Zdroje
1. ThomasAJEricksonCA 2008 The making of a melanocyte: the specification of melanoblasts from the neural crest. Pigment Cell Melanoma Res 21 598 610
2. GeisslerENRyanMAHousmanDE 1988 The dominant-white spotting (W) locus of the mouse encodes the c-kit proto-oncogene. Cell 55 185 192
3. HodgkinsonCAMooreKJNakayamaASteingrímssonECopelandNG 1993 Mutations at the mouse microphthalmia locus are associated with defects in a gene encoding a novel basic-helix-loop-helix-zipper protein. Cell 74 395 404
4. SilverDLHouLSomervilleRYoungMEApteSS 2008 The secreted metalloprotease ADAMTS20 is required for melanoblast survival. PLoS Genet 4 e1000003 doi:10.1371/journal.pgen.1000003
5. ThomasAJEricksonCA 2009 FOXD3 regulates the lineage switch between neural crest-derived glial cells and pigment cells by repressing MITF through a non-canonical mechanism. Development 136 1849 1858
6. SchollFAKamarashevJMurmannOVGeertsenRDummerR 2001 PAX3 is expressed in human melanomas and contributes to tumor cell survival. Cancer Res 61 823 826
7. BaxterLLHouLLoftusSKPavanWJ 2004 Spotlight on spotted mice: a review of white spotting mouse mutants and associated human pigmentation disorders. Pigment Cell Res 17 215 224
8. RiederSHaggerCObexer-RuffGLeebTPoncetPA 2008 Genetic analysis of white facial and leg markings in the Swiss Franches-Montagnes Horse breed. J Hered 99 130 136
9. SponenbergDP 2009 Equine color genetics, 3rd edition Ames Wiley-Blackwell 277
10. BrooksSABaileyE 2005 Exon skipping in the KIT gene causes a Sabino spotting pattern in horses. Mamm Genome 16 893 902
11. BrooksSALearTLAdelsonDLBaileyE 2007 A chromosome inversion near the KIT gene and the Tobiano spotting pattern in horses. Cytogenet Genome Res 119 225 230
12. HaaseBBrooksSASchlumbaumAAzorPJBaileyE 2007 Allelic heterogeneity at the equine KIT locus in dominant white (W) horses. PLoS Genet 3 e195 doi:10.1371/journal.pgen.0030195
13. HaaseBBrooksSATozakiTBurgerDPoncetPA 2009 Seven novel KIT mutations in horses with white coat colour phenotypes. Anim Genet 40 623 629
14. HaaseBRiederSTozakiTHasegawaTPenedoMC 2011 Five novel KIT mutations in horses with white coat colour phenotypes. Anim Genet 42 337 339
15. MetallinosDLBowlingATRineJ 1998 A missense mutation in the endothelin-B receptor gene is associated with Lethal White Foal Syndrome: an equine version of Hirschsprung disease. Mamm Genome 9 426 431
16. SantschiEMPurdyAKValbergSJVrotsosPDKaeseH 1998 Endothelin receptor B polymorphism associated with lethal white foal syndrome in horses. Mamm Genome 9 306 309
17. YangGCCroakerDZhangALManglickPCartmillT 1998 A dinucleotide mutation in the endothelin-B receptor gene is associated with lethal white foal syndrome (LWFS); a horse variant of Hirschsprung disease. Hum Mol Genet 7 1047 1052
18. PingaultVEnteDDastot-Le MoalFGoossensMMarlinS 2010 Review and update of mutations causing Waardenburg syndrome. Hum Mutat 31 391 406
19. TachibanaMKobayashiYMatsushimaY 2003 Mouse models for four types of Waardenburg syndrome. Pigment Cell Res 16 448 454
20. XuHERouldMAXuWEpsteinJAMaasRL 1999 Crystal structure of the human Pax6 paired domain-DNA complex reveals specific roles for the linker region and carboxy-terminal subdomain in DNA binding. Genes Dev 13 1263 1275
21. FuseNYasumotoKSuzukiHTakahashiKShibaharaS 1996 Identification of a melanocyte-type promoter of the microphthalmia-associated transcription factor gene. Biochem Biophys Res Commun 219 702 707
22. BondurandNPingaultVGoerichDELemortNSockE 2000 Interaction among SOX10, PAX3 and MITF, three genes altered in Waardenburg syndrome. Hum Mol Genet 9 1907 1917
23. HallssonJHHaflidadóttirBSSchepskyAArnheiterHSteingrímssonE 2007 Evolutionary sequence comparison of the Mitf gene reveals novel conserved domains. Pigment Cell Res 20 185 200
24. KarlssonEKBaranowskaIWadeCMSalmon HillbertzNHZodyMC 2007 Efficient mapping of mendelian traits in dogs through genome-wide association. Nat Genet 39 1321 1328
25. SmithSDKelleyPMKenyonJBHooverD 2000 Tietz syndrome (hypopigmentation/deafness) caused by mutation of MITF. J Med Genet 37 446 448
26. EpsteinDJVekemansMGrosP 1991 Splotch (Sp2H), a mutation affecting development of the mouse neural tube, shows a deletion within the paired homeodomain of Pax-3. Cell 67 767 774
27. HodgkinsonCAMooreKJNakayamaASteingrímssonECopelandNG 1993 Mutations at the mouse microphthalmia locus are associated with defects in a gene encoding a novel basic-helix-loop-helix-zipper protein. Cell 74 395 404
28. WoolfCM 1991 Common white facial markings in bay and chestnut Arabian horses and their hybrids. J Hered 82 167 169
29. SteingrímssonECopelandNGJenkinsNA 2004 Melanocytes and the microphthalmia transcription factor network. Annu Rev Genet 38 365 411
30. PurcellSNealeBTodd-BrownKThomasLFerreiraMA 2007 PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 81 559 575
31. AbecasisGRChernySSCooksonWOCardonLR 2002 Merlin - rapid analysis of dense genetic maps using sparse gene flow trees. Nat Genet 30 97 101
32. Wilkinson-WhiteLGamsjaegerRDastmalchiSWienertBStokesPH 2011 Structural basis of simultaneous recruitment of the transcriptional regulators LMO2 and FOG1/ZFPM1 by the transcription factor GATA1. Proc Natl Acad Sci USA 108 14443 14448
Štítky
Genetika Reprodukční medicína
Článek A Genome-Wide Screen for Genetic Variants That Modify the Recruitment of REST to Its Target GenesČlánek Population Structure of Hispanics in the United States: The Multi-Ethnic Study of AtherosclerosisČlánek Differing Requirements for RAD51 and DMC1 in Meiotic Pairing of Centromeres and Chromosome Arms inČlánek Transcriptional Regulation of Rod Photoreceptor Homeostasis Revealed by NRL Targetome AnalysisČlánek Cell Contact–Dependent Outer Membrane Exchange in Myxobacteria: Genetic Determinants and MechanismČlánek Formation of Rigid, Non-Flight Forewings (Elytra) of a Beetle Requires Two Major Cuticular Proteins
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2012 Číslo 4- Akutní intermitentní porfyrie
- Farmakogenetické testování pomáhá předcházet nežádoucím efektům léčiv
- Růst a vývoj dětí narozených pomocí IVF
- Pilotní studie: stres a úzkost v průběhu IVF cyklu
- Intrauterinní inseminace a její úspěšnost
-
Všechny články tohoto čísla
- Runs of Homozygosity Implicate Autozygosity as a Schizophrenia Risk Factor
- Modifier Genes and the Plasticity of Genetic Networks in Mice
- The DSIF Subunits Spt4 and Spt5 Have Distinct Roles at Various Phases of Immunoglobulin Class Switch Recombination
- A Genome-Wide Screen for Genetic Variants That Modify the Recruitment of REST to Its Target Genes
- Population Structure of Hispanics in the United States: The Multi-Ethnic Study of Atherosclerosis
- Deep Sequencing of Plant and Animal DNA Contained within Traditional Chinese Medicines Reveals Legality Issues and Health Safety Concerns
- Differing Requirements for RAD51 and DMC1 in Meiotic Pairing of Centromeres and Chromosome Arms in
- Insulin Signaling Mediates Sexual Attractiveness in
- Progressive Telomere Dysfunction Causes Cytokinesis Failure and Leads to the Accumulation of Polyploid Cells
- Long-Range Chromosome Organization in : A Site-Specific System Isolates the Ter Macrodomain
- Regulation of Budding Yeast Mating-Type Switching Donor Preference by the FHA Domain of Fkh1
- Polyglutamine Toxicity Is Controlled by Prion Composition and Gene Dosage in Yeast
- Patterns of Regulatory Variation in Diverse Human Populations
- Sequence-Specific Targeting of Dosage Compensation in Favors an Active Chromatin Context
- Whole-Exome Sequencing and Homozygosity Analysis Implicate Depolarization-Regulated Neuronal Genes in Autism
- Replication Fork Reversal after Replication–Transcription Collision
- Common Variants at 9p21 and 8q22 Are Associated with Increased Susceptibility to Optic Nerve Degeneration in Glaucoma
- Coordinate Regulation of Lipid Metabolism by Novel Nuclear Receptor Partnerships
- Epigenome-Wide Scans Identify Differentially Methylated Regions for Age and Age-Related Phenotypes in a Healthy Ageing Population
- A Coordinated Interdependent Protein Circuitry Stabilizes the Kinetochore Ensemble to Protect CENP-A in the Human Pathogenic Yeast
- Budding Yeast Dma Proteins Control Septin Dynamics and the Spindle Position Checkpoint by Promoting the Recruitment of the Elm1 Kinase to the Bud Neck
- , a Homolog of a Deaf-Blindness Gene, Regulates Circadian Output and Slowpoke Channels
- Transcriptional Regulation of Rod Photoreceptor Homeostasis Revealed by NRL Targetome Analysis
- Cell Contact–Dependent Outer Membrane Exchange in Myxobacteria: Genetic Determinants and Mechanism
- Defective Membrane Remodeling in Neuromuscular Diseases: Insights from Animal Models
- Formation of Rigid, Non-Flight Forewings (Elytra) of a Beetle Requires Two Major Cuticular Proteins
- SPE-44 Implements Sperm Cell Fate
- A Shared Role for RBF1 and dCAP-D3 in the Regulation of Transcription with Consequences for Innate Immunity
- A Companion Cell–Dominant and Developmentally Regulated H3K4 Demethylase Controls Flowering Time in via the Repression of Expression
- The HEN1 Ortholog, HENN-1, Methylates and Stabilizes Select Subclasses of Germline Small RNAs
- Improved Statistics for Genome-Wide Interaction Analysis
- The Probability of a Gene Tree Topology within a Phylogenetic Network with Applications to Hybridization Detection
- Context-Dependent Dual Role of SKI8 Homologs in mRNA Synthesis and Turnover
- Mu Insertions Are Repaired by the Double-Strand Break Repair Pathway of
- Competition between Replicative and Translesion Polymerases during Homologous Recombination Repair in Drosophila
- An Unbiased Assessment of the Role of Imprinted Genes in an Intergenerational Model of Developmental Programming
- Type 2 Diabetes Risk Alleles Demonstrate Extreme Directional Differentiation among Human Populations, Compared to Other Diseases
- Mutations in and Cause “Splashed White” and Other White Spotting Phenotypes in Horses
- Fine-Scale Mapping of Natural Variation in Fly Fecundity Identifies Neuronal Domain of Expression and Function of an Aquaporin
- Dynamics of Brassinosteroid Response Modulated by Negative Regulator LIC in Rice
- Genetic Inhibition of Solute-Linked Carrier 39 Family Transporter 1 Ameliorates Aβ Pathology in a Model of Alzheimer's Disease
- The Functions of Mediator in Support a Role in Shaping Species-Specific Gene Expression
- Patterns of Ancestry, Signatures of Natural Selection, and Genetic Association with Stature in Western African Pygmies
- Dissection of Pol II Trigger Loop Function and Pol II Activity–Dependent Control of Start Site Selection
- PIWI Associated siRNAs and piRNAs Specifically Require the HEN1 Ortholog
- Genome-Wide Patterns of Gene Expression in Nature
- Hypoxia Disruption of Vertebrate CNS Pathfinding through EphrinB2 Is Rescued by Magnesium
- A New Role for Translation Initiation Factor 2 in Maintaining Genome Integrity
- Sex Reversal in C57BL/6J XY Mice Caused by Increased Expression of Ovarian Genes and Insufficient Activation of the Testis Determining Pathway
- The Rac GTP Exchange Factor TIAM-1 Acts with CDC-42 and the Guidance Receptor UNC-40/DCC in Neuronal Protrusion and Axon Guidance
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- A Coordinated Interdependent Protein Circuitry Stabilizes the Kinetochore Ensemble to Protect CENP-A in the Human Pathogenic Yeast
- Coordinate Regulation of Lipid Metabolism by Novel Nuclear Receptor Partnerships
- Defective Membrane Remodeling in Neuromuscular Diseases: Insights from Animal Models
- Formation of Rigid, Non-Flight Forewings (Elytra) of a Beetle Requires Two Major Cuticular Proteins
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



