-
Články
- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Coordinate Regulation of Lipid Metabolism by Novel Nuclear Receptor Partnerships
Mammalian nuclear receptors broadly influence metabolic fitness and serve as popular targets for developing drugs to treat cardiovascular disease, obesity, and diabetes. However, the molecular mechanisms and regulatory pathways that govern lipid metabolism remain poorly understood. We previously found that the Caenorhabditis elegans nuclear hormone receptor NHR-49 regulates multiple genes in the fatty acid beta-oxidation and desaturation pathways. Here, we identify additional NHR-49 targets that include sphingolipid processing and lipid remodeling genes. We show that NHR-49 regulates distinct subsets of its target genes by partnering with at least two other distinct nuclear receptors. Gene expression profiles suggest that NHR-49 partners with NHR-66 to regulate sphingolipid and lipid remodeling genes and with NHR-80 to regulate genes involved in fatty acid desaturation. In addition, although we did not detect a direct physical interaction between NHR-49 and NHR-13, we demonstrate that NHR-13 also regulates genes involved in the desaturase pathway. Consistent with this, gene knockouts of these receptors display a host of phenotypes that reflect their gene expression profile. Our data suggest that NHR-80 and NHR-13's modulation of NHR-49 regulated fatty acid desaturase genes contribute to the shortened lifespan phenotype of nhr-49 deletion mutant animals. In addition, we observed that nhr-49 animals had significantly altered mitochondrial morphology and function, and that distinct aspects of this phenotype can be ascribed to defects in NHR-66 – and NHR-80–mediated activities. Identification of NHR-49's binding partners facilitates a fine-scale dissection of its myriad regulatory roles in C. elegans. Our findings also provide further insights into the functions of the mammalian lipid-sensing nuclear receptors HNF4α and PPARα.
Published in the journal: . PLoS Genet 8(4): e32767. doi:10.1371/journal.pgen.1002645
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1002645Summary
Mammalian nuclear receptors broadly influence metabolic fitness and serve as popular targets for developing drugs to treat cardiovascular disease, obesity, and diabetes. However, the molecular mechanisms and regulatory pathways that govern lipid metabolism remain poorly understood. We previously found that the Caenorhabditis elegans nuclear hormone receptor NHR-49 regulates multiple genes in the fatty acid beta-oxidation and desaturation pathways. Here, we identify additional NHR-49 targets that include sphingolipid processing and lipid remodeling genes. We show that NHR-49 regulates distinct subsets of its target genes by partnering with at least two other distinct nuclear receptors. Gene expression profiles suggest that NHR-49 partners with NHR-66 to regulate sphingolipid and lipid remodeling genes and with NHR-80 to regulate genes involved in fatty acid desaturation. In addition, although we did not detect a direct physical interaction between NHR-49 and NHR-13, we demonstrate that NHR-13 also regulates genes involved in the desaturase pathway. Consistent with this, gene knockouts of these receptors display a host of phenotypes that reflect their gene expression profile. Our data suggest that NHR-80 and NHR-13's modulation of NHR-49 regulated fatty acid desaturase genes contribute to the shortened lifespan phenotype of nhr-49 deletion mutant animals. In addition, we observed that nhr-49 animals had significantly altered mitochondrial morphology and function, and that distinct aspects of this phenotype can be ascribed to defects in NHR-66 – and NHR-80–mediated activities. Identification of NHR-49's binding partners facilitates a fine-scale dissection of its myriad regulatory roles in C. elegans. Our findings also provide further insights into the functions of the mammalian lipid-sensing nuclear receptors HNF4α and PPARα.
Introduction
Modern day lifestyle and diet dramatically increase the threat of chronic diseases including obesity, diabetes and atherosclerosis. These metabolic disorders have been consistently linked to the imbalance between energy consumption and expenditure. Growing evidence suggests that faulty regulation of fat metabolism promotes metabolic diseases [1]. The control of fat metabolism is often mediated by nuclear receptors (NR), which are ligand-regulated transcription factors that play a central role in the cell's ability to sense, transduce and respond to lipophilic signals by modulating the appropriate target genes [2]. Dissecting the role of nuclear receptors in fat metabolism is therefore essential to our understanding of how energy homeostasis is maintained in an organism.
Nuclear receptors typically exhibit highly conserved modular domains including a zinc-finger DNA binding domain (DBD) and a ligand-binding domain (LBD) [2]. Ligand binding affects nuclear receptor activity by inducing structural changes within the LBD, which then alters the receptor's affinity to different co-factor proteins such as co-regulators and binding partner(s). Co-regulators include both co-activators and co-repressors, and are critical in mediating transcriptional responses. Alteration in co-regulator–NR binding can thus lead to a change in the transcriptional response. For instance, co-repressors can be replaced with co-activators to promote transcription of target genes, or NRs can form distinct homo - or heterodimers to activate or repress a specific set of target genes [3]. Thus, binding of distinct co-factors determines how nuclear receptors influence different gene networks.
The Hepatocyte Nuclear Factor 4 - alpha (HNF4α) is an example of a lipid sensing nuclear receptor in mammals. HNF4α orchestrates the regulation of a diverse range of target genes and is especially important in the control of genes involved in glucose and fatty acid homeostasis [4], and is mainly expressed in the liver, pancreas, kidney and small intestine [5]. Consistent with its role in metabolism, mutations in HNF4α are associated with both maturity onset diabetes of the young (MODY) and type 2 diabetes [6]–[9]. Although it is not entirely clear how HNF4α protects against diabetes, it has been reported that mutations in HNF4α can lead to early death of pancreatic beta-cells resulting in pancreatic dysfunction and a subsequent decrease in insulin production [10], [11]. In addition, knock-out of HNF4α in the adult mouse liver leads to increased lipid accumulation in hepatocytes and to the misregulation of genes involved in glucose and lipid metabolism [12], [13].
HNF4 receptors are evolutionarily conserved. Whereas mammals have two paralogs of this receptor, HNF4α and HNF4γ, the HNF4 family has expanded enormously in the nematode C. elegans to include 269 HNF4-like members [14], [15]. One of these 269 HNF4-like members is NHR-49, which is remarkably similar to the mammalian Peroxisome Proliferator-Activated Receptors (PPARs) in its overall biological effects on metabolism, fat storage, and life span [16]. The PPARs are global modulators of fat metabolism, controlling fat storage, expenditure, distribution and transport [17]. Perhaps most striking is the finding that NHR-49 and the PPARs, particularly PPARα and PPARδ, positively influence similar genes in multiple metabolic processes, including fatty acid β-oxidation, fatty acid desaturation, and fatty acid binding/transport [17], [18]. Moreover, knockout of PPARα or PPARδ can lead to high-fat phenotypes [18], [19] that are similar to those observed in nhr-49 animals, demonstrating the common physiological effects of these receptors on fat storage.
In this study, we set out to elucidate the genome-wide regulatory network of NHR-49 and to characterize its target genes to better understand the impact of NHR-49 mediated transcriptional regulation on worm physiology. We identified NHR-66 and NHR-80 as physical NHR-49 co-factors and were able to delineate their specific contribution to distinct phenotypes of nhr-49 mutants, namely selective effects of individual co-factors on life span and on mitochondrial function. We also uncovered novel roles for NHR-49 in the regulation of lipid metabolism including sphingolipid breakdown and lipid remodeling. Taken together, our findings support a model whereby NHR-49 heterodimerizes with other nuclear receptors to mediate the activation or repression of genes involved in distinct aspects of lipid metabolism.
Results
NHR-49 activates and represses genes in multiple lipid metabolism pathways
We previously found that NHR-49 promotes two distinct aspects of lipid metabolism, fatty acid desaturation and fatty acid β-oxidation [16]. However, the complete list of NHR-49's regulatory targets was still not known. Thus, we used whole genome C. elegans oligonucleotide microarrays to define the transcriptional profiles in an nhr-49(nr2041) deletion strain (compared to N2 wild-type worms). Table 1 lists genes that exhibit statistically significant changes in expression with (|log2(ratio)|≥0.848 and p-value≤0.001), in nhr-49 animals ([20] and Materials and Methods). The genes with negative values of logFC (fold change) are down regulated in the nhr-49 mutant (i.e. activated by NHR-49), whereas the genes with positive values of logFC are up regulated in the nhr-49 mutant (repressed by NHR-49). The finding that acs-2, which participates in mitochondrial β-oxidation and was previously demonstrated to be an NHR-49 target gene [16] had reduced expression in nhr-49 mutants validated the experimental approach. However, some of the previously reported genes like fat-5, fat-6, fat-7, cpt-2 and ech-1[16], were not found to be significant in our microarray analysis. This is likely due to the cut-off that we employed. We also uncovered new targets that are repressed by NHR-49 that include genes involved in sphingolipid breakdown, lipid remodeling and xenobiotic detoxification.
Tab. 1. List of differentially regulated genes in <i>nhr-49</i> compared to wild-type animals. 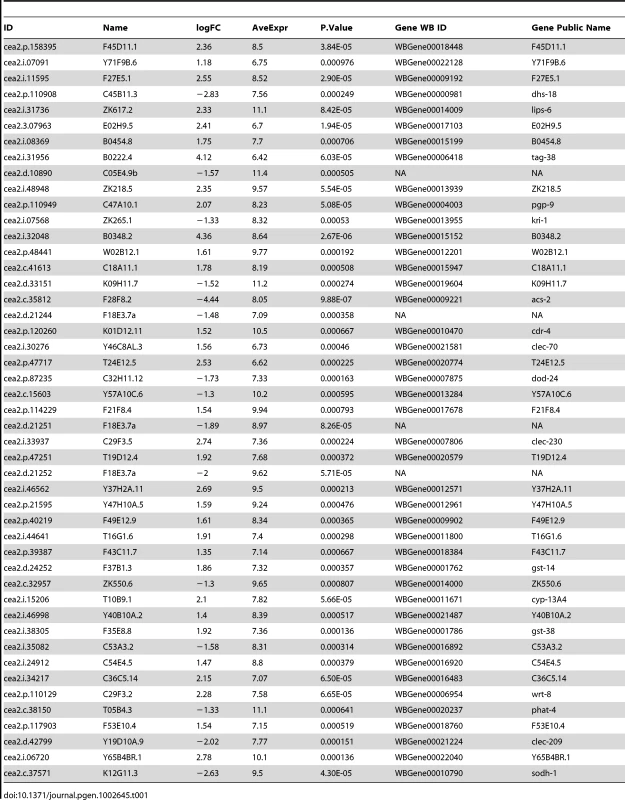
To validate these candidate NHR-49 targets, we employed quantitative RT-PCR to analyze their mRNA levels (Figure 1). In nhr-49 mutants, the expression of the sphingolipid processing genes acid ceramidase, glycosyl hydrolase, sphingosine-phosphate lyase (SPL), as well as lipid remodeling genes like phospholipases, TAG lipase and O-acyltransferase, was up regulated, as expected (Figure 1A and Table S1). Consistent with previously published data, nhr-49 mutants also showed a decrease in the expression of the fatty acid desaturase genes fat-7, fat-5 and fat-6, and the fatty acid beta-oxidation genes acs-2, cpt-5 and ech-1 (Figure 1B). Together, the gene expression data confirm that NHR-49 activates fatty acid β-oxidation and desaturase genes, and for the first time identifies target genes repressed by NHR-49 that include those involved in sphingolipid processing and lipid remodeling.
Fig. 1. NHR-49 collaborates with NHR-66 to regulate sphingolipid and lipid remodeling genes and with NHR-80 and NHR-13 to regulate fatty acid desaturase genes. 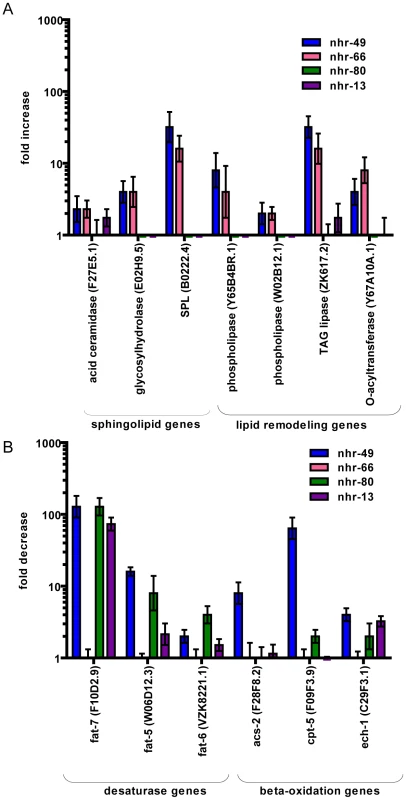
(A) qRT-PCR measurement of genes involved in sphingolipid metabolism including fatty acid ceramidase, sphingosine phosphate lyase, glycosyl hydrolase and genes involved in lipid remodeling namely phospholipases, TAG lipase and O-acyltransferease expression in nhr-49(nr2041) (blue bars), nhr-66(ok940) (pink bars), nhr-80(tm1011) (green bars) and nhr-13(gk796) (purple bars) relative to wild-type animals. Values represent overall fold change of nhr-49(nr2041), nhr-66(ok940), nhr-80(tm1011) and nhr-13(gk796) animals with respect to wild-type worms. Expression was measured in L4 stage of development. Error bars represent SEM from 3 independent experiments. (B) qRT-PCR measurement of genes involved in fatty acid desaturases including fat-5, fat-6 and fat-7 and genes involved in fatty acid beta-oxidation including acs-2, cpt-5 and ech-1 in nhr-49(nr2041) (blue bars), nhr-66(ok940) (pink bars), nhr-80(tm1011) (green bars) and nhr-13(gk796) (purple bars) relative to wild-type animals. Values represent overall fold change in nhr-49(nr2041), nhr-66(ok940), nhr-80(tm1011) and nhr-13(gk796) animals relative to wild-type worms. Expression was measured at L4 stage of development. Error bars represent SEM from 3 independent experiments. To identify pathways and molecular functions common to the genes observed by microarray analysis, we employed the gene ontology (GO) enrichment analysis using GOrilla [21]. As expected due to NHR-49's known role in lipid biology, there was a significant overrepresentation of GO-terms for functions related to fat metabolism (Figure 2 and Table 2). We also found that pathways regulating protein processing, maturation and proteolysis were overrepresented.
Fig. 2. Functional classification summary for the nhr-49 mutant are represented as a scatter plot using the GO visualization tool REViGO. 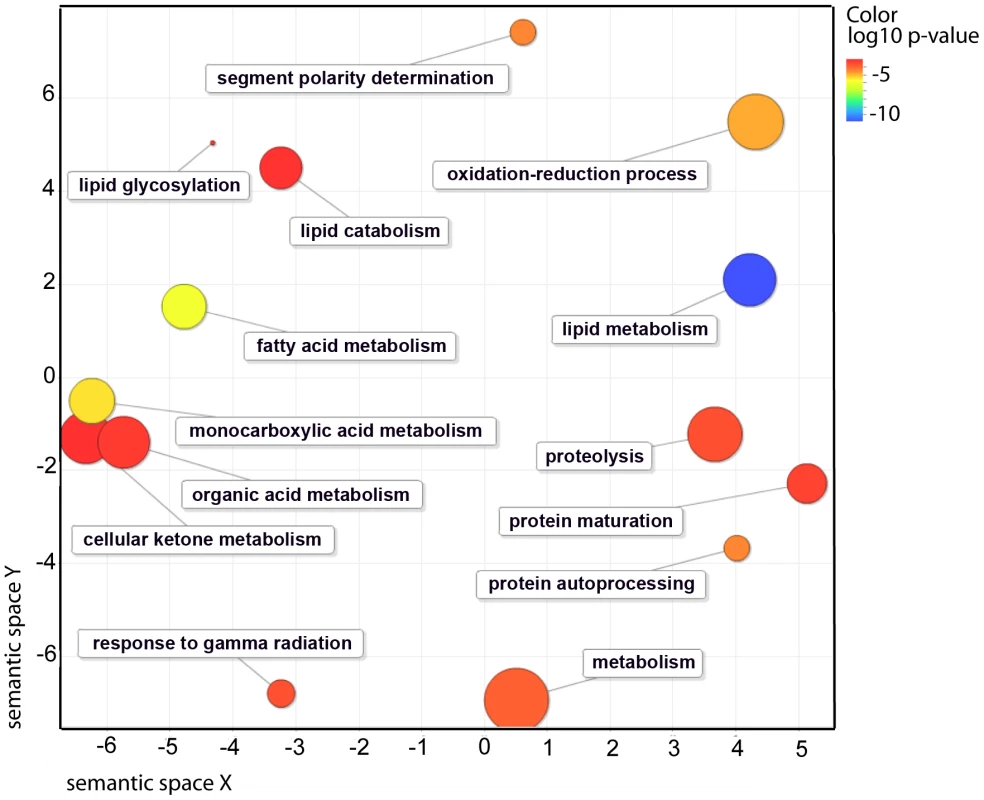
X- and Y-axes represent a two-dimensional annotation space derived from a multi-dimensional scaling procedure used on a matrix of GO terms' semantic similarities (see reference 51 for details). By employing this visualization method, similar functional categories will cluster together. Bubble color represents the p-value obtained from GO term enrichment analysis (GOrilla, see Materials and Methods) and bubble size relates to the frequency of GO terms in the Gene Ontology Annotation Database. Tab. 2. Occurrence of gene families in microarray results for NHR-49, based on GO-terms. 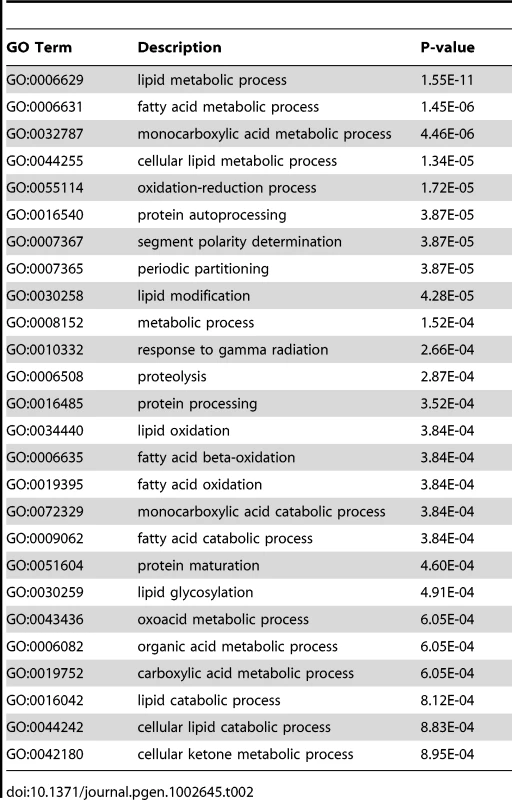
Identification of candidate NHR-49 co-factors
Because NRs form homo and/or heterodimers and associate with co-regulators, we hypothesized that NHR-49 might differentially regulate its distinct target genes by interacting with specific transcriptional co-factors [22]. To identify such factors, we performed a yeast two-hybrid screen using the NHR-49-LBD as bait. In addition to the Mediator subunit MDT-15 [23], we identified six NHRs as candidate NHR-49-LBD partners (Table S2a). Notably, all NHR prey clones identified included their respective LBDs, suggesting that dimerization occurs via the LBD, as has been described for other NRs. We estimated the relative binding strength of these preys using a LacZ reporter, and found that five of the six NHRs interacted strongly and specifically with the NHR-49 LBD (Table S2b). In parallel, a large-scale yeast-two-hybrid screen identified a set of 13 additional candidates to interact with full-length NHR-49 (Table S2a) [24]. Because they were identified in both yeast-two-hybrid screens, we deemed NHR-13, MDT-15, NHR-22, NHR-66, and NHR-105 as most likely to represent NHR-49 cofactors.
Specific co-factors regulate distinct NHR-49 pathways
To determine whether any of these cofactors regulate the newly identified NHR-49 pathways of sphingolipid processing, lipid remodeling or the β-oxidation and fatty acid desaturation pathways, we quantified the mRNA levels of the NHR-49 activated and repressed genes in these pathways (see primer pairs, Tables S3 and S4). We chose to analyze the nhr-13(gk796) and nhr-66(ok940) mutants, and also included nhr-80(tm1011) because it regulates the fatty acid desaturase genes [25]. Our qRT-PCR analyses revealed a striking separation of gene regulation by the different candidate co-factors. The deletion of nhr-66(ok940) resulted in the up regulation of most of the genes that are repressed by NHR-49, including the sphingolipid and lipid remodeling genes (Figure 1A and Table S1). The levels of derepression of these genes observed in nhr-66 mutants were comparable to that seen in nhr-49 animals, suggesting that the two NRs act together. In contrast, nhr-66 mutants did not show any change in the expression of nhr-49 activated genes, like those involved in β-oxidation and desaturation (Figure 1B and Table S1). These results strongly suggest that NHR-49 acts with NHR-66 to specifically repress the transcription of sphingolipid and lipid remodeling genes.
In contrast, nhr-80 and nhr-13 deletion mutants do not affect the sphingolipid, lipid remodeling, or β-oxidation genes. Instead, these mutants exhibited a decreased expression of the fatty acid desaturase genes fat-7, fat-5 and fat-6 (Figure 1B), confirming previous analyses of nhr80 mutants by the Watts laboratory [25]. The fat-5, fat-6 and fat-7 genes are members of the Δ9 fatty acid desaturases and are key enzymes in fatty acid metabolism [26]. Their function is to introduce a double bond in saturated fatty acid chains to generate monounsaturated fatty acids (MUFAs) that are important in membrane fluidity and energy storage [27]. These results suggest that NHR-49 associates with NHR-80 and NHR-13 to regulate the ratio of saturated and unsaturated fat in lipid membranes. However, the fold changes observed in fatty acid desaturation regulation by NHR-80 and NHR-13 did not parallel those observed in nhr-49 mutants, suggesting the involvement of other regulatory factors. Consistent with this, the C. elegans ortholog of the sterol-regulatory-element-binding protein (SREBP) transcription factor SBP-1 and the Mediator subunit MDT-15 are also implicated in the regulation of fatty acid desaturases [23], [28]–[30].
NHR-66 and NHR-80 regulate additional targets independently of NHR-49
Our data strongly suggest that NHR-66 regulates genes in the sphingolipid and lipid remodeling pathways whereas NHR-80 regulates the fatty acid desaturase genes. To determine whether these NRs regulate other genes in addition to these NHR-49 targets, we performed a genome-wide microarray analysis on nhr-66 and nhr-80 mutants. The list of differentially regulated nhr-66 and nhr-80 genes is presented in tables S5 and S7, respectively, and the corresponding GO term analyses of NHR-66 and NHR-80 is presented in Tables S6 and S8, respectively. As expected, the microarray data confirmed that NHR-66 regulates several genes involved in the sphingolipid and lipid remodeling pathways, and that NHR-80 regulates genes involved in fatty acid desaturation. In addition, Figure 3A and Table S9 show the genes commonly regulated by NHR-49 & NHR-66 and NHR-49 & NHR-80, using our microarray analysis. We also found evidence that these nuclear receptors regulate additional target genes. For example, NHR-66 is involved in GDP-mannose metabolic processes and regulates gmd-2, a GDP-mannose dehydratase. The NHR-80 GO analysis indicates that it regulates genes involved in several different processes including embryonic development and cell death. In conclusion, our analyses suggest that NHR-66 and -80 are not only transcriptional partners required for NHR-49, but can independently act with other binding partners to regulate unique pathways (Figure 3A).
Fig. 3. NHR-49 shares target genes and physically interacts with NHR-66 and NHR-80. 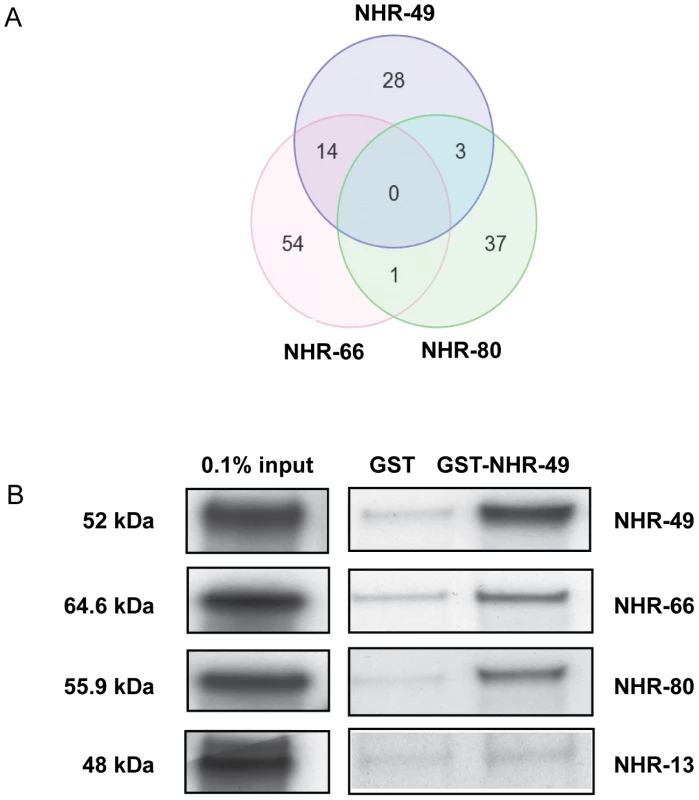
(A) Venn diagram showing the comparison of genes regulated by NHR-49, NHR-66 and NHR-80. Gene identities are presented in Table S9. (B) NHR-49 physically interacts with NHR-49, NHR-66 and NHR-80 in vitro. Pull-down assays showing in vitro translated 35S-methionine labeled NHR-49, NHR-66, NHR-80 and NHR-13 binding purified GST-NHR-49 fusion protein versus purified GST-alone control. The protein mixtures of 35S-methionine-labeled NHRs were incubated with glutathione-sepharose 4B beads and analyzed by SDS-PAGE (4–12%) followed by autoradiography. The input represents 0.1% of the labeled proteins used for the pull down assays. NHR-49 physically interacts with NHR-66 and NHR-80
Our yeast-two-hybrid and gene expression data suggested that NHR-49 might directly interact with NHR-66, NHR-80 and NHR-13. To test this, we performed in vitro GST-NHR-49 pull-down assays with full-length NHR-66, NHR-80 and NHR-13 proteins. NHR-49 was a positive control since it exhibits a two-hybrid interaction with itself [24]. Purified glutathione-S-transferase (GST)-tagged NHR-49 was incubated with in vitro translated 35S-methionine-labeled NHR-49, NHR-66, NHR-80 and NHR-13. As expected, GST-NHR-49 successfully formed a homodimer with radiolabeled NHR-49 (Figure 3B). In addition, it was also able to interact directly with NHR-66 and NHR-80. NHR-13 did not bind to NHR-49 in vitro above background levels, suggesting that additional factors may contribute to an interaction with NHR-49 in vivo. Alternately, NHR-13 may regulate fatty acid desaturation via another unknown mechanism.
NHR-49's regulation of fatty acid desaturases contributes to lifespan
The physical interaction and functional studies support a model whereby the control of distinct NHR-49 regulatory modules is based on NHR-49's association with distinct partner proteins. In this case, mutation of individual NHR-49 partners should delineate the contributions of each co-factor to the phenotypes observed in nhr-49 mutants. For example, knocking out NHR-66 could reveal NHR-49-dependent sphingolipid and lipid remodeling gene mediated phenotypes, whereas knocking out NHR-80 and NHR-13 could reveal NHR-49 dependent fatty acid desaturase regulated phenotypes. We note that although we did not obtain clear evidence that NHR-13 is a direct physical partner of NHR-49, we continued to characterize it because its gene expression profile suggests it plays a role in fatty acid desaturation.
Among the many phenotypes exhibited by nhr-49 mutants, we chose to focus on its reduced lifespan. To determine what pathway of NHR-49 is important in regulating its lifespan, we analyzed the individual co-factor mutants for effects on lifespan. The nhr-66 mutant animals had a lifespan of 17.16+/−0.41 days compared to wild-type lifespan of 17.35+/−0.34 days, suggesting that sphingolipid and lipid remodeling genes do not play a role in the reduced lifespan phenotype of nhr-49 mutants (Figure 4). In contrast, at 20°C, nhr-80 and nhr-13 mutants had significantly shorter lifespans of 13.19+/−0.38 days and 14.17+/−0.4 days, respectively (Figures 4A and 4B and Table S10). Moreover, an nhr-80; nhr-13 double mutant had a lifespan of 12.29+/−0.37 days, which approaches the nhr-49 mutant lifespan of 9.52+/−0.23 days. Together, these results suggest that the reduced expression of fatty acid desaturases observed in the nhr-80 and nhr-13 mutants might contribute to the shortened lifespan of nhr-49 animals.
Fig. 4. nhr-80; nhr-13 double mutants have a reduced lifespan comparable to that of nhr-49 animals. 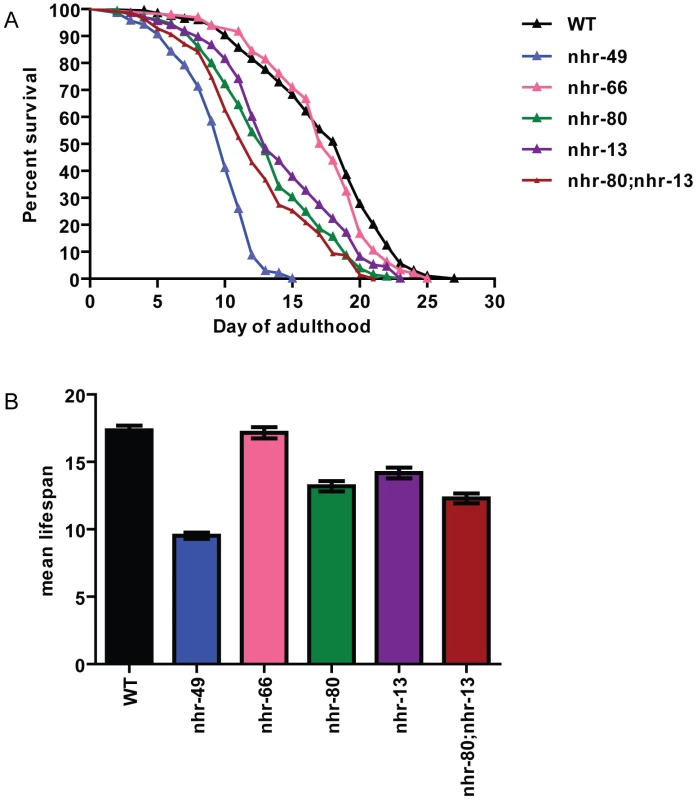
(A) The percentage of animals remaining alive is plotted against animal age. Adult lifespan survival curves of WT (black), nhr-49(nr2041) (blue), nhr-66(ok940) (pink), nhr-80(tm1011) (green), nhr-13(gk796) (purple) and double mutant nhr-80; nhr-13 (brown) animals. (B) The bar graph represents the combined mean (+/−SEM) adult lifespan (days) from at least four independent experiments. These observations are in accordance with previously published work demonstrating that shortened lifespan is correlated with lowered expression of the fat-7 SCD (Stearoyl CoA Desaturase) [16]. In addition, both nhr-49 mutants and worms where FAT-7 was knocked down using RNA-interference, exhibit a concomitant increase in the levels of saturated fat. More specifically, the reduction in desaturase levels strongly alter the ratio between stearic acid and oleic acid (C18 : 0/C18 : 1n9) to approximately 3.74+/−0.33 in nhr-49 animals (0.98+/−0.06 in wild-type animals). We thus quantified the abundance of individual fatty acids species in nhr-13, nhr-80, and nhr-80; nhr-13 mutants using Gas Chromatography-Mass Spectrometry (GC-MS). The C18 : 0/C18 : 1n9 ratio in nhr-80; nhr-13 double mutants (2.99+/−0.22) was similar to the ratio observed in nhr-49 mutants, and higher than the ratio in wild-type worms (0.98+/−0.06) (Figure 5A and Table S11). Strikingly, there is a strong inverse correlation (r2 = 0.86) between the level of saturated fat and the duration of mean worm lifespan (Figure 5B), supporting the notion that excess saturated fat plays a role in the early death of the nhr mutants. These results also suggest that NHR-49's interaction with NHR-80 and possibly NHR-13 contributes to the shortened lifespan phenotype that is observed in nhr-49 mutants.
Fig. 5. Higher levels of saturated fat in nhr-80;nhr-13 and nhr-49 animals correlate with shortened lifespans. 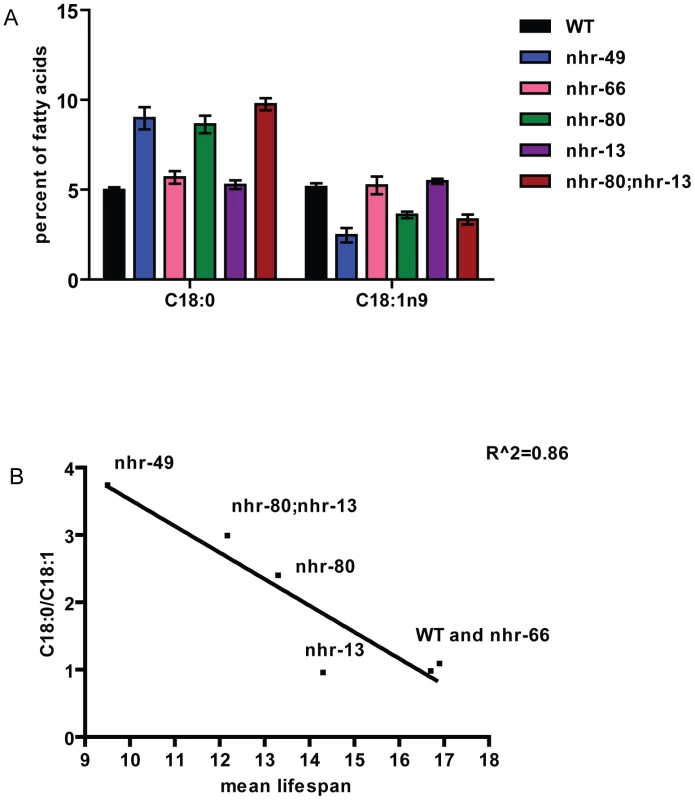
(A) Relative abundance of individual fatty acid species expressed as percentage of total measured fatty acid. Fatty acids were isolated and quantified by GC/MS from WT (black bars), nhr-49 (blue bars), nhr-66 (pink bars), nhr-80 (green bars), nhr-13 (purple bars) and nhr-80; nhr-13 (brown) animals. Error bars represent SEM. (B) Correlation between the ratio of C18:0 to C18:1n9 and mean lifespan. NHR-49 is important for normal mitochondrial morphology
To understand why the imbalance of lipid saturation observed in nhr-49 mutants could be detrimental to the animal, we hypothesized that the excess pool of saturated fat might get incorporated into membranes, thus affecting their function. Because NHR-49 functionally resembles the PPAR family of mammalian nuclear receptors that promote mitochondrial biogenesis [31], [32], we employed high pressure-transmission electron microscopy (HP-TEM) to visualize mitochondria in nhr-49 animals (Figure 6). This ultrastructural analysis revealed multiple morphological defects in the intestinal mitochondria of one-day old adult nhr-49 mutants (compared to wild-type worms), although mitochondria in nhr-49 mutants were comparable in size to wild-type mitochondria (Figure 7A). Strikingly, when we measured the average fractional area occupied by mitochondria per total intestinal area in nhr-49 mutants, we found that there was a considerable variation in the distribution, although the average was not statistically different from wild-type animals (Figure 7B). Moreover, about 25% of the intestinal mitochondria of the nhr-49 mutants were uncharacteristically irregular in shape and appeared to have more turns compared to wild-type animals (Figure 6A). The average number of turns exhibited by the intestinal mitochondria in nhr-49 animals was greater than the more rounded wild-type mitochondria (Figure 7C). Taken together, these results suggested that mitochondria in nhr-49 animals exhibit an altered shape when compared to those in wild-type animals of the same age.
Fig. 6. nhr-49 mutants show abnormal mitochondrial morphology. 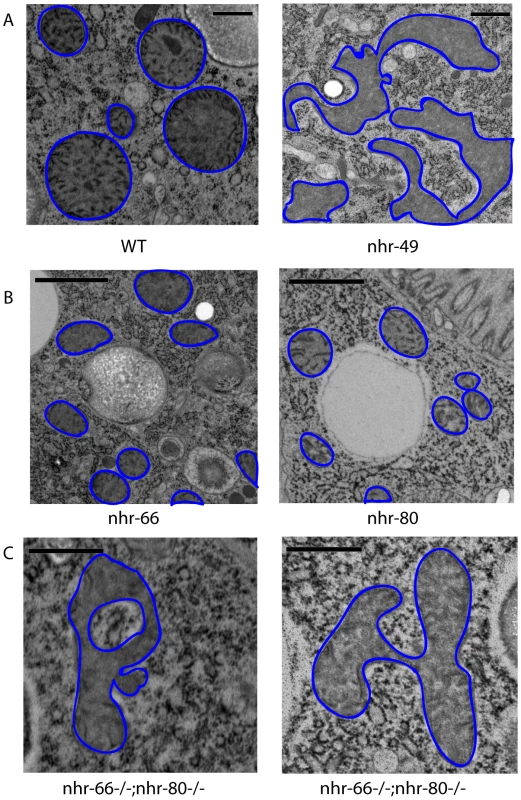
(A) High pressure freezing transmission electron microscopy (HP-TEM) images of wild-type animals and nhr-49 mutants at the same magnification. Note that mitochondria are outlined in blue. Bar, 0.5 µm. (B) HP-TEM images of nhr-66 and nhr-80 mutants at the same magnification. Note that the mitochondria are outlined in blue. Bar, 1.0 µm. (C) Two HP-TEM images of nhr-66; nhr-80 animals at the same magnification. Note that the mitochondria are outlined in blue. Bar, 1.0 µm. Fig. 7. nhr-49, nhr-66, and nhr-80 animals have abnormal mitochondrial phenotypes. 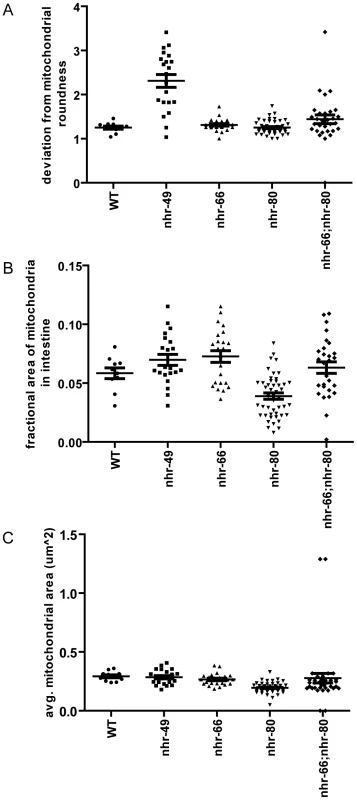
(A) Quantification of mitochondrial size in WT, nhr-49, nhr-80, nhr-66 and nhr-66; nhr-80 animals. Each point represents the average size of all the intestinal mitochondria examined per section. P-values were obtained using the unpaired t-test with Welch's correction for unequal variance. This analysis showed that the difference between nhr-80 animals and wild-type is statistically significant with a p-value<0.0001. (B) Quantification of mitochondrial fractional area in WT, nhr-49, nhr-80, nhr-66 and nhr-66; nhr-80 animals. Each point represents the average fractional area (µm2) of the examined mitochondria per section. P-values were obtained using the unpaired t-test with Welch's correction for unequal variance. This analysis showed that the difference between nhr-66 and nhr-80 animals compared to wild-type was statistically significant with p-values of 0.0440 and 0.0025 respectively. (C) Quantification of mitochondrial shape in WT, nhr-49, nhr-80, nhr-66 and nhr-66; nhr-80 animals. Each point represents the average number of mitochondrial turns in the section examined. P-values were obtained using the unpaired t-test with Welch's correction for unequal variance. This analysis showed that the difference between nhr-49animals compared to wild-type was statistically significant with a p-value<0.0001. Partner co-factors contribute to NHR-49's regulation of mitochondrial morphology
We next asked if the NHR-49 partners NHR-66 and NHR-80 contribute to the observed NHR-49 mitochondrial abnormalities. In contrast to nhr-49 mutants, the mitochondria in one-day old adult nhr-66 and nhr-80 mutants were comparable in shape to those observed in wild-type worms (Figure 6B and 7A). However, the average fractional area occupied by the mitochondria in the intestine was significantly higher in nhr-66 animals and significantly lower in nhr-80 animals (compared to wild-type worms; Figure 7B). Lastly, even though the average size of the intestinal mitochondria in the nhr-66 animals was similar to the size of mitochondria in wild-type animals, the nhr-80 animals showed significantly smaller mitochondria (Figure 7C).
Because these data suggested that NHR-66 and NHR-80 contribute to mitochondrial morphology, we next examined intestinal mitochondria in one-day old adult nhr-66; nhr-80 double mutants. The fractional area of mitochondria in these animals was not statistically different from wild-type when averaged, but the distribution was much broader, similar to nhr-49 animals (Figure 7B). In addition, 11.4% of the intestinal mitochondria in the double mutants were highly irregular in shape, as was observed in 25% of the mitochondria in nhr-49 mutants (Figure 6C and Figure 7C). Although the mitochondrial phenotypes in nhr-66; nhr-80 mutants are not as strong as those in the nhr-49 animals, these data do suggest that NHR-66 and NHR-80 contribute to normal mitochondria morphology. Together, these data suggest that NHR-49 maintains mitochondrial morphology via multiple pathways, including NHR-66 and NHR-80 dependent regulation as well as additional, unknown mechanisms.
Impaired mitochondrial morphology affects physiological function
Because mitochondrial morphology was altered in the nhr mutants, we assayed mitochondrial function using two metabolic assays. First, we monitored the basal oxygen consumption rates in live animals (normalized to worm count). Consistent with the mitochondrial abnormalities observed by electron microscopy, nhr-49 animals consumed oxygen at significantly reduced rates of 5.22 pmoles/min/worm compared to 9.625 pmoles/min/worm in wild-type animals (Figure 8A). The single mutants nhr-66 and nhr-80 also had reduced basal oxygen consumption rates of 6.12 and 7.44 pmoles/min/worm, respectively. Interestingly, the respiration rate for the nhr-66; nhr-80 double mutant was lower than that of nhr-49 with a rate of 3.87 pmoles/min/worm but higher than the electron transport chain (ETC) complex I defective mutant gas-1(fc21), which showed rates of 2.55 pmoles/min/worm.
Fig. 8. nhr-49 animals have reduced basal oxygen consumption rates and reduced beta-oxidation. 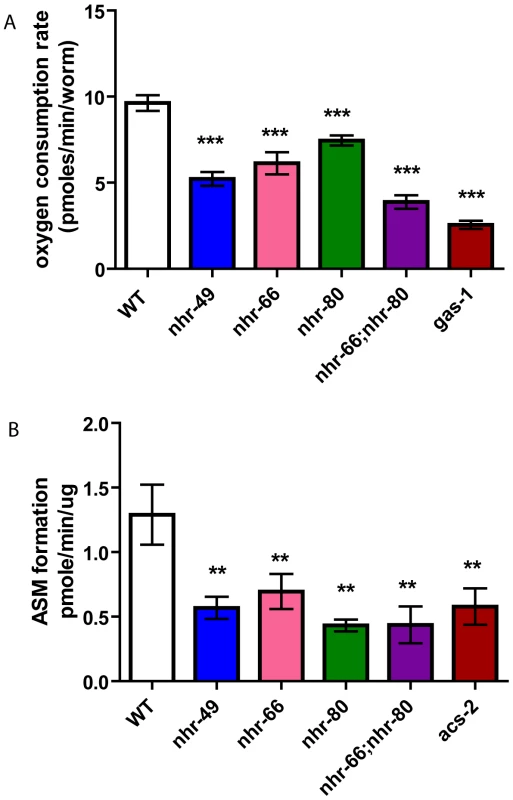
(A) Basal oxygen consumption rates of WT, nhr-49, nhr-80, nhr-66, nhr-66; nhr-80 and gas-1 animals. P-values were obtained using the unpaired t-test. *** p<0.001. (B) Evaluating β-oxidation by measuring amounts of radiolabeled acid-soluble metabolites in WT, nhr-49, nhr-80, nhr-66, nhr-66; nhr-80 and acs-2 animals. The data represents the average results from two independent experiments. The results are expressed as means +/− SEM (n = 6) normalized to the specific activity of the radioactive palmitate. One-way ANOVA was used to compare with the Neuman-Keuls multiple comparison tests. **<p<0.01, *** p<0.001. In a second assay for mitochondrial function, we indirectly measured β-oxidation by feeding the animals radiolabeled palmitic acid and then measuring the rates of production of an acid soluble metabolites, a byproduct of lipid oxidation. As the mitochondrial β-oxidation gene acs-2 is down regulated in nhr-49 mutants and contributes to their increased fat content, we used the acs-2(ok2457) mutant as a control [16]. Just like the acs-2 mutants, the nhr-49 animals also had a significant reduction in acid soluble metabolite production with a rate of 0.56 pmole/min/µg protein compared to 1.29 observed in wild-type animals (Figure 8B). Surprisingly, although the qRT-PCR analysis did not predict NHR-66 and NHR-80 to regulate genes in the β-oxidation pathway, the nhr-66 and nhr-80 single mutants also exhibited reduced oxidation rates (0.69 and 0.43 pmole/min/µg protein, respectively). This is consistent with our EM data that nhr-66 and nhr-80 mitochondria differ from wild-type, which may be explained by the reduced β-oxidation in these mutants. The β-oxidation rate for the nhr-66; nhr-80 double mutants was also significantly reduced relative to wild-type animals (0.43 pmole/min/ug protein), although the rate in the double mutant was identical to the rate in the single mutant with the stronger effect (nhr-80), suggesting that the two NRs may act in the same pathway. Together, our data show that altered mitochondrial morphology strongly correlates with aberrant mitochondrial physiology, and they further suggest that several NRs play a significant role in maintaining normal mitochondrial function.
Discussion
NHR-49 regulates genes involved in fatty acid β-oxidation and fatty acid desaturation [16], but its full influence on lipid metabolism remained obscure. Here, we reveal a previously uncharacterized group of NHR-49 targets, namely genes involved in the repression of sphingolipid processing and lipid remodeling. We also identify several NHR-49 interacting partner receptors and show that at least two of these, NHR-66 and NHR-80 directly bind to NHR-49 and modulate separable NHR-49 dependent pathways. In characterizing NHR-49's transcriptional network and studying its influence on physiology, our findings inform us on the evolutionary history of HNF4-related receptors across species and on their conserved metabolic pathways and physiological functions.
NHR-49's transcriptional network
HNF4α plays an important role in mammalian physiology and despite the identification of several HNF4 bound and regulated genes, it is still unclear what the relevant targets are in vivo. The number of genes estimated to be regulated by HNF4α varies greatly and depends on the experimental approaches [33], [34]. For example, Odom et al. performed ChIP on chip to identify promoters occupied by HNF4α in the human liver and pancreas and identified 1575 potential HNF4 target genes [35]. In contrast, gene expression analysis of pancreatic cells in an HNF4α conditional knockout model identified only 133 genes as HNF4α regulated [36]. This highlighted that revealing the regulatory targets and processes of metabolic nuclear receptors needed further characterization. We therefore performed genome-wide transcriptional profiling to identify the targets of the PPAR/HNF4 like nuclear receptor NHR-49 in C. elegans. Our analysis revealed a previously unrecognized role of NHR-49 in the regulation of genes involved in membrane lipid metabolism, particularly glycosphingolipid processing. It will be important to determine whether mammalian HNF4α also regulates these genes.
It has also been unclear what co-factors of mammalian HNF4α influence distinct aspects of gene expression. We took advantage of C. elegans genetics to identify multiple partners of NHR-49 and confirmed that NHR-66 and NHR-80 physically interact with NHR-49 using in vitro binding assays. We further characterized the transcript level changes observed for genes regulated by NHR-66, NHR-80 and NHR-13. Notably, knockout of nhr-66 affected only a subset of NHR-49 targets, sphingolipid and lipid remodeling target genes, whereas knockout of nhr-80 and nhr-13 affected the expression of NHR-49-regulated fatty acid desaturases. We therefore propose that NHR-49 and NHR-66 heterodimerize to regulate genes that mediate the breakdown of glycolipids and remodeling of lipid membranes, whereas NHR-49 cooperates with NHR-80 and possibly NHR-13 to sense and regulate the balance between saturated and unsaturated fat (Figure 9). Even though we did not detect a direct interaction between NHR-13 and NHR-49, NHR-13 clearly affects fatty acid desaturation. In the future, it will be important to determine if the different target genes are direct NR targets in vivo. To date, it has been difficult to identify a consensus binding sequence for NHR-49 and other NRs given the degeneracy of response elements often observed in C. elegans, and the lack of antibodies suitable for chromatin immunoprecipitation to define in vivo NR binding sites. GFP reporters of NHR-49/NHR-66 and NHR-49/NHR-80 regulated genes will be useful to determine whether these distinct dimers may selectively drive gene expression in individual tissues. Although HNF4 is not yet known to dimerize with other partners, it will be critical to determine if there are similar cofactors and/or it carries out similar mechanistic roles.
Fig. 9. Model of NHR-49–dependent regulation of lipid metabolism. 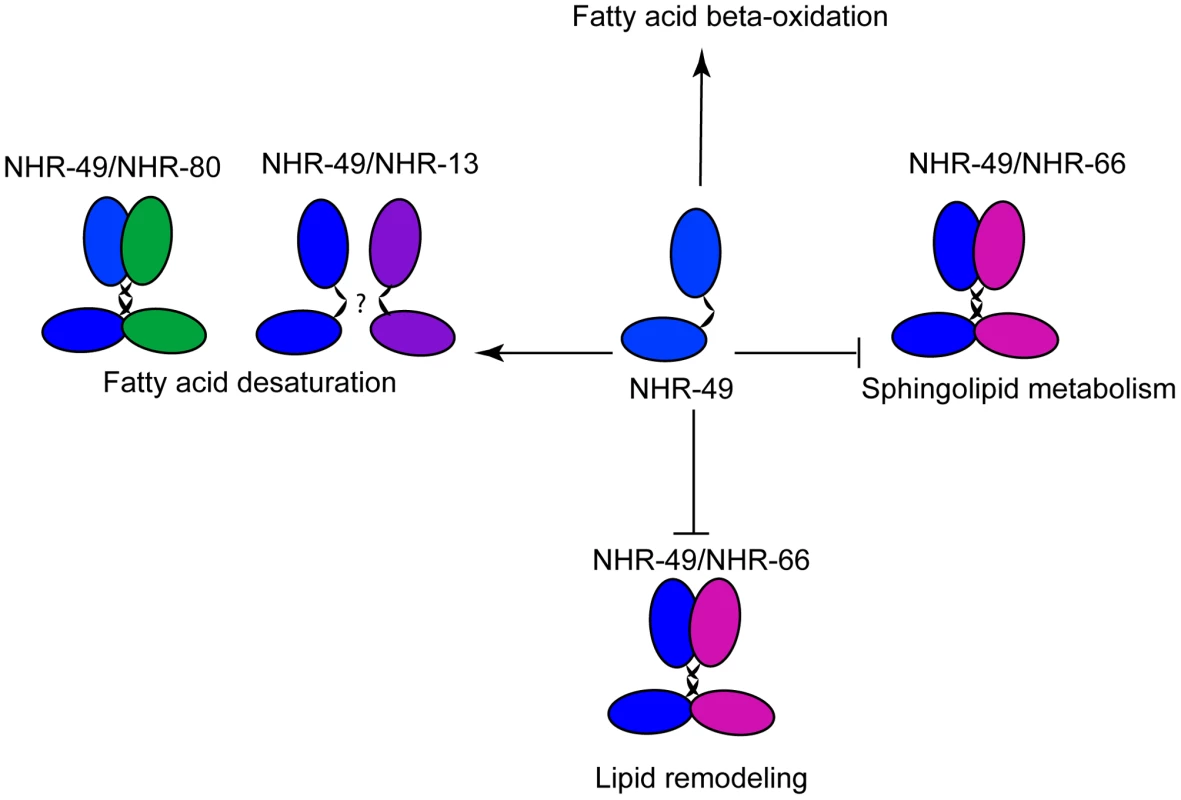
We propose that NHR-49 (blue) interacts with NHR-66 (pink) to repress genes involved in sphingolipid processing and lipid remodeling, whereas NHR-49 (blue) binds to NHR-80 (green) to activate the fatty acid desaturase genes. Another interesting finding of this study is the complexity of the NHR-49 regulatory network. For instance, both NHR-66 and NHR-80 are NHR-49 co-factors, but there are nuances in their co - regulation of NHR-49 genes. In the case of NHR-66, the fold changes observed in sphingolipid and lipid remodeling genes were comparable to those seen in nhr-49 animals. However, in the case of NHR-80 and NHR-13, the levels of activation in fatty acid desaturation regulation did not completely parallel the levels we observed in nhr-49 mutants. This suggests that additional regulation of these genes may involve NHR-49 hetero and/or homodimers or NHR-49 independent mechanisms, consistent with our observation that NHR-13 influences NHR-49 regulated desaturases but may not directly bind to NHR-49. Adding to the complexity, our gene expression data suggest that, in addition to the genes jointly regulated by NHR-49/NHR-66 and NHR-49/NHR-80, NHR-66 and NHR-80 regulate additional genes independently of NHR-49. In fact, our studies on mitochondrial morphology and function do not clearly differentiate between the possibilities that the observed defects are the result of NHR-49-dependent pathways or the result of multiple independent pathways regulated by NHR-66 and NHR-80. Our data also does not exclude the possibility that NHR-66 and NHR-80 act together for some functions. Future work will be needed to address these different scenarios and elucidate the complexity of this transcriptional network.
Influence of NHR-49's target genes on physiology
Our lifespan analysis revealed that NHR-80 and NHR-13 regulated pathways appear to contribute to the early death of nhr-49 deletion mutants. We propose that NHR-49's interaction with NHR-80 and possibly with NHR-13 modulates the conversion of saturated to unsaturated fat, and contributes to the shortened lifespan phenotype of nhr-49 mutants. Supporting this idea, there is a significant correlation between the C18 : 0/C18 : 1n9 ratio and mean lifespan. This fits well with a recent study that showed the correlation between several fatty acid metabolic parameters to longevity, including the ratio of C18 to C18 : 1n9 [37]. Our data my also provide insight into the mechanism by which mammalian HNF4α protects against diabetes. Both in vitro and in vivo studies showed that pancreatic β-cells are highly susceptible to saturated fat-induced apoptosis or lipotoxicity [38], [39], whereas overexpression of the fat-6/fat-7 homologue Stearoyl CoA Desaturase (SCD) protects against this lipotoxicity [40], [41]. It is intriguing to speculate that the shortened lifespan could be related to roles of HNF4 in protection against premature cell death.
Although NHR-49 is derived from the ancestral HNF4-like receptors [42], NHR-49 functionally resembles the PPARα. Our EM data suggest that mitochondrial morphology was abnormal in nhr-49 mutants, which led us to test mitochondrial function in these worms. We observed defects in oxygen consumption and fatty acid β-oxidation, which may be the result of altered membrane lipids affecting the function of intestinal mitochondria. Because the co-factor mutants also exhibit mitochondrial defects, we propose that multiple pathways regulate mitochondrial function.
It has been shown that PPARs regulate the density (number) of mitochondria. In fact, PPARγ and PPARδ promote mitochondrial biogenesis in a cell-type specific manner, specifically increasing biogenesis in white adipose tissue and skeletal muscle, respectively [31], [32]. PPARs have also been shown to influence mitochondrial function by mostly regulating genes encoding mitochondrial fatty acid β -oxidation enzymes and mitochondrial uncouplers [43]. Our data suggest that nhr-49 mutants differ from wild-type animals in the shape but not the fractional area (number) of their intestinal mitochondria. Given that some mitochondria in the nhr-49 mutants are irregular in shape, it would be interesting to determine if the mitochondria in mammalian PPAR mutant mice exhibit similar phenotypes. Alternatively, a set of genes not commonly regulated by NHR-49 and PPAR may contribute to the altered morphology of the mitochondria in nhr-49 mutants only. In line with this notion, our study revealed novel NHR-49 dependent targets and pathways, particularly glycosphingolipid processing and lipid remodeling, which mammalian PPARs are not known to regulate.
Insights on evolution of the HNF4 family
When HNF4 and HNF4-like NRs are compared across species (Table 3), a common theme that emerges is the conservation of their roles in lipid metabolism pathways. It is possible that one ancestral role of HNF4–the mobilization of stored energy during nutrient deprivation and the regulation of β-oxidation–was adopted by PPARα, whereas other functions of HNF4 were either retained or outsourced to other NRs. In C. elegans, HNF4-like NRs underwent a massive expansion and it is likely that various functions were allocated to different nuclear receptors [44]. HNF4 paralogs in nematodes could then have been selected for divergent functions that included sphingolipid metabolism and lipid remodeling.
Tab. 3. Insights on the evolution of the HNF4 family across species. 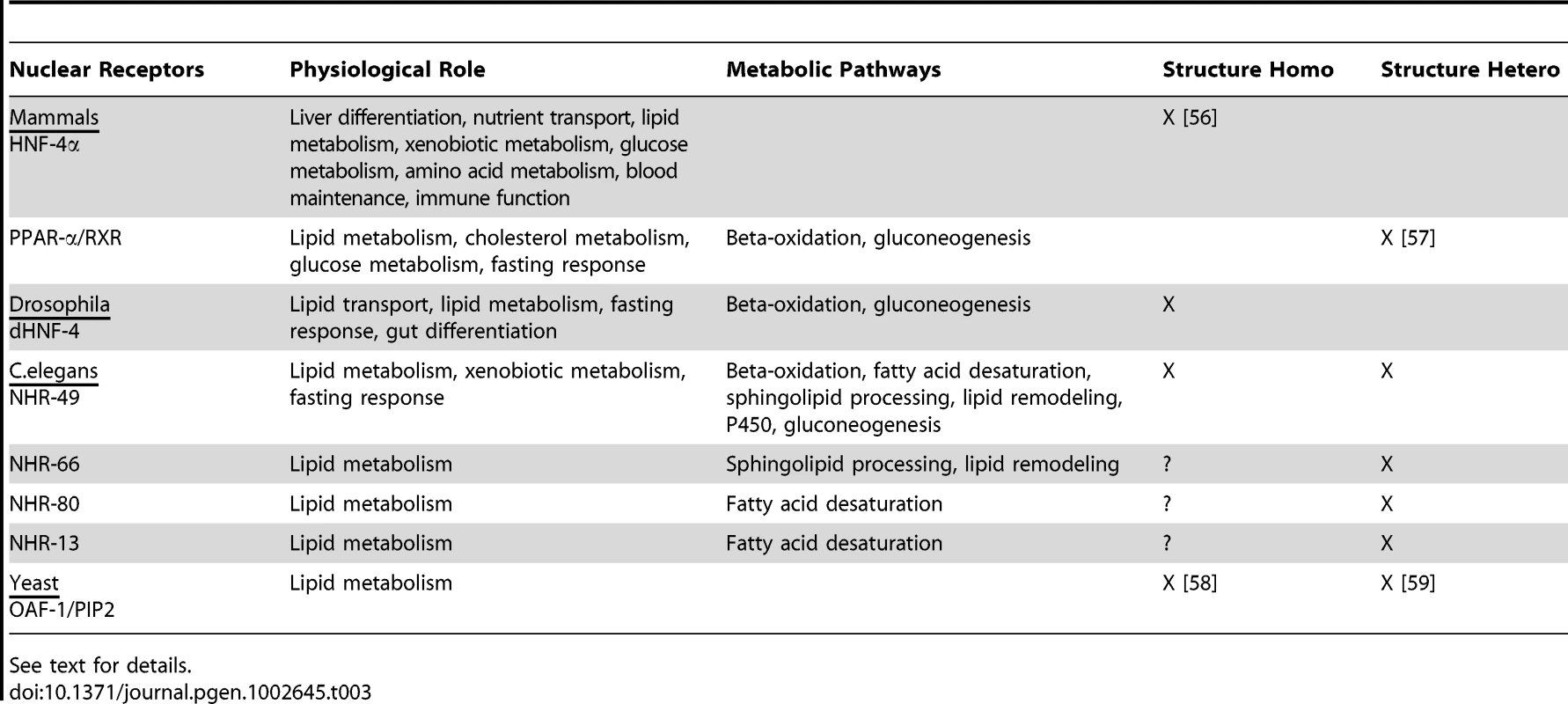
See text for details. Mammalian RXR serves as a heterodimer partner for numerous NRs, including PPARs [45]. We thus speculate that like PPAR and its heterodimeric partner, RXR adopted specific HNF4 roles in mammals. It is possible that NHR-49 heterodimerized with other binding partners to adopt individual tasks that encompassed the numerous functions of the precursor nematode HNF4. This is particularly intriguing given the striking absence of any C. elegans orthologs of RXR-like molecules that in other organisms heterodimerize with a range of interacting proteins. NHR-49's action is promiscuous as it is clearly assuming different function with different partners. NHR-49 may therefore serve as an ancestral binding partner reminiscent of the mammalian RXR. C. elegans thus represents a class of animals that switched from solely using HNF4 receptors to regulate lipid metabolism to those that began to employ different heterodimeric partners to regulate distinct metabolic pathways.
In closing, our study has identified distinct binding partners that modulate specific pathways of NHR-49 activity. Additionally, we have also shown that these NRs play a role in mitochondrial physiology and function. Given the conservation of HNF4 structure, function and regulatory modules across species, it is possible that mammalian HNF4α utilizes similar mechanisms as that of C. elegans NHR-49, thus informing us of ways to selectively modulate its activity. Exploring these possibilities can assist in pharmacological efforts to manipulate mammalian HNF4α for specific desired outcomes without any detrimental side-effects.
Materials and Methods
Nematode strains and growth conditions
C. elegans strains N2 Bristol (wild-type), nhr-49(nr2041), nhr-66(ok940), nhr-80(tm1011), nhr-13(gk796), gas-1(fc21) and acs-2(ok2457) were grown at 20°C on high-growth plates seeded with OP50 bacteria and maintained as described [46]. The mutant strains were obtained from the CGC and were outcrossed at least five times with wild-type N2 worms. Single-worm PCR was used to determine the genotype of worms during crossing to generate nhr-80; nhr-13 and nhr-66; nhr-80 double-mutant lines. For mRNA and GC/MS analysis, worm embryos were allowed to hatch on unseeded nematode growth media (NGM)-lite plates overnight at 20°C. The next day, synchronized L1 larvae were plated onto NGM-lite plates seeded with Escherichia coli strain OP50. Worms were grown to early L4s at 20°C, harvested, washed three times with M9, and flash-frozen in liquid N2.
Preparation of total nematode mRNA
C. elegans strains N2-Bristol (wild-type), nhr-49(nr2041), nhr-66(ok940), nhr-80(tm1011) and nhr-13(gk796) were grown at 20°C on high-growth plates seeded with OP50 bacteria and maintained as described [46]. Gravid adults from 10 10-cm plates were bleached, and embryos were dispersed onto 15-cm nematode growth media (NGM)-lite plates seeded with OP50. Worms at L4 stage were harvested, washed twice with M9, and frozen in liquid nitrogen. For RNA preparation, worms were thawed at 65°C for 10 min, and RNA was isolated using the Tri-Reagent Kit (Molecular Research Center, Cincinnati, Ohio, United States). Isolated total RNA was subjected to DNAase treatment and further purification using RNAeasy (Qiagen, Valencia, California).
Yeast two-hybrid screen
For the NHR-49-LBD yeast two-hybrid screen [23], the strain AH109/pGBK-Leu2-NHR-49-LBD was used to probe a C. elegans oligo(dT)-primed cDNA library cloned into pPC86 (kindly provided by the Vidal laboratory, Dana Farber Cancer Institute, Boston). In short, ∼4.3×106 independent transformants were screened for growth on medium-stringency plates (lacking tryptophan, leucine, histidine, and uracil containing 7.5 mM 3-amino-1,2,4-triazole (3-AT; Sigma, H-8056). Candidate clones were retested twice for their ability to grow on high-stringency medium (i.e. medium-stringency plates additionally lacking adenine), allowing recovery of 141 candidates. Following PCR and AluI digestion to identify redundant clones, 49 candidate plasmids were extracted and sequenced, yielding 24 independent cDNAs of 13 genes (Table S1c). To estimate the relative interaction strength of these candidate proteins with the NHR-49-LBD, plasmid pairs were transformed into strain Y187 (Clontech) and liquid β-galactosidase assays were performed as described in the manufacturer's protocol.
qRT–PCR and microarray analysis
cDNA was prepared from 5 µg of total RNA in a 100-µl reaction using the Protoscript cDNA preparation kit (New England Biolabs, Beverly, Massachusetts, United States). Primer pairs were diluted into 96-well cell culture plates at a concentration of 3 µM. Next, 30-µl PCR reactions were prepared in 96-well plates. Each PCR reaction was carried out with TaqDNA Polymerase (Invitrogen, Carlsbad, California, United States) and consisted of the following reaction mixture: 0.3 µM primers, 1/500th of the cDNA reaction (corresponds to cDNA derived from 10 ng of total RNA), 125 µM dNTPs, 1.5 mM MgCl2, and 1× reaction buffer (20 mM Tris pH 8.4, 50 mM KCl), 0.15 µl (0.75 units) of TaqDNA Polymerase was used for each reaction. Formation of double-stranded DNA product was monitored using SYBR-Green (Molecular Probes, Eugene, Oregon, United States). Data were collected using RNA from at least three independent C. elegans growths. To determine the relationship between mRNA abundance and PCR cycle number, all primer sets were calibrated using serial dilutions of cDNA preparations. qRT-PCR primers were designed using Primer3 software [47]. qPCR was performed using a BioRad iCycler (MyiQ Single color).
For microarray analysis, total RNA was assayed for quality using an UV-Vis spectrophotometer and an Agilent 2100 Bioanalyzer (Agilent Technologies, Inc., Santa Clara, CA). Cy5 - and Cy3-labeled cDNA targets were generated from 30 µg of total RNA using a reverse transcription, amino-allyl based labeling protocol [48]. Mutant and wild-type targets were co-hybridized to Washington University/Genome Sequencing Center C. elegans 23K spotted oligo arrays for 16 hours at 42°C, followed by a series of stringency washings at 42°C for 5 cycles in 2× SSC and 3 cycles in 0.1× SSC using an automated GeneTac Hybridization Station (Genomic Solutions, Ann Arbor, MI, USA). Post-hybridized arrays were scanned using a GenePix 4000B scanner (Axon Instruments, Union City, CA) and image analysis was performed using GenePix Pro software. Independent datasets were generated for each mutant vs. wild-type comparison: nhr-49 (n = 2, with dye swap), nhr-66 (n = 3), nhr-80 (n = 4).
Array data was pre-processed through a custom-built application used to filter data based on criteria associated with spot-level signal quality (e.g., signal-to-noise, lower bound intensity threshold, spot dimension). Each array was background subtracted and the ratios (mutant/wt) were log2 transformed. Intra-array normalization was performed using a loess algorithm to correct for intensity-dependent ratio biasing [49]. For each comparison, the dataset was filtered by applying a lower-bound signal intensity cutoff, followed by the application of a variance filter using the ‘shorth’ function in the Bioconductor package genefilter. Pair-wise significance testing (mutant vs. wild-type) was performed using the Bioconductor package limma [20] and p-values were initially corrected for multiple testing using the false discovery rate (FDR) method of Benjamini and Hochberg [50]. We attempted to define differential expression as |log2(ratio)|≥0.848 with the FDR set to 5%, however the multiple testing penalty was too high to produce a gene list in some cases and as such we slightly relaxed our criteria by employing |log2(ratio)|≥0.848 and p-value≤0.001.
Gene ontology (GO) enrichment analysis was performed using GOrilla [21]. Each list from the limma analysis was ranked from smallest to largest p-value and analyzed for enriched biological process ontology terms found near the top of the list. Functional classification summary for the nhr-49 mutant were presented as a scatter plot using the GO visualization tool REViGO [51].
Fatty acid analysis
Fatty acids were isolated from 10,000 L4 animals grown on a single 15-cm NGM-Lite plate. Total lipids were extracted and converted to fatty acid methyl esters (FAMEs) as described [52]. After incubation at 80°C for 1 hour the samples were cooled and the FAMEs were extracted by adding water and hexane. FAMEs were analyzed for fatty acid composition by gas chromatography/mass spectrometry (GC/MS) (Agilent 5975GC, 6920MS). Peaks were assigned using fatty acid standards.
Plasmids
The PCR products of the full length NHR-49, NHR-66, NHR-80 and NHR-13 were cloned into the expression vector pCS2+ that was generously provided by Dr. B. Eisenmann (Fred Hutchinson Cancer Research Center, Seattle). The PCR product for NHR-49 was flanked by BamH1/EcoR1, NHR-66 by Stu1/Xba1, NHR-80 by BamH1/EcoR1 and NHR-13 by BamH1/EcoR1 and then cloned into the corresponding sites in pCS2+. For bacterial expression, GST-NHR-49 was constructed by inserting BamH1-NHR-49 cDNA-EcoR1 into PGEX-2T vector. All clones were confirmed by sequence analysis. All plasmid requests should be directed to Stefan Taubert.
GST–pull down assays
GST-fused to NHR-49 (GST-NHR-49) was induced with 1 mM IPTG for 2 hours at 23°C, expressed in the Escherichia coli BL21 (DE3) strain and purified using glutathione-Sepharose 4B beads. The IVTs were transcribed from the SP6 promoter and translated with the wheat germ system (Promega: Madison, WI), in the presence of unlabeled (Promega: Madison, WI) or 35S - labeled methionine (Perkin Elmer) according to the manufacturer's instructions. For the in vitro protein-protein interaction assays, the IVT protein was incubated with GST-NHR-49 for 90 minutes at 37°C and washed in binding buffer. The samples were then run on SDS-PAGE and the radiolabeled protein detected by autoradiography.
Lifespan assays
Approximately 30–40 L1 worms were transferred to 6-cm plates seeded with L4440 RNAi bacteria and life-span assays were carried out at 20°C as described previously [53]. The L4 stage was counted as day 0. Adults were transferred to new plates daily until progeny production had ceased. Animals were considered dead when they no longer responded to a gentle tap with a worm pick. Life span curves and statistical data including p-values from Log-rank (Mantel-Cox) test were generated using GraphPad Prism version 5 software (GraphPad Software, San Diego, CA).
High pressure transmission electron microscopy
Day 1 adults were placed into a 20% BSA/PBS buffer solution and prepared in a Leica-Impact-2 high-pressure freezer. In short, animals were kept for 60 hours in 100% acetone and uranyl acetate at −90°C. The temperature was then ramped from −90°C to −25°C over the course of 32.5 hours. Next, samples were incubated at −25°C for 13 hours. Finally, the temperature was brought from −25°C to 27°C in a 13 hour temperature ramp. Serial sections were post-stained in uranyl acetate followed by lead citrate. Thin cross sections were taken from resin-embedded clusters of young adults. Sections for adult animals were obtained from 3 different animals.
Quantification of mitochondria
Ten micron transverse sections typically spanning the length of the worm from the pharynx to the vulva were examined per animal. Mitochondria from each section were individually outlined and the average measurements were obtained using Image J software. At least two animals were examined per mutant strain. Statistical data including p-values from unpaired t-tests using Welch's correction for unequal variance were generated using GraphPad Prism version 5 (GraphPad Software). Irregularity in mitochondrial shape was measured by counting the number of times the mitochondrial outer membrane changed from convex to concave. A turn was measured every time the mitochondria changed from convex to concave and vice versa.
Determination of β-oxidation by acid-soluble metabolite production
The production of acid-soluble metabolites was used as an index of the β-oxidation of fatty acids based on an assay that was originally developed for cell lines [54]. We further modified the protocol for C. elegans based on the protocol reported by Mullaney et al, 2010 [55]. Synchronized L4 animals were washed off plates with 1× M9 medium and counted. The animals were then rinsed three times in 0.9% NaCl. An aliquot was stored at −80 C for subsequent protein determination. The worms were resuspended in S-Basal medium with 14C-palmitic acid (40–60 mCi/mmol) to a final concentration of 1 µCi /ml complexed to 25% fatty-acid-free BSA (all from GE Healthcare Life Sciences) per well. Samples were incubated on an orbital shaker for 2 hours at room temperature. Subsequently, 70% perchloric acid was added to precipitate the BSA-bound fatty acid. The samples were centrifuged for 10 minutes at 14,000 g and the radioactivity of the supernatant was determined by liquid scintillation. Samples without animals were included as background controls. These experiments were performed in triplicates two independent times. Statistical data was analyzed using one-way ANOVA and p-values were generated using GraphPad Prism version 5 (GraphPad Software).
Measuring oxygen consumption rates by Seahorse XF-24 analyzer
Between 50 and 100 synchronized L4 worms were washed with 1XM9 and seeded into triplicates of the Seahorse XF-24 cell culture plates (Seahorse Bioscience, North Billerica, MA) in M9. Oxygen consumption rates were measured at least five times using the Seahorse XF-24 Analyzer (Seahorse Bioscience). Measurements were taken under basal conditions and were normalized to the number of worms counted per well. The Seahorse software was used to plot the results. The experiment was repeated two times under these conditions. Statistical data including p-values from unpaired t-test were generated using GraphPad Prism version 5 (GraphPad Software).
Supporting Information
Zdroje
1. EvansRMBarishGDWangYX 2004 PPARs and the complex journey to obesity. Nat Med 10 355 361
2. ChawlaARepaJJEvansRMMangelsdorfDJ 2001 Nuclear receptors and lipid physiology: opening the X-files. Science 294 1866 1870
3. GlassCKRosenfeldMG 2000 The coregulator exchange in transcriptional functions of nuclear receptors. Genes & Development 14 121 141
4. StoffelMDuncanSA 1997 The maturity-onset diabetes of the young (MODY1) transcription factor HNF4α regulates expression of genes required for glucose transport and metabolism. Proceedings of the National Academy of Sciences 94 13209 13214
5. MiquerolLLopezSCartierNTulliezMRaymondjeanM 1994 Expression of the L-type pyruvate kinase gene and the hepatocyte nuclear factor 4 transcription factor in exocrine and endocrine pancreas. Journal of Biological Chemistry 269 8944 8951
6. YamagataKFurutaHOdaNKaisakiPJMenzelS 1996 Mutations in the hepatocyte nuclear factor-4[alpha] gene in maturity-onset diabetes of the young (MODY1). Nature 384 458 460
7. HaniEHSuaudLBoutinPChèvreJCDurandE 1998 A missense mutation in hepatocyte nuclear factor-4 alpha, resulting in a reduced transactivation activity, in human late-onset non-insulin-dependent diabetes mellitus. The Journal of Clinical Investigation 101 521 526
8. Love-GregoryLDWassonJMaJJinCHGlaserB 2004 A Common Polymorphism in the Upstream Promoter Region of the Hepatocyte Nuclear Factor-4α Gene on Chromosome 20q Is Associated With Type 2 Diabetes and Appears to Contribute to the Evidence for Linkage in an Ashkenazi Jewish Population. Diabetes 53 1134 1140
9. SilanderKMohlkeKLScottLJPeckECHollsteinP 2004 Genetic Variation Near the Hepatocyte Nuclear Factor-4α Gene Predicts Susceptibility to Type 2 Diabetes. Diabetes 53 1141 1149
10. GuptaRKGaoNGorskiRKWhitePHardyOT 2007 Expansion of adult β-cell mass in response to increased metabolic demand is dependent on HNF-4α. Genes & Development 21 756 769
11. RhodesCJ 2005 Type 2 Diabetes-a Matter of {beta}-Cell Life and Death? Science 307 380 384
12. HayhurstGPLeeY-HLambertGWardJMGonzalezFJ 2001 Hepatocyte Nuclear Factor 4{alpha} (Nuclear Receptor 2A1) Is Essential for Maintenance of Hepatic Gene Expression and Lipid Homeostasis. Molecular and Cellular Biology 21 1393 1403
13. Weissglas-VolkovDHuertas-VazquezASuviolahtiELeeJPlaisierC 2006 Common Hepatic Nuclear Factor-4α Variants Are Associated With High Serum Lipid Levels and the Metabolic Syndrome. Diabetes 55 1970 1977
14. SluderAEMainaCV 2001 Nuclear receptors in nematodes: themes and variations. Trends in Genetics 17 206 213
15. BertrandSBrunetFGEscrivaHParmentierGLaudetV 2004 Evolutionary Genomics of Nuclear Receptors: From Twenty-Five Ancestral Genes to Derived Endocrine Systems. Molecular Biology and Evolution 21 1923 1937
16. Van GilstMRHadjivassiliouHJollyAYamamotoKR 2005 Nuclear hormone receptor NHR-49 controls fat consumption and fatty acid composition in C. elegans. PLoS Biol 3 e53 doi:10.1371/journal.pcbi.0030053
17. DesvergneBWahliW 1999 Peroxisome Proliferator-Activated Receptors: Nuclear Control of Metabolism. Endocr Rev 20 649 688
18. WangY-XLeeC-HTiepSYuRTHamJ 2003 Peroxisome-Proliferator-Activated Receptor [delta] Activates Fat Metabolism to Prevent Obesity. Cell 113 159 170
19. CostetPLCMoreJEdgarAGaltierP 1998 Peroxisome proliferator-activated receptor alpha-isoform deficiency leads to progressive dyslipidemia with sexually dimorphic obesity and statosis. . J Biol Chem 273 29577 29585
20. SmythGK 2005 397 420 Limma: linear models for microarray data. Springer, NY
21. EdenENavonRSteinfeldILipsonDYakhiniZ 2009 GOrilla: a tool for discovery and visualization of enriched GO terms in ranked gene lists. BMC Bioinformatics 10 48
22. ArandaAPascualA 2001 Nuclear Hormone Receptors and Gene Expression. Physiological Reviews 81 1269 1304
23. TaubertSVan GilstMRHansenMYamamotoKR 2006 A Mediator subunit, MDT-15, integrates regulation of fatty acid metabolism by NHR-49-dependent and -independent pathways in C. elegans. Genes Dev 20 1137 1149
24. LiSArmstrongCMBertinNGeHMilsteinS 2004 A Map of the Interactome Network of the Metazoan C. elegans. Science 303 540 543
25. BrockTJBrowseJWattsJL 2006 Genetic regulation of unsaturated fatty acid composition in C. elegans. PLoS Genet 2 e108 doi:10.1371/journal.pgen.0020108
26. WattsJLBrowseJ 2000 A Palmitoyl-CoA-Specific Δ9 Fatty Acid Desaturase from Caenorhabditis elegans. Biochemical and Biophysical Research Communications 272 263 269
27. NtambiJM 1995 The regulation of stearoyl-CoA desaturase (SCD). Progress in Lipid Research 34 139 150
28. KniazevaMCrawfordQTSeiberMWangC-YHanM 2004 Monomethyl Branched-Chain Fatty Acids Play an Essential Role in Caeonorhabditis elegans Development. PLoS Biol 2 e257 doi:10.1371/journal.pbio.0020257
29. AshrafiKChangFYWattsJLFraserAGKamathRS 2003 Genome-wide RNAi analysis of Caenorhabditis elegans fat regulatory genes. Nature 421 268 272
30. McKayRMMcKayJPAveryLGraffJM 2003 C. elegans: A Model for Exploring the Genetics of Fat Storage. Developmental Cell 4 131 142
31. BogackaIXieHBrayGASmithSR 2005 Pioglitazone Induces Mitochondrial Biogenesis in Human Subcutaneous Adipose Tissue In Vivo. Diabetes 54 1392 1399
32. TanakaTYamamotoJIwasakiSAsabaHHamuraH 2003 Activation of peroxisome proliferator-activated receptor delta induces fatty acid beta-oxidation in skeletal muscle and attenuates metabolic syndrome. Proc Natl Acad Sci U S A 100 15924 15929
33. NaikiTNagakiMShidojiYKojimaHImoseM 2002 Analysis of Gene Expression Profile Induced by Hepatocyte Nuclear Factor 4α in Hepatoma Cells Using an Oligonucleotide Microarray. Journal of Biological Chemistry 277 14011 14019
34. LucasBGrigoKErdmannSLausenJKlein-HitpassL 2005 HNF4[alpha] reduces proliferation of kidney cells and affects genes deregulated in renal cell carcinoma. Oncogene 24 6418 6431
35. OdomDTZizlspergerNGordonDBBellGWRinaldiNJ 2004 Control of Pancreas and Liver Gene Expression by HNF Transcription Factors. Science 303 1378 1381
36. GuptaRKGaoNGorskiRKWhitePHardyOT 2007 Expansion of adult beta-cell mass in response to increased metabolic demand is dependent on HNF-4{alpha}. Genes & Development 21 756 769
37. Shmookler Reis1, 3RobertJXu2LuluLee2HoonyongChae2,*MinhoThaden2JohnJBharill1,3PuneetTazearslan3,†CagdasSiegel4EricAlla1RamaniZimniak1,5PiotrAyyadevara1,2Srinivas 2011 Modulation of lipid biosynthesis contributes to stress resistance and longevity of C. elegans mutants. Aging 3 2 125 147
38. UngerRHZhouYT 2001 Lipotoxicity of beta-cells in obesity and in other causes of fatty acid spillover. Diabetes 50 S118
39. ShimabukuroMHigaMZhouY-TWangM-YNewgardCB 1998 Lipoapoptosis in Beta-cells of Obese Prediabeticfa/fa Rats. Journal of Biological Chemistry 273 32487 32490
40. PeterAWeigertCStaigerHRittigKCeganA 2008 Induction of stearoyl-CoA desaturase protects human arterial endothelial cells against lipotoxicity. American Journal of Physiology - Endocrinology And Metabolism 295 E339 E349
41. ListenbergerLLHanXLewisSECasesSFareseRV 2003 Triglyceride accumulation protects against fatty acid-induced lipotoxicity. Proceedings of the National Academy of Sciences 100 3077 3082
42. Robinson-RechaviMMainaCVGissendannerCRLaudetVSluderA 2005 Explosive Lineage-Specific Expansion of the Orphan Nuclear Receptor HNF4 in Nematodes. Journal of Molecular Evolution 60 577 586
43. PuigserverPWuZParkCWGravesRWrightM 1998 A Cold-Inducible Coactivator of Nuclear Receptors Linked to Adaptive Thermogenesis. Cell 92 829 839
44. TaubertSWardJDYamamotoKR 2011 Nuclear hormone receptors in nematodes: Evolution and function. Molecular and Cellular Endocrinology 334 49 55
45. MangelsdorfDJEvansRM 1995 The RXR heterodimers and orphan receptors. Cell 83 841 850
46. BrennerS 1974 THE GENETICS OF CAENORHABDITIS ELEGANS. Genetics 77 71 94
47. SkaletskySRaHJ 2000 Primer3 on the www for general users and for biologist programmers. Methods Mol Biol 132 365 386
48. FazzioTGKooperbergCGoldmarkJPNealCBasomR 2001 Widespread Collaboration of Isw2 and Sin3-Rpd3 Chromatin Remodeling Complexes in Transcriptional Repression. Molecular and Cellular Biology 21 6450 6460
49. YangYHDudoitSLuuPLinDMPengV 2002 Normalization for cDNA microarray data: a robust composite method addressing single and multiple slide systematic variation. Nucleic Acids Research 30 e15
50. BenjamininiYaHY 1995 Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Statistical Soc Ser B-Methodological 57 289 300
51. SupekFBošnjakMŠkuncaNŠmucT 2011 REVIGO Summarizes and Visualizes Long Lists of Gene Ontology Terms. PLoS ONE 6 e21800 doi:10.1371/journal.pone.0021800
52. WattsJLBrowseJ 2002 Genetic dissection of polyunsaturated fatty acid synthesis in Caenorhabditis elegans. Proc Natl Acad Sci U S A 99 5854 5859
53. HansenMHsuA-LDillinAKenyonC 2005 New Genes Tied to Endocrine, Metabolic, and Dietary Regulation of Lifespan from a Caenorhabditis elegans Genomic RNAi Screen. PLoS Genet 1 e17 doi:10.1371/journal.pgen.0040017
54. GolejDLAskariBKramerFBarnhartSVivekanandan-GiriA 2011 Long-chain acyl-CoA synthetase 4 modulates prostaglandin E2 release from human arterial smooth muscle cells. Journal of Lipid Research 52 782 793
55. MullaneyBCBlindRDLemieuxGAPerezCLElleIC 2010 Regulation of C. elegans Fat Uptake and Storage by Acyl-CoA Synthase-3 Is Dependent on NR5A Family Nuclear Hormone Receptor nhr-25. Cell Metabolism 12 398 410
56. JiangGNepomucenoLHopkinsKSladekF 1995 Exclusive homodimerization of the orphan receptor hepatocyte nuclear factor 4 defines a new subclass of nuclear receptors. Mol Cell Biol 15 5131 5143
57. SchulmanIGShaoGHeymanRA 1998 Transactivation by Retinoid X Receptor-Peroxisome Proliferator-Activated Receptor gamma (PPARgamma ) Heterodimers: Intermolecular Synergy Requires Only the PPARgamma Hormone-Dependent Activation Function. Mol Cell Biol 18 3483 3494
58. Trzcinska-DanielewiczJIshikawaTMicialkiewiczAFronkJ 2008 Yeast transcription factor Oaf1 forms homodimer and induces some oleate-responsive genes in absence of Pip2. Biochemical and Biophysical Research Communications 374 763 766
59. RottensteinerHKalAJHamiltonBRuisHTabakHF 1997 A Heterodimer of the Zn2Cys6 Transcription Factors Pip2p and Oaf1p Controls Induction of Genes Encoding Peroxisomal Proteins in Saccharomyces Cerevisiae. European Journal of Biochemistry 247 776 783
Štítky
Genetika Reprodukční medicína
Článek A Genome-Wide Screen for Genetic Variants That Modify the Recruitment of REST to Its Target GenesČlánek Population Structure of Hispanics in the United States: The Multi-Ethnic Study of AtherosclerosisČlánek Differing Requirements for RAD51 and DMC1 in Meiotic Pairing of Centromeres and Chromosome Arms inČlánek Transcriptional Regulation of Rod Photoreceptor Homeostasis Revealed by NRL Targetome AnalysisČlánek Cell Contact–Dependent Outer Membrane Exchange in Myxobacteria: Genetic Determinants and MechanismČlánek Formation of Rigid, Non-Flight Forewings (Elytra) of a Beetle Requires Two Major Cuticular Proteins
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2012 Číslo 4- Akutní intermitentní porfyrie
- Souvislost haplotypu M2 genu pro annexin A5 s opakovanými reprodukčními ztrátami
- Příjem alkoholu a menstruační cyklus
- Doporučení pro diagnostiku a léčbu akutních jaterních porfyrií
- Doc. Miloš Kubánek: Nemocní se srdeční amyloidózou jsou často skryti a sledováni pod jinými diagnózami
-
Všechny články tohoto čísla
- Runs of Homozygosity Implicate Autozygosity as a Schizophrenia Risk Factor
- Modifier Genes and the Plasticity of Genetic Networks in Mice
- The DSIF Subunits Spt4 and Spt5 Have Distinct Roles at Various Phases of Immunoglobulin Class Switch Recombination
- A Genome-Wide Screen for Genetic Variants That Modify the Recruitment of REST to Its Target Genes
- Population Structure of Hispanics in the United States: The Multi-Ethnic Study of Atherosclerosis
- Deep Sequencing of Plant and Animal DNA Contained within Traditional Chinese Medicines Reveals Legality Issues and Health Safety Concerns
- Differing Requirements for RAD51 and DMC1 in Meiotic Pairing of Centromeres and Chromosome Arms in
- Insulin Signaling Mediates Sexual Attractiveness in
- Progressive Telomere Dysfunction Causes Cytokinesis Failure and Leads to the Accumulation of Polyploid Cells
- Long-Range Chromosome Organization in : A Site-Specific System Isolates the Ter Macrodomain
- Regulation of Budding Yeast Mating-Type Switching Donor Preference by the FHA Domain of Fkh1
- Polyglutamine Toxicity Is Controlled by Prion Composition and Gene Dosage in Yeast
- Patterns of Regulatory Variation in Diverse Human Populations
- Sequence-Specific Targeting of Dosage Compensation in Favors an Active Chromatin Context
- Whole-Exome Sequencing and Homozygosity Analysis Implicate Depolarization-Regulated Neuronal Genes in Autism
- Replication Fork Reversal after Replication–Transcription Collision
- Common Variants at 9p21 and 8q22 Are Associated with Increased Susceptibility to Optic Nerve Degeneration in Glaucoma
- Coordinate Regulation of Lipid Metabolism by Novel Nuclear Receptor Partnerships
- Epigenome-Wide Scans Identify Differentially Methylated Regions for Age and Age-Related Phenotypes in a Healthy Ageing Population
- A Coordinated Interdependent Protein Circuitry Stabilizes the Kinetochore Ensemble to Protect CENP-A in the Human Pathogenic Yeast
- Budding Yeast Dma Proteins Control Septin Dynamics and the Spindle Position Checkpoint by Promoting the Recruitment of the Elm1 Kinase to the Bud Neck
- , a Homolog of a Deaf-Blindness Gene, Regulates Circadian Output and Slowpoke Channels
- Transcriptional Regulation of Rod Photoreceptor Homeostasis Revealed by NRL Targetome Analysis
- Cell Contact–Dependent Outer Membrane Exchange in Myxobacteria: Genetic Determinants and Mechanism
- Defective Membrane Remodeling in Neuromuscular Diseases: Insights from Animal Models
- Formation of Rigid, Non-Flight Forewings (Elytra) of a Beetle Requires Two Major Cuticular Proteins
- SPE-44 Implements Sperm Cell Fate
- A Shared Role for RBF1 and dCAP-D3 in the Regulation of Transcription with Consequences for Innate Immunity
- A Companion Cell–Dominant and Developmentally Regulated H3K4 Demethylase Controls Flowering Time in via the Repression of Expression
- The HEN1 Ortholog, HENN-1, Methylates and Stabilizes Select Subclasses of Germline Small RNAs
- Improved Statistics for Genome-Wide Interaction Analysis
- The Probability of a Gene Tree Topology within a Phylogenetic Network with Applications to Hybridization Detection
- Context-Dependent Dual Role of SKI8 Homologs in mRNA Synthesis and Turnover
- Mu Insertions Are Repaired by the Double-Strand Break Repair Pathway of
- Competition between Replicative and Translesion Polymerases during Homologous Recombination Repair in Drosophila
- An Unbiased Assessment of the Role of Imprinted Genes in an Intergenerational Model of Developmental Programming
- Type 2 Diabetes Risk Alleles Demonstrate Extreme Directional Differentiation among Human Populations, Compared to Other Diseases
- Mutations in and Cause “Splashed White” and Other White Spotting Phenotypes in Horses
- Fine-Scale Mapping of Natural Variation in Fly Fecundity Identifies Neuronal Domain of Expression and Function of an Aquaporin
- Dynamics of Brassinosteroid Response Modulated by Negative Regulator LIC in Rice
- Genetic Inhibition of Solute-Linked Carrier 39 Family Transporter 1 Ameliorates Aβ Pathology in a Model of Alzheimer's Disease
- The Functions of Mediator in Support a Role in Shaping Species-Specific Gene Expression
- Patterns of Ancestry, Signatures of Natural Selection, and Genetic Association with Stature in Western African Pygmies
- Dissection of Pol II Trigger Loop Function and Pol II Activity–Dependent Control of Start Site Selection
- PIWI Associated siRNAs and piRNAs Specifically Require the HEN1 Ortholog
- Genome-Wide Patterns of Gene Expression in Nature
- Hypoxia Disruption of Vertebrate CNS Pathfinding through EphrinB2 Is Rescued by Magnesium
- A New Role for Translation Initiation Factor 2 in Maintaining Genome Integrity
- Sex Reversal in C57BL/6J XY Mice Caused by Increased Expression of Ovarian Genes and Insufficient Activation of the Testis Determining Pathway
- The Rac GTP Exchange Factor TIAM-1 Acts with CDC-42 and the Guidance Receptor UNC-40/DCC in Neuronal Protrusion and Axon Guidance
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- A Coordinated Interdependent Protein Circuitry Stabilizes the Kinetochore Ensemble to Protect CENP-A in the Human Pathogenic Yeast
- Coordinate Regulation of Lipid Metabolism by Novel Nuclear Receptor Partnerships
- Defective Membrane Remodeling in Neuromuscular Diseases: Insights from Animal Models
- Formation of Rigid, Non-Flight Forewings (Elytra) of a Beetle Requires Two Major Cuticular Proteins
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



