-
Články
- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Translational Regulation of the Post-Translational Circadian Mechanism
article has not abstract
Published in the journal: . PLoS Genet 10(9): e32767. doi:10.1371/journal.pgen.1004628
Category: Perspective
doi: https://doi.org/10.1371/journal.pgen.1004628Summary
article has not abstract
Much of our behavior and physiology exhibits daily oscillations driven by a circadian rhythm. While the phase of these oscillations is typically set by the daily light–dark cycle, the oscillations themselves are actually produced by an internal circadian clock. A summary of the Drosophila circadian clock mechanism, which exhibits evolutionary conservation with the human one, is outlined in Figure 1. Daily oscillations of the transcription factor period protein (PER) lead to elevated nuclear PER levels in the late night and early morning, when PER binds to a positively acting CLOCK/CYCLE (CLK/CYC) transcription factor to repress transcription of genes with a CLK/CYC-responsive promoter. One of these genes is the per gene, so PER regulates its own transcription in a transcriptional negative feedback loop (colored red in Figure 1). Delays in the negative feedback loop allow per mRNA to accumulate to its daily peak and PER protein to persist as a repressor even as its mRNA levels fall. The delays are thought to arise principally from a post-translational feedback loop (colored blue in Figure 1) in which PER is phosphorylated by the casein kinase I ortholog doubletime (DBT), resulting in PER degradation throughout the daytime. During the night, PER accumulates because it is no longer degraded in response to light signals (transduced by the CRY photoreceptor and the TIM protein) and represses CLK/CYC-dependent transcription, including that of per, tim, and many genes leading to the physiological consequences (outputs) of the clock [1]. Now, in this issue of PLOS Genetics, Yanmei Huang and coauthors demonstrate translational regulation that in turn regulates the post-translational regulatory loop of the Drosophila circadian clock (green loop in Figure 1) [2].
Fig. 1. The molecular mechanism for the Drosophila circadian clock. 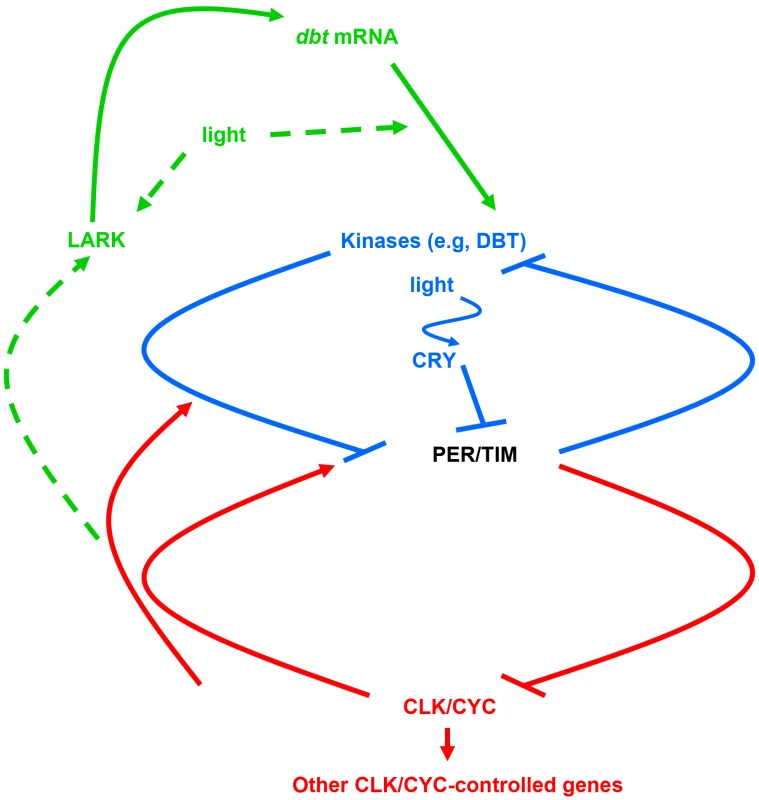
This mechanism has been proposed to consist of a transcriptional feedback loop (red) and a post-translational feedback loop (blue). This paper adds a translational regulatory loop (green) in which the LARK protein binds to dbt mRNAs to promote translation of DBT, the principal circadian kinase; tentative pathways are marked with dashed arrows. Some earlier work did implicate a role for translational regulation in clock-related processes. For instance, the circadian bioluminescence rhythm in Gonyaulax (a single-celled dinoflagellate) has been shown to arise from circadian control of luciferin-binding protein translation [3]. In Neurospora (a bread mold), differential translation initiation at two different AUG codons of the circadian protein FRQ extends the effective temperature range for circadian rhythmicity [4]. But there was little evidence that translational control was necessary to support the underlying oscillator mechanism.
However, recently regulation of protein translation has been increasingly moved to a place in the oscillator mechanism itself, as has been reviewed extensively elsewhere [1] and is briefly summarized here. Work in Drosophila has identified per RNA-binding proteins that stimulate PER translation, demonstrating a role for translational regulation in the transcriptional feedback loop [5], [6]. Work in a number of systems has shown that regulation of translation through circadian changes in polyA tail length [7], Tor signaling [8], [9], ribosome biogenesis [8], and miRNA [1] contributes to the daily oscillations of many proteins whose mRNAs do not oscillate [10], thereby demonstrating a significant role for translational regulation in circadian output pathways [1].
The current work of Huang and coauthors [2] builds on their previous findings that the circadian RNA-binding and translational regulator LARK binds to RNA encoding the circadian kinase DBT [11] and that circadian changes in translation are common for many mRNAs [12]. In the current study, the authors show that LARK binds to each of the four alternatively spliced dbt transcripts. By recovering transcripts that co-immunoprecipitate with a tagged ribosomal protein expressed specifically in circadian neurons, it is shown that LARK promotes the translation of these dbt mRNAs, because lower levels are associated with ribosomes in the absence of LARK and higher levels with LARK overexpression. Moreover, translation of one of these transcripts undergoes circadian changes in constant darkness (but curiously, not in light–dark cycles), while the translation of another transcript is light inducible with a requirement for LARK (i.e., its translation is not light induced with lark knock-down, and the induction by light is increased with LARK overexpression.). Since DBT is involved in setting the circadian period, altered translation of DBT would be predicted to alter circadian period, and in fact, altered LARK abundance does produce changes in circadian period. Knock-down of LARK in the brain neurons that produce rhythms of behavior in constant darkness shortens circadian period, while overexpression of LARK in these neurons lengthens the period. These changes are modified by changes in the dbt genotype. For instance, they are not produced in the presence of catalytically inactive DBT, suggesting that the period-altering effects of LARK are mediated through DBT and require DBT activity. Increased expression of LARK delays PER degradation at the beginning of the day in behaviorally relevant brain neurons, suggesting that it reduces DBT-dependent degradation at these times. This might seem counterintuitive, since translation of DBT (which targets PER for degradation) is increased with LARK overexpression. But immunoblot analysis shows that the increased DBT from LARK overexpression comes in a number of atypical molecular weight ranges that include a lower molecular weight form unlikely to retain catalytic activity, as well as some very high molecular weight forms. The authors hypothesize that the lower molecular weight form may interact with full-length DBT to regulate circadian period, so that the ratio of the two determines period. One way this could happen would be if the short isoform and the full-length DBT isoform associate to promote preferential phosphorylation of a PER domain that has previously been shown to slow the pace of the clock rather than accelerate it [13], [14].
The authors present a very creative and provocative model that makes a number of predictions. The short DBT isoform is proposed as a regulator of DBT activity, so it will be important to determine if it is expressed and exhibits circadian oscillations under normal conditions (i.e., without LARK or DBT overexpression, which presumably raises its levels above the current lower limit of detection). Moreover, what exactly are its sequence and role, does it associate with full-length DBT as proposed to target a specific domain in PER and does it interact with another recently described regulator of DBT [15]?
The findings also raise additional questions. Both the levels of the small DBT isoform detected with LARK overexpression and the translation of the light-inducible mRNA exhibit a diurnal oscillation that peaks during the day, and the basis for this oscillation could be the previously demonstrated LARK oscillation, since LARK is also higher during the day than at night. But how does the circadian clock (and/or light) control these oscillations (dashed arrow to LARK in Figure 1)? Moreover, it is not yet known if LARK is acutely induced by light to mediate the effects of light on translation, or alternatively if light induces translation through a LARK-independent pathway, with LARK increasing the magnitude of the effect (dashed arrows from light in Figure 1). It is also not known whether the circadian and diurnal changes in translation occur throughout the circadian system or only in specific circadian cells. Finally, does a similar translational mechanism exist for the mammalian circadian clock, in which mammalian DBT orthologs subserve a similar role [16] and in which a LARK ortholog has also been implicated in circadian translation of PER [17]? If so, the work has revealed an evolutionarily conserved translational mechanism for the regulation of the post-translational loop of the circadian clock.
Zdroje
1. LimC, AlladaR (2013) Emerging roles for post-transcriptional regulation in circadian clocks. Nat Neurosci 16 : 1544–1550.
2. HuangY, McNeilGP, JacksonFR (2014) Translational Regulation of the Doubletime/CKIdelta/epsilon Kinase by LARK Contributes to Circadian Period Modulation. PLoS Genet 10: e1004536 doi: 10.1371/journal.pgen.1004536
3. MorseD, MilosPM, RouxE, HastingsJW (1989) Circadian regulation of bioluminescence in Gonyaulax involves translational control. Proc Natl Acad Sci U S A 86 : 172–176.
4. LiuY, GarceauNY, LorosJJ, DunlapJC (1997) Thermally regulated translational control of FRQ mediates aspects of temperature responses in the Neurospora circadian clock. Cell 89 : 477–486.
5. LimC, AlladaR (2013) ATAXIN-2 activates PERIOD translation to sustain circadian rhythms in Drosophila. Science 340 : 875–879.
6. ZhangY, LingJ, YuanC, DubruilleR, EmeryP (2013) A role for Drosophila ATX2 in activation of PER translation and circadian behavior. Science 340 : 879–882.
7. KojimaS, Sher-ChenEL, GreenCB (2012) Circadian control of mRNA polyadenylation dynamics regulates rhythmic protein expression. Genes Dev 26 : 2724–2736.
8. JouffeC, CretenetG, SymulL, MartinE, AtgerF, et al. (2013) The circadian clock coordinates ribosome biogenesis. PLoS Biol 11: e1001455.
9. CaoR, AndersonFE, JungYJ, DziemaH, ObrietanK (2011) Circadian regulation of mammalian target of rapamycin signaling in the mouse suprachiasmatic nucleus. Neuroscience 181 : 79–88.
10. ReddyAB, KarpNA, MaywoodES, SageEA, DeeryM, et al. (2006) Circadian orchestration of the hepatic proteome. Curr Biol 16 : 1107–1115.
11. HuangY, GenovaG, RobertsM, JacksonFR (2007) The LARK RNA-binding protein selectively regulates the circadian eclosion rhythm by controlling E74 protein expression. PLoS ONE 2: e1107.
12. HuangY, AinsleyJA, ReijmersLG, JacksonFR (2013) Translational profiling of clock cells reveals circadianly synchronized protein synthesis. PLoS Biol 11: e1001703.
13. ChiuJC, KoHW, EderyI (2011) NEMO/NLK phosphorylates PERIOD to initiate a time-delay phosphorylation circuit that sets circadian clock speed. Cell 145 : 357–370.
14. KivimaeS, SaezL, YoungMW (2008) Activating PER repressor through a DBT-directed phosphorylation switch. PLoS Biol 6: e183.
15. FanJY, AgyekumB, VenkatesanA, HallDR, KeightleyA, et al. (2013) Noncanonical FK506-binding protein BDBT binds DBT to enhance its circadian function and forms foci at night. Neuron 80 : 984–996.
16. LowreyPL, ShimomuraK, AntochMP, YamazakiS, ZemenidesPD, et al. (2000) Positional syntenic cloning and functional characterization of the mammalian circadian mutation tau. Science 288 : 483–492.
17. KojimaS, MatsumotoK, HiroseM, ShimadaM, NaganoM, et al. (2007) LARK activates posttranscriptional expression of an essential mammalian clock protein, PERIOD1. Proc Natl Acad Sci U S A 104 : 1859–1864.
Štítky
Genetika Reprodukční medicína
Článek An Evolutionarily Conserved Role for the Aryl Hydrocarbon Receptor in the Regulation of MovementČlánek Requirement for Drosophila SNMP1 for Rapid Activation and Termination of Pheromone-Induced ActivityČlánek Co-regulated Transcripts Associated to Cooperating eSNPs Define Bi-fan Motifs in Human Gene NetworksČlánek Identification of a Regulatory Variant That Binds FOXA1 and FOXA2 at the Type 2 Diabetes GWAS LocusČlánek tRNA Modifying Enzymes, NSUN2 and METTL1, Determine Sensitivity to 5-Fluorouracil in HeLa CellsČlánek Derlin-1 Regulates Mutant VCP-Linked Pathogenesis and Endoplasmic Reticulum Stress-Induced ApoptosisČlánek A Genetic Assay for Transcription Errors Reveals Multilayer Control of RNA Polymerase II FidelityČlánek The Proprotein Convertase KPC-1/Furin Controls Branching and Self-avoidance of Sensory Dendrites inČlánek Regulation of p53 and Rb Links the Alternative NF-κB Pathway to EZH2 Expression and Cell SenescenceČlánek BMPs Regulate Gene Expression in the Dorsal Neuroectoderm of and Vertebrates by Distinct MechanismsČlánek Unkempt Is Negatively Regulated by mTOR and Uncouples Neuronal Differentiation from Growth Control
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2014 Číslo 9- Akutní intermitentní porfyrie
- Farmakogenetické testování pomáhá předcházet nežádoucím efektům léčiv
- Růst a vývoj dětí narozených pomocí IVF
- Pilotní studie: stres a úzkost v průběhu IVF cyklu
- Intrauterinní inseminace a její úspěšnost
-
Všechny články tohoto čísla
- Translational Regulation of the Post-Translational Circadian Mechanism
- An Evolutionarily Conserved Role for the Aryl Hydrocarbon Receptor in the Regulation of Movement
- Eliminating Both Canonical and Short-Patch Mismatch Repair in Suggests a New Meiotic Recombination Model
- Requirement for Drosophila SNMP1 for Rapid Activation and Termination of Pheromone-Induced Activity
- Co-regulated Transcripts Associated to Cooperating eSNPs Define Bi-fan Motifs in Human Gene Networks
- Targeted H3R26 Deimination Specifically Facilitates Estrogen Receptor Binding by Modifying Nucleosome Structure
- Role for Circadian Clock Genes in Seasonal Timing: Testing the Bünning Hypothesis
- The Tandem Repeats Enabling Reversible Switching between the Two Phases of β-Lactamase Substrate Spectrum
- The Association of the Vanin-1 N131S Variant with Blood Pressure Is Mediated by Endoplasmic Reticulum-Associated Degradation and Loss of Function
- Identification of a Regulatory Variant That Binds FOXA1 and FOXA2 at the Type 2 Diabetes GWAS Locus
- Regulation of Flowering by the Histone Mark Readers MRG1/2 via Interaction with CONSTANS to Modulate Expression
- The Actomyosin Machinery Is Required for Retinal Lumen Formation
- Plays a Conserved Role in Assembly of the Ciliary Motile Apparatus
- Hidden Diversity in Honey Bee Gut Symbionts Detected by Single-Cell Genomics
- Ribosome Rescue and Translation Termination at Non-Standard Stop Codons by ICT1 in Mammalian Mitochondria
- tRNA Modifying Enzymes, NSUN2 and METTL1, Determine Sensitivity to 5-Fluorouracil in HeLa Cells
- Causal Variation in Yeast Sporulation Tends to Reside in a Pathway Bottleneck
- Tissue-Specific RNA Expression Marks Distant-Acting Developmental Enhancers
- WC-1 Recruits SWI/SNF to Remodel and Initiate a Circadian Cycle
- Clonal Expansion of Early to Mid-Life Mitochondrial DNA Point Mutations Drives Mitochondrial Dysfunction during Human Ageing
- Methylation QTLs Are Associated with Coordinated Changes in Transcription Factor Binding, Histone Modifications, and Gene Expression Levels
- Differential Management of the Replication Terminus Regions of the Two Chromosomes during Cell Division
- Obesity-Linked Homologues and Establish Meal Frequency in
- Derlin-1 Regulates Mutant VCP-Linked Pathogenesis and Endoplasmic Reticulum Stress-Induced Apoptosis
- Stress-Induced Nuclear RNA Degradation Pathways Regulate Yeast Bromodomain Factor 2 to Promote Cell Survival
- The MAPK p38c Regulates Oxidative Stress and Lipid Homeostasis in the Intestine
- Widespread Genome Reorganization of an Obligate Virus Mutualist
- Trans-kingdom Cross-Talk: Small RNAs on the Move
- The Vip1 Inositol Polyphosphate Kinase Family Regulates Polarized Growth and Modulates the Microtubule Cytoskeleton in Fungi
- Myosin Vb Mediated Plasma Membrane Homeostasis Regulates Peridermal Cell Size and Maintains Tissue Homeostasis in the Zebrafish Epidermis
- GLD-4-Mediated Translational Activation Regulates the Size of the Proliferative Germ Cell Pool in the Adult Germ Line
- Genome Wide Association Studies Using a New Nonparametric Model Reveal the Genetic Architecture of 17 Agronomic Traits in an Enlarged Maize Association Panel
- Translational Regulation of the DOUBLETIME/CKIδ/ε Kinase by LARK Contributes to Circadian Period Modulation
- Positive Selection and Multiple Losses of the LINE-1-Derived Gene in Mammals Suggest a Dual Role in Genome Defense and Pluripotency
- Out of Balance: R-loops in Human Disease
- A Genetic Assay for Transcription Errors Reveals Multilayer Control of RNA Polymerase II Fidelity
- Altered Behavioral Performance and Live Imaging of Circuit-Specific Neural Deficiencies in a Zebrafish Model for Psychomotor Retardation
- Nipbl and Mediator Cooperatively Regulate Gene Expression to Control Limb Development
- Meta-analysis of Mutations in Autism Spectrum Disorders: A Gradient of Severity in Cognitive Impairments
- The Proprotein Convertase KPC-1/Furin Controls Branching and Self-avoidance of Sensory Dendrites in
- Hydroxymethylated Cytosines Are Associated with Elevated C to G Transversion Rates
- Memory and Fitness Optimization of Bacteria under Fluctuating Environments
- Regulation of p53 and Rb Links the Alternative NF-κB Pathway to EZH2 Expression and Cell Senescence
- Interspecific Tests of Allelism Reveal the Evolutionary Timing and Pattern of Accumulation of Reproductive Isolation Mutations
- PRO40 Is a Scaffold Protein of the Cell Wall Integrity Pathway, Linking the MAP Kinase Module to the Upstream Activator Protein Kinase C
- Low Levels of p53 Protein and Chromatin Silencing of p53 Target Genes Repress Apoptosis in Endocycling Cells
- SPDEF Inhibits Prostate Carcinogenesis by Disrupting a Positive Feedback Loop in Regulation of the Foxm1 Oncogene
- RRP6L1 and RRP6L2 Function in Silencing Regulation of Antisense RNA Synthesis
- BMPs Regulate Gene Expression in the Dorsal Neuroectoderm of and Vertebrates by Distinct Mechanisms
- Unkempt Is Negatively Regulated by mTOR and Uncouples Neuronal Differentiation from Growth Control
- Atkinesin-13A Modulates Cell-Wall Synthesis and Cell Expansion in via the THESEUS1 Pathway
- Dopamine Signaling Leads to Loss of Polycomb Repression and Aberrant Gene Activation in Experimental Parkinsonism
- Histone Methyltransferase MMSET/NSD2 Alters EZH2 Binding and Reprograms the Myeloma Epigenome through Global and Focal Changes in H3K36 and H3K27 Methylation
- Bipartite Recognition of DNA by TCF/Pangolin Is Remarkably Flexible and Contributes to Transcriptional Responsiveness and Tissue Specificity of Wingless Signaling
- The Olfactory Transcriptomes of Mice
- Muscular Dystrophy-Associated and Variants Disrupt Nuclear-Cytoskeletal Connections and Myonuclear Organization
- Interplay of dFOXO and Two ETS-Family Transcription Factors Determines Lifespan in
- Evidence for Widespread Positive and Negative Selection in Coding and Conserved Noncoding Regions of
- Genome-Wide Association Meta-analysis of Neuropathologic Features of Alzheimer's Disease and Related Dementias
- Rejuvenation of Meiotic Cohesion in Oocytes during Prophase I Is Required for Chiasma Maintenance and Accurate Chromosome Segregation
- Admixture in Latin America: Geographic Structure, Phenotypic Diversity and Self-Perception of Ancestry Based on 7,342 Individuals
- Local Effect of Enhancer of Zeste-Like Reveals Cooperation of Epigenetic and -Acting Determinants for Zygotic Genome Rearrangements
- Differential Responses to Wnt and PCP Disruption Predict Expression and Developmental Function of Conserved and Novel Genes in a Cnidarian
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Admixture in Latin America: Geographic Structure, Phenotypic Diversity and Self-Perception of Ancestry Based on 7,342 Individuals
- Nipbl and Mediator Cooperatively Regulate Gene Expression to Control Limb Development
- Genome Wide Association Studies Using a New Nonparametric Model Reveal the Genetic Architecture of 17 Agronomic Traits in an Enlarged Maize Association Panel
- Histone Methyltransferase MMSET/NSD2 Alters EZH2 Binding and Reprograms the Myeloma Epigenome through Global and Focal Changes in H3K36 and H3K27 Methylation
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



