-
Články
- Vzdělávání
- Časopisy
Top články
- Témata
- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Epstein-Barr Virus Down-Regulates Tumor Suppressor Expression
Many oncogenic viruses exhibit cellular transforming properties, often involving oncogenes activation and tumor suppressor genes inactivation. The DOK1 gene is a newly identified tumor suppressor gene with altered expression via hypermethylation of its promoter in a variety of human cancers, including head and neck, lung, gastric and others. In addition, a correlation has been reported between DOK1 aberrant hypermethylation and the presence of oncogenic viruses such as hepatitis B virus (HBV) in hepatocellular carcinoma (HCC) and Epstein-Barr virus (EBV) in Burkitt's lymphoma-derived cell lines. Here we demonstrate for the first time that EBV is directly involved in the inhibition of DOK1 expression in B-cells. We show that EBV leads to epigenetic repression of DOK1 through increased DNA methylation of its promoter and H3K27 tri-methylation. The LMP1 oncoprotein plays a key role in the repression of DOK1 expression. It promotes the formation and the recruitment to the DOK1 promoter of transcriptionally inhibitory complexes composed of E2F1/pRB/DNMT1 and of EZH2 which is part of the polycomb repressive complex 2. Interestingly, one or more additional EBV protein(s) cooperate(s) with LMP1 in inducing massive DNA methylation at the DOK1 promoter, leading to the loss of E2F1 complexes recruitment and even stronger repression of DOK1 expression.
Published in the journal: . PLoS Pathog 10(5): e32767. doi:10.1371/journal.ppat.1004125
Category: Research Article
doi: https://doi.org/10.1371/journal.ppat.1004125Summary
Many oncogenic viruses exhibit cellular transforming properties, often involving oncogenes activation and tumor suppressor genes inactivation. The DOK1 gene is a newly identified tumor suppressor gene with altered expression via hypermethylation of its promoter in a variety of human cancers, including head and neck, lung, gastric and others. In addition, a correlation has been reported between DOK1 aberrant hypermethylation and the presence of oncogenic viruses such as hepatitis B virus (HBV) in hepatocellular carcinoma (HCC) and Epstein-Barr virus (EBV) in Burkitt's lymphoma-derived cell lines. Here we demonstrate for the first time that EBV is directly involved in the inhibition of DOK1 expression in B-cells. We show that EBV leads to epigenetic repression of DOK1 through increased DNA methylation of its promoter and H3K27 tri-methylation. The LMP1 oncoprotein plays a key role in the repression of DOK1 expression. It promotes the formation and the recruitment to the DOK1 promoter of transcriptionally inhibitory complexes composed of E2F1/pRB/DNMT1 and of EZH2 which is part of the polycomb repressive complex 2. Interestingly, one or more additional EBV protein(s) cooperate(s) with LMP1 in inducing massive DNA methylation at the DOK1 promoter, leading to the loss of E2F1 complexes recruitment and even stronger repression of DOK1 expression.
Introduction
Cellular transformation induced by oncogenic viruses often involves the activation of growth-promoting signaling pathways and the inactivation of tumor suppressor genes. The downstream of tyrosine kinase 1gene (DOK1) has emerged as a newly identified tumor suppressor gene that encodes a multi-domain adapter protein and acts as a negative regulator of signaling pathways involved in several cellular functions. DOK1 inhibits cell proliferation, down regulates MAP kinase activity, and has an opposing role in leukemogenesis and promotes cell spreading, motility, and apoptosis [1], [2]. Functional studies showed that mice lacking the DOK1 and/or DOK2 genes have a high susceptibility to the development of lung adenocarcinomas [3] and exhibit significant defects in their immune responses and immune cell development, often developing myelo-proliferative and autoimmune diseases, e.g. lupus-like renal disease [4], [5]. The DOK1 gene locus is located in the human chromosome 2p13 region, which is frequently rearranged in a number of human tumors [6]. Oncogenic tyrosine kinases such as p210BCR-ABL, the causative mutation in chronic myelogenous leukemia (CML), and Src target DOK1 for ubiquitin-mediated proteasomal degradation [7], therefore promoting cell proliferation. We have reported a frameshift mutation of the DOK1 gene in chronic lymphoid leukemia (CLL) resulting in the expression of truncated DOK1 that is exclusively localized in the nucleus and loses its tumor suppressive activities, in contrast with the cytoplasmic wild type protein [8]. We also showed that DOK1 gene expression is repressed in a large proportion of head and neck cancer (HNC), lung cancer and Burkitt's lymphoma [9], as a result of aberrant hypermethylation of its promoter region. The inactivation of DOK1 through promoter methylation also occurred in liver and gastric cancers [10], [11]. Thus, DOK1 emerged as a tumor suppressor frequently altered in a variety of human cancers, making it a potential marker and therapeutic target in cancer control.
Epstein-Barr virus (EBV) is a γ-herpes-virus that is widespread in 90% of human populations. In the majority of individuals, EBV persists as a permanent, asymptomatic infection of the lymphocytes B-lymphocyte pool [12]. EBV occasionally causes infectious mononucleosis in adolescents [13] and is considered a human carcinogenic infectious agent. Indeed, EBV is associated with the development of different types of B-cell lymphoma such as Burkitt's lymphoma (BL), Hodgkin disease, lympho-proliferative disorders in immuno-deficient individuals, and nasopharyngeal carcinoma [14], [15], [16]. EBV is also associated with gastric cancer [17]. The oncogenic potential of EBV has been further demonstrated by its ability to immortalize efficiently the primary human B-cells in vitro in lymphoblastoid cell lines (LCLs) [18]. LCLs carry the EBV genome in an extra-chromosomal episome state and express nine latent viral proteins: three trans-membrane proteins (LMP1, LMP2A and 2B) and six nuclear antigens (EBNAs 1, 2, 3A, 3B, 3C and LP), along with other non-translated RNA products [12]. These viral products enhance the proliferation of quiescent B-cells and maintain the viral genome in its episomal form. However, only EBNA1, 2, 3A, 3C, LP, and LMP1 are essential for the transformation of primary B-cells into LCLs [19]. The latent membrane protein 1 (LMP1) is crucial for EBV-induced B-cell immortalization. It is the only EBV latent protein that displays transforming properties in vitro [20].
LMP1 protein is thought to alter cell growth transformation by mimicking the activated forms of tumor necrosis factor receptor (TNFR), CD40 and CD30 receptors [21], [22], [23]. Through its long C-terminal cytosolic domain, LMP1 has the ability to induce several signaling pathways, including the MAP kinase (both ERK/MAPK and p38/MAPK), nuclear factor kappa B (NF-κB) and c-Jun N-terminal kinase (JNK) [24], [25], [26], [27]. The alteration of these signaling pathways by LMP1 is essential for the oncogenicity of EBV.
The presence of the EBV genome in several lymphomas, and its ability to induce B-cell immortalization, and alter host-cell expression profiles and epigenome (i.e. DNA methylation patterns) strongly support an etiological role for EBV in these cancers. We recently reported that the expression of DOK1 gene is repressed through DNA hypermethylation in BL cell lines, it became of interest to investigate the possible role of EBV in the inhibition of DOK1 expression in infected B-cells. To date, very little is known about the regulation of DOK1 expression by oncogenic viruses.
In the present study, we demonstrate a strong association between EBV infection and DOK1 gene silencing via hypermethylation of its promoter in EBV-infected cell lines. We show that EBV infection in B-cells leads to epigenetic repression and CpG methylation of the DOK1 gene and that LMP1 expression inhibits DOK1 promoter activity via the recruitment of inhibitory complexes including E2F1, pRB, DNMT1 and EZH2.
Results
EBV infection of primary human B-cells in vitro leads to down-regulation of DOK1 expression
Based on our previous results that showed the down-regulation of DOK1 expression in BL cell-lines [9], we evaluate whether this event was linked to infection with EBV, a key risk factor for this malignancy. Primary human B-cells, isolated from different healthy donors, were infected in independent experiments with recombinant EBV virus expressing the green fluorescent protein (GFP-EBV). The infection efficiency was evaluated by flow cytometry to monitor GFP expression (data not shown). The expression of EBV genes EBNA1 and LMP1, as well as DOK1 was determined by real-time PCR and western blot at different time points post-infection (Figure 1A and B). EBV infection resulted in a strong reduction of DOK1 mRNA and protein levels, which was evident at 16 hours post-infection (Figure 1A). Similarly, DOK1 mRNA and protein levels were strongly down-regulated by EBV in three cancers B-cell lines (RPMI, BJAB and Louckes) infected by EBV, as well as in EBV-immortalized lymphoblastoid cells lines (LCLs) (Figure 1C and D). Together, these findings highlight a role for EBV in down-regulating DOK1 gene expression.
Fig. 1. EBV infection in vitro inhibits DOK1 gene expression. 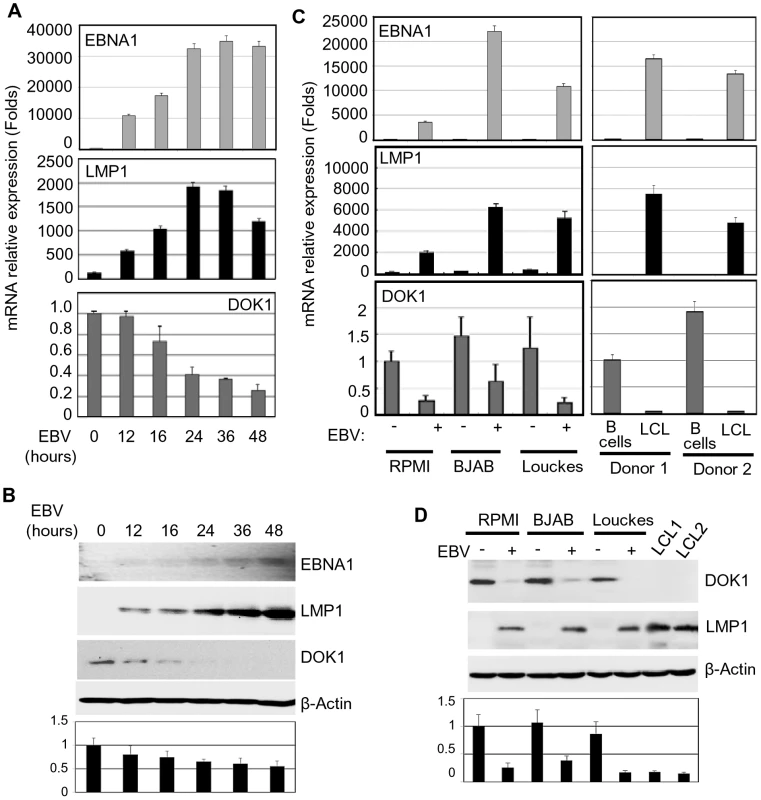
Primary B-cells were isolated from healthy donor blood using negative selection, and then infected with GFP-EBV recombinant virus. (A) mRNA levels of EBNA1, LMP1, and DOK1 were measured using real time PCR at different time points 12, 16, 24, 36, and 48 hours post infection and normalized to GAPDH expression. The isolated primary B-cells were used as a control (time point 0). Data are average of three independent experiments. (B) DOK1, LMP1, EBNA1, and β-actin protein levels were determined by western blotting. (C) Total mRNA was extracted from RPMI, BJAB and Louckes cells two weeks after infection with GFP-EBV recombinant virus. The respective non-infected cells were used as control. Similarly, mRNA was extracted from two LCLs and their original primary B-cells. The expression levels of EBNA1, LMP1, and DOK1 were measured by real time PCR and normalized to GAPDH expression. (D) DOK1, LMP1 and β-actin protein levels were determined by western blotting. DOK1 protein levels were quantified from two independent immunoblots and normalized to the corresponding β-actin level (bottom of B and D). LMP1 plays a key role in the inhibition of DOK1 expression
The EBV oncoprotein LMP1 is essential for EBV-induced B-cell immortalization by altering cellular gene expression via the activation of several signaling pathways [28]. To determine whether LMP1 can affect the expression of DOK1, we infected the RPMI cells with wild-type GFP-EBV or GFP-EBV lacking the LMP1 gene (EBVΔLMP1). The infection efficiency was monitored using flow cytometry for GFP expression (Figure 2A). In contrast to wild-type GFP-EBV, EBVΔLMP1 infection in primary B cells and in RPMI cells did not significantly decrease DOK1 mRNA or protein levels (Figure 2B and C). Re-expression of LMP1 in EBVΔLMP1 RPMI cells by retroviral transduction restored the ability of EBV to down-regulate DOK1 expression, while transduction of the same cells with empty retrovirus (pLXSN) did not affect DOK1 mRNA or protein levels (Figure 2D and E), highlighting the key role of LMP1 in this event. Accordingly, expression of LMP1 alone in RPMI cells was sufficient to reduce DOK1 mRNA and protein expression (Figure 2D and E), whereas expression of other viral proteins, such as EBNA1, 2, 3A, 3B, and 3C, did not lead to down-regulation of DOK1 protein levels (supplementary Figure S1A–C) In addition, transient transfection of RPMI with increasing concentrations of LMP1 expressing vector resulted in the decrease of DOK1 expression is a dose dependent manner (Figure 2F and G). Together, these data underline the key role of LMP1 in EBV-mediated DOK1 down-regulation in infected B-cells.
Fig. 2. LMP1 plays a key role in EBV-mediated DOK1 silencing. 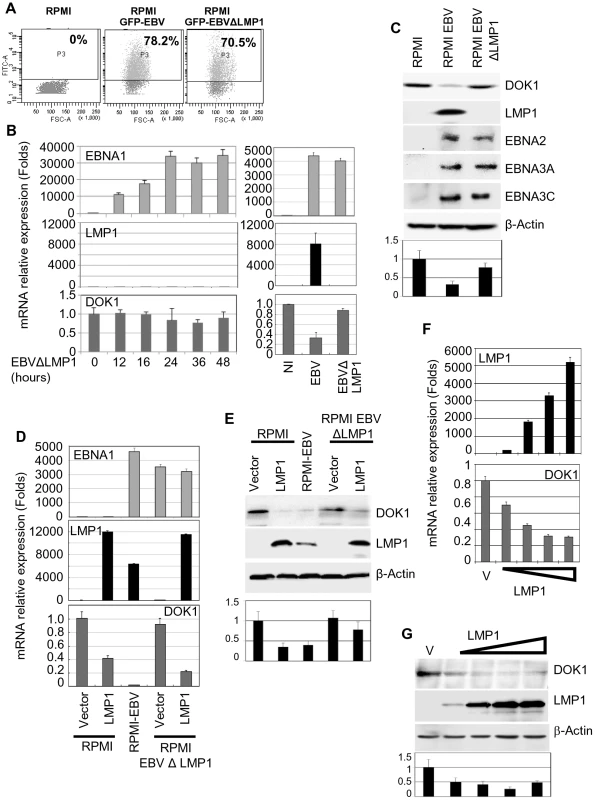
RPMI cells were infected with GFP recombinant EBV wild type (GFP-EBV) or lacking LMP1 (EBVΔLMP1). (A) The infection was monitored using flow cytometry for GFP expression. (B and C) mRNA levels of EBNA1, LMP1, GAPDH and DOK1 in these cells were determined using real time PCR and the indicated proteins expression were analyzed using western blotting. Both RPMI and RPMI-EBVΔLMP1 cells were transduced using retroviral vector pLXSN empty (Vector) or expression vector pLXSN-LMP1. The cells were collected for mRNA and protein analysis. (D and E) The mRNA levels of EBNA1, LMP1, and DOK1 in these cells were determined using real time PCR and normalized to GAPDH expression, while the indicated proteins expression were analyzed using western blot. RPMI cells were transiently transfected with increasing amounts of pcDNA3 empty plasmid (Vector) or expression vector pcDNA3-LMP1. (F) Cells were collected for mRNA and protein analysis. LMP1and DOK1 gene expressions were measured using real time PCR for RNA levels and normalized to GAPDH expression. (G) The indicated protein levels were detected using western blotting. DOK1 protein levels were quantified from two independent immunoblots and normalized to the corresponding β-actin level (bottom of C, E and G). LMP1 down-regulates DOK1 expression by altering the composition of the E2F transcription complex
We recently showed that the E2F1 transcription factor has a key role in activation of DOK1 transcription [29]. The 500 nucleotide upstream of the start site of the DOK1 promoter contains three E2F1 responsive elements (RE) which appear to have a role in transcription activation; in particular the one at position −498/−486 (ERE1) [29]. Transient transfection experiments showed that LMP1 was able to efficiently inhibit the activity of −500/+33 DOK1 promoter cloned in front of the luciferase reporter gene (Figure 3A). The addition of upstream regions (−1000/−500 or −2000/−500) did not modify the pattern of LMP1 inhibition (Figure 3A). In addition, LMP1 was not able to further decrease the activity of DOK1 promoter harboring point mutations in ERE1 (Figure 3A). Together, these results suggest that LMP1 may exert its inhibitory activity targeting the regulatory complexes able to bind ERE1 within the −500/+33 region of the DOK1 promoter. Chromatin immuno-precipitation (ChIP) experiments using an anti-E2F1 antibody showed that infection with wild-type GFP-EBV significantly decreases the recruitment of E2F1 to ERE1 in RPMI and two independent LCLs (Figure 3B), while EBVΔLMP1 did not have any impact on this event in RPMI (Figure 3B). Interestingly, LMP1 alone did not prevent the recruitment of E2F1 to the DOK1 promoter in RPMI cells (Figure 3B), although it is able to efficiently down-regulate DOK1 expression (Figures 2D, 2E, and 3A).
Fig. 3. LMP1 represses DOK1 promoter activity through the recruitment of E2F1/pRB/DNMT1 inhibitory complex. 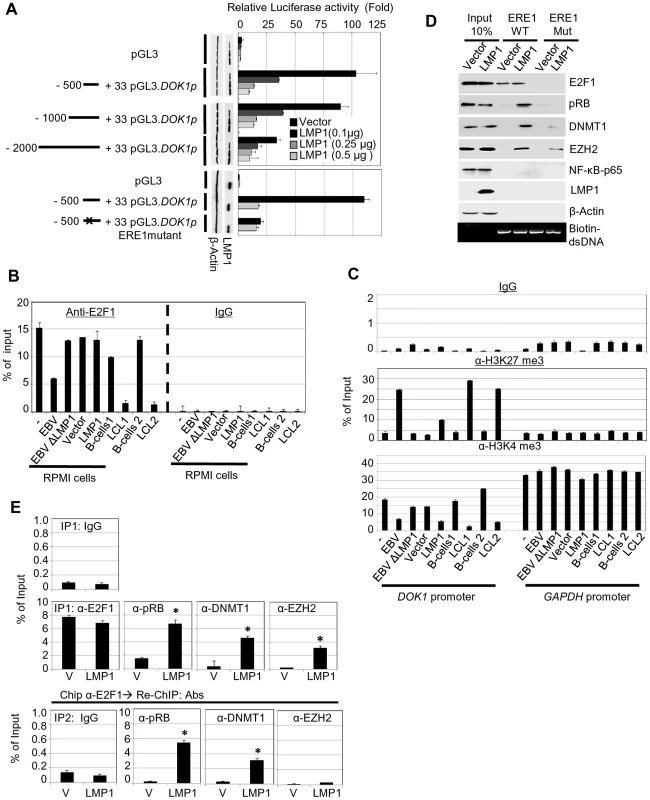
(A) RPMI cells were transfected with the indicated firefly luciferase reporter pGL3-DOK1 promoter constructs along with increasing amounts of pcDNA3 LMP1. Renilla luciferase was used as an internal control for the reporter assay. After 48 hours, cells were collected and processed for luciferase activity measurement. The data are average of three independent experiments. (B) RPMI cells, RPMI cells infected with GFP-EBV recombinant virus, or GFP-EBVΔLMP1, RPMI cells transduced with empty pLXSN (V) or expression vector pLXSN-LMP1, and LCLs and their original primary B-cells were subjected to quantitative ChIP assay using anti-E2F1 (KH 95) antibody or IgG. The DOK1 promoter was amplified by real-time PCR using specific primers flanking the E2F-response element located at (−498/−486). Data were calculated as percentages of enrichment of input. Error bars indicate the standard deviation from three independent experiments performed in triplicate. (C) The same cells from (B) were subjected to ChIP assay using the anti-H3K27 trimethylation antibody, anti-H3K4 trimethylation antibody or IgG. The DOK1 promoter and GAPDH promoter were amplified by real-time PCR. (D) In vitro DNA pull-down assay. The DOK1 promoter region containing the original E2F-response element located at (−498/−486) or a mutated one (obtained by replacing the core GGCG of the consensus sequence with AAAA), was amplified by PCR using specific 5′biotinylated primers. The PCR products (agarose gel, bottom panel) were incubated with total lysate from RPMI cells transduced with empty pLXSN (Vector) or expression vector pLXSN-LMP1, and then pulled down using streptavidin-agarose beads. Immunoblotting was used to check the recruitment of E2F1, pRB, DNMT1, EZH2 and p65 to the different PCR fragments. β-Actin was used as a negative control of binding to DNA. (E) Transduced RPMI cells with empty pLXSN (Vector) or expression vectorpLXSN-LMP1 were used for quantitative ChIP Re-ChIP assay. To assess the individual recruitment of E2F1, pRB, DNMT1, and EZH2 to the DOK1 promoter, chromatin was immuno-precipitated (IP 1) with the indicated antibodies and IgG was used as a negative control (top). To determine the E2F1 association with the indicated factors, E2F1 chromatin complex (IP 1) was subjected to Re-ChIP (IP 2) using the indicated antibodies (bottom). The DOK1 promoter was amplified using real time PCR and data from IP 1 and IP 2 were calculated as percentage of total input. We next analyzed the chromatin organization within the DOK1 promoter in the same cells by monitoring the tri-methylation of histone H3 at lysine 4 (H3K4me3) or at lysine 27 (H3K27me3) which are events associated with transcriptionally active or inactive chromatin, respectively. According to their ability to repress DOK1 expression, wild-type GFP-EBV or LMP1 alone induced an increase of H3K27me3 and a decrease of H3K4me3 within the DOK1 promoter compared with mock cells (Figure 3C). However, LMP1 was less efficient than the entire virus in promoting these epigenetic changes (Figure 3C). In summary, although LMP1 alone is not able to prevent the recruitment of E2F1 to the DOK1 promoter, it is capable of inducing epigenetic changes and inhibition of DOK1 transcription.
Based on these findings, we hypothesized that LMP1 mediates DOK1 down-regulation by altering the composition of the E2F1 complex. To explore this possibility, we performed oligo pull-down experiments using biotinylated DNA probes which contain a region of the DOK1 promoter encompassing the wild-type or mutated ERE1. Biotinylated DNA probes were incubated with protein extracts from RPMI cells transduced with empty retrovirus or with retrovirus expressing LMP1. In both extracts and as expected, E2F1 was found associated with the DNA, while only in the presence of LMP1 were three additional cellular proteins, which are usually part of negative regulatory complexes of transcription found associated with the DOK1 promoter fragment: (i) the E2F1 inhibitor retinoblastoma (pRB), (ii) the DNA methyl-transferase DNMT1 and (iii) the polycomb-group (PcG) 2 member EZH2 (Figure 3D). Deletion of ERE1 prevented the association of E2F1 in both cellular extracts. In addition, in LMP1-containing extracts, mutation of ERE1 also significantly decreased the pRB and DNMT1 protein levels precipitated with DNA (Figure 3D), suggesting that both proteins are recruited in the same complex as E2F1. With regard to EZH2, its binding to the DOK1 promoter was less affected by the ERE1 mutation, indicating that it is recruited by a different complex. Although LMP1 is able to activate the NF-κB pathway, no binding of the p65 transcription factor was found in both cellular extracts (Figure 3D). ChIP Re-ChIP experiments in mock and LMP1-expressing cells confirmed the data obtained in the pull-down assay. Indeed, Re-ChIP showed that a significant proportion of E2F1 complexes recruited to the DOK1 promoter contains pRB and DNMT1 proteins (80% and 40% respectively), but not EZH2 (Figure 3E), which appears to be associated with an independent complex.
Finally, the events occurring at DOK1 promoter were determined at early stages post-infection with EBV. We observed a significant enrichment of pRB, DNMT1 and EZH2 recruitment to DOK1 promoter in primary naive B cells infected with recombinant GFP-EBV virus for 48 hours. Consequently, an increase of H3K27 trimethylation (∼5 folds) and CpG methylation (∼10%) was detected (Supplementary Figure S3). Thus, early stage of EBV infection mimics the scenario observed in LMP1-expressing cells.
In summary, these data show that LMP1 initiates the repression of DOK1 expression by inducing the formation of transcriptional inhibitory complexes.
LMP1-mediated NF-κB activation is required for DOK1 down-regulation
LMP1 has the ability to activate different signaling pathways, such as NF-κB, MAPK p38, JNK, and MAPK/ERK [28]. To explore the potential role of these pathways in DOK1 down-regulation, RPMI cells infected with recombinant GFP-EBV were treated with different chemical inhibitors specific for these signaling pathways. No change was observed in mock or GFP-EBV cells treated with the chemical inhibitors of the MAPK p38, JNK, and MAPK/ERK pathways (SB203580, S600125 and PD98059, respectively) (data not shown). However, DOK1 mRNA and protein levels were found to be considerably increased in GFP-EBV-infected cells treated with a specific inhibitor of NF-κB (Bay11), but not in mock cells (Figure 4A and B). Similarly, Bay11 treatment of LMP1-expressing cells increased the DOK1 mRNA and protein levels (Figure 4A and B). To further demonstrate the role of NF-κB signaling in EBV-mediated DOK1 down-regulation, we inhibited the NF-κB canonical pathway by expressing a non-degradable deletion mutant of IκBα (Δ-IκBα) that lacks the first 36 amino acids at the N-terminus containing the IKK-phosphorylated amino acid. Similarly to Bay 11, Δ-IκBα expression in GFP-EBV RPMI cells led to an increase of transcript and protein levels of DOK1 (Figure 4C and D). Accordingly, transient transfection experiments using a plasmid containing the DOK1 promoter cloned upstream of the luciferase gene showed that Δ-IκBα antagonized LMP1 in inhibiting the DOK1 promoter (Supplementary Figure S2).
Fig. 4. LMP1-mediated NF-κB activation is required for EBV-related DOK1 down-regulation. 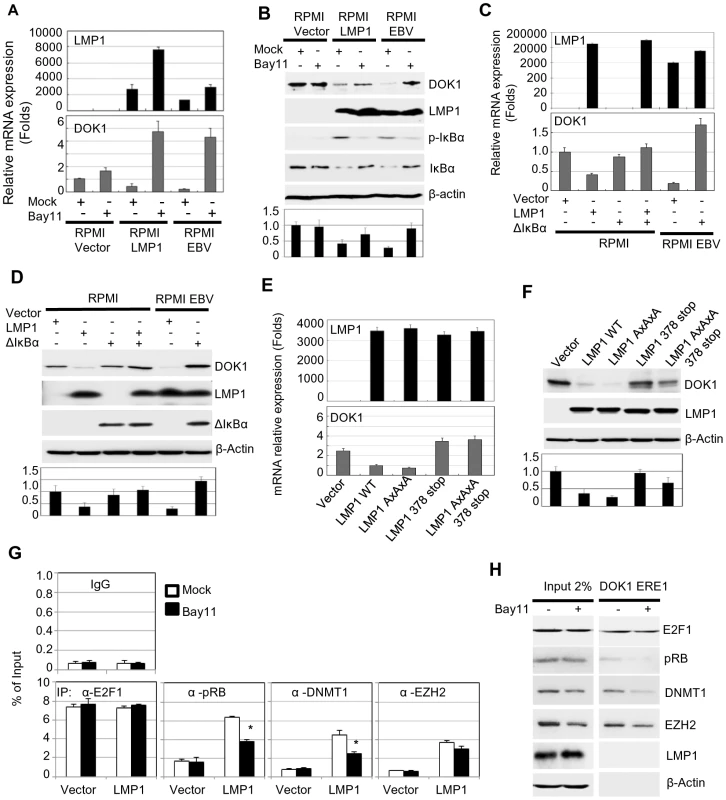
RPMI cells transduced with empty retroviral pLXSN (Vector), expression vector pLXSN-LMP1, or infected with GFP-EBV recombinant virus were treated with Bay11 or the equivalent volume of DMSO (Mock). (A) mRNA levels of LMP1 and DOK1 were measured by real time PCR, and normalized to GAPDH expression. (B) The indicated proteins were detected using western blotting. RPMI cells were transfected with pcDNA3 empty plasmid (Vector), expression vector pcDNA3-LMP1 and/or expressing the super-repressor IκBα (ΔIκBα), while RPMI cells infected with GFP-EBV recombinant virus were transfected only with pcDNA3 empty (Vector) or expression vector of the super-repressor IκBα (ΔIκBα). After 48 hours, cells were collected for analysis. (C) mRNA levels of LMP1 and DOK1 were measured by real time PCR, and normalized to GAPDH expression. (D) The indicated proteins were detected using western blotting. RPMI cells were transfected with empty pLXSN (Vector), or expression vector pLXSN-LMP1 wild type (WT), LMP1 mutant for the CTAR1 domain (AxAxA), and CTAR2 domain (378 stop), or both CRAT1 and 2 domains (AxAxA/378 stop). After 48 hours, cells were harvested for expression analysis. (E) mRNA levels of LMP1, GAPDH and DOK1 were measured using real time PCR. (F) The indicated proteins were detected using western blotting. DOK1 protein levels were quantified from two independent immunoblots and normalized to the corresponding β-actin level (bottom of B, D and F). Stable RPMI cells with empty pLXSN (Vector), or expression vector pLXSN-LMP1, were treated with Bay11 or the equivalent volume of DMSO (Mock). (G) Cells were subjected to quantitative ChIP assay using the indicated antibody or IgG. The DOK1 promoter was amplified by real-time PCR using specific primers flanking the E2F-response element located at −498/−486. Data were calculated as percentages of enrichment of total input. Error bars indicate the standard deviation from two independent experiments performed in triplicate. (H) In vitro DNA pull-down assay. The DOK1 promoter region containing ERE1 was incubated with total lysate from RPMI cells expressing LMP1 treated with Mock or Bay11, and then pulled down using streptavidin-agarose beads. Immunoblotting was used to check the recruitment of E2F1, pRB, DNMT1, EZH2. β-Actin was used as a negative control of binding to DNA. The LMP1 protein has two important C-terminal cytosolic domains named C-terminal activation region 1 (CTAR-1) (residues 194–232) and 2 (CTAR-2) (residues 351–386). Both the CTAR1 and CTAR2 domains have the ability to activate the NF-κB pathway through their interactions with tumor necrosis factor receptor (TNFR)-associated factors (TRAFs) [30], and TNFR-associated death domain protein (TRADD) [31], respectively. In particular, the CTAR2 domain is required for the activation of the canonical NF-κB pathway, while the CTAR1 domain is critical for the stimulation of the non-canonical NF-κB pathway [32]. The LMP1 mutants AxAxA (mutated CTAR1), 378 stop (deleted in CTAR2) and AxAxA/378 stop (mutated CTAR1 and deleted CTAR2) were expressed in RPMI cells. Both LMP1 378 stop and AxAxA/378 stop mutants failed to down-regulate the DOK1 gene, but not the LMP1 AxAxA mutant, which still retained its ability to suppress DOK1 expression at similar levels of wild-type LMP1 (Figure 4E and F). Therefore, LMP1 down-regulates DOK1 expression through its CTAR2 domain. In addition, we investigated whether the LMP1-mediated NF-κB activation plays a role in the formation of inhibitory complexes and their recruitment to the DOK1 promoter. LMP1-expressing RPMI cells were cultured in the presence of NF-κB inhibitor Bay11. No significant change in pRB and DMNT1 intracellular levels was observed, whereas EZH2 levels were slightly decreased (Figure 4H). However, oligo pull-down experiments clearly showed that inhibition of the NF-κB signaling affected the binding efficiency of pRB and DNMT1 to DOK1 promoter, while E2F1 and EZH2 continued to be associated with the DNA (Figure 4H). ChIP assay confirmed that inhibition of NF-κB significantly decreased the recruitment of pRB and DNMT1 to the DOK1 promoter (Figure 4G). Together, the data show that activation of the canonical NF-κB pathway by LMP1 is an important event for the down-regulation of DOK1 expression.
DOK1 gene silencing through DNA methylation is associated with EBV infection in B-cells
In our previous study [9], we reported that DOK1 expression is repressed in 64% of Burkitt's lymphoma cell lines through DNA hypermethylation of its promoter. These findings prompted us to assess whether hypermethylation of the DOK1 promoter could be ascribed to the presence of EBV. Using pyrosequencing and real-time PCR, respectively, DOK1 methylation and expression levels were measured in our experimental model. EBV infection of RPMI cells led to hypermethylation of DOK1 promoter (Figure 5A). This phenomenon was even more evident in LCLs (Figure 5A). EBVΔLMP1 was unable to promote DOK1 promoter methylation, further underlining the importance of the viral oncoprotein in this event. However, in agreement with the fact that LMP1 alone is unable to displace E2F1 from the DOK1 promoter, low DNA methylation was detected in RPMI cells expressing LMP1 alone (Figure 5A). In addition, no further methylation was observed in RPMI cells co-expressing LMP1 with other EBV proteins, i.e. EBNA3A, 3B, or 3C (data not shown), suggesting that a more complex pattern of viral gene expression is required to induce hypermethylation of DOK1 promoter. Treatments with the methyl-transferase inhibitor 5-Aza-2′-deoxycytidine (5-Aza) significantly affected DOK1 promoter methylation in LCLs and RPMI cells infected with EBV (Figure 5A). As expected, 5-Aza treatment rescued the recruitment of E2F1 to the DOK1 promoter in GFP-EBV infected cells, while no change was observed in LMP1-expressing cells (Figure 5B). However, an increase of DOK1 mRNA and protein levels was observed upon exposure to 5-Aza in GFP-EBV-infected cells (RPMI or LCLs) as well as in LMP1-expressing RPMI cells (Figure 5C and D). This event correlates with the decrease of H3K27me3 and the increase of H3K4me3 levels (Figure 5E).
Fig. 5. 5-Aza treatment rescue DOK1 expression in EBV infected cells. 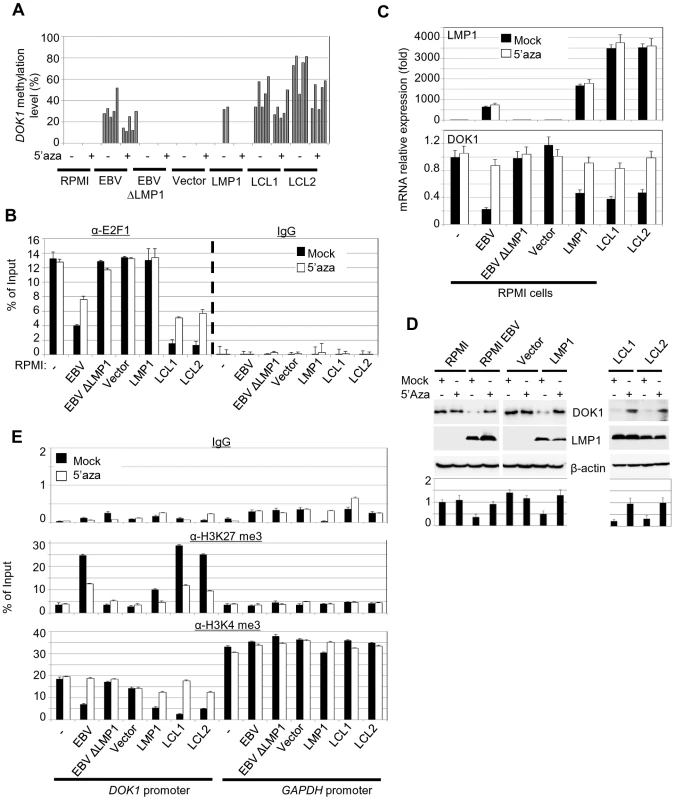
Cells were treated with 1 µM methyl-transferase inhibitor 5-Aza-2′deoxycytidine (5-Aza) for 4 days or equivalent volume of DMSO (Mock), then collected for analysis. (A) DNA methylation levels of the DOK1 promoter were measured using pyrosequencing. Each bar represents the percentage of methylation for individual CpG sites. (B) Quantitative ChIP assay using anti-E2F1 (KH 95) antibody or IgG. The DOK1 promoter was amplified by real-time PCR using specific primers flanking the E2F-response element located at (−498/−486). Data were calculated as percentages of enrichment of input. Error bars indicate the standard deviation (SD) from two independent experiments performed in triplicate. (C) The mRNA expression levels of LMP1, GAPDH and DOK1 were determined using real time PCR. (D) The indicated proteins were analyzed using western blotting. DOK1 protein levels were quantified from two independent immunoblots and normalized to the corresponding β-actin level (bottom). (E) ChIP assays were carried out using anti-H3K27 trimethylation antibody, anti-H3K4 trimethylation antibody or IgG. The DOK1 promoter and GAPDH promoter were amplified by real-time PCR. Data were calculated as percentages of enrichment of input. Together, these data show that EBV induces hypermethylation of the DOK1 promoter. Although expression of LMP1 alone marginally promotes DNA methylation, deletion of its gene in the EBV genome prevents the occurrence of this event. Thus, LMP1 appears to be essential, but not sufficient for hypermethylation of the DOK1 promoter.
Re-expression of DOK1 in LCLs decreased cellular proliferation and induced apoptosis
To understand the biological significance of EBV-induced DOK1-down-regulation, we re-expressed DOK1 in LCLs. We observed that ectopic DOK1 levels decreased LCL proliferation in a dose-dependent manner (Figure 6A). Consistently with these observations, DOK1 induced a significance decrease of cell populations in G0/G1 and G2/M phases (Figure 6B). In addition, high levels of DOK1 led to a significant increase of subG0 population and AnnexinV-positive cells (Figure 6B and C).Together, these data demonstrate the role of DOK1 in inhibiting cell proliferation induced by EBV and promoting both cell growth arrest and apoptosis.
Fig. 6. (A) LCL cells were transfected with the indicated amounts of empty pcDNA3 (Vector) or expression vector pcDNA3-Flag-DOK1. 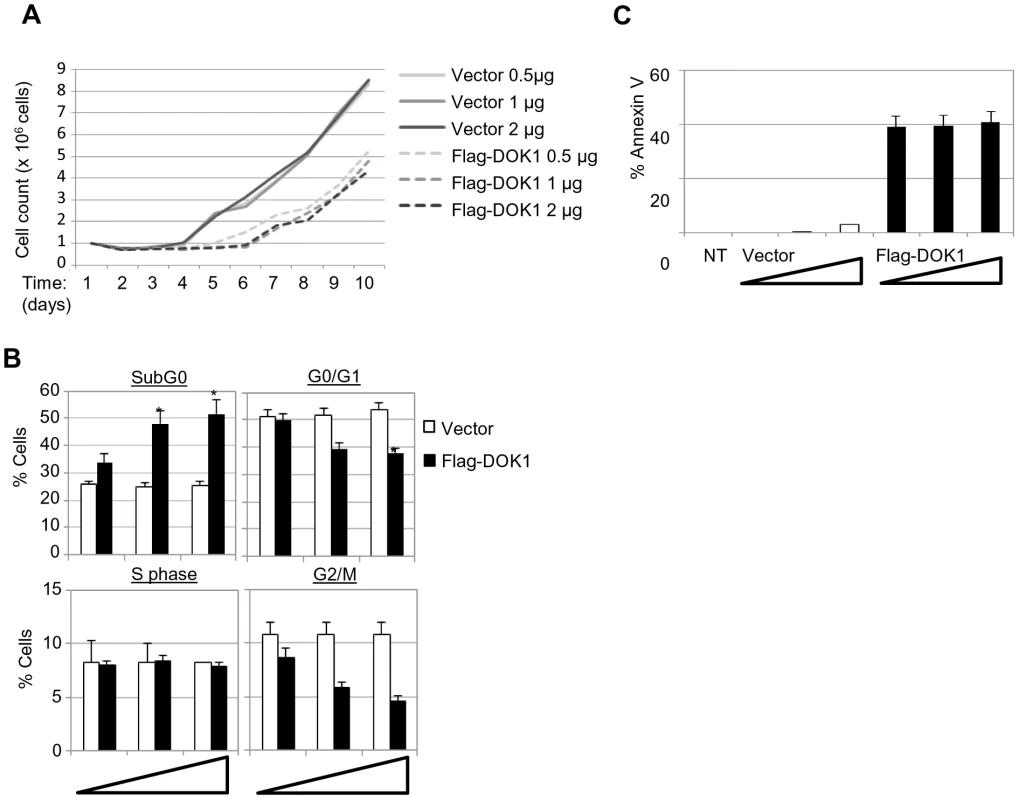
After 24(B) LCL cells were monitored for cell cycle analysis 48 hours after being transfected with the indicated amounts of pcDNA3 empty (Vector) or expression vector pcDNA3-Flag-DOK1. Cells in different cycle phases (SubG0, G0/G1, S, or G2/M) are represented as percentage of total cells. (C) The same cells from (B) were monitored for apoptosis using Annexin V staining. Non transfected cells were used as control (NT). Error bars indicate the SD from two independent experiments. Data were analyzed using Student's t test (*, P<0.05). Discussion
Several studies have demonstrated that the loss of DOK1 function is a key event in human carcinogenesis [1], [3], [4], [9], [33]. Indeed several mechanisms of DOK1 inactivation have been characterized so far DOK1 expression was found to be silenced by hypermethylation of its promoter in a variety of human cancers, including, head and neck, lung, gastric and liver cancer as well as in Burkitt's lymphoma-derived cell lines [9], [10], [11]. In addition, DOK1 was found to be mutated in chronic lymphocytic leukemia (CLL) [8]. At the protein level, DOK1 is targeted for proteasome degradation triggered by oncoprotein kinases (OTKs) such as p210bcr-abl and oncogenic forms of Src [7].
A recent study has provided evidence that DOK1 inactivation also occurs in virus-induced cancers [10]. Indeed, a correlation between DOK1 aberrant hypermethylation and the presence of hepatitis B virus (HBV) has been reported in hepatocellular carcinoma (HCC) [10]. Similarly, the expression of DOK1 mRNA was found to be down-regulated in cell lines derived from Burkitt's lymphoma [34], a pathological condition associated with EBV infection. However, these initial findings do not provide evidence about whether the down-regulation of DOK1 expression is directly induced by the viral proteins or is a consequence of the chromosomal alterations occurring during the carcinogenic processes. In this study, we demonstrate for the first time that EBV is directly involved in the inhibition of DOK1 expression. Our data show that the EBV LMP1 oncoprotein plays a key role in this event. Indeed, an EBV mutant lacking the entire LMP1 gene was unable to inhibit DOK1 transcription, while re-expression of LMP1 in cells infected with the EBVΔLMP1 mutant fully restored the ability of EBV to decrease DOK1 mRNA and protein levels. Expression of LMP1 alone in human cancer B-cells was sufficient to efficiently inhibit DOK1 transcription by promoting the formation of a transcriptional repressor complex containing E2F1, pRB, and the DNA methyl-transferase DNMT1. In addition, deletion of the E2F1-binding element (ERE1) strongly affected the binding of three cellular proteins to the DOK1 promoter, and a Re-ChIP assay confirmed that E2F1 is the carrier of pRB and DNMT1. We also observed that LMP1 promotes the recruitment of the histone-lysine N-methyl-transferase EZH2 independently of E2F1, leading to an increase in the level of H3K27me3. In agreement with the recruitment of the two epigenetic enzymes, an increase in H3K27me3 and DNA methylation levels was detected at the DOK1 promoter.
It has previously been shown that LMP1 is able to increase the expression and activity of DNA methyl-transferases (DNMT 1, 3a, and 3b), which could explain the increase of the DOK1 promoter methylation. Interestingly, DNA methylation was strongly enhanced in B-cells infected by the entire virus compared with cells expressing only LMP1. Thus, it is likely that additional viral products may cooperate with LMP1 in promoting DOK1 silencing via DNA methylation. No down-regulation of DOK1 was observed when EBNA1, 2, 3A, 3B, and 3C are expressed in RPMI cells. In addition, none of these viral proteins further stimulate DNA methylation at DOK1 promoter when co-expressed with LMP1 (data not shown). Thus, a more complex pattern of viral gene expression may be involved in the hyper-methylation of DOK1 promoter. Most importantly, we show that in EBV-infected B-cells the DNA methylation extends over a large region of the DOK1 promoter including ERE1 that loses the ability to recruit the active form of E2F1. Inhibition of DNA methylation significantly increases DOK1 transcription in LMP1-expressing cells as well as EBV-infected cells.
In summary, based on our findings, a two-step model can be proposed for EBV in the inhibition of DOK1 expression (Figure 7). In the first step, LMP1 favors the formation and recruitment of transcriptional repressor complexes containing E2F1/pRB/DNMT1 and EZH2. These complexes induce epigenetic changes in the DOK1 promoter region, leading to its inhibition. In the second step, LMP1 in collaboration with other EBV proteins leads to further increase of DNA methylation which in turn results in a loss of all transcriptional regulatory complexes and a strong repression of the DOK1 promoter. These data corroborate our previous studies that highlighted the key role of E2F1 and DNA methylation in the regulation of DOK1 expression [29]. Our data also show that the LMP1-induced DOK1 down-regulation is linked to activation of the NF-κB canonical pathway. Indeed, NF-κB activation by LMP1 plays a role in the formation and recruitment of inhibitory complex E2F1/pRB/DNMT1 to the DOK1 promoter. Although we did not observe any recruitment of p65 to the DOK1 promoter, neither by DNA-pull-down assay nor by chromatin immuno-precipitation (data not shown), we cannot exclude the involvement of other NF-κB transcription factors.
Fig. 7. Schematic model of DOK1 gene regulation in EBV-infected cells. 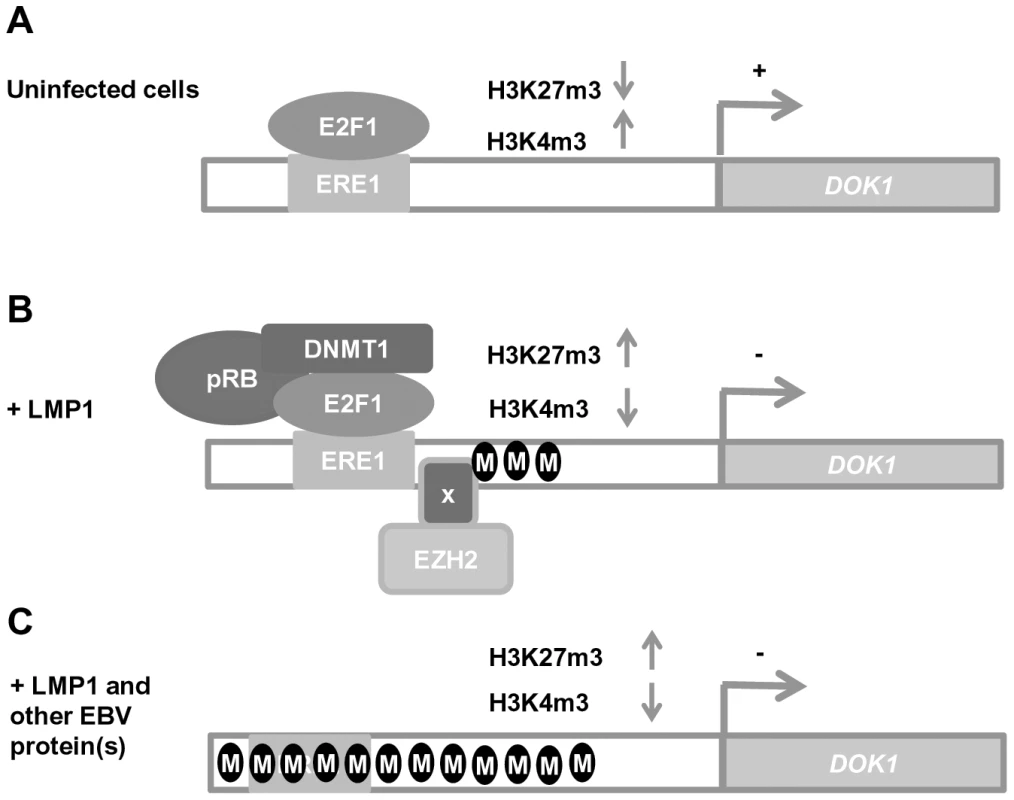
(A) In uninfected cells, DOK1 expression is activated via the recruitment of the active form of the E2F1 transcription factor to its response element located at (−498/−486) on the DOK1 promoter. (B) In cells expressing the oncoprotein LMP1, DOK1 is down-regulated through the recruitment of the inhibitory complexes E2F1/pRB/DNMT1 and EZH2 to its promoter region. These complexes lead to the induction of partial DNA methylation and the increase of H3K27 trimethylation levels, respectively. (C) In EBV-infected cells, DOK1 is repressed through heavy DNA methylation of its promoter region and the increase in H3K27 trimethylation level. These events likely induce conformational changes in the chromatin, which become less permissive to E2F1 transcription factor recruitment. Until now, several studies reported that DNA methylation patterns were higher in EBV positive tumors compared to the EBV-negative ones and that EBV infection was clearly demonstrated to induce specific methylation epigenotypes that lead to silencing of multiple tumor suppressor genes such as BIM, p16INK4A, p14ARF, p73, E-cadherin and PTEN in EBV–associated nasopharyngeal and gastric cancers [17], [35], [36], [37], [38]. While these events are believed to be caused by elevated levels of DNMTs induced by LMP1 and 2, the mechanisms establishing the methylation patterns themselves are unknown. As DNA methyl-transferases have little specificity in vitro, we propose the notion that LMP1 triggers DOK1 gene repression through the recruitment of DNMT1 to its promoter in a specific manner via E2F1-binding to its response element, and this event might be an early step for EBV-induced DNA methylation. As some of the genes listed above are targets of E2F1 [39], [40], it will be interesting to see whether their methylation patterns are also specific to the recruitment of the inhibitory complex E2F1/pRB/DNMT1. Moreover, EBV appears to have an initiator role of epigenetic alterations and therefore inducing oncogenesis, however, the latency expression patterns of EBV genes differ in different cancers, which make unclear the contribution of the virus to some types. One explanation would be that EBV-induced epigenetic changes, such as EBV-mediated DNA methylation of DOK1 promoter, are stable events and could also persist even after the changes in EBV latent gene expression. As DOK1 gene silencing was found to be related to its promoter hypermethylation in gastric cancer [11], it will be important to investigate whether these events are associated with the presence of EBV in these cancers and others.
In conclusion, the present study sheds light on the association between EBV infection and DOK1 down-regulation in B-cells. It provides novel insights into the regulation of DOK1 in viral-related carcinogenesis, and could define it as a potential cancer biomarker and an attractive target for epigenetic-based therapy.
Materials and Methods
Expression vectors
Cellular and viral genes were expressed using the retroviral vector pLXSN (Clontech, Palo Alto, CA) or the expression vector pcDNA-3 (Invitrogen). The pLXSN-LMP-1 and the mutants LMP-1AxAxA, LMP-1 378 stop, and LMP-1AxAxA/378 stop constructs have been previously described [41]. The pGL3 basic luciferase reporter (Promega) and pGL3 containing the DOK1 promoter constructs have been described previously [29], The NF-κB super-repressor Δ-IκBα, which lacks the coding sequence of the first 36 N-terminal amino-acids, was kindly provided by Dr Elliot Kieff (Harvard Medical School, Boston, Massachusetts, USA). The expression plasmids pDEST-myc-EBNA1, pSG5-EBNA2, pDEST-myc-EBN3A1, pDEST-myc-EBNA3B, pDEST-myc-EBNA3C were kindly provided by Dr Evelyne Manet (ENS, Lyon, France).
Cells, transfection, and chemicals
RPMI 8226 cells were kindly provided by Dr Christophe Caux (Centre Léon Bérard, Lyon, France). The EBV-negative immortalized B-cells, BJAB were previously described [42], and the Louckes cells were kindly provided by Dr Evelyne Manet (ENS, Lyon, France). The primary B-cells were isolated from total blood of healthy donors using negative selection EasySep or RosetteSep (StemCell Technologies). Primary naive B cells and RPMI cells were infected with recombinant GFP-EBV, and GFP-EBVΔLMP-1 as described in [43], [44], [45], RPMI pLXSN-empty or pLXSN-LMP1 cell lines were generated as described previously [41]. Expression of LMP-1 wild-type, LMP-1 AxAxA , LMP1 378 stop, and LMP1 AxAxA/378 stop mutants in RPMI was achieved by transduction with recombinant retroviruses [41]. The EBV-immortalized lymphoblastoid cell lines (LCLs) were generated in this study by infecting primary B-cells isolated from different donors with recombinant EBV expressing GFP, as described previously [41]. Primary and immortalized B-cells were cultured in RPMI 1640 medium (GIBCO, Invitrogen life Technologies, Cergy-Pontoise, France) supplemented with 10% FBS, 100 U/ml penicillin G, 100 mg/ml streptomycin, 2 mM L-glutamine, and 1 mM sodium pyruvate (PAA, Pasching, Austria). Expression plasmids were transiently transfected in cells using Xtreme gene 9 reagents (Roche) according to the manufacturer's protocol.
For treatment, cells were incubated in media containing different reagents: with a final concentration of 1 µM of the NF-κB pathway inhibitor Bay11 in dimethyl sulfoxide (DMSO) for 6 hours. Inhibition of DNA methylation was performed by incubation for 4 days with 5-aza-2′-deoxycytidine (5-aza) at 1 µM (Sigma) dissolved in DMSO. Cells were then harvested for analysis.
Quantitative RT-PCR
Total RNA was extracted using TRIzol reagent (Life Technologies). Reverse transcription was performed using the RevertAid H Minus First Strand cDNA synthesis kit (Fermentas) according to the manufacturer's protocol. Real-time PCR was performed using the following gene-specific primers:
DOK1: Fw ATGGACGGAGCAGTGATGGA, Rev CCCAGGTCTTCCTCCACCTC
LMP1: Fw CCCCCTCTCCTCTTCCATAG, Rev GCCAAAGATGAACAGCACAA
EBNA1: Fw GGACCCGGCCCACAACCTG, Rev CTCCTGCCCTTCCTCACCCTCATC
GAPDH: Fw GAAGGTGAAGGTCGGAGTC, Rev AAGATGGTGATGGGATTTC. Data were analyzed using the ΔΔCT method.
Antibodies and immunoblotting
The following antibodies were used: anti-DOK1 (ab8112, Abcam), anti-E2F1 (KH-95; Santa Cruz Biotechnology), anti - β-Actin C4 (MP Biomedicals), anti-LMP1 (S12), anti - phosphor IκBα (#9246, Cell Signaling Technology), anti-total IκBα (#9242, Cell Signaling Technology), mouse IgG, rabbit IgG (Santa Cruz Biotechnology), anti-p65 (#3034, Cell Signaling Technology), anti-H3K4me3, and anti-H3K27me3 (Epigentek), anti-EZH2 (AC22; Cell Signaling Technology), anti-pRB (4H1, Cell Signaling Technology), anti-DNMT1 (60B1220, Abnova), anti-EBNA1 (1EB12, Santa Cruz Biotechnology), anti-EBNA2 (Novocastra), anti-EBNA3A (Exalpha), anti-EBNA3C (ab16128, Abcam). Immunoblotting was performed as described previously [29].
Reporter assays
Cells were transfected with 0.250 µg of pGL3 or DOK1 promoter constructs along with other experimental plasmids using X-tremeGENE 9 (Roche Diagnostics). The Renilla construct was included for normalization of transfection efficiency. At 48 hours after transfection, cells were harvested and the enzyme activities of firefly and Renilla luciferases were measured using the Dual-Luciferase reporter assay system (Promega). The luminescence signal was quantified using an Optocomp I luminometer (MGM Instruments). Each condition was used in triplicate and replicated in different independent experiments.
Chromatin immuno-precipitation (ChIP)
For each reaction, 106 cells were cross-linked with 1% formaldehyde, harvested and subjected to sonication to shear the chromatin into fragments of 0.2 kb, immuno-precipitated with 2 µg of appropriate antibody, and then processed according to the standard protocol for ChIP analysis from Cell Signaling Technology.
Low cell ChIP kit (Diagenode) was used for primary B cells and infected with EBV for 48 hours. 50 000 cells per reaction were processed according to the manufacturer's protocol.
The input and immuno-precipitated DNA from both methods (standard and low cell) were then analyzed by real-time PCR using primers flanking the E2F-response element (−498/−486) of the DOK1 promoter: Fw GCCAAAACCGAGGACTTTCG, Rev CATCACTGCTCCGTCCATGG, or primers for GAPDH promoter: Fw GACGGCCGCATCTTCTTGT, Rev CCTGGTGACCAGGCGC. Data were calculated as a percentage of enrichment of input.
Re-ChIP assay
Following the initial anti-E2F1 ChIP (performed as above using 107 cells and 10 µg of anti E2F1 KH-95 antibody), up to the final wash step with TE buffer, E2F1–chromatin complexes were eluted by the addition of 10 mM dithiothreitol (DTT) and incubated for 30 minutes at 37°C. Supernatants were diluted 1∶20 with re-ChIP buffer (1% Triton X-100; 20 mM Tris–HCl, pH 8.1; 2 mM EDTA; 150 mM NaCl; supplemented with protease inhibitors), and immuno-precipitated a second time (IP 2) using 4 µg of antibody against pRB, DNMT1, and EZH2. IgG was used as negative control. The Re-ChIP mixtures were incubated overnight at 4°C with rotation. Isolation and purification of associated DNA were carried out as described for the standard ChIP experiment. The binding of each factor was determined by real-time PCR as previously described. Data were calculated as a percentage of enrichment of total input.
DNA pull-down assay
Cells were lysed by sonication in HKMG buffer (10 mM HEPES, pH 7.9; 100 mM KCl; 5 mM MgCl2; 10% glycerol; 1 mM dithiothreitol (DTT); and 0.5% NP-40) containing protease and phosphatase inhibitors. Cellular debris was removed by centrifugation. Then, 1 mg of total lysate was pre-cleared with 40 µl of streptavidin-agarose beads (Thermo Scientific) for 1 hour at 4°C, with rotation, and incubated with 2 µg of biotinylated PCR product oligonucleotides and 20 µg of poly (dI-dC) for 16 hours at 4°C, with rotation. Biotin-oligonucleotide-protein complexes were collected with 60 µl of streptavidin-agarose beads for 1 hour at 4°C, with rotation, washed twice with HKMG buffer, separated on SDS-PAGE, and detected by western blotting. The biotinylated double-stranded oligonucleotides were amplified using the same primers as for ChIP with 5′ biotin.
DNA extraction and pyrosequencing
Genomic DNA was extracted using the QIAamp DNA minikit (Qiagen) and bisulfite converted using the EZ DNA Methylation-Gold kit (Zymo Research). Converted DNA was then subjected to Pyrosequencing (Qiagen) as previously described [46]. The primers used to measure the methylation of DOK1 promoter were: Fw GAGGTGGAGGAAGATTTG, Rev BIOTIN-CCACACTCACACACTCAA, and sequencing primer AGTTTTGGGGGTGGT. The percentage of methylation was evaluated as the mean of each CpG analyzed.
Flow cytometry analysis
To determine cell cycle profile, cells were collected 48 hours post-transfection with empty pCDNA3 (Vector) or expression vector pCDNA3-Flag-DOK1, washed twice with PBS 1×, and then cell pellets were re-suspended in 70% ethanol while vortexing, in order to prevent cell clumps. After ethanol fixation (30 minutes at 4°C) the cells were rewashed in PBS 1× and finally re-suspended in PBS 1×+ 100 µg/mL RNAse (Roche) + 25 µg/mL of Propidium iodide (Sigma).
Apoptotic cells were detected using the PE Annexin V apoptosis detection kit I (BD Pharmingen) according to the manufacturer's instructions.
Stained cells for cell cycle and for apoptosis were detected using the BD FACSCanto II flow cytometer (BD Biosciences) and analyzed using FACSDiva software.
Ethics statement
Blood samples from healthy donors were provided by the Etablissement Français du Sang (EFS, Lyon, France) after being anonymized. All participants signed a written informed consent.
Supporting Information
Zdroje
1. MashimaR, HishidaY, TezukaT, YamanashiY (2009) The roles of Dok family adapters in immunoreceptor signaling. Immunol Rev 232 : 273–285.
2. Di CristofanoA, NikiM, ZhaoM, KarnellFG, ClarksonB, et al. (2001) p62(dok), a negative regulator of Ras and mitogen-activated protein kinase (MAPK) activity, opposes leukemogenesis by p210(bcr-abl). J Exp Med 194 : 275–284.
3. BergerAH, NikiM, MorottiA, TaylorBS, SocciND, et al. (2010) Identification of DOK genes as lung tumor suppressors. Nature Genetics 42 : 216–U227.
4. YasudaT, ShirakataM, IwamaA, IshiiA, EbiharaY, et al. (2004) Role of Dok-1 and Dok-2 in myeloid homeostasis and suppression of leukemia. J Exp Med 200 : 1681–1687.
5. YasudaT, BundoK, HinoA, HondaK, InoueA, et al. (2007) Dok-1 and Dok-2 are negative regulators of T cell receptor signaling. Int Immunol 19 : 487–495.
6. NelmsK, SnowAJ, Noben-TrauthK (1998) Dok1 encoding p62(dok) maps to mouse chromosome 6 and human chromosome 2 in a region of translocation in chronic lymphocytic leukemia. Genomics 53 : 243–245.
7. JanasJA, Van AelstL (2011) Oncogenic tyrosine kinases target Dok-1 for ubiquitin-mediated proteasomal degradation to promote cell transformation. Mol Cell Biol 31 : 2552–2565.
8. LeeS, RoyF, GalmariniCM, AccardiR, MichelonJ, et al. (2004) Frameshift mutation in the Dok1 gene in chronic lymphocytic leukemia. Oncogene 23 : 2287–2297.
9. SaulnierA, VaissiereT, YueJ, SioudaM, MalfroyM, et al. (2012) Inactivation of the putative suppressor gene DOK1 by promoter hypermethylation in primary human cancers. Int J Cancer 130 : 2484–2494.
10. LambertMP, PaliwalA, VaissiereT, CheminI, ZoulimF, et al. (2011) Aberrant DNA methylation distinguishes hepatocellular carcinoma associated with HBV and HCV infection and alcohol intake. J Hepatol 54 : 705–715.
11. BalassianoK, LimaS, JenabM, OvervadK, TjonnelandA, et al. (2011) Aberrant DNA methylation of cancer-associated genes in gastric cancer in the European Prospective Investigation into Cancer and Nutrition (EPIC-EURGAST). Cancer Lett 311 : 85–95.
12. YoungLS, RickinsonAB (2004) Epstein-Barr virus: 40 years on. Nat Rev Cancer 4 : 757–768.
13. StrausSE (1988) The chronic mononucleosis syndrome. J Infect Dis 157 : 405–412.
14. EpsteinMA, AchongBG, BarrYM (1964) Virus Particles in Cultured Lymphoblasts from Burkitt's Lymphoma. Lancet 1 : 702–703.
15. FlavellKJ, MurrayPG (2000) Hodgkin's disease and the Epstein-Barr virus. Mol Pathol 53 : 262–269.
16. CarboneA, GloghiniA, DottiG (2008) EBV-associated lymphoproliferative disorders: classification and treatment. Oncologist 13 : 577–585.
17. MatsusakaK, KanedaA, NagaeG, UshikuT, KikuchiY, et al. (2011) Classification of Epstein-Barr virus-positive gastric cancers by definition of DNA methylation epigenotypes. Cancer Res 71 : 7187–7197.
18. FarrellPJ (1995) Epstein-Barr virus immortalizing genes. Trends Microbiol 3 : 105–109.
19. BornkammGW, HammerschmidtW (2001) Molecular virology of Epstein-Barr virus. Philos Trans R Soc Lond B Biol Sci 356 : 437–459.
20. KayeKM, IzumiKM, KieffE (1993) Epstein-Barr virus latent membrane protein 1 is essential for B-lymphocyte growth transformation. Proc Natl Acad Sci U S A 90 : 9150–9154.
21. Cahir McFarlandED, IzumiKM, MosialosG (1999) Epstein-barr virus transformation: involvement of latent membrane protein 1-mediated activation of NF-kappaB. Oncogene 18 : 6959–6964.
22. EliopoulosAG, YoungLS (2001) LMP1 structure and signal transduction. Semin Cancer Biol 11 : 435–444.
23. MosialosG, BirkenbachM, YalamanchiliR, VanArsdaleT, WareC, et al. (1995) The Epstein-Barr virus transforming protein LMP1 engages signaling proteins for the tumor necrosis factor receptor family. Cell 80 : 389–399.
24. RobertsML, CooperNR (1998) Activation of a ras-MAPK-dependent pathway by Epstein-Barr virus latent membrane protein 1 is essential for cellular transformation. Virology 240 : 93–99.
25. EliopoulosAG, GallagherNJ, BlakeSM, DawsonCW, YoungLS (1999) Activation of the p38 mitogen-activated protein kinase pathway by Epstein-Barr virus-encoded latent membrane protein 1 coregulates interleukin-6 and interleukin-8 production. J Biol Chem 274 : 16085–16096.
26. HuenDS, HendersonSA, Croom-CarterD, RoweM (1995) The Epstein-Barr virus latent membrane protein-1 (LMP1) mediates activation of NF-kappa B and cell surface phenotype via two effector regions in its carboxy-terminal cytoplasmic domain. Oncogene 10 : 549–560.
27. EliopoulosAG, BlakeSM, FloettmannJE, RoweM, YoungLS (1999) Epstein-Barr virus-encoded latent membrane protein 1 activates the JNK pathway through its extreme C terminus via a mechanism involving TRADD and TRAF2. J Virol 73 : 1023–1035.
28. DawsonCW, PortRJ, YoungLS (2012) The role of the EBV-encoded latent membrane proteins LMP1 and LMP2 in the pathogenesis of nasopharyngeal carcinoma (NPC). Semin Cancer Biol 22 : 144–153.
29. SioudaM, YueJ, ShuklaR, GuillermierS, HercegZ, et al. (2012) Transcriptional regulation of the human tumor suppressor DOK1 by E2F1. Mol Cell Biol 32 : 4877–4890.
30. DevergneO, HatzivassiliouE, IzumiKM, KayeKM, KleijnenMF, et al. (1996) Association of TRAF1, TRAF2, and TRAF3 with an Epstein-Barr virus LMP1 domain important for B-lymphocyte transformation: role in NF-kappaB activation. Mol Cell Biol 16 : 7098–7108.
31. IzumiKM, KieffED (1997) The Epstein-Barr virus oncogene product latent membrane protein 1 engages the tumor necrosis factor receptor-associated death domain protein to mediate B lymphocyte growth transformation and activate NF-kappaB. Proc Natl Acad Sci U S A 94 : 12592–12597.
32. AtkinsonPG, CoopeHJ, RoweM, LeySC (2003) Latent membrane protein 1 of Epstein-Barr virus stimulates processing of NF-kappa B2 p100 to p52. J Biol Chem 278 : 51134–51142.
33. NikiM, Di CristofanoA, ZhaoM, HondaH, HiraiH, et al. (2004) Role of Dok-1 and Dok-2 in leukemia suppression. J Exp Med 200 : 1689–1695.
34. LeeS, HuangH, NiuY, TommasinoM, LenoirG, et al. (2007) Dok1 expression and mutation in Burkitt's lymphoma cell lines. Cancer Lett 245 : 44–50.
35. PaschosK, SmithP, AndertonE, MiddeldorpJM, WhiteRE, et al. (2009) Epstein-barr virus latency in B cells leads to epigenetic repression and CpG methylation of the tumour suppressor gene Bim. PLoS Pathog 5: e1000492.
36. HinoR, UozakiH, MurakamiN, UshikuT, ShinozakiA, et al. (2009) Activation of DNA methyltransferase 1 by EBV latent membrane protein 2A leads to promoter hypermethylation of PTEN gene in gastric carcinoma. Cancer Res 69 : 2766–2774.
37. SakumaK, ChongJM, SudoM, UshikuT, InoueY, et al. (2004) High-density methylation of p14ARF and p16INK4A in Epstein-Barr virus-associated gastric carcinoma. Int J Cancer 112 : 273–278.
38. PaschosK, AlldayMJ (2010) Epigenetic reprogramming of host genes in viral and microbial pathogenesis. Trends Microbiol 18 : 439–447.
39. UristM, TanakaT, PoyurovskyMV, PrivesC (2004) p73 induction after DNA damage is regulated by checkpoint kinases Chk1 and Chk2. Genes Dev 18 : 3041–3054.
40. ZhangHJ, LiWJ, YangSY, LiSY, NiJH, et al. (2009) 8-Chloro-adenosine-induced E2F1 promotes p14ARF gene activation in H1299 cells through displacing Sp1 from multiple overlapping E2F1/Sp1 sites. J Cell Biochem 106 : 464–472.
41. AccardiRFI, GruffatH, MariggiòG, Le Calvez-KelmF, VoegeleC, BartoschB, Hernandez-VargasH, McKayJ, SyllaBS, ManetE, TommasinoM (2013 Mar) Epstein - Barr virus transforming protein LMP-1 alters B cells gene expression by promoting accumulation of the oncoprotein ΔNp73α. PLoS Pathog 9(3).
42. LeeS, AndrieuC, SaltelF, DestaingO, AuclairJ, et al. (2004) IkappaB kinase beta phosphorylates Dok1 serines in response to TNF, IL-1, or gamma radiation. Proc Natl Acad Sci U S A 101 : 17416–17421.
43. DelecluseHJ, HilsendegenT, PichD, ZeidlerR, HammerschmidtW (1998) Propagation and recovery of intact, infectious Epstein-Barr virus from prokaryotic to human cells. Proc Natl Acad Sci U S A 95 : 8245–8250.
44. DelecluseHJ, PichD, HilsendegenT, BaumC, HammerschmidtW (1999) A first-generation packaging cell line for Epstein-Barr virus-derived vectors. Proc Natl Acad Sci U S A 96 : 5188–5193.
45. DirmeierU, NeuhierlB, KilgerE, ReisbachG, SandbergML, et al. (2003) Latent membrane protein 1 is critical for efficient growth transformation of human B cells by Epstein-Barr virus. Cancer Research 63 : 2982–2989.
46. FrommerM, McDonaldLE, MillarDS, CollisCM, WattF, et al. (1992) A genomic sequencing protocol that yields a positive display of 5-methylcytosine residues in individual DNA strands. Proc Natl Acad Sci U S A 89 : 1827–1831.
Štítky
Hygiena a epidemiologie Infekční lékařství Laboratoř
Článek Combined Systems Approaches Reveal Highly Plastic Responses to Antimicrobial Peptide Challenge inČlánek Two Novel Human Cytomegalovirus NK Cell Evasion Functions Target MICA for Lysosomal Degradation
Článek vyšel v časopisePLOS Pathogens
Nejčtenější tento týden
2014 Číslo 5- Stillova choroba: vzácné a závažné systémové onemocnění
- Choroby jater v ordinaci praktického lékaře – význam jaterních testů
- Diagnostický algoritmus při podezření na syndrom periodické horečky
- Perorální antivirotika jako vysoce efektivní nástroj prevence hospitalizací kvůli COVID-19 − otázky a odpovědi pro praxi
- Diagnostika virových hepatitid v kostce – zorientujte se (nejen) v sérologii
-
Všechny články tohoto čísla
- Surveillance for Emerging Biodiversity Diseases of Wildlife
- The Emerging Role of Urease as a General Microbial Virulence Factor
- PARV4: An Emerging Tetraparvovirus
- Epigenetic Changes Modulate Schistosome Egg Formation and Are a Novel Target for Reducing Transmission of Schistosomiasis
- The Human Adenovirus E4-ORF1 Protein Subverts Discs Large 1 to Mediate Membrane Recruitment and Dysregulation of Phosphatidylinositol 3-Kinase
- A Multifactorial Role for Malaria in Endemic Burkitt's Lymphoma Pathogenesis
- Structural Basis for the Ubiquitin-Linkage Specificity and deISGylating Activity of SARS-CoV Papain-Like Protease
- Cathepsin-L Can Resist Lysis by Human Serum in
- Epstein-Barr Virus Down-Regulates Tumor Suppressor Expression
- BCA2/Rabring7 Targets HIV-1 Gag for Lysosomal Degradation in a Tetherin-Independent Manner
- The Evolutionarily Conserved Mediator Subunit MDT-15/MED15 Links Protective Innate Immune Responses and Xenobiotic Detoxification
- Suppressor of Cytokine Signaling 4 (SOCS4) Protects against Severe Cytokine Storm and Enhances Viral Clearance during Influenza Infection
- T Cell Inactivation by Poxviral B22 Family Proteins Increases Viral Virulence
- Dynamics of HIV Latency and Reactivation in a Primary CD4+ T Cell Model
- HIV and HCV Activate the Inflammasome in Monocytes and Macrophages via Endosomal Toll-Like Receptors without Induction of Type 1 Interferon
- Virus and Autoantigen-Specific CD4+ T Cells Are Key Effectors in a SCID Mouse Model of EBV-Associated Post-Transplant Lymphoproliferative Disorders
- Severe Acute Respiratory Syndrome Coronavirus Envelope Protein Ion Channel Activity Promotes Virus Fitness and Pathogenesis
- Squalene Synthase As a Target for Chagas Disease Therapeutics
- The Contribution of Viral Genotype to Plasma Viral Set-Point in HIV Infection
- Combined Systems Approaches Reveal Highly Plastic Responses to Antimicrobial Peptide Challenge in
- Anthrax Lethal Factor as an Immune Target in Humans and Transgenic Mice and the Impact of HLA Polymorphism on CD4 T Cell Immunity
- Ly49C-Dependent Control of MCMV Infection by NK Cells Is -Regulated by MHC Class I Molecules
- Two Novel Human Cytomegalovirus NK Cell Evasion Functions Target MICA for Lysosomal Degradation
- A Large Family of Antivirulence Regulators Modulates the Effects of Transcriptional Activators in Gram-negative Pathogenic Bacteria
- Broad-Spectrum Anti-biofilm Peptide That Targets a Cellular Stress Response
- Malaria Parasite Infection Compromises Control of Concurrent Systemic Non-typhoidal Infection via IL-10-Mediated Alteration of Myeloid Cell Function
- A Role for in Higher Order Structure and Complement Binding of the Capsule
- Hip1 Modulates Macrophage Responses through Proteolysis of GroEL2
- CD8 T Cells from a Novel T Cell Receptor Transgenic Mouse Induce Liver-Stage Immunity That Can Be Boosted by Blood-Stage Infection in Rodent Malaria
- Phosphorylation of KasB Regulates Virulence and Acid-Fastness in
- HIV-Infected Individuals with Low CD4/CD8 Ratio despite Effective Antiretroviral Therapy Exhibit Altered T Cell Subsets, Heightened CD8+ T Cell Activation, and Increased Risk of Non-AIDS Morbidity and Mortality
- A Novel Mechanism Inducing Genome Instability in Kaposi's Sarcoma-Associated Herpesvirus Infected Cells
- Structural and Biochemical Characterization Reveals LysGH15 as an Unprecedented “EF-Hand-Like” Calcium-Binding Phage Lysin
- Hepatitis C Virus Cell-Cell Transmission and Resistance to Direct-Acting Antiviral Agents
- Different Modes of Retrovirus Restriction by Human APOBEC3A and APOBEC3G
- TNFα and IFNγ but Not Perforin Are Critical for CD8 T Cell-Mediated Protection against Pulmonary Infection
- Large Scale RNAi Reveals the Requirement of Nuclear Envelope Breakdown for Nuclear Import of Human Papillomaviruses
- The Cytoplasmic Domain of Varicella-Zoster Virus Glycoprotein H Regulates Syncytia Formation and Skin Pathogenesis
- A New Class of Multimerization Selective Inhibitors of HIV-1 Integrase
- Are We There Yet? The Smallpox Research Agenda Using Variola Virus
- High-Efficiency Targeted Editing of Large Viral Genomes by RNA-Guided Nucleases
- Dynamic Functional Modulation of CD4 T Cell Recall Responses Is Dependent on the Inflammatory Environment of the Secondary Stimulus
- Bacterial Superantigens Promote Acute Nasopharyngeal Infection by in a Human MHC Class II-Dependent Manner
- Follicular Helper T Cells Promote Liver Pathology in Mice during Infection
- A Nasal Epithelial Receptor for WTA Governs Adhesion to Epithelial Cells and Modulates Nasal Colonization
- Unexpected Role for IL-17 in Protective Immunity against Hypervirulent HN878 Infection
- Human Cytomegalovirus Fcγ Binding Proteins gp34 and gp68 Antagonize Fcγ Receptors I, II and III
- Expansion of Murine Gammaherpesvirus Latently Infected B Cells Requires T Follicular Help
- Venus Kinase Receptors Control Reproduction in the Platyhelminth Parasite
- Molecular Signatures of Hemagglutinin Stem-Directed Heterosubtypic Human Neutralizing Antibodies against Influenza A Viruses
- The Downregulation of GFI1 by the EZH2-NDY1/KDM2B-JARID2 Axis and by Human Cytomegalovirus (HCMV) Associated Factors Allows the Activation of the HCMV Major IE Promoter and the Transition to Productive Infection
- Inactivation of Fructose-1,6-Bisphosphate Aldolase Prevents Optimal Co-catabolism of Glycolytic and Gluconeogenic Carbon Substrates in
- New Insights into Rotavirus Entry Machinery: Stabilization of Rotavirus Spike Conformation Is Independent of Trypsin Cleavage
- Prophenoloxidase Activation Is Required for Survival to Microbial Infections in
- SslE Elicits Functional Antibodies That Impair Mucinase Activity and Colonization by Both Intestinal and Extraintestinal Strains
- Timed Action of IL-27 Protects from Immunopathology while Preserving Defense in Influenza
- HIV-1 Envelope gp41 Broadly Neutralizing Antibodies: Hurdles for Vaccine Development
- The PhoP-Dependent ncRNA Mcr7 Modulates the TAT Secretion System in
- Cellular Superspreaders: An Epidemiological Perspective on HIV Infection inside the Body
- The Inflammasome Pyrin Contributes to Pertussis Toxin-Induced IL-1β Synthesis, Neutrophil Intravascular Crawling and Autoimmune Encephalomyelitis
- Papillomavirus Genomes Associate with BRD4 to Replicate at Fragile Sites in the Host Genome
- Integrative Functional Genomics of Hepatitis C Virus Infection Identifies Host Dependencies in Complete Viral Replication Cycle
- Co-assembly of Viral Envelope Glycoproteins Regulates Their Polarized Sorting in Neurons
- Targeting Membrane-Bound Viral RNA Synthesis Reveals Potent Inhibition of Diverse Coronaviruses Including the Middle East Respiratory Syndrome Virus
- Dual-Site Phosphorylation of the Control of Virulence Regulator Impacts Group A Streptococcal Global Gene Expression and Pathogenesis
- PLOS Pathogens
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Venus Kinase Receptors Control Reproduction in the Platyhelminth Parasite
- Dual-Site Phosphorylation of the Control of Virulence Regulator Impacts Group A Streptococcal Global Gene Expression and Pathogenesis
- Severe Acute Respiratory Syndrome Coronavirus Envelope Protein Ion Channel Activity Promotes Virus Fitness and Pathogenesis
- High-Efficiency Targeted Editing of Large Viral Genomes by RNA-Guided Nucleases
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



