-
Články
- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Bmp and Nodal Independently Regulate Expression to Maintain Unilateral Nodal Activity during Left-Right Axis Specification in Zebrafish
In vertebrates, left-right (LR) axis specification is determined by a ciliated structure in the posterior region of the embryo. Fluid flow in this ciliated structure is responsible for the induction of unilateral left-sided Nodal activity in the lateral plate mesoderm, which in turn regulates organ laterality. Bmp signalling activity has been implied in repressing Nodal expression on the right side, however its mechanism of action has been controversial. In a forward genetic screen for mutations that affect LR patterning, we identified the zebrafish linkspoot (lin) mutant, characterized by cardiac laterality and mild dorsoventral patterning defects. Mapping of the lin mutation revealed an inactivating missense mutation in the Bmp receptor 1aa (bmpr1aa) gene. Embryos with a mutation in lin/bmpr1aa and a novel mutation in its paralogue, bmpr1ab, displayed a variety of dorsoventral and LR patterning defects with increasing severity corresponding with a decrease in bmpr1a dosage. In Bmpr1a-deficient embryos we observed bilateral expression of the Nodal-related gene, spaw, coupled with reduced expression of the Nodal-antagonist lefty1 in the midline. Using genetic models to induce or repress Bmp activity in combination with Nodal inhibition or activation, we found that Bmp and Nodal regulate lefty1 expression in the midline independently of each other. Furthermore, we observed that the regulation of lefty1 by Bmp signalling is required for its observed downregulation of Nodal activity in the LPM providing a novel explanation for this phenomenon. From these results we propose a two-step model in which Bmp regulates LR patterning. Prior to the onset of nodal flow and Nodal activation, Bmp is required to induce lefty1 expression in the midline. When nodal flow has been established and Nodal activity is apparent, both Nodal and Bmp independently are required for lefty1 expression to assure unilateral Nodal activation and correct LR patterning.
Published in the journal: . PLoS Genet 7(9): e32767. doi:10.1371/journal.pgen.1002289
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1002289Summary
In vertebrates, left-right (LR) axis specification is determined by a ciliated structure in the posterior region of the embryo. Fluid flow in this ciliated structure is responsible for the induction of unilateral left-sided Nodal activity in the lateral plate mesoderm, which in turn regulates organ laterality. Bmp signalling activity has been implied in repressing Nodal expression on the right side, however its mechanism of action has been controversial. In a forward genetic screen for mutations that affect LR patterning, we identified the zebrafish linkspoot (lin) mutant, characterized by cardiac laterality and mild dorsoventral patterning defects. Mapping of the lin mutation revealed an inactivating missense mutation in the Bmp receptor 1aa (bmpr1aa) gene. Embryos with a mutation in lin/bmpr1aa and a novel mutation in its paralogue, bmpr1ab, displayed a variety of dorsoventral and LR patterning defects with increasing severity corresponding with a decrease in bmpr1a dosage. In Bmpr1a-deficient embryos we observed bilateral expression of the Nodal-related gene, spaw, coupled with reduced expression of the Nodal-antagonist lefty1 in the midline. Using genetic models to induce or repress Bmp activity in combination with Nodal inhibition or activation, we found that Bmp and Nodal regulate lefty1 expression in the midline independently of each other. Furthermore, we observed that the regulation of lefty1 by Bmp signalling is required for its observed downregulation of Nodal activity in the LPM providing a novel explanation for this phenomenon. From these results we propose a two-step model in which Bmp regulates LR patterning. Prior to the onset of nodal flow and Nodal activation, Bmp is required to induce lefty1 expression in the midline. When nodal flow has been established and Nodal activity is apparent, both Nodal and Bmp independently are required for lefty1 expression to assure unilateral Nodal activation and correct LR patterning.
Introduction
In vertebrates the internal organs are positioned asymmetrically along the left-right (LR) axis. For example, in humans, the heart is positioned on the left side, as is the stomach whilst the liver is positioned on the right side. Within organs LR asymmetry also exists. For example, the two lungs appear identical however they are divided into lobes with 3 on the right lung and 2 on the left. Animals with situs inversus totalis (a LR reversal of all organs) have no pathological features [1] however severe medical problems occur in infants with a partial reversal in a subset of organs (situs ambigious or heterotaxia). These heterotaxic phenotypes occur during early embryonic development and can have both genetic as well as environmental causes [2], [3].
A ciliated organ at the posterior end of the embryo is required for LR-axis specification in the embryo. In this LR organ, the node in mouse or Kupffer's vesicle (KV) in zebrafish, cilia rotate and create a directional fluid flow from the right to left side of the embryo. This directional nodal flow induces a unilateral and asymmetric expression of Nodal in the left lateral plate mesoderm (LPM) directing organ laterality. Unilateral expression of Nodal is essential for correct LR-axis specification, a function that has been highly conserved from human to snails [2], [4], [5]. Although unilateral expression of Nodal is highly conserved and essential for LR–axis specification, there is still very little understanding of how this unilateral Nodal expression is initiated by nodal flow and maintained in the LPM.
Nodal is a member of the Tgf-ß superfamily of secreted growth factors. Nodal signaling is activated by the interaction of Nodal ligands with the type I and II Activin receptors and the Cripto coreceptor (reviewed by A.F. Schier [6]). Upon Nodal interaction with its receptor, intracellular Smad2 protein is phosphorylated, which after associating with Smad4 protein is translocated to the nucleus to activate transcription of downstream target genes. Extracellular antagonists such as Lefty and Cerberus can inhibit Nodal signalling either by direct interaction with Nodal or by competing with Nodal for binding to the receptor. The activity of Lefty proteins, Lefty1 and Lefty2, is controlled at the level of transcription. In most tissues Lefty expression is dependent on Nodal signalling [6]. During LR-axis formation in mouse embryos Lefty1 and Lefty2 have reciprocal expression patterns. While Lefty1 is expressed strongly in the presumptive floor plate and only weakly in the left LPM, Lefty2 is expressed strongly in the left LPM and only weakly in the presumptive floorplate [7]. During LR axis formation in zebrafish embryos lefty1 is expressed in the notochord. Only after LR patterning has been established are lefty1 and lefty2 expressed in the left cardiac field [8]. Nodal likely activates its own expression via a positive feedback loop while it also activates expression of its own antagonists Lefty1 and Lefty2. Genetic experiments in mouse demonstrated that Lefty1 is the more important antagonist and is essential for LR-axis formation [9]. It is believed that Lefty1 expression in the midline prevents Nodal from crossing the midline, blocking activation of Nodal signalling in the right LPM. Indeed loss of Lefty1 expression caused the ectopic expression of Nodal and other left-sided genes in the right LPM and resulted in various laterality defects. It has been suggested that Nodal and Lefty maintain the L/R asymmetry by a self-enhancement and lateral-inhibition (SELI) mechanism [10]. With the SELI model it is possible to explain how a small difference between two separated regions is converted into a robust difference through local activation and long-range inhibition [11].
Bmps have been implicated in LR patterning but data on their precise role has been contradictory [12]–[22]. This is partly due to Bmp ligands acting in opposite fashions, depending on the time and place of action during LR-axis specification [13], [16]. Bmp proteins are members of the Tgf-ß superfamily of growth factors. Extracellular antagonists of Bmp signalling are Noggin, Chordin and Follistatin. Upon interaction with their serine/threonine kinase type I and II Bmp receptors, Bmp ligands induce intracellular phosphorylation of Smad1, 5 or 8 proteins [23]. Mouse embryos deficient for the type I Bmp receptor Bmpr1a/Alk3 or Acvr1/Alk2 fail to form mesoderm, which has hampered the study of their role during LR-axis specification [24]–[26].
In the current work we describe the identification of the linkspoot (lin) mutant from a forward genetic screen for laterality mutants. A missense mutation in the bmpr1aa gene is responsible for the LR defect of lin mutant embryos. Due to a genome duplication event, there is a second gene encoding a Bmpr1a (bmpr1ab) in the zebrafish genome. By screening an ENU-mutagenized zebrafish library we identified a nonsense allele in the bmpr1ab gene. Genetic analysis reveals that a reduction in Bmpr1a activity results in left isomerism of the viscera, demonstrating an essential and early role in LR-axis specification. Previous genetic data has provided evidence that Bmp signalling is required to repress Nodal activation in the right LPM but various direct and indirect models have been proposed to explain this activity [12]–[22], [27]. Here we provide evidence that Bmp signalling via Bmpr1a inhibits Nodal activation in the right LPM indirectly by inducing lefty1 expression in the midline, offering a new model of the interactions between Nodal, Bmp and Lefty in induction and maintenance of LR asymmetry.
Results
Identification of the laterality mutant, linkspoot, in a forward genetic screen
From an ENU-mutagenesis screen, we identified a unique mutant, linkspoot (linhu4087), that displayed a reduced ventral tail fin in combination with a heart-specific laterality defect (Figure 1A, 1B, 1E and 1F). At 30 hours post fertilization (hpf), 24.6% (n = 464) of the embryos derived from an incross of two lin heterozygous carriers displayed the small but noticeable reduction of the ventral tail fin (Figure 1A and 1E). Whilst the majority of lin mutant embryos with the ventral tail fin reduction had no other obvious morphological defects and survived to adulthood, 29% (33 out of 114 lin mutant embryos) showed cardiac defects resulting in cardiac failure and death at around 5 days post fertilization (dpf) (Figure 1F, 1G). Examination of the cardiac defect in lin mutant embryos revealed a midline positioning of the heart in contrast to a leftward positioning in wild-type siblings at 28 hpf. Furthermore at 48 hpf, when the heart in wild-type sibling embryos has completed looping toward the right, heart looping in these lin mutant embryos was incomplete (n = 6/8) (data not shown). Despite the aberrant heart looping in almost 30% of the lin mutant embryos, patterning of the myocardium and endocardium was grossly normal. Expression of tbx2b and has2 in the atrioventricular canal myocardium and endocardium, respectively, was comparable between lin mutant and sibling embryos (Figure S1). In addition, bmp4 expression was still restricted (although slightly expanded) to the venous pole, atrioventricular canal and arterial pole (Figure S1).
Fig. 1. Dorsoventral and laterality defects in zygotic and maternal zygotic lin mutant embryos. 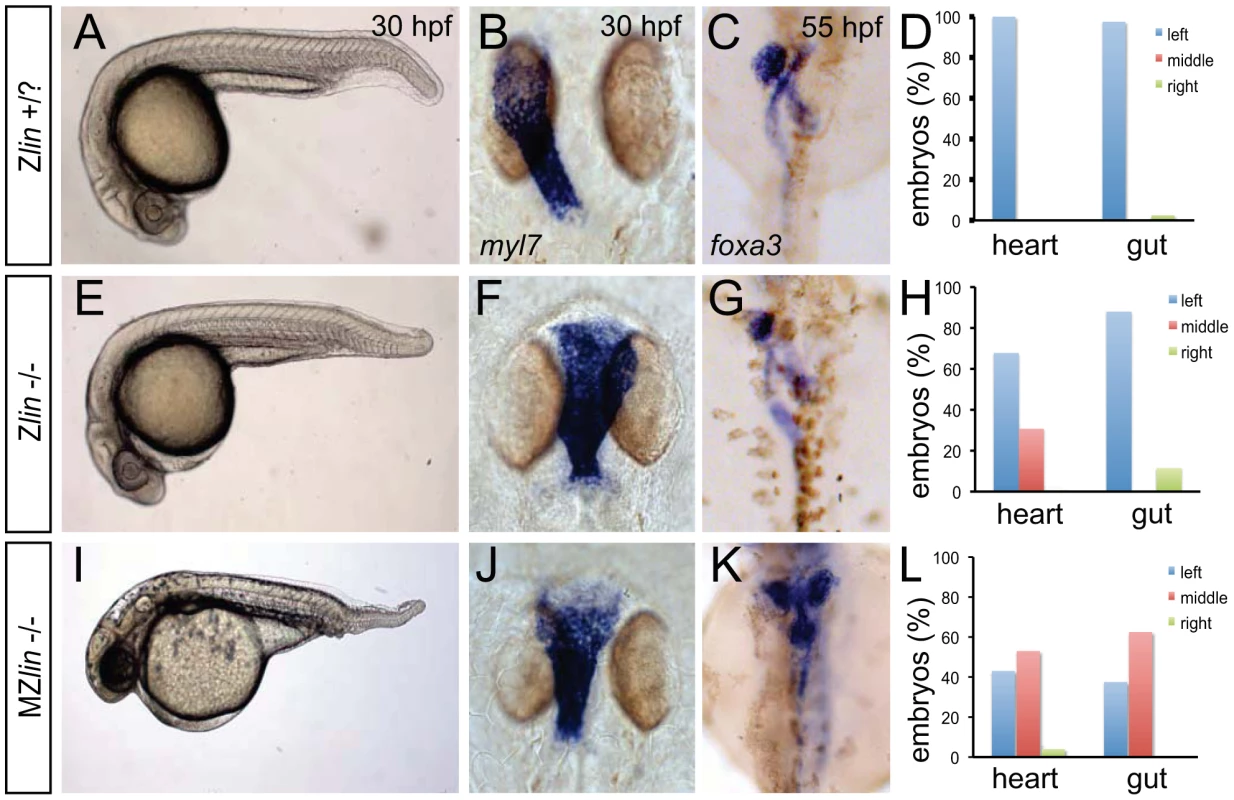
(A–D) Wild-type zygotic lin siblings with normal ventral tail fin (A), left-positioned heart tube (B) and normal organ situs with liver on the left, pancreas on the right and left looped gut tube (C). (D) Quantification of heart position (n = 14) and direction of gut looping (n = 40). (E–H) Zygotic lin (Zlin) mutant embryos displayed a mild reduction of the ventral tail fin (n = 100/108) (E). In addition, in almost 30% of Zlin mutant embryos, the heart tube was positioned at the midline (F). Gut laterality was unaffected in Zlin mutant embryos (G). Quantification of heart position (n = 108) and direction of gut looping (n = 100). (I–L) Maternal zygotic lin (MZlin) mutant embryos derived from a cross of a homozygous lin mutant female and male showing the more severe posterior truncation (I) compared to a Zlin mutant embryo (E). In addition, most MZlin mutant embryos displayed a laterality defect in the heart (J), liver (bilateral, K) and in looping of the gut (K). (L) Quantification of heart positioning (n = 151) and direction of gut looping (n = 16). We observed that the laterality of the other visceral organs (direction of gut looping, positioning of the liver and pancreas) was unaffected in lin mutant embryos (30 out of 34) (Figure 1C, 1D, 1G and 1H). Since lin mutant embryos that did not display the cardiac defects described above survived up to adulthood we crossed homozygous lin mutant females with heterozygous lin carrier males. The resulting maternal and zygotic (MZ) lin mutant embryos displayed a reduction of ventral structures such as the tail fin and blood islands (Figure 1I). Such phenotypes have been associated with aberrant dorsoventral patterning of the embryo [28]. In addition, we observed in MZlin mutants, uncoordinated laterality defects in the viscera (Figure 1J–1L). Aberrant positioning of the heart and other viscera can be caused by defects in formation or function of the Kupffer's vesicle, resulting in disrupted LR patterning. We therefore examined cilia rotation in the KV, and found that both in Zlin and MZlin mutant embryos with a midline positioning of the heart, cilia rotation in the Kupffer's vesicle was unaffected (Figure S2 and Videos S1, S2, S3), suggesting that the laterality defect was not due to a disruption of Kupffer's vesicle function. Together, these results suggest that the affected gene product in lin mutants is required for dorsal-ventral and left-right axis specification.
linkspoot encodes Bmpr1aa
To better understand the molecular nature of the lin mutant phenotype, we positionally cloned the gene that is disrupted in the lin mutant. Using bulk segregant analysis with SSLP markers we placed the lin mutation onto chromosome 13. Mapping of the lin locus using 570 mutant embryos resulted in the identification of a chromosomal region containing a zebrafish orthologue of the mammalian Bmpr1a/Alk3 gene, encoding a Bmp Type I receptor (Figure 2A). Since Bmp signalling is instructive for cardiac laterality as well as ventral tail fin formation [13], [29], [30], we sequenced the coding region of the bmpr1aa gene for mutations. We identified a base pair substitution (T > G) at position 1538 resulting in a leucine to arginine substitution at position 337 (L337R) in the kinase domain of the Bmpr1aa protein (Figure 2B). The T1538G polymorphism was invariably linked with the mutant phenotype (n = 570). No other non-synonymous substitutions were identified in the coding region of bmpr1aa that were linked with the mutant phenotype. Modelling of the corresponding region of human BMPR1A suggested that the L312R (corresponding to zebrafish L337R) substitution is incompatible with proper folding of this region and thereby likely destabilizes the entire kinase domain (Figure 2C, 2D).
Fig. 2. Genetic variations found in zebrafish bmpr1aa and bmpr1ab genes. 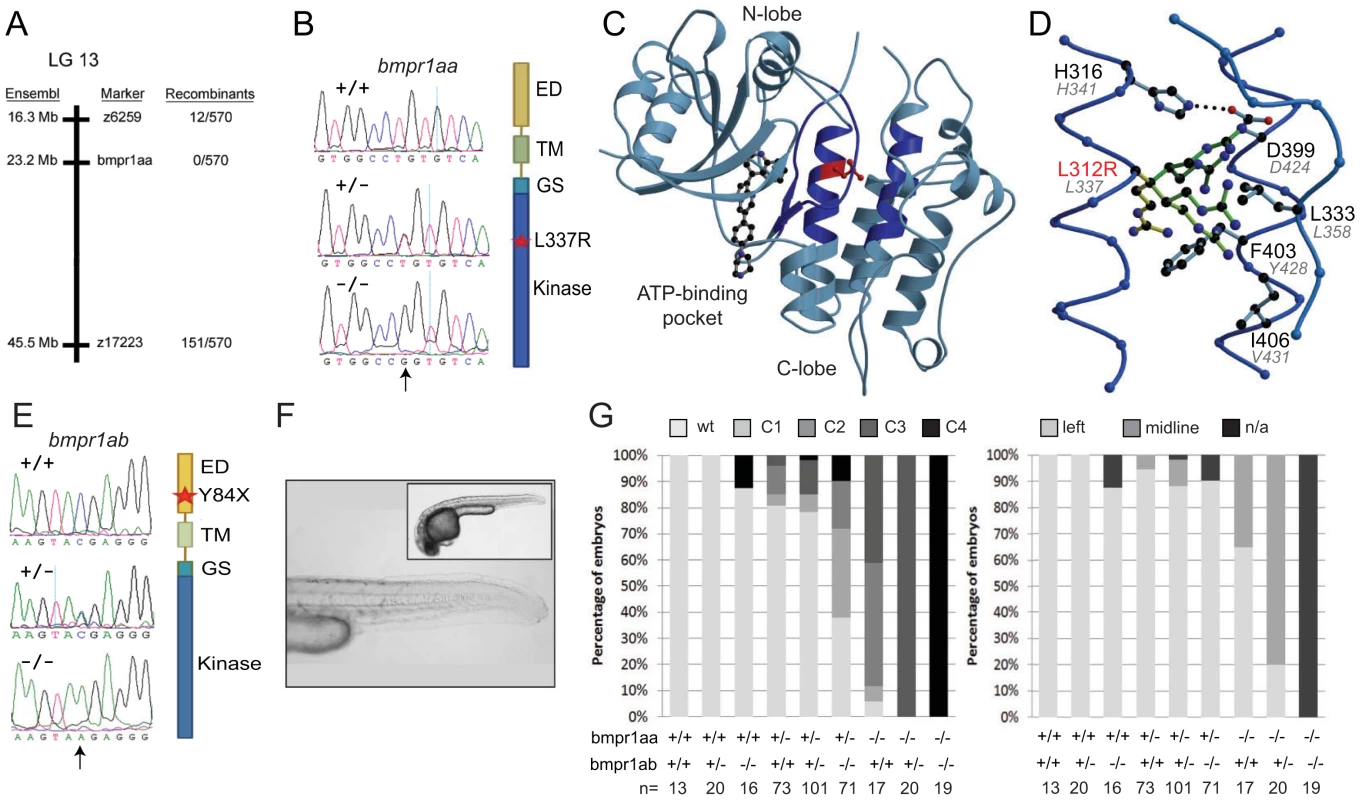
(A) The lin mutation was mapped to a region on chromosome 13 that includes the bmpr1aa gene. (B) T>A basepair change that was found in all lin mutant embryos results in a Leu to Arg change at position 337 (L337R). (C) Crystal structure of human BMPR1B. The kinase domain from the human BMPR1B with the kinase inhibitor LDN-193189 (ball-and-stick representation) bound to the ATP binding site (pdb entry 3MDY). Leu 312 (corresponding to Leu 337 in fish) is shown in red. Structural elements providing residues to the hydrophobic core surrounding Leu 312 are highlighted in dark blue. (D) Detailed view of the hydrophobic core surrounding Leu 312 (in red). Black labels refer to the structure of human BMPR1A, the corresponding residues in fish are indicated by grey italic labels. Consequences of the L312R mutation are analyzed by replacing the leucine side chain in the structure model with arginine, of which five typical rotamers are shown (yellow to green). All rotamers cause serious clashes with surrounding residue, which are highly conserved in fish. (E) C>A basepair change in the bmpr1ab gene that results in a premature stop codon at position 84 in the extracellular domain of the receptor. (F) A MZbmpr1ab mutant embryo at 2 dpf with no obvious phenotypes in the heart or tail region (magnified). (G) Bmpr1a dose-dependent effect on dorsoventral and left-right patterning. Embryos derived from an incross of bmpr1aa+/−;bmpr1ab+/− double carrier fish was analyzed and quantified for the dorsoventral phenotypes (classified as C1 (mild) to C4 (strong)) and position of the heart (left or midline) if present. n/a, not applicable since no heart tissue was present. To address the functional consequence of the L337R substitution, we introduced the lin mutation in the bmpr1aa gene and generated synthetic mRNA for injection into embryos. Surprisingly, injection of wild-type bmpr1aa mRNA into wild-type 1-cell stage embryos resulted in a loss of the ventral tail fin (Table 1). Injection of bmpr1aa L337R mRNA had a stronger inhibition of Bmp signalling since more of the injected embryos displayed a dorsalised phenotype, which was also stronger in its effect (Table 1). These results suggest that increasing wild-type Bmpr1a beyond physiological levels has a negative effect on Bmp signalling, possibly by titrating out other components of the signalling pathway. The dominant-negative effect is stronger for the Bmpr1aa L337R most likely because Bmpr1aa L337R is still able to form a receptor complex and interact with Bmp but it can no longer phosphorylate the receptor Smad protein due to its mutation in the kinase domain. To test this hypothesis we injected a lower dose of the wild-type bmpr1aa mRNA into embryos derived from an incross of two lin heterozygous carriers to determine whether we could rescue the tail fin defects of lin mutant embryos. Indeed we observed that injection of low levels of wild-type bmpr1aa was able to rescue the ventral tail fin defects in almost 50% of lin mutant embryos (Table 1). Consistent with our model that Bmpr1aa L337R has reduced signalling activity, we never observed a rescue of the tail fin defects of lin mutant embryos when we injected the bmpr1aa L337R mRNA. From these results we conclude that the gene that is disrupted in lin mutants encodes the Bmp receptor, Bmpr1aa, and that the lin mutation inactivates Bmpr1aa activity.
Tab. 1. Injection studies. 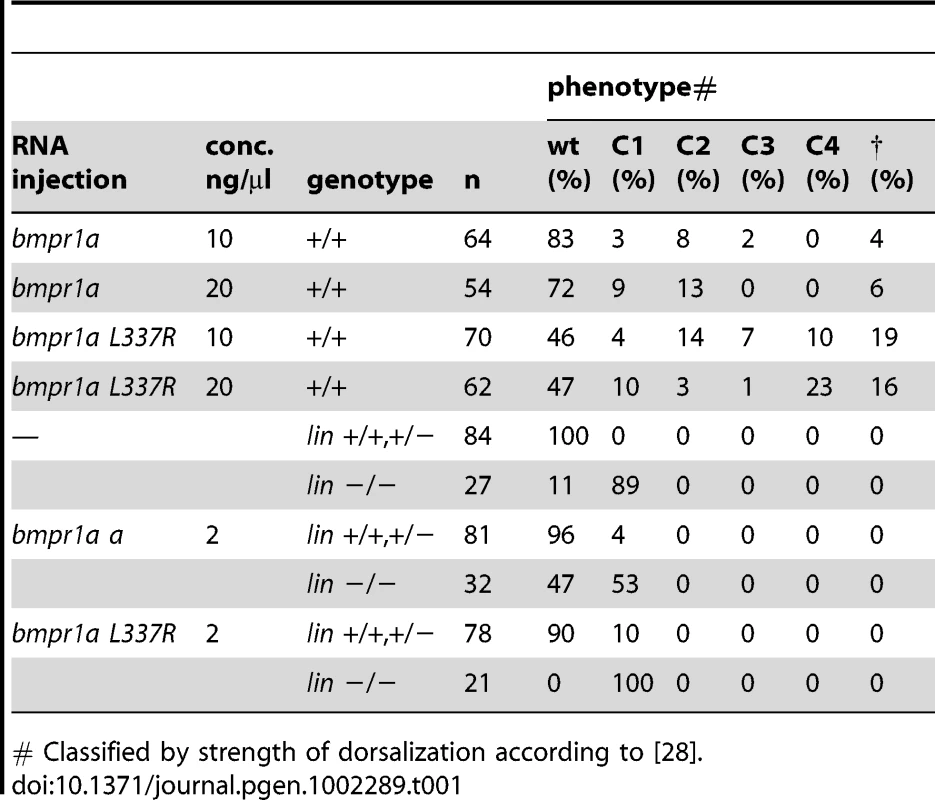
# Classified by strength of dorsalization according to [28]. Bmpr1aa and Bmpr1ab are partially redundant during dorsal-ventral and left-right axis formation
To further characterise the requirement for bmpr1a during zebrafish development, we analysed its expression. Interestingly, database searches revealed that due to a genome duplication event, a paralogue of bmpr1aa existed in the form of bmpr1ab/alk3b (exhibiting 80% identity at the protein level). We, therefore, simultaneously analysed the expression pattern of these two closely related genes. ISH analysis revealed that both bmpr1a paralogues are expressed from the 2-cell stage, indicating maternal deposition of the transcripts (Figure S3). Each paralogue was expressed in a ubiquitous fashion up until the 10-somite stage, however the signal for bmpr1aa was more intense compared to the signal of bmpr1ab suggesting different levels of expression. From 20-somites onwards, the expression of both paralogues became progressively restricted to anterior regions.
The similar expression patterns observed for bmpr1aa and bmpr1ab suggest comparable functions for the paralogues. To analyse this possibility further we screened a mutagenesis library for a bmpr1ab mutant. We identified a mutant harbouring a stop codon (TAC>TAA) in the second exon of the gene, truncating the protein 84 amino acids into the ligand-binding domain (Y84X) (Figure 2E). Although a DV patterning defect was reported upon morpholino knockdown of bmpr1ab [31], we observed no morphological phenotype in the majority of bmpr1ab zygotic mutants. Furthermore, maternal zygotic bmpr1ab mutants exhibited no observable phenotype (Figure 2F).
We next tested for possible redundancy between the two paralogues. By incrossing double heterozygous carriers for the two mutations (bmpr1aa+/−;bmpr1ab+/−), we observed a spectrum of dorsalised embryonic phenotypes, ranging from wild-type phenotypes to C4 dorsalisation in the most severe instances (categorisation according to Mullins et al., [28]) (Figure 2G). Genotyping revealed that the severity of the dorsalisation phenotype correlates with decreasing gene dosage of bmpr1aa and bmpr1ab, with double mutant embryos always exhibiting a C4 dorsalised phenotype. Importantly, this gene dosage effect was also observed on LR patterning, with 80% of embryos of genotype bmpr1aa−/−;bmpr1ab+/ − presenting with a cardiac laterality defect (Figure 2H). Interestingly, loss of the bmpr1aa paralogue affected phenotypic severity more robustly than loss of the bmpr1ab. Unfortunately, we were unable to score the cardiac laterality phenotype of double mutant embryos as no cardiac field was detected in these embryos (Figure S4), consistent with previous observations that Bmp signalling is required for cardiac specification [32], [33]). These results demonstrate that the bmpr1a paralogues play partially redundant roles in both dorsoventral and LR patterning.
Bmp acts upstream of Nodal signalling during left-right patterning
Since no role for Bmp1a in LR axis formation has been reported thus far, we further investigated how Bmp signalling via Bmpr1a regulates LR patterning. We analysed the expression pattern of marker genes whose expression is controlled by LR patterning in embryos derived from an incross of bmpr1aa+/−;bmpr1ab+/ − parental fish. Expression analysis of the Nodal-related gene spaw revealed that embryos that retained at least one wild-type copy of bmpr1aa, displayed normal spaw expression (Figure 3A). However, embryos that had lost both wild-type copy of bmpr1aa and retained at least one wild-type copy of bmpr1ab displayed strong and bilateral expression of spaw in the entire LPM (Figure 3B). On the contrary, in embryos that had lost all wild-type copies of bmpr1aa and bmpr1ab we observed a reduction of spaw expression in the LPM by in situ hybridization (Figure 3C) and quantitative RT-PCR (Figure S5). The bmpr1aa/bmpr1ab double mutant embryos displayed a strong (C4) dorsalised phenotype resulting in a curling of the tail region. Although a Kupffer's vesicle was present in these embryos (data not shown), the structure of the tail is suspected to have physically intervened with the potential of the Kupffer's vesicle to activate and/or propagate spaw expression in the posterior LPM.
Fig. 3. Dose-dependent effect of Bmpr1a on the expression of laterality genes. 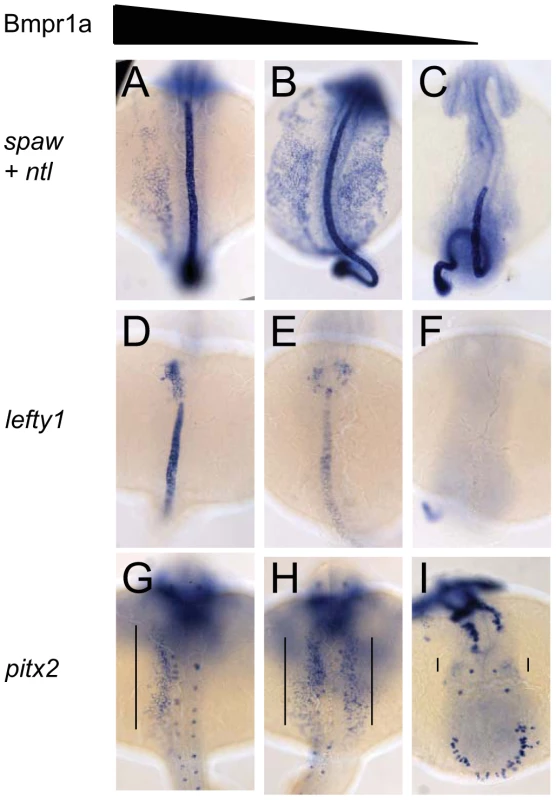
In situ hybridisation at 18-somites for spaw (in LPM) and no tail (ntl) (in midline) (A–C), lefty1 at 23-somites (heart field and midline) (D–F) and pitx2 at 23-somites (in LPM) (G–I). (A,D,G) Embryos selected for normal ventral tail fin or C1 dorsalization (genotypes: bmpr1aa +/+ or +/−; bmpr1ab +/+ or +/− or −/−). (B,E,H) Embryos selected for C3 dorsalization (genotype bmpr1aa−/−;bmp1ab+/−). (C,F,I) Embryos selected for C4 dorsalization (genotype bmpr1aa−/−;bmpr1ab−/−). All embryos are shown as dorsal views with anterior to the top and left to the left. Number of embryos examined is presented in Table 2. Tab. 2. Expression pattern of <i>spaw, lefty1</i>, and <i>pitx2</i> in <i>bmpr1aa/ab</i> genotypes. 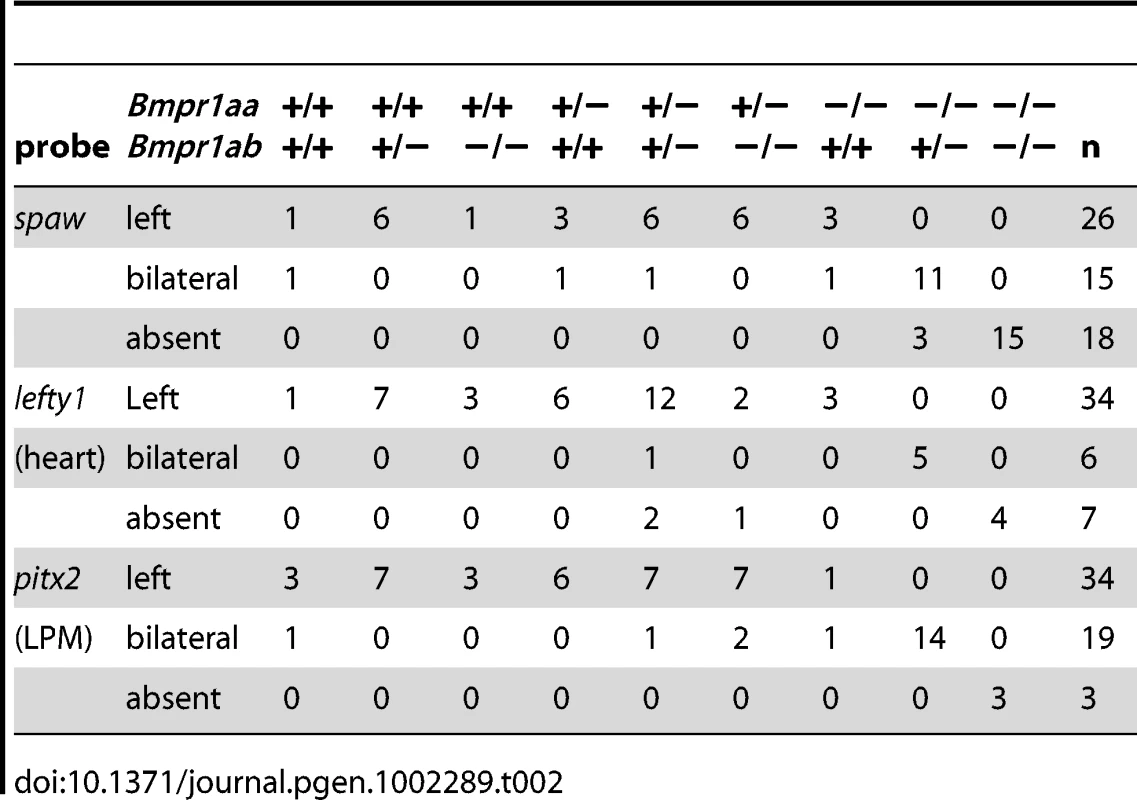
Similar disruptions to asymmetric gene expression were observed upon analysis of lefty1 expression in the cardiac field and pitx2 expression in the gut region. Expression of lefty1 was restricted to the left cardiac field in embryos that retained at least one wild-type copy of bmpr1aa (Figure 3D). Embryos that had lost both wild-type copy of bmpr1aa and retained at least one wild-type copy of bmpr1ab, however, displayed a clear bilateral expression of lefty1 in the cardiac field (Figure 3E). Since embryos without any wild-type bmpr1a gene lack the entire cardiac field, no lefty1 expression was observed in the cardiac region of these embryos (Figure 3F). Furthermore, pitx2 is expressed in the posterior LPM and its expression is regulated by Nodal activity; this expression was unaltered in embryos that still possessed at least one wild-type copy of bmpr1aa (Figure 3G). Consistent with the observed spaw and lefty1 expression, pitx2 expression was also bilateral in the LPM of embryos that had lost both wild-type copy of bmpr1aa and retained at least one wild-type copy of bmpr1ab and was compromised in embryos that had lost all 4 copies of the wild-type bmpr1a gene (Figure 3H, 3I).
These results suggest that during LR patterning Bmp signalling via Bmpr1a regulates Nodal activity. To address the interrelation between Bmp and Nodal signalling we tested the possibility that Nodal acts downstream of Bmp signalling during cardiac laterality. Therefore we attempted to rescue the Bmp-dependent cardiac laterality defect by implanting Nodal-soaked beads in the anterior LPM (ALPM), in order to induce ectopic Nodal signalling. To block Bmp signalling, Tg(hsp70l:nog3) embryos were heat-shocked at 16 hpf which resulted in a cardiac laterality defect in almost all embryos (6 out of 7; Figure 4A–4C). Interestingly, when a Nodal bead was placed in the right ALPM of non-heat-shocked embryos the heart tube was displaced from the left side towards the midline in approximately 50% of the embryos (Figure 4C, 4F). This effect of the Nodal bead was even stronger when the bead was placed in heat-shocked Tg(hsp70l:nog3) embryos. The cardiac tube in such embryos with reduced Bmp signalling was directed towards the right-sided bead in nearly 70% of cases (Figure 4D–4G). In a similar experiment using MZbmpr1aa mutant embryos we again observed that the cardiac tube was directed towards the Nodal bead in 75% of embryos examined (Figure 4H–4J). Together these results suggest that during generation of cardiac laterality Bmp and Bmpr1a act upstream of, or in parallel with, Nodal.
Fig. 4. Rescue of Bmp-related cardiac laterality defects by Nodal beads. 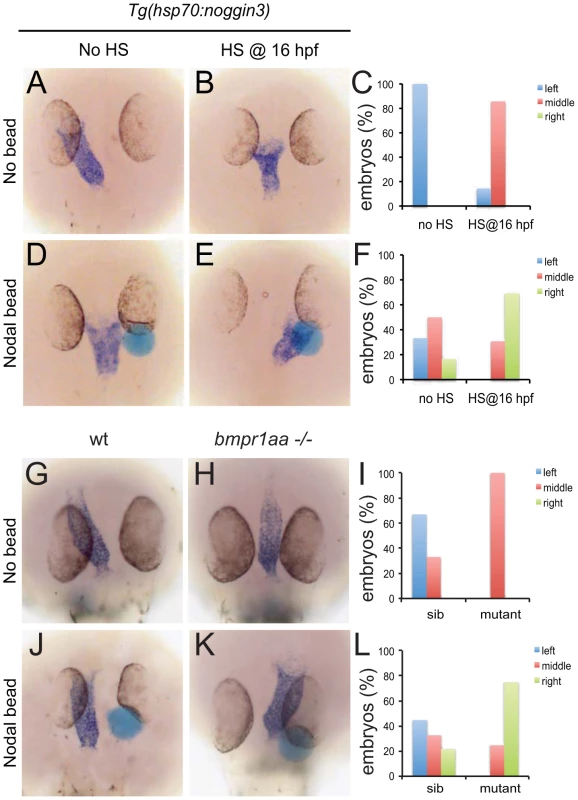
In situ hybridisation for myl7 to highlight the position of the linear heart tube at 30 hpf. Tg(hsp70l:nog3) embryos with no heat-shock (A,D) or heat-shocked at 16 hpf (B,E). Beads (blue) preincubated with recombinant Nodal protein placed in the right ALPM of non-heat-shocked (D) or heat-shocked (E) Tg(hsp70l:nog3) embryos at 17–18 hpf. Control siblings (G,J) or MZbmpr1aa mutant embryos (H,K). Beads (blue) preincubated with Nodal protein placed in the right ALPM of siblings (J) or MZbmpr1aa mutant embryos (K). Position of the inflow pole of the linear heart tube was determined for embryos without a Nodal bead (C) and for embryos in which a Nodal bead was placed on the right side (F). Embryos are shown as dorsal views with anterior to the top and left to the left. Expression of lefty1 in the midline is regulated by Bmp
Our observation that spaw is ectopically expressed in the right LPM mesoderm in embryos that had lost 2–3 copies of their wild-type bmpr1a indicated that Bmpr1a is normally required to repress spaw expression in the right LPM. For this to be a direct effect of Bmp signalling it is expected that Bmp signalling is elevated in the right LPM, as recently reported studying mouse embryos [17]. Although we previously reported on elevated Bmp activity in the left anterior LPM before the cardiac tube is formed (22-somite stage)[34], we never observed enhanced Bmp activity in the right posterior LPM using an anti phospho-Smad1,5,8 antibody (data not shown). An alternative to the model in which Bmp activity directly regulates spaw expression in the right LPM is a model in which Bmp activity regulates spaw expression in an indirect manner. It is well established that Lefty1 in the midline is required to prevent Nodal protein produced in the left LPM from crossing the midline and inducing Nodal expression ectopically in the right LPM [9]. We, therefore, systematically analysed lefty1 expression in the midline of embryos with a gradual loss of Bmpr1a signalling. Doing so, we observed that embryos with 4 or 3 copies of the wild-type bmpr1a gene displayed normal and robust lefty1 expression in the embryonic midline (Figure 5A). Analysis of lefty1 expression in embryos that had lost 2 or 3 copies of the wild-type receptor gene, we observed an increase in the number of embryos with reduced lefty1 expression levels in the midline. Embryos that had lost all 4 copies of the wild-type bmpr1a gene consistently showed a near loss of all lefty1 expression (Figure 5A).
Fig. 5. Bmp via Bmpr1a regulates lefty1 expression in the midline. 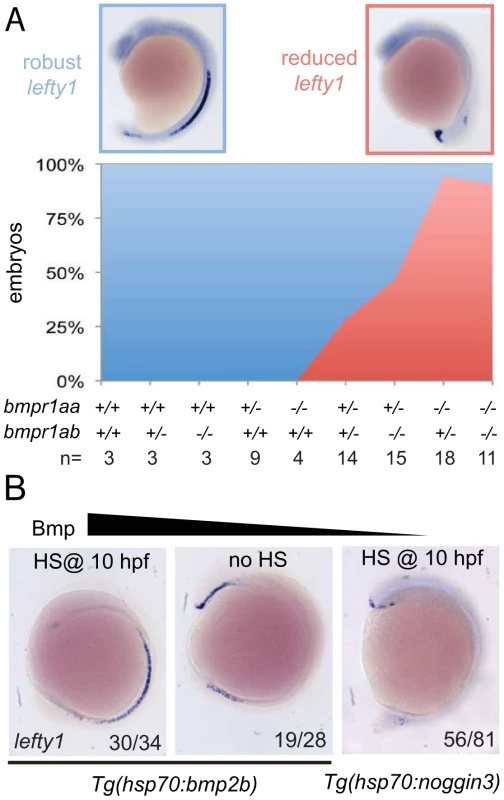
(A) In situ hybridisation for lefty1 at 15-somites on embryos from an incross of bmpr1aa+/−;bmpr1ab+/− double carrier fish. Embryos were analysed for lefty1 expression and classified as robust (blue boxed panel) or reduced (red boxed panel) expression after which the embryos were genotyped. Quantification of the results is shown in the stacked area graph (blue, robust lefty1; red reduced lefty1). (B) In situ hybridisation for lefty1 at 10-somite stage. Embryos shown are Tg(hsp70l:bmp2b) embryos either heat-shocked at 10 hpf to induce bmp2b expression (left panel) or without heat-shock (middle panel) and Tg(hsp70l:nog3) embryos heat-shocked at 10 hpf to inhibit Bmp signalling (right panel). Lateral view of 10-somite stage embryos with dorsal to the right and anterior up. To address whether a disruption of fluid flow in Kupffer's vesicle might explain the reduced lefty1 expression we analyzed the lrrc50hu255h mutant, a loss-of-function allele of a conserved cilia protein that is required for cilia motility [35]. We observed that in the majority of lrrc50hu255h mutant embryos lefty1 was robustly expressed in the midline (Figure S6), suggesting that the observed reduction of lefty1 expression in bmpr1a mutant embryos was not a consequence of a disruption in Kupffer's vesicle function.
To test whether Bmp activity can regulate lefty1 expression in the midline we analysed lefty1 expression in embryos with increased or reduced Bmp activity. We manipulated levels of Bmp signalling by performing heat-shock experiments on embryos carrying the Tg(hsp70l:bmp2b) or Tg(hsp70l:nog3) transgenes, allowing temporally controlled upregulation or downregulation of Bmp signalling, respectively. The embryos were heat-shocked after gastrulation to prevent strong effects on dorsal-ventral patterning due to altered Bmp signalling levels and lefty1 expression was analysed at somitogenesis stages. Consistent with the data from the bmpr1a mutant analysis, we observed an upregulation of lefty1 expression in embryos with ectopic Bmp activity, intermediate levels of lefty1 in wild-type embryos and reduced lefty1 expression in embryos with reduced Bmp activity (Figure 5B). These results demonstrate that Bmp signalling is both required and sufficient for lefty1 expression in the midline.
Bmp and Nodal regulate lefty1 expression independently
Thus far it has been proposed that lefty1 expression in the midline is directly regulated by Nodal protein produced in the LPM [36]. Importantly, our observations demonstrate that lefty1 expression also requires Bmp signalling. Next, we wanted to address whether the regulation of lefty1 by Bmp is Nodal (in)-dependent. The suggestion that Nodal activity itself is not sufficient to induce lefty1 expression in the midline arises from our observation that upon reduction of Bmp signalling, lefty1 expression was reduced while spaw was still strongly expressed in the LPM. In addition, our observation that upon ectopic expression of bmp2b, lefty1 expression is induced while spaw expression is lost goes further to suggests that Bmp signalling can induce lefty1 expression in the absence of Nodal activity. We, therefore, wanted to address whether Bmp signalling regulates lefty1 expression in the midline independent from its regulation by Nodal activity. To investigate further the possibility that Bmp induces lefty1 expression in a Nodal-independent manner, we incubated embryos with the Nodal inhibitor SB431542 from tail-bud stage until the time point of analysis (18 hpf) [37]. As expected, we observed that in wild-type embryos treated with SB431542, spaw expression was compromised in the LPM, indicating the efficiency of the SB431542 treatment in blocking Nodal signalling (data not shown). When wild-type embryos or Tg(hsp70l:bmp2b) that were not subjected to heat-shock (both exhibiting wild-type Bmp levels) were treated with SB431542, we observed a loss of lefty1 expression from the midline. These results demonstrate that Nodal activity is indeed required for lefty1 expression at this stage, which is consistent with previous reports [5], [38]. However, when heat-shock induced Tg(hsp70l:bmp2b) embryos were directly treated with SB431542, lefty1 expression was induced in the midline. From these results we can conclude that in embryos with wild-type Bmp activity, Nodal is essential to drive robust expression of lefty1 in the midline. In addition, these results demonstrate that when Bmp signalling is ectopically activated, lefty1 expression is induced independently of Nodal. When comparing the level of lefty1 induction in heat-shocked Tg(hsp70l:bmp2b) embryos with or without the SB treatment we observed less ectopic lefty1 expression in the presence of the SB inhibitor (comparing Figure 6B and 6D). This result suggests a synergistic effect of Nodal and Bmp on lefty1 expression.
Fig. 6. Bmp and Nodal induce lefty1 independently. 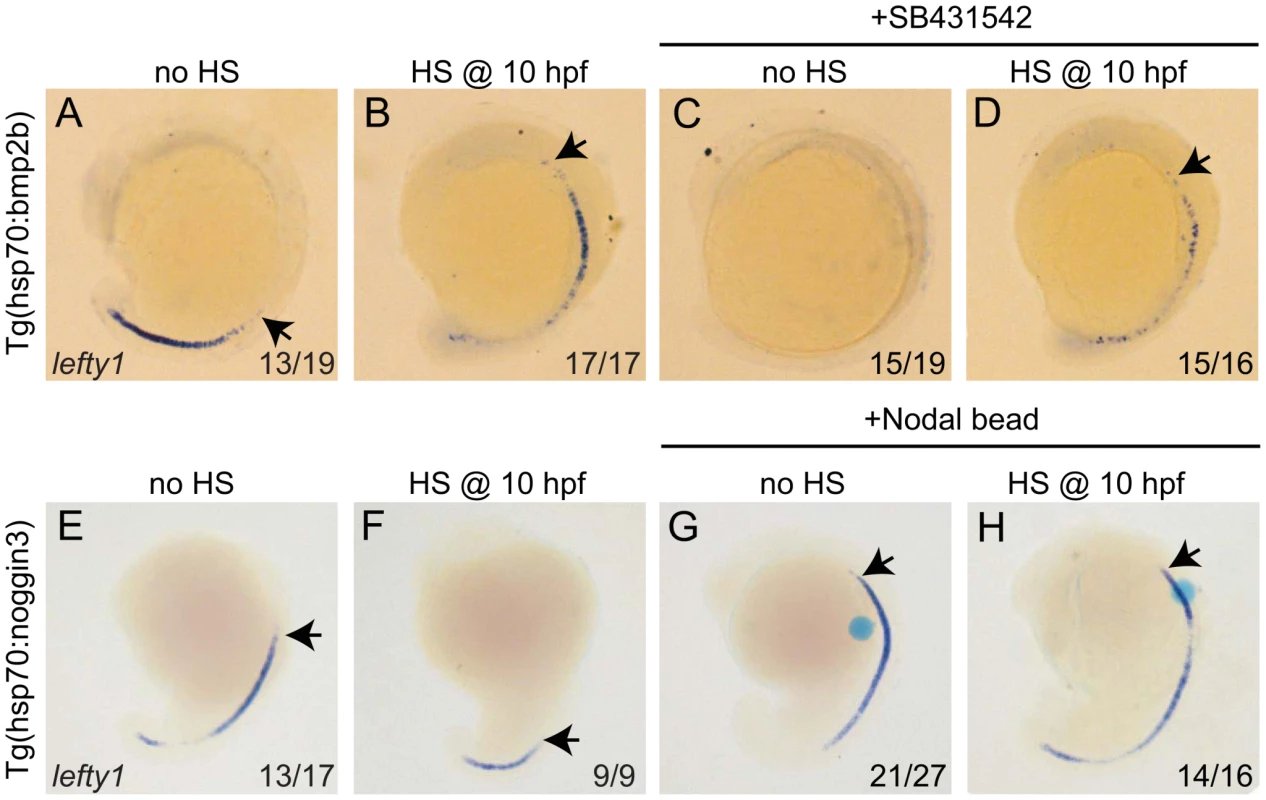
(A–D) Tg(hsp70l:bmp2b) embryos were left at 28°C (A,C) or heat-shocked at 10 hpf to induce bmp2b expression (B,D). A subset of embryos were incubated in the presence of the Nodal inhibitor SB431542 directly after the heat-shock. Embryos were analysed by in situ hybridisation for lefty1 expression at 15-somites. (E–H) Tg(hsp70l:nog3) embryos were left at 28°C (E,G) or heat-shocked at 10 hpf to induce noggin3 expression (F,H). In a subset of embryos a bead preincubated with recombinant Nodal was placed in the ALPM. Embryos were analysed by in situ hybridisation for lefty1 expression at 18-somites. All embryos are shown as lateral views with dorsal to the right and anterior to the top. Arrows point to most anterior lefty1 expression domain. Numbers in lower right represents the number of embryos that displayed the phenotype represented in the panels. Thus far our results suggest that Nodal and Bmp regulate lefty1 expression in the midline independent from each other. To confirm such an independent regulation we tested whether Nodal can regulate lefty1 expression independent from Bmp signalling. To block Bmp activity Tg(hsp70l:nog3) embryos were heat-shocked at tail-bud stage (10 hpf), which resulted in reduced expression of lefty1 in the anterior midline at 18 hpf (Figure 6E, 6F). To induce Nodal in Bmp-depleted embryos, a Nodal bead was placed in the ALPM. As a consequence of Nodal bead implantation we observed restoration of the anterior lefty1 expression even in the absence of Bmp signalling (Figure 6G, 6H). These results demonstrate that Nodal can activate lefty1 expression independent from Bmp and confirm that Bmp and Nodal regulate lefty1 expression independent from each other. Together these results indicate that lefty1 expression is regulated by at least two parallel pathways involving Nodal and Bmp.
Lefty1 is required for the suppression of spaw expression by Bmp
Finally, we wanted to address whether the observed effect of Bmp on spaw expression in the LPM is direct or indirect via its proposed role in regulating lefty1 expression. In Tg(hsp70l:bmp2b) embryos that were heat-shocked at the tail bud stage, we observed a strong down-regulation of spaw expression in the LPM (Figure 7A, 7B), which was coupled with ectopic lefty1 expression (Figure 5B). To test whether the upregulation of lefty1 in the midline was responsible for the downregulation of spaw expression in the LPM, we performed lefty1 knock-down by injecting embryos with a previously published morpholino that effectively targets lefty1 [39]. Interestingly, injection of the lefty1 MO in heat-shock induced Tg(hsp70l:bmp2b) embryos resulted in restoration of spaw expression in the left LPM, with ectopic expression also observed in the right LPM (Figure 7D), similar to non-heat-shocked embryos (Figure 7C). These results demonstrate that lefty1 expression in the midline is required for Bmp to repress spaw expression in the LPM and acts as an intermediary between Bmp signalling and spaw expression.
Fig. 7. Lefty1 is required for Bmp induced repression of spaw. 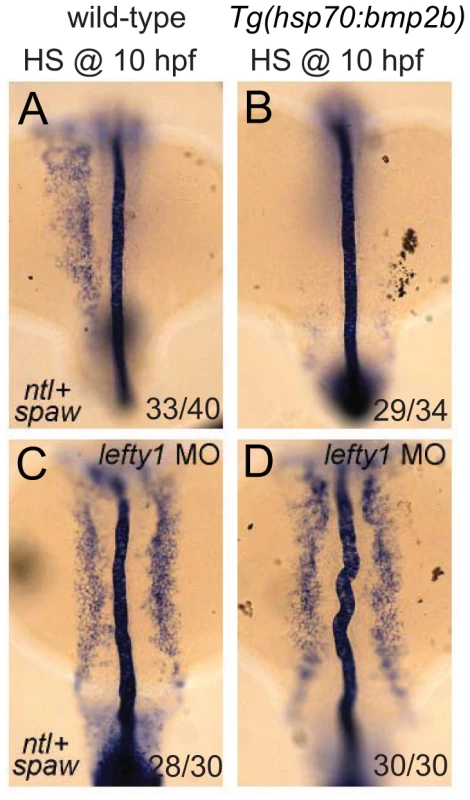
In situ hybridisation of spaw (in LPM) and ntl (in midline) at 18-somites. Wild-type (A,C) or Tg(hsp70l:bmp2b) (B,D) embryos were heat-shocked at 10 hpf to induce bmp2b expression (B,D). Ectopic expression of bmp2b resulted in the loss of spaw expression in the LPM (B). A subset of embryos were injected with a lefty1 MO (C,D), which resulted in bilateral spaw expression even in the presence of ectopic bmp2b (D). Embryos are shown as dorsal views with anterior to the top and left to the left. Discussion
We describe here the identification of two novel zebrafish bmpr1a mutants; a bmpr1aa mutant allele from a forward genetic screen for laterality mutants and a bmpr1ab mutant allele by screening a mutagenized library. By generating and analyzing compound heterozygous and double mutant embryos for bmpr1aa and bmpr1ab, we observed a strong correlation between the number of wild-type bmpr1a gene copies being lost and the severity of the LR patterning defects observed. Most strikingly we observed a shift from the normal unilateral expression of the Nodal-related spaw gene in the left LPM to a bilateral spaw expression in both the left and the right LPM. This shift was accompanied by a reduction in the expression of lefty1 at the midline. This demonstrates that Bmp signalling regulates normal unilateral Nodal activation in the LPM, an observation supported by Nodal bead implantation in the LPM that restored cardiac laterality in Bmp-deficient embryos. Mechanistically our data suggests that there are two parallel pathways, a Bmp and a Nodal dependent pathway, to promote lefty1 expression in the midline and regulate LR patterning (see Figure 8 for proposed model). This model also explains the observation made in several animal models that ectopic Bmp signalling downregulates Nodal activation, suggesting that Bmp signalling is required on the right side to repress Nodal activation [13], [20], [22], [38]. Our data now demonstrates that, at least in zebrafish, this regulation of Nodal activity by Bmp is indirect and depends on the activation of lefty1 expression, as was demonstrated by knock-down of lefty1 in embryos with elevated Bmp signalling (Figure 7). Expanding the previous reaction-diffusion model of an agonist (Nodal) and antagonist (Lefty1), we can now include an additional level of regulation, in which Bmp induces Lefty1, which is required to establish unilateral Nodal activity in the LPM.
Fig. 8. Schematic representation of lefty1 regulation during LR axis specification. 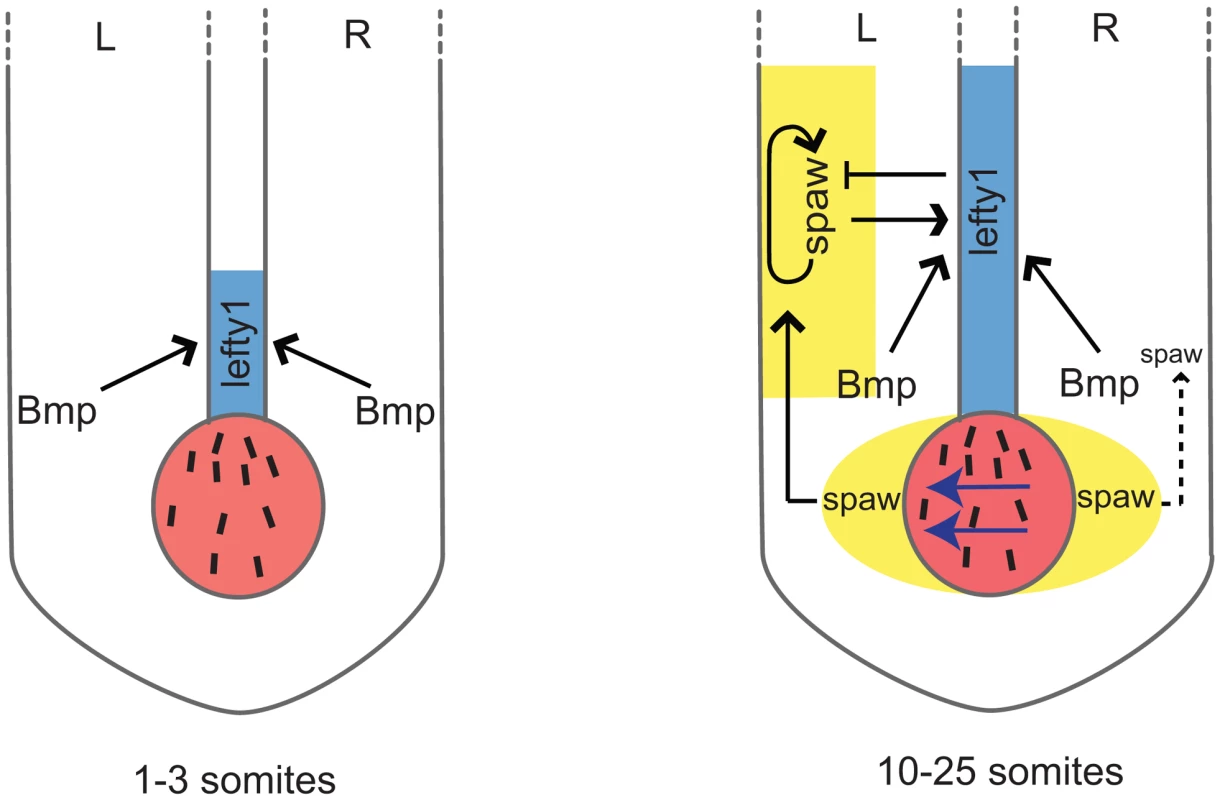
Two phases of lefty1 regulation can be distinguished. i) At the 1–3 somite stage the KV (shown in red) is formed but the nodal flow (indicated by blue arrows) has not yet been initiated. While at this stage lefty1 is already expressed in midline (shown in blue) spaw expression is still absent from the embryo. Thus, this early lefty1 expression is induced independent of Nodal but does depend on Bmp activity. Most likely, robust lefty1 expression is required prior to the initiation of LR axis specification to prevent ectopic activation of spaw in the right LPM later on. ii) At the 5-somite stage spaw expression becomes apparent in the perinode region (yellow area flanking the KV) and from the 10-somite stage onward (up to the 25-somite stage) spaw is expressed unilateral in the left LPM (yellow-boxed area). Our results demonstrate that at this second phase both Spaw/Nodal and Bmp activity are required independently to maintain lefty1 expression in the midline. Lefty1 in the midline antagonises Spaw and prevents it from crossing the midline where it would induce its own expression in the right LPM. Lefty1 is essential for formation of the LR axis [9]. Loss of Lefty1 in mouse embryos results in a left-isomerism, whereby left-sided genes become expressed bilaterally. These described effects are very similar to those observed upon reducing Bmpr1a levels or Bmp signalling in the zebrafish embryo shown here. Others have reported that expression of Lefty1 in the midline is dependent on Nodal activity from the LPM in both zebrafish and mouse embryos [5], [36], [38]. Detailed analysis of the Lefty1 promoter region by Saijoh and colleagues identified a 1.2 kb upstream region of the Lefty1 gene that was sufficient to drive its midline expression [40]. In addition, it was reported that although Foxh1 binding sites are present in this upstream promoter region, these were not required to drive Lefty1 expression in the midline [36]. This suggests that, besides Nodal, additional factors are required for inducing midline Lefty1 expression. Indeed our data demonstrate that during zebrafish LR axis formation, Bmp signalling is required and sufficient to drive lefty1 expression in the midline. Firstly, we found that in mutants with reduced copies of the wild-type bmpr1a gene, lefty1 expression is gradually lost from the midline. Secondly, in transgenic embryos that ectopically express noggin3, a potent Bmp antagonist, lefty1 expression is diminished from the midline while Nodal signalling is still active (indicated by bilateral spaw expression). Thirdly, ectopic activation of the Bmp signalling pathway using a Tg(hsp70l:bmp2b) transgenic results in elevated and ectopic expression of lefty1 in the midline.
Our experiment using ectopic Bmp signalling in the absence of Nodal activity demonstrated that under conditions where Bmp signalling is sufficiently high, Nodal is not required to induce lefty1 in the midline. This might be important during early stages of LR axis formation. Based on the following observations, we hypothesize that at the initiation of LR axis formation, lefty1 expression in the midline is initiated by Bmp signalling independently of Spaw activity. Firstly, in zebrafish embryos lefty1 expression in the midline was observed at the 1–3 somite stage while spaw expression is initiated only at the 5-somite stage in the perinode region and at the 10-somite stage in the LPM ([5] and unpublished observations M. Verhoeven, E. Noël and J. Bakkers). Secondly, this initial lefty1 expression was unaffected by the injection of MOs that efficiently targeted spaw [5]. Thirdly, at these early somite stages expression of Bmp ligands is very strong in the tail bud region [41]. When blocking all Bmp signalling at this early stage in heat-shocked Tg(hsp70l:nog3) embryos, lefty1 expression was indeed not initiated in the midline. Together these results suggest that at the initiation of LR axis formation, lefty1 expression in the midline is initiated by Bmp while the maintenance of lefty1 expression in the midline requires both Nodal and Bmp (see model in Figure 8).
Non-redundant roles for Bmp1a and Acvr1l during LR patterning
Although the zebrafish has been used extensively to identify new regulators by conducting forward genetic screens, there has been very limited success identifying novel mutants displaying LR patterning defects [42], [43]. This might be due to the variability and mixture of the phenotypes that can be observed (situs inversus, situs ambigious or situs solitus) as well as the natural occurrence of these phenotypes in the commonly used wild-type strains. Alternatively, an earlier and essential function of the gene product in embryo development masking any LR defects would hamper the identification of such LR genes. In addition, redundancy with paralogous genes often present in the zebrafish genome can mask the full loss-of-function phenotype. In lin mutant embryos, two copies of the wild-type bmp1aa gene are lost while the two wild-type bmpr1ab copies are still present. The lin/bmpr1aa mutant embryos displayed heart-specific laterality defects (although not fully penetrant) without displaying any gut laterality defects. Previously, we showed temporally distinct requirements for Bmp signalling functions during both LR axis formation and heart morphogenesis [13], [34]. The heart-specific laterality defect of lin/bmpr1aa mutant embryos (eg. loss of leftward cardiac jogging and rightward cardiac looping) is very similar to the cardiac laterality defect previously observed in the lost-a-fin/alk8 mutant. This suggests that during these processes Bmpr1a/Alk3 and Acvrl1/Alk8 play non-redundant functions similar to those described for these receptors during dorsoventral patterning [31]. These results also imply that either the regulation of heart laterality is more sensitive to reducing Bmp signalling activity than the digestive system or that this process is less compensated by wild-type maternal bmpr1aa RNA present in the oocyte. In agreement with the latter suggestion are the observations that bmpr1aa is maternally provided in the oocyte and that maternal zygotic (MZ)lin/bmpr1aa mutant embryos (from surviving lin/bmpr1aa homozygous females) displayed an increase in the strength of the LR patterning defects, including gut laterality defects.
Conservation of the Nodal-Bmp-Lefty1 pathway
To our knowledge, this is the first report describing the requirement for Bmpr1a in regulating LR axis formation. Mouse Bmpr1a mutant embryos do not form mesoderm at embryonic day 7.5 and subsequently die before embryonic day 9.5, preventing the study of LR axis formation in these mutants [26]. Interestingly, the closely related mouse Acvr1/Alk2 gene has been implicated in LR patterning [16]. Since the Acvr1 mutant mouse embryos also die early due to severe gastrulation defects, chimeric embryos were produced and analysed for LR patterning. Depending on the relative contribution of mutant cells to the chimeric embryos, a variety of laterality defects were described. In chimeric embryos with a relative high contribution of Acvr1 mutant cells, bilateral expression of Nodal and Pitx2 in the LPM was observed in combination with reduced expression of Lefty1 in the midline. The phenotypes described for the chimeric embryos with Acvr1 mutant cells corroborate our observations in the Bmpr1a compound heterozygous/mutant embryos, suggesting a conserved role for Bmp type I receptors during LR axis formation.
The Bmp signal that regulates Lefty1 expression in the midline does so independent of Smad1, one of the three Bmp-specific Smad proteins. Although Smad1 inactivation in mouse embryos resulted in the activation of Nodal expression in the right LPM, Lefty expression in the midline was unaffected in such embryos [15]. Alternatively, Smad5 could be responsible for transducing the Bmp signal. Embryos lacking Smad5 no longer express Lefty1 in the midline, which is accompanied by bilateral Nodal and Pitx2 expression in the LPM [12]. Several observations in mouse suggest that during LR axis specification, Bmp signalling can also repress Nodal activation in the right LPM more directly and independently from its regulation of Lefty1 in the midline. As mentioned above, Smad1-deficient embryos showed bilateral Nodal expression while Lefty expression in the midline was reported to be unaffected [15]. In a study by Mine and co-workers, elevated phospho-Smad1,5,8 levels in the right LPM compared with the left LPM of mouse embryos was reported [17]. In addition, an increase on the left side of phospho-Smad1,5,8 levels was observed in Chordin and Noggin double mutant embryos, combined with a loss of Nodal and Lefty1,2 expression. However in Chordin;Noggin double mutant embryos, perinodal Nodal was also reduced and defects in the morphology of the node and the density of cilia were described, suggesting an additional defect in the transduction of a signal from the node to the LPM in such embryos. This defect in communication between the node and the LPM most likely also explains why we observed a complete lack of spaw expression in the LPM of bmpr1aa;bmp1ab double mutant embryos. In zebrafish embryos, we did not observe a stronger phospho-Smad1,5,8 level in the right LPM compared to the left side during LR specification. However, at later stages we did observe the opposite in the anterior LPM where phospho-Smad1,5,8 levels were increased on the left side [34]. In addition, our observation that ectopic Bmp signalling in the Tg(hsp70l:bmp2b) embryos can no longer repress spaw activation in the LPM when Lefty1 is absent makes it very unlikely that such a direct repression of Bmp signalling on spaw expression exists in the zebrafish embryo. Together this indicates that the regulation of lefty1 by Nodal and Bmp during LR axis specification is conserved amongst various vertebrate species. However there are species-specific differences as to what other activities Bmp signalling has during this process. Possibly, differences in geometry or scale of the embryos and speed of their development might require additional regulatory mechanisms to maintain the crucial but very unstable unilateral Nodal activation during LR axis specification.
Materials and Methods
Zebrafish strains and screen
All zebrafish strains were maintained in the Hubrecht Institute using standard husbandry conditions. Animal experiments were approved by the Animal Experimentation Committee (DEC) of the Royal Netherlands Academy of Arts and Sciences. The bmpr1ahu4087 mutant was identified during a forward genetic screen performed at the Hubrecht institute. ENU mutagenesis was performed as previously described for the creation of the Hubrecht Institute target selected mutagenesis library [44]. F1 progeny of mutagenised males were outcrossed to create approximately 300 F2 families, which were then incrossed. F3 progeny were screened for cardiac laterality defects at 28–34 hpf. The bmpr1ahu4087 mutant can be identified using nested PCR with the following primers:
PCR1
Forward primer: AGCTCATCCGGAGAAGTATG
Reverse primer: TCCACTTCATTTGTGTCACTG
PCR2
Forward primer: TGTAAAACGACGGCCAGT ATATGTACCCAGCCCTGATG
Reverse primer: AGGAAACAGCTATGACCAT AGCTTCAGATTCAGATCAACAC
The bmpr1absa0028 mutant was identified from the mutagenesis library at the Sanger institute by screening finclip DNA using nested PCR with the following primers:
PCR1:
Forward primer: CCAGACTACATGCTTCATG
Reverse Primer: ATTGTGACAGGCCTACAATG
PCR2:
Forward primer: TGTAAAACGACGGCCAGT CAGAAGATGCCACAAACAAC
Reverse primer: AGGAAACAGCTATGACCATGGTCACACCGAGTAATTTCC
Products were then sequenced with M13F or M13R primers.
Published transgenic lines used were Tg(hsp70ll:nog3)fr14 and Tg(hsp70ll:bmp2b)fr13 [13].
Genetic mapping and genotyping
Meiotic mapping of the linkspoot mutation was performed using standard simple sequence length polymorphisms. The primers used for SSLP can be found on www.ensembl.org.
Morpholino oligo and RNA synthesis
The lefty1 morpholino was described previously [39].
The coding region of the bmpr1aa gene was cloned into pCS2+ by PCR amplification. The lin mutation was introduced in the pCS2+ bmpr1aa construct using the QuickChange kit (Stratagene). In vitro transcription was performed from Acc65I digested template using the SP6 mMessage mMachine kit for all injected mRNA (Ambion).
SB431542 treatment
SB431542 (Sigma) was resuspended in DMSO to a concentration of 10 mM, and subsequently diluted to a working concentration of 150 µM in embryo medium. Control embryos were treated with an equal volume of DMSO. 30 embryos were treated per 5 ml of SB/DMSO solution.
In situ hybridization
In situ hybridization was carried out as previously described [45]. Embryos were cleared in MetOH and mounted in benzylbenzoate/benzylalcohol (2 : 1) before pictures were taken. Riboprobes were generated by transcription from a linearized template in the presence of 11-UTP.
Bead implants
Agarose beads (Affigel blue, BioRad) were rinsed twice in PBS and incubated for 1 hr at 37°C with 50 µg/ml recombinant mouse Nodal protein (R&D systems). Implants were performed as previously described [46].
Supporting Information
Zdroje
1. RamsdellAF 2005 Left-right asymmetry and congenital cardiac defects: getting to the heart of the matter in vertebrate left-right axis determination. Dev Biol 288 1 20
2. BamfordRNRoesslerEBurdineRDSaplakoğluUdela CruzJ 2000 Loss-of-function mutations in the EGF-CFC gene CFC1 are associated with human left-right laterality defects. Nat Genet 26 365 369
3. KuehlKSLoffredoC 2002 Risk factors for heart disease associated with abnormal sidedness. Teratology 66 242 248
4. GrandeCPatelNH 2009 Nodal signalling is involved in left-right asymmetry in snails. Nature 457 1007 1011
5. LongSAhmadNRebagliatiM 2003 The zebrafish nodal-related gene southpaw is required for visceral and diencephalic left-right asymmetry. Development 130 2303 2316
6. SchierAF 2009 Nodal morphogens. Cold Spring Harb Perspect Biol 1 a003459
7. MenoCItoYSaijohYMatsudaYTashiroK 1997 Two closely-related left-right asymmetrically expressed genes, lefty-1 and lefty-2: their distinct expression domains, chromosomal linkage and direct neuralizing activity in Xenopus embryos. Genes Cells 2 513 524
8. ThisseBPflumioSFurthauerMLoppinBHeyerV 2001 Expression of the zebrafish genome during embryogenesis. ZFIN Direct Data Submission
9. MenoCShimonoASaijohYYashiroKMochidaK 1998 lefty-1 is required for left-right determination as a regulator of lefty-2 and nodal. Cell 94 287 297
10. NakamuraTMineNNakaguchiEMochizukiAYamamotoM 2006 Generation of robust left-right asymmetry in the mouse embryo requires a self-enhancement and lateral-inhibition system. Dev Cell 11 495 504
11. MeinhardtHGiererA 2000 Pattern formation by local self-activation and lateral inhibition. Bioessays 22 753 760
12. ChangHZwijsenAVogelHHuylebroeckDMatzukMM 2000 Smad5 is essential for left-right asymmetry in mice. Dev Biol 219 71 78
13. ChocronSVerhoevenMCRentzschFHammerschmidtMBakkersJ 2007 Zebrafish Bmp4 regulates left-right asymmetry at two distinct developmental time points. Dev Biol 305 577 588
14. FujiwaraTDehartDBSulikKKHoganBLM 2002 Distinct requirements for extra-embryonic and embryonic bone morphogenetic protein 4 in the formation of the node and primitive streak and coordination of left-right asymmetry in the mouse. Development 129 4685 4696
15. FurtadoMBSollowayMJJonesVJCostaMWBibenC 2008 BMP/SMAD1 signaling sets a threshold for the left/right pathway in lateral plate mesoderm and limits availability of SMAD4. Genes & Development 22 3037 3049
16. KishigamiSYoshikawaS-ICastranioTOkazakiKFurutaY 2004 BMP signaling through ACVRI is required for left-right patterning in the early mouse embryo. Dev Biol 276 185 193
17. MineNAndersonRMKlingensmithJ 2008 BMP antagonism is required in both the node and lateral plate mesoderm for mammalian left-right axis establishment. Development 135 2425 2434
18. Monsoro-BurqALe DouarinNM 2001 BMP4 plays a key role in left-right patterning in chick embryos by maintaining Sonic Hedgehog asymmetry. Mol Cell 7 789 799
19. PiedraMERosMA 2002 BMP signaling positively regulates Nodal expression during left right specification in the chick embryo. Development 129 3431 3440
20. Rodríguez EstebanCCapdevilaJEconomidesANPascualJOrtizA 1999 The novel Cer-like protein Caronte mediates the establishment of embryonic left-right asymmetry. Nature 401 243 251
21. SchlangeTArnoldH-HBrandT 2002 BMP2 is a positive regulator of Nodal signaling during left-right axis formation in the chicken embryo. Development 129 3421 3429
22. YokouchiYVoganKJPearseRVTabinCJ 1999 Antagonistic signaling by Caronte, a novel Cerberus-related gene, establishes left-right asymmetric gene expression. Cell 98 573 583
23. ShiYMassaguéJ 2003 Mechanisms of TGF-ß signaling from cell membrane to the nucleus. Cell 113 685 700
24. GuZReynoldsEMSongJLeiHFeijenA 1999 The type I serine/threonine kinase receptor ActRIA (ALK2) is required for gastrulation of the mouse embryo. Development 126 2551 2561
25. MishinaYCrombieRBradleyABehringerRR 1999 Multiple roles for activin-like kinase-2 signaling during mouse embryogenesis. Dev Biol 213 314 326
26. MishinaYSuzukiAUenoNBehringerRR 1995 Bmpr encodes a type I bone morphogenetic protein receptor that is essential for gastrulation during mouse embryogenesis. Genes & Development 9 3027 3037
27. ZhuLMarvinMJGardinerALassarABMercolaM 1999 Cerberus regulates left-right asymmetry of the embryonic head and heart. Curr Biol 9 931 938
28. MullinsMCHammerschmidtMKaneDAOdenthalJBrandM 1996 Genes establishing dorsoventral pattern formation in the zebrafish embryo: the ventral specifying genes. Development 123 81 93
29. BauerHLeleZRauchGJGeislerRHammerschmidtM 2001 The type I serine/threonine kinase receptor Alk8/Lost-a-fin is required for Bmp2b/7 signal transduction during dorsoventral patterning of the zebrafish embryo. Development 128 849 858
30. SchillingTFConcordetJPInghamPW 1999 Regulation of left-right asymmetries in the zebrafish by Shh and BMP4. Dev Biol 210 277 287
31. LittleSCMullinsMC 2009 Bone morphogenetic protein heterodimers assemble heteromeric type I receptor complexes to pattern the dorsoventral axis. Nat Cell Biol 11 637 643
32. KishimotoYLeeKHZonLHammerschmidtMSchulte-MerkerS 1997 The molecular nature of zebrafish swirl: BMP2 function is essential during early dorsoventral patterning. Development 124 4457 4466
33. SchultheissTMBurchJBLassarAB 1997 A role for bone morphogenetic proteins in the induction of cardiac myogenesis. Genes & Development 11 451 462
34. SmithKAChocronSvon der HardtSde PaterESoufanA 2008 Rotation and asymmetric development of the zebrafish heart requires directed migration of cardiac progenitor cells. Dev Cell 14 287 297
35. van RooijenEGilesRHVoestEEvan RooijenCSchulte-MerkerS 2008 LRRC50, a conserved ciliary protein implicated in polycystic kidney disease. J Am Soc Nephrol 19 1128 1138
36. YamamotoMMineNMochidaKSakaiYSaijohY 2003 Nodal signaling induces the midline barrier by activating Nodal expression in the lateral plate. Development 130 1795 1804
37. InmanGJNicolásFJCallahanJFHarlingJDGasterLM 2002 SB-431542 is a potent and specific inhibitor of transforming growth factor-beta superfamily type I activin receptor-like kinase (ALK) receptors ALK4, ALK5, and ALK7. Mol Pharmacol 62 65 74
38. WangXYostHJ 2008 Initiation and propagation of posterior to anterior (PA) waves in zebrafish left-right development. Dev Dyn 237 3640 3647
39. FeldmanBConchaMLSaúdeLParsonsMJAdamsRJ 2002 Lefty antagonism of Squint is essential for normal gastrulation. Curr Biol 12 2129 2135
40. SaijohYOkiSOhishiSHamadaH 2003 Left-right patterning of the mouse lateral plate requires nodal produced in the node. Dev Biol 256 161 173
41. Martínez-BarberáJPToressonHDa RochaSKraussS 1997 Cloning and expression of three members of the zebrafish Bmp family: Bmp2a, Bmp2b and Bmp4. Gene 198 53 59
42. ChenJNvan BebberFGoldsteinAMSerlucaFCJacksonD 2001 Genetic steps to organ laterality in zebrafish. Comp Funct Genomics 2 60 68
43. ChenJNvan EedenFJWarrenKSChinANüsslein-VolhardC 1997 Left-right pattern of cardiac BMP4 may drive asymmetry of the heart in zebrafish. Development 124 4373 4382
44. WienholdsE 2003 Efficient Target-Selected Mutagenesis in Zebrafish. Genome Research 13 2700 2707
45. ThisseCThisseB 2008 High-resolution in situ hybridization to whole-mount zebrafish embryos. Nat Protoc 3 59 69
46. von der HardtSBakkersJInbalACarvalhoLSolnica-KrezelL 2007 The Bmp gradient of the zebrafish gastrula guides migrating lateral cells by regulating cell-cell adhesion. Curr Biol 17 475 487
Štítky
Genetika Reprodukční medicína
Článek Produces a Long Noncoding RNA and Has General Functions in the Germline, Stem Cells, and BrainČlánek Inference of Relationships in Population Data Using Identity-by-Descent and Identity-by-StateČlánek A Genome-Wide Meta-Analysis of Six Type 1 Diabetes Cohorts Identifies Multiple Associated LociČlánek Retrotransposon-Induced Heterochromatin Spreading in the Mouse Revealed by Insertional PolymorphismsČlánek Essential Roles of BCCIP in Mouse Embryonic Development and Structural Stability of ChromosomesČlánek The Receptor Tyrosine Kinase Alk Controls Neurofibromin Functions in Drosophila Growth and LearningČlánek PCNA Ubiquitination Is Important, But Not Essential for Translesion DNA Synthesis in Mammalian CellsČlánek Genome-Wide Association Study Identifies Four Loci Associated with Eruption of Permanent Teeth
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2011 Číslo 9- Růst a vývoj dětí narozených pomocí IVF
- Délka menstruačního cyklu jako marker ženské plodnosti
- Vztah užívání alkoholu a mužské fertility
- Intrauterinní inseminace a její úspěšnost
- Šanci na úspěšný průběh těhotenství snižují nevhodné hladiny progesteronu vznikající při umělém oplodnění
-
Všechny články tohoto čísla
- Temporal Trends in Results Availability from Genome-Wide Association Studies
- Association of eGFR-Related Loci Identified by GWAS with Incident CKD and ESRD
- Genetic Variants at Chromosomes 2q35, 5p12, 6q25.1, 10q26.13, and 16q12.1 Influence the Risk of Breast Cancer in Men
- Large-Scale Gene-Centric Analysis Identifies Novel Variants for Coronary Artery Disease
- Genetic Association for Renal Traits among Participants of African Ancestry Reveals New Loci for Renal Function
- Transcriptome Kinetics Is Governed by a Genome-Wide Coupling of mRNA Production and Degradation: A Role for RNA Pol II
- Conserved Regulation of p53 Network Dosage by MicroRNA–125b Occurs through Evolving miRNA–Target Gene Pairs
- Heterozygous Mutations of Are Associated with an Increased Risk of Isolated Metopic Craniosynostosis in Humans and Mice
- Study of FoxA Pioneer Factor at Silent Genes Reveals Rfx-Repressed Enhancer at and a Potential Indicator of Esophageal Adenocarcinoma Development
- Cholesterol Metabolism Is Required for Intracellular Hedgehog Signal Transduction
- Produces a Long Noncoding RNA and Has General Functions in the Germline, Stem Cells, and Brain
- Age-Dependent Recombination Rates in Human Pedigrees
- Sequence Conservation and Functional Constraint on Intergenic Spacers in Reduced Genomes of the Obligate Symbiont
- Sex Chromosome Mosaicism and Hybrid Speciation among Tiger Swallowtail Butterflies
- A Negative Feedback Loop That Limits the Ectopic Activation of a Cell Type–Specific Sporulation Sigma Factor of
- Phased Whole-Genome Genetic Risk in a Family Quartet Using a Major Allele Reference Sequence
- Mutations in or near the Transmembrane Domain Alter PMEL Amyloid Formation from Functional to Pathogenic
- Inactivation of Alters Melanosome Shape But Has Only a Subtle Effect on Visible Pigmentation
- Novel Interactions between Actin and the Proteasome Revealed by Complex Haploinsufficiency
- Germline Genetic Variants Disturbing the /LIN28 Double-Negative Feedback Loop Alter Breast Cancer Susceptibility
- Separation of Recombination and SOS Response in RecA Suggests LexA Interaction Sites
- Inference of Relationships in Population Data Using Identity-by-Descent and Identity-by-State
- Misregulation of Scm3p/HJURP Causes Chromosome Instability in and Human Cells
- A Noncoding Point Mutation of Causes Multiple Developmental Malformations and Obesity in Twirler Mice
- The Evolutionarily Conserved Longevity Determinants HCF-1 and SIR-2.1/SIRT1 Collaborate to Regulate DAF-16/FOXO
- A Genome-Wide Metabolic QTL Analysis in Europeans Implicates Two Loci Shaped by Recent Positive Selection
- Bacterial Communities of Diverse Species: Ecological Context of a Host–Microbe Model System
- A Genome-Wide Meta-Analysis of Six Type 1 Diabetes Cohorts Identifies Multiple Associated Loci
- Elongator Complex Influences Telomeric Gene Silencing and DNA Damage Response by Its Role in Wobble Uridine tRNA Modification
- Elevated Proteasome Capacity Extends Replicative Lifespan in
- Genome-Wide Analysis of Heteroduplex DNA in Mismatch Repair–Deficient Yeast Cells Reveals Novel Properties of Meiotic Recombination Pathways
- MicroRNA Predictors of Longevity in
- An Iterative Genetic and Dynamical Modelling Approach Identifies Novel Features of the Gene Regulatory Network Underlying Melanocyte Development
- Atypical AT Skew in Firmicute Genomes Results from Selection and Not from Mutation
- Retrotransposon-Induced Heterochromatin Spreading in the Mouse Revealed by Insertional Polymorphisms
- Genomic Analysis of QTLs and Genes Altering Natural Variation in Stochastic Noise
- The Abnormal Phenotypes of Cartilage and Bone in Calcium-Sensing Receptor Deficient Mice Are Dependent on the Actions of Calcium, Phosphorus, and PTH
- Cell Type–Specific Transcriptome Analysis Reveals a Major Role for and miR-200b in Mouse Inner Ear Morphogenesis
- Essential Roles of BCCIP in Mouse Embryonic Development and Structural Stability of Chromosomes
- IAP1-Mediated Ubiquitylation Controls Activation of the Initiator Caspase DRONC Independent of Protein Degradation
- VANG-1 and PRKL-1 Cooperate to Negatively Regulate Neurite Formation in
- The Receptor Tyrosine Kinase Alk Controls Neurofibromin Functions in Drosophila Growth and Learning
- Comparative and Functional Genomics of PD630 for Biofuels Development
- Identification of Type 1 Diabetes–Associated DNA Methylation Variable Positions That Precede Disease Diagnosis
- PCNA Ubiquitination Is Important, But Not Essential for Translesion DNA Synthesis in Mammalian Cells
- Genetic Effects at Pleiotropic Loci Are Context-Dependent with Consequences for the Maintenance of Genetic Variation in Populations
- Genome-Wide Association Study Identifies Four Loci Associated with Eruption of Permanent Teeth
- Bmp and Nodal Independently Regulate Expression to Maintain Unilateral Nodal Activity during Left-Right Axis Specification in Zebrafish
- Inter-Allelic Prion Propagation Reveals Conformational Relationships among a Multitude of [] Strains
- Emergence and Modular Evolution of a Novel Motility Machinery in Bacteria
- Histone Methyltransferase MET-2 Shields the Male X Chromosome from Checkpoint Machinery and Mediates Meiotic Sex Chromosome Inactivation
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Retrotransposon-Induced Heterochromatin Spreading in the Mouse Revealed by Insertional Polymorphisms
- The Evolutionarily Conserved Longevity Determinants HCF-1 and SIR-2.1/SIRT1 Collaborate to Regulate DAF-16/FOXO
- Genome-Wide Analysis of Heteroduplex DNA in Mismatch Repair–Deficient Yeast Cells Reveals Novel Properties of Meiotic Recombination Pathways
- Association of eGFR-Related Loci Identified by GWAS with Incident CKD and ESRD
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



