-
Články
- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Whole-Genome Sequencing of a Single Proband Together with Linkage Analysis Identifies a Mendelian Disease Gene
Although more than 2,400 genes have been shown to contain variants that cause Mendelian disease, there are still several thousand such diseases yet to be molecularly defined. The ability of new whole-genome sequencing technologies to rapidly indentify most of the genetic variants in any given genome opens an exciting opportunity to identify these disease genes. Here we sequenced the whole genome of a single patient with the dominant Mendelian disease, metachondromatosis (OMIM 156250), and used partial linkage data from her small family to focus our search for the responsible variant. In the proband, we identified an 11 bp deletion in exon four of PTPN11, which alters frame, results in premature translation termination, and co-segregates with the phenotype. In a second metachondromatosis family, we confirmed our result by identifying a nonsense mutation in exon 4 of PTPN11 that also co-segregates with the phenotype. Sequencing PTPN11 exon 4 in 469 controls showed no such protein truncating variants, supporting the pathogenicity of these two mutations. This combination of a new technology and a classical genetic approach provides a powerful strategy to discover the genes responsible for unexplained Mendelian disorders.
Published in the journal: . PLoS Genet 6(6): e32767. doi:10.1371/journal.pgen.1000991
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1000991Summary
Although more than 2,400 genes have been shown to contain variants that cause Mendelian disease, there are still several thousand such diseases yet to be molecularly defined. The ability of new whole-genome sequencing technologies to rapidly indentify most of the genetic variants in any given genome opens an exciting opportunity to identify these disease genes. Here we sequenced the whole genome of a single patient with the dominant Mendelian disease, metachondromatosis (OMIM 156250), and used partial linkage data from her small family to focus our search for the responsible variant. In the proband, we identified an 11 bp deletion in exon four of PTPN11, which alters frame, results in premature translation termination, and co-segregates with the phenotype. In a second metachondromatosis family, we confirmed our result by identifying a nonsense mutation in exon 4 of PTPN11 that also co-segregates with the phenotype. Sequencing PTPN11 exon 4 in 469 controls showed no such protein truncating variants, supporting the pathogenicity of these two mutations. This combination of a new technology and a classical genetic approach provides a powerful strategy to discover the genes responsible for unexplained Mendelian disorders.
Introduction
Elucidation of the molecular bases of Mendelian disease has provided a rich resource for understanding genetic mechanisms, protein functions, the behavior of biological systems and mechanisms of disease [1]–[3]. Despite intense efforts with a variety of approaches, however, human geneticists have so far identified only ∼2,400 genes responsible for Mendelian phenotypes, or about 11% of the total number of protein coding genes in our genome. Currently, OMIM, a catalog of Mendelian disorders [4], lists >1,500 mapped Mendelian disorders for which the gene has yet to be identified, and practicing clinical geneticists know that there is an untold number of families with Mendelian disorders for which a molecular explanation or even clear mapping information has yet to be accomplished. Challenges that prevent harvesting this trove of biomedical information include the rarity of each disorder, small family sizes, reduced reproductive fitness of affected individuals, locus heterogeneity and diagnostic tools that query only a fraction of all biological systems [1]–[3], [5], [6].
The recent development of massively parallel DNA sequencing technologies has reduced the cost and increased the throughput of large-scale sequencing (LSS) and provides a new and potentially powerful way to identify virtually all of the mutations responsible for Mendelian disorders [7]. Indeed, at least two groups have used LSS coupled with hybridization strategies to “capture” the majority of known exons (the “exome”) for protein coding genes to identify genes responsible for three Mendelian disorders [6], [8], [9]. While there are many good reasons to use whole-exome sequencing (WES), including the lower cost (currently ∼5-fold) and the fact that exon variation is the most readily interpreted, it is also clear that WES will miss mutations of interest, including those variants that are either in exons that are not captured, are in non-exonic regulatory regions, or are structural variants. At least 1.4% of the disease variants listed in the Human Gene Mutation Database <http://www.hgmd.cf.ac.uk/ac/index.php> are in regulatory sequences and this is likely to be an underestimate given the traditional strategies used for mutation detection (largely PCR amplification and sequencing of exons). The same database lists about 7.5% of disease variants as structural variants. WES also requires higher average coverage levels than WGS, both because of the variable success of capturing different regions and because of “allelic imbalance” where one allele is preferentially captured over the other. For these reasons we elected to employ a WGS strategy.
Both WES and WGS identify a large number of sequence variants when compared to the reference sequence, making it important to prioritize variants. This can be based upon both the assessment of the likelihood of the variants being functional, especially in the WGS setting (Shianna et al. submitted), and on their frequency in healthy control populations. These approaches were used in the WES study that discovered the gene for Miller syndrome (OMIM 263750) by identifying functional mutations in the same gene in each of four unrelated patients and no controls (6). In addition to these approaches, it also is possible to utilize classical genetic strategies that depend on family structure and inheritance patterns to prioritize certain genomic regions. In our case, we show that it is possible to combine partial linkage information with other criteria for prioritizing variants to identify the genetic basis of a rare autosomal dominant disorder (metachondromatosis, OMIM 156250).
The condition we have studied, metachondromatosis (MC, OMIM 156250), is an autosomal dominant condition characterized by exostoses, commonly of the bones of the hands and feet, and enchondromas of the metaphyses of long bones and iliac crest. It was first described by Maroteaux in 1971, based on clinical observation of the six affected individuals from two families [10]. Shortly thereafter, Lachman reported a young male with enlarging, painless, hard lumps on multiple fingers with concurrent long bone metaphyseal and iliac irregularities, both proven by histopathology to be classic exostoses and enchondroma, respectively [11]. The enchondromas often have a “striated” appearance in radiographs, and in MC both types of lesions typically appear in childhood and may regress or even resolve over several years [10], [11] (Figure 1). This phenomenon likely contributes to the incomplete penetrance that has been described in MC families [11], [12]. The exostoses of MC differ in location, orientation and duration from those observed in a related set of phenotypes known as the hereditary multiple exostoses syndromes (MES I and II, OMIM 133700 and 133701). The exostoses of MESs rarely resolve and can cause permanent deformity [11]–[13]. The osteochondromas of MESs typically point away from the adjacent epiphysis and rarely affect the hands or feet, while those of MC point toward the epiphyses and usually present on the hands and feet [11], [13]. Though palpable, the exostoses of MC may not be calcified and therefore may be radiolucent [14], in part depending on the timing of the clinical exam and radiography in the lifespan of a given lesion. The enchondromas of MC are similar to those of Ollier disease (OMIM 166000, also known as multiple enchondromatosis) but the latter disorder usually lacks exostoses. Mutations in EXT1 and EXT2, located respectively at 8q24 and 11p11–p12, have been identified in 70% of MES cases [15].
Fig. 1. Manifestations of metachondromatosis. 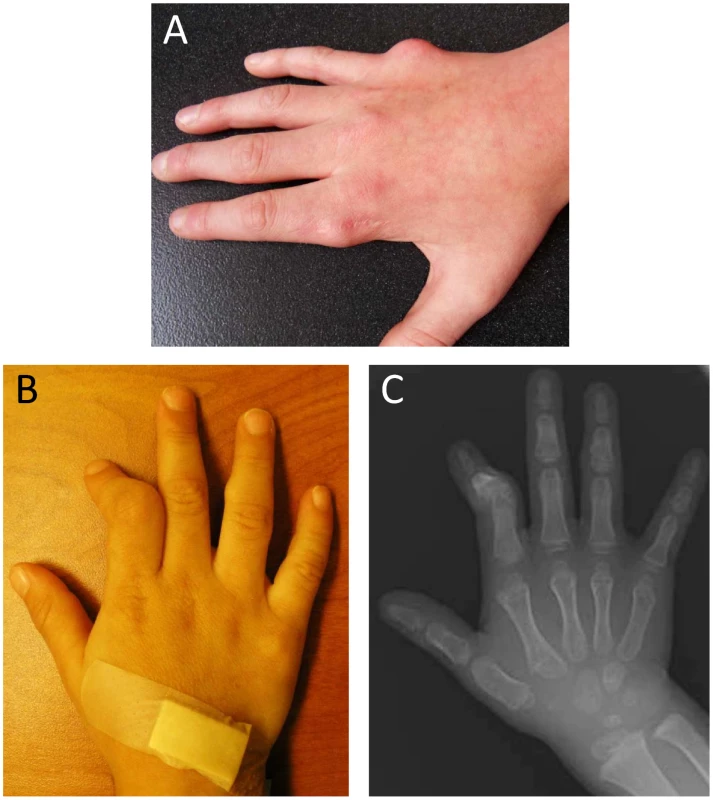
(A) Dorsal view of the right hand of individual III-2 in Pedigree 2 at age 12 years, with residual exostoses following partial surgical removal over the metacarpophalangeal (MCP) joint of the second digit and primary exostoses of MCP of the fifth digit. (B) Dorsal view of the right hand of individual V-1 in Pedigree 1 at three years, showing deformity of the second digit secondary to exostoses of the middle phalanx and an exostoses over the middle phalanx of the fourth digit. (C) Radiograph of the dorsal view of the right hand of individual V-1 in Pedigree 1 at three years. Results
Linkage data
At the outset of our studies, we had access to a single MC family (Pedigree 1) and had not personally examined individual III-1, who was reported to be unaffected. We performed linkage analysis using multiple individuals from Pedigree 1 (Figure 2A) and found six regions with positive LOD scores. Three of these had LOD scores of 1.8 or higher: one at 7p14.1 (39.5–43.1 Mb) showed complete linkage and a LOD of 2.5 (the maximum possible in this family, resulting from perfect co-segregation); two more, 8q24.1 (129.3–141.2 Mb) and 12q23 (106.0–116.4 Mb), were each consistent with one non-penetrant individual and a LOD score of 1.8. Three other regions had LOD scores between 1.0 and 1.5; one was at 2p25 (3.9–10.0 Mb), one at 5q12.1 (60.6–62.5 Mb) and one at 9q31.1–q33.1 (111.1–119.3). We considered these six regions as showing suggestive evidence for the presence of a causal variant, and therefore concentrated attention on the approximately 42 Mb (767 Kb exonic) of included sequence.
Fig. 2. Pedigrees. 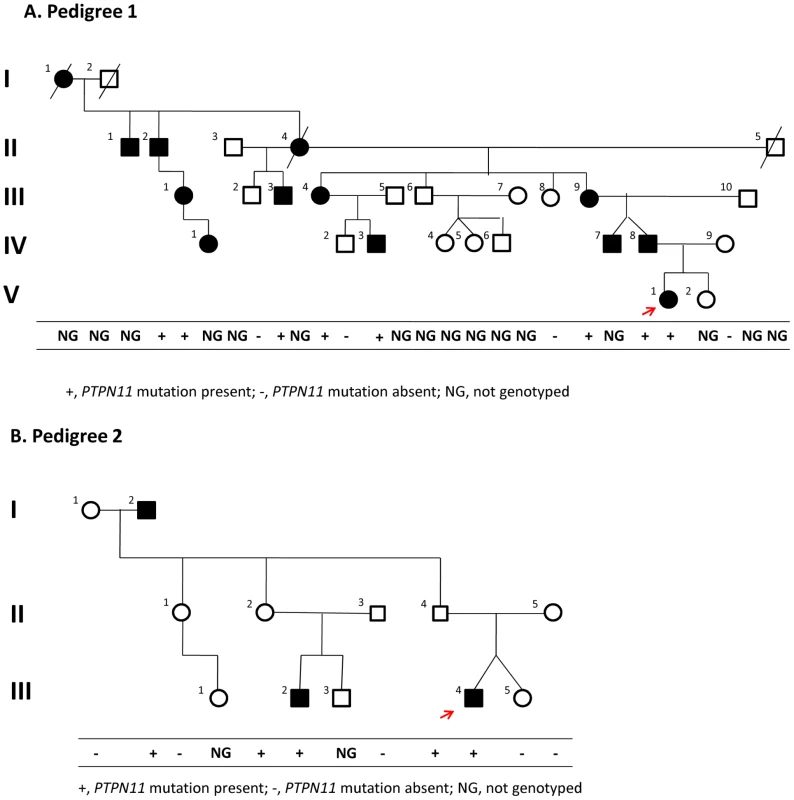
Of families 1 (A) and 2 (B). The red arrow indicates the proband in each family. Segregation of the causal variant with the disease is shown in each family. Although III-1 and IV-1 in pedigree 1 were originally reported to be unaffected, we had the opportunity to examine them during the preparation of this manuscript and found that both had exostoses. Evaluation of sequence data
We performed WGS on a single patient (V-1) from Pedigree 1, to an average of 31.8× coverage. In the 42 Mb of candidate regions defined by our linkage results, 95% of the exonic sequence was covered at a depth of >10×. We also sequenced eight unrelated controls and used data available from dbSNP. In the 7p14.1 (14 RefSeq genes), 8q24.1 region (27 RefSeq genes), 2p25 region (20 RefSeq genes), 5q12.1 region (7 RefSeq genes), and 9q31.1–q33.1 (71 RefSeq genes) regions we found no variants unique to the patient genome with a high likelihood of functional significance (stops gained and frameshifting indels). However, in the 12q23 region (105 RefSeq genes), we identified one frameshifting indel, an 11 bp deletion extending to but not including the 3′ base in exon 4 of PTPN11 (c.514_524del11, see Supplementary Figure 4 in Text S1). This deletion shifts the reading frame, leading to a new sequence of 12 codons followed by a premature stop codon. We verified these results by direct Sanger sequencing of the PTPN11 exon 4 amplicon.
PTPN11 mutations
We confirmed the segregation of the PTPN11 deletion with the affected status in members of Pedigree 1 using a PCR assay (Supplementary Figure 3 in Text S1); all affected members carried the deletion. Additionally, one apparently unaffected individual (III-1), the same individual that was scored as non-penetrant in the linkage analysis, was also heterozygous for the PTPN11 deletion. After obtaining these results, we had the opportunity to examine III-1 and found that she was affected with bilateral internal exostoses of her mandible and that her daughter (IV-1) was also affected with an exostoses of her right proximal tibia.
These WGS results together with the segregation analysis suggested that the PTPN11 deletion was responsible for MC in Pedigree 1. To test this hypothesis, we identified a second family segregating MC as an autosomal dominant trait (Pedigree 2) (Figure 2B). We sequenced all PTPN11 exons and flanking splice sites in individual III-4 of Pedigree 2 and found a heterozygous nonsense mutation, p.R138X, resulting from a C to T transition at position 111,375,382 in exon 4. This mutation abolishes the recognition site for RsaI, and we used PCR amplification of PTPN11 exon 4, followed by restriction of the amplicon with RsaI, to follow the segregation of p.R138X in members of Pedigree 2. All affected individuals were p.R138X heterozygotes, as were two apparently unaffected individuals, II-2 and II-4, who were adults when examined. These results are consistent with the conclusion that this nonsense mutation is responsible for MC in affected individuals in pedigree 2, with non-penetrance in individuals II-2 and II-4, both of whom have affected children.
As a further test of the significance of the mutations we identified, we amplified and successfully Sanger sequenced PTPN11 exon 4 in 469 control, unrelated individuals of whom 60% were of European descent similar to that of our two affected families, 11% were African-American, 11% were East Asian and 18% were of other ethnicities. We found no examples of either of these mutations, nor any other variants in exon 4 predicted to result in the loss of PTPN11 function.
Discussion
PTPN11 encodes the protein tyrosine phosphatase SHP-2, which is an src homology-2 (SH2)-containing protein tyrosine phosphate (PTP) that is highly conserved among metazoans and plays a central role in RAS/MAPK signaling downstream of several receptor tyrosine kinases including EGFR and FGFR [16]. The N-terminal half of SHP-2 contains two SH2 domains (N-SH2 and C-SH2), while the C-terminal half contains the catalytic PTP domain. SHP-2 is ubiquitously expressed. Activation of SHP-2 has a positive effect on RAS/MAPK signal transduction in most contexts. Germline gain of function missense mutations in PTPN11 cause an overlapping but distinct group of dominant disorders with involvement of the face, heart, skeleton, skin and brain, including Noonan syndrome (OMIM 163950), Noonan-like disorder with multiple giant cell lesion syndrome (OMIM 163955) and LEOPARD syndrome (OMIM 151100) [17], [18]. Nonsense mutations in PTPN11 have not been described in humans, but in mice a gene-targeted mutant Ptpn11 allele that deletes codons 46–110 is an early developmental recessive lethal [19], [20]. No phenotype was described in the heterozygotes, but the murine counterpart of MC could easily be overlooked and it would be useful to re-examine Ptpn11+/− mice [20]. The loss of function PTPN11 mutations we report here (c.514_524del11 and p.R138X) are the first to be described in human disease. Not surprisingly, MC has essentially no phenotypic overlap with the other disorders caused by PTPN11 mutations, with the possible exception that one affected individual in Pedigree 2, I-2, a 71-year-old male, has multiple truncal lentigenes (Supplementary Figure 1 in Text S1). How, or if, this is explained by a pathophysiologic overlap with the more extensive and earlier age of onset lentigenes characteristic of the LEOPARD syndrome is not clear.
Incomplete penetrance is a well-described feature of MC and many other dominant disorders and complicates co-segregation tests of candidate causative mutations. Interestingly, in Pedigree 1, individual III-1 was initially reported as unaffected and her status influenced our linkage analysis. However, during the preparation of this manuscript, we had our first opportunity to examine III-1. She has bilateral, internal exostoses of her mandible. Thus, her original classification as non-penetrant was incorrect.
Our results suggest that PTPN11 and other members of the RAS/MAPK pathway should be examined in related and as yet unexplained Mendelian phenotypes such as Ollier's disease, Mafucci syndrome (OMIM 166000) and the trichorhinophalangeal syndrome type II (OMIM150230). Additionally, it is interesting to speculate on the focal nature and limited duration of the enchondromas and exostoses in MC. The local nature and childhood onset of these lesions suggests the possibility that a second mutational event is required for their appearance, a possibility that can be tested by examining PTPN11 in these tumors. The reason for spontaneous regression of some of these lesions is unclear, but may be due to the developmental maturation of the affected tissue.
Our study adds to a small but growing list of examples where genome-wide sequencing approaches have successfully identified rare, high-penetrant risk factors for disease. Ours is one of the first to take the whole-genome approach, although the variant we identified would have been found using either WGS or WES. However, two key distinguishing features of this study are that discovery of the disease-causing variant resulted from the initial sequencing of only a single patient genome and that weak linkage evidence helped to identify regions most likely to harbor the causative variant. The linkage evidence present in this family restricted our search for the causal variant to 42 Mb, which contained only one protein-truncating variant unique to our sequenced case. Without this linkage evidence, sifting through the 109 protein-truncating variants unique to this genome (19 stops gained, 90 frameshifting indels) to find the causal variant would have been a difficult task. It would also have been difficult to sift through the many unique variants in the linked regions if it were not possible to assess the function of all identified variants, whether previously known or novel as the one we found. We also note that generating the necessary sequence data for all genes in the 42 Mb implicated region in the first family would be a daunting task using traditional sequencing approaches. This paradigm of WGS with the prioritization of variants by predicted functional consequence and frequency in controls combined with information gleaned by classical genetic approaches should prove effective for other unexplained Mendelian phenotypes and may also prove effective in the study of more common diseases that show modest linkage evidence.
Materials and Methods
Informed consent was obtained through a Johns Hopkins Medical Institutions IRB approved protocol and a Duke University IRB approved protocol.
Individuals with MC from two families were identified and recruited from the Johns Hopkins Hospital Genetics Clinic. Informed consent was obtained through a Johns Hopkins Medical Institutions IRB approved protocol. Family 1 was a four-generation family comprised of 12 individuals for whom we had DNA. Seven individuals were classified as “affected” and five were “unaffected”. Family 2 was a three-generation family of three “affected” individuals and seven “unaffecteds” for whom we had DNA.
In preparation for interpreting the results of our WGS, we performed linkage analysis on Pedigree 1 (Figure 2A) by genotyping 7 family members using the Illumina HumanHap 550 Genotyping BeadChip v1.0 and 5 family members with Illumina Human 610-Quad v1.0 Genotyping BeadChip. We selected ∼2% of the genotyped SNPs (10,763 autosomal SNPs), requiring that they were heterozygous in at least 4 genotyped individuals, and used them for parametric linkage as calculated with the Merlin linkage analysis software [21] assuming a dominant model with reduced penetrance (set at 0.8). We also analyzed allele sharing and determined non-penetrant individuals using the tools pediSNP and SNPduo (Supplementary Figure 2 in Text S1). Figure 3 shows the results of the linkage analysis across all autosomes.
Fig. 3. Plot of linkage results from Pedigree 1. 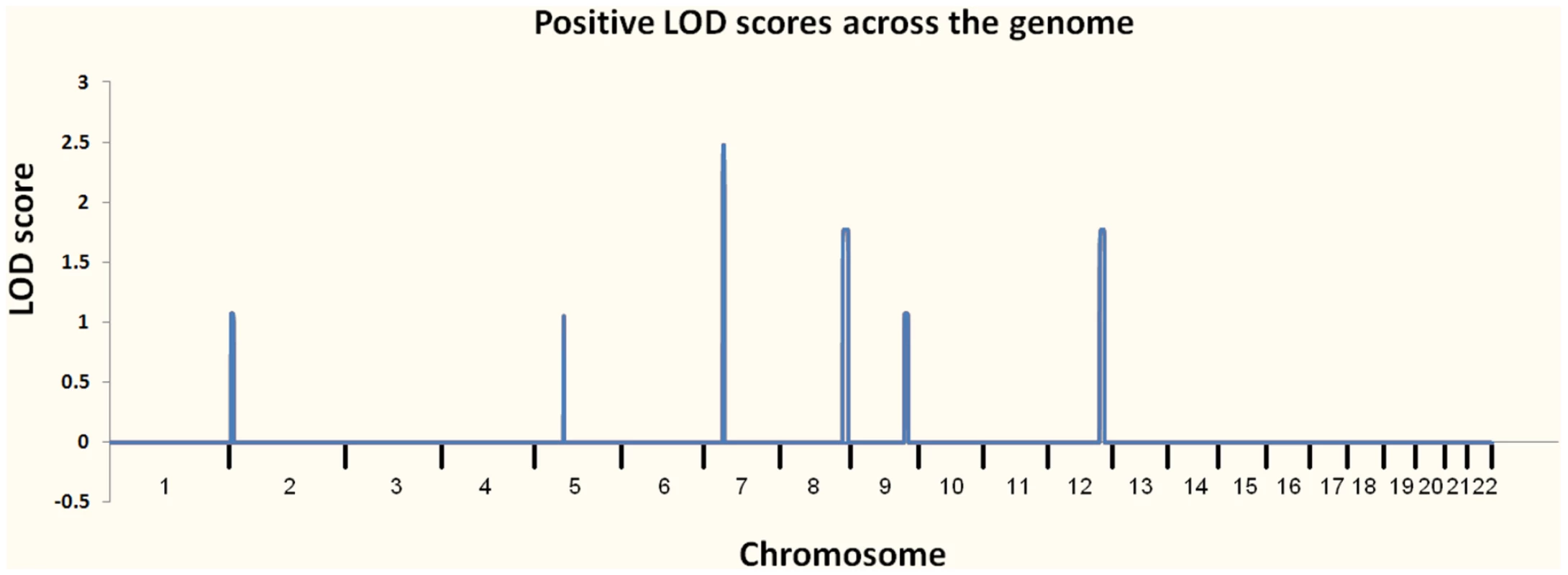
The regions investigated for causal variants were 7p14.1, 8q24.1, 12q23, 2p25, 5q12.1, and 9q31.1–q33.1. We performed WGS on a single individual (V-1) from MC pedigree 1 with an Illumina Genome Analyzer II. The sequence runs were paired-end 75 base pair reads, with 110 billion bases passing the Illumina analysis filter. To assess overall coverage, all gaps (stretches of N's) in the reference genome (Ensembl build 36 release 50) were excluded, resulting in the reference having 2,855,996,286 bases. After accounting for PCR duplicates and reads that did not align to the reference genome, our genomic coverage was 31.8×. We defined a “covered” base as one with at least five reads with a Phred-like consensus score greater than zero. Using these criteria, we covered approximately 97.8% of the reference genome. We then used data from eight control genomes that we had sequenced to an average coverage of 35.9× and dbSNP as filters to remove common variants. To identify candidate pathogenic mutations, we used SVA (http://www.svaproject.org/) to group all variants into functional categories. We expected that the variant responsible for MC would be rare and would not be found in dbSNP or in the control sequenced genomes. We also expected that it would be a severe functional variant, and thus concentrated on those variants that resulted in protein truncation: nonsense mutations and frameshifting indels.
Supporting Information
Zdroje
1. BotsteinD
RischN
2003
Discovering genotypes underlying human phenotypes: Past successes for mendelian disease, future approaches for complex disease.
Nat Genet
S33
228
237
2. AntonarakisSE
BeckmannJS
2006
Mendelian disorders deserve more attention.
Nat Rev Genetics
7
277
282
3. RopersH-H
2007
New perspectives for the elucidation of genetic disorders.
Am J Hum Genet
81
199
207
4. AmbergerJ
BocchiniCA
ScottAF
HamoshA
2009
McKusick's Online Mendelian Inheritance in Man (OMIM).
Nucl Acids Res
37
D793
D796
5. GohKI
CusickME
ValleD
ChildsB
VidalM
2007
The human disease network.
PNAS
104
8685
8690
6. NgSB
BuckinghamKJ
LeeC
BighamAW
TaborHK
2010
Exome sequencing identifies the cause of a mendelian disorder.
Nat Genet
42
30
35
7. MardisER
2008
Next-generation DNA sequencing methods.
Annu Rev Genomics Hum Genet
9
387
402
8. NgSB
TurnerEH
RobertsonPD
FlygareSD
BighamAW
2009
Targeted capture and massively parallel sequencing of 12 human exomes.
Nature
461
272
276
9. ChoiM
SchollUI
JiW
LiuT
TikhonovaIR
2009
Genetic diagnosis by whole exome capture and massively parallel DNA sequencing.
PNAS
106
19096
19101
10. MaroteauxP
1971
Metachondromatosis.
Z Kinderheilkd
109
246
261
11. LachmanRS
CohenA
HollisterD
RimoinDL
1974
Metachondromatosis.
Birth Defects Orig Artic Ser
10
171
178
12. KoslowskiK
ScougallJS
1975
Aust Paediatri J
11
42
45
13. BassettGS
CowellHR
1985
Metachondromatosis: A report of four cases.
J Bone Joint Surg Am
67
811
814
14. KennedyLA
1983
Metachondromatosis.
Radiology
148
117
118
15. BoveeJVMG
HameetmanL
KroonHM
AignerT
HogendoomPCW
2006
EXT-related pathways are not involved in the pathogenesis of dysplasia epipysealis hemimelica and metachondromatosis.
J Pathol
209
411
419
16. TidymanWE
RauenKA
2009
The RASopathies: Developmental syndromes of Ras/MAPK pathway dysregulation.
Curr Opin Genet Devel
19
230
236
17. TartagliaM
GelbBD
2005
Noonan syndrome and related disorders: Genetics and pathogenesis.
Annu Rev Genomics Hum Genet
6
45
68
18. JorgeAAL
MalaquiasAC
ArnholdIJP
MendoncaBB
2009
Noonan syndrome and related disorders: A review of clinical features and mutations in genes of the RAS/MAPK pathway.
Horm Res
71
185
193
19. OishiK
ZhangH
GaultWJ
WangCJ
TanCC
2009
Phosphatase-defective LEOPARD syndrome mutations in PTPN11 gene have gain-of-function effects during Drosophila development.
Hum Mol Genet
18
193
201
20. SaxtonTM
HenkemeyerM
GascaS
ShenR
RossiDJ
1997
Abnormal mesoderm patterning in mouse embryos mutant for the SH2 tyrosine phosphatase Shp-2.
EMBO J
16
2352
2364
21. AbecasisGR
ChernySS
CooksonWO
CardonLR
2002
Merlin - rapid analysis of dense genetic maps using spare gene flow trees.
Nat Genet
30
3
4
Štítky
Genetika Reprodukční medicína
Článek The Met 470 Allele Is Associated with Lower Birth Rates in Fertile Men from a Population IsolateČlánek Copy Number Variation and Transposable Elements Feature in Recent, Ongoing Adaptation at the Locus
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2010 Číslo 6- Růst a vývoj dětí narozených pomocí IVF
- Délka menstruačního cyklu jako marker ženské plodnosti
- Vztah užívání alkoholu a mužské fertility
- Šanci na úspěšný průběh těhotenství snižují nevhodné hladiny progesteronu vznikající při umělém oplodnění
- Vliv melatoninu a cirkadiálního rytmu na ženskou reprodukci
-
Všechny články tohoto čísla
- Translational Selection Is Ubiquitous in Prokaryotes
- Whole-Genome Sequencing of a Single Proband Together with Linkage Analysis Identifies a Mendelian Disease Gene
- Web-Based, Participant-Driven Studies Yield Novel Genetic Associations for Common Traits
- The Met 470 Allele Is Associated with Lower Birth Rates in Fertile Men from a Population Isolate
- Contributions of Status and Allele Expression, But Not Copy Number Variation, to the Control of SIVmac251 Replication in Indian-Origin Rhesus Monkeys
- A Genome-Wide Association Study of Optic Disc Parameters
- Amplification of a Cytochrome P450 Gene Is Associated with Resistance to Neonicotinoid Insecticides in the Aphid
- The Transcription Factor REST Is Lost in Aggressive Breast Cancer
- 14-3-3 Mediates Histone Cross-Talk during Transcription Elongation in
- Copy Number Variation and Transposable Elements Feature in Recent, Ongoing Adaptation at the Locus
- Use of Genome-Wide Expression Data to Mine the “Gray Zone” of GWA Studies Leads to Novel Candidate Obesity Genes
- Genome-Wide RNAi Screen Identifies Multiple Regulators of HIF–Dependent Transcription in Hypoxia
- The CYCLIN-A CYCA1;2/TAM Is Required for the Meiosis I to Meiosis II Transition and Cooperates with OSD1 for the Prophase to First Meiotic Division Transition
- Inactivation of hnRNP K by Expanded Intronic AUUCU Repeat Induces Apoptosis Via Translocation of PKCδ to Mitochondria in Spinocerebellar Ataxia 10
- Mice with Alopecia, Osteoporosis, and Systemic Amyloidosis Due to Mutation in , a Gene Coding for Palmitoyl Acyltransferase
- siRNA–Mediated Methylation of Telomeres
- Chromosome 4 Replicates in Two Phases That Correlate with Chromatin State
- Dynamic Switch of Negative Feedback Regulation in Akt–TOR Signaling
- On the Use of Variance per Genotype as a Tool to Identify Quantitative Trait Interaction Effects: A Report from the Women's Genome Health Study
- and Deficiency Cooperate in the Progression of Mouse Prostate Tumourigenesis
- An Integration of Genome-Wide Association Study and Gene Expression Profiling to Prioritize the Discovery of Novel Susceptibility Loci for Osteoporosis-Related Traits
- Consent and Internet-Enabled Human Genomics
- Understanding Adaptation in Large Populations
- Identification of a Functional Genetic Variant at 16q12.1 for Breast Cancer Risk: Results from the Asia Breast Cancer Consortium
- Evidence that Adaptation in Is Not Limited by Mutation at Single Sites
- The IG-DMR and the -DMR at Human Chromosome 14q32.2: Hierarchical Interaction and Distinct Functional Properties as Imprinting Control Centers
- Cushing's Syndrome and Fetal Features Resurgence in Adrenal Cortex–Specific Knockout Mice
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Web-Based, Participant-Driven Studies Yield Novel Genetic Associations for Common Traits
- The IG-DMR and the -DMR at Human Chromosome 14q32.2: Hierarchical Interaction and Distinct Functional Properties as Imprinting Control Centers
- Amplification of a Cytochrome P450 Gene Is Associated with Resistance to Neonicotinoid Insecticides in the Aphid
- Copy Number Variation and Transposable Elements Feature in Recent, Ongoing Adaptation at the Locus
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



