-
Články
- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
South Asia as a Reservoir for the Global Spread of Ciprofloxacin-Resistant : A Cross-Sectional Study
Stephen Baker and colleagues use whole-genome sequencing and phylogenetic charactarization of a global collection of ciprofloxacin-resistant Shigella sonnei isolates to explore the origins of the recent spread of the antimicrobial-resistant infection.
Published in the journal: . PLoS Med 13(8): e32767. doi:10.1371/journal.pmed.1002055
Category: Research Article
doi: https://doi.org/10.1371/journal.pmed.1002055Summary
Stephen Baker and colleagues use whole-genome sequencing and phylogenetic charactarization of a global collection of ciprofloxacin-resistant Shigella sonnei isolates to explore the origins of the recent spread of the antimicrobial-resistant infection.
Introduction
Diarrheal disease is the second most common cause of mortality in children under the age of 5 y worldwide, equating to approximately 800,000 deaths per year [1]. The recent Global Enteric Multicentre Study (GEMS), a large, prospective, case-control study focusing on mild and severe paediatric diarrheal illnesses in sub-Saharan Africa and South Asia, found that Shigella (a genus of Gram-negative enteric bacteria) were amongst the top four most prevalent diarrhoeal pathogens in these settings [2]. The most recent estimates suggest that Shigella infections account for around 125 million cases of diarrhoea annually, with the majority occurring in children in low-income countries [3]. There are four Shigella species (dysenteriae, boydii, flexneri, and sonnei), but the overwhelming majority of the current global burden is presently caused by S. sonnei and S. flexneri. Present-day international epidemiology of the various Shigella species is particularly intriguing, as S. sonnei is replacing S. flexneri as the most common cause of shigellosis worldwide; this pattern is accentuated in regions undergoing rapid economic development [4,5], where S. flexneri dominated as recently as a decade ago.
Shigella infections are characterised by the invasion and disruption of the epithelial cells lining the gastrointestinal mucosa, resulting in mucous and/or bloody diarrhoeal discharge. Although shigellosis is typically self-limiting, antimicrobial treatment is used to prevent complications, reduce dysenteric discharge, and curb post-symptomatic faecal shedding [6,7]. Consequently, resistance to antimicrobials restricts treatment options, placing vulnerable individuals suffering from shigellosis at increased risk of complications and increasing the likelihood of protracted faecal shedding. One of the current recommended first-line treatments for shigellosis is the fluoroquinolone ciprofloxacin [8]. The fluoroquinolones target the DNA gyrase, a type II topoisomerase that is essential for bacterial DNA replication and transcription [9].
Antimicrobial resistance is an emerging global issue in S. sonnei, with a specific multidrug-resistant (MDR) lineage (III) now dominating internationally. Furthermore, organisms belonging to lineage III appear to be highly proficient at acquiring resistance to additional antimicrobials (including third-generation cephalosporins) when they are introduced into new locations [10]. However, given their common usage and broad spectrum of activity, resistance against the fluoroquinolones is the most concerning. Since the first isolation of S. sonnei with reduced susceptibility to ciprofloxacin in Japan in 1993 [11], ciprofloxacin-resistant S. sonnei have been increasingly reported throughout Asia [12–14]. Furthermore, public health laboratories in several non-Asian countries with low incidences of shigellosis have reported the isolation of ciprofloxacin-resistant S. sonnei, often from individuals reporting recent travel to locations with a high risk of shigellosis [15–17].
Whole-genome sequencing has proven to be the gold standard for tracking the international dissemination of clonal bacterial pathogens [18,19], and we have previously exploited this method to study the phylogenetic structure and spread of S. sonnei at both national and intercontinental levels [10,20]. Hypothesising that Asia was a hub for the recent international spread of ciprofloxacin-resistant S. sonnei, we performed whole-genome sequencing and phylogenetic characterisation of a collection of ciprofloxacin-resistant S. sonnei isolated from within and outside Asia, aiming to explore the origins of this growing international epidemic.
Methods
Ethics Statement
S. sonnei isolates from Bhutan, Thailand, and Vietnam were collected during diarrheal surveillance studies [14]. IRB approval was granted for these studies (including organism characterization) from the Research Ethics Board of Health (REBH), Ministry of Health, Bhutan (Bhutan study), the Walter Reed Army Institute of Research (WRAIR) Institutional Review Board, USA (Bhutan and Thailand studies), the Oxford Tropical Research Ethics Committee (OxTREC), UK, and the Hospital for Tropical Diseases Ho Chi Minh City, Vietnam (Vietnam study). Written (Vietnam) or oral (Thailand and Bhutan) consent was obtained from a parent or guardian at the time of enrolment into the study. The target patient groups for these studies were generally hospitalised children aged less than 5 y residing in close proximity to the study centres. S. sonnei isolates from Cambodia were collected at the Angkor Hospital for Children (AHC) in Siem Reap province from the routine diagnostic laboratory; no patient data were collected, and these organisms were not subject to local or international IRB approval. The ciprofloxacin-resistant S. sonnei from countries outside Asia were collected and characterized by the National Salmonella, Shigella, and Listeria monocytogenes Reference Laboratory, Galway, Ireland, and the Microbiological Diagnostic Unit Public Health Laboratory, Melbourne, Australia. These isolates were generally, but not exclusively, obtained from patients reporting recent travel to countries with a high incidence of shigellosis in Asia (Table 1). As these isolates were from anonymous sources and collected at local public health laboratories, these were not subject to IRB approval, and informed consent was not required.
Tab. 1. The origins of the Shigella sonnei isolates and sequences used in this study. 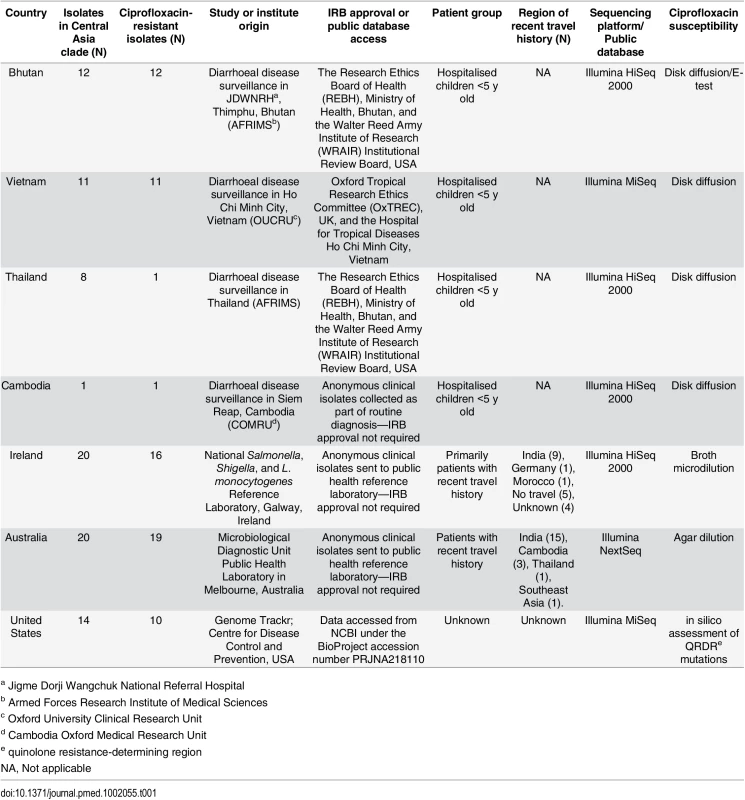
a Jigme Dorji Wangchuk National Referral Hospital Strain Collection
Aiming to investigate the current international upsurge in ciprofloxacin-resistant S. sonnei in detail, we gathered a collection of 60 contemporary ciprofloxacin-resistant S. sonnei from six countries for whole-genome sequencing. The isolates originated from Asian countries with a high incidence of shigellosis (Vietnam, n = 11; Bhutan, n = 12; Thailand, n = 1; Cambodia, n = 1), as well as countries with a low incidence of shigellosis (Australia, n = 19; Ireland, n = 16). Twelve additional ciprofloxacin-susceptible S. sonnei sequences from these settings were also included for phylogenetic context. All strains were isolated independently between 2010 and 2015; details of the isolates used in this study are shown in Table 1.
Susceptibility to ciprofloxacin was determined by disk diffusion, E-test, agar dilution, or broth microdilution, depending on the collaborating institution, and susceptibility breakpoints were interpreted according to the European Committee on Antimicrobial Susceptibility Testing (http://www.eucast.org/clinical_breakpoints). Namely, resistance was determined as strains with a zone of inhibition <19 mm (5 μg disc) and/or a minimum inhibitory concentration (MIC) >1 μg/ml against ciprofloxacin; the various location-specific methods and resulting data are described in Table 1.
Genome Sequencing and Analysis
All isolated S. sonnei were subcultured and subjected to DNA extraction prior to whole-genome sequencing on various Illumina platforms to produce pair-ended short read sequences; the specific sequencing system and the resulting public database numbers are shown in Table 1. We additionally included 14 S. sonnei sequences from the NCBI database under the Bioproject accession number PRJNA218110. These organisms were isolated in the US and deposited as part of the GenomeTrackr Project. All sequences were mapped to the S. sonnei Ss046 reference sequence (Accession number: NC_007384) using SMALT (version 0.7.4), and SNPs were called against the reference and filtered using SAMtools [21]. To contextualize all ciprofloxacin-resistant S. sonnei within the global phylogeny, we appended our collection to include 133 publicly available sequences from a previous global analysis (accession ERP000182) [20]. Previously characterized mobile genetic elements and putative recombination (predicted using Gubbins) were removed [20], resulting in a gap-free alignment of 211 non-duplicate pseudo-whole genome sequences of 4,738 SNPs. A whole-genome phylogeny was inferred from this alignment using RAxML v8.1.3 under the GTRGAMMA substitution model, and sufficient bootstrap replicates were determined automatically using the extended majority rule (MRE) bootstrap convergence criterion. Bootstrap values >75% signify strong statistical support for a node within a maximum likelihood phylogeny, thus indicating that all isolates falling within that lineage are highly likely to be linked in evolutionary history at a more recent time than those falling outside of that lineage. In order to obtain a refined phylogenetic structure of the Central Asia clade, we applied the aforementioned approach to a set of 97 S. sonnei sequences (86 novel sequences and 11 historical sequences) belonging to this clade. This resulted in an alignment of 1,121 SNPs, which was used for phylogenetic inference. De novo assemblies were generated for each read set using Velvet and VelvetOptimiser, and read sets were mapped back to each assembly [22]. A manually curated database based on ResFinder [23] was mapped against each assembly to identify mobile resistance genetic determinants in all ciprofloxacin-resistant strains.
Results
Fluoroquinolone-Resistant S. sonnei in a Global Context
We constructed a whole genome phylogeny of S. sonnei, incorporating sequences from 133 globally representative isolates and 86 novel isolates from Vietnam, Cambodia, Thailand, Bhutan, Australia, Ireland, and the US. The novel sequences included 60 from ciprofloxacin-resistant (MIC >1 μg/ml or zone of inhibition <19 mm) organisms and 26 from ciprofloxacin-susceptible organisms (or of unknown ciprofloxacin susceptibility isolated in the US). The related metadata for the bacterial isolates are shown in S1 Table. The overall tree topology reflected the previously described global phylogenetic structure [20], confirming the presence of four distinct lineages (I, II, III, and IV); lineage III was the most commonly represented and the most widely geographically distributed (Fig 1A). All ciprofloxacin-resistant S. sonnei formed a single, well-supported monophyletic clade within the Central Asian expansion of lineage III (Central Asia III), an MDR group that is closely related but distinct from the Global III clade (Fig 1A and 1B).
Fig. 1. The phylogenetic structure of ciprofloxacin-resistant Shigella sonnei in an international context. 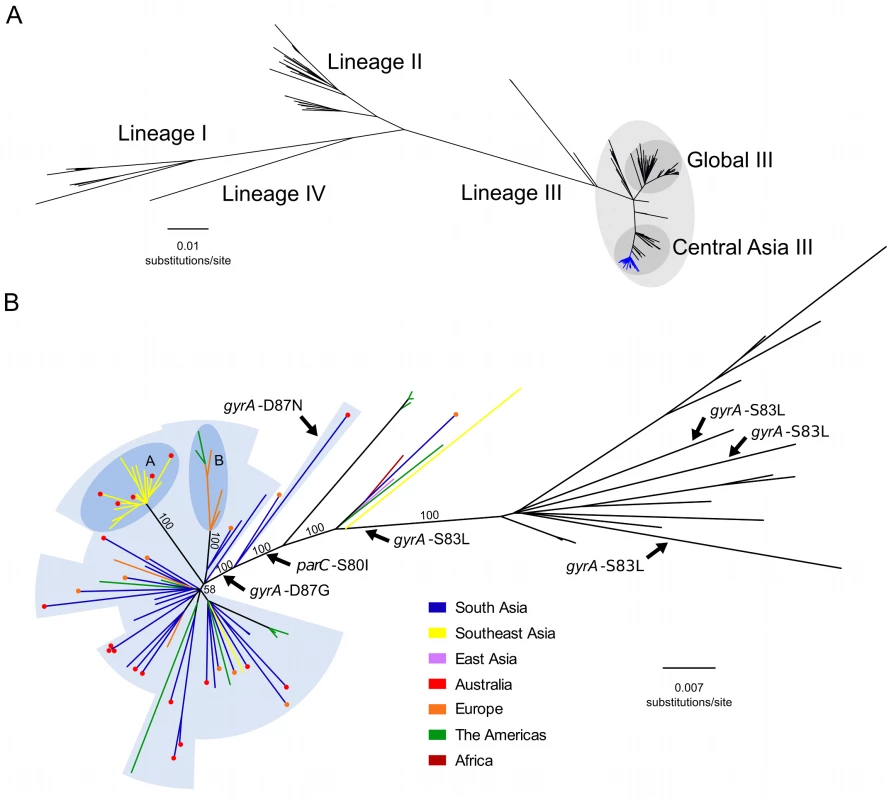
A) Unrooted maximum likelihood phylogeny of 211 globally representative S. sonnei, including 60 sequences from ciprofloxacin-resistant isolates (highlighted by the blue branches). Major lineages are indicated by numbers (I, II, III, and IV) as defined in Holt et al. 2012, with clades Global III and Central Asia III within lineage III highlighted. Horizontal scale bar indicates the number of nucleotide substitutions per site. B) Unrooted maximum likelihood phylogeny of Central Asia III, composed of 97 S. sonnei sequences. Branch colours indicate region of isolation (where no travel history is confirmed) or region of recent travel (where travel history was confirmed) according to the keys. For isolates with confirmed recent travel, a coloured circle at the tip indicates the region where the isolate was collected (multiple coloured circles are indicative of multiple isolates). Labelled arrows indicate branches where the mutations gyrA-S83L, gyrA-D87N, gyrA-D87G, and parC-S80I have arisen. Blue background shading denotes isolates exhibiting ciprofloxacin resistance conferred by triple mutations (gyrA-S83L, parC-S80I, and gyrA-D87G [or gyrA-D87N]). Subpopulations A and B are highlighted in the darker blue shaded areas, denoting clonal expansions in Southeast Asia and Europe/America, respectively. Numbers above major branches indicate bootstrap support values, and horizontal scale bar denotes the number of nucleotide substitutions per site. The Emergence of a Fluoroquinolone-Resistant S. sonnei Clone
We next performed a more detailed phylogenetic reconstruction of the Central Asia III clade, incorporating sequence data from the 60 phenotypically ciprofloxacin-resistant isolates and 26 others (ciprofloxacin-susceptible or of unknown ciprofloxacin susceptibility), along with 11 historical Central Asia III sequences sourced from our previous global study (Fig 1B) [20]. The majority of the Central Asia III isolates carried more than three antimicrobial resistance genes, encoding resistance to a wide range of first-line antimicrobials including tetracycline (tetA), streptomycin (strAB), and co-trimoxazole (dfrA1 and sul2), as previously described [20]. No plasmid-mediated quinolone resistance genes (qnr, qepA, oxqAB, aac(6’)lb-cr) were detected in the sequences of any of the ciprofloxacin-resistant S. sonnei. We additionally examined the genome sequences for mutations in the quinolone resistance-determining region (QRDR) within the DNA gyrase gene (gyrA) and the topoisomerase IV gene (parC), the regions encoding the target residues for fluoroquinolones. Overlaying these mutations on the phylogenetic tree indicated that the gyrA-S83L mutation, the first sequential mutation that confers reduced susceptibility against fluoroquinolones, has arisen independently within the Central Asia III clade on at least four separate occasions (Fig 1B). Amongst the isolates examined here for the first time, extensive resistance to ciprofloxacin can be attributed to a single clonal emergence event, via the sequential accumulation of gyrA-S83L followed by parC-S80I and gyrA-D87G, except for a single outlier isolated in Australia (Fig 1B). Strong bootstrap support at each of these nodes suggests that these mutations were sequential lineage-defining events, with the final gyrA-D87G mutation preceding the expansion and intercontinental dissemination of the Central Asia III clade. These three QRDR mutations were also shared by ten phenotypically uncharacterized S. sonnei from the US, thus providing genotypic evidence for ciprofloxacin resistance. The single outlier isolate shares the gyrA-S83L and parC-S80I QRDR mutations of the other ciprofloxacin-resistant isolates, but harbours gyrA-D87N rather than a gyrA-D87G, and is within a closely related out-group of the major ciprofloxacin-resistant clone (Fig 1B).
South Asia as a Hub of Fluoroquinolone-Resistant S. sonnei
We additionally mapped the country of isolation and patient travel history onto the Central Asia III phylogeny to investigate the geographical structure of the clade (Fig 1B). For the ciprofloxacin-resistant S. sonnei isolated from countries with a low incidence of shigellosis (Ireland, Australia, and US), and for which data on recent travel history was confirmed (27/45; 60%), India was the most commonly reported travel destination (21/27; 78%). The majority of the isolates associated with travel to India clustered closely with strains isolated in neighbouring Bhutan. These data suggest that South Asia is a primary source of ciprofloxacin-resistant S. sonnei that have increasingly been isolated both within and outside of Asia in recent years. Furthermore, greater genetic diversity was observed within the South Asian S. sonnei than within the other sampled countries (Fig 1B), suggesting that this region currently acts as the most likely geographical source population.
Our data also show evidence of regional diversification of ciprofloxacin-resistant S. sonnei within Asia. The phylogenetic structure is highly suggestive of a clonal expansion of ciprofloxacin-resistant S. sonnei in Southeast Asia, specifically within Vietnam, as indicated by a long branch with 100% bootstrap support (Fig 1B). We additionally noted that S. sonnei nested within this clonal expansion were also isolated from travellers returning from countries including Cambodia and Thailand, indicating that isolates from this lineage have spread widely across Southeast Asia, as well as having been introduced into Australia on at least five separate occasions. An additional well-supported subpopulation of ciprofloxacin-resistant S. sonnei, isolated in Ireland (five individuals with no recent history of travel and one individual returning from Germany) and the US, likely represents an extended chain of local transmission within Europe and the US (Fig 1B). Alternatively, although less likely, this subpopulation could represent multiple importations and short-term local transmission of S. sonnei strains originating from an unsampled source population. Whilst it was not possible to identify the geographical source or extent of local transmission definitively, the isolates most closely related to this European/US subpopulation originated in India and Bhutan, again suggesting South Asia was the most likely origin of this subpopulation. These two expansion events in Southeast Asia and Europe/US indicate that this clone of ciprofloxacin-resistant S. sonnei is also capable of sustained circulation upon introduction into new locations.
Discussion
Here we provide direct evidence for the ongoing global expansion of S. sonnei exhibiting new and clinically relevant antimicrobial resistance profiles. What is more, as we can use phylogeography in high resolution, we can link the reservoir of these organisms to a specific region. Therefore, this study has significant implications for understanding the international trafficking of antimicrobial-resistant bacterial pathogens from Asia. Furthermore, we suggest that, as a single-serotype, human-adapted pathogen with a clonal population structure, S. sonnei serves as a tractable model for understanding how Gram-negative antimicrobial resistant pathogens are being regularly mobilised around the globe.
To our knowledge, this is the first study that has used whole-genome sequencing to examine the emergence and global spread of ciprofloxacin-resistant S. sonnei. Our data show that all sequenced extant ciprofloxacin-resistant S. sonnei, though sourced from disparate geographical locations, belonged to a single clonal expansion of lineage III, with South Asia being the most likely hub for its origin and spread. Our findings support previous hypotheses suggesting that ciprofloxacin-resistant S. sonnei in industrialised countries is being imported from South Asia [15,16]. A recent estimation of worldwide antimicrobial usage reported that India was the largest consumer of antimicrobials in 2010 [24]. Additionally, the fluoroquinolones are ranked as the most common antimicrobial prescribed for acute enteric diseases in India and Bangladesh [25,26]. The intensive use of fluoroquinolones in a region where there are foci of high population density and inconsistent access to good sanitation is likely to have contributed to emergence of ciprofloxacin-resistant enteric bacteria, such as S. sonnei and Salmonella Typhi, on the Indian subcontinent [19]. Global dissemination of these organisms is likely facilitated by the volume of travel between these regions and other areas of the world.
Our new data highlight the limitations of current typing protocols for tracking S. sonnei. It had been previously observed that some of the ciprofloxacin-resistant S. sonnei isolates in this study (originating from Bhutan and Ireland) shared a similar XbaI pulsed field gel electrophoresis (PFGE) pattern [14,15]. This pulsotype has been observed previously in India and Bangladesh [12,13,27–29], as well as in Canada [30], Belgium [31], and Japan [32], where the association with ciprofloxacin resistance was inconsistent. However, PFGE in this context did not offer sufficient granularity to link all of the isolates or provide sufficient resolution into the regional evolution of S. sonnei. Our phylogenetic analyses show that this pulsotype is associated with a phylogenetic lineage, supporting the notion that this pulsotype actually represents a widespread and pervasive subclade of Central Asia III.
This work has limitations. First, the lack of historical organisms from South Asia restricts our inference to only the current situation. Furthermore, additional contemporary organisms from other settings would have improved our understanding of the current geographical spread of this clonal group. Although the data included in this analysis was generated from organisms sampled in diverse geographic locations, the majority of sequences were retrieved from South Asia, Southeast Asia, Europe, Australia, and the Americas. It is possible, albeit unlikely, that our study might have overlooked an undersampled population that may ultimately act as an additional reservoir. This issue is inherent to all phylogeographical investigations and can potentially be overcome by global-scale prospective sampling to capture and characterize maximal global diversity. Notwithstanding these limitations, whole-genome sequencing of these geographically disparate organisms, together with plausible epidemiological links, has provided data at the highest resolution for deciphering the emergence and international spread of ciprofloxacin-resistant S. sonnei. Future studies interrogating extensive spatial and temporal collections of ciprofloxacin-resistant S. sonnei, as well as the S. sonnei diversity specific to South Asia prior to and during the emergence of antimicrobial resistance, are essential to further elucidate the origins and epidemiological dynamics of these populations. These supplementary investigations will greatly aid our efforts in controlling the spread of the current ciprofloxacin-resistant clone and to prevent future emergent antimicrobial-resistant bacterial populations.
In conclusion, the international surge of ciprofloxacin-resistant S. sonnei clone poses a substantial global health challenge, and our data show this threat is not only manifested in sporadic cases from returning travellers but also the establishment of endemic transmission in new settings. The latter is already evident in high shigellosis incidence areas such as Southeast Asia. Therefore, integrative efforts from both the research community and public health authorities should be prioritised to track, monitor, and prevent the international spread of this key enteric pathogen.
Supporting Information
Zdroje
1. Liu L, Johnson HL, Cousens S, Perin J, Scott S, Lawn JE, et al. Global, regional, and national causes of child mortality: an updated systematic analysis for 2010 with time trends since 2000. Lancet. Elsevier Ltd; 2012;379 : 2151–61.
2. Kotloff KL, Nataro JP, Blackwelder WC, Nasrin D, Farag TH, Panchalingam S, et al. Burden and aetiology of diarrhoeal disease in infants and young children in developing countries (the Global Enteric Multicenter Study, GEMS): a prospective, case-control study. Lancet. 2013;382 : 209–22. doi: 10.1016/S0140-6736(13)60844-2 23680352
3. Bardhan P, Faruque ASG, Naheed A, Sack DA. Decrease in shigellosis-related deaths without Shigella spp.-specific interventions, Asia. Emerg Infect Dis. 2010/10/30 ed. 2010;16 : 1718–23. doi: 10.3201/eid1611.090934 21029529
4. Vinh H, Nhu NT, Nga T V, Duy PT, Campbell JI, Hoang N V, et al. A changing picture of shigellosis in southern Vietnam: shifting species dominance, antimicrobial susceptibility and clinical presentation. BMC Infect Dis. 2009/12/17 ed. 2009;9 : 204. doi: 10.1186/1471-2334-9-204 20003464
5. Thompson CN, Duy PT, Baker S. The Rising Dominance of Shigella sonnei: An Intercontinental Shift in the Etiology of Bacillary Dysentery. PLoS Negl Trop Dis. 2015;9 : 1–13.
6. Bennish ML. Potentially lethal complications of shigellosis. Rev Infect Dis. 1991;13 Suppl 4: S319–S324. 2047657
7. Vinh H, Main J, Chinh M, Tam C, Trang P, Nga D, et al. Treatment of bacillary dysentery in Vietnamese children: two doses of ofloxacin versus 5-days nalidixic acid. Trans R Soc Trop Med Hyg. 2000;94 : 323–326. 10975012
8. WHO. Guidelines for the control of shigellosis, including epidemics due to Shigella dysenteriae type 1. World Health Organization. 2005.
9. Ruiz J. Mechanisms of resistance to quinolones: Target alterations, decreased accumulation and DNA gyrase protection. J Antimicrob Chemother. 2003;51 : 1109–1117. 12697644
10. Holt KE, Vu T, Nga T, Pham D, Vinh H, Wook D, et al. Tracking the establishment of local endemic populations of an emergent enteric pathogen. PNAS. 2013;110.
11. Horiuchi S, Inagaki Y, Yamamoto N, Okamura N, Imagawa Y, Nakaya R. Reduced susceptibilities of Shigella sonnei strains isolated from patients with dysentery to fluoroquinolones. Antimicrob Agents Chemother. 1993;37 : 2486–2489. 8285640
12. Nandy S, Dutta S, Ghosh S, Ganai A, Rajahamsan J, Theodore RBJ, et al. Foodborneassociated Shigella sonnei, India, 2009 and 2010. Emerg Infect Dis. 2011;17 : 2072–2073. 8 doi: 10.3201/eid1711.110403 22099103
13. Ud-Din AIMS, Wahid SUH, Latif H a, Shahnaij M, Akter M, Azmi IJ, et al. Changing trends in the prevalence of Shigella species: emergence of multi-drug resistant Shigella sonnei biotype g in Bangladesh. PLoS ONE. 2013;8: e82601. doi: 10.1371/journal.pone.0082601 24367527
14. Ruekit S, Wangchuk S, Dorji T, Tshering KP, Pootong P, Nobthai P, et al. Molecular characterization and PCR-based replicon typing of multidrug resistant Shigella sonnei isolates from an outbreak in Thimphu, Bhutan. BMC Res Notes. BMC Research Notes; 2014;7 : 1–9.
15. De Lappe N, Connor JO, Garvey P, Mckeown P, Cormican M. Ciprofloxacin-Resistant Shigella sonnei Associated with Travel to India. Emerg Infect Dis. 2015;21 : 2013–2015.
16. Bowen A, Hurd J, Hoover C, Khachadourian Y, Traphagen E, Harvey E, et al. Importation and Domestic Transmission of Shigella sonnei Resistant to Ciprofloxacin—United States, May 2014-February 2015. Morb Mortal Wkly Rep. 2015;64 : 318–320.
17. Kim JS, Kim JJ, Kim SJ, Jeon S, Seo KY, Choi J, et al. Shigella sonnei Associated with Travel to Vietnam, Republic of Korea. Emerg Infect Dis. 2015;21 : 1247–1250. doi: 10.3201/eid2107.150363 26079171
18. He M, Miyajima F, Roberts P, Ellison L, Pickard DJ, Martin MJ, et al. Emergence and global spread of epidemic healthcare-associated Clostridium difficile. Nat Genet.; 2013;45 : 109–13. doi: 10.1038/ng.2478 23222960
19. Wong VK, Baker S, Pickard DJ, Parkhill J, Page AJ, Feasey N a, et al. Phylogeographical analysis of the dominant multidrug-resistant H58 clade of Salmonella Typhi identifies inter - and intracontinental transmission events. Nat Genet. 2015;
20. Holt KE, Baker S, Weill FX, Holmes EC, Kitchen A, Yu J, et al. Shigella sonnei genome sequencing and phylogenetic analysis indicate recent global dissemination from Europe. Nat Genet. 2012/08/07 ed. 2012;44 : 1056–1059. doi: 10.1038/ng.2369 22863732
21. Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25 : 2078–9. doi: 10.1093/bioinformatics/btp352 19505943
22. Zerbino DR, Birney E. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 2008;18 : 821–9. doi: 10.1101/gr.074492.107 18349386
23. Zankari E, Hasman H, Cosentino S, Vestergaard M, Rasmussen S, Lund O, et al. Identification of acquired antimicrobial resistance genes. J Antimicrob Chemother. 2012;67 : 2640–4. doi: 10.1093/jac/dks261 22782487
24. Van Boeckel TP, Gandra S, Ashok A, Caudron Q, Grenfell BT, Levin S a., et al. Global antibiotic consumption 2000 to 2010: An analysis of national pharmaceutical sales data. Lancet Infect Dis. Elsevier Ltd; 2014;14 : 742–750.
25. Fahad BM, Matin A, Shill MC, Asish KD. Antibiotic usage at a primary health care unit in Bangladesh. Australas Med J. 2010;3 : 414–421.
26. Kotwani A, Chaudhury RR, Holloway K. Antibiotic-prescribing practice of primary care prescribers for acute diarrhea in New Delhi, India. Value Heal. 2012;15: S16–S119.
27. Pazhani GP, Niyogi SK, Singh AK, Sen B, Taneja N, Kundu M, et al. Molecular characterization of multidrug-resistant Shigella species isolated from epidemic and endemic cases of shigellosis in India. J Med Microbiol. 2008;57 : 856–863. d doi: 10.1099/jmm.0.2008/000521-0 18566144
28. Ghosh S, Pazhani GP, Chowdhury G, Guin S, Dutta S, Rajendran K, et al. Genetic characteristics and changing antimicrobial resistance among Shigella spp. isolated from hospitalized diarrhoeal patients in Kolkata, India. J Med Microbiol. 2011;60 : 1460–1466. 0 doi: 10.1099/jmm.0.032920-0 21659504
29. Talukder K a., Islam Z, Dutta DK, Aminul Islam M, Khajanchi BK, Azmi IJ, et al. Antibiotic resistance and genetic diversity of Shigella sonnei isolated from patients with diarrhoea between 1999 and 2003 in Bangladesh. J Med Microbiol. 2006;55 : 1257–1263. 16914657
30. Gaudreau C, Ratnayake R, Pilon P a, Gagnon S, Roger M, Lévesque S. CiprofloxacinResistant Shigella sonnei among Men Who Have Sex with Men, Canada, 2010. Emerg Infect Dis. 2011;17 : 1747–1750. doi: 10.3201/eid1709.102034 21888811
31. Vrints M, Mairiaux E, Van Meervenne E, Collard JM, Bertrand S. Surveillance of antibiotic susceptibility patterns among Shigella sonnei strains isolated in Belgium during the 18-year period 1990 to 2007. J Clin Microbiol. 2009;47 : 1379–1385. doi: 10.1128/JCM.02460-08 19321731
32. Uchimura M, Kishida K, Koiwai K. Increasing Incidence and the Mechanism of Resistance of Nalidixic Acid Resistant Shigella sonnei. Kansenshogaku Zasshi. 2001;75 : 923–930. 11766375
Štítky
Interní lékařství
Článek vyšel v časopisePLOS Medicine
Nejčtenější tento týden
2016 Číslo 8- Není statin jako statin aneb praktický přehled rozdílů jednotlivých molekul
- Magnosolv a jeho využití v neurologii
- Moje zkušenosti s Magnosolvem podávaným pacientům jako profylaxe migrény a u pacientů s diagnostikovanou spazmofilní tetanií i při normomagnezémii - MUDr. Dana Pecharová, neurolog
- Biomarker NT-proBNP má v praxi široké využití. Usnadněte si jeho vyšetření POCT analyzátorem Afias 1
- Antikoagulační léčba u pacientů před operačními výkony
-
Všechny články tohoto čísla
- Dementia across the Lifespan and around the Globe—Pathophysiology, Prevention, Treatment, and Societal Impact: A Call for Papers
- Progress and Challenges in Scaling Up Laboratory Monitoring of HIV Treatment
- Facility-Based Delivery during the Ebola Virus Disease Epidemic in Rural Liberia: Analysis from a Cross-Sectional, Population-Based Household Survey
- Availability and Use of HIV Monitoring and Early Infant Diagnosis Technologies in WHO Member States in 2011–2013: Analysis of Annual Surveys at the Facility Level
- Duration of Adulthood Overweight, Obesity, and Cancer Risk in the Women’s Health Initiative: A Longitudinal Study from the United States
- Improving Clinical Risk Stratification at Diagnosis in Primary Prostate Cancer: A Prognostic Modelling Study
- Genetically Predicted Body Mass Index and Breast Cancer Risk: Mendelian Randomization Analyses of Data from 145,000 Women of European Descent
- Social Dancing and Incidence of Falls in Older Adults: A Cluster Randomised Controlled Trial
- Genetic and Environmental Risk for Chronic Pain and the Contribution of Risk Variants for Major Depressive Disorder: A Family-Based Mixed-Model Analysis
- Long-Term Outcomes Associated with Traumatic Brain Injury in Childhood and Adolescence: A Nationwide Swedish Cohort Study of a Wide Range of Medical and Social Outcomes
- On Risk Estimation versus Risk Stratification in Early Prostate Cancer
- Make Data Sharing Routine to Prepare for Public Health Emergencies
- Assessment of Adverse Events in Protocols, Clinical Study Reports, and Published Papers of Trials of Orlistat: A Document Analysis
- Concussions and Repercussions
- South Asia as a Reservoir for the Global Spread of Ciprofloxacin-Resistant : A Cross-Sectional Study
- Building from the HIV Response toward Universal Health Coverage
- Transitioning to Country Ownership of HIV Programs in Rwanda
- Accelerating the Uptake and Timing of Antiretroviral Therapy Initiation in Sub-Saharan Africa: An Operations Research Agenda
- Glycemic Control and the Risk of Tuberculosis: A Cohort Study
- Adjuvant Trastuzumab in HER2-Positive Early Breast Cancer by Age and Hormone Receptor Status: A Cost-Utility Analysis
- Uptake of Home-Based HIV Testing, Linkage to Care, and Community Attitudes about ART in Rural KwaZulu-Natal, South Africa: Descriptive Results from the First Phase of the ANRS 12249 TasP Cluster-Randomised Trial
- An Audit and Feedback Intervention for Reducing Antibiotic Prescribing in General Dental Practice: The RAPiD Cluster Randomised Controlled Trial
- Multidrug-Resistant Tuberculosis Treatment in North Korea: Is Scale-Up Possible?
- Associations between Mental Health and Ebola-Related Health Behaviors: A Regionally Representative Cross-sectional Survey in Post-conflict Sierra Leone
- Pancreatic Cancer Surgical Resection Margins: Molecular Assessment by Mass Spectrometry Imaging
- Integrated Delivery of Antiretroviral Treatment and Pre-exposure Prophylaxis to HIV-1–Serodiscordant Couples: A Prospective Implementation Study in Kenya and Uganda
- Measuring Burden of Unhealthy Behaviours Using a Multivariable Predictive Approach: Life Expectancy Lost in Canada Attributable to Smoking, Alcohol, Physical Inactivity, and Diet
- Comparison of Outcomes before and after Ohio's Law Mandating Use of the FDA-Approved Protocol for Medication Abortion: A Retrospective Cohort Study
- Core Outcomes for Colorectal Cancer Surgery: A Consensus Study
- Adverse Renal, Endocrine, Hepatic, and Metabolic Events during Maintenance Mood Stabilizer Treatment for Bipolar Disorder: A Population-Based Cohort Study
- PLOS Medicine
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Glycemic Control and the Risk of Tuberculosis: A Cohort Study
- Transitioning to Country Ownership of HIV Programs in Rwanda
- Dementia across the Lifespan and around the Globe—Pathophysiology, Prevention, Treatment, and Societal Impact: A Call for Papers
- Social Dancing and Incidence of Falls in Older Adults: A Cluster Randomised Controlled Trial
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



