-
Články
- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Sox11 Is Required to Maintain Proper Levels of Hedgehog Signaling during Vertebrate Ocular Morphogenesis
Ocular coloboma is a condition in which tissue is missing from a portion of the eye due to its abnormal development. Coloboma is also frequently associated with additional anomalies, including microphthalmia (abnormally small eye) and cataracts. Although some of the genes that cause coloboma have been identified, in the majority of cases the underlying genetic cause has not been determined. One pathway that has been implicated in coloboma is the Hedgehog (Hh) signaling pathway. In this study, we have taken advantage of the ability to titrate levels of gene expression in zebrafish to demonstrate for the first time that the transcription factor Sox11 is required to limit levels of Hedgehog (Hh) signaling during ocular development. We show that in the absence of Sox11, levels of the Sonic Hedgehog (Shh) ligand are greatly elevated, which disrupts the proper patterning of the optic stalk and optic vesicle, resulting in coloboma. We also provide evidence that SOX11 dosage changes or mutations contribute to human coloboma, microphthalmia, and rod photoreceptor dysfunction. Thus, our work establishes a novel link between Sox11 and Hh signaling, and suggests that mutations in SOX11 contribute to pediatric eye disorders such as coloboma.
Published in the journal: . PLoS Genet 10(7): e32767. doi:10.1371/journal.pgen.1004491
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1004491Summary
Ocular coloboma is a condition in which tissue is missing from a portion of the eye due to its abnormal development. Coloboma is also frequently associated with additional anomalies, including microphthalmia (abnormally small eye) and cataracts. Although some of the genes that cause coloboma have been identified, in the majority of cases the underlying genetic cause has not been determined. One pathway that has been implicated in coloboma is the Hedgehog (Hh) signaling pathway. In this study, we have taken advantage of the ability to titrate levels of gene expression in zebrafish to demonstrate for the first time that the transcription factor Sox11 is required to limit levels of Hedgehog (Hh) signaling during ocular development. We show that in the absence of Sox11, levels of the Sonic Hedgehog (Shh) ligand are greatly elevated, which disrupts the proper patterning of the optic stalk and optic vesicle, resulting in coloboma. We also provide evidence that SOX11 dosage changes or mutations contribute to human coloboma, microphthalmia, and rod photoreceptor dysfunction. Thus, our work establishes a novel link between Sox11 and Hh signaling, and suggests that mutations in SOX11 contribute to pediatric eye disorders such as coloboma.
Introduction
Ocular coloboma arises when the embryonic choroid fissure in the ventral optic cup fails to close. It can cause significant pediatric visual impairment [1], and is often associated with other ocular abnormalities such as microphthalmia or anophthalmia (collectively referred to as MAC). Coloboma may also be observed in conjunction with dysgenesis of the anterior segment (front portion of the eye) or optic nerve, lenticular defects (such as cataract), or systemic congenital malformation syndromes [2]. In addition to phenotypic heterogeneity, coloboma is genetically heterogeneous, exhibiting differing patterns of inheritance, variable expressivity, and reduced penetrance [2].
Among the signaling pathways that converge to regulate ocular morphogenesis, Hedgehog (Hh) signaling has a critical role and acts reiteratively during eye development [3]. Hh signaling from the midline promotes the segregation of the single eye field into two optic primordia, and is required for the correct proximodistal and dorsoventral patterning of the optic vesicle [3]–[5]. Once the optic cup has formed, intraretinal Hh signaling regulates the differentiation of retinal progenitor cells [3]. Given its central role in eye development, it is unsurprising that mutations in genes encoding Hh pathway ligands (SHH) or targets (PAX2, VAX1) are associated with congenital ocular malformations in humans [6]–[9]. However, these mutations account for only a minority of patients; for the majority of MAC cases, the molecular defect has yet to be identified. Because of their potency, the spatiotemporal levels of Hh ligands must be tightly regulated throughout eye development; yet, very little is known about the factors that restrict their expression during oculogenesis. Such factors would represent excellent candidate genes for human coloboma and associated ocular defects, and potentially could be used to influence Hh signaling.
Here, we focus on the role of the SRY-box transcription factor Sox11 during eye development. Sox11 is a member of the group C family of SOX proteins, which also includes Sox4 and Sox12 [10]. Sox11 is required for a variety of processes, including organogenesis and neurogenesis, craniofacial and skeletal development [10], as well as being implicated in carcinogenesis (including mantle cell lymphoma, medulloblastoma, and glioblastoma) [10], [11]. Expression and functional studies support a role for Sox11 during several stages of eye development. In the mouse, Sox11 is expressed in the optic cup and periocular mesenchyme during early eye development, and in the developing lens and retina at later stages [12], [13]. In the zebrafish retina, we previously found that Sox11 is upregulated in rod progenitor cells during rod photoreceptor regeneration [14]. Sox11−/− mice exhibit ocular abnormalities such as anterior segment dysgenesis, microphthalmia, a persistent lens stalk, delayed lens formation, and coloboma [13]. Finally, some human chromosomal rearrangements resulting in ocular abnormalities have been mapped to the vicinity of the SOX11 locus at chromosome 2p25.2 [15]–[18]. These data together suggested intriguing roles for Sox11 in ocular morphogenesis and rod photoreceptor differentiation, however the underlying mechanisms were undefined.
In this study, we inhibited Sox11 activity in zebrafish embryos, and based on the resultant phenotypes demonstrate that the function of Sox11 in regulating lens development and choroid fissure closure is evolutionarily conserved, and that Sox11 is required for rod photoreceptor differentiation. We demonstrate that elevated Hh signaling causes the ocular phenotypes in Sox11-deficient zebrafish, and that Sox11 is required to repress expression of the Sonic hedgehog gene (shha). Finally, we identify SOX11 variants with reduced transactivation ability in MAC patients, and in parallel demonstrate that decreased SOX11 gene dosage results in congenital ocular abnormalities. In revealing a previously uncharacterized role for Sox11 upstream of Hh signaling, these studies may substantially extend our understanding of additional Sox11-dependent developmental and pathologic processes.
Results
Expression of sox11a/b during ocular development
Zebrafish possess two orthologs of mammalian Sox11, which are expressed in overlapping and distinct domains ([19]–[21], this study). Previous studies have shown that both sox11a and sox11b are maternally expressed prior to the midblastula transition, and are expressed in the region of the anterior neural plate that gives rise to the diencephalon at the onset of the segmentation period [20]. Using in situ hybridization with paralog-specific probes, we investigated the expression of sox11a and sox11b both within the forebrain during optic cup formation, and in the eye at later stages of retinal development. At 18 hours post fertilization (hpf), we detected expression of sox11a and sox11b in the telencephalon, and in the dorsal “corner” formed by the diencephalon and the evaginated optic stalk/optic vesicle (top panel arrows and second row closed asterisks, Figure 1A). We also detected faint expression of sox11a and sox11b at the ventral hinge of the optic stalk/optic vesicle axis (open asterisks, Figure 1A). However, we did not detect expression of sox11a or sox11b within the optic vesicle itself (Figure 1A). At 24 hpf, sox11a/b expression persisted in the diencephalon adjacent to the retina and in the telencephalon, and both paralogs were also expressed in the hypothalamus (Figures 1A, C). Within the developing retina at 24 hpf, sox11b was expressed diffusely across the lens and retinal neuroepithelium, and was distinctly visible in a small cluster of cells in the ventro-nasal retina (arrow, bottom right panel, Figure 1A), corresponding to the location at which retinal neurogenesis initiates [22]. As retinal development proceeded, sox11a expression was observed in the ganglion cell layer (GCL) at 48 hpf, whereas the expression of sox11b was detected in a few scattered cells across the central retina but was mostly restricted to the undifferentiated peripheral retina (Figure 1B; [14]). By 72 hpf, when retinal neurogenesis was mostly complete, both sox11a and sox11b were predominantly expressed in the persistently neurogenic ciliary marginal zone (Figure 1B; [14]); expression of sox11a also persisted in the GCL and in some cells in the inner half of the inner nuclear later (INL). Interestingly, the expression domains of both sox11 paralogs were adjacent to regions of shha expression in the ventral diencephalon at 18 and 24 hpf (Figure 1C), whereas at 48 hpf sox11a expression overlapped with the previously described location of shha in the GCL [23], [24].
Fig. 1. Developmental expression of sox11. 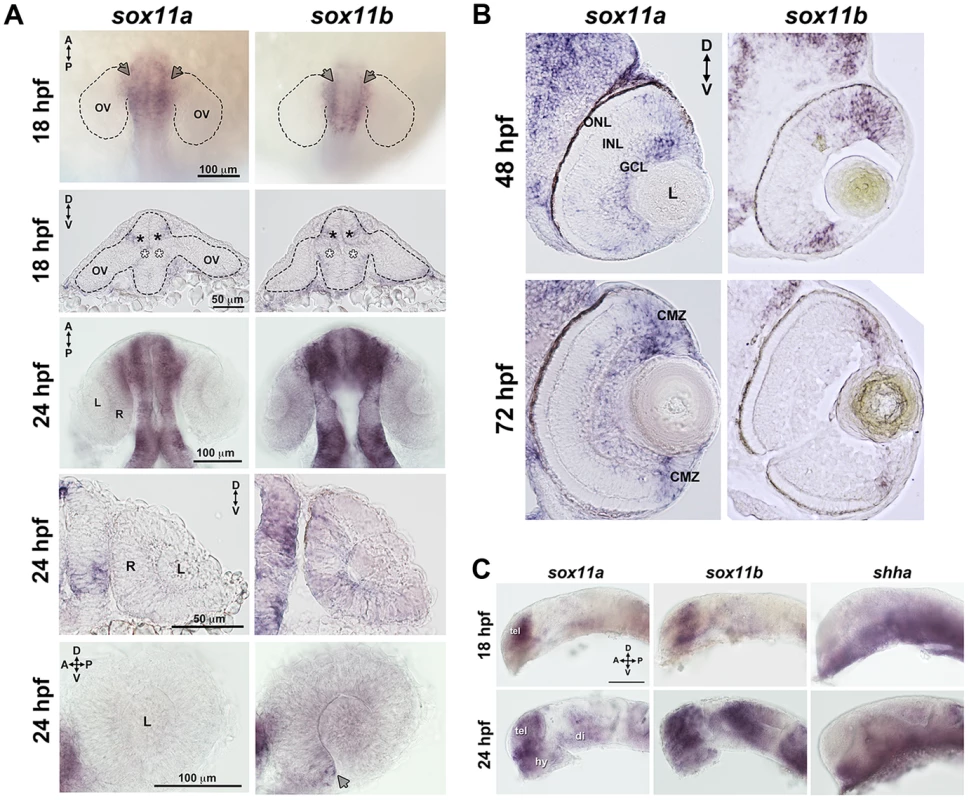
In situ hybridization with antisense probes for sox11a, sox11b, and shha was performed on whole embryos or on tissue sections at the indicated time points. (A) Sox11a and sox11b were expressed in the diencephalon adjacent to the optic vesicle (arrows in top row and asterisks in second row) at 18 hpf (top two rows) and 24 hpf (third and fourth rows). Sox11a expression was not detected in the lens or retina at 24 hpf (bottom left). Sox11b was expressed in a patch of cells in the ventronasal retina (arrow, bottom right) and more diffusely across the rest of the retina and lens. Top and third rows are dorsal views of flat-mounted embryos. Second and fourth rows are frontal sections through the head. Bottom panels are lateral views of dissected eyes; (n = 20 embryos examined per time point, 3 independent repeats). (B) Transverse sections through the eye at 48 hpf (top) and 72 hpf (bottom). Sox11a expression was detected in the ganglion cell layer (GCL) and in few sporadic cells in the inner nuclear layer (INL); sox11b expression was observed in scattered cells across the central retina and in the peripheral retina. At 72 hpf, sox11a expression persisted in the GCL and in some cells in the INL; sox11a and sox11b were also expressed in the persistently neurogenic ciliary marginal zone (CMZ); n = 20 embryos examined per time point, 3 independent repeats. (C) Expression patterns of sox11a (left), sox11b (center), and shha (right) in the developing brain at 18 hpf (top) and 24 hpf (bottom). The eye was removed to better image the brain. Expression of sox11a and sox11b, but not shha, was observed in the telencephalon. Expression of all three genes was detected in the hypothalamus and ventral diencephalon at 24 hpf (n = 20 embryos examined per time point, 3 independent repeats). Scale bar = 100 µm; D, dorsal; V, ventral; A, anterior; P, posterior; hpf, hours post fertilization; OV, optic vesicle; L, lens; R, retina; GCL, ganglion cell layer; INL, inner nuclear layer; ONL, outer nuclear layer; CMZ, ciliary marginal zone; tel, telencephalon; hy, hypothalamus; di, diencephalon. Knockdown of sox11a/b causes abnormal ocular morphogenesis
To investigate the function of Sox11 paralogs during eye development, translation of sox11a and sox11b was blocked with morpholino oligonucleotides (MOs), whose efficiency and specificity were confirmed using a second sox11 MO and a GFP reporter assay, respectively (Figures S1A, C). Zebrafish embryos were injected with sox11a and sox11b MOs simultaneously (hereafter referred to as sox11 morphants), as co-inhibition of both paralogs induced the highest prevalence of ocular phenotypes (Figure S1B). At 24 hpf, 72.9±7.4% of sox11 morphants displayed a misshapen, rudimentary, or absent lens (Figures 2A, S1B). Sox11 morphant lenses mostly recovered to a spherical shape by 2 days post fertilization (dpf), however at this stage a similar proportion of morphants (70.0±7.7%) displayed coloboma (Figures 2A, S1B). Sox11 morphant eyes were also hypopigmented ventrally, and microphthalmic compared to controls (Figure 2A, B). Histological sections revealed that the colobomatous retinas in sox11 morphants frequently extruded through the open choroid fissure into the brain (Figure 2C). Approximately 54% (15 of 28 individuals examined) of sox11 morphant retinas with coloboma also exhibited poor or reduced retinal lamination, suggesting a delay in retinal differentiation. In contrast, of the sox11 morphant retinas that did not display coloboma, only 14% were poorly laminated (4 of 29). This suggests that similar mechanisms may underlie the ocular morphogenesis and retinal developmental defects observed in sox11 morphants with coloboma. The presence of the coloboma prevented the retinal pigmented epithelium (RPE) from completely enclosing the posterior eye (Figure 2C), which likely accounts for the hypopigmented appearance of the ventral portion of the eye when viewed laterally (Figure 2A). This coloboma phenotype was reminiscent of the zebrafish blowout mutant, which has a mutation in patched2 (formerly named patched1), a negative regulator of Hh signaling [25], [26]. In addition to the ocular phenotypes, sox11 morphants also frequently displayed a downward kink of the tail, as well as brain abnormalities such as widened ventricles, likely reflecting Sox11's expression and function in the posterior somites and developing brain, respectively [10], [20]. All of the morphant phenotypes were rescued by injection of wild type sox11a and sox11b mRNA, consistent with the morpholinos being specific for Sox11 (Figure 2A, D). Importantly, these phenotypes were also rescued by injection of human SOX11 mRNA, indicating that the function of Sox11 in regulating early eye development is evolutionarily conserved (Figure 2D).
Fig. 2. Sox11 knockdown disrupts ocular morphogenesis and causes coloboma in zebrafish. 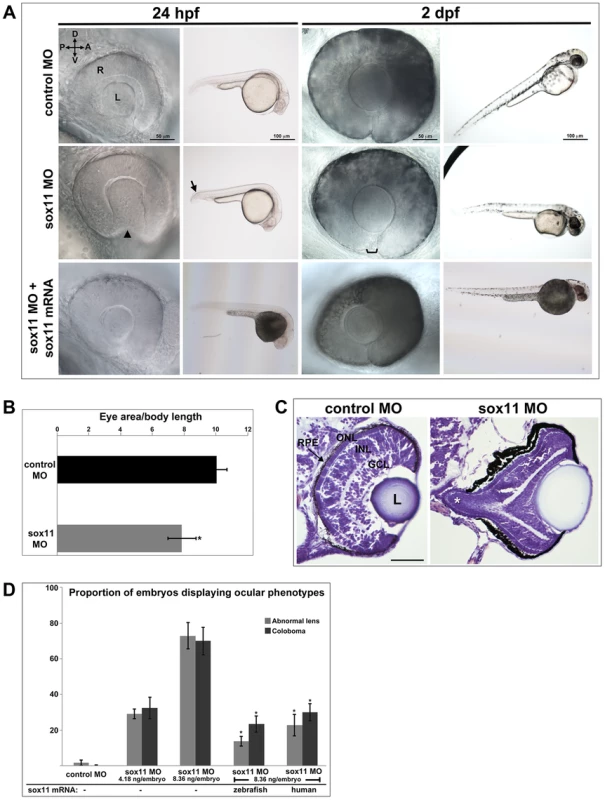
(A) Representative eye and body images of control and sox11 morphants (taken from the set of embryos analyzed in (D). At 24 hpf, approximately 70% of sox11 morphants displayed a malformed lens (arrowhead) and a posterior kink in the tail (arrow). At 2 dpf, a similar proportion of sox11 morphants displayed coloboma (bracket), and had a hypopigmented and underdeveloped ventral retina. Both the abnormal lens and coloboma phenotypes were rescued with co-injection of wild type zebrafish sox11 mRNA (bottom row). (B) Sox11 morphants were microphthalmic at 24 hpf. Eye area was normalized to body length (*p<0.0001, Student's t-test; control MO: n = 10 embryos examined; sox11 MO: n = 12 embryos examined, 3 independent repeats). (C) Sections of 72 hpf control (left) and sox11 morphant eyes (right) stained with cresyl violet revealed the extrusion of the retina into the brain through the open choroid fissure of sox11 morphants (asterisk); n = 6 individuals examined per group. The thickened appearance of the dorsal RPE in the sox11 morphant retina is a staining artifact and was not observed in fresh tissue sections. Scale bar = 50 µm. (D) Injection of zebrafish and human sox11 mRNA rescued the ocular phenotypes in sox11 morphants. Number of embryos analyzed: 24 hpf control MO, 4.18 ng/embryo, n = 1007; 2 dpf control MO, 4.18 ng/embryo, n = 1001; 24 hpf sox11 MO, 4.18 ng/embryo, n = 309; 2 dpf sox11 MO, 4.18 ng/embryo, n = 294; 24 hpf sox11 MO, 8.36 ng/embryo, n = 559; 2 dpf sox11 MO, 8.36 ng/embryo, n = 392; 24 hpf sox11 MO 8.36 ng/embryo plus 2.0 ng/embryo zebrafish sox11 mRNA, n = 185; 2 dpf sox11 MO, 8.36 ng/embryo plus 2.0 ng/embryo zebrafish sox11 mRNA, n = 167; 24 hpf sox11 MO, 8.36 ng/embryo plus 0.3 ng/embryo human SOX11 mRNA, n = 130; 2 dpf sox11 MO, 8.36 ng/embryo plus 0.3 ng/embryo human SOX11 mRNA, n = 125. Three biological replicates were performed for all experiments. (*p<0.001, Student's t- test). D, dorsal; V, ventral; A, anterior; P, posterior; L, lens; R, retina; hpf, hours post fertilization; dpf, days post fertilization; MO, morpholino; GCL, ganglion cell layer; INL, inner nuclear layer; ONL, outer nuclear layer; RPE, retinal pigmented epithelium. One mechanism that has been suggested to contribute to optic fissure closure defects is overproliferation of progenitor cells within the presumptive neural retina [27]. To determine whether changes in mitotic activity underlie the lens and coloboma phenotypes in sox11 morphants, we immunolabeled retinal sections from control and sox11 morphants with an antibody to phosphohistone H3 (PH3). We observed a modest but significant increase in the number of PH3-positive cells in sox11 morphant optic vesicle and retinas at 18 and 24 hpf, and a larger increase in proliferation relative to controls at 48 and 72 hpf (Figure S2C, D). However, the excess PH3-positive cells were not clustered in the ventral retina, optic stalk, or lens at 24 hpf (Figure S2D), by which time the abnormal ocular phenotypes are already apparent. Therefore, we conclude that overproliferation likely does not underlie the early ocular phenotypes of sox11 morphants. We also performed TUNEL staining on sections from control and sox11 morphant retinas (Figure S2A, B). This analysis revealed a variable but significant increase in TUNEL-positive cells in the optic vesicle of sox11 morphants at 18 hpf. At 24 hpf, we did not detect elevated apoptosis in the retina or optic stalk of Sox11-deficient embryos. However, we did observe a significant increase in TUNEL-positive cells in the anterior lens of sox11 morphants, which persisted through 72 hpf (Figure S2A, B). This increase in apoptotic cells in the lens may be related to the abnormal lens morphology we observed by light microscopy (Figure 2A). Finally, we observed an increase in TUNEL-positive cells in the colobomatous tissue of sox11 morphant retinas at 48 hpf (Figure S2B), indicating that this abnormal ocular structure negatively impacted the survival of the cells within it.
Sox11 morphants possess fewer mature rod photoreceptors
Given that expression of sox11a/b is upregulated in adult zebrafish rod progenitor cells during rod photoreceptor regeneration [14], we investigated whether Sox11-deficient embryos displayed altered rod development. Since we found that a significant proportion of sox11 morphant retinas with coloboma also displayed poor lamination, indicating a potential delay in retinal development, for analysis we divided the sox11 morphants into those with and without coloboma. This approach minimized the potential secondary effects on retinal development from the ocular morphogenetic defect masking any additional role for Sox11 in retinal neurogenesis. Using immunohistochemistry with cell-type specific antibodies, we found that sox11 morphants without coloboma (approximately 30% of morphant embryos) possessed well-laminated retinas with normal numbers of ganglion, amacrine, horizontal, and bipolar cells, Müller glia, and cone photoreceptors at 72 hpf (Figure S3A, B). In contrast, when control and sox11 MOs were injected into a rod photoreceptor-GFP transgenic reporter line [28], we observed a significant reduction in mature rod photoreceptors in sox11 morphant retinas without coloboma at 3 dpf (control embryos, 34.9±7.4rods/section; sox11 morphants, 8.7±8.9 rods/section; p<0.00001; Figure 3A, B). Furthermore, several retinal sections from sox11 morphants contained no detectable GFP-positive rods at 3 dpf. The reduction in mature rod photoreceptors in sox11 morphant retinas was confirmed by immunolabeling with the rod-specific antibody 4C12 (not shown), by fluorescent in situ hybridization (FISH) of retinal sections with a probe for rhodopsin (rho), and by quantitative RT-PCR (qPCR) for the rho transcript at 3 dpf (Figure 3C, D). Rod photoreceptor number could be rescued by injection of wild type sox11 mRNA (Figure 3B), demonstrating that the reduction in rods was due to Sox11 deficiency. To determine whether depletion of Sox11 blocks specification of the rod photoreceptor fate, we conducted FISH on 3 dpf retinal sections from control and sox11 morphants using probes for three genes associated with the rod photoreceptor lineage: neuroD, crx, and nr2e3 [29]–[31]. Interestingly, we found that expression of all three rod lineage genes was qualitatively normal in sox11 morphant retinas, even those with coloboma and poor lamination (Figure 3C). We also verified by qPCR that nr2e3 transcript levels were not significantly different in sox11 morphants and controls (Figure 3D) Therefore, these data suggest that Sox11 is required for the terminal differentiation, but not the specification, of rod photoreceptor cells. Because the window of rod photoreceptor differentiation is longer than that of cones or other retinal neurons [32], [33] we investigated whether rod photoreceptor number remained reduced in sox11 morphants later in development. The number of rods in sox11 morphant retinas was higher at 4 dpf than at 3 dpf, but remained significantly reduced relative to controls (sox11 morphants, 15.9±2.9 rods/section; controls, 57.9±5.4 rods/section; p<0.001; Figure S3C). Taken together, these data suggest that terminal differentiation of rods requires Sox11.
Fig. 3. Sox11 morphants lack mature rod photoreceptors. 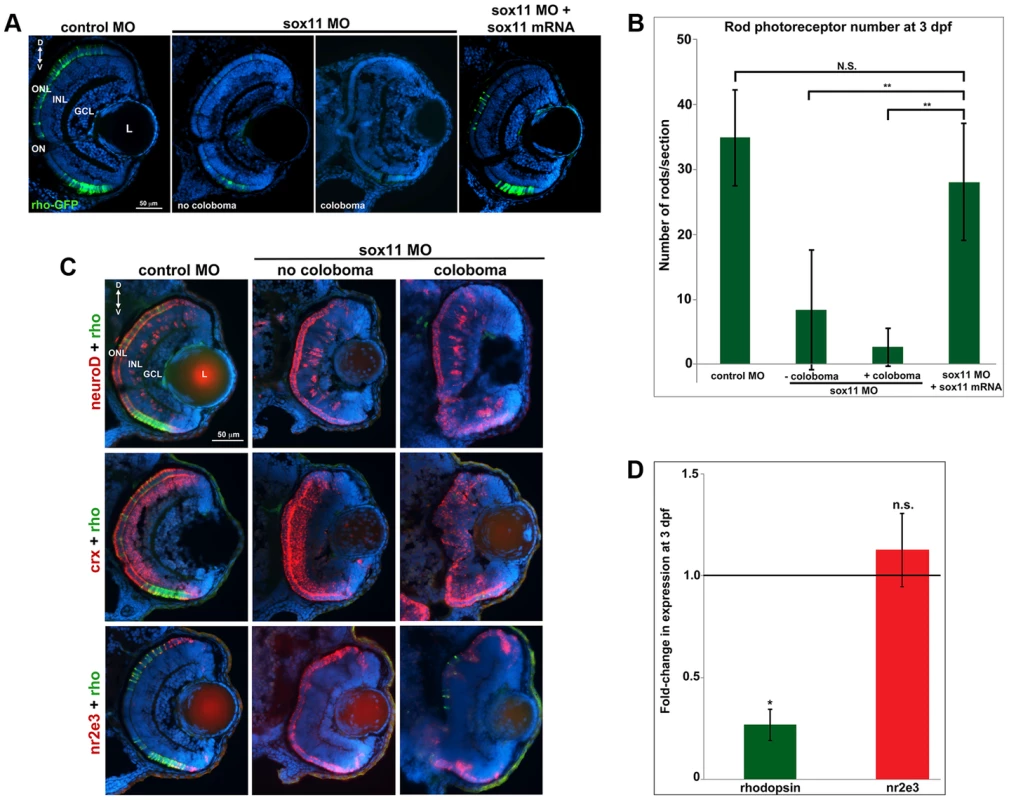
(A) Representative transverse retinal sections from XOPS-GFP zebrafish injected with control, sox11 MO, or sox11 MO plus zebrafish sox11 mRNA at 3 dpf (from the set of individuals analyzed in (B). Even sox11 morphants with well-laminated retinas and no evidence of coloboma (second panel) displayed greatly reduced numbers of mature rods compared to controls (left panel). Co-injection of wild type zebrafish sox11 mRNA (right panel) rescued the rod deficiency at 3 dpf. (B) Quantification of the number of rod photoreceptors/section. Number of embryos analyzed: control MO, n = 25; sox11 MO without coloboma, n = 25; sox11 MO with coloboma, n = 25; sox11 MO plus zebrafish sox11 mRNA, n = 17 (**p<0.00001; n.s., p>0.05, Student's t- test). (C) Two-color fluorescent in situ hybridization (FISH) for neuroD, crx, nr2e3, and rhodopsin expression in control and sox11 morphants with and without coloboma at 3 dpf. Expression of the rod lineage genes neuroD (top), crx (middle), and nr2e3 (bottom) was qualitatively normal in sox11 morphants with or without coloboma. However, rhodopsin expression (green) was greatly reduced compared to control morphants (left column). Number of embryos analyzed: n = 14 per group, 3 independent biological replicates. (D) Quantitative RT-PCR (qPCR) performed on mRNA from control and sox11 morphant heads at 3 dpf revealed a significant decrease in rhodopsin expression in sox11 morphants compared to controls. However, nr2e3 transcript levels were not significantly different between control and sox11 morphants. Relative transcript abundance was normalized to atp5h levels and is presented as the mean fold-change in expression relative to controls (n = 30 embryos per group, 3 independent biological replicates). *p<0.003; n.s, p>0.05, Student's t-test. D, dorsal; V, ventral; L, lens; dpf, days post fertilization; MO, morpholino; GCL, ganglion cell layer; INL, inner nuclear layer; ONL, outer nuclear layer; ON, optic nerve. Sox11 negatively regulates Hedgehog signaling
As mentioned above, the coloboma phenotype of sox11 morphants is similar to the zebrafish blowout mutant, in which increased Hedgehog signaling results in altered proximodistal patterning of the optic vesicle [25]. To determine whether a similar defect was present in sox11 morphants, we performed FISH on retinal sections to examine the expression of pax2a and pax6a, which mark optic stalk and retinal territories, respectively. This revealed expansion of the pax2a domain in approximately 50% of sox11 morphant embryos at 18 and 36 hpf, while at later stages (48 hpf), expression persisted around the open choroid fissure, whereas it was barely detectable in controls (Figure 4A). These expression changes were verified by qPCR at 18 and 24 hpf. Although we did not observe a concomitant decrease in pax6a expression in the optic vesicle at 18 hpf, there was a significant reduction in transcript levels detected by qPCR at 24 hpf (Figure 4B). To further test whether Hh signaling was elevated in sox11 morphants, we made use of a recently described Hh signaling reporter line of zebrafish, which expresses GFP under the control of the patched2 (ptc2) promoter [34]. Sections through the head of 24 hpf control and sox11 morphants on the ptc2:GFP background revealed both an increase in GFP expression and an expansion of the GFP-positive domains in the brain, retina, and RPE of sox11 morphants (Figure 5A). Taken together, these data strongly suggest that Hh signaling is indeed elevated in sox11 morphants.
Fig. 4. Pax2.1 and pax6a expression is altered in sox11 morphants. 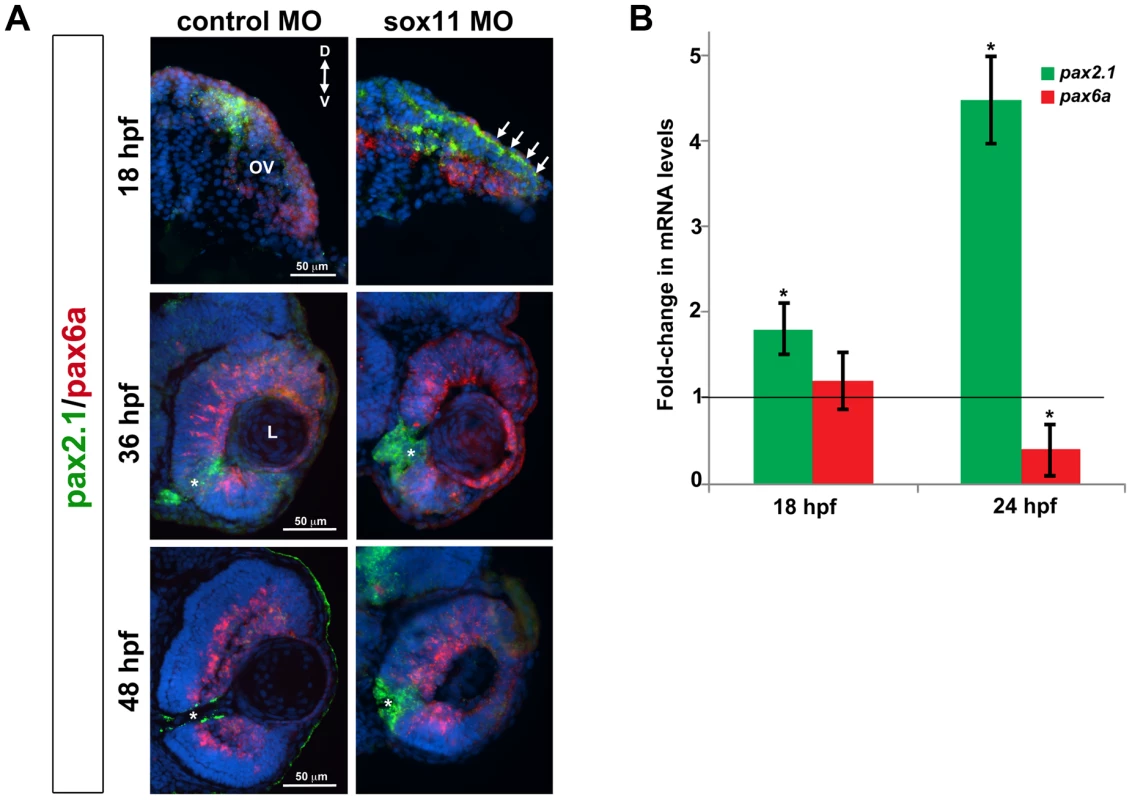
(A) Fluorescent in situ hybridization on transverse sections from control and sox11 morphants with probes for pax2.1 and pax6a. The expression domain of f pax2.1 was expanded into the optic vesicle of sox11 morphants at 18 hpf (top right, arrows), and there was a modest retraction of pax6a expression compared to controls (top left; number of embryos analyzed: control MO, n = 14; sox11 MO, n = 13). At 36 and 48 hpf, in control retinas pax2.1 expression decreased and was only observed lining the optic nerve (asterisk; left middle and bottom rows); in contrast, pax2.1 expression was expanded and persisted around the open choroid fissure in sox11 morphant retinas (asterisks, right middle and bottom rows). Pax6a expression in the retina of sox11 morphants at 36 and 48 hpf appeared comparable to the control morphant retinas at this stage (number of embryos analyzed: 36 hpf control MO, n = 7; 36 hpf sox11 MO, n = 12; 48 hpf control MO, n = 8; 48 hpf sox11 MO, n = 14). (B) QPCR performed on mRNA from control and sox11 morphant heads at 18 and 24 hpf revealed a significant increase in pax2.1 expression at both 18 and 24 hpf, and a downregulation of pax6a expression at 24 hpf, in sox11 morphants compared to controls. Relative transcript abundance was normalized to atp5h (18 hpf) or gapdh (24 hpf) levels and is presented as the mean fold-change in expression relative to controls (n = 50 embryos per group, 3 independent biological replicates. *p<0.05. D, dorsal; V, ventral; OV, optic vesicle; L, lens; hpf, hours post fertilization; MO, morpholino. Fig. 5. Sox11 negatively regulates Hedgehog (Hh) signaling. 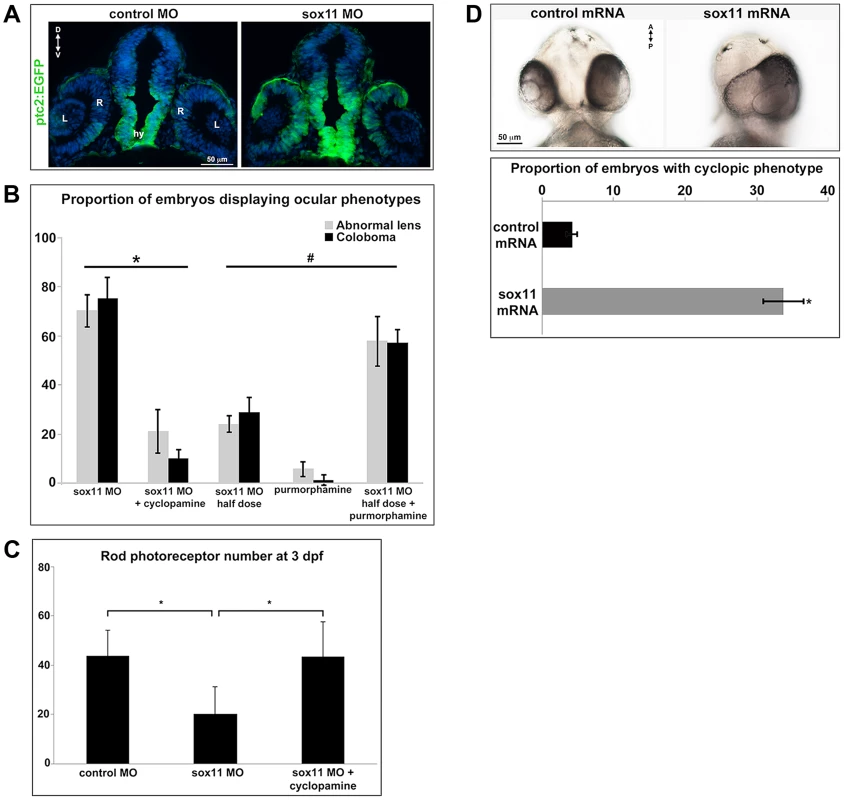
(A) Transverse retinal sections from 24 hpf ptc2:EGFP zebrafish embryos injected with control or sox11 MO. Sox11 morphants displayed elevated GFP expression in the brain as well as in the central and dorsal retina, and the dorsal RPE (number of embryos analyzed: control MO, n = 8; sox11 MO, n = 10). (B) Treatment with the Hh inhibitor cyclopamine rescued the ocular phenotypes in sox11 morphants. In contrast, treatment with the Hh agonist purmorphamine increased the prevalence of ocular phenotypes in embryos injected with a half dose of sox11 MO. Number of embryos analyzed: 24 hpf sox11MO (plus 100% ethanol), n = 393; 2 dpf sox11 MO (plus 100% ethanol), n = 319; 24 hpf sox11 MO plus cyclopamine, n = 276; 2 dpf sox11 MO plus cyclopamine, n = 263; 24 hpf half dose sox11 MO (plus DMSO), n = 258; 2 dpf sox11 MO half dose (plus DMSO), n = 241; 24 hpf uninjected plus purmorphamine, n = 83; 2 dpf uninjected plus purmorphamine, n = 81; 24 hpf half dose sox11 MO plus purmorphamine, n = 291; 2 dpf half dose sox11 MO plus purmorphamine, n = 270; 3 independent biological replicates. * and # p<0.0001, Fisher's exact test. (C) Treatment with cyclopamine rescued rod photoreceptor number in sox11 morphants. Rods were counted in retinal cryosections from 3 dpf embryos. Number of embryos analyzed: control MO, n = 17; sox11 MO, n = 20; sox11 MO plus cyclopamine, n = 18; 3 independent replicates. *p = 0.02, Student's t-test). (D) Overexpression of zebrafish sox11 increased the proportion of embryos with a cyclopic phenotype (right) compared to embryos injected with equimolar amounts of control td-tomato mRNA (left). Number of embryos analyzed: control mRNA, n = 168; sox11 mRNA, n = 202, 3 independent biological replicates.*p<0.001, Fisher's exact test. D, dorsal; V, ventral; A, anterior; P, posterior; hpf, hours post fertilization; dpf, days post fertilization R, retina; hy, hypothalamus; L, lens; MO, morpholino. To directly test this hypothesis, control and sox11 morphant embryos were treated from 5.5–13 hpf with the Hh inhibitor cyclopamine. This treatment window was chosen because it resulted in maximal rescue of colobomas in the blowout mutant [25]. The proportion of embryos displaying a malformed lens at 24 hpf (21.3±8.8%) or coloboma at 2 dpf (10.1±3.8%) was significantly reduced after cyclopamine treatment, compared to vehicle-treated sox11 morphants (>70% for both phenotypes; p<0.0001; Figures 5B and S4A). Moreover, cyclopamine treatment significantly increased the number of rods at 72 hpf (sox11 MO: 20.3±11.1 rods/section; sox11 MO + cyclopamine: 43.8±10.5 rods/section; p = 0.02; Figures 5C and S4B), and corrected the lamination and differentiation defects that were associated with colobomatous retinas (Figure S4C). In a reciprocal experiment, embryos were injected with half the full dose of each sox11 MO, and treated with either a Hh agonist (purmorphamine) or vehicle control (DMSO) from 5.5–24 hpf. We used a sub-threshold dose of purmorphamine (75 µM), which did not cause coloboma when given alone (Figure 5B). In contrast, when the half dose of sox11 MO was combined with purmorphamine, the prevalence of lens malformations at 24 hpf and coloboma at 2 dpf significantly increased (sox11 MO half dose + purmorphamine: 57.9±10.2% malformed lens, 57.2±5.3% coloboma; sox11 MO half dose + DMSO: 24.2±3.5% malformed lens, 28.9±5.9% coloboma; p<0.0001; Figures 5B and S4A). Together, these data demonstrate that deficiency of Sox11 increases Hh signaling, resulting in defects in ocular morphogenesis and reduced rod photoreceptor number.
Finally, we injected sox11a and sox11b mRNA into wildtype zebrafish embryos and evaluated the prevalence of a cyclopic phenotype, which is classically associated with reduced Hh pathway activity [3], at 24 hpf. Injection of sox11a and sox11b mRNA caused a cyclopic phenotype in 33.8±2.9% of the embryos, whereas only 4.2±0.7% of embryos had cyclopia when injected with a control td-tomato mRNA (p<0.001; Figure 5D). Taken together, these results demonstrate that Sox11 is required to limit Hh signaling during zebrafish ocular development.
Transcription of sonic hedgehog a (shha) is strongly upregulated in sox11 morphants
Zebrafish possess five Hedgehog ligands (Sonic hedgehog a and b, Indian hedgehog a and b, and Desert hedgehog), two Patched and one Smoothened receptor, and four Gli effectors. To determine whether expression of any of these pathway members was altered in sox11 morphants, we performed qPCR on mRNA prepared from 18 and 24 hpf control and sox11 morphant heads. At 18 hpf, no significant gene expression changes were observed, except for gli2a and gli3, which were both slightly elevated in sox11 morphants (Figure S5A). In contrast, at 24 hpf we observed a very strong increase (189-fold) in the expression of shha in sox11 morphants relative to controls, as well as a modest decrease in ihhb, ptc1, and gli2b expression, and a 2-fold increase in expression of ptc2 (Figure 6A). The increase in shha expression in sox11 morphants appeared to be dose-dependent, as injection of one-half the full dose of sox11 MOs resulted in only a 65-fold elevation in shha (Figure S5B). In situ hybridization revealed greatly increased shha signal intensity in regions of the sox11 morphant embryo that normally express shha, such as the ventral forebrain and the notochord (Figure 6B), with ectopic expression observed in the dorsal midbrain and telencephalon (Figure 6B). These results suggest that the ocular phenotypes in sox11 morphants are caused by elevated levels of Shha. However, we were puzzled that shha transcript levels were not significantly increased at 18 hpf (Figure S5A), and yet cyclopamine treatment from 5.5–13 hpf rescued the ocular defects of sox11 morphants. Therefore, we asked whether shha levels were elevated in sox11 morphants at earlier time points. We performed qPCR analysis on mRNA from control and sox11 morphants at 8, 10, and 12 hpf and found that shha levels are elevated approximately 2-fold in sox11 morphants at these time points (Figure 6C). Moreover, using the ptc2:GFP line, we detected increased GFP levels in the ventral midline of sox11 morphants at 12 hpf, confirming that Hh signaling was elevated at this stage (Figure 6D). Taken together, these results suggest that knockdown of sox11 results in elevated expression of shha, and an increase in Hh signaling, as early as 8–12 hpf when the optic vesicle is evaginating from the midline.
Fig. 6. Shha expression is upregulated in sox11 morphants. 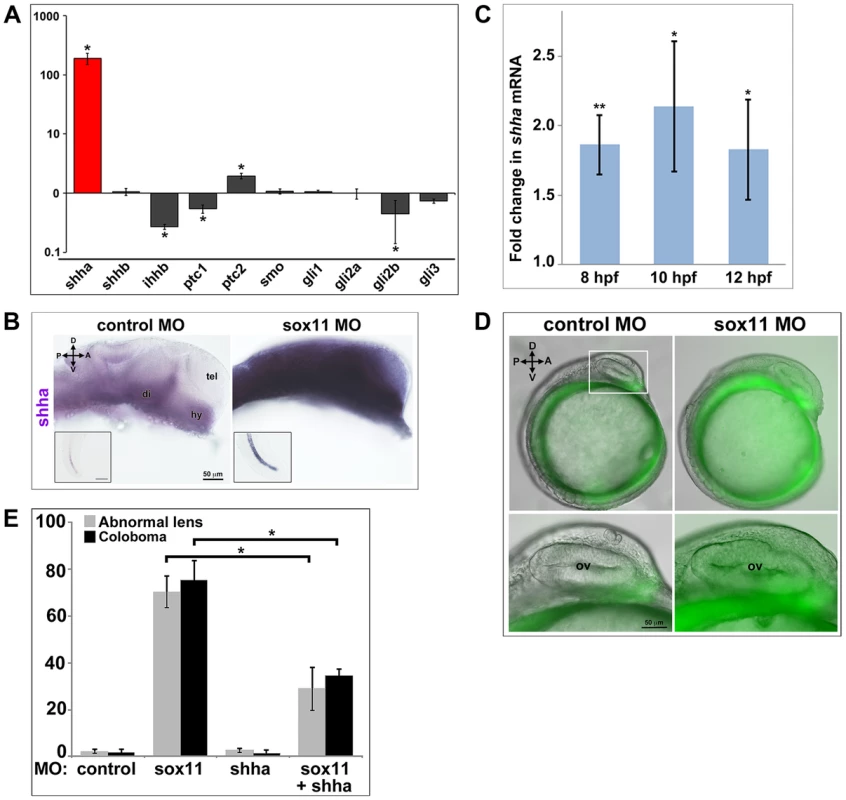
(A) QPCR performed on mRNA from control and sox11 morphant heads at 24 hpf revealed a dramatic upregulation of shha expression, and a small but significant increase in ptc2 expression, in sox11 morphants compared to controls (n = 70 embryos per group, 3 independent biological replicates). Relative transcript abundance was normalized to gapdh levels. The Y-axis (log-scale) represents the mean ratio of sox11 morphant to control expression for three biological and three technical replicates. *p<0.01, Student's t-test. (B) In situ hybridization with a shha probe on control (left) and sox11 morphant (right) embryos at 24 hpf revealed expanded shha expression in sox11 morphants throughout the brain and also in the notochord (inset). Numbers of embryos analyzed: n = 15 embryos per group, 3 independent repeats. (C) QPCR performed on mRNA from control and sox11 morphant heads at 8, 10 and 12 hpf demonstrated an upregulation of shha expression in sox11 morphants compared to controls. Relative transcript abundance was normalized to gapdh levels and is presented as the mean fold-change in expression relative to controls (n = 60 embryos per group, 3 independent biological repeats). **p<0.001, *p = 0.01, Student's t-test. (D) Sox11 morphants (right) on the ptc2:EGFP background displayed elevated GFP expression in the midline at 12 hpf compared to control morphants (left). The bottom panels are an enlargement of the boxed area indicated in the top left panel. Number of embryos analyzed: control MO, n = 34; sox11 MO, n = 41, 3 independent biological replicates. (E) Co-knockdown of shha and sox11 reduced the proportion of embryos displaying abnormal lens and coloboma phenotypes at 24 hpf and 2 dpf, respectively. Number of embryos analyzed: 24 hpf control MO, n = 186; 2 dpf control MO, n = 165; 24 hpf sox11 MO, n = 199, 2 dpf sox11 MO, n = 182; 24 hpf shha MO, n = 249; 2 dpf shha MO, n = 231; 24 hpf sox11+ shha MO, n = 207; 2 dpf sox11+ shha MO, n = 190; 3 independent biological replicates. *p<0.0001, Student's t-test. D, dorsal; V, ventral; A, anterior; P, posterior; hpf, hours post fertilization; dpf, days post fertilization; R, retina; di; diencephalon, tel, telencephalon; hy, hypothalamus; MO, morpholino. To test the hypothesis that elevated Shha levels cause the ocular phenotypes of sox11 morphants, we knocked down both shha and sox11 simultaneously (using our sox11 MOs and a previously described shha MO [35]) and scored embryos at 24 hpf and 2 dpf for malformed lens and coloboma phenotypes, respectively. We used a low dose of the shha MO (3.14 ng/embryo), which by itself did not produce lens defects, coloboma, or rod photoreceptor defects (Figures 6E and S5C). The prevalence of ocular phenotypes was significantly reduced in the double morphants (sox11 MO: 70.3%±6.7% malformed lens, 75.4%±8.3% coloboma; sox11 + shha MOs: 28.9%±9.2% malformed lens; 34.7%±2.7% coloboma; p<0.0001; Figures 6E and S5C). Rod photoreceptor number was also significantly increased at 3 dpf in the double shha/sox11 morphants, however it did not reach the levels observed in controls (sox11 MO: 5.8±7.1 rods/section; sox11 + shha MOs: 14.6±2.3 rods/section; p<0.05; Figure S5D). We performed qPCR analysis on sox11 morphants treated with cyclopamine and purmorphamine and confirmed that these treatments caused a decrease and an increase in shha transcript levels, respectively (Figure S5E, F). Cyclopamine treatment of sox11 morphants also restored expression of the Hh target gene ptc2 to control levels (data not shown). Moreover, qPCR analysis of embryos injected with control or sox11 mRNA confirmed that overexpression of sox11 resulted in a concomitant decrease in shha expression (Figure S5G). Finally, we determined that there was not a reciprocal regulation of sox11 by shha, because injection of the shha morpholino alone did not result in a change in expression of sox11a or sox11b (Figure S5H). Taken together, these results demonstrate that Sox11 controls levels of Hh signaling primarily through negative regulation of shha expression, and that limiting shha expression is essential for proper ocular morphogenesis.
Bmp7b can rescue the ocular phenotypes in sox11 morphants
Thus far, our data strongly suggest that Sox11 is required to limit levels of shha expression during ocular development. However, Sox11 and other members of the SoxC family have previously been shown to function as transcriptional activators rather than repressors [36]–[38]. Furthermore, a scan of the shha promoter revealed no perfect consensus binding sequences for Sox factors (not shown; [37]), and the expression domains of sox11 and shha only partially overlap in the ventral midline during ocular morphogenesis. Therefore, we hypothesized that Sox11 negatively regulates Shha indirectly and perhaps non-cell autonomously, by activating the expression of an upstream inhibitor of Shha. We searched the literature to identify candidate Shha repressors that are expressed in the forebrain during development, and then asked whether expression of any of these factors was reduced in sox11 morphant heads at 24 hpf (Figure 7A). We analyzed five candidate genes: bmp7b, fgfr2, tbx2a, tbx2b, and kras, which had been shown previously to negatively regulate shha expression during development [39]–[44]. Of these five, bmp7b showed significantly decreased expression in sox11 morphants compared to controls (Figure 7A). Bmp7b represents a good candidate intermediary between Sox11 and shha for several reasons. First, Bmp7 null mice display microphthalmia and optic fissure defects, similar to sox11 null mice [45]. Second, bmp7 is expressed in the ventral midline and proximal optic vesicle in the mouse [45], and bmp7b is expressed in the forebrain adjacent to the optic vesicle in zebrafish at 18 hpf in a similar pattern to sox11 [41]. Third, bmp7 expression was reported to be reduced in Sox11−/− mice [13]. And finally, a scan of the bmp7b promoter revealed two perfect Sox consensus binding sites [37] located approximately 950 bp upstream of the transcription start site (not shown).
Fig. 7. Bmp7b expression is reduced in sox11 morphants. 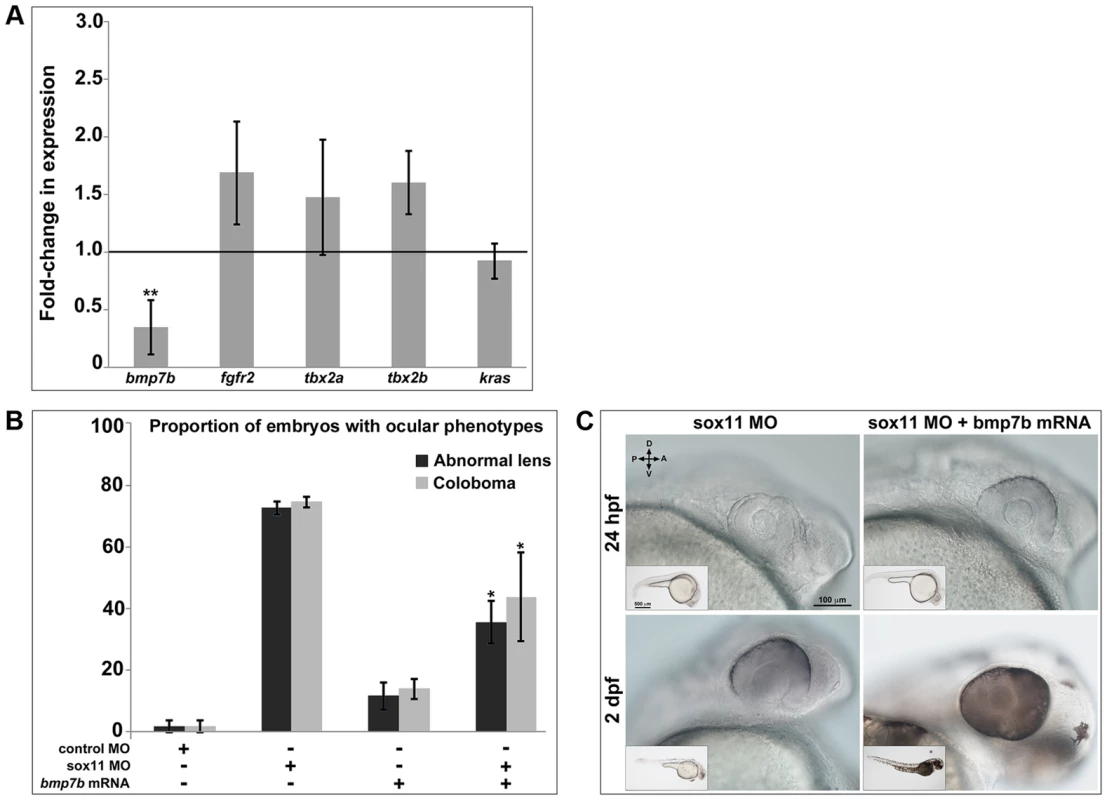
(A) QPCR was performed on mRNA from control and sox11 morphant heads at 24 hpf for known repressors of shha transcription. A significant downregulation of bmp7b was observed in sox11 morphants compared to controls. Relative transcript abundance was normalized to gapdh levels and is presented as the mean fold-change in expression relative to controls (n = 50 embryos per group, 3 independent biological repeats). **p<0.01, Student's t –test. (B) Injection of bmp7b mRNA significantly reduced the proportion of sox11 morphants displaying abnormal lens and coloboma phenotypes at 24 hpf and 2 dpf, respectively. Number of embryos analyzed: 24 hpf control MO, n = 127; 2 dpf control MO, n = 123; 24 hpf sox11 MO, n = 282; 2 dpf sox11 MO, n = 274; 24 hpf bmp7b mRNA, n = 95; 2 dpf bmp7b mRNA, n = 91; 24 hpf sox11 MO + bmp7b mRNA, n = 140, 2 dpf sox11 MO + bmp7b mRNA, n = 134; 3 independent biological replicates. *p<0.006. (C) Brightfield images of a representative sox11 morphant and a sox11 morphant rescued with bmp7b mRNA, taken from the set of embryos analyzed in (B). D, dorsal; V, ventral; A, anterior; P, posterior; hpf, hours post fertilization; dpf, days post fertilization; MO, morpholino. Because we had detected elevated shha levels as early as 8 hpf in sox11 morphants, we asked whether bmp7b expression is also downregulated at that time. qPCR analysis revealed that bmp7b transcript levels were significantly reduced at 8, 10, and 12 hpf in sox11 morphants when compared to controls (Figure S6). Interestingly, bmp7b expression increased to just above control levels at 18 hpf, before declining significantly again at 24 hpf. This rebound in bmp7b expression at 18 hpf precisely mirrors the normal levels of shha expression in sox11 morphants at this time (Figure S5A). Taken together, these data suggest that the initial decrease in bmp7b expression (and corresponding elevation of shha) caused by knockdown of sox11 induces a compensatory pathway that works to bring transcriptional levels back to normal, but that the continued knockdown of sox11 results in renewed dysregulation of bmp7b and shha.
We reasoned that if Bmp7b functions downstream of Sox11 and upstream of Shha, then expression of bmp7b in sox11 morphants should rescue the ocular phenotypes caused by elevated Hh signaling. To test this hypothesis, we injected bmp7b mRNA into control and sox11 morphant embryos, and determined the proportion of embryos that displayed lens defects and coloboma at 24 hpf and 2 dpf, respectively. We found that co-injection of bmp7b mRNA into sox11 morphants significantly reduced the number of embryos displaying ocular phenotypes (sox11 MO: 72.6±2.22% malformed lens, 74.5±1.8% coloboma; sox11 MO + bmp7b mRNA: 35.6±6.9% malformed lens; 43.8±14.4% coloboma; p<0.001; Figure 7B, C), although the rescue was not as large as that observed with cyclopamine treatment. These data suggest that Sox11 negatively regulates shha at least in part through Bmp7b.
Sox4 can compensate for the loss of Sox11
As functional redundancy between SoxC family members has been observed in mouse models [10],[36],[46], we investigated whether another SoxC factor could compensate for the loss of Sox11 during zebrafish ocular morphogenesis. By in situ hybridization and qPCR, we observed elevated expression of the SoxC factor sox4a in sox11 morphants at 24 and 36 hpf, suggesting that sox11 deficiency induces a compensatory increase in sox4 expression (Figure S7A, B). We then injected sox4 mRNA into sox11 morphants and found that this significantly reduced the proportion of embryos with lens and coloboma phenotypes (Figure S7C). This result suggests that increased Sox4 expression may buffer the effects of Sox11 deficiency. Consistent with this hypothesis, we observed a significantly greater proportion of embryos with coloboma in sox4/sox11 double morphants than when either gene was knocked down alone (data not shown).
Identification of SOX11 variants in patients with coloboma
To investigate whether SOX11 mutations contribute to patient phenotypes, the coding region was sequenced in DNA samples from 79 MAC patients [47]. These DNA samples had been previously screened for mutations in two other coloboma-related genes, GDF3 and GDF6 [47]–[49]. We identified heterozygous sequence changes in two probands (Figure 8A), both of whom are Canadians of white European ancestry. The first, a c.488G→T missense mutation in a coloboma patient, is predicted to result in a G145C amino acid alteration, considered damaging by SIFT analysis (http://sift.jcvi.org/). The second variant, a 12-nucleotide duplication (c.1106–1117) in a patient with bilateral iris and retino-choroidal coloboma (Figure 8B), is predicted to result in an in-frame, four amino acid duplication (S351–354dup). The affected amino acid residues are located outside previously defined functional domains and are conserved in chimp and macaque SOX11 (Figure 8A). These variants were absent from dbSNP and the 1000 Genomes databases, and from the NHLBI database comprising more than ten thousand exomes (Figure S8A). Sequencing of SOX11 from the probands' family members revealed that the S351–354dup alteration was present in the proband's mother, who did not exhibit a phenotype clinically (Figure 8C). In light of the rod photoreceptor phenotype in zebrafish sox11 morphants, an electroretinogram (ERG) was performed on the mother carrying the S351–354dup alteration. This analysis demonstrated a reduction in scotopic b-wave amplitude, indicating reduced rod photoreceptor function (Figure S8B). In addition, her 10Hz dim white flicker response was appreciably reduced, and was associated with a change in latency. The mother was asymptomatic at the time the ERG was performed, which may reflect her young age (37 years). Her cone flicker response was normal.
Fig. 8. Association of SOX11 locus with MAC. 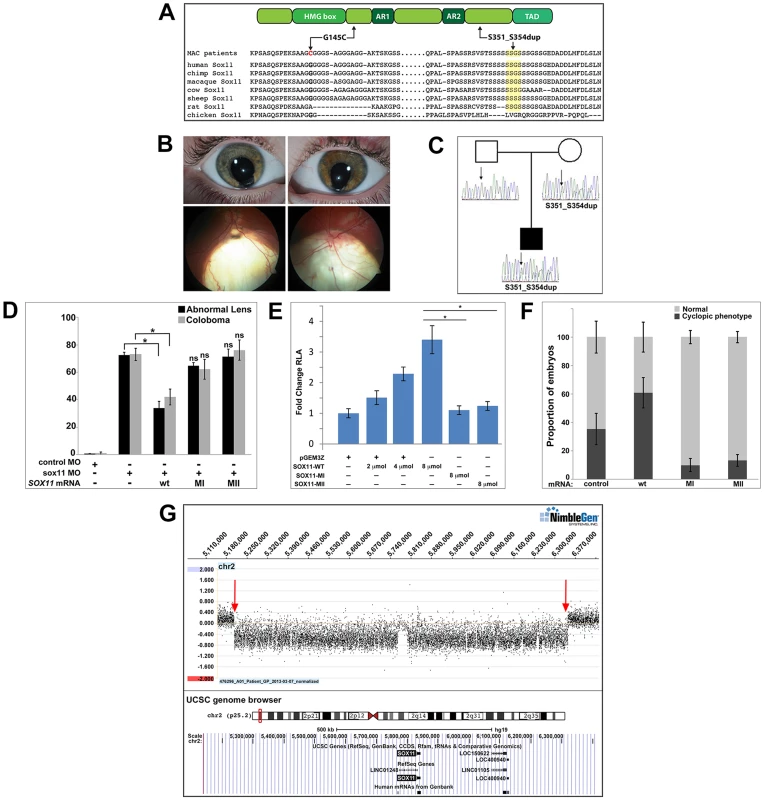
(A) Schematic representation of SOX11, indicating the positions of the two MAC sequence variants, and an alignment of the SOX11 protein sequence encompassing the two affected regions. (B) Photographs of the S351–354dup proband, indicating bi-lateral iris coloboma (top) and retino-choroidal coloboma (bottom). (C) Pedigree showing the S351–354dup proband and his parents. The proband's mother also carries the S351–354dup mutation, but does not have coloboma. (D) SOX11 mRNA containing G145C (MI) or S351–354dup (MII) did not rescue the abnormal lens or coloboma phenotypes of sox11 morphants. Number of embryos analyzed: 24 hpf control MO, n = 202; 2 dpf control MO, n = 174; 24 hpf sox11 MO, n = 148; 2 dpf sox11 MO, n = 133; 24 hpf sox11 MO +wild type SOX11 mRNA, n = 177; 2 dpf sox11 MO + wild type SOX11 mRNA , n = 159; 24 hpf sox11 MO + MI SOX11 mRNA, n = 203, 2 dpf sox11 MO + MI SOX11 mRNA, n = 188; 24 hpf sox11 MO + MII SOX11 mRNA, n = 219, 2 dpf sox11 MO + MII SOX11 mRNA, n = 201; average of three independent biological replicates. *p<0.0001, Fisher's exact test. (E) GDF5-luciferase reporter activity. Transfection of either SOX11 G145C (MI) or S351–354dup (MII) did not significantly enhance luciferase levels. Firefly luciferase activity was normalized to Renilla luciferase and is represented as mean fold change over the empty vector (pGEM3Z) from three biological and six technical replicates (*p<0.0001, Student's t-test). (F) Whereas overexpression of wild type (WT) human SOX11 mRNA increased the proportion of embryos with a cyclopic phenotype compared to injection of control (td-Tomato) mRNA, human SOX11 mRNA containing G145C (MI) or S351–354dup (MII) did not cause cyclopia. Number of embryos analyzed: control mRNA, n = 67; WT SOX11 mRNA, n = 62; MI SOX11 mRNA, n = 165; MII SOX11 mRNA, n = 128, 3 independent biological repeats. *p<0.006 (G) Array CGH data for 2p25.2 demonstrating deletion breakpoints (red arrows) in a patient with agenesis of the optic nerve, microphthalmia, and developmental delay. The corresponding annotated genomic region, modified from the UCSC genome browser (http://genome.ucsc.edu/), is shown below; SOX11 is the only protein-coding gene within the deleted region. MAC, microphthalmia, anophthalmia, and coloboma; MO, morpholino. Intrigued by the presence of phenotypic effects in a heterozygote only on targeted testing, 384 DNA samples derived from patients undergoing screening for hemochromatosis were sequenced, which detected the S351–354dup variant in three individuals, whilst the G145C variant was absent. Unfortunately, these three carriers could not be recalled for clinical examination.
To determine whether the two SOX11 sequence variants had functional consequences, their ability to rescue the lens and coloboma phenotypes of zebrafish sox11 morphants was compared to wild type human SOX11 mRNA. Whereas wild type SOX11 mRNA significantly reduced the proportion of sox11 morphants displaying lens defects and coloboma, no significant rescue was observed with mRNA containing either SOX11 variant (Figures 8D and S8D), suggesting that both sequence changes compromise SOX11 function. Next, we utilized a luciferase reporter containing the promoter region of the SOX11 target gene GDF5 [50] to further analyze the functional consequences of the two mutations. Expression of increasing amounts of wild type SOX11 in COS-7 cells produced a dose-dependent increase in luciferase activity from the GDF5 reporter (Figure 8E). In contrast, transfection of equivalent amounts of either SOX11 variant did not enhance luciferase activity over the empty vector control (Figure 8E), although the variants showed comparable levels of protein expression by Western blot (Figure S8C). Equivalent results were obtained with the luciferase assay in two additional cell lines (HEK293 and HeLa; data not shown). To further confirm that the two SOX11 sequence variants are functionally compromised, we overexpressed them in zebrafish and quantified the proportion of embryos that exhibited a cyclopic phenotype at 24 hpf. Whereas injection of WT human SOX11 mRNA caused a significant increase in the proportion of cyclopic embryos compared to injection of control td-Tomato mRNA (61.16±10.7% in WT SOX11 injected vs. 35.3±11% in control injected; p<0.05), neither of the SOX11 sequence variants produced elevated levels of cyclopia (G145C, 10.0±4.8%, S351–354dup, 13.1±4.11%; Figures 8F and S8E). Taken together, these data suggest that the two variants compromise SOX11's transactivation ability.
Finally, array comparative genomic hybridization (array CGH) was performed on DNA from a patient with microphthalmia, unilateral optic nerve agenesis, and a de novo chromosome 2p25 deletion [18]. This defined a 1.14 Mb segmental deletion (5,206,155–6,343,906; chromosome build GRCh37), encompassing an interval within which SOX11 is the only protein-coding gene (Figures 8G and S8F). Taken together, these data demonstrate that perturbed SOX11 function, either through mutation or decreased gene dosage, contributes to structural (microphthalmia/coloboma) or functional (rod photoreceptor) phenotypes.
Discussion
This study reveals a novel role for Sox11 in maintaining the correct level of Hedgehog (Hh) signaling during ocular morphogenesis. We demonstrate that knockdown of Sox11 in zebrafish perturbs lens formation, induces coloboma, and reduces the number of differentiated rod photoreceptors – phenotypes that can be rescued by pharmacological inhibition of the Hh pathway (cyclopamine) or morpholino inhibition of shha. Comparable lenticular and coloboma phenotypes have also been observed in murine mutants [13], demonstrating that Sox11's function in vertebrate ocular development is evolutionarily conserved. However, the perinatal lethality of Sox11 null mice has precluded a thorough in vivo assessment of rod photoreceptor differentiation, which mostly occurs postnatally. Expression of the rod photoreceptor genes Nrl, Nr2e3, and Sag (Rod arrestin) is significantly reduced in E16 retinas from Sox11−/− mice [51], suggesting that Sox11 does regulate aspects of rod photoreceptor differentiation in mammals. However, in retinal explants derived from Sox11 null mice and cultured for several days, reduced rod photoreceptor number was not observed [51]. Our data suggesting that early, midline-derived Shh influences rod photoreceptor differentiation (see below), indicates that retinal explants, being removed from the source of extra-retinal Shh, may not accurately reflect the in vivo response of retinal progenitor cells to their environment. In this context, the external embryogenesis, rapid pace of retinal development, and continual rod photoreceptor genesis in the zebrafish have benefitted our studies and permitted us to uncover for the first time both the mechanism of Sox11's action during early ocular development, as well as a role for Sox11 in regulating rod photoreceptor differentiation.
A second key finding of our study is that Sox11 acts upstream of Hh signaling specifically by negatively regulating transcription of the ligand shha. In Sox11-deficient embryos, we observed a strong increase in shha expression in the ventral forebrain, as well as an expansion of the shha territory into the dorsal diencephalon and the telencephalon. Therefore, in addition to regulating expression of shha expression in the ventral midline, our data suggest that Sox11 is also required to prevent activation of shha in the more dorsal regions of the brain. Within the retina, the expression of sox11a in the GCL at 48 hpf suggests that Sox11 continues to regulate Hh signaling during retinal neurogenesis.
The magnitude of the increase in shha expression in the absence of sox11 (over 180-fold) at 24 hpf suggests that loss of shha transcriptional repression is accompanied by a significant positive transcriptional feedback loop. However, the Hh target gene ptc2 demonstrated a much smaller increase in expression (2-fold) at this time, raising the question of why the dramatic upregulation in shha did not produce a correspondingly large transcriptional response. One possible explanation is that post-transcriptional mechanisms narrow the range of Shha protein expression in sox11 morphants. Moreover, additional feedback mechanisms may work to attenuate the transcriptional response of Hh target genes such as patched. In any case, the elevated and expanded GFP expression in the Hh reporter line ptc2:GFP, as well as the rescue by cyclopamine and shha co-knockdown, strongly argue that the rise in shha transcription induced by sox11 deficiency has functional consequences.
In the absence of Sox11, we observed an early expansion of the optic stalk marker pax2.1 in the optic vesicle, and a later reduction in the retinal marker pax6a. Such altered proximodistal patterning of the optic vesicle has been observed in several models of elevated Hh signaling [4], [5], [25], [52], [53]. The increased apoptosis in the lens and its abnormal development may be attributable to reduced pax6a expression in sox11 morphants, since similar phenotypes were observed in a lens-specific Pax6 conditional mutant mouse model [54]. In parallel, we suggest the expansion of pax2.1 expression due to elevated levels of Shh enlarged the area of the optic vesicle that was specified as optic stalk, hindering closure of the choroid fissure and thus causing coloboma. Elevated Hh signaling could also account for the increase in mitotic cells in the retina, as this pathway is known to be mitogenic [55].
Previous studies in zebrafish have shown that blocking early Hh signaling, either with shha and shhb morpholinos or by cyclopamine treatment, caused a reduction in rhodopsin expression in the retina, suggesting that Hh signaling promotes rod photoreceptor differentiation [56]. However, murine studies have found that activation of the Hh pathway results in a non-cell autonomous inhibition of rhodopsin expression [57], which is consistent with our results. Moreover, loss of Shh was shown to cause accelerated differentiation of rods and cones in a conditional mouse model [58]. The seemingly paradoxical response to increased and decreased Shh levels is potentially explained by the requirement for precise Shh dosage, with either alteration resulting in reduced photoreceptor number. This accords with a comparable model for Shh's effect on reactive astrocytes [59], and is a well-recognized feature of transcription factors, as exemplified by the effects of altered Pax6 dosage in inducing microphthalmia [60].
Interestingly, we observed a significant increase in shha expression at 8–12 hpf, when the optic vesicle is evaginating from the midline, and we confirmed that Hh signaling was increased at this time using a ptc2:GFP reporter line (Figure 6). Furthermore, treatment of sox11 morphants with cyclopamine during this developmental window was sufficient to restore rod photoreceptor number at 72 hpf. Thus, taken together, these data indicate that early, midline-derived Shh influences rod photoreceptor differentiation. This is not the first demonstration that early midline Hh signals influence later neurogenesis in the retina. It has been shown previously that the timely progression of ath5 expression in the retina, which coincides with the activation of neurogenesis, depends on axial Shh [61]. As ath5-positive cells contribute significantly to the rod photoreceptor lineage [62], it is plausible that elevated Shh coming from the midline in sox11 morphants delays rod photoreceptor differentiation by influencing the cell-intrinsic neurogenic program of retinal progenitor cells.
Although the phenotypes of zebrafish blowout (blw) mutants and sox11 morphants are similar with respect to coloboma, blw mutants do not appear to have a defect in the differentiation of rod photoreceptors or any other retinal cell types. This is surprising, given the well-described influence of Hh signaling on retinal neurogenesis [3], [4], [33], [56], [61], [63]–[66]. Moreover, patients with elevated Hh signaling due to heterozygous loss of function mutations in PTCH exhibit retinal abnormalities, and PtchlacZ+/− mice display a delay in photoreceptor and horizontal cell maturation at P5, all of which is consistent with our data [67]. One possible explanation as to why sox11 morphants and blw mutants differ in this aspect of their phenotype is that the mutation in ptc2 may be a partial loss of function allele, which is supported by the observation that ptc2 morphants display more severe phenotypes than blw mutants [25].
Since SoxC factors are generally considered to function as transcriptional activators rather than repressors [36], [38], we hypothesized that Sox11 regulates Shha indirectly, through the induction of a repressor. Indeed, we found that bmp7b expression was significantly reduced in sox11 morphants, and that injection of bmp7b mRNA into sox11 morphants could rescue the lens and coloboma phenotypes (Figure 7). As Bmp7 has previously been shown to antagonize Shh signaling [42], [68], our results are compatible with a model whereby Bmp7 functions downstream of Sox11 to limit Shh expression during ocular morphogenesis. However, since the magnitude of the bmp7b rescue was not as large as that observed with cyclopamine treatment, additional mechanisms linking Sox11 with the regulation of Hh signaling are likely.
So far, more than 27 genes are associated with coloboma in humans [2], however mutations in these account for less than 20% of cases. Consequently, it is important to define additional causative genes, both to extend understanding of pathogenesis and define pathways that may be amenable to therapeutic modulation. Our work, in combination with previous studies [13], strongly supports a contribution from SOX11 to coloboma phenotypes, however our data indicate that the relationship is complex. With a 50% reduction in gene dosage (2p25 segmental deletion; Figure 8G), a profound phenotype was observed. In contrast, milder coding changes (S351–354dup) with a low prevalence in the general population, resulted in incompletely penetrant phenotypes, with the unaffected carrier exhibiting a sub-clinical phenotype, only detectable on ERG testing. Since the variants had significantly reduced function on in vitro and in vivo assays, this suggests that such mild alleles contribute to MAC but may be insufficient to induce phenotypes alone in all cases. Coloboma, like many developmental defects, exhibits extensive phenotypic variability, suggesting complex relationships between disease genes and modifying alleles that complicate simple genotype-phenotype correlations. It is also possible that oligogenic inheritance is a factor in coloboma, in which individuals in non-penetrant families carry a combination of pathogenic alleles at two or more disease loci, as has been described for other genetically heterogeneous developmental disorders such as the ciliopathies [69]. Furthermore, functional redundancy between Sox subgroup family members is also commonly observed [36], [46], [70], suggesting that one SoxC family member may buffer the effects of mutation in a second. Consistent with this model, we observed elevated expression of the SoxC factor sox4 in sox11 morphants at 24 and 36 hpf, and found that the lens and coloboma phenotypes of sox11 morphants could be rescued by injection of sox4 mRNA (Figure S7). Finally, in light of the incompletely penetrant phenotypes evident with multiple other MAC-causing genes [47], [49], [71]–[73], a similar additive contribution from other SOX gene variants is highly plausible.
In summary, we describe here a novel role for Sox11 in regulating levels of Shh during ocular morphogenesis. It will be interesting to determine whether dysregulated Hh signaling underlies any of the additional developmental defects observed in Sox11−/− mice, such as congenital cardiac malformations and craniofacial anomalies. Future studies will continue to explore the mechanisms of how Sox11 regulates Hh signaling and Shh transcription, as well as the identification of direct molecular targets of Sox11 transcriptional control.
Materials and Methods
Zebrafish
The Tg (XlRho:EGFP)fl1 transgenic line has been previously described [28], and was generously provided by J.M. Fadool (Florida State University, Tallahassee, FL). The Tg (gfap:GFP)mi2001 line has been previously described [74] and was obtained from the Zebrafish International Resource Center (Eugene, OR). The Tg (3.2TαC-EGFP) line has been previously described [75], and was generously provided by S.E. Brockerhoff (University of Washington, Seattle, WA). Tg(GBS-ptch2:nlsEGFP) has been previously described [34] and was kindly provided by R. Karlstrom (University of Massachusetts, Amherst, MA). Zebrafish (Danio rerio) were reared, bred, and staged according to standard protocols [76], [77]. All animal procedures were carried out in accordance with the policies established by the University of Kentucky Institutional Animal Care and Use Committee (IUCAC).
Morpholino (MO) injection and analysis
Morpholinos (MOs) were obtained from Gene Tools, LLC (Philomath, OR) and were prepared and injected as previously described [78]. The following MOs were used in this study: standard control MO, 5′-CCTCTTACCTCAGTTACAATTTATA-3′; sox11a MO1, 5′ –GTGCGTTGTCAGTCCAAAATATCAA-3′; sox11b MO1, 5′ –CATGTTCAAACACACTTTTCCCTCT; shha-MO: 5′CAGCACTCTCGTCAAAAGCCGCATT [35]. The specificity of the sox11 morphant phenotype was confirmed using a second sox11 morpholino placed further downstream of the first set (completely non-overlapping with sox11a MO1, and overlapping by only 4 nucleotides with sox11b MO1). Because the target site for this morpholino extended into the coding region (which is highly similar in sequence for both genes) it simultaneously targets both sox11a and sox11b (sox11 MO2, 5′ –TCCGTTTGCPGCACCATG-3′; the “P” indicates a photo-cleavable moiety that was not used in this study). The sox11 MO2 produced the same coloboma phenotype as the first set of MOs (Figure S1C). All data presented in this study are from embryos injected with sox11a MO1 and sox11b MO1. Unless stated otherwise, embryos were injected with 4.18 ng each of sox11a MO1 and sox11b MO1, 4.18 ng of the standard control MO, or with 3.14 ng of shha MO. We also confirmed that no abnormal phenotypes were observed when embryos were injected with 8 ng of standard control MO. To determine the efficiency of the sox11 MOs, PCR fragments corresponding to the 5′UTRs of sox11a and sox11b encompassing the morpholino target sequences were amplified (using primers listed in Table S1) and cloned upstream and in frame with the EGFP gene in the pEF1α:GFP plasmid (Addgene plasmid 11154). One-cell stage zebrafish embryos were injected with 100 pg/embryo of pEF1α:GFP plasmid containing the MO binding site in the presence or absence of the sox11 MOs. GFP expression in injected embryos was analyzed by fluorescence microscopy at 24 hpf.
mRNA synthesis and injection
Zebrafish sox11a and sox11b or human wild type and variant SOX11 coding sequences were PCR amplified (using primers listed in Table S1) and cloned into the pGEMT-easy vector (Promega). The pCRII-bmp7b plasmid has been previously described [41] and was a kind gift from Dr. S. Fabrizio (The Novartis Institutes for Biomedical Research, Cambridge, MA). The constructs were linearized and mRNA was prepared using the mMESSAGE mMACHINE kit (Ambion) according to manufacturer's instructions. Zebrafish sox11a and sox11b mRNAs (1.0 ng each), human SOX11 mRNA (0.3 ng), zebrafish bmp7b mRNA (1.0 ng) or zebrafish sox4a and sox4b (0.5 ng each) were injected into zebrafish embryos at the one-cell stage. For mRNA rescue experiments, the mRNAs were either co-injected with sox11 MOs, or were injected sequentially after injection of the MOs. As both methods produced similar results, the data presented here are for co-injection of mRNA and morpholino. Injections were always performed in triplicate, and a minimum of 55 injected embryos were analyzed in each experiment. For mRNA overexpression experiments, embryos were injected with either a control (tdTomato) mRNA, zebrafish sox11a/b mRNA, or human WT, G145C (MI), or S351–354dup (MII) SOX11 mRNA, all at equimolar concentrations. The control mRNA was synthesized from pRSET-B-td-Tomato (kindly provided by Dr. D.A. Harrison, University of Kentucky, Lexington, KY). To compare control versus sox11a/b mRNA, 0.003 pmol of each mRNA was injected. To compare control versus human WT and variant SOX11 mRNA, 0.0133 pmol of each mRNA was injected. Zebrafish embryos were injected at the one-cell stage, and embryos were scored for cyclopic phenotypes (one single eye in the center of the head, two eyes that were almost fused at the midline, or one normal eye and one vestigial eye) at 24 hpf.
Patient analysis
To screen for mutations in human SOX11, PCR was performed using three sets of overlapping primers that spanned the entire coding region of the single-exon SOX11 gene. The amplicons were sequenced on an ABI Prism 3100 capillary sequencer (Applied Biosystems), analyzed using DNABaser v.3.1.5 and sequence alignments were performed using ClustalW. Mutations were confirmed by bi-directional Sanger sequencing and RFLP analysis of the SOX11 amplicons. Half of the 384 control DNA samples were screened by RFLP analysis, using TseI (NEB) for the G145C variant and SfcI (NEB) for the S351–S354dup variant, and the other half were screened by direct Sanger sequencing of the SOX11 coding region. Array CGH analysis was performed using a custom designed Nimblegen 4×72 whole human genome array. Oligonucleotide probes were spaced approximately every 75 bp across a 2.65 Mb region at 2p25.2, and backbone probes covered the rest of the genome. Four technical replicates were performed on the proband's DNA, and two replicate hybridizations were performed for each parental DNA sample. Array hybridization and scanning were performed by the Roy Carver Center for Genomics at the University of Iowa (Iowa City, IA). Array data were analyzed using the segMNT analysis program (Nimblegen). Informed consent was obtained from all participants. Study approval was provided by the University of Alberta Hospital Health Research Ethics Board and the Ethics Committee of the IRCCS Oasi Maria SS Onlus, Troina, Italy.
Pharmacological manipulations
Cyclopamine (Sigma) was resuspended at 1 mM concentration in 100% ethanol and diluted in fish water for exposure. A dose response curve was generated by exposing wild type embryos to 0.5, 1.0, and 2.0 µM of cyclopamine from 5.5–13 hpf, and the dose (2.0 µM) at which no abnormal phenotype and negligible toxicity was observed was used for control and sox11 morphants. Purmorphamine (Calbiochem) was resuspended at 50 mM concentration in DMSO and diluted in fish water for exposures. Wild type embryos were exposed to 10–100 µM of purmorphamine from 5.5–24 hpf, and the dose (75 µM) at which no ocular phenotypes were observed was used to treat control and sox11 morphants.
Whole mount in situ hybridization, two-color fluorescent in situ hybridization (FISH) and immunohistochemistry
Whole mount in situ hybridization (WISH) and immunohistochemistry were performed essentially as previously described [78]. For FISH embryos were manually dechorionated and fixed in 4% paraformaldehyde (PFA) made with diethyl pyrocarbonate (DEPC)-treated PBS at 4°C overnight. The fixed embryos were sequentially cryoprotected in 10% sucrose-DEPC and 30% sucrose-DEPC at 4°C overnight. Embryos were then embedded in OCT (Ted Pella, Redding, CA) and frozen at −80°C. Ten-micron sections were collected using a cryostat (Leica CM1900, Leica Biosystems, Buffalo Grove, IL), placed on Superfrost plus glass slides (Fisher Scientific, Waltham, MA) and air dried at room temperature overnight. The sections were post-fixed in 1% PFA-DEPC and rehydrated in PBST-DEPC. The sections were permeabilized for 10 minutes with 1 µg/ml proteinase K. Sections were acetylated in triethanolamine buffer plus 0.25% acetic anhydride (Sigma-Aldrich, Saint Louis, MO), and then rinsed in DEPC treated water. Sections were hybridized with digoxigenin (DIG) and fluorescein (FITC) labeled probes (2.5 ng/µl) in hybridization buffer (0.25% SDS, 10% dextran sulfate, 1× Denhardt's solution , 200 µg/ml torula yeast tRNA, 50% de-ionized formamide, 1 mM EDTA, 600 mM NaCl, and 10 mM Tris pH 7.5 in DEPC-treated water) at 65°C in a sealed humidified chamber for a minimum of 16 hours. Following hybridization, the slides were rinsed in 5× SSC and then with pre-warmed 1× SSC/50% formamide. Endogenous peroxidase activity was quenched with 1% H2O2 for 30 minutes. Sections were blocked using 0.5% PE blocking solution (Perkin Elmer Inc, Waltham, MA) for at least 1 hour. For two-color FISH, sections were incubated first with anti-DIG-POD Fab fragment (Roche, Indianapolis, IN) at 4°C overnight. Subsequently, probe signal was detected using the TSA plus Cy3 kit (Perkin Elmer Inc, Waltham, MA) following the manufacturer's instructions. For the second color detection, the sections were treated with 1% H2O2 for 30 minutes and then incubated with anti-FITC-POD Fab fragment (Roche, Indianapolis, IN) at 4°C overnight. Subsequently, the FITC-labeled probe signal was revealed using TSA plus Fluorescein (Perkin Elmer Inc, Waltham, MA). Finally, sections were counterstained with 4′, 6-diamidino-2-phenylindole (DAPI; Sigma-Aldrich, Saint Louis, MO), mounted in 40% glycerol, and imaged on an inverted fluorescent microscope (Nikon Eclipse Ti-U; Nikon Instruments, Melville, NY) using a 40× objective.
The sox11a, sox11b and shha cDNAs were amplified (using primers listed in Table S1) and cloned from 48 hpf whole embryo cDNA. The sox11b and NeuroD antisense probes have been previously described [14]. The pax6a , crx, and nr2e3 probes have been previously described, and were kindly provided by Y.F. Leung (Purdue University, Indiana). The pax2.1 probe has been described previously [25] and was a gift from J.M. Gross (University of Texas, Austin, TX). The following primary antibodies and dilutions were used: Zpr-1 (1∶20; ZIRC), which labels red-green cones; Zn-8 (1∶10; ZIRC), which labels ganglion cells; anti-Prox-1 (1∶2000; Millipore), which recognizes horizontal cells; anti-PH3 (1∶500; Millipore), which marks cells in G2/M phase; 5E11 (1∶10; J.M. Fadool, Florida State University), which labels amacrine cells; and anti-PKCα (1∶300; Santa Cruz Biotechnology), which labels bipolar cells. Alexa Fluor secondary antibodies (Molecular Probes, Invitrogen) and Cy-conjugated secondary antibodies (Jackson ImmunoResearch) were all used at 1∶200 dilution. Sections from the same region of the eye were analyzed for quantification purposes. One section was quantified per individual embryo (for both control and sox11 morphants).
TUNEL Assay
Terminal deoxynucleotide transferase (TdT)-mediated dUTP nick end labeling (TUNEL) was performed on retinal cryosections using the ApopTag Fluorescein Direct In Situ Apoptosis Detection Kit (Millipore) according to the manufacturer's instructions. Sections from the same region of the eye were analyzed for quantification purposes. One section was quantified per individual embryo (for both control and sox11 morphants).
Real-time quantitative RT-PCR
RNA extracted from the heads of control, sox11, and shha morphant embryos at various time points was used to perform first-strand cDNA synthesis (GoScript Reverse Transcriptase System; Promega). Real time PCR was performed using either Maxima SYBR Green qPCR master mix (Thermo Scientific) or FastStart SYBR Green Master (Roche) on an iCycler iQ Real Time PCR Detection system (Bio-Rad) or LightCycler 96 (Roche) with primers listed in Table S1. Three biological replicates were performed for each experiment. The gene expression change was determined using a relative standard curve quantification method with gapdh, atp5h, or 18s rRNA [79] expression as the normalization control.
Statistics
Statistical analysis was performed on all data using the GraphPad Prism 6.02 software. Continuous data were analyzed using Student's-t-test and Fisher's exact test. For all graphs, data are represented as the mean ± the standard deviation (s.d.).
Dual luciferase assays
COS-7 cells were transfected with the pcDNA3 expression vector (Invitrogen) containing the coding region of wild type, G145C, or S351–354dup SOX11; the pGL3 Firefly Luciferase reporter vector (Promega) containing the GDF5 core promoter was a kind gift from Akinori Kan (Harvard Medical School, Boston, MA) [80]; and the pRL-TK vector (Promega) containing Renilla luciferase driven by a ubiquitous tyrosine kinase promoter to control for transfection efficiency. Transfections were performed using Fugene 6 (Promega), following manufacturer's instructions. The total mass of DNA and molar ratios of pGL3 and pRL-TK were held constant across transfections, which were repeated a minimum of 6 times. Dose response curves were generated using wild type SOX11 at 0∶100, 1∶20, 1∶10, and 1∶5 molar ratios to the GDF5 reporter. The mutant SOX11 variants were transfected at a 1∶5 molar ratio to the GDF5 reporter. Firefly and Renilla luciferase activity were measured 24–36 hours post transfection using the DualGlo Luciferase Assay System (Promega). Data was analyzed as follows: Firefly luciferase (FFLuc) was baselined against untransfected control (UTC) samples ( = FFLuc – UTC) and normalized using the Renilla luciferase (RLuc). The Relative Luciferase Activity (RLA) was calculated as (FFLuc-UTC)/RLuc and compared between experimental and control transfections.
Supporting Information
Zdroje
1. ShahSP, TaylorAE, SowdenJC, RaggeN, Russell-EggittI, et al. (2012) Anophthalmos, Microphthalmos, and Coloboma in the United Kingdom: Clinical Features, Results of Investigations, and Early Management. Ophthalmology 119 : 362–368.
2. ChangL, BlainD, BertuzziS, BrooksBP (2006) Uveal coloboma: clinical and basic science update. Current Opinion in Ophthalmology 17 : 447–470 410.1097/1001.icu.0000243020.0000282380.f0000243026
3. AmatoMA, BoyS, PerronM (2004) Hedgehog signaling in vertebrate eye development: a growing puzzle. Cellular and Molecular Life Sciences CMLS 61 : 899–910.
4. EkkerSC, UngarAR, GreensteinP, von KesslerDP, PorterJA, et al. (1995) Patterning activities of vertebrate hedgehog proteins in the developing eye and brain. Current Biology 5 : 944–955.
5. ZhangX-M, YangX-J (2001) Temporal and Spatial Effects of Sonic Hedgehog Signaling in Chick Eye Morphogenesis. Developmental Biology 233 : 271–290.
6. BakraniaP, Ugur IseriSA, WyattAW, BunyanDJ, LamWWK, et al. (2010) Sonic hedgehog mutations are an uncommon cause of developmental eye anomalies. American Journal of Medical Genetics Part A 152A: 1310–1313.
7. SanyanusinP, SchimmentiLA, McNoeLA, WardTA, PierpontME, et al. (1995) Mutation of the PAX2 gene in a family with optic nerve colobomas, renal anomalies and vesicoureteral reflux. Nature genetics 9 : 358–364.
8. SchimmentiLA, de la CruzJ, LewisRA, KarkeraJD, ManligasGS, et al. (2003) Novel mutation in sonic hedgehog in non-syndromic colobomatous microphthalmia. American Journal of Medical Genetics Part A 116A: 215–221.
9. SlavotinekAM, ChaoR, VacikT, YahyaviM, AbouzeidH, et al. (2012) VAX1 mutation associated with microphthalmia, corpus callosum agenesis, and orofacial clefting: The first description of a VAX1 phenotype in humans. Human mutation 33 : 364–368.
10. Penzo-MéndezAI (2010) Critical roles for SoxC transcription factors in development and cancer. The International Journal of Biochemistry & Cell Biology 42 : 425–428.
11. de BontJM, KrosJM, PassierMM, ReddingiusRE, Sillevis SmittPA, et al. (2008) Differential expression and prognostic significance of SOX genes in pediatric medulloblastoma and ependymoma identified by microarray analysis. Neuro-oncology 10 : 648–660.
12. SockE, RettigSD, EnderichJ, BöslMR, TammER, et al. (2004) Gene Targeting Reveals a Widespread Role for the High-Mobility-Group Transcription Factor Sox11 in Tissue Remodeling. Molecular and cellular biology 24 : 6635–6644.
13. WurmA, SockE, FuchshoferR, WegnerM, TammER (2008) Anterior segment dysgenesis in the eyes of mice deficient for the high-mobility-group transcription factor Sox11. Experimental Eye Research 86 : 895–907.
14. MorrisAC, Forbes-OsborneMA, PillaiLS, FadoolJM (2011) Microarray Analysis of XOPS-mCFP Zebrafish Retina Identifies Genes Associated with Rod Photoreceptor Degeneration and Regeneration. Investigative Ophthalmology & Visual Science 52 : 2255–2266.
15. HeathcoteJG, SholdiceJ, WaltonJC, WillisNR, SergovichFR (1991) Anterior segment mesenchymal dysgenesis associated with partial duplication of the short arm of chromosome 2. Canadian journal of ophthalmology Journal canadien d'ophtalmologie 26 : 35–43.
16. Aviram-GoldringA, FritzB, BartschC, SteuberE, DanielyM, et al. (2000) Molecular cytogenetic studies in three patients with partial trisomy 2p, including CGH from paraffin-embedded tissue. American Journal of Medical Genetics 91 : 74–82.
17. TiradoCA, HendersonS, UddinN, StewartE, IyerS, et al. (2009) Cytogenetic and molecular characterization of a partial trisomy 2p arising from inverted duplication of 2p with terminal deletion of 2pter. American Journal of Medical Genetics Part A 149A: 2507–2512.
18. Lo-CastroA, GianaG, FicheraM, CastigliaL, GrilloL, et al. (2009) Deletion 2p25.2: A cryptic chromosome abnormality in a patient with autism and mental retardation detected using aCGH. European Journal of Medical Genetics 52 : 67–70.
19. RiminiR, BeltrameM, ArgentonF, SzymczakD, CotelliF, et al. (1999) Expression patterns of zebrafish sox11A, sox11B and sox21. Mechanisms of Development 89 : 167–171.
20. De MartinoS, YanY-L, JowettT, PostlethwaitJH, VargaZM, et al. (2000) Expression of sox11 gene duplicates in zebrafish suggests the reciprocal loss of ancestral gene expression patterns in development. Developmental Dynamics 217 : 279–292.
21. NavratilovaP, FredmanD, LenhardB, BeckerTS (2010) Regulatory divergence of the duplicated chromosomal loci sox11a/b by subpartitioning and sequence evolution of enhancers in zebrafish. Molecular genetics and genomics : MGG 283 : 171–184.
22. HuM, EasterSSJr (1999) Retinal Neurogenesis: The Formation of the Initial Central Patch of Postmitotic Cells. Developmental Biology 207 : 309–321.
23. NeumannCJ, Nuesslein-VolhardC (2000) Patterning of the Zebrafish Retina by a Wave of Sonic Hedgehog Activity. Science 289 : 2137–2139.
24. VinothkumarS, RastegarS, TakamiyaM, ErtzerR, SträhleU (2008) Sequential and cooperative action of Fgfs and Shh in the zebrafish retina. Developmental Biology 314 : 200–214.
25. LeeJ, WillerJR, WillerGB, SmithK, GreggRG, et al. (2008) Zebrafish blowout provides genetic evidence for Patched1-mediated negative regulation of Hedgehog signaling within the proximal optic vesicle of the vertebrate eye. Developmental Biology 319 : 10–22.
26. LeeJ, CoxBD, DalyCMS, LeeC, NuckelsRJ, et al. (2012) An ENU Mutagenesis Screen in Zebrafish for Visual System Mutants Identifies a Novel Splice-Acceptor Site Mutation in patched2 that Results in Colobomas. Investigative Ophthalmology & Visual Science 53 : 8214–8221.
27. KimT-H, GoodmanJ, AndersonKV, NiswanderL (2007) Phactr4 Regulates Neural Tube and Optic Fissure Closure by Controlling PP1-, Rb-, and E2F1-Regulated Cell-Cycle Progression. Developmental Cell 13 : 87–102.
28. FadoolJM (2003) Development of a rod photoreceptor mosaic revealed in transgenic zebrafish. Developmental Biology 258 : 277–290.
29. ShenY-c, RaymondPA (2004) Zebrafish cone-rod (crx) homeobox gene promotes retinogenesis. Developmental Biology 269 : 237–251.
30. ChenJ, RattnerA, NathansJ (2005) The Rod Photoreceptor-Specific Nuclear Receptor Nr2e3 Represses Transcription of Multiple Cone-Specific Genes. The Journal of Neuroscience 25 : 118–129.
31. OchocinskaMJ, HitchcockPF (2009) NeuroD regulates proliferation of photoreceptor progenitors in the retina of the zebrafish. Mechanisms of Development 126 : 128–141.
32. BruhnS, CepkoC (1996) Development of the pattern of photoreceptors in the chick retina. The Journal of Neuroscience 16 : 1430–1439.
33. Stenkamp DL (2007) Neurogenesis in the Fish Retina. In: Kwang WJ, editor. International Review of Cytology: Academic Press. pp. 173–224.
34. ShenM-C, OzacarAT, OsgoodM, BoerasC, PinkJ, et al. (2013) Heat-shock–mediated conditional regulation of hedgehog/gli signaling in zebrafish. Developmental Dynamics 242 : 539–549.
35. NaseviciusA, EkkerSC (2000) Effective targeted gene ‘knockdown’ in zebrafish. Nature genetics 26 : 216–220.
36. BergslandM, RamskoldD, ZaouterC, KlumS, SandbergR, et al. (2011) Sequentially acting Sox transcription factors in neural lineage development. Genes & development 25 : 2453–2464.
37. WiebeMS, NowlingTK, RizzinoA (2003) Identification of Novel Domains within Sox-2 and Sox-11 Involved in Autoinhibition of DNA Binding and Partnership Specificity. Journal of Biological Chemistry 278 : 17901–17911.
38. ChewL-J, GalloV (2009) The Yin and Yang of Sox proteins: Activation and repression in development and disease. Journal of Neuroscience Research 87 : 3277–3287.
39. MukhopadhyayA, KrishnaswamiSR, Cowing-ZitronC, HungN-J, Reilly-RhotenH, et al. (2013) Negative regulation of Shh levels by Kras and Fgfr2 during hair follicle development. Developmental Biology 373 : 373–382.
40. ManningL, OhyamaK, SaegerB, HatanoO, WilsonSA, et al. (2006) Regional Morphogenesis in the Hypothalamus: A BMP-Tbx2 Pathway Coordinates Fate and Proliferation through Shh Downregulation. Developmental Cell 11 : 873–885.
41. ShawiM, SerlucaFC (2008) Identification of a BMP7 homolog in zebrafish expressed in developing organ systems. Gene Expression Patterns 8 : 369–375.
42. BastidaMF, ShethR, RosMA (2009) A BMP-Shh negative-feedback loop restricts Shh expression during limb development. Development 136 : 3779–3789.
43. FarinHF, LüdtkeTHW, SchmidtMK, PlaczkoS, Schuster-GosslerK, et al. (2013) Tbx2 Terminates Shh/Fgf Signaling in the Developing Mouse Limb Bud by Direct Repression of Gremlin1. PLoS Genet 9: e1003467.
44. TakabatakeY, TakabatakeT, SasagawaS, TakeshimaK (2002) Conserved expression control and shared activity between cognate T-box genes Tbx2 and Tbx3 in connection with Sonic hedgehog signaling during Xenopus eye development. Development, Growth & Differentiation 44 : 257–271.
45. MorcilloJ, Martínez-MoralesJR, TrousseF, FerminY, SowdenJC, et al. (2006) Proper patterning of the optic fissure requires the sequential activity of BMP7 and SHH. Development 133 : 3179–3190.
46. DyP, Penzo-MéndezA, WangH, PedrazaCE, MacklinWB, et al. (2008) The three SoxC proteins—Sox4, Sox11 and Sox12—exhibit overlapping expression patterns and molecular properties. Nucleic Acids Research 36 : 3101–3117.
47. YeM, Berry-WynneKM, Asai-CoakwellM, SundaresanP, FootzT, et al. (2010) Mutation of the bone morphogenetic protein GDF3 causes ocular and skeletal anomalies. Human Molecular Genetics 19 : 287–298.
48. Asai-CoakwellM, FrenchCR, BerryKM, YeM, KossR, et al. (2007) GDF6, a Novel Locus for a Spectrum of Ocular Developmental Anomalies. The American Journal of Human Genetics 80 : 306–315.
49. Asai-CoakwellM, FrenchCR, YeM, GarchaK, BigotK, et al. (2009) Incomplete penetrance and phenotypic variability characterize Gdf6-attributable oculo-skeletal phenotypes. Human Molecular Genetics 18 : 1110–1121.
50. KanA, IkedaT, FukaiA, NakagawaT, NakamuraK, et al. (2013) SOX11 contributes to the regulation of GDF5 in joint maintenance. BMC developmental biology 13 : 4.
51. UsuiA, MochizukiY, IidaA, MiyauchiE, SatohS, et al. (2013) The early retinal progenitor-expressed gene Sox11 regulates the timing of the differentiation of retinal cells. Development 140 : 740–750.
52. MacdonaldR, BarthKA, XuQ, HolderN, MikkolaI, et al. (1995) Midline signalling is required for Pax gene regulation and patterning of the eyes. Development 121 : 3267–3278.
53. PerronM, BoyS, AmatoMA, ViczianA, KoebernickK, et al. (2003) A novel function for Hedgehog signalling in retinal pigment epithelium differentiation. Development 130 : 1565–1577.
54. ShahamO, SmithAN, RobinsonML, TaketoMM, LangRA, et al. (2009) Pax6 is essential for lens fiber cell differentiation. Development 136 : 2567–2578.
55. MartíE, BovolentaP (2002) Sonic hedgehog in CNS development: one signal, multiple outputs. Trends in Neurosciences 25 : 89–96.
56. StenkampDL, FreyRA, PrabhudesaiSN, RaymondPA (2000) Function for Hedgehog Genes in Zebrafish Retinal Development. Developmental Biology 220 : 238–252.
57. YuC, MazerolleCJ, ThurigS, WangY, PacalM, et al. (2006) Direct and indirect effects of hedgehog pathway activation in the mammalian retina. Molecular and cellular neurosciences 32 : 274–282.
58. WangY, DakuboGD, ThurigS, MazerolleCJ, WallaceVA (2005) Retinal ganglion cell-derived sonic hedgehog locally controls proliferation and the timing of RGC development in the embryonic mouse retina. Development 132 : 5103–5113.
59. SirkoS, BehrendtG, Johansson PiaA, TripathiP, CostaMR, et al. (2013) Reactive Glia in the Injured Brain Acquire Stem Cell Properties in Response to Sonic Hedgehog. Cell Stem Cell 12 : 426–439.
60. SchedlA, RossA, LeeM, EngelkampD, RashbassP, et al. (1996) Influence of PAX6 Gene Dosage on Development: Overexpression Causes Severe Eye Abnormalities. Cell 86 : 71–82.
61. KayJN, LinkBA, BaierH (2005) Staggered cell-intrinsic timing of ath5 expression underlies the wave of ganglion cell neurogenesis in the zebrafish retina. Development 132 : 2573–2585.
62. BrzezinskiJAt, PrasovL, GlaserT (2012) Math5 defines the ganglion cell competence state in a subpopulation of retinal progenitor cells exiting the cell cycle. Developmental Biology 365 : 395–413.
63. KayJN, Finger-BaierKC, RoeserT, StaubW, BaierH (2001) Retinal ganglion cell genesis requires lakritz, a zebrafish atonal homolog. Neuron 30 : 725–736.
64. StenkampDL, FreyRA (2003) Extraretinal and retinal hedgehog signaling sequentially regulate retinal differentiation in zebrafish. Developmental Biology 258 : 349–363.
65. LevineEM, RoelinkH, TurnerJ, RehTA (1997) Sonic Hedgehog Promotes Rod Photoreceptor Differentiation in Mammalian Retinal Cells In Vitro. The Journal of Neuroscience 17 : 6277–6288.
66. WangYP, DakuboG, HowleyP, CampsallKD, MazarolleCJ, et al. (2002) Development of normal retinal organization depends on Sonic hedgehog signaling from ganglion cells. Nature neuroscience 5 : 831–832.
67. BlackGCM, MazerolleCJ, WangY, CampsallKD, PetrinD, et al. (2003) Abnormalities of the vitreoretinal interface caused by dysregulated Hedgehog signaling during retinal development. Human Molecular Genetics 12 : 3269–3276.
68. LiemKFJr, TremmlG, RoelinkH, JessellTM (1995) Dorsal differentiation of neural plate cells induced by BMP-mediated signals from epidermal ectoderm. Cell 82 : 969–979.
69. DavisEE, KatsanisN (2012) The ciliopathies: a transitional model into systems biology of human genetic disease. Current Opinion in Genetics & Development 22 : 290–303.
70. BhattaramP, Penzo-MendezA, SockE, ColmenaresC, KanekoKJ, et al. (2010) Organogenesis relies on SoxC transcription factors for the survival of neural and mesenchymal progenitors. Nature communications 1 : 9.
71. ReisL, TylerR, SchilterK, Abdul-RahmanO, InnisJ, et al. (2011) BMP4 loss-of-function mutations in developmental eye disorders including SHORT syndrome. Human Genetics 130 : 495–504.
72. BakraniaP, EfthymiouM, KleinJC, SaltA, BunyanDJ, et al. (2008) Mutations in BMP4 Cause Eye, Brain, and Digit Developmental Anomalies: Overlap between the BMP4 and Hedgehog Signaling Pathways. The American Journal of Human Genetics 82 : 304–319.
73. MihelecM, AbrahamP, GibsonK, KrowkaR, SusmanR, et al. (2009) Novel SOX2 partner-factor domain mutation in a four-generation family. European journal of human genetics : EJHG 17 : 1417–1422.
74. BernardosRL, RaymondPA (2006) GFAP transgenic zebrafish. Gene Expression Patterns 6 : 1007–1013.
75. KennedyBN, AlvarezY, BrockerhoffSE, StearnsGW, Sapetto-RebowB, et al. (2007) Identification of a Zebrafish Cone Photoreceptor–Specific Promoter and Genetic Rescue of Achromatopsia in the nof Mutant. Investigative Ophthalmology & Visual Science 48 : 522–529.
76. Westerfield M, 1995 (Westerfield, M.,1995) The zebrafish book. A guide for the laboratory use of zebrafish (Danio rerio). 4th ed, Univ of Oregon Press, Eugene.
77. KimmelCB, BallardWW, KimmelSR, UllmannB, SchillingTF (1995) Stages of embryonic development of the zebrafish. Developmental Dynamics 203 : 253–310.
78. Forbes-OsborneMA, WilsonSG, MorrisAC (2013) Insulinoma-associated 1a (Insm1a) is required for photoreceptor differentiation in the zebrafish retina. Developmental Biology 380 : 157–171.
79. McCurleyAT, CallardGV (2008) Characterization of housekeeping genes in zebrafish: male-female differences and effects of tissue type, developmental stage and chemical treatment. BMC molecular biology 9 : 102.
80. KanA, IkedaT, FukaiA, NakagawaT, NakamuraK, et al. (2013) SOX11 contributes to the regulation of GDF5 in joint maintenance. BMC Dev Biol 13 : 4 10.1186/1471-1213X-1113-1184
Štítky
Genetika Reprodukční medicína
Článek Comparative Phylogenomics Uncovers the Impact of Symbiotic Associations on Host Genome EvolutionČlánek Distribution and Medical Impact of Loss-of-Function Variants in the Finnish Founder PopulationČlánek Common Transcriptional Mechanisms for Visual Photoreceptor Cell Differentiation among PancrustaceansČlánek Integrative Genomics Reveals Novel Molecular Pathways and Gene Networks for Coronary Artery DiseaseČlánek An ARID Domain-Containing Protein within Nuclear Bodies Is Required for Sperm Cell Formation inČlánek Knock-In Reporter Mice Demonstrate that DNA Repair by Non-homologous End Joining Declines with Age
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2014 Číslo 7- Akutní intermitentní porfyrie
- Farmakogenetické testování pomáhá předcházet nežádoucím efektům léčiv
- Růst a vývoj dětí narozených pomocí IVF
- Pilotní studie: stres a úzkost v průběhu IVF cyklu
- Vliv melatoninu a cirkadiálního rytmu na ženskou reprodukci
-
Všechny články tohoto čísla
- Cuba: Exploring the History of Admixture and the Genetic Basis of Pigmentation Using Autosomal and Uniparental Markers
- Clonal Architecture of Secondary Acute Myeloid Leukemia Defined by Single-Cell Sequencing
- Mechanisms of Functional Variants That Impair Regulated Bicarbonate Permeation and Increase Risk for Pancreatitis but Not for Cystic Fibrosis
- Nucleosomes Shape DNA Polymorphism and Divergence
- Functional Diversification of Hsp40: Distinct J-Protein Functional Requirements for Two Prions Allow for Chaperone-Dependent Prion Selection
- Comparative Phylogenomics Uncovers the Impact of Symbiotic Associations on Host Genome Evolution
- Activation of the Immune System by Combinations of Common Alleles
- Age-Associated Sperm DNA Methylation Alterations: Possible Implications in Offspring Disease Susceptibility
- Muscle-Specific SIRT1 Gain-of-Function Increases Slow-Twitch Fibers and Ameliorates Pathophysiology in a Mouse Model of Duchenne Muscular Dystrophy
- MDRL lncRNA Regulates the Processing of miR-484 Primary Transcript by Targeting miR-361
- Hypersensitivity of Primordial Germ Cells to Compromised Replication-Associated DNA Repair Involves ATM-p53-p21 Signaling
- Intrapopulation Genome Size Variation in Reflects Life History Variation and Plasticity
- SlmA Antagonism of FtsZ Assembly Employs a Two-pronged Mechanism like MinCD
- Distribution and Medical Impact of Loss-of-Function Variants in the Finnish Founder Population
- Determinative Developmental Cell Lineages Are Robust to Cell Deaths
- DELLA Protein Degradation Is Controlled by a Type-One Protein Phosphatase, TOPP4
- Wnt Signaling Interacts with Bmp and Edn1 to Regulate Dorsal-Ventral Patterning and Growth of the Craniofacial Skeleton
- Common Transcriptional Mechanisms for Visual Photoreceptor Cell Differentiation among Pancrustaceans
- UVB Induces a Genome-Wide Acting Negative Regulatory Mechanism That Operates at the Level of Transcription Initiation in Human Cells
- The Nesprin Family Member ANC-1 Regulates Synapse Formation and Axon Termination by Functioning in a Pathway with RPM-1 and β-Catenin
- Combinatorial Interactions Are Required for the Efficient Recruitment of Pho Repressive Complex (PhoRC) to Polycomb Response Elements
- Recombination in the Human Pseudoautosomal Region PAR1
- Microsatellite Interruptions Stabilize Primate Genomes and Exist as Population-Specific Single Nucleotide Polymorphisms within Individual Human Genomes
- An Intronic microRNA Links Rb/E2F and EGFR Signaling
- An Essential Nonredundant Role for Mycobacterial DnaK in Native Protein Folding
- Integrative Genomics Reveals Novel Molecular Pathways and Gene Networks for Coronary Artery Disease
- The Genomic Landscape of the Ewing Sarcoma Family of Tumors Reveals Recurrent Mutation
- Evolution and Genetic Architecture of Chromatin Accessibility and Function in Yeast
- An ARID Domain-Containing Protein within Nuclear Bodies Is Required for Sperm Cell Formation in
- Stage-Dependent and Locus-Specific Role of Histone Demethylase Jumonji D3 (JMJD3) in the Embryonic Stages of Lung Development
- Genome Wide Association Identifies Common Variants at the Locus Influencing Plasma Cortisol and Corticosteroid Binding Globulin
- Regulation of Feto-Maternal Barrier by Matriptase- and PAR-2-Mediated Signaling Is Required for Placental Morphogenesis and Mouse Embryonic Survival
- Apomictic and Sexual Germline Development Differ with Respect to Cell Cycle, Transcriptional, Hormonal and Epigenetic Regulation
- Functional EF-Hands in Neuronal Calcium Sensor GCAP2 Determine Its Phosphorylation State and Subcellular Distribution , and Are Essential for Photoreceptor Cell Integrity
- Comparison of Methods to Account for Relatedness in Genome-Wide Association Studies with Family-Based Data
- Knock-In Reporter Mice Demonstrate that DNA Repair by Non-homologous End Joining Declines with Age
- Cis and Trans Effects of Human Genomic Variants on Gene Expression
- 8.2% of the Human Genome Is Constrained: Variation in Rates of Turnover across Functional Element Classes in the Human Lineage
- Novel Approach Identifies SNPs in and with Evidence for Parent-of-Origin Effect on Body Mass Index
- Hypoxia Adaptations in the Grey Wolf () from Qinghai-Tibet Plateau
- A Loss of Function Screen of Identified Genome-Wide Association Study Loci Reveals New Genes Controlling Hematopoiesis
- Unraveling Genetic Modifiers in the Mouse Model of Absence Epilepsy
- DNA Topoisomerase 1α Promotes Transcriptional Silencing of Transposable Elements through DNA Methylation and Histone Lysine 9 Dimethylation in
- The Coding and Noncoding Architecture of the Genome
- A Novel Locus Is Associated with Large Artery Atherosclerotic Stroke Using a Genome-Wide Age-at-Onset Informed Approach
- Brg1 Loss Attenuates Aberrant Wnt-Signalling and Prevents Wnt-Dependent Tumourigenesis in the Murine Small Intestine
- The PTK7-Related Transmembrane Proteins Off-track and Off-track 2 Are Co-receptors for Wnt2 Required for Male Fertility
- The Co-factor of LIM Domains (CLIM/LDB/NLI) Maintains Basal Mammary Epithelial Stem Cells and Promotes Breast Tumorigenesis
- Essential Genetic Interactors of Required for Spatial Sequestration and Asymmetrical Inheritance of Protein Aggregates
- Meiosis-Specific Cohesin Component, Is Essential for Maintaining Centromere Chromatid Cohesion, and Required for DNA Repair and Synapsis between Homologous Chromosomes
- Silencing Is Noisy: Population and Cell Level Noise in Telomere-Adjacent Genes Is Dependent on Telomere Position and Sir2
- The Two Cis-Acting Sites, and , Contribute to the Longitudinal Organisation of Chromosome I
- A Broadly Conserved G-Protein-Coupled Receptor Kinase Phosphorylation Mechanism Controls Smoothened Activity
- Requirements for Acute Burn and Chronic Surgical Wound Infection
- LIN-42, the PERIOD homolog, Negatively Regulates MicroRNA Transcription
- WAPL Is Essential for the Prophase Removal of Cohesin during Meiosis
- Expression in Planarian Neoblasts after Injury Controls Anterior Pole Regeneration
- Sox11 Is Required to Maintain Proper Levels of Hedgehog Signaling during Vertebrate Ocular Morphogenesis
- Accumulation of a Threonine Biosynthetic Intermediate Attenuates General Amino Acid Control by Accelerating Degradation of Gcn4 via Pho85 and Cdk8
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Wnt Signaling Interacts with Bmp and Edn1 to Regulate Dorsal-Ventral Patterning and Growth of the Craniofacial Skeleton
- Novel Approach Identifies SNPs in and with Evidence for Parent-of-Origin Effect on Body Mass Index
- Hypoxia Adaptations in the Grey Wolf () from Qinghai-Tibet Plateau
- DNA Topoisomerase 1α Promotes Transcriptional Silencing of Transposable Elements through DNA Methylation and Histone Lysine 9 Dimethylation in
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



