-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaThe Myc-Mondo/Mad Complexes Integrate Diverse Longevity Signals
Transcription factors are essential proteins that regulate the expression of genes and play an important role in most biological processes. The results of our study presented here demonstrate for the first time a role in aging for a small family of transcription factors in the nematode worm Caenorhabditis elegans. Importantly, these proteins have close relatives in higher organisms, including humans that influence metabolism, cell replication, and have been implicated in the development of cancer. Moreover, the loss of one homologue has also been implicated in Williams-Beuren syndrome, a disease characterized in part by signs of premature aging. Our data demonstrate that these transcription factors function within insulin/IGF-1 signaling and dietary restriction, two highly conserved pathways that link nutrient sensing to longevity. Taken together, our findings provide exciting new insight into a family of proteins that may be essential for linking nutrient sensing to longevity and have implications for the improvement of human healthspan.
Published in the journal: . PLoS Genet 10(4): e32767. doi:10.1371/journal.pgen.1004278
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1004278Summary
Transcription factors are essential proteins that regulate the expression of genes and play an important role in most biological processes. The results of our study presented here demonstrate for the first time a role in aging for a small family of transcription factors in the nematode worm Caenorhabditis elegans. Importantly, these proteins have close relatives in higher organisms, including humans that influence metabolism, cell replication, and have been implicated in the development of cancer. Moreover, the loss of one homologue has also been implicated in Williams-Beuren syndrome, a disease characterized in part by signs of premature aging. Our data demonstrate that these transcription factors function within insulin/IGF-1 signaling and dietary restriction, two highly conserved pathways that link nutrient sensing to longevity. Taken together, our findings provide exciting new insight into a family of proteins that may be essential for linking nutrient sensing to longevity and have implications for the improvement of human healthspan.
Introduction
A large body of evidence indicates that sensors and regulators of cell metabolism modulate organismal aging through evolutionarily conserved pathways. Research on the nematode C. elegans has been instrumental in the characterization of many of these pathways, including the insulin/IGF (ILS), Target of Rapamycin (TOR), AMP protein kinase (AMPK), sirtuin (Sir2), and dietary restriction (DR) pathways [1]–[5]. For example, the ILS and DR signaling pathways target the forkhead transcription factors DAF-16 (FoxO) and PHA-4 (FoxA), respectively, suggesting that these two regulatory inputs function independently [6]–[8]. Although much is known about the structure and function of these pathways, many questions remain about how those signals are integrated, the extent of their functional overlap, and how these pathways determine the progression of aging.
The large Myc super-family is comprised of basic helix-loop-helix leucine zipper (bHLHZip) containing transcription factors that are important regulators of cell growth, proliferation, and energy metabolism. In mammals, there are over 100 bHLH transcription factors [9]. Within this larger super-family, the core mammalian “Myc interaction network” consists of 11 proteins: 3 Myc; 4 Mad/Mxi; 2 Mnt/Mga; and 2 Mondo proteins. These proteins can heterodimerize in a complex manner with either Max or Mlx to regulate transcription by binding to a range of enhancer box (E-box) DNA sequences. C. elegans possesses 42 bHLH transcription factors, including a simplified Myc interaction network [10] comprised of four genes: mml-1 (T20B12.6, Myc and Mondo like), mxl-2 (F40G9.11, Mlx), mxl-1 (T19B10.11, Max), and mdl-1 (R03E9.1, Mad) (Figure 1A). While C. elegans lacks a true Myc orthologue, mml-1 contains a region that is highly similar to the N-terminal region of c-Myc, and presumably incorporates both Myc and Mondo functions [11]. Similar to their mammalian counterparts, members of the C. elegans Myc interaction network form obligate heterodimers that bind to a range of E-box sequences and either activate or repress transcription (respectively: MML-1:MXL-2, hereafter referred to as the Myc-Mondo complex and MDL-1:MXL-1, referred to as the Mad complex) (Figure 1A). Unlike mammalian Mlx and Max, these protein-protein interactions are specific, and no evidence exists for cross-heterodimerization. In C. elegans, null mutations in any of the four genes produces no overt developmental phenotype, although a subtle cell migration defect has been reported in mxl-2(tm1516) mutant animals [11]. From a comprehensive genome-wide RNAi screen, we had previously discovered that mxl-2 is essential for the extended longevity of the daf-2(e1370) insulin/IGF1 receptor loss of function mutants [12]. Additionally, mdl-1 has been putatively identified as a target of ILS based on microarray analysis, and loss of mdl-1 has been reported to produce a subtle negative effect on the extension of lifespan in ILS mutants [13].
Fig. 1. The C. elegans Myc-Mondo/Mad complexes have opposing functions in longevity. 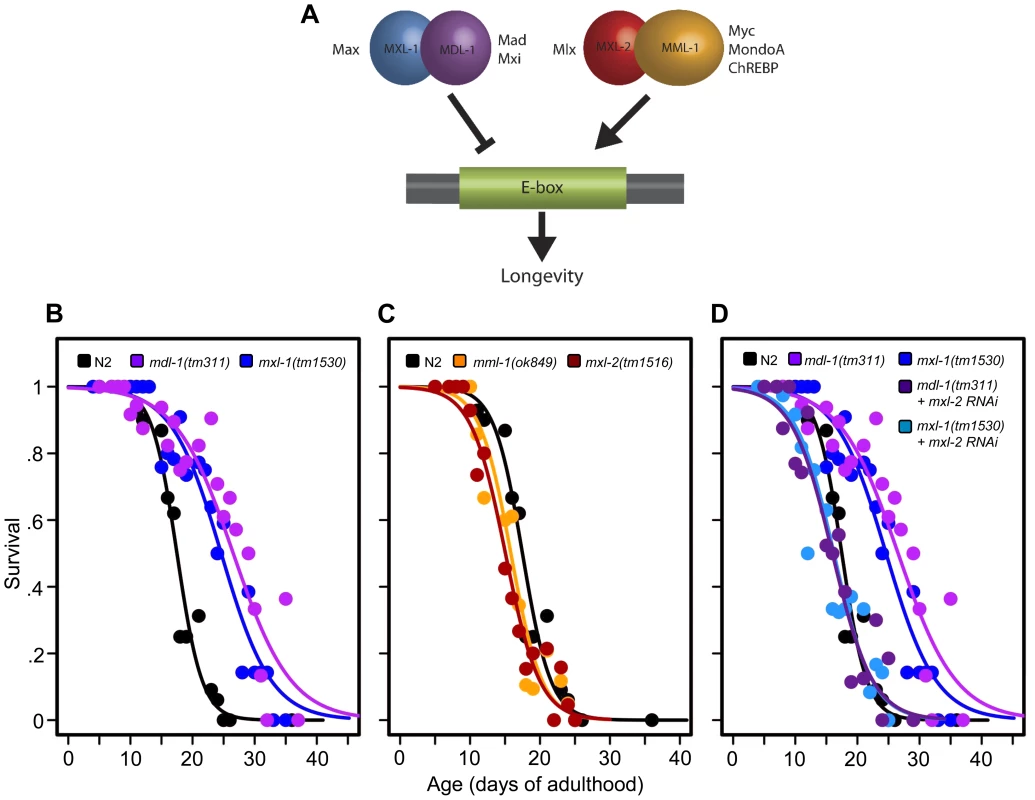
(A) The C. elegans Myc-Mondo/Mad interaction network consists of four proteins that form two heterodimeric complexes, with opposing effects on transcription and longevity. The Myc-Mondo transcriptional activation complex consists of MML-1 and MXL-2, and the Mad transcriptional repression complex consists of MDL-1 and MXL-1 [11]. The mammalian homologues are listed beside each protein. (B) The mdl-1(tm311) and mxl-1(tm1530) null mutations significantly increase lifespan (compare black (wild-type) to purple and blue, respectively). (C) The mml-1(ok849) and mxl-2(tm1516) null mutations significantly shorten lifespan (compare to wild-type (black)). (D) RNAi inactivation of mxl-2 abolishes the lifespan extension conferred by mdl-1(tm311) and mxl-1(tm1530) null mutations (dark purple and dark blue, versus purple and blue traces respectively). Similar results were obtained using mml-1 RNAi (Dataset S1). Graphs illustrate the combined data from multiple trials. Complete information regarding the number of trials, total number of animals examined, and statistical significance for each experiment can be found in Dataset S1. In mammals, two Mondo paralogs (MondoA and ChREBP) function as intracellular sensors of carbohydrate availability and regulators of glycolytic and lipogenic gene expression. MondoA (MLXIP) is predominately expressed in skeletal muscle. When activated by glucose-6-phosphate (G6P), it translocates from the mitochondrial outer membrane to the nucleus where it upregulates the expression of glycolytic genes, by DNA binding to E-box sequences. ChREBP (Carbohydrate Response Element Binding Protein/MLXIPL/MondoB/WBSCR14) is expressed in the liver and is also activated by carbohydrate intermediates, most likely G6P (reviewed in [14]). Activated ChREBP exhibits cytoplasmic-to-nuclear shuttling similar to MondoA and upregulates the expression of both glycolytic and lipogenic genes (triglyceride synthesis) through binding of carbohydrate response elements (ChORE sequences) within the genome [15]–[17]. In mammals, the absence of ChREBP ablates glucose-induced target gene activation [18], [19]. ChREBP –/ – mice display marked physiological changes: large glycogen-laden livers; smaller fat deposits; decreased plasma free fatty acid levels and lipogenic enzyme expression; signs of insulin resistance; reduced glycolytic flux; and enhanced glycogen accumulation [20]. While the specific roles of ChREBP in humans have not been elucidated, the gene encoding ChREBP is one of 27 genes deleted in Williams-Beuren Syndrome (WBS), a complex developmental disorder. Interestingly, WBS patients present signs of premature aging including: premature graying of hair; glucose intolerance; diabetes; and hearing loss that commonly develops during adolescence or young adulthood, concomitant with declining memory skills or dementia [21].
Results
C. elegans Myc-Mondo and Mad Complexes have Opposing Roles in Longevity
Prompted by our previous identification of mxl-2 as an effector of daf-2 mutants in the extension of C. elegans lifespan [12], we asked whether Myc-Mondo and Mad complexes might affect longevity. Thus we obtained null mutant animals for mml-1, mxl-2, mdl-1, and mxl-1, then assessed their lifespan. Loss of either component of the transcriptional repressor complex resulted in a robust extension of lifespan: mdl-1(tm311) and mxl-1(tm1530) single null mutant animals had a 44.5% and 33.2% (p<0.0001) increase in median lifespan compared to wild-type (N2) animals, which had a median lifespan of 18.4+/–0.4 days (Figure 1B, Dataset S1). Using traditional and replica set lifespan approaches, similar results were obtained when either mdl-1 or mxl-1 were inactivated by RNAi (Figure S6B, S6C, Dataset S1). Furthermore, simultaneous inactivation of mxl-1 and mdl-1 resulted in lifespans comparable to the single mutant animals (Figure S1A, S1B, Dataset S1), consistent with published observations in C. elegans that these two proteins only function as an obligate heterodimer [10], [11]. In contrast, loss of either component of the transcriptional activation complex shortened lifespan; mml-1(ok849) and mxl-2(tm1516) single null mutant animals had a 14.1% and 22.8% reduction in median lifespan compared to wild-type animals (Figure 1C, Dataset S1). Inactivation of mml-1 or mxl-2 by RNAi produced a similar result using both traditional and replica set lifespan analyses (Figure S6, Dataset S1). Analogous to what was observed with mdl-1 and mxl-1, simultaneous inactivation of mml-1 and mxl-2 resulted in lifespans comparable to the single mutant animals (Figure S1C, S1D, Dataset S1). Thus loss of the Mad (MDL-1:MXL-1) transcriptional repressor complex extends lifespan, while loss of the Myc-Mondo (MML-1:MXL-2) activator complex shortens lifespan.
We conducted an epistasis analysis to determine whether the lifespan extension conferred by loss of the Mad complex (MDL-1:MXL-1) was dependent on the function of the Myc-Mondo complex (MML-1:MXL-2). Inactivation of either mml-1 or mxl-2 via RNAi fully suppressed the extended longevity of both mdl-1(tm311) and mxl-1(tm1530) mutant animals (Figure 1D, Dataset S1). Therefore, the extended longevity conferred by the loss of the Mad transcriptional repression complex is dependent upon an intact Myc-Mondo complex. Interestingly, C. elegans possess a second Max like protein, MXL-3 (F46G10.6) [22]. Both MXL-2 and MXL-3 share similar sequence homology to mammalian Max; however, MXL-3 likely functions as a homodimer unlike MXL-2 and mammalian Max [10]. Moreover, mxl-3 links lipolysis and autophagy to nutrient availability [10], [23], and mxl-3(ok1947) null mutant animals are also long-lived (Figures S1E, S1F, Dataset S1 and [23]). In contrast to mdl-1 or mxl-1 mutants, inactivation of either mml-1 or mxl-2 had a negligible effect on mxl-3(ok1947) longevity (Figures S1E, S1F, Dataset S1). Thus the Myc-Mondo transactivation complex is required for the extension of lifespan in the absence of the Mad complex. In contrast, Myc-Mondo complex function is dispensable for the extension of lifespan conferred by loss of the Max paralog mxl-3. Collectively, this implies that the Myc-Mondo and Mad complexes have similar functions that influence longevity, which are separate from those of the Max paralog mxl-3.
The Myc-Mondo and Mad Complexes Functionally Overlap with ILS-Mediated Longevity
We next determined whether the Myc-Mondo and Mad complexes functioned in insulin/IGF1 signaling, which is known to depend on the FoxO homologue daf-16. First we asked whether daf-16 was necessary for loss of the Mad complex (MDL-1:MXL-1) to extend longevity; second, we asked whether inactivation of daf-16 further shortened longevity in the absence of the Myc-Mondo complex (MML-1:MXL-2). Inactivation of daf-16 by RNAi completely suppressed the extended lifespan of mxl-1(tm1530) mutant animals (Figure 2A, Dataset S1). Similarly, inactivation of either mdl-1 or mxl-1 failed to confer longevity in daf-16(mgDf47) null mutant animals (Dataset S1). Conversely, daf-16(RNAi) did not further shorten the lifespan of mxl-2(tm1516) null mutant animals (Figure 2A, Dataset S1) and neither mml-1(RNAi) nor mxl-2(RNAi) further shortened the longevity of daf-16(mgDf47) null mutant animals (Dataset S1). Thus daf-16 (FoxO) is essential for loss of the Mad complex to extend longevity, and loss of the Myc-Mondo complex has no further negative effect on longevity in the absence of daf-16.
Fig. 2. The Myc-Mondo/Mad complexes intersect with ILS in longevity. 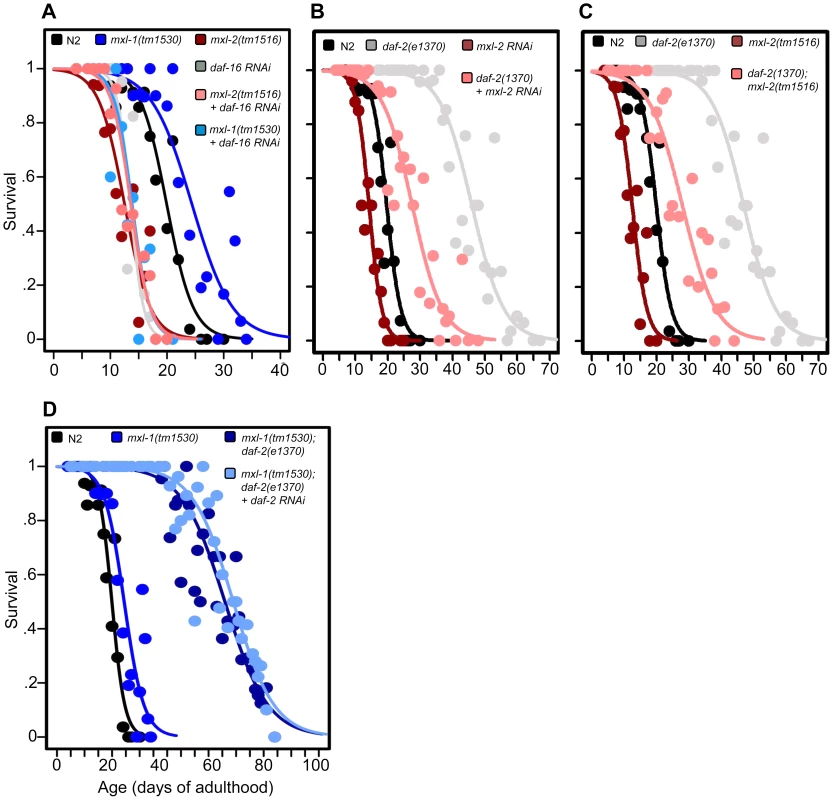
(A) RNAi inactivation of daf-16 ablates the lifespan extension conferred by mxl-1(tm1530) null mutation (dark blue versus blue traces), and does not further shorten the lifespan of mxl-2(tm1516) null mutants (pink versus red traces). Reciprocal experiments show that RNAi inactivation of mxl-1 or mdl-1 do not extend the lifespan of daf-16(mgDf47) null mutant animals, nor does RNAi inactivation of mxl-2 or mml-1 further shorten the lifespan of daf-16(mgDf47) null mutant animals (Dataset S1). (B) RNAi inactivation of mxl-2 partially suppresses the lifespan of daf-2(e1370) mutant animals (pink versus gray traces). Similar results were obtained with mml-1 RNAi (Figure S2A). (C) The mxl-2(tm1516) null mutation partially suppresses the extended lifespan observed in daf-2(e1370) mutant animals (pink versus gray traces). (D) RNAi inactivation of daf-2 does not further extend the lifespan of mxl-1(tm1530); daf-2(e1370) mutant animals (darker blue versus light blue traces). Traces represent the combined data from multiple separate trials. Complete information regarding the number of trials, total number of animals examined, and statistical significance for each experiment can be found in Dataset S1. Experiments shown within this figure were performed simultaneously with those shown in Figure 3 and were split into multiple figures for readability. To further study the role of the Myc proteins in the longevity effects of ILS, we asked whether the transcription-activating Myc-Mondo complex (MML-1:MXL-2) was required for daf-2(e1370) extension of lifespan. daf-2(e1370) mutant animals fed control RNAi had a median and maximum lifespan of 46.0 +/ – 1.2 days and 60.5 +/–2.4 at 20°C, respectively (Figure 2B, Dataset S1). Inactivation of either mxl-2 or mml-1 by RNAi reduced the median lifespan of daf-2(e1370) mutant animals, to 27.6+/–0.9 days and 27.2+/–1.3 days, respectively; a result consistent with our previous findings (Figures 2B, S2A, Dataset S1, and [12]). Similarly, daf-2(e1370);mxl-2(tm1516) double mutant animals had a median lifespan of 28.0+/–1.4 days (Figure 2C, Dataset S1). Thus loss of Myc-Mondo complex function by null mutation reduces daf-2(e1370) median lifespan by 18.0+/–1.4 days and wild-type lifespan by 4.2+/–0.5 days. The reduction caused by loss of mxl-2 is significantly greater (p = 0.001) in the daf-2(e1370) background (39.1%) than in the wild-type background (22.8%). This implies that the effect of the loss of the Myc-Mondo complex on lifespan has specificity to insulin/IGF-1 signaling. This is consistent with the results of a global analysis of synthetic genetic interactions [24], which predicted a relationship between mml-1 and daf-2.
If, as suggested by the above experiments, daf-2 and the Myc proteins are part of a common lifespan regulating system, they would not be expected to act independently. We sought to confirm this hypothesis by testing whether there is additivity between loss of the Mad complex and decreased ILS signaling. However, interpreting an epistasis analysis with ILS mutants is complicated, as even strong temperature-sensitive (ts) alleles of daf-2 [25] retain some level of ILS. For example, placing daf-2(e1370) mutant animals onto daf-2(RNAi) results in a 21.8% increase in median longevity from 46.0+/–1.2 to 56.0+/–1.6 days at 20°C (p = 0.0032) (Figure S2B). In order to perform daf-2 pathway analysis with the lowest sub-lethal amount of insulin-like signaling we examined the lifespan of daf-2(e1370) and daf-2(e1370);mxl-1(tm1530) mutants in the presence of daf-2 RNAi. Unlike daf-2(e1370) single mutant animals, daf-2(e1370);mxl-1(tm1530) double mutant animals show no significant further increase in lifespan on daf-2(RNAi) (p = 0.1164) (Figure 2D), implying that the C. elegans Mad complex and decreased ILS have similar functions in longevity.
The Myc-Mondo and Mad Complexes Contribute to DR-Mediated Longevity
As mammalian MondoA and ChREBP are known sensors of carbohydrate availability [14], we hypothesized that the Myc-Mondo (MML-1:MXL-2) and Mad (MDL-1:MXL-1) complexes may function in DR signaling. The pha-4 (FoxA) transcription factor is a major transcriptional output of DR signaling [8]. Analogous to our analysis with daf-16(RNAi) and ILS, we conducted epistasis experiments with pha-4(RNAi) to determine whether the Myc-Mondo and Mad complexes function in DR signaling. Inactivation of pha-4 completely suppressed the extended lifespan of both mxl-1(tm1530) (p = 0.0001) and mdl-1(tm311) (p = 0.0002) mutant animals, similar to our results with daf-16(RNAi) (Figure 3A, Dataset S1). Additionally, pha-4(RNAi) did not further shorten the lifespan of mxl-2(tm1516) mutant animals (p = 0.5835) (Figure 3A, Dataset S1). Thus pha-4 is required for the extension of lifespan conferred by loss of the Mad complex, and has similar pro-longevity functions as the Myc-Mondo complex.
Fig. 3. The Myc-Mondo/Mad complexes intersect with dietary restriction in longevity. 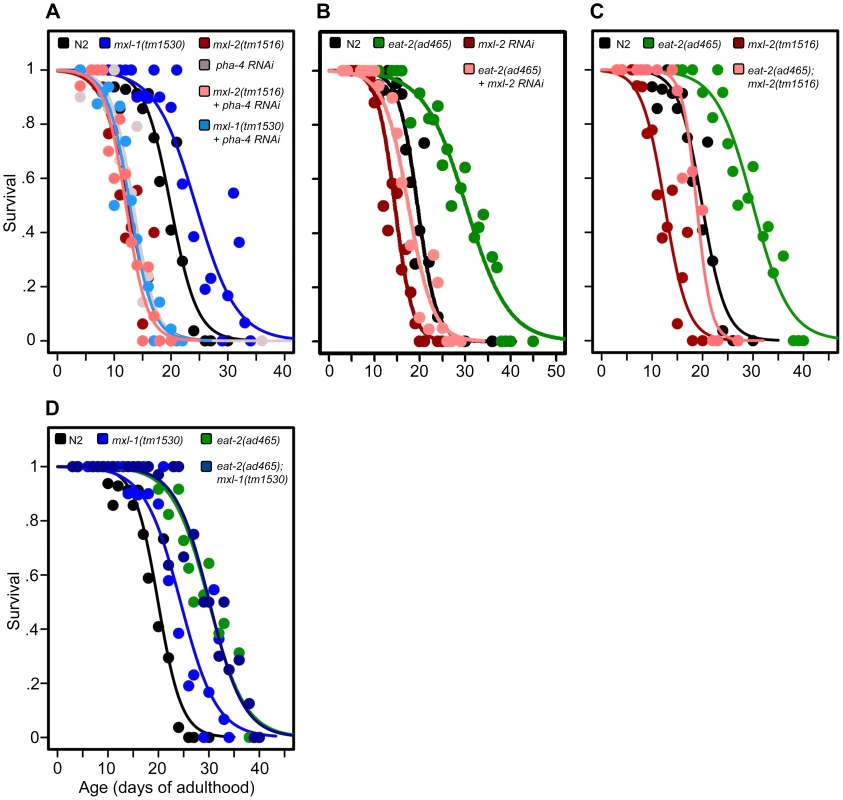
(A) RNAi inactivation of pha-4 ablates the lifespan extension conferred by the mxl-1(tm1530) null mutation (light blue versus blue traces), and does not further shorten the lifespan of mxl-2(tm1516) null mutant animals (pink versus red traces). (B) RNAi inactivation of mxl-2 partially suppresses the extended lifespan observed in eat-2(ad465) mutants (pink versus green traces). Similar results were obtained with mml-1 RNAi (Dataset S1). (C) The mxl-2(tm1516) null mutation partially suppresses the extended lifespan observed in eat-2(ad465) mutant animals (pink versus green traces). (D) The mxl-1(tm1530) null mutation does not further extend the lifespan of eat-2(ad465) mutant animals (dark blue versus green traces). Similar results were obtained with mxl-1 and mdl-1 RNAi (Dataset S1). Traces represent the combined data from multiple separate trials. Complete information regarding the number of trials, total number of animals examined, and statistical significance for each experiment can be found in Dataset S1. Experiments shown within this figure were performed simultaneously with those shown in Figure 2 and were split into multiple figures for readability. Next, we tested whether the Myc-Mondo (MML-1:MXL-2) complex is required for lifespan extension by DR. To do this, we conducted a lifespan analysis in eat-2(ad465) mutant animals, which harbor a mutation in a nicotinic acetylcholine receptor, resulting in drastically decreased pharyngeal pumping and increased lifespan [26], [27]. Notably, eat-2(ad465);mxl-2(tm1516) and eat-2(ad465);mxl-1(tm1530) mutant animals have pumping rates identical to those seen in eat-2(ad465) mutants (Figure S7). Loss of the Myc-Mondo complex by mxl-2(RNAi), or eat-2(ad465);mxl-2(tm1516) double mutant partially suppressed the DR-mediated lifespan extension (Figure 3B, 3C, Dataset S1) by 11.8+/–1.3 days (40.5%), and 10.3+/–1.3 days (35.4%), respectively. This reduction is significantly greater than the 4.1+/–0.5 day decrease (22.7% reduction) in lifespan that results from the loss of mxl-2 in an otherwise wild-type background (p = 0.00055). Thus the Myc-Mondo complex is required for the increased lifespan conferred by DR, similar to what we found with ILS.
Lastly, we tested whether there would be additivity to overall lifespan in the absence of the Mad complex (MDL-1:MXL-1) under conditions of DR. Inactivation of either mxl-1 or mdl-1 did not significantly extend eat-2(ad465) lifespan, with median lifespans of 29.1+/–1.1; 29.3+/–1.1 (p = 0.398); 32.0+/–2.2 days (p = 0.28), respectively (Dataset S1). Additionally, eat-2(ad465) and eat-2(ad465);mxl-1(tm1530) mutant animals have similar lifespans (median lifespans of 29.1+/–1.1, and 30.1+/–1.3 (p = 0.565), Figure 3D, Dataset S1). Collectively, these results indicate that, in addition to ILS, C. elegans Myc-Mondo and Mad complexes function in DR longevity signaling.
TGF-β Signaling Does Not Influence Lifespan by Suppressing Mad Complex Function
The TGF-β pathway functions in parallel with ILS to regulate energy-balance, thereby affecting dauer formation, fat metabolism, egg laying, feeding behavior, and lifespan [13],[28]–[31]. daf-7 encodes a member of the TGF-β superfamily, which inactivates the co-SMAD DAF-3 via the TGF-β Type I and II receptors DAF-1 and DAF-4. Loss of daf-7 extends longevity dependent upon daf-3 [32]. Interestingly, DAF-3 binds the mdl-1 promoter, and mdl-1 expression in the pharynx is increased in daf-3 RNAi-treated animals, suggesting that DAF-3 negatively regulates the expression of mdl-1 [33]. Thus the decreased longevity of daf-3(tm4940) null mutant animals might, at least in part, be explained by upregulation of the Mad complex, and we predicted that loss of the Mad complex might rescue the shortened longevity of daf-3(tm4940) animals. To test this hypothesis, wild-type and daf-3(tm4940) null mutant animals were treated with control and mxl-1 RNAi, and lifespan was assessed. mxl-1 RNAi robustly increased the lifespan of wild-type animals by 5.0+/–0.9 days (27.3% increase, p<0.0001, Figure S3, Dataset S1), which is comparable to the effect of the mxl-1 null mutation (Figure 1B, Dataset S1). In contrast, inactivation of mxl-1 failed to extend the lifespan of daf-3(tm4940) null mutant animals (Figure S3, p = 0.24). Therefore, we conclude that TGF-β signaling does not influence lifespan through regulation of the Mad complex.
Decreased ILS or Conditions of DR Promote Nuclear Accumulation of MML-1 (Myc-Mondo), Dependent on pha-4
In mammals, the MondoA/ChREBP:Mlx complexes exhibit translocation from cytoplasm to nucleus in response to changes in carbohydrate availability [34], [35]. We hypothesized that changes in metabolic status, as relayed through decreased ILS or conditions of DR, might alter MML-1 subcellular localization. To determine whether the C. elegans Myc-Mondo complex (MML-1:MXL-2) might be regulated by a similar translocation, we generated an MML-1::GFP translational reporter under control of the endogenous mml-1 gene promoter. As has been previously reported, MML-1::GFP is broadly expressed (Figure 4A and [11]). However, contrary to previous reports, MML-1::GFP was observed to reside in both the nucleus and the cytoplasm under basal conditions (Figure 4A). The MML-1::GFP fusion was functional, as it rescued the thermotolerance defect of mml-1(ok849) (data not shown). In control animal populations, the relative distribution of MML-1::GFP varied between animals, ranging from entirely nuclear to entirely cytoplasmic; however, subpopulations showed predominately either nuclear with some cytoplasmic (56.1%), or approximately equal distribution between the nucleus and cytoplasm (28.3%) (Figure 4A, File S1). In contrast, decreased ILS (daf-2(e1370)) or conditions of DR (eat-2(ad465)) significantly increased the prevalence of the subpopulation with entirely nuclear MML-1::GFP from 8.3% to 36.1% and 40.0%, respectively (Figure 4A, compare columns 1 to 4 and 1 to 7). The increased nuclear accumulation of MML-1::GFP in response to reduced ILS and conditions of DR supports the idea that this aspect of MML-1/MondoA/ChREBP biology is conserved in nematodes, and provides a potential mechanistic explanation for the requirement of the Myc-Mondo complex in ILS and DR-mediated longevity; specifically that Myc-Mondo complex function is activated by decreased ILS or by conditions of DR to increase the expression of genes that extend longevity.
Fig. 4. ILS and DR pathways differentially regulate mml-1 gene expression and MML-1 localization. 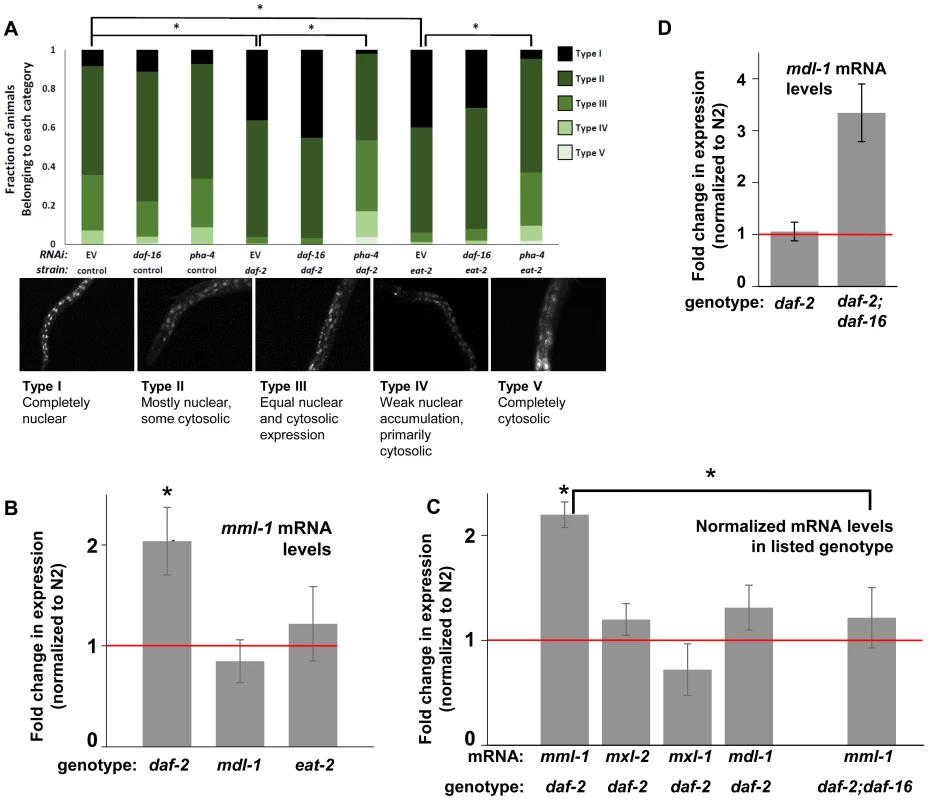
(A) Nuclear accumulation of MML-1::GFP is increased in daf-2(e1370) and eat-2(ad465) mutant animals. Loss of pha-4, but not daf-16, blocks increases in MML-1::GFP nuclear accumulation in daf-2(e1370) and eat-2(ad465), but does not affect MML-1::GFP localization under basal conditions. Representative images of Types I-V are found below the graph. Additional high-resolution images of MML-1::GFP expressing animals can be found in File S1 (* denotes p < 0.05). (B) mml-1 mRNA levels are significantly increased in daf-2(e1370), but not in mdl-1(tm311) or eat-2(ad465) mutant animals, as compared to N2 (* denote p<0.05). (C) mxl-2, mxl-1, and mdl-1 mRNA levels are unaffected in daf-2(e1370) mutants. A daf-16 null mutation suppresses the increases in mml-1 mRNA observed in daf-2(e1370) mutants. (* denotes p<0.01) (D) mdl-1 mRNA levels are significantly increased in daf-2(e1370);daf-16(mgDf47) mutants (* denotes p<0.01). We hypothesized that the nuclear translocation of MML-1 in conditions of decreased ILS might be dependent upon daf-16, while increased nuclear accumulation in conditions of DR would require pha-4. Inactivation of either daf-16 or pha-4 failed to significantly alter the basal spectrum of MML-1::GFP nuclear accumulation in control animals (Figure 4A compare columns 2 and 3 to 1). To our surprise, inactivation of daf-16 failed to alter the increased nuclear accumulation of MML-1::GFP conferred by decreased ILS or conditions of DR (Figure 4A compare column 4 to 5 and column 7 to 8). In contrast, inactivation of pha-4 completely abolished the increased nuclear accumulation of MML-1::GFP conferred by decreased ILS or conditions of DR back to the distributions found in control populations (Figure 4A, compare column 4 to 6 and column 7 to 9). Thus we conclude that pha-4, but not daf-16, is essential for the increased nuclear accumulation of MML-1 under conditions of DR or decreased ILS.
DAF-16 Activity Regulates the Expression of Myc Family Members
We next asked whether ILS or DR influenced the expression of Myc family members. Interestingly, according to the modENCODE database (www.modENCODE.org) only the mml-1 promoter is directly bound by DAF-16, PHA-4, and MDL-1, making it the most likely target for transcriptional regulation in response to aging-relevant signals, such as ILS and DR [36]. Thus we first assessed whether decreased ILS, DR, or loss of mdl-1 altered expression of mml-1 by qRT-PCR. In daf-2(e1370) mutant animals, mml-1 mRNA levels were significantly increased by 2-fold (Figure 4B, p = 0.0027). In contrast, a significant change in mml-1 mRNA levels was not detected in mdl-1(tm311), or eat-2(ad465), mutant animals (Figure 4B, p = 0.3817 and 0.6033). To test whether the increased mRNA expression under conditions of decreased ILS was unique to mml-1, we also used qRT-PCR to measure the relative mRNA levels of mxl-2, mxl-1, and mdl-1 in daf-2(e1370) compared to wild-type animals. Only mml-1 was significantly upregulated under conditions of low ILS (Figure 4C, p = 0.0024); this implies that insulin-like signaling only transcriptionally regulates Myc-Mondo complex function at the level of mml-1 expression. Consistent with that notion, mxl-2 and mxl-1 are not direct targets of DAF-16 as measured by ChIP-seq (www.modENCODE.org), although mxl-1 is the last gene in an operon that is a target of DAF-16. Therefore, it is possible that ILS regulates mxl-1 expression within the context of the operon at a level that cannot be distinguished by qRT-PCR.
Since decreased ILS, but not conditions of DR, regulated the Myc family at the mRNA level, we explored the role of DAF-16 in this regulation. We first determined in qRT-PCR experiments whether the upregulation of mml-1 by decreased ILS was daf-16 dependent. The increased mml-1 mRNA levels observed in daf-2(e1370) mutant animals is daf-16 dependent, as daf-2(e1370);daf-16(mgDf47) double mutant animals expressed mml-1 at levels comparable to wild-type control animals (Figure 4C). Thus we conclude that decreased ILS activates DAF-16 to induce expression of mml-1, perhaps as a feed-forward mechanism to amplify DAF-16 signaling. Since MML-1 (Mondo) and MDL-1 (Mad) have opposing roles in transcription and longevity control, we hypothesized that loss of daf-16 might induce expression of mdl-1. Indeed, daf-2(e1370);daf-16(mgDf47) double mutant animals have a 3-fold increase in mdl-1 mRNA levels (Figure 4D, p = 0.0013). DAF-16 binds in the promoter region of mdl-1 (http://www.modENCODE.org and [37]). Thus we conclude that DAF-16 activity regulates expression of the key components of the Myc-Mondo and Mad complexes in a manner that correlates with their role in longevity. Specifically, decreased ILS activates DAF-16, which in turn results in increased mml-1 expression and extension of longevity; in contrast, loss of daf-16 results in increased mdl-1 expression and shortened lifespan.
DAF-16, PHA-4, and MDL-1 Genomic Binding Overlaps at Metabolic and Cytoprotective Target Genes
We found that the Myc family members are regulated by DR and ILS signaling and have overlapping functions in longevity control, implying that the C. elegans Myc-Mondo/Mad complexes may cooperate with PHA-4 and DAF-16 at shared target gene promoters. Through an informatics analysis of available ChIP-seq data provided by the modENCODE project (www.modENCODE.org) [38], we sought to identify the overlap in genomic binding sites for DAF-16, PHA-4, and MDL-1. We compared the global frequency of the MDL-1 and PHA-4 binding midpoints to the position of the transcription start site (TSS), as has been done for DAF-16 [37]. MDL-1 and PHA-4 had enriched binding frequency within a region almost identical to what has been reported for DAF-16 (between –700 to +100 of the TSS, Figure 5A). We used this metric to distinguish genes more likely to be regulated by MDL-1, and identified 4605 possible target genes. The Myc family of bHLH transcription factors bind to E box sequences, therefore we asked whether the MDL-1 binding regions identified by ChIP-seq also contained E boxes, and found this to be true in 3555 cases (77.2%, Dataset S2). Next, we found that there is significant overlap of 2879 target genes that are bound by DAF-16, PHA-4, and MDL-1 (Figure 5B, p = 2.2e−16). Of 963 genes that have been identified as influencing longevity (based on published studies, gene ontology and description), 299 are bound by DAF-16, PHA-4, and MDL-1. In contrast, only 13 previously identified longevity genes are bound by MDL-1 and not DAF-16 or PHA-4, a significant enrichment (p = 2.2e−16). We next asked whether the actual binding sites of the three transcription factors overlapped. For 234 of 299 putative longevity genes the MDL-1 binding peak is 75% or more overlapped by DAF-16 and PHA-4, suggesting that binding occurs in common regulatory regions. We conclude that DAF-16, PHA-4, and MDL-1 bind at many common target genes, in common regions.
Fig. 5. ChIP-seq data shows significant overlap amongst MDL-1, DAF-16, and PHA-4 promoter binding. 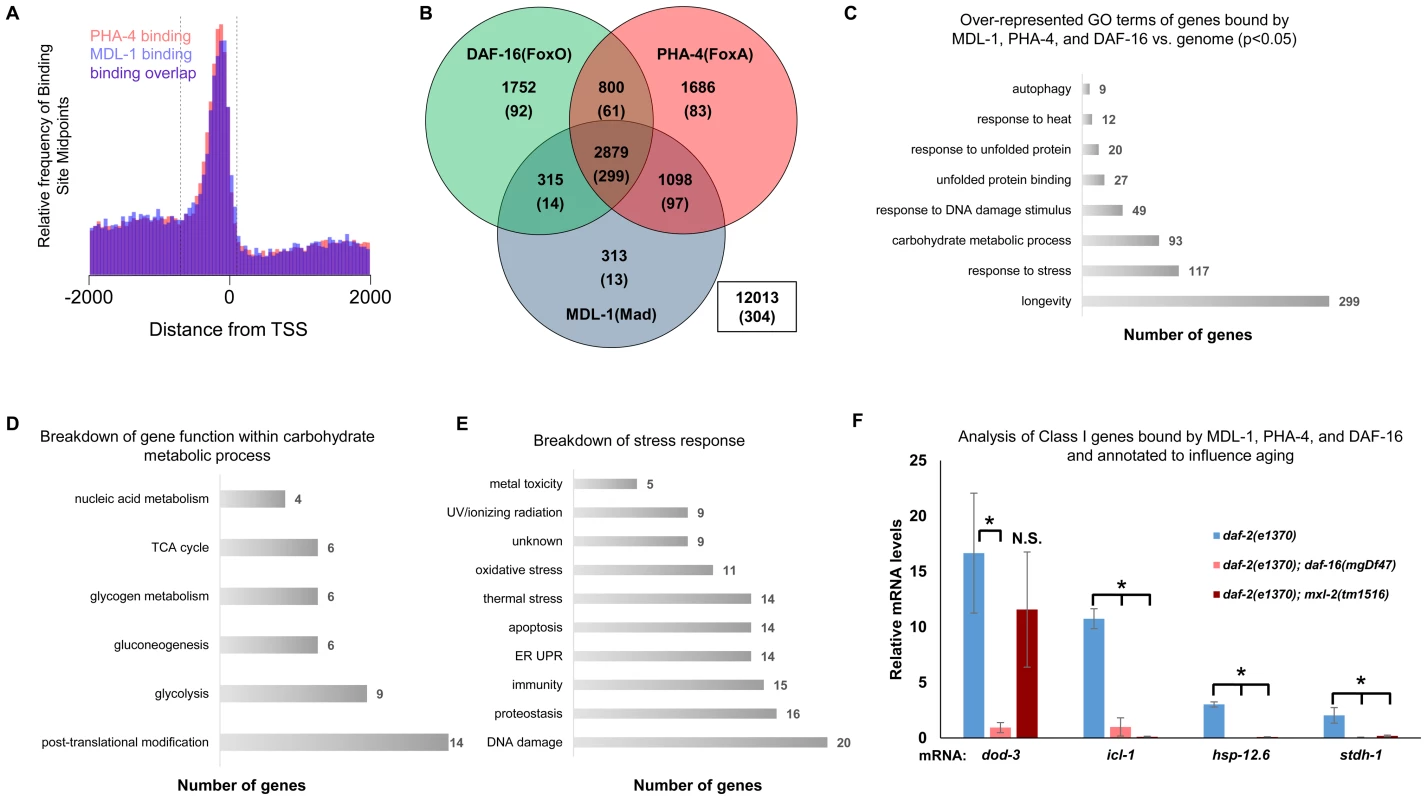
(A) Analysis of binding sites throughout the genome indicates that MDL-1 (blue) and PHA-4 (red) bind predominantly in a region located –700 to +100 bp (dashed lines) relative to the transcriptional start site (TSS). (B) Venn diagram based on ChIP-Seq data showing gene promoters bound by MDL-1, DAF-16, and PHA-4. Numbers indicate the total number of genes belonging to each group. Parenthesis denote the number of genes belonging to each group that have previously been annotated to be involved in aging (based on published studies, gene ontology and description). The inset box shows the total number of genes in the genome that are not bound by MDL-1, DAF-16, and PHA-4. (C) GO terms related to aging that are enriched in genes bound by MDL-1, DAF-16, and PHA-4 versus the entire genome (p <0.05). The total number of common target genes annotated with each GO term is shown to the right of each bar. Enrichment was determined as described in Materials and Methods. Details of this analysis can be found in Dataset S2 (D) Analysis of metabolic pathways contained within the GO term “carbohydrate metabolic process” (GO: 0005975). The total number of genes annotated within each pathway is shown. Some genes belong to more than one category, notably several genes function in glycolysis and gluconeogenesis. Details can be found in Dataset S2 (E) An analysis of stress response pathways contained with the GO term “response to stress” (GO: 0006950). The total number of genes annotated to be involved in each pathway is shown. Many genes are important for resistance to multiple stresses. Details can be found in Dataset S2. (F) qRT-PCR analysis of Class I genes [37] known to be bound by MDL-1, DAF-16, and PHA-4 in daf-2(e1370), daf-2(e1370);daf-16(mgDf47), and daf-2(e1370);mxl-2(tm1516) animals. Class I genes were significantly upregulated (* denotes p<0.01) in daf-2(e1370) in a daf-16 dependent manner as previously described [13], [37]. Additionally, icl-1, hsp-12.6, and stdh-1 failed to be induced in daf-2(e1370);mxl-2(tm1516) mutants as well. To gain mechanistic insight into the potential transcriptional programs co-regulated by DAF-16, PHA-4, and MDL-1, we determined whether there was over-representation of aging related gene ontology terms at putative common target genes. Comparing gene ontology of genes bound by DAF-16, PHA-4, and MDL-1 versus all genes revealed that determination of adult lifespan, carbohydrate metabolism, autophagy, unfolded protein binding, and stress response were significantly over-represented (Figure 5C and Dataset S2). An analysis of all potential MDL-1 targets (an additional 1726 genes) revealed similar classes of over-represented gene functions. Further, comparing MDL-1 target genes to previously identified putative targets of mammalian Mad1 [39], [40] reveals a large degree of overlap, which implies evolutionary conservation (Dataset S2). This suggests that DAF-16, PHA-4, and the Myc family converge on a significant subset of common target genes that have functions relevant to aging, carbohydrate metabolism, proteostasis, and stress response.
An examination of the specific carbohydrate metabolic pathways putatively regulated by MDL-1, DAF-16, and PHA-4 revealed a number of genes involved in glycolysis, gluconeogenesis, and glycogen metabolism (Figure 5D, Dataset S2). This is consistent with the reported function of the mammalian Mondo/ChREBP complexes, which are essential for the expression of genes in the same metabolic pathways [18]–[20], [41], [42]. Additionally, our analysis identified a number of stress response pathways that may be influenced by MDL-1 in conjunction with DAF-16 and PHA-4, including DNA damage response, proteostasis, unfolded protein response (UPR), oxidative stress, and thermotolerance (Figure 5E, Dataset S2).
If DAF-16 and Myc family members converge on common target genes, then they should have similar functions in longevity control. We tested this hypothesis by assessing whether overexpression of a DAF-16::GFP translational fusion could rescue the shortened lifespan that occurs in the absence of the Myc-Mondo activation complex. Compared to wild-type control, daf-16 overexpression increased lifespan by 6 days (median lifespan of 17 and 23 days, respectively a 35.3% increase, p<0.0001) consistent with published findings [43]. Interestingly, overexpression of daf-16 completely rescued the shortened lifespan of the mxl-2(tm1516) null mutation (Figure S4). Additionally, the mxl-1(tm1530)) null mutation did not further extend lifespan when daf-16 was over-expressed (Figure S4). Thus over-expression of daf-16 is sufficient to compensate for the lack of Myc-Mondo transactivation, and loss of the Mad transcriptional repression complex does not further extend lifespan when daf-16 is overexpressed, which is consistent with the notion that these transcription factors may act at common target genes.
We next asked whether Myc-Mondo function was necessary for the activation of DAF-16 target genes under conditions of decreased ILS. To this end, we measured mRNA expression levels of several candidates by qRT-PCR, which were chosen based on the criteria that they:are bound by DAF-16, PHA-4, and MDL-1 (modENCODE and [37]); are upregulated under conditions of decreased ILS in a daf-16 dependent manner (i.e. are “Class 1” DAF-16 target genes, [37]); and have been implicated in the determination of adult longevity (GO term: 0008340). Based on these criteria five candidate genes were chosen: dod-3, an apparently nematode-specific gene previously shown to be “downstream of daf-16” [13]; icl-1, encoding isocitrate lyase and malate synthetase, which together form the glyoxylate shunt, an alternative metabolic pathway known to be favored in daf-2 mutants, dietarily restricted worms, and Mit mutants [13], [44], [45]; sodh-1 which encodes a sorbitol dehydrogenase; hsp-12.6, a small heat-shock protein; and stdh-1, a sterol dehydrogenase. Consistent with published findings, all but sodh-1 were induced by decreased ILS in a daf-16 dependent manner [37]. Of these four genes, the expression of three (icl-1, hsp-12.6, and stdh-1) were also suppressed in daf-2(e1370);mxl-2(tm1516) double mutant animals (Figure 5F). Additionally, we discovered that Myc-Mondo complex was not necessary for the increased nuclear accumulation of DAF-16 under conditions of decreased ILS, as GFP was solely nuclear in daf-2(e1370);DAF-16::GFP and daf-2(e1370);mxl-2(tm1516);DAF-16::GFP mutant animals (data not shown). We conclude that the Myc-Mondo transcriptional activation complex is necessary for the activation of at least a subset of DAF-16 target genes under conditions of decreased ILS.
The C. elegans Myc-Mondo Complex (MML-1:MXL-2) Is Required for Resistance to Oxidative and Thermal Stress
Stress resistance is intimately connected to aging. For instance, reactive oxygen species cause the accumulation of oxidative damage to proteins and other cellular constituents as an organism ages [46]. Thermal stress also challenges the molecular chaperone network, leading to the collapse of proteostasis and a shortened lifespan [47]. Indeed, many of the genes known to influence longevity also function in stress response; the most studied example being ILS and DAF-16 (reviewed in [48]). DAF-16 functions in numerous forms of stress response including: heat shock, oxidative damage, exposure to heavy metals, radiation, anoxia, osmotic, and pathogen exposure [49]–[54]. Thus sensors of metabolic status or environmental damage may also regulate the expression of cytoprotective genes that influence aging. Based on the functional overlap between DAF-16 and Myc family members, we hypothesized that the Myc family influences longevity through altered stress response.
To assess whether loss of the Myc-Mondo or Mad complex altered C. elegans oxidative stress resistance, N2, mxl-2(tm1516), and mxl-1(tm1530) mutants were grown until day 2 of adulthood and survival was measured following oxidative stress imposed by exposure to tert-butylhydroperoxide (tBOOH; an organic peroxide). While loss of mxl-1 failed to improve survival under any conditions (data not shown), loss of the Myc-Mondo complex via deletion of mxl-2 impaired survival to oxidative stress (Figure 6A, Dataset S3). Specifically, following exposure to 7.7mM tBOOH, median survival was reduced from 13.6+/–0.8 hours in N2 animals, to 7.5+/–0.5 hours in mxl-2(tm1516) animals, a reduction of 45% (Figure 6A, Dataset S3). As expected, daf-2(e1370) mutant animals were much more resistant to tBOOH treatment; median survival was 25.0+/–1.0 hours. However, the median survival of daf-2(e1370);mxl-2(tm1516) double mutant animals was only 15.5+/–0.6 hours, a reduction of 38% (Figure 6A, Dataset S3). Thus loss of mxl-2 does not reduce daf-2(e1370) tBOOH survival to a greater extent when compared to N2 (p = 0.986). To our surprise, eat-2(ad465) mutant animals were significantly more sensitive to tBOOH treatment than wild-type animals (data not shown), precluding a requirement to test the effects of mxl-2 deletion in this background. Thus the Myc-Mondo complex is essential for resistance to oxidative stress and loss of the Mad complex is insufficient to increase stress resistance. Our bioinformatic analysis of oxidative stress response genes found that 12 out of 50 genes annotated as responding to oxidative stress are bound by DAF-16 and MDL-1 (i.e. no overrepresentation vs. the genome, p = 0.088). In contrast, 20 out of 50 oxidative stress response genes are bound by DAF-16, a significant enrichment (p = 0.049). Taken together, these data suggest that the C. elegans Myc-Mondo complex is generally required for resistance to oxidative stress, rather than specifically required for the enhanced resistance of the daf-2(e1370) mutant.
Fig. 6. Loss of the Myc-Mondo complex impairs resistance to oxidative and thermal stress. 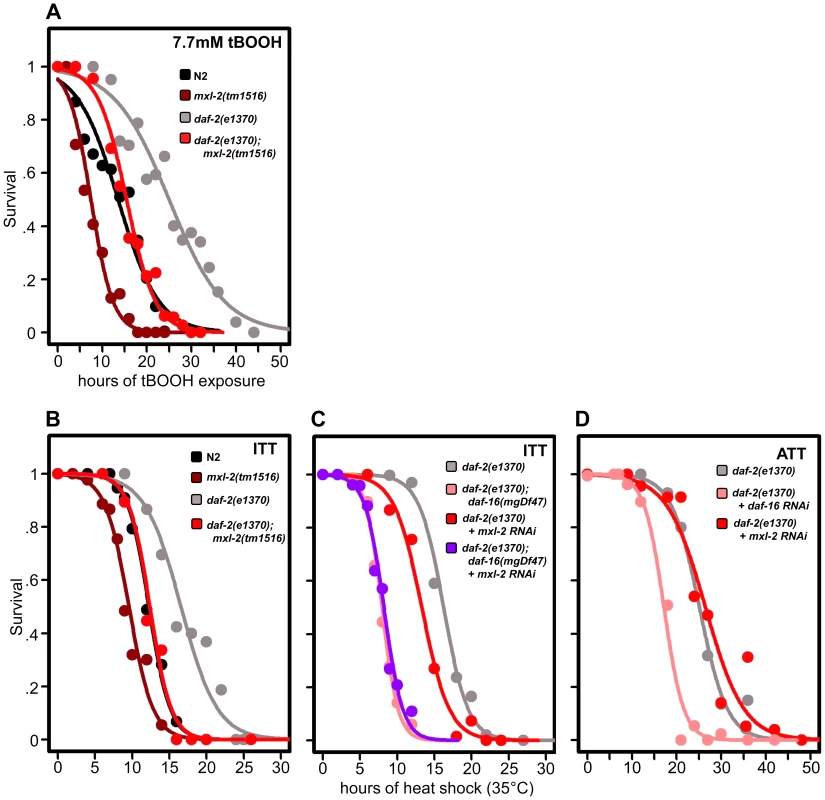
(A) Loss of mxl-2 impairs resistance to oxidative stress imposed by the exposure to tert-butylhydroperoxide (tBOOH) in wild-type and daf-2(e1370) mutant animals. Animals were exposed to 7.7mM tBOOH for the denoted time and survival was assessed. Loss of mxl-2 in wild-type and daf-2(e1370) mutant animals impaired tBOOH survival to a similar extent. (B) Loss of mxl-2 significantly impairs intrinsic thermotolerance (ITT) in wild-type and daf-2(e1370) mutant animals. (C) Loss of mxl-2 does not further impair ITT in daf-2(e1370);daf-16(mgDf47) mutants. (D) mxl-2 is dispensable for acquired thermotolerance in daf-2(e1370) mutants. Graphs were generated from the combined data from multiple experiments performed as described in Materials and Methods. Statistical analyses and experimental details can be found in Dataset S3. We next assessed whether loss of the Myc-Mondo or Mad complexes altered C. elegans thermotolerance (i.e. intrinsic thermal tolerance, ITT). N2, mxl-2(tm1516), and mxl-1(tm1530) mutants were grown until day 2 of adulthood and survival at 35°C was measured. Loss of mxl-2 significantly shortened the median survival time of wild-type animals by 22.0%, from 12.3+/–0.4 hours to 9.6+/–0.3 hours at 35°C (Figure 6B, Dataset S3, p<0.0001). Similar results were obtained in the mml-1(ok849) mutant (data not shown). Nearly identical results were obtained in the eat-2(ad465) and eat-2(ad465);mxl-2(tm1516) animals (Figure S5) confirming previous reports that eat-2 mutations do not enhance thermal stress resistance [55]. Conversely, loss of mxl-1 failed to alter survival to thermal stress (Dataset S3) in all three genetic backgrounds.
As previously reported, daf-2(e1370) mutant animals had increased thermal stress resistance compared to N2 animals (median survival of 16.6+/–0.8 hours versus 12.3+/–0.4 hours, Figure 6B, 6C, Dataset S3), and daf-16 was essential for this increased resistance (Figure 6C, Dataset 3, and [56]). daf-2(e1370);mxl-2(tm1516) had thermal stress resistance comparable to wild-type control animals (median survival of 12.4+/–0.5 and 12.3+/–0.4 hours, respectively) (Figure 6B, 6C). Thus loss of Myc-Mondo function suppresses the enhanced survival of daf-2(e1370) mutant animals and wild-type survival to similar extents (25.3% and 22.2%, respectively).
We sought to better understand the relationship between ILS, DAF-16, the Myc family of transcription factors, and thermal stress resistance. To this end we first tested whether inactivating mxl-2 in the absence of daf-16 would further impair survival to thermal stress. Inactivating mxl-2 in daf-2(e1370);daf-16(mgDf47) resulted in no significant further reduction in thermal stress survival when compared to control (median survival 8.0+/–0.11 and 7.6+/–0.10, Figure 6C, p = 0.313). We next asked whether mxl-2 was required for the acquired thermotolerance (ATT) of daf-2(e1370) mutant animals. daf-2(e1370) mutant animals grown at 25°C become extremely tolerant to thermal stress. By day 3 of adulthood daf-2(e1370) animals subsequently exposed to 35°C have a median survival of 25.1+/–0.6 hours (Figure 6D), which is significantly greater than daf-2(e1370) intrinsic thermotolerance (ITT) (p<0.0001). Similar to ITT, the acquired thermotolerance of daf-2(e1370) is dependent upon daf-16, as median survival is reduced from 25.1+/–0.6 to 17.2+/–0.53 hours (Figure 6D). In contrast, mxl-2 is dispensable for the acquired thermotolerance of daf-2(e1370) as median survival is unchanged (Figure 6D, p = 0.17). A comparison of genes that have promoter regions bound by DAF-16, PHA-4, and MDL-1 against all genes in the genome revealed a 2.17-fold enrichment (p = 0.006) for genes associated with a “response to heat”; which includes chaperones such sti-1, hsp16.2, and hsp-16.41. Collectively, we conclude that the Myc-Mondo complex: is required for intrinsic but not acquired thermotolerance; has overlapping genetic requirements with daf-16; and that MDL-1 and DAF-16 bind at the promoters of molecular chaperones implicated in the heat shock response.
C. elegans Mad and Myc-Mondo Complexes Have Opposing Roles in Proteostasis
We determined whether loss of C. elegans Myc-Mondo or Mad complexes might alter the decline/progressive collapse of protein homeostasis (‘proteostasis’), a hallmark of normal aging [57]. One way to assess whether a genetic perturbation affects C. elegans proteostasis is by monitoring the solubility of a polyglutamine repeat fused to YFP (Punc-54::Q35::YFP). During normal aging transgenic animals expressing such poly Gln-YFP fusion proteins in body wall muscle cells accumulate protein aggregates that can be visualized as fluorescent foci [58]. Later in life, these toxic foci overwhelm the chaperone network that maintains proper protein folding, resulting in the collapse of proteostasis within the body wall muscle cells, thus causing paralysis in the animal [59].
To test whether the Myc-Mondo and Mad complexes might function in proteostasis we inactivated mml-1, mxl-2, mdl-1, or mxl-1 in Pmyo-3::Q35::YFP animals and quantified the accumulation of fluorescent foci (protein aggregates) over time. Loss of the Myc-Mondo complex (either mml-1 or mxl-2) resulted in a significantly premature accumulation of protein aggregates (mxl-2: day 1 p = 7.6×10−4, day 2 p = 4.21×10−6, day 3 p = 9.66×10−6; mml-1: day 1 p = 5.68×10−10, day 2 p = 2.89×10−9, day 3 p = 5.86×10−8). For instance, by day 3 of adulthood mml-1(RNAi) or mxl-2(RNAi) treated Q35::YFP animals showed on average 113+/–16.7 and 108+/–22.1 foci protein aggregates, respectively. In contrast, control RNAi animals had an average of 72+/–13.6 foci per animal (Figure 7A, Dataset 3). This is consistent with the notion that loss of the Myc-Mondo complex compromises the protein chaperone network. In contrast, inactivation of the Mad complex (mdl-1 or mxl-1) had no effect on polyglutamine foci formation (Figure 7A, Dataset 3). For example, at day 3 adulthood, Q35::YFP animals treated with mdl-1 or mxl-1 RNAi had on average 70+/–11.7 and 75+/–10.4 foci, which is not significantly different from animals fed control RNAi (mdl-1 p = 0.711, mxl-1 p = 0.458). Of note, inactivation of daf-2 also failed to appreciably delay the accumulation of foci (Figure 5A): the average number of foci at day 3 adulthood is 70+/–18.0 (p = 0.791). Thus loss of the Mad complex or decreased ILS does not appreciably alter the acute accumulation of protein aggregates in Q35::YFP animals, but loss of the Myc-Mondo complex hastens the collapse of proteostasis as measured by the accumulation of fluorescent aggregates.
Fig. 7. Myc-Mondo/Mad transcription factors influence proteostasis. 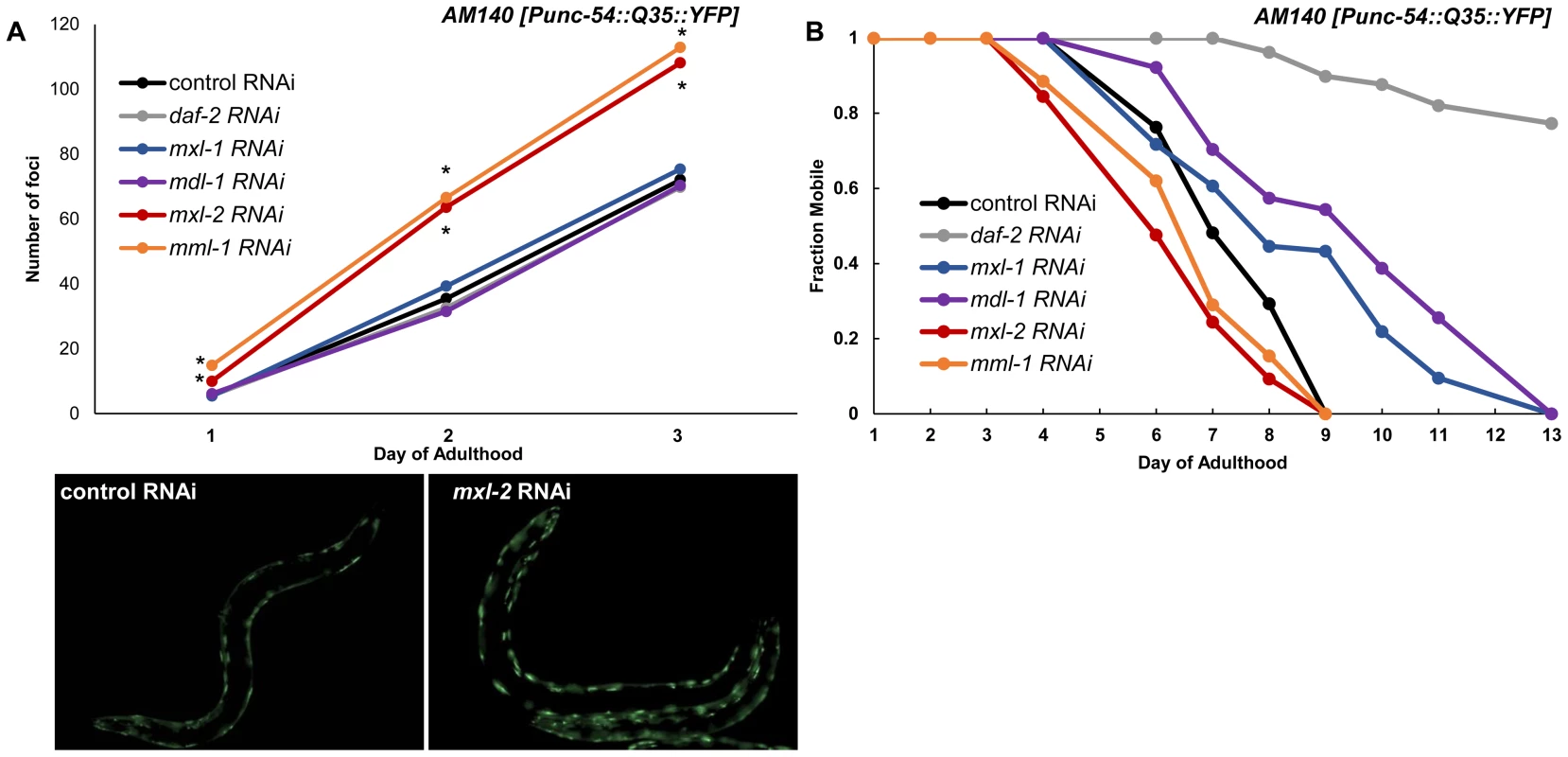
(A) RNAi inactivation of mxl-2 and mml-1 leads to early accumulation of Q35::YFP foci (red and orange versus black). RNAi inactivation of mxl-1, mdl-1, and daf-2 fail to delay accumulation of Q35::YFP foci (compare vector control RNAi treated animals (black) to blue, purple, and gray respectively. Traces are the combined data from three trials (*denotes p<0.01, standard deviations and statistical analysis can be found in Dataset S3). Images are from empty vector control and mxl-2 RNAi treated animals of Day 2 of adulthood. (B) RNAi inactivation of mxl-2 and mml-1 leads to premature paralysis (compare red and orange to vector RNAi control (black)). RNAi inactivation of mxl-1(blue), mdl-1(purple), and daf-2(gray) delays paralysis induced by Q35::YFP expression in the body wall muscles in comparison to animals treated with empty vector control RNAi (black). Traces are representative of data from two trials performed in duplicate. Complete statistical analysis for data represented in this figure can be found in Dataset S3. We assessed the long term consequences of loss of Myc-Mondo or Mad complex function in Punc-54::Q35::YFP animals as measured by the onset of paralysis. daf-2(RNAi) served as a positive control and completely negated the onset of paralysis over the course of our analysis, consistent with published results [60]. In contrast, the average age of paralysis of Q35::YFP animals treated with control RNAi was day 7 of adulthood, and all animals were paralyzed by day 9. Inactivation of mml-1 or mxl-2 resulted in a significant premature onset of paralysis (mxl-2 p = 0.0000 and mml-1 p = 4.93×10−11, Figure 7B). Conversely, inactivation of mdl-1 or mxl-1 resulted in a significant (mxl-1 p = 0.0187; mdl-1 p = 0.0384) delay in the onset of paralysis (Figure 7B). Collectively, our findings indicate that the C. elegans Myc-Mondo and Mad complexes have opposing effects on proteostasis, which parallel their effect on longevity and known roles in transcriptional control [11].
Discussion
The Myc family of transcription factors have well established functions in growth control and metabolic regulation; processes that are known to influence lifespan. Yet, this is the first study to identify a role for the Myc interaction network in longevity control and proteostasis. We have identified a role for the Myc-family of bHLH transcription factors: mml-1, mdl-1, mxl-1, and mxl-2 in the regulation of C. elegans lifespan. We show that the Myc-Mondo (MML-1:MXL-2) and Mad (MDL-1:MXL-1) transcriptional complexes have opposing roles in longevity control and proteostasis analogous to their known functions as transcriptional activators and repressors in C. elegans [11]. Specifically, loss of the Myc-Mondo complex leads to premature aging, while loss of the Mad complex delays aging. Lifespan extension by loss of the Mad complex is dependent upon the Myc-Mondo complex, daf-16, and pha-4, suggesting a common mechanism in longevity control. Conversely, Myc-Mondo is required for lifespan extension by decreased ILS or conditions of DR. Both decreased ILS and conditions of DR promote nuclear accumulation of MML-1, but do so through distinct mechanisms; altered DAF-16 activity regulates expression of Myc family members, while pha-4 is essential for the increased nuclear accumulation of MML-1. In contrast, the Myc-Mondo complex is dispensable for the nuclear accumulation of DAF-16 by decreased ILS, and overexpression of DAF-16::GFP rescues the shortened lifespan of mxl-2 null mutant animals, which suggests that DAF-16 and the Myc-Mondo complex may co-regulate transcription at overlapping target genes. From a candidate approach, we find that Myc-Mondo is required for the induction of longevity genes by decreased ILS. DAF-16, PHA-4, and MDL-1 bind within the genome at many overlapping target genes involved in unfolded protein binding, carbohydrate metabolism, autophagy, and stress response. Finally, loss of the Myc-Mondo complex impairs oxidative and thermal stress survival. Collectively, our results suggest that Myc family members are regulated by diverse signals of metabolic status and converge with DAF-16 and PHA-4 at metabolic and cytoprotective transcriptional programs that influence aging (Figure 8).
Fig. 8. Model for Myc-Mondo/Mad transcription factors in longevity control under basal conditions the Myc-Mondo activation complex (MML-1:MXL-2) is largely inactive, and transcription of genes encoding functions related to aging is limited by the Mad transcriptional repression complex (MDL-1:MXL-1). 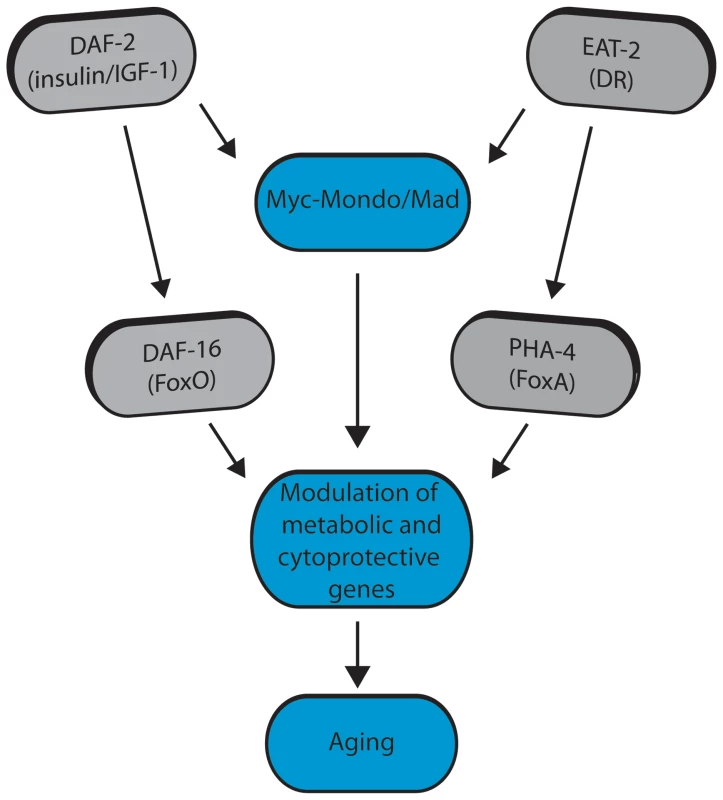
Conditions of reduced ILS and DR promote Myc-Mondo complex activity, likely by regulating cellular localization and transcription of mml-1. Myc-Mondo/Mad transcription factors may cooperate with DAF-16 and PHA-4 to modulate the expression of key metabolic and cytoprotective genes to influence aging. The Myc interaction network in mammals has been defined based on the combinations of known protein-protein interactions between Myc/Mnt/Mad/Mondo and either Max or Mlx, which distinguishes them from the larger super-family of bHLH transcription factors [61]–[64]. The discovery that the Myc-Mondo and Mad complexes have antagonistic functions in both transcription and longevity control sets them apart from other transcription factors that are relevant to aging, such as DAF-16, PHA-4, or SKN-1. The related bHLH protein, MXL-3, has recently been implicated in lifespan [23], [65]. mxl-3 encodes a paralog of mxl-1 (Max), which is unique to nematodes [22]. Furthermore, MXL-3 homodimerizes and does not interact with other C. elegans Myc family members [10]. This is in contrast to mammalian Max and C. elegans MXL-1 (Max), which function as heterodimers. Additionally, there is little overlap between predicted MXL-3 and MDL-1::MXL-1 target genes [10]. In C. elegans, MXL-3 functions in conjunction with HLH-30 to regulate the expression of a family of lipases, which mobilize lysosomal fat stores, and function independent of ILS and DR to influence longevity [23], [65]. We find no genetic interaction between the C. elegans Myc-Mondo or Mad complexes and mxl-3, implying that MXL-3 functions are distinct from the function of the Myc-Mondo/Mad interaction network.
MML-1, (for ““Myc and Mondo-like”), has a high degree of sequence identity to c-Myc, MondoA, and MondoB/ChREBP. The C. elegans genome does not encode a clear Myc ortholog [22]. Therefore, it is likely that C. elegans MML-1 incorporates both mammalian c-Myc and Mondo functions. Myc proteins have been implicated in ribosomal and mitochondrial biogenesis, energy metabolism, biosynthesis, cell growth, and cell cycle regulation. For example, many glucose metabolism genes are directly regulated by Myc [66]–[71]. In this way, Myc promotes glucose transport and catabolism. C. elegans MML-1 has an overall domain structure and size similar to mammalian Mondo/ChREBP. Work in mammals has identified MondoA:MLX and ChREBP:MLX complexes as non-hormonal sensors of glucose and key regulators of glycolytic metabolism, fat storage, and energy sensing [20], [72]. Recently, ChREBP has been shown in adipose tissue to regulate systemic glucose metabolism, fatty acid synthesis, and insulin sensitivity [73]. While the genomic binding patterns of MML-1 remain to be determined, we find an overrepresentation of genes involved in carbohydrate metabolism bound by MDL-1, DAF-16, and PHA-4. Thus the Myc-Mondo/Mad complex functions relevant to longevity may be at least partially linked to evolutionarily conserved alterations in carbohydrate metabolism.
Surprisingly, there have been few large-scale efforts to identify Mad targets in mammalian systems. Individual Mad target genes have largely been identified by the examination of genes previously identified as Myc targets [40]. However, one study overexpressed Mad1 in T-lymphocytes and identified reduced expression in 57 genes. [39]. Collectively, of 64 putative Mad1 target genes we found 53 clear C. elegans homologues, of which 41 are MDL-1 targets (Figure 5, Dataset S2). Many of these homologues are components of important cellular processes such as protein translation/ribosome function (23), metabolism (7), DNA transcription (2), and cell cycle control (2). Moreover, 11 have been implicated in C. elegans longevity: the eukaryotic initiation factors eif-1/eIF1, ifg-1/eIFGI, ife-2/eIF4E, and inf-1/eIF4AI; elongation factors eef-1a.2/EF1 and eef-2/EF2; the ribosomal protein rpl-18/Rpl18; the PTEN homologue daf-18; the glucose-6-phosphate isomerase gpi-1; the mitochondrial stress protein hsp-6; and the cytochrome b-c1 complex subunit ucr-1. Thus while the full extent of bona fide Mad target genes in mammals is relatively unknown, we find good overlap at conserved putative target genes, suggesting that the expression of Mad target genes relevant to longevity may be conserved.
We have found that decreased ILS and conditions of DR regulate C. elegans Myc family activity. Both decreased ILS and conditions of DR promote nuclear accumulation of MML-1, presumably to alter gene expression. In this respect MML-1 resembles its mammalian homologues, MondoA and ChREBP, which are also regulated by cytoplasmic-to-nuclear shuttling under changing conditions of nutrient availability. In contrast to mammalian Mondo complexes, Mad complexes are reported to be constitutively nuclear [74], [75]. Furthermore, we find that DAF-16 activity regulates Myc family expression. This is reminiscent of the reported function of mammalian MAD1, which is upregulated at the transcriptional level by several different signaling pathways, including activated PI3K/Akt downstream of the granulocyte-colony stimulating factor receptor (G-CSFR) [76]. This contrasts with findings that identified mdl-1 as an upregulated DAF-16 target gene by microarray analysis [13], [77]. Furthermore, a previous study found that mdl-1 inactivation slightly shortened daf-2 mutant longevity [13]. We find no effect of loss of mdl-1 on daf-2(e1370) longevity (data not shown). One possible explanation is that the published lifespan analysis was conducted at 25°C, which we find is a slightly stressed state that alters the genetic requirements for lifespan extension by decreased ILS (A.V.S. unpublished observations).
We found that the Myc-Mondo and Mad complexes function in both DR and ILS signaling, implying that the Myc-Mondo/Mad complexes may act as a molecular convergence point to integrate cues of metabolic status, and coordinate the transcriptional response to these diverse signals. A number of metabolic signals are widely accepted as being intimately linked to aging, yet how these signals are integrated to “fine-tune” the appropriate transcriptional response to maximize longevity under adverse environmental conditions is unknown. Interestingly, Myc has recently been shown to function as a “universal amplifier” of transcription; instead of having distinct target genes, Myc amplifies the output of the existing gene expression [78], . Thus the Myc-Mondo and Mad complexes’ antagonistic effect on transcription may function as a rheostat to modulate gene expression in response to multiple inputs of metabolic status, which is a possibility mutually exclusive from regulating transcription of distinct target genes.
Our results raise a number of questions of how Myc family members integrate diverse signals of metabolic status. The roles of ILS and DR in longevity control are genetically separable, yet there is evidence of overlap between the transcriptional programs that are activated by decreased ILS and conditions of DR (Reviewed in [1]). For instance, overexpression of pha-4 has the greater influence on lifespan in the absence of daf-16 [8], which suggests either an inherent competition between daf-16 and pha-4 in wild-type animals, or that the role of daf-16 and pha-4 may be partially redundant. However, pha-4 is dispensable for the increased lifespan of daf-2 mutants [8], yet wefind that pha-4 is necessary for the increased nuclear accumulation of MML-1::GFP by decreased ILS. One relevant finding is that loss of pha-4 has no detrimental effect on the distribution of MML-1::GFP localization in wild-type animals, which still have substantial amounts of nuclear MML-1. Thus the basal distribution of MML-1 may be sufficient to potentiate insulin/IGF1 signals either through altered patterns of DNA binding, recruitment of co-factors, or changes of stability at DNA. However, our results predict that there would be no additivity of lifespan extension by the combination of DR and decreased ILS in the absence of pha-4. The mechanism through which PHA-4 potentiates DR signals through increased nuclear accumulation of MML-1 remains to be determined. Nuclear localization of ChREBP, the mammalian homologue of MML-1, is regulated by changes in carbohydrate availability through post-translational modifications [80]. Thus identifying whether MML-1 is similarly regulated will be an important step to further understand how DR signaling alters Myc family function.
We find that overexpression of DAF-16 can rescue the shortened lifespan caused by the loss of Myc-Mondo, yet Myc-Mondo is required for the increased expression of DAF-16 target genes under conditions of decreased ILS. How might these two paradoxical findings be resolved? One possibility is that DAF-16 activity is not maximally induced by daf-2(e1370). Our discovery that the daf-16 dependent acquired thermotolerance of daf-2(e1370) is independent of mxl-2 is consistent with the possibility that mild heat stress in daf-2(e1370) maximally potentiates DAF-16 activity, thereby removing the need for the Myc-Mondo transcriptional activation complex. A second possibility is that DAF-16::GFP overexpression is not equivalent to the activation of DAF-16 in daf-2(e1370) mutant animals.
We sought to identify putative common target genes between DAF-16, PHA-4, and MDL-1 to begin to elucidate the molecular mechanisms through which the Myc family may influence longevity. In contrast to DAF-16/PHA-4, MDL-1 and the Mad mammalian homologues are only known to function as transcriptional repressors [11], [40]. Importantly, MDL-1 genomic binding patterns were identified in wild-type animals; thus under normal conditions the MDL-1 complex limits the expression of DAF-16/PHA-4 target genes relevant to aging. We favor a model where under conditions of reduced ILS or DR, increased nuclear accumulation of MML-1 results in the replacement of MDL-1 binding at target genes. This idea is consistent with the model of complex switching that has been proposed to describe the antagonistic relationship between Myc and Mad complexes in mammals [40]. We found significant overlap of genomic binding between these transcription factors at many target genes, consistent with the possibility of shared transcriptional programs. Assessing whether these putative common target genes were enriched over the genome for any specific biological function by gene ontology revealed a significant enrichment for genes that function in lifespan (299), stress response (117), carbohydrate metabolism (93), unfolded protein binding (27), and autophagy (9). Specific forms of stress response that were over-represented included response to heat (12), unfolded protein response (20), immunity/pathogen resistance (15), and DNA damage response (49). Surprisingly, response to oxidative stress was not over-represented. However, we found no evidence that heat or oxidative stress altered the expression of Myc family members, nor the nuclear accumulation of MML-1 (D.W.J., unpublished observations). Thus despite their requirement for stress response survival, and the finding that MDL-1 binds at many genes involved in survival to stress, Myc family members do not seem to be stress response genes, which is in contrast to their regulation by altered signals of metabolic status. Rather, these data support a model where the Myc family converges with DAF-16 and PHA-4 at overlapping metabolic and cytoprotective transcriptional programs that set the progression of aging.
Given their close ties to metabolism it is not surprising that DAF-16 and PHA-4 bind extensively in the promoter regions of key metabolic genes. Our analysis shows that MDL-1, DAF-16, and PHA-4 binding is significantly enriched at genes involved in carbohydrate metabolism (Dataset S2), including nine glycolytic genes (Y77E11A.1, H25P06.1, enol-1, pyk-2, ZK632.4, gpi-1, aldo-2, F57B10.3, pgk-1), and five gluconeogenesis genes (gpi-1, aldo-2, F57B10.3, pgk-1, pyc-1), which is strongly reminiscent of mammalian Mondo-ChREBP/Mlx complexes [18]-[20], [42]. Interestingly, it has been found that gluconeogenesis is upregulated in conditions of low ILS and DR [44], [81]; therefore, it is possible that one role for Myc-Mondo complexes under these conditions is to promote the expression of gluconeogenic genes, which results in the increase of intracellular glucose. Other metabolic genes with known roles in longevity included icl-1 and stdh-1. Both are metabolic enzymes that are upregulated by decreased ILS, in a daf-16 dependent manner [37]. We discovered their upregulation by decreased ILS is also dependent on the Myc-Mondo complex (Figure 5F). stdh-1 encodes a steroid dehydrogenase, which is involved in hormone biosynthesis and suggests that the Myc family may contribute to cell non-autonomous signaling. icl-1 encodes a dual enzyme isocitrate lyase/malate synthase essential for the glyoxylate cycle, a variation of the citric acid cycle that allows the production of sugar from fat. Like gluconeogenesis, the expression of icl-1 is also upregulated in conditions of low ILS and DR [13], [44], and increased glyoxylate cycle activity is required for the increased longevity of C. elegans insulin signaling mutants [13].
Proteostasis is the ability of the cell’s proteome to maintain a proper folding environment. The predominant theory is that during aging, post-mitotic cells accumulate protein damage (e.g. through oxidative damage and DNA mutations), which in turn produces misfolded proteins that rapidly form highly stable protein aggregates that ultimately overwhelm the ability of the chaperone network, protein degradation machinery, and sequestration strategies to maintain a proper folding environment. This collapse may be the underlying basis for age-associated proteotoxic diseases [82]. We find that the Myc-Mondo and Mad complexes have opposite effects on proteostasis, analogous to their effect on transcription and longevity. Since many putative target genes of MDL-1 are involved in either protein folding, unfolded protein response, or heat-shock response, we favor a model where changes in Myc family activity alter the expression of genes involved in protein homeostasis, which in turn influences the progression of aging.
Putative target genes of MDL-1 and DAF-16 were enriched over genes bound only by DAF-16 in two major gene ontology terms that are highly relevant for the maintenance of proteostasis: unfolded protein binding and the unfolded protein response. These somewhat mutually exclusive categories cover de novo protein folding and the response to proteotoxic stress, respectively. Twenty-eight of 52 genes annotated to function in unfolded protein binding were discovered and include the gene classes: cct (Chaperonin Containing TCP-1; 6 of 8), pfd (PreFolDin; 4 of 6), and dnj (DNaJ domain; 11 of 29) (Dataset 3). The cct genes encode the components of the TCP-1 ring complex (TRiC), which complete de novo protein folding and interacts with prefoldin co-chaperones. The TRiC complex has been implicated in longevity control and an ILS-mediated soma to germline transformation gene expression program [83], [84]. In addition, 20 of 40 genes annotated to function in the unfolded protein response are bound by DAF-16, PHA-4, and MDL-1. Genes involved in the mitochondrial and ER unfolded protein responses were represented (3 and 9 genes respectively, Dataset S2). Lifespan extension in daf-2 mutants is dependent on both ire-1 and xbp-1, suggesting that genes involved in ER proteostasis may contribute to daf-2 mutant lifespan [85]. We found several small heat shock proteins (hsp-16.2, hsp-16.41, and hsp-12.6) to be bound by DAF-16, PHA-4, and MDL-1. The latter is not stress responsive, but is upregulated in daf-2 mutant animals dependent on daf-16 [13], [86], [87], and we find this upregulation is also mxl-2 dependent (Figure 5D). Interestingly, hsp-12.6 is required for the increased lifespan of ILS mutants, and hsp-12.6(RNAi) results in the premature collapse of proteostasis [88].
This is the first study to identify a role of the Myc-Mondo/Mad interaction network in longevity. Significantly, a homologue of one of these transcription factors may also influence aging in mammals. The mml-1 mammalian homologue ChREBP is also known as William-Beuren Syndrome Chromosomal Region 14 (WBSCR14). Williams-Beuren Syndrome (WBS) is a multisystem disorder resulting from the deletion of 26 to 28 genes, including ChREBP, on human chromosome 7. WBS has been characterized as a disorder of mild accelerated aging. Patients display premature gray hair, diverticulosis, diabetes, and hearing loss that develops during adolescence or young adulthood, in combination with instances of declining memory skills or dementia [21]. Thus our findings collectively imply that the Myc-Mondo/Mad interaction network of basic-helix transcription factors function as a molecular convergence point: integrating diverse signals of metabolic status that converge at evolutionarily conserved metabolic and cytoprotective transcriptional programs to influence the progression of aging.
Materials and Methods
Strains, Plasmid Construction, RNAi, and Nematode Culture
Standard culture techniques were used to maintain nematodes [89]. The wild-type strain is Bristol N2 and all other strains used in this work were backcrossed into Bristol N2 a minimum of six times before use. The mxl-2(tm1516), mxl-1(tm1530), and mdl-1(tm311) strains were created by and obtained from Dr. Shohei Mitani with the Japanese Bioresource Project; the mml-1(ok849) and mxl-3(ok1947) were generated by Dr. Robert Barstead with the Oklahoma Medical Research Foundation Knockout Group, and were generously provided by the C. elegans Genetics Center (University of Minnesota).
The Pmml-1::mml-1::gfp construct was generated through PCR sewing [90]. The mml-1 genomic fragment was PCR amplified from the WRM0615CE07 fosmid using the following primers: 5′-AGTCGACCTGCAGGCATGCAAGCTaaatggatttttgagttgttgcat-3′ and 5′-TAGAGGGTCGCGTTAGAGGT-3′. The GFP open-reading frame was PCR amplified from pPD95.75 (Fire Lab vector kit) using the following primers: 5′-AGCTTGCATGCCTGCAGGTCG-3′ and 5′-AAGGGCCCGTACGGCCGACTA-3′. The two fragments were then PCR sewn together using the following primers: 5′-TTCCACTCGTTTCTCCGTCC-3′ and 5′-GGAAACAGTTATGTTTGGTATA-3′. Eight sewing reactions were simultaneously performed. Each reaction generated a single PCR product, which were combined to form a single pool of DNA which was injected into mml-1(ok849) young adult hermaphrodites at a concentration of 5 ng/μL along with 2.5 ng/μL pCFJ90 as a co-injection marker, and 142.5 ng/μL 2-log DNA ladder (New England Biolabs). The transgene was maintained as a high-transmission rate extrachromosomal array. Three independent mml-1(ok849) lines expressing this array were subsequently mated into daf-2(e1370) and eat-2(ad465) mutant to generate independent lines for examination. Three independently generated lines were examined per background.
The strains used in this work are: N2 Bristol, eat-2(ad465), daf-2(e1370), mxl-2(tm1516), mxl-1(tm1530), mdl-1(tm311), mml-1(ok849), mxl-3(tm1947), mxl-2(tm1516);daf-2(e1370), mxl-1(tm1530); daf-2(e1370), mxl-2(tm1516);eat-2(ad465), mxl-1(tm1530);eat-2(ad465), AM140 rmIs132 [Punc-54::Q35::YFP] [58]; AVS317-319 mml-1(ok849);artEx6-8 [Pmml-1::mml-1::gfp; pCFJ90] (three independent lines), AVS329-331 daf-2(e1370);artEx6-8 [Pmml-1::mml-1::gfp; pCFJ90] (three independent lines), AVS332-334 eat-2(ad465);artEx6-8artEx3 [Pmml-1::mml-1::gfp; pCFJ90] (three independent lines). RNAi clones were grown overnight in Luria broth and seeded onto plates containing 5 mM isopropylthiogalatoside, to induce dsRNA expression overnight at room temperature.
Lifespan Assays
Lifespan and survival assays were performed using either a replica set protocol, as previously described [12], or through traditional methods. Briefly, for replica set protocol: rather than following a single population of animals over time, animals were cultured in 24-well format on HT115 bacteria expressing either empty vector or the indicated RNAi. A sufficient number of plates were created for each condition (combination of strain and RNAi) so that each could be counted at least every other day and then disposed. On average, 25 animals were counted per well. In all cases animals were cultured from L1 to L4 stage of development at 15°C, at which point FuDR was added to a final concentration of 400 μM and the animals were transferred to 20°C for the remainder of their lifespan. For traditional survival assays, proportional survival was charted over time using the Kaplan-Meier estimator. Replicate survival assays were fit via generalized linear modeling to a logit curve, according to the equation y = 1/(1+e∧(a*x-b)). The LD50 for a fit survival curve was then a function of the coefficients: LD50 = b/a. For two sample comparison, p-values were calculated through resampling and refitting, where the difference in resampled LD50s was compared to the difference in the observed LD50s, see [12]. Similar analyses were used on stress survival assays. For each comparison of either a mutant or RNAi treatment compared to control, between two and four independent trials were conducted to measure alterations in lifespan or survival (see figure legends and Dataset S1). Raw mortality data from replicate set lifespan has been deposited into the Dryad repository (doi:10.5061/dryad.pj0p3 Data files: Raw Data of Mortality Observations of Replica Set Experiments).
MML-1::GFP Localization
Synchronized L1 transgenic animals were grown to the L4 stage of development on HT115 bacteria, expressing: empty vector control, daf-16, or pha-4 RNAi at 15°C on 6 cm plates. FuDR was added to a final concentration of 400 μM and the animals were transferred to 20°C. GFP localization was examined on day 2 of adulthood. Worms were mounted on 2% agarose pads and anaesthetized with a mixture of 15 mM tricaine mesylate and 15 mM tetramisole hydrochloride prior to imaging. Microscopy was performed on a Zeiss AxioImager.M2m equipped with an AxioCam MRm camera using high NA Zeiss EC-Plan-NEOFLUAR 20X and 40X objectives, and a Semrock Brightline GFP filter (488 nm/535 nm Ex/Em). Scoring was performed in three separate trials examined under blinded conditions, and each animal was scored according to the rubric shown in (Figure 4).
qRT-PCR
Synchronized populations were grown to young adulthood at 15°C on 10 cm RNA plates containing HT115 bacteria expressing vector control RNAi. Animals were then rinsed from the plates using M9 and washed three times to remove bacteria. RNA was isolated using Trizol reagent (Life Sciences) following the manufacturer’s protocol. Isolated RNA was converted to cDNA using the Bio-Rad iScript reverse transcriptase kit with oligo dT primers. Negative samples were prepared by omitting the reverse transcriptase enzyme from the reaction.
Analysis of transcript levels via qRT-PCR was performed on three distinct biological replicates per condition, and performed in triplicate in each instance. The total cDNA concentration for each sample was normalized to the levels of act-1 and rpl-32 transcript. mml-1, mxl-2, mxl-1, and mdl-1 transcript levels were normalized to N2 control samples. For analysis of Class I genes: act-1, rpl-32, and pat-10 were used to normalize total cDNA concentration. dod-3, hsp-12.6, stdh-1, and icl-1 transcript levels were normalized to N2 control samples. Statistical analyses of fold-changes from three independent trials were accomplished using Student’s paired t-test.
The primers used are as follows:
act-1:forward 5′-CCA TCA TGA AGT GCG ACA TTG-3′
reverse 5′-CAT GGT TGA TGG GGC AAG AG-3′
rpl-32:forward 5′-AGG GAA TTG ATA ACC GTG TCC GCA-3′
reverse 5′-TGT AGG ACT GCA TGA GGA GCA TGT-3′
mml-1:forward 5′-GGA GAA ATC CGG AAG GCA TTA CTT AC-3′
reverse 5′-CGC CAG AAT ACA AAT CGG GTA TAA TAT C-3′
mxl-2:forward 5′-TCA GAG CCT GCG ACT TCA TG-3′
reverse 5′-GTC GAG GAG AAG TTG GAG CAT-3′
mxl-1:forward 5′-GAC ATG AGT GAC CTC GAA GAT GAC-3′
reverse 5′-CAG GCG AGC TAT CTC TTC TCT G-3′
mdl-1:forward 5′-CGA TCT TTC AAA TGA GTC CGA ACT CC-3′
reverse 5′-GTC TTG CAA TTC AAT GAT GTG ATC TCG-3′
pat-10: forward 5′-GACGGAAAGCTTCACGAAGTTC-3′
reverse 5′-CCTTCGTAAACTGATCCGCAAG-3′
dod-3: forward 5′-AAAAAGCCATGTTCCCGAAT-3′
reverse 5′-GCTGCGAAAAGCAAGAAAAT-3′
hsp-12.6: forward 5′-GGAGTTGTCAATGTCCTCGACG-3′
reverse 5′-GAAGTTCTCCAATGTTCTTGAC-3′
stdh-1: forward 5′-ACAGGATGTCTTCAAAAGGAATATGG-3′
reverse 5′-TTGCTGGGGTGATAGCTTGG-3′
icl-1: forward 5′-GACTACGAGGCTGGAAGAACGATTG-3′
reverse 5′ - GTAGGCGAACATCTTGTCTGGGTAC-3′
Statistical and Informatic Analysis
Statistical analysis of traditional lifespan analysis was performed by two sample tests, which used the Mantel-Cox log-rank statistic. For replica set lifespan and stress survival experiments, data were fit to a two parameter logit survival curve, a generalized linear model with a binomial link function. This curve models the probability of a worm's survival at a given time point t, according to the equation p = 1/(1+e∧(+ax-b)). The parameters a and b, which represent logit slope and intercept, are optimized by iteratively reweighted least squares. For a fit logit curve, the LD50 is b/a, or intercept divided by slope. For maximum life, we took the value of the logit curve where it dropped below 5% survival. We estimate 95% confidence intervals on the LD50 non-parametrically, through resampling of observations (K = 10,000), with replacement. For two sample tests comparing condition LD50s to control, we again used a resampling approach, where labels were swapped between conditions compared, then curves refit, and the resampled difference in LD50 was compared to the observed difference, (K = 10,000).
To compare the effect loss that mxl-2 had in different backgrounds, we compared the ratio of mxl-2 loss of function LD50 over control LD50 between backgrounds. To put confidence intervals on this ratio, and make two sample comparisons, we again used resampling to estimate the distribution (K = 10,000 for each of the four conditions).
The binding site locations for PHA-4, MDL-1, or DAF-16 were identified based on ChIP-seq data generated by the modENCODE project (www.modencode.org). ChIP-seq binding peaks for DAF-16, PHA-4, and MDL-1 were validated in [38]. Binding sites were assigned to specific genes based on location data from WormBase (www.wormbase.org) and then PHA-4 and MDL-1 binding sites were mapped relative to the transcriptional start site (TSS) of nearby open reading frames. Examination of genome-wide binding patterns revealed that MDL-1 and PHA-4 binding was most highly enriched between –700bp and +100bp relative to the TSS. Based on this we assessed the binding of PHA-4 and MDL-1 within this window relative to each gene in the genome. Genes bound by DAF-16 were identified in [37].
For characterizing ontologies among genes bound by MDL-1, we used a list of GO terms relevant to aging mined from WormBase version 210. To evaluate GO term enrichment between a set of bound genes and the genome we used the chi-squared test.
Thermotolerance, Oxidative Stress, and Proteostasis Assays
For stress response assays, synchronized L1 animals were cultured on HT115 bacteria expressing the identified RNAi vector at 15°C until the L4 stage of development. FuDR was then added to a final concentration of 400 μM, and the animals were then cultured at 20°C unless otherwise stated. For thermotolerance assays, two day old adult animals were exposed to 35°C for the indicated time period, then removed and allowed to recover for 1–2 hours at room temperature before survival was then scored. For oxidative stress survival assays, two day old adult animals were treated with 7.7 mM tert-butylhydroperoxide (Alfa Aesar) for the indicated time period, then survival was scored. For proteostasis assays, foci formation in AM140 animals was scored on days one, two, and three of adulthood. Scoring was performed on a Zeiss M2BIO upright fluorescence microscope using a 10X objective and an EGFP filter set. Worms were mounted on 2% agarose pads and anaesthetized with a mixture of 15 mM tricaine mesylate and 15 mM tetramisole hydrochloride prior to imaging. For paralysis assays animals were scored in their ability to retreat from gentle nose touch. Paralyzed animals were removed, and scoring continued until the entire population had become paralyzed. Foci accumulation and paralysis were measured in three separate trials each. For stress response assays two separate trials were performed in duplicate.
Supporting Information
Zdroje
1. Greer EL, Brunet A (2009) Different dietary restriction regimens extend lifespan by both independent and overlapping genetic pathways in C. elegans. Aging Cell. pp. 113–127.
2. GuarenteL (2007) Sirtuins in aging and disease. Cold Spring Harb Symp Quant Biol 72 : 483–488.
3. LapierreLR, HansenM (2012) Lessons from C. elegans: signaling pathways for longevity. Trends Endocrinol Metab 23 : 637–644.
4. MairW (2013) Tipping the energy balance toward longevity. Cell Metab 17 : 5–6.
5. Mair W, Dillin A (2008) Aging and survival: the genetics of life span extension by dietary restriction. Annu Rev Biochem. pp. 727–754.
6. KenyonC, ChangJ, GenschE, RudnerA, TabtiangR (1993) A C. elegans mutant that lives twice as long as wild type. Nature 366 : 461–464.
7. GottliebS, RuvkunG (1994) daf-2, daf-16 and daf-23: genetically interacting genes controlling Dauer formation in Caenorhabditis elegans. Genetics 137 : 107–120.
8. PanowskiSH, WolffS, AguilaniuH, DurieuxJ, DillinA (2007) PHA-4/Foxa mediates diet-restriction-induced longevity of C. elegans. Nature 447 : 550–555.
9. SimionatoE, LedentV, RichardsG, Thomas-ChollierM, KernerP, et al. (2007) Origin and diversification of the basic helix-loop-helix gene family in metazoans: insights from comparative genomics. BMC Evol Biol 7 : 33.
10. GroveCA, De MasiF, BarrasaMI, NewburgerDE, AlkemaMJ, et al. (2009) A multiparameter network reveals extensive divergence between C. elegans bHLH transcription factors. Cell 138 : 314–327.
11. PickettCL, BreenKT, AyerDE (2007) A C. elegans Myc-like network cooperates with semaphorin and Wnt signaling pathways to control cell migration. Dev Biol 310 : 226–239.
12. SamuelsonAV, CarrCE, RuvkunG (2007) Gene activities that mediate increased life span of C. elegans insulin-like signaling mutants. Genes Dev 21 : 2976–2994.
13. MurphyCT, McCarrollSA, BargmannCI, FraserA, KamathRS, et al. (2003) Genes that act downstream of DAF-16 to influence the lifespan of Caenorhabditis elegans. Nature 424 : 277–283.
14. HavulaE, HietakangasV (2012) Glucose sensing by ChREBP/MondoA-Mlx transcription factors. Semin Cell Dev Biol 23 : 640–647.
15. IizukaK, HorikawaY (2008) Regulation of lipogenesis via BHLHB2/DEC1 and ChREBP feedback looping. Biochem Biophys Res Commun 374 : 95–100.
16. MaL, TsatsosNG, TowleHC (2005) Direct role of ChREBP.Mlx in regulating hepatic glucose-responsive genes. J Biol Chem 280 : 12019–12027.
17. StoeckmanAK, MaL, TowleHC (2004) Mlx is the functional heteromeric partner of the carbohydrate response element-binding protein in glucose regulation of lipogenic enzyme genes. J Biol Chem 279 : 15662–15669.
18. DentinR, PegorierJP, BenhamedF, FoufelleF, FerreP, et al. (2004) Hepatic glucokinase is required for the synergistic action of ChREBP and SREBP-1c on glycolytic and lipogenic gene expression. J Biol Chem 279 : 20314–20326.
19. IshiiS, IizukaK, MillerBC, UyedaK (2004) Carbohydrate response element binding protein directly promotes lipogenic enzyme gene transcription. Proc Natl Acad Sci U S A 101 : 15597–15602.
20. UyedaK, RepaJJ (2006) Carbohydrate response element binding protein, ChREBP, a transcription factor coupling hepatic glucose utilization and lipid synthesis. Cell Metab 4 : 107–110.
21. PoberBR (2010) Williams-Beuren syndrome. N Engl J Med 362 : 239–252.
22. McFerrinLG, AtchleyWR (2011) Evolution of the Max and Mlx networks in animals. Genome Biol Evol 3 : 915–937.
23. O'RourkeEJ, RuvkunG (2013) MXL-3 and HLH-30 transcriptionally link lipolysis and autophagy to nutrient availability. Nat Cell Biol 15 : 668–676.
24. ByrneAB, WeirauchMT, WongV, KoevaM, DixonSJ, et al. (2007) A global analysis of genetic interactions in Caenorhabditis elegans. J Biol 6 : 8.
25. GemsD, SuttonAJ, SundermeyerML, AlbertPS, KingKV, et al. (1998) Two pleiotropic classes of daf-2 mutation affect larval arrest, adult behavior, reproduction and longevity in Caenorhabditis elegans. Genetics 150 : 129–155.
26. LakowskiB, HekimiS (1998) The genetics of caloric restriction in Caenorhabditis elegans. Proc Natl Acad Sci U S A 95 : 13091–13096.
27. McKayJP, RaizenDM, GottschalkA, SchaferWR, AveryL (2004) eat-2 and eat-18 are required for nicotinic neurotransmission in the Caenorhabditis elegans pharynx. Genetics 166 : 161–169.
28. GreerER, PérezCL, Van GilstMR, LeeBH, AshrafiK (2008) Neural and molecular dissection of a C. elegans sensory circuit that regulates fat and feeding. Cell Metab 8(2): 118–31.
29. LuoS, KleemannGA, AshrafJM, ShawWM, MurphyCT (2010) TGF-β and insulin signaling regulate reproductive aging via oocyte and germline quality maintenance. Cell 143(2): 299–312.
30. VowelsJJ, ThomasJH (1992) Genetic analysis of chemosensory control of dauer formation in Caenorhabditis elegans. Genetics 130 : 105–123.
31. YouYJ, KimJ, RaizenDM, AveryL (2008) Insulin, cGMP, and TGF-beta signals regulate food intake and quiescence in C. elegans: a model for satiety. Cell Metab 7 : 249–257.
32. ShawWM, LuoS, LandisJ, AshrafJ, MurphyCT (2007) The C. elegans TGF-beta Dauer pathway regulates longevity via insulin signaling. Current Biology 17(19): 1635–45.
33. DeplanckeB, MukhopadhyayA, AoW, ElewaAM, GroveCA, et al. (2006) A gene-centered C. elegans protein-DNA interaction network. Cell 125 : 1193–1205.
34. BillinAN, EilersAL, CoulterKL, LoganJS, AyerDE (2000) MondoA, a novel basic helix-loop-helix-leucine zipper transcriptional activator that constitutes a positive branch of a max-like network. Mol Cell Biol 20 : 8845–8854.
35. KawaguchiT, TakenoshitaM, KabashimaT, UyedaK (2001) Glucose and cAMP regulate the L-type pyruvate kinase gene by phosphorylation/dephosphorylation of the carbohydrate response element binding protein. Proc Natl Acad Sci U S A 98 : 13710–13715.
36. CelnikerSE, DillonLA, GersteinMB, GunsalusKC, HenikoffS, et al. (2009) Unlocking the secrets of the genome. Nature 459 : 927–930.
37. TepperRG, AshrafJ, KaletskyR, KleemannG, MurphyCT, et al. (2013) PQM-1 complements DAF-16 as a key transcriptional regulator of DAF-2-mediated development and longevity. Cell 154 : 676–690.
38. NiuW, LuZJ, ZhongM, SarovM, MurrayJI, et al. (2011) Diverse transcription factor binding features revealed by genome-wide ChIP-seq in C. elegans. Genome Res 21 : 245–254.
39. IritaniBM, DelrowJ, GrandoriC, GomezI, KlackingM, et al. (2002) Modulation of T-lymphocyte development, growth and cell size by the Myc antagonist and transcriptional repressor Mad1. EMBO J 21 : 4820–4830.
40. LuscherB (2012) MAD1 and its life as a MYC antagonist: an update. Eur J Cell Biol 91 : 506–514.
41. IizukaK, BruickRK, LiangG, HortonJD, UyedaK (2004) Deficiency of carbohydrate response element-binding protein (ChREBP) reduces lipogenesis as well as glycolysis. Proc Natl Acad Sci U S A 101 : 7281–7286.
42. PedersenKB, ZhangP, DoumenC, CharbonnetM, LuD, et al. (2007) The promoter for the gene encoding the catalytic subunit of rat glucose-6-phosphatase contains two distinct glucose-responsive regions. Am J Physiol Endocrinol Metab 292: E788–801.
43. HendersonST, JohnsonTE (2001) daf-16 integrates developmental and environmental inputs to mediate aging in the nematode Caenorhabditis elegans. Curr Biol 11 : 1975–1980.
44. CasteleinN, HoogewijsD, De VreeseA, BraeckmanBP, VanfleterenJR (2008) Dietary restriction by growth in axenic medium induces discrete changes in the transcriptional output of genes involved in energy metabolism in Caenorhabditis elegans. Biotechnol J 3 : 803–812.
45. GalloM, ParkD, RiddleDL (2011) Increased longevity of some C. elegans mitochondrial mutants explained by activation of an alternative energy-producing pathway. Mech Ageing Dev 132 : 515–518.
46. BokovA, ChaudhuriA, RichardsonA (2004) The role of oxidative damage and stress in aging. Mech Ageing Dev 125 : 811–826.
47. MorimotoRI (2008) Proteotoxic stress and inducible chaperone networks in neurodegenerative disease and aging. Genes Dev 22 : 1427–1438.
48. ShoreDE, RuvkunG (2013) A cytoprotective perspective on longevity regulation. Trends Cell Biol 23 : 409–420.
49. BarsyteD, LovejoyDA, LithgowGJ (2001) Longevity and heavy metal resistance in daf-2 and age-1 long-lived mutants of Caenorhabditis elegans. Faseb J 15 : 627–634.
50. GarsinDA, VillanuevaJM, BegunJ, KimDH, SifriCD, et al. (2003) Long-lived C. elegans daf-2 mutants are resistant to bacterial pathogens. Science 300 : 1921.
51. HondaY, HondaS (1999) The daf-2 gene network for longevity regulates oxidative stress resistance and Mn-superoxide dismutase gene expression in Caenorhabditis elegans. Faseb J 13 : 1385–1393.
52. Lamitina ST, Strange K (2005) Transcriptional targets of DAF-16 insulin signaling pathway protect C. elegans from extreme hypertonic stress. Am J Physiol Cell Physiol 288: C467–474. Epub 2004 Oct 2020.
53. Mendenhall AR, LaRue B, Padilla PA (2006) Glyceraldehyde-3-phosphate dehydrogenase mediates anoxia response and survival in Caenorhabditis elegans. Genetics. pp. 1173–1187.
54. MurakamiS, JohnsonTE (1996) A genetic pathway conferring life extension and resistance to UV stress in Caenorhabditis elegans. Genetics 143 : 1207–1218.
55. HouthoofdK, BraeckmanBP, LenaertsI, BrysK, De VreeseA, et al. (2002) Axenic growth up-regulates mass-specific metabolic rate, stress resistance, and extends life span in Caenorhabditis elegans. Exp Gerontol 37 : 1371–1378.
56. WalkerGA, WhiteTM, McCollG, JenkinsNL, BabichS, et al. (2001) Heat shock protein accumulation is upregulated in a long-lived mutant of Caenorhabditis elegans. J Gerontol A Biol Sci Med Sci 56: B281–287.
57. Ben-Zvi A, Miller EA, Morimoto RI (2009) Collapse of proteostasis represents an early molecular event in Caenorhabditis elegans aging. Proc Natl Acad Sci USA. pp. 14914–14919.
58. SatyalSH, SchmidtE, KitagawaK, SondheimerN, LindquistS, et al. (2000) Polyglutamine aggregates alter protein folding homeostasis in Caenorhabditis elegans. Proc Natl Acad Sci USA 97(11): 5750–5.
59. MorleyJF, BrignullHR, WeyersJJ, MorimotoRI (2002) The threshold for polyglutamine-expansion protein aggregation and cellular toxicity is dynamic and influenced by aging in Caenorhabditis elegans. Proc Natl Acad Sci USA 99(16): 10417–22.
60. MorleyJF, BrignullHR, WeyersJJ, MorimotoRI (2002) The threshold for polyglutamine-expansion protein aggregation and cellular toxicity is dynamic and influenced by aging in Caenorhabditis elegans. Proc Natl Acad Sci U S A 99 : 10417–10422.
61. LuscherB, VervoortsJ (2012) Regulation of gene transcription by the oncoprotein MYC. Gene 494 : 145–160.
62. PetersonCW, AyerDE (2011) An extended Myc network contributes to glucose homeostasis in cancer and diabetes. Front Biosci (Landmark Ed) 16 : 2206–2223.
63. WahlstromT, HenrikssonM (2007) Mnt takes control as key regulator of the myc/max/mxd network. Adv Cancer Res 97 : 61–80.
64. BillinAN, AyerDE (2006) The Mlx network: evidence for a parallel Max-like transcriptional network that regulates energy metabolism. Curr Top Microbiol Immunol 302 : 255–278.
65. PaekJ, LoJY, NarasimhanSD, NguyenTN, Glover-CutterK, et al. (2012) Mitochondrial SKN-1/Nrf mediates a conserved starvation response. Cell Metab 16 : 526–537.
66. KimJW, GaoP, LiuYC, SemenzaGL, DangCV (2007) Hypoxia-inducible factor 1 and dysregulated c-Myc cooperatively induce vascular endothelial growth factor and metabolic switches hexokinase 2 and pyruvate dehydrogenase kinase 1. Mol Cell Biol 27 : 7381–7393.
67. KimJW, ZellerKI, WangY, JeggaAG, AronowBJ, et al. (2004) Evaluation of myc E-box phylogenetic footprints in glycolytic genes by chromatin immunoprecipitation assays. Mol Cell Biol 24 : 5923–5936.
68. LewisBC, ShimH, LiQ, WuCS, LeeLA, et al. (1997) Identification of putative c-Myc-responsive genes: characterization of rcl, a novel growth-related gene. Mol Cell Biol 17 : 4967–4978.
69. OsthusRC, ShimH, KimS, LiQ, ReddyR, et al. (2000) Deregulation of glucose transporter 1 and glycolytic gene expression by c-Myc. J Biol Chem 275 : 21797–21800.
70. ShimH, DoldeC, LewisBC, WuCS, DangG, et al. (1997) c-Myc transactivation of LDH-A: implications for tumor metabolism and growth. Proc Natl Acad Sci U S A 94 : 6658–6663.
71. ZellerKI, ZhaoX, LeeCW, ChiuKP, YaoF, et al. (2006) Global mapping of c-Myc binding sites and target gene networks in human B cells. Proc Natl Acad Sci U S A 103 : 17834–17839.
72. SloanEJ, AyerDE (2010) Myc, mondo, and metabolism. Genes Cancer 1 : 587–596.
73. HermanMA, PeroniOD, VilloriaJ, SchonMR, AbumradNA, et al. (2012) A novel ChREBP isoform in adipose tissue regulates systemic glucose metabolism. Nature 484 : 333–338.
74. GrinbergAV, HuCD, KerppolaTK (2004) Visualization of Myc/Max/Mad family dimers and the competition for dimerization in living cells. Mol Cell Biol 24 : 4294–4308.
75. YinX, LandayMF, HanW, LevitanES, WatkinsSC, et al. (2001) Dynamic in vivo interactions among Myc network members. Oncogene 20 : 4650–4664.
76. JiangK, HeinN, EckertK, Luscher-FirzlaffJ, LuscherB (2008) Regulation of the MAD1 promoter by G-CSF. Nucleic Acids Res 36 : 1517–1531.
77. McElwee JJ, Schuster E, Blanc E, Thomas JH, Gems D (2004) Shared transcriptional signature in Caenorhabditis elegans Dauer larvae and long-lived daf-2 mutants implicates detoxification system in longevity assurance. J Biol Chem 279 : 44533–44543. Epub 42004 Aug 44511.
78. NieZ, HuG, WeiG, CuiK, YamaneA, et al. (2012) c-Myc is a universal amplifier of expressed genes in lymphocytes and embryonic stem cells. Cell 151 : 68–79.
79. LinCY, LovenJ, RahlPB, ParanalRM, BurgeCB, et al. (2012) Transcriptional amplification in tumor cells with elevated c-Myc. Cell 151 : 56–67.
80. IizukaK (2013) Recent progress on the role of ChREBP in glucose and lipid metabolism. Endocr J 60 : 543–555.
81. FuchsS, BundyJG, DaviesSK, VineyJM, SwireJS, et al. (2010) A metabolic signature of long life in Caenorhabditis elegans. BMC Biol 8 : 14.
82. BalchWE, MorimotoRI, DillinA, KellyJW (2008) Adapting proteostasis for disease intervention. Science 319(5865): 916–9.
83. CurranSP, RuvkunG (2007) Lifespan regulation by evolutionarily conserved genes essential for viability. PLoS Genet 3: e56.
84. CurranSP, WuX, RiedelCG, RuvkunG (2009) A soma-to-germline transformation in long-lived Caenorhabditis elegans mutants. Nature 459 : 1079–1084.
85. Henis-KorenblitS, ZhangP, HansenM, McCormickM, LeeSJ, et al. (2010) Insulin/IGF-1 signaling mutants reprogram ER stress response regulators to promote longevity. Proc Natl Acad Sci U S A 107 : 9730–9735.
86. Halaschek-WienerJ, KhattraJS, McKayS, PouzyrevA, StottJM, et al. (2005) Analysis of long-lived C. elegans daf-2 mutants using serial analysis of gene expression. Genome Res 15 : 603–615.
87. ZhaoJ, LiL, LeissringM (2009) Molecular Neurodegeneration. Mol Neurodegener. 4 : 39 doi:10.1186/1750-1326-4-39
88. HsuAL, MurphyCT, KenyonC (2003) Regulation of aging and age-related disease by DAF-16 and heat-shock factor. Science 300 : 1142–1145.
89. BrennerS (1974) The genetics of Caenorhabditis elegans. Genetics 77 : 71–94.
90. EtchbergerJF, HobertO (2008) Vector-free DNA constructs improve transgene expression in C. elegans. Nat Methods 5 : 3.
Štítky
Genetika Reprodukční medicína
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2014 Číslo 4- Akutní intermitentní porfyrie
- Růst a vývoj dětí narozených pomocí IVF
- Farmakogenetické testování pomáhá předcházet nežádoucím efektům léčiv
- Pilotní studie: stres a úzkost v průběhu IVF cyklu
- IVF a rakovina prsu – zvyšují hormony riziko vzniku rakoviny?
-
Všechny články tohoto čísla
- The Challenges of Mitochondrial Replacement
- Concocting Cholinergy
- Genome-Wide Diet-Gene Interaction Analyses for Risk of Colorectal Cancer
- Statistical Power to Detect Genetic (Co)Variance of Complex Traits Using SNP Data in Unrelated Samples
- Mouse Pulmonary Adenoma Susceptibility 1 Locus Is an Expression QTL Modulating -4A
- Transcription-Associated R-Loop Formation across the Human CGG-Repeat Region
- Epigenetic Regulation by Heritable RNA
- Protein Quantitative Trait Loci Identify Novel Candidates Modulating Cellular Response to Chemotherapy
- Genome-Wide Profiling of Yeast DNA:RNA Hybrid Prone Sites with DRIP-Chip
- The Mechanism of Gene Targeting in Human Somatic Cells
- A LINE-1 Insertion in DLX6 Is Responsible for Cleft Palate and Mandibular Abnormalities in a Canine Model of Pierre Robin Sequence
- Interaction between Two Timing MicroRNAs Controls Trichome Distribution in
- DNA Glycosylases Involved in Base Excision Repair May Be Associated with Cancer Risk in and Mutation Carriers
- The Myc-Mondo/Mad Complexes Integrate Diverse Longevity Signals
- Evolutionarily Diverged Regulation of X-chromosomal Genes as a Primal Event in Mouse Reproductive Isolation
- Mutations in Conserved Residues of the microRNA Argonaute ALG-1 Identify Separable Functions in ALG-1 miRISC Loading and Target Repression
- Genetic Predisposition to In Situ and Invasive Lobular Carcinoma of the Breast
- Isl1 Directly Controls a Cholinergic Neuronal Identity in the Developing Forebrain and Spinal Cord by Forming Cell Type-Specific Complexes
- A Synthetic Community Approach Reveals Plant Genotypes Affecting the Phyllosphere Microbiota
- The Sequence-Specific Transcription Factor c-Jun Targets Cockayne Syndrome Protein B to Regulate Transcription and Chromatin Structure
- Determining the Control Circuitry of Redox Metabolism at the Genome-Scale
- DNA Repair Pathway Selection Caused by Defects in , , and Telomere Addition Generates Specific Chromosomal Rearrangement Signatures
- Methylome Diversification through Changes in DNA Methyltransferase Sequence Specificity
- Folliculin Regulates Ampk-Dependent Autophagy and Metabolic Stress Survival
- Fine Mapping of Dominant -Linked Incompatibility Alleles in Hybrids
- Unexpected Role of the Steroid-Deficiency Protein Ecdysoneless in Pre-mRNA Splicing
- Three Groups of Transposable Elements with Contrasting Copy Number Dynamics and Host Responses in the Maize ( ssp. ) Genome
- Sox5 Functions as a Fate Switch in Medaka Pigment Cell Development
- Synergistic Interactions between the Molecular and Neuronal Circadian Networks Drive Robust Behavioral Circadian Rhythms in
- Chromatin Landscapes of Retroviral and Transposon Integration Profiles
- Widespread Use of Non-productive Alternative Splice Sites in
- Ras GTPase-Like Protein MglA, a Controller of Bacterial Social-Motility in Myxobacteria, Has Evolved to Control Bacterial Predation by
- Cell Type-Specific Functions of Genes Revealed by Novel Adipocyte and Hepatocyte Circadian Clock Models
- Phenotype Ontologies and Cross-Species Analysis for Translational Research
- Embryogenesis Scales Uniformly across Temperature in Developmentally Diverse Species
- In Pursuit of the Gene: An Interview with James Schwartz
- Molecular Mechanisms of Hypoxic Responses via Unique Roles of Ras1, Cdc24 and Ptp3 in a Human Fungal Pathogen
- Analysis of the Genome and Transcriptome of var. Reveals Complex RNA Expression and Microevolution Leading to Virulence Attenuation
- Genotypic and Functional Impact of HIV-1 Adaptation to Its Host Population during the North American Epidemic
- RNA Editome in Rhesus Macaque Shaped by Purifying Selection
- Proper Actin Ring Formation and Septum Constriction Requires Coordinated Regulation of SIN and MOR Pathways through the Germinal Centre Kinase MST-1
- Interplay of the Serine/Threonine-Kinase StkP and the Paralogs DivIVA and GpsB in Pneumococcal Cell Elongation and Division
- A Quality Control Mechanism Coordinates Meiotic Prophase Events to Promote Crossover Assurance
- CNNM2 Mutations Cause Impaired Brain Development and Seizures in Patients with Hypomagnesemia
- The RNA-Binding Protein QKI Suppresses Cancer-Associated Aberrant Splicing
- Uncoupling Transcription from Covalent Histone Modification
- Rad51–Rad52 Mediated Maintenance of Centromeric Chromatin in
- FRA2A Is a CGG Repeat Expansion Associated with Silencing of
- A General Approach for Haplotype Phasing across the Full Spectrum of Relatedness
- A Novel Highly Divergent Protein Family Identified from a Viviparous Insect by RNA-seq Analysis: A Potential Target for Tsetse Fly-Specific Abortifacients
- A Central Role for in Regulation of Islet Function in Man
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- The Sequence-Specific Transcription Factor c-Jun Targets Cockayne Syndrome Protein B to Regulate Transcription and Chromatin Structure
- The Mechanism of Gene Targeting in Human Somatic Cells
- Genetic Predisposition to In Situ and Invasive Lobular Carcinoma of the Breast
- Widespread Use of Non-productive Alternative Splice Sites in
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



