-
Články
- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
New Models of Microsporidiosis: Infections in Zebrafish, , and Honey Bee
article has not abstract
Published in the journal: . PLoS Pathog 7(2): e32767. doi:10.1371/journal.ppat.1001243
Category: Pearls
doi: https://doi.org/10.1371/journal.ppat.1001243Summary
article has not abstract
Microsporidia Are Minimalist Parasites That Invade and Replicate inside a Wide Range of Hosts
Microsporidia comprise a large phylum of fungal-related pathogens that have been studied since the time of Louis Pasteur, who in 1870 found that they were responsible for silkworm disease that was decimating the silkworm industry [1], [2]. These obligate intracellular microbes are ubiquitous, but have remained enigmatic because of the difficulties of culturing them in the lab. In the 1990s there was a surge of interest in microsporidia when it was found that they were responsible for severe diarrhea and death in AIDS patients, but most research on these parasites has been conducted in fish and insects [3]. A recent PLoS Pathogens Pearl focused on the phylogenetic placement of microsporidia and the compactness of their genomes [4]. In this review we will consider the interaction between microsporidia and their hosts, with a focus on three non-mammalian hosts: zebrafish, Caenorhabditis elegans, and honey bee (Figure 1). These hosts are relatively new systems for the study of microsporidia, with distinct reasons motivating interest in each of them as described below. These systems provide exciting new opportunities to obtain insights into the mechanisms of microsporidia pathogenesis.
Fig. 1. Three new systems for study of infection by microsporidia. 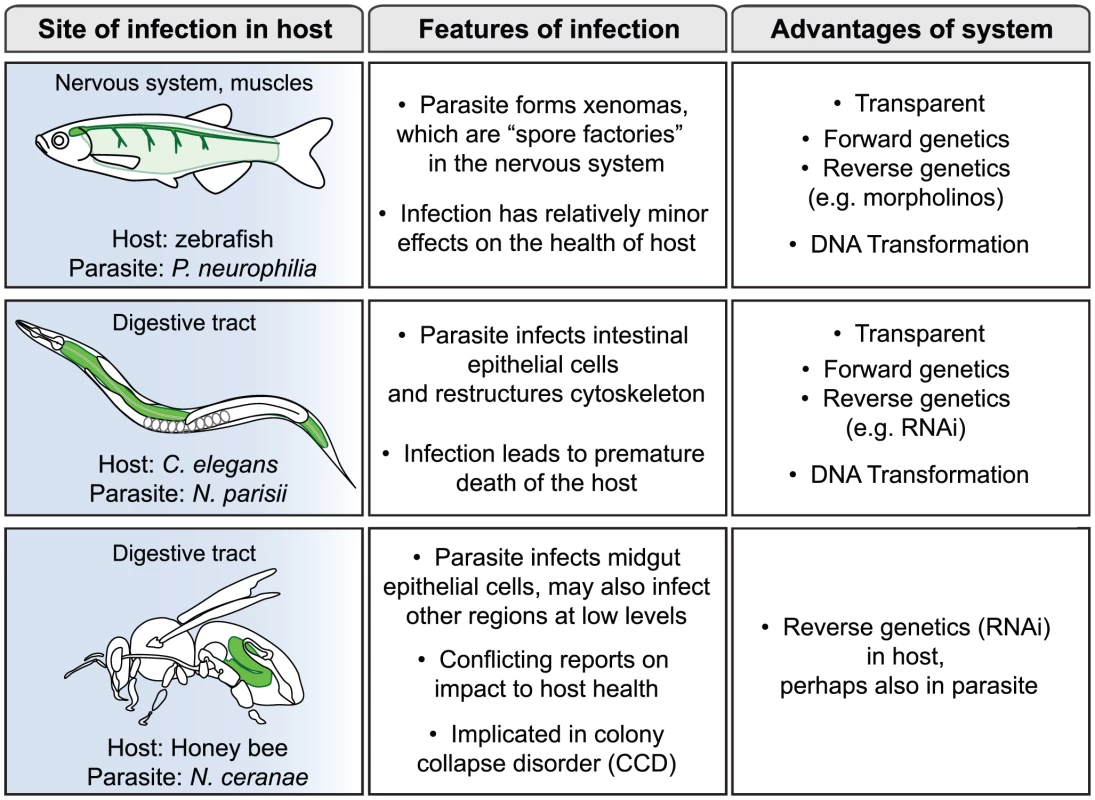
Zebrafish, C. elegans, and honey bee are infected by distinct species of microsporidia. Site of infection is highlighted in green for each host. Transmission in each system likely occurs via a fecal-oral route and the host is infected by ingesting infectious spores. Diagram by Malina Bakowski. Microsporidia as a group are able to infect an astonishingly wide range of hosts, including all animal phyla as well as a few protists. But how specific is the host range for a particular species of microsporidia? Roughly 1,200 species of microsporidia have been described, and it is difficult to make a blanket statement about their host range. On one end of the spectrum is Antonospora locustae, which appears to have a very narrow host range. A. locustae infects and eventually kills locusts, and has been approved by the United States Environmental Protection Agency as a “green pesticide” for locust control. In the process of obtaining approval for agricultural use, it was shown that A. locustae did not infect mammals, birds, fish, aquatic invertebrates, or honey bees (http://www.epa.gov/). On the other end of the spectrum is Anncallia (formerly Brachiola or Nosema) algerae, which was originally isolated from mosquitoes, but can replicate in several different species of invertebrate and vertebrate cell lines [5]. Interestingly, A. algerae appears to have caused death in an immunocompromised patient, possibly from a mosquito bite [6]. Thus microsporidia may have narrow or wide host range, depending on the species. The determinants of this host range are likely to encompass both host and pathogen factors that allow microsporidia infection.
How do microsporidia infect their hosts? Many microsporidia use a fecal-oral route of transmission, with some species restricted to intestinal cells and other species disseminating to a variety of tissues and organs. Very generally, microsporidia have two life stages: the actively replicating meront that develops inside of host cells, and the dormant spore form, which is the transmissible form that survives outside the host cell. In order to invade cells, spores contain a highly specialized infection apparatus called a polar tube. This tube is coiled inside the spore and then “fires” outside the spore to directly inject the parasite into host cells, although the exact details of this process are poorly understood in vivo. A variety of cues have been shown to induce polar tube firing in vitro, but no cue has been found that is universal [7]. Presumably the host environment encountered by each microsporidia species during infection differs, and thus each species may respond to a distinct set of conditions.
Once inside the host cell, microsporidia are very dependent occupants. Microsporidian genomes are extremely reduced in size, having jettisoned functions that can be accomplished by the host cell. An example of this dependence is that microsporidia cannot make ATP (except perhaps through substrate level phosphorylation) and instead appear to steal this vital energy currency from the host. This strategy of dramatic invasion followed by extreme dependence on the host cell appears to be evolutionarily successful, since there are a large number of species and hosts for microsporidia. A lifestyle such as this may represent the “bare bones” of a eukaryotic pathogen and the closest lifestyle to a virus of any eukaryote. Thus, study of microsporidia may provide insight into what represents the minimal arsenal for a eukaryotic parasite.
The Zebrafish Danio rerio Is a Host for Microsporidia
About 100 species of microsporidia have been shown to infect fish, including agriculturally relevant fish, such as salmon and rainbow smelt [8]. Indeed, the decline of entire fisheries has been attributed to microsporidiosis on several occasions. With the advent of zebrafish as a genetic model system, the infection of zebrafish by microsporidia has been increasingly observed. In fact, analyses from the University of Oregon diagnostic service indicated that microsporidia are the most common cause of disease for laboratory zebrafish [9], making this pathogen a serious concern for zebrafish researchers. The most commonly observed species that infects zebrafish is Pseudoloma neurophilia [10]. As is often the case in fish microsporidia infections, P. neurophilia form complexes called xenomas, which are essentially spore factories that generate vast quantities of spores. P. neurophilia xenomas are found in the nervous system, such as the hind brain, spinal cord, nerve roots, and occasionally in muscle (Figure 1). Despite the substantial size and frequency of these xenomas, they appear to be a relatively minor burden on the health and lifespan of the fish. Therefore, in this system microsporidia appear well-adapted to exploit their hosts by maximizing spore production but minimizing impact on the host.
The Nematode C. elegans Is a Host for Microsporidia
The nematode C. elegans is another genetic model organism that has recently been a focus for the study of microsporidia infections. C. elegans has been an extremely useful system for addressing many biological questions since 1970, including host defense and pathogenesis more recently [11]. However, most studies in the field of C. elegans pathogenesis have involved clinically relevant human pathogens that were not known to be natural pathogens of this animal. In a search for natural pathogens of C. elegans, a new genus and species of microsporidia was found in a wild-caught strain of C. elegans isolated from a compost pit near Paris [12]. This new species was named Nematocida parisii, or nematode-killer from Paris. In addition to this species, several other wild-caught Caenorhabditis nematodes have been isolated that harbor microsporidia ([12]; M-A. Félix, personal communication). Infection with N. parisii eventually leads to premature death of the host, but nematodes can carry a substantial parasite burden and still feed and move relatively normally for some time. One interesting aspect of the infection is that N. parisii appears to restructure the cytoskeleton of C. elegans host cells, perhaps as part of a non-damaging exit strategy (Figure 1). This restructuring may again be an example of microsporidia maximizing spore production and transmission, but minimizing impact on the host.
The Honey Bee Apis mellifera Is a Host for Microsporidia
In addition to the two genetic model hosts described above, microsporidia are also common in agriculturally relevant organisms, such as Apis mellifera, the Western honey bee. Precipitous drops in honey bee numbers have been observed in recent years, a phenomenon referred to as honey bee colony collapse disorder (CCD) [13]. Because honey bees are responsible for pollinating crops of economic importance, such as almonds, berries, fruits, and vegetables, this die-off is of great concern. The reason for this die-off is controversial, and some have even questioned whether it is significantly different from episodic declines in the past. In any case, there has been an active search for pathogens that could be responsible for CCD. The microsporidian species Nosema apis has long been known to afflict Western honey bee colonies, and in recent years, a new species of microsporidia called Nosema ceranae has increasingly been found in Western honey bee colonies around the world [14]. Study of N. ceranae has been an active area of interest as a possible cause of CCD. Some reports have indicated that N. ceranae is more pathogenic to honey bees than N. apis and could be a cause of CCD [15], although other studies have not found a difference in pathogenicity between the two species [16]. A recent report may provide a reconciliation of these conflicting results: this study indicates that CCD requires infection by both N. ceranae and a virus [17]. Clearly, N. ceranae warrants further study for its potential contribution to CCD, and for its general effects on the health of honey bee colonies.
Why Do These Systems Provide Fertile Ground for Study, and What Can Be Learned from Them?
After microsporidia invade host cells they undergo elaborate development, which has been described in rich detail through decades of electron microscopy (EM) in a variety of host/parasite pairs. However, these studies lack kinetic information, since EM provides only a single snapshot and requires labor-intensive fixation, sectioning, and staining to visualize the infection. For this reason, the transparent hosts C. elegans and zebrafish provide excellent systems for analysis of microsporidia development, since infections can be visualized inside living, intact animals. C. elegans and zebrafish provide advantages over other hosts like mammals and insects, which require dissection to analyze infection, even for standard microscopy studies. Individual C. elegans and zebrafish animals can repeatedly be analyzed microscopically throughout infection, making it possible to track the kinetics of parasite development inside these transparent and hardy hosts.
Another limitation of previous microsporidia studies is the relative dearth of molecular information. The field of microsporidia pathogenesis is full of rich cell biology questions, which have been underexplored molecularly due to a lack of tools. In particular, how do microsporidia exploit and restructure their hosts in order to minimize their impact, such as for xenomas in the fish and for cytoskeletal restructuring in C. elegans? Do microsporidia secrete enzymes to perform this work, or do they intersect pathways further upstream and instruct the host to do its own restructuring? Again, answers to these questions may come from imaging and genetic studies in genetically accessible hosts such as C. elegans and zebrafish. Both the zebrafish and C. elegans communities have generated strains that express fluorescently tagged proteins to allow for analysis of host molecules in live animals. In our own studies we have used such strains to reveal dramatic changes in host cytoskeletal proteins during infection (unpublished observations). In addition, C. elegans and zebrafish provide powerful genetic tools that allow for unbiased identification of host proteins involved in microsporidia infection through forward genetic screens. Proteins identified in these studies can then be examined in a more directed manner for their roles in less tractable hosts such as mice and humans.
In contrast to the above systems, the honey bee has not traditionally been a strong genetic system, but reverse genetics have recently been developed in the honey bee with the use of RNA interference [18]. This technique should allow for directed investigations into the roles of honey bee proteins in microsporidia infection. A tissue culture system has also been developed for N. ceranae that will facilitate study [19]. Intriguingly, RNAi may also be possible in the parasite itself. The genome sequence of N. ceranae indicates that the RNAi pathway is intact in this parasite, and an exciting report suggests that N. ceranae genes can be silenced by feeding dsRNA to the honeybee host [20]. This may be the first example of genetic manipulation in microsporidia, and opens up enormous potential for doing functional analysis of microsporidian genes.
Analysis of other microsporidian species will be facilitated by the Microsporidian Genomes Consortium effort at the Broad Institute (http://www.broadinstitute.org/files/shared/genomebio/Microsporidia_wp.pdf). The genomes of several species are being sequenced, including N. parisii and P. neurophilia. Sequence data from these small genomes will allow for molecular analysis, perhaps through RNAi or misexpression studies in the host. Comparative studies such as these will likely yield insight into what endows each species with its own characteristics and ability to interact and exploit its specific host. They may also provide insight into the general strategies used by microsporidia, which are some of the most streamlined eukaryotic parasites.
Zdroje
1. TexierC
VidauC
ViguesB
El AlaouiH
DelbacF
2010 Microsporidia: a model for minimal parasite-host interactions. Curr Opin Microbiol 13 443 449
2. KeelingPJ
FastNM
2002 Microsporidia: biology and evolution of highly reduced intracellular parasites. Annu Rev Microbiol 56 93 116
3. DidierES
WeissLM
2006 Microsporidiosis: current status. Curr Opin Infect Dis 19 485 492
4. KeelingP
2009 Five questions about microsporidia. PLoS Pathog 5 e1000489 doi:10.1371/journal.ppat.1000489
5. Becnel JJAT
1999 Microsporidia in insects.
WittnerM
The microsporidia and microsporidiosis Washington (D.C.) ASM 447 501
6. CoyleCM
WeissLM
RhodesLV3rd
CaliA
TakvorianPM
2004 Fatal myositis due to the microsporidian Brachiola algerae, a mosquito pathogen. N Engl J Med 351 42 47
7. Keohane EMWL
1999 The structure, function, and composition of the microsporidian polar tube.
WittnerM
The microsporidia and microsporidiosis Washington (D.C.) ASM
8. Shaw RWKM
1999 Fish microsporidia.
WittnerM
The microsporidia and microsporidiosis Washington (D.C.) ASM 418 446
9. RamsayJM
WatralV
SchreckCB
KentML
2009 Pseudoloma neurophilia infections in zebrafish Danio rerio: effects of stress on survival, growth, and reproduction. Dis Aquat Organ 88 69 84
10. MatthewsJL
BrownAM
LarisonK
Bishop-StewartJK
RogersP
2001 Pseudoloma neurophilia n. g., n. sp., a new microsporidium from the central nervous system of the zebrafish (Danio rerio). J Eukaryot Microbiol 48 227 233
11. IrazoquiJE
UrbachJM
AusubelFM
Evolution of host innate defence: insights from Caenorhabditis elegans and primitive invertebrates. Nat Rev Immunol 10 47 58
12. TroemelER
FelixMA
WhitemanNK
BarriereA
AusubelFM
2008 Microsporidia are natural intracellular parasites of the nematode Caenorhabditis elegans. PLoS Biol 6 e309 doi:10.1371/journal.pbio.0060309
13. VanengelsdorpD
EvansJD
SaegermanC
MullinC
HaubrugeE
2009 Colony collapse disorder: a descriptive study. PLoS ONE 4 e6481 doi:10.1371/journal.pone.0006481
14. KleeJ
BesanaAM
GenerschE
GisderS
NanettiA
2007 Widespread dispersal of the microsporidian Nosema ceranae, an emergent pathogen of the western honey bee, Apis mellifera. J Invertebr Pathol 96 1 10
15. HigesM
Martin-HernandezR
BotiasC
BailonEG
Gonzalez-PortoAV
2008 How natural infection by Nosema ceranae causes honeybee colony collapse. Environ Microbiol 10 2659 2669
16. ForsgrenE
FriesI
Comparative virulence of Nosema ceranae and Nosema apis in individual European honey bees. Vet Parasitol 170 212 217
17. BromenshenkJJ
HendersonCB
WickCH
StanfordMF
ZulichAW
2010 Iridovirus and microsporidian linked to honey bee colony decline. PLoS ONE 5 e13181 doi:10.1371/journal.pone.0013181
18. Honeybee Genome Sequencing Consortium 2006 Insights into social insects from the genome of the honeybee Apis mellifera. Nature 443 931 949
19. GisderS
MockelN
LindeA
GenerschE
2010 A cell culture model for Nosema ceranae and Nosema apis allows new insights into the life cycle of these important honey bee-pathogenic microsporidia. Environ Microbiol E-pub ahead of print. doi: 10.1111/j.1462-2920.2010.02346.x
20. PaldiN
GlickE
OlivaM
ZilberbergY
AubinL
2010 Effective gene silencing in a microsporidian parasite associated with honeybee (Apis mellifera) colony declines. Appl Environ Microbiol 76 5960 5964
Štítky
Hygiena a epidemiologie Infekční lékařství Laboratoř
Článek Compensatory Evolution of Mutations Restores the Fitness Cost Imposed by β-Lactam Resistance inČlánek The C-Terminal Domain of the Arabinosyltransferase EmbC Is a Lectin-Like Carbohydrate Binding ModuleČlánek A Viral microRNA Cluster Strongly Potentiates the Transforming Properties of a Human Herpesvirus
Článek vyšel v časopisePLOS Pathogens
Nejčtenější tento týden
2011 Číslo 2- Stillova choroba: vzácné a závažné systémové onemocnění
- Jak souvisí postcovidový syndrom s poškozením mozku?
- Perorální antivirotika jako vysoce efektivní nástroj prevence hospitalizací kvůli COVID-19 − otázky a odpovědi pro praxi
- Diagnostika virových hepatitid v kostce – zorientujte se (nejen) v sérologii
- Infekční komplikace virových respiračních infekcí – sekundární bakteriální a aspergilové pneumonie
-
Všechny články tohoto čísla
- A Fresh Look at the Origin of , the Most Malignant Malaria Agent
- In Situ Photodegradation of Incorporated Polyanion Does Not Alter Prion Infectivity
- Highly Efficient Protein Misfolding Cyclic Amplification
- Positive Signature-Tagged Mutagenesis in : Tracking Patho-Adaptive Mutations Promoting Airways Chronic Infection
- Charge-Surrounded Pockets and Electrostatic Interactions with Small Ions Modulate the Activity of Retroviral Fusion Proteins
- Whole-Body Analysis of a Viral Infection: Vascular Endothelium is a Primary Target of Infectious Hematopoietic Necrosis Virus in Zebrafish Larvae
- Inhibition of Nox2 Oxidase Activity Ameliorates Influenza A Virus-Induced Lung Inflammation
- STAT2 Mediates Innate Immunity to Dengue Virus in the Absence of STAT1 via the Type I Interferon Receptor
- Uropathogenic P and Type 1 Fimbriae Act in Synergy in a Living Host to Facilitate Renal Colonization Leading to Nephron Obstruction
- Elite Suppressors Harbor Low Levels of Integrated HIV DNA and High Levels of 2-LTR Circular HIV DNA Compared to HIV+ Patients On and Off HAART
- DC-SIGN Mediated Sphingomyelinase-Activation and Ceramide Generation Is Essential for Enhancement of Viral Uptake in Dendritic Cells
- Short-Lived IFN-γ Effector Responses, but Long-Lived IL-10 Memory Responses, to Malaria in an Area of Low Malaria Endemicity
- Induces T-Cell Lymphoma and Systemic Inflammation
- The C-Terminus of RON2 Provides the Crucial Link between AMA1 and the Host-Associated Invasion Complex
- Critical Role of the Virus-Encoded MicroRNA-155 Ortholog in the Induction of Marek's Disease Lymphomas
- Type I Interferon Signaling Regulates Ly6C Monocytes and Neutrophils during Acute Viral Pneumonia in Mice
- Atypical/Nor98 Scrapie Infectivity in Sheep Peripheral Tissues
- Innate Sensing of HIV-Infected Cells
- BosR (BB0647) Controls the RpoN-RpoS Regulatory Pathway and Virulence Expression in by a Novel DNA-Binding Mechanism
- Compensatory Evolution of Mutations Restores the Fitness Cost Imposed by β-Lactam Resistance in
- Expression of Genes Involves Exchange of the Histone Variant H2A.Z at the Promoter
- The RON2-AMA1 Interaction is a Critical Step in Moving Junction-Dependent Invasion by Apicomplexan Parasites
- Epstein-Barr Virus Nuclear Antigen 3C Facilitates G1-S Transition by Stabilizing and Enhancing the Function of Cyclin D1
- Transcription and Translation Products of the Cytolysin Gene on the Mobile Genetic Element SCC Regulate Virulence
- Phosphatidylinositol 3-Monophosphate Is Involved in Apicoplast Biogenesis
- The Rubella Virus Capsid Is an Anti-Apoptotic Protein that Attenuates the Pore-Forming Ability of Bax
- Episomal Viral cDNAs Identify a Reservoir That Fuels Viral Rebound after Treatment Interruption and That Contributes to Treatment Failure
- Genetic Mapping Identifies Novel Highly Protective Antigens for an Apicomplexan Parasite
- Relationship between Functional Profile of HIV-1 Specific CD8 T Cells and Epitope Variability with the Selection of Escape Mutants in Acute HIV-1 Infection
- The Genotype of Early-Transmitting HIV gp120s Promotes αβ –Reactivity, Revealing αβ/CD4 T cells As Key Targets in Mucosal Transmission
- Small Molecule Inhibitors of RnpA Alter Cellular mRNA Turnover, Exhibit Antimicrobial Activity, and Attenuate Pathogenesis
- The bZIP Transcription Factor MoAP1 Mediates the Oxidative Stress Response and Is Critical for Pathogenicity of the Rice Blast Fungus
- Entrapment of Viral Capsids in Nuclear PML Cages Is an Intrinsic Antiviral Host Defense against Varicella-Zoster Virus
- NS2 Protein of Hepatitis C Virus Interacts with Structural and Non-Structural Proteins towards Virus Assembly
- Measles Outbreak in Africa—Is There a Link to the HIV-1 Epidemic?
- New Models of Microsporidiosis: Infections in Zebrafish, , and Honey Bee
- The C-Terminal Domain of the Arabinosyltransferase EmbC Is a Lectin-Like Carbohydrate Binding Module
- A Viral microRNA Cluster Strongly Potentiates the Transforming Properties of a Human Herpesvirus
- Infections in Cells: Transcriptomic Characterization of a Novel Host-Symbiont Interaction
- Secreted Bacterial Effectors That Inhibit Host Protein Synthesis Are Critical for Induction of the Innate Immune Response to Virulent
- Genital Tract Sequestration of SIV following Acute Infection
- Functional Coupling between HIV-1 Integrase and the SWI/SNF Chromatin Remodeling Complex for Efficient Integration into Stable Nucleosomes
- DNA Damage and Reactive Nitrogen Species are Barriers to Colonization of the Infant Mouse Intestine
- The ESCRT-0 Component HRS is Required for HIV-1 Vpu-Mediated BST-2/Tetherin Down-Regulation
- Targeted Disruption of : Invasion of Erythrocytes by Using an Alternative Py235 Erythrocyte Binding Protein
- Trivalent Adenovirus Type 5 HIV Recombinant Vaccine Primes for Modest Cytotoxic Capacity That Is Greatest in Humans with Protective HLA Class I Alleles
- PLOS Pathogens
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Genetic Mapping Identifies Novel Highly Protective Antigens for an Apicomplexan Parasite
- Type I Interferon Signaling Regulates Ly6C Monocytes and Neutrophils during Acute Viral Pneumonia in Mice
- Infections in Cells: Transcriptomic Characterization of a Novel Host-Symbiont Interaction
- The ESCRT-0 Component HRS is Required for HIV-1 Vpu-Mediated BST-2/Tetherin Down-Regulation
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



