-
Články
- Vzdělávání
- Časopisy
Top články
- Témata
- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Chromosomal Organization and Segregation in
The study of chromosomal organization and segregation in a handful of bacteria has revealed surprising variety in the mechanisms mediating such fundamental processes. In this study, we further emphasized this diversity by revealing an original organization of the Pseudomonas aeruginosa chromosome. We analyzed the localization of 20 chromosomal markers and several components of the replication machinery in this important opportunistic γ-proteobacteria pathogen. This technique allowed us to show that the 6.3 Mb unique circular chromosome of P. aeruginosa is globally oriented from the old pole of the cell to the division plane/new pole along the oriC-dif axis. The replication machinery is positioned at mid-cell, and the chromosomal loci from oriC to dif are moved sequentially to mid-cell prior to replication. The two chromosomal copies are subsequently segregated at their final subcellular destination in the two halves of the cell. We identified two regions in which markers localize at similar positions, suggesting a bias in the distribution of chromosomal regions in the cell. The first region encompasses 1.4 Mb surrounding oriC, where loci are positioned around the 0.2/0.8 relative cell length upon segregation. The second region contains at least 800 kb surrounding dif, where loci show an extensive colocalization step following replication. We also showed that disrupting the ParABS system is very detrimental in P. aeruginosa. Possible mechanisms responsible for the coordinated chromosomal segregation process and for the presence of large distinctive regions are discussed.
Published in the journal: . PLoS Genet 9(5): e32767. doi:10.1371/journal.pgen.1003492
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1003492Summary
The study of chromosomal organization and segregation in a handful of bacteria has revealed surprising variety in the mechanisms mediating such fundamental processes. In this study, we further emphasized this diversity by revealing an original organization of the Pseudomonas aeruginosa chromosome. We analyzed the localization of 20 chromosomal markers and several components of the replication machinery in this important opportunistic γ-proteobacteria pathogen. This technique allowed us to show that the 6.3 Mb unique circular chromosome of P. aeruginosa is globally oriented from the old pole of the cell to the division plane/new pole along the oriC-dif axis. The replication machinery is positioned at mid-cell, and the chromosomal loci from oriC to dif are moved sequentially to mid-cell prior to replication. The two chromosomal copies are subsequently segregated at their final subcellular destination in the two halves of the cell. We identified two regions in which markers localize at similar positions, suggesting a bias in the distribution of chromosomal regions in the cell. The first region encompasses 1.4 Mb surrounding oriC, where loci are positioned around the 0.2/0.8 relative cell length upon segregation. The second region contains at least 800 kb surrounding dif, where loci show an extensive colocalization step following replication. We also showed that disrupting the ParABS system is very detrimental in P. aeruginosa. Possible mechanisms responsible for the coordinated chromosomal segregation process and for the presence of large distinctive regions are discussed.
Introduction
Members of the genus Pseudomonas (γ-proteobacteria) show remarkable metabolic and physiological diversity and versatility, enabling colonization of diverse terrestrial and aquatic habitats. These species are of great interest because of their importance in plant and human diseases and their growing potential in biotechnological applications. One of these bacteria, Pseudomonas aeruginosa, is an aquatic and soil bacterium that can infect a range of organisms, including plants, invertebrates and various mammals [1]. In humans, P. aeruginosa is an opportunistic pathogen that causes serious infections in immunocompromised patients, and it is the leading cause of morbidity in cystic fibrosis patients [2]. These infections are particularly challenging because of P. aeruginosa's broad intrinsic antimicrobial resistance [3].
The PAO1 genome was the first P. aeruginosa genome to be sequenced [4]. It comprises 6.3 Mbp encoding 5,570 genes. Its size results from genetic complexity rather than gene duplications, which may suggest that it adapts to colonize a diverse range of ecological niches [1], [4]. A comparison of five P. aeruginosa genomes revealed a relatively large set of 5021 conserved genes, which constitute the core genome. Beside this set, insertions containing blocks of strain-variable genes are found in a limited number of chromosomal locations, termed Regions of Genomic Plasticity (RGPs). The P. aeruginosa oriC sequence has been characterized and is adjacent to dnaA and dnaN [5], [6]. The dif site, a sequence that is required for chromosome dimer resolution by the XerCD recombinases upon activation by the DNA translocase FtsK, was identified opposite of oriC, whereas no system dedicated to replication termination (such as replication fork trap) has yet been characterized in P. aeruginosa.
To fit inside bacterial cells, bacterial chromosomes need to be organized into a compact structure called the nucleoid. DNA compaction is thought to result from the interplay between macromolecular crowding, DNA supercoiling and the specific action of DNA binding proteins [7]. In all organisms in which this process has been examined, the nucleoid is oriented in a specific way inside the cell, which preserves the linear order of genes in the DNA [8]. This global orientation is longitudinal in the case of Bacillus subtilis, Caulobacter crescentus and Vibrio cholerae chromosomes and transversal in E. coli [9], [10]. In E. coli, large-scale organization in several domains is also present [11], [12].
The compaction of the chromosome must be compatible with various processes of DNA metabolism, such as gene expression and replication and the segregation of genetic information. Different hypotheses have been proposed to explain chromosomal segregation. For instance, studies in B. subtilis led to the development of the capture extrusion model, which suggests that the energy required for initial chromosomal segregation could come from the replication process that occurs at mid-cell in a static replication factory [13]. However, it was later shown that in E. coli, the two replication forks appear to follow chromosomal arms, rendering the capture extrusion model irrelevant for E. coli chromosomes [14], [15]. It was proposed instead that loss of cohesion between replicated origin regions could trigger global chromosomal movement and mediate chromosomal segregation [14]. It has also been proposed that entropic exclusion of replicated chromosomes might participate in the segregation process [16].
At the molecular level, a mitotic-like apparatus composed of a DNA binding protein (ParB) that binds to a specific sequence (parS) and a Walker-type ATPase (ParA) is present in a number of bacteria, including P. aeruginosa, but absent in E. coli. This apparatus is involved in chromosomal segregation in V. cholera, C. crescentus (where it is essential), B. subtilis and Streptococcus pneumonia [17]–[20]. It was also demonstrated in B. subtilis and S. pneumonia that the recruitment of bacterial condensins to the origin region by ParB-parS complex contributes to chromosomal segregation [20]–[22]. Two bacterial condensins have been identified in P. aeruginosa, and both the ParABS system and these two condensins appear to be important for chromosomal segregation, as the deletion of either of these factors leads to an increase in anucleated cells [23]–[25].
As a first step in the study of chromosomal organization and segregation in P. aeruginosa, we used a fluorescent microscopy approach to investigate chromosomal localization during growth. This approach allowed us to show that the global orientation of the P. aeruginosa chromosome is longitudinal and that the replication forks are mostly colocalized near mid-cell. Chromosomal loci appear to be relocated closer to mid-cell prior to replication, and the two replicated copies are then progressively segregated into opposite cell halves. This process is surprisingly different from what is observed in E. coli, another gamma-proteobacterium, but closer to what has been described for the gram-positive bacterium B. subtilis. Moreover, we could discern two distinctive regions in the P. aeruginosa chromosome, one surrounding oriC and the other surrounding the dif site. The origin region was defined by the fact that approximately 1.4 Mbp of DNA centered on oriC segregate to the same position around the 0.2/0.8 relative cell length. The Ter region encompasses at least 800 kbp that remain close to the new pole of the cell before being relocated to near mid-cell prior to replication. Loci in the Ter region show a high level of colocalization following replication. Chromosomal arms linking these two regions are longitudinally distributed. These features allow us to propose P. aeruginosa chromosomal organization as an original model, combining longitudinal organization, as observed in C. crescentus, with large distinctive regions that might be suggestive of long-range organization, as observed in E. coli. We also provide evidence that the ParABS system plays a major role in chromosomal organization and segregation in P. aeruginosa.
Results
Visualizing 20 chromosomal loci in live P. aeruginosa cells
To study chromosomal organization in P. aeruginosa, we used a fluorescent microscopy approach. The intracellular position of chromosomal loci was visualized using two different reporter systems based on the binding of a fluorescently labeled protein to a target sequence inserted at twenty specific locations in the P. aeruginosa chromosome (see Figure 1A and Text S1). We used both the TetR-CFP chimera, which recognizes tetO arrays, and the yGFP-ParBpMT1 chimera, which binds to the parSpMT1 sequence [26], [27]. We created a plasmid allowing expression of both yGFP-parBpMT1 and tetR-CFP from an IPTG inducible promoter. The visualization of two chromosomal loci by fluorescent microscopy was thus possible upon growth in IPTG-containing medium. The use of either the parSpMT1 sequence or the tetO arrays to label a specific chromosomal locus gives similar results regarding foci number and positions inside the cell (data not shown).
Fig. 1. Progressive segregation of P. aeruginosa chromosomal loci. 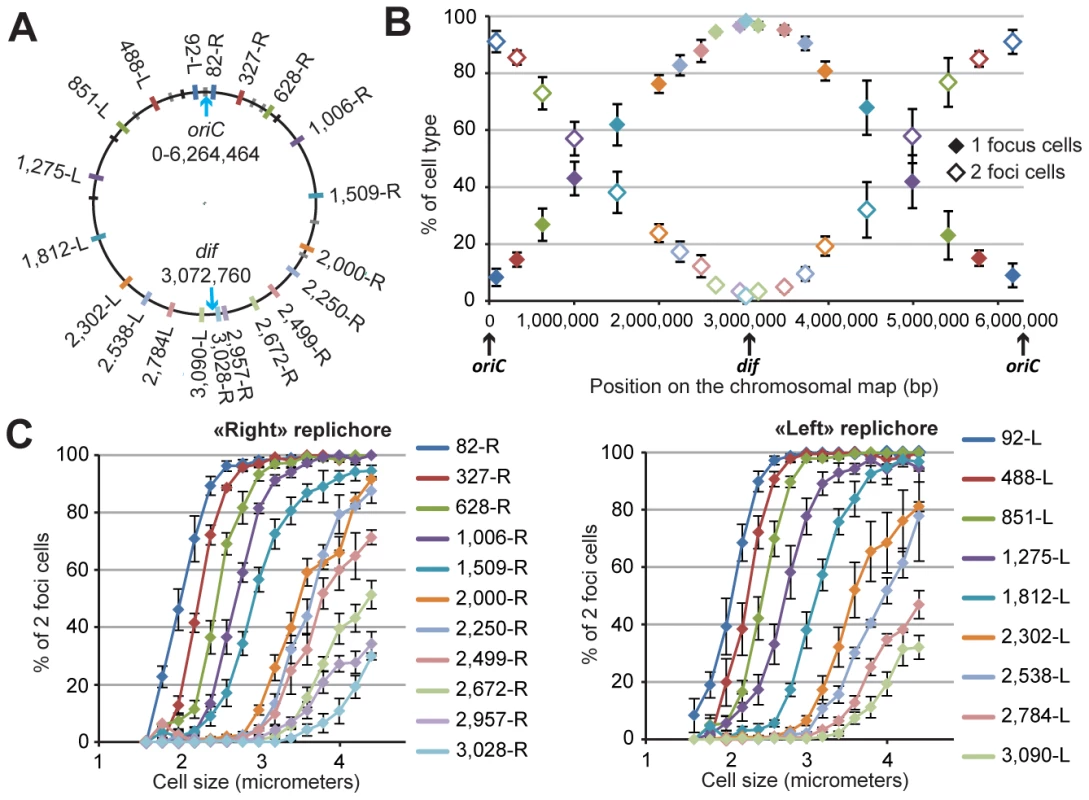
(A) Position of each chromosomal locus on the PAO1 chromosomal map. Each locus is represented by a colored tag on the PAO1 chromosome according to its position. The color code is used for all figures. Black tags indicate rRNA operons. Grey tags represent parS sequence positions. Four of these parS sites are clustered close to oriC, four more are dispersed between positions 851-L and 628-R, and two are localized in the “right” replichore. (B) Average percentage of one-focus cells (solid diamonds) and two-foci cells (open diamonds) in a bacterial population grown in minimal medium supplemented with citrate. Cells with no visible focus were removed from this analysis (they constituted between 5 to 10% of the cells). The proportion of cells exhibiting more than two foci was always smaller than 0.5 %. The x-axis represents the positions of the loci on the chromosomal map. Values from four to eleven experiments were averaged, and the error bars represent standard deviations. (C) The average percentage of two-foci cells in a bacterial population grown in minimal medium supplemented with citrate, according to cell size, for each chromosomal locus of the “right” replichore (left panel) or of the “left” replichore (right panel). Values from four to eleven experiments were averaged, and the error bars represent standard deviations. Chromosomal loci are sequentially segregated
We tested different growth conditions and chose to perform extensive studies in minimal medium supplemented with citrate at 30 degrees. Under these growth conditions, the P. aeruginosa doubling time is approximately 45 to 50 minutes, with cell sizes ranging from 2 to 5 µm (mean size of approximately 2.9 µm). We predicted, based on the number of foci corresponding to chromosomal loci or replisome proteins (see below), that most of the cells would contain a single replicating chromosome. Indeed, each chromosomal locus is visualized as one or two foci. Figure 1B shows the number of cells exhibiting one or two foci for each of the 20 positions. Loci close to oriC are mostly visualized as two foci (>90% of two-foci cells), indicating that they are replicated and segregated upon cell birth (focus duplication corresponds to the segregation of the two copies of a chromosomal locus). In contrast, loci located close to the dif site (presumably in the Ter region where the replication ends) are mostly visualized as one focus (>90% of one-focus cells), indicating that they are segregated immediately prior to cell division. For loci located in between, a linear increase in the proportion of one-focus cells (and therefore a decrease in the proportion of two-foci cells) is observed. Figure 1C shows the percentage of two-foci cells according to cell size for each position. This percentage indicates that the foci duplication process is sequential, proceeding from the origin of replication to the terminus. For each locus, the number of two-foci cells increases with cell size. The curves are remarkably parallel for loci close to oriC, and their slope decreases as the distance from oriC increases. Very interestingly, for positions close to the dif site (2,672-R, 2,957-R and 3,028-R on the right replichore, and 2,784-L and 3,090-L on the left replichore), the proportion of two-foci cells never exceeds 50%, even in larger cells. This finding indicates that the separation of the two copies of these loci occurs concomitantly with cell division in more than 50% of cells.
The P. aeruginosa chromosome shows two distinctive regions surrounding oriC and dif and is longitudinally organized in between
We studied foci localization inside the cells. To orientate bacterial cells, we took advantage of the fact that loci located near dif exhibit a striking pattern: in one-focus cells, they are localized close to one pole (closer than the 0.2 relative cell length) in small cells and close to mid-cell in larger cells. In two-foci cells, the 2 foci stay close to the division plane, suggesting that upon division, the pole where the dif region is localized is the new pole of the cell. We thus labeled two different loci: one close to dif, which we used as a marker of the new pole, and another elsewhere on the chromosome, whose position we were interested in. The data for each locus are presented in Figure S1, and the results are summarized in Figure 2. The relative positions of foci inside bacterial cells are represented for small cells (smaller than 2.8 µm, Figure 2A), medium cells (between 2.8 and 3.5 µm, Figure 2B) and large cells (larger than 3.5 µm, Figure 2C). The proportions of one-focus cells for each cell type are indicated in Table S3. Moreover, Figure S2 presents the same data differently: the relative positions of the foci in cells within 0.2 µm size intervals are presented according to cell size, yielding curves that represent the segregation profiles for each chromosomal locus.
Fig. 2. Position of chromosomal loci inside bacterial cells, oriented relative to the new pole of the cell. 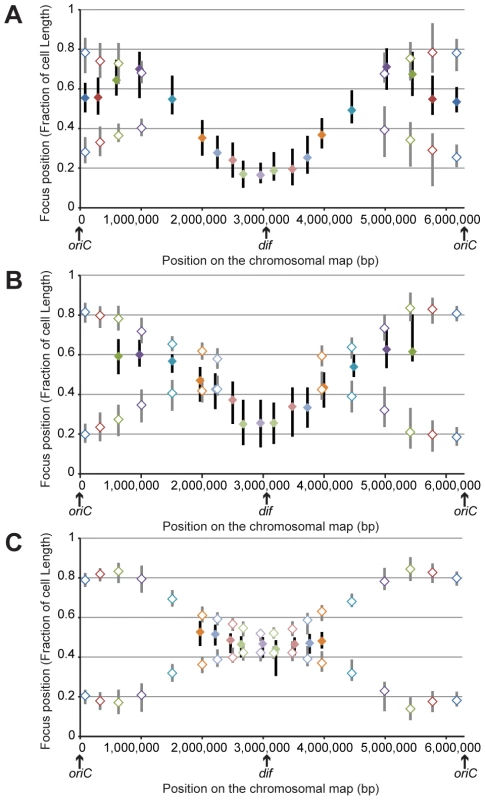
Positions in cells smaller than 2.8 μm (A), in cells between 2.8 and 3.5 μm (B) and in cells larger than 3.5 μm (C). The y-axis represents the relative position of the focus in bacterial cells, 0 being the new pole and 1 the old pole. The markers represent the median values of relative positions, and the vertical bars represent the 25-75th percentiles of the relative positions. The positions of the single focus in one-focus cells are represented with a solid diamond and a black vertical bar, whereas the positions of the two foci in two-foci cells are indicated with an open diamond and a gray vertical bar. From our results, we inferred the existence of two specific regions in the P. aeruginosa chromosome. The first region surrounds oriC. Indeed, in large cells (when most of the chromosome is replicated, before cell division), it is clearly observed that the two copies of the three loci closest to oriC on each replichore (loci 82-R, 327-R, 628-R and 92-L, 488-L, 851-L) are located at approximately the same position, close to the 0.2/0.8 relative cell length (blue, red and green markers, Figure 2C). Remarkably, eight of the ten parS sites found in P. aeruginosa are located in between these chromosomal loci (Figure 1A): four of them are very close to oriC (between 4 to 15kb from oriC), two are found between loci 327-R and 628-R, one is found between loci 92-L and 488-L, and one is found between loci 488-L and 851-L. From these data, we proposed that the three loci closer to oriC on each replichore (blue, red and green markers) define a distinctive region that we called the Ori region. Loci in this region segregate sequentially (Figure 1, Figure 2A and 2B) but reach the same final position inside the cell (around the 0.2/0.8 relative cell length).
Loci closest to oriC are not the most polarly localized (the red and green markers are farther away from the new pole than the blue marker, Figure 2C), but they are precisely positioned at the 0.2/0.8 relative cell length. This finding is not expected from the position of the loci on the chromosomal map and is not consistent with a strictly longitudinal organization (as in C. crescentus chromosomal organization, for instance [28]).
The second region that we discerned in the P. aeruginosa chromosome surrounds the dif site. Indeed, another striking pattern is observed for the four loci closer to dif (positions 2,672-R, 2,957-R, 3,090-L and 2,784-L). As indicated above, they are mostly visualized as a single focus, even in large cells (more than 75% of one-focus cells, Table S3 and Figure 1C). They are located close
to the new pole (below the 0.2 relative cell length) in small cells (Figure 2A); in medium cells, their position is still quite polar (Figure 2B and Figure S2), whereas they are located near mid-cell in large cells (Figure 2C and Figure S2). Because of their polar localization in small cells and the fact that the proportion of cells exhibiting two foci never exceeds 50%, even in large cells, we proposed that these 4 loci belong to another distinctive region that we called the Ter region. Chromosomal locus 3,028-R also belongs to this Ter region, but because it was used to orientate cells, it is not included in Figure 2. Once again, loci in this region segregate sequentially (Figure 1), but their positioning inside the cell is similar.
Chromosomal loci located in between these Ori and Ter regions are spread between the 0.7 and the 0.2 relative cell length when they are visualized as a single focus and between the 0.2/0.8 relative cell length and mid-cell when they are visualized as two foci. This suggests a longitudinal organization of these parts of the P. aeruginosa chromosome. To confirm this hypothesis, we measured distances between pairs of loci. We observed that loci located at equivalent distances from oriC on different replichores are indeed more frequently colocalized than loci on the same replichore (Figure S3).
Looking more specifically at one-focus cells, most chromosomal loci are relocated close to mid-cell (0.5 relative cell length) before they are resolved into two elements (Figure 2 and Figure S2). This relocation could suggest a sequential repositioning of each chromosomal locus before replication.
P. aeruginosa replisomes are mostly colocalized near mid-cell
To investigate where replication takes place in P. aeruginosa, DNA polymerase was visualized in living cells using fusions between replisome components and green fluorescent protein (GFP). This approach has been successfully used in B. subtilis [29], C. crescentus [30] and E. coli [15] to study DNA polymerase localization. Chromosomal genes encoding DnaX (PA1532), HolB (PA2961) and HolA (PA3989) were replaced with genes encoding GFP tagged versions of these proteins (see Text S1 for details). We also engineered a Dronpa-tagged version of DnaX, as Dronpa is a fluorescent protein that has been reported not to cause aberrant foci formation [31]. The results using this DnaX-Dronpa fusion were quite similar to those obtained with DnaX-eGFP (Figure S5).
In minimal medium supplemented with citrate, these GFP fusions were mostly observed as a single spot localized near mid-cell (Figure 3A and Figure S4A). Some cells had no visible focus (about 10%), and although we cannot exclude experimental faults, the highest proportion was observed in smaller cells or larger cells, which is consistent with replication starting immediately upon cell birth and ending before cell division. More than 90% of the cells with foci possessed only one visible focus, indicating that the two replication forks are mostly colocalized. However, a small number of cells presenting two GFP foci were observed for each cell size. In small cells, these foci were found close to mid-cell and relatively close to each other (distance <0.35 of the relative cell length), which suggests that the two forks can separate during the replication process. In the largest cells, however, the two foci were localized far apart, around the 0.2/0.8 relative cell length. These foci coincide with the position of foci close to oriC, which could indicate that another round of replication is starting in a few cells, before division is achieved.
Fig. 3. Localization of P.aeruginosa DNA polymerase. 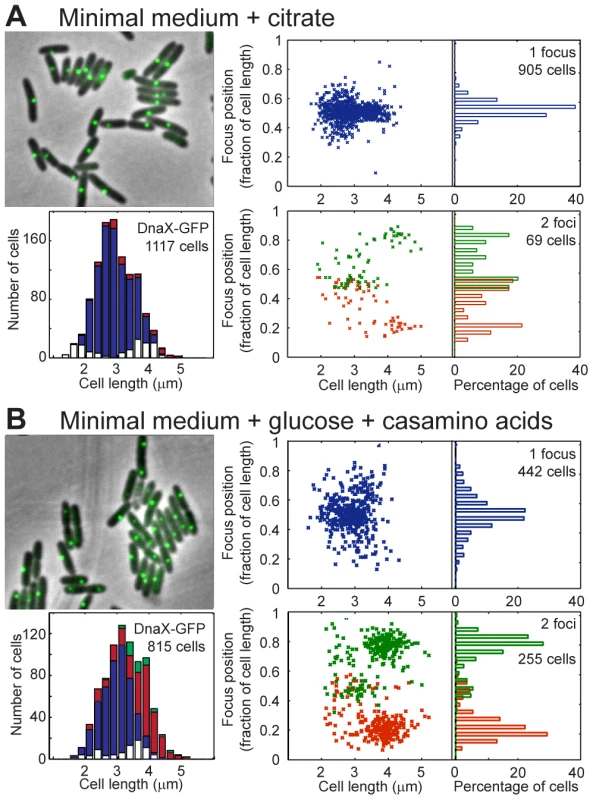
EGFP-labeled replisome protein DnaX was observed in minimal medium supplemented with citrate (A) or glucose and casamino acids (B). For each panel, the upper left area shows a sample of representative cells; the lower left area presents the amount of cells exhibiting zero (white), one (blue), two (red) or three (green) fluorescent foci according to cell size. The upper right area presents the relative positions of the focus in one-focus cells, and the lower left area presents the relative positions of the foci in two-foci cells. Overall, these results show that replication forks are mostly colocalized near mid-cell in P. aeruginosa. Therefore, the repositioning of all chromosomal loci close to mid-cell prior to their duplication could indeed be linked to the replication process.
Global chromosomal organization is conserved in different growth conditions
We next wondered what would happen to chromosomal organization if the growth conditions were changed. In minimal medium supplemented with glucose and casamino acids, 2 to 4 copies of chromosomal loci close to oriC were observed (Figure S6), implying that a new round of replication starts before cell division more often in this medium than in citrate medium. The cells are slightly larger in this minimal medium than in minimal medium supplemented with citrate (Figure 3), and the doubling time is similar (approximately 45 minutes, data not shown).
Observing DnaX-eGFP (as well as HolB-eGFP, Figure S4B, and DnaX-Dronpa, Figure S5B) revealed that in one-focus cells, the focus is localized close to mid-cell, as observed in minimal medium supplemented with citrate. The proportion of two-foci cells is higher in MM supplemented with glucose and casamino acids than in MM supplemented with citrate (approximately one third of cells with visible foci presented two foci, Figure 3B). The proportion of two-foci cells increased with cell size: a large majority of cells (approximately 65%) larger than 3.5 µm exhibited two foci. Moreover, a clear difference was observed between the position of the two foci in cells smaller than approximately 3.5 µm, in which the two foci were close together (distance <0.35 of the relative cell length), and the position of the two foci in cells larger than approximately 3.5 µm, where they were located near the one quarter/three quarter position in the cells. These results suggest that in cells smaller than approximately 3.5 µm, a replication round is finishing near mid-cell, most often with the two forks colocalized. In contrast, in cells larger than approximately 3.5 µm, another round of replication started on the two duplicated chromosomes, close to the mid-cell of the future daughter cells. The two replication forks that reproduce the same chromosome are also mostly colocalized.
In contrast to loci close to oriC (which are present in two to four copies), intermediate chromosomal loci (1,812-L and 1,509-R) are observed as only one or two foci (Figure S6), which might suggest that cell division occurs before the replication forks reach these positions. Loci located near dif (3,090-L and 2,499-R) show the same localization pattern in minimal medium supplemented with glucose and casamino acids as in minimal medium supplemented with citrate: they are localized near the cell pole in small cells and near mid-cell in larger cells. A single focus near mid-cell can be observed in large cells despite the fact that the chromosome is fully replicated (as can be inferred from visualizing replisome proteins). When a locus in the Ori region and a locus in the Ter region are visualized in the same cells, the proportion of cells exhibiting four foci of the Ori locus and one focus of the Ter locus is twice the proportion of cells exhibiting four foci of the Ori locus and two focus of the Ter locus (data not shown). This finding suggests an extensive colocalization step following replication for the Ter region.
Together, these results suggest that in minimal medium supplemented with glucose and casamino acids, the P. aeruginosa chromosome is organized similarly to how it is organized in minimal medium supplemented with citrate, except that two chromosomes are being replicated instead of one (see discussion).
Disruption of the ParABS system has a major impact on chromosomal organization
As a first step to study molecular mechanisms involved in P. aeruginosa chromosomal organization and segregation, we disrupted the ParABS system by deleting either the parA gene or the parB gene. Consistent with previously reported work [24], the ΔparA mutant showed an increase in its doubling time (approximately 100 minutes in minimal medium supplemented with citrate, as opposed to approximately 50 minutes for the wild-type strain, data not shown). In the genetic context we used, the ΔparB mutant showed the same growth defect, in contrast to previous observations reporting a growth rate only 5 to 10% lower than that of the wild type strain [25]. Both mutants produced more than 20% anucleated cells when grown in minimal medium supplemented with citrate (data not shown), suggesting a major defect in chromosomal segregation. To further investigate the impact of these mutations on chromosomal organization, we analyzed the positioning of chromosomal loci located in the Ter and Ori regions during growth in minimal medium supplemented with citrate. The results are shown in Figure 4. Consistent with the large number of anucleated cells, more than half the cells exhibited no visible foci for both the Ter and the Ori loci. Additionally, the proportion of cells containing a given number of foci in the Δpar mutants is strikingly different from the wild type strain (Figure 4A). For instance, only 15% of the cells have one visible Ter focus and two visible Ori foci in these mutants, compared to more than 80% of the wild type cells.
Fig. 4. Extensive chromosomal disorganization in mutants of the ParABS system. 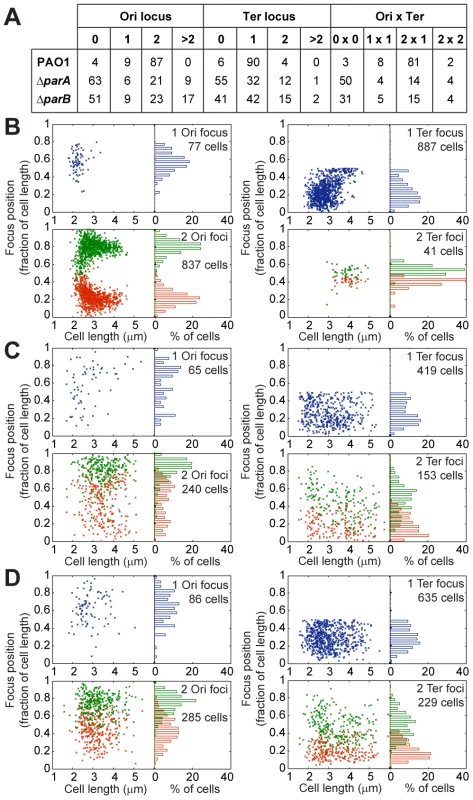
(A) Percentages of the population presenting a given number of foci corresponding to the Ter locus or the Ori locus. Positioning of chromosomal loci located in the Ori region (left panels) and in the Ter region (right panels) in a wild type PAO1 strain (B), the ΔparA mutant (C) and the ΔparB mutant (D) grown in minimal medium supplemented with citrate. The position of the foci in cells containing 1 (upper panels) or 2 (bottom panels) foci are presented. Ori locus: 82-R in (A), (B) and (C). Ter loci: 2,957-R in (A), 3,090-L in (B) and (C). Moreover, the localization of these foci inside the cells also completely changed (Figure 4B–4D). A strong segregation defect is observed for the Ori locus in both the ΔparA and the ΔparB mutants: the two copies of this locus are not segregated to the 0.2/0.8 relative cell length. Furthermore, the typical localization pattern of the Ter locus is completely lost in the ΔparA and ΔparB mutants, which indicates that the whole chromosomal organization is altered when the ParABS system is disrupted.
Discussion
In this study, we showed that the P. aeruginosa chromosome is longitudinally organized between 2 large regions in which loci are localized at a similar position inside the cell upon segregation, one surrounding oriC (approximately 1.4 Mbp) and the other surrounding dif (approximately 800 kb). Segregation occurs sequentially along the Ori/Ter axis and follows replication, which occurs near mid-cell (Figure 5). Segregation and positioning of the P. aeruginosa chromosome rely on a functional ParABS system.
Fig. 5. Proposed model for P. aeruginosa chromosomal organization. 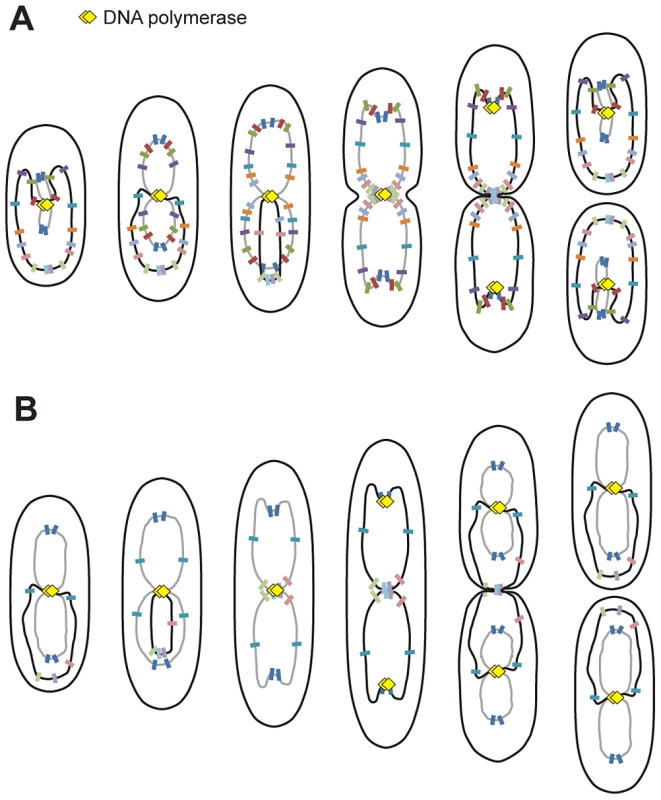
Organization in minimal medium supplemented with citrate (A) or glucose and casamino acids (B). Black lines represent fully replicated chromosomes, whereas grey lines represent partially replicated chromosomes. Colored markers represent chromosomal loci, and yellow diamonds represent replisomes. P. aeruginosa's chromosome is longitudinally organized between 2 distinctive regions
The longitudinal organization of the P. aeruginosa chromosome is similar to chromosomal organization in C. crescentus. However, in C. crescentus, a linear correspondence exists between the position of any given chromosomal locus and its position inside the bacterial cell [28]; however, this is not always the case in P. aeruginosa for loci that belong to the Ori or Ter regions. Indeed, loci belonging to these regions are localized around the same position before and after segregation: loci from the Ori region are near the 0.2/0.8 relative cell length in large cells and loci from the Ter region are below the 0.2 relative cell length in small cells. Large domains have only been described thus far in E. coli [11], [32], which exhibits a transversal organization of its chromosome: oriC and the Ter region are localized near mid-cell, while the replichores are localized in different halves of the cell [27], [33]. P. aeruginosa is thus the first bacterium in which a unique combination of properties could coexist: longitudinal positioning and the presence of large distinctive regions. A systematic study of the positioning of chromosomal loci in B. subtilis or V. cholera has yet to be published.
The 1.4 Mb region surrounding oriC could constitute a specific region
In P. aeruginosa, the three loci closest to oriC on each replichore (encompassing approximately 1.4 Mbp around oriC) appear to be localized at the same position upon segregation (See model, Figure 5). Interestingly, this region includes the eight parS sites located around oriC [34], suggesting a putative role of the ParABS system in organizing this specific region, in addition to its global role in the segregation of the P. aeruginosa chromosome. In C. crescentus, the most polar region of the chromosome contains two parS sites (which are located 8 kb away from oriC in the wild type chromosomal configuration). In P. aeruginosa, the whole Ori region is localized near the 0.2/0.8 relative cell length position. The two loci closest to oriC (82-R and 92-L) are precisely positioned, whereas loci 327-R, 628-R, 488-L and 851-L can be localized slightly more towards the cell pole (Figure 2C). Even if parS sites are dispersed in this Ori region, four of them are located between +4 kb and +15 kb from oriC, which could account for the precise positioning of the loci closest to oriC. Deletion or displacement of parS sites will be necessary to determine their role in the positioning and existence of the Ori region as we defined it.
It is worth noting that in C. crescentus, sporulating B. subtilis and V. cholerae, oriC is more polarly localized than what we observed in P. aeruginosa. Proteins anchoring oriC to the cell pole via the ParABS system have been identified in these bacteria [35]–[38] and are not found in P. aeruginosa. The FimV protein, a homolog of HubP that is responsible for pole anchorage in V. cholerae, lacks the specific domain responsible for interacting with the ParA protein [38]. It is thus very unlikely that it plays a role in positioning the Ori region of P. aeruginosa at the 0.2/0.8 relative cell length position. The mechanisms responsible for this precise localization have yet to be identified.
In B. subtilis, ParB (Spo0J) binds to parS and recruits the SMC complex, which plays a major role in organizing the origin region and promotes chromosomal segregation [21], [22]. Two condensins have been identified in P. aeruginosa, the SMC complex and the MksB complex, which could also be involved in chromosomal segregation [23]. The role of these proteins in organizing P. aeruginosa's chromosome has yet to be carefully investigated, as well as their interplay with the ParABS system. We might imagine that the strong detrimental effect of the deletion of parB is linked to the inability of the condensins to be recruited to the Ori region, as observed in B. subtilis [21], [22]. However, the deletion of parA in P. aeruginosa is also very detrimental to the cell, in contrast to what is observed in B. subtilis.
Considering the major impact on chromosomal organization and segregation of the impairment of the ParABS system in P. aeruginosa, the development of specific tools will be required to more specifically analyze the role of ParA and ParB in organizing the Ori region, as well as their interplay with Smc and MksB during chromosome segregation.
Properties of the Ter region in P. aeruginosa
Another striking observation in P. aeruginosa is that loci located in the approximately 800 kb surrounding the dif site show a characteristic localization during the cell cycle. We arbitrarily define the Ter region as containing these loci, which are positioned below the 0.2 relative cell length in small cells and are sequentially repositioned near mid-cell prior to their duplication. Interestingly, loci in the Ter region are mostly visualized as one single focus, even when they are obviously already duplicated. This distinctive feature is clearly observed when cells are grown in minimal medium supplemented with glucose and casamino acids. In these conditions, large cells possess 2 complete chromosomes that are being replicated (see model, Figure 5), and nevertheless, loci close to dif are frequently visualized as 1 focus. In most cases, the separation of these Ter loci is concomitant with cell division and might require septum formation. Further analyses will be required to elucidate the nature of this Ter region and to identify processes that specifically control the segregation of loci belonging to that region.
A specific domain surrounding the dif site was characterized in E. coli. This domain, called the Ter domain, was first identified using a cellular biological approach [11], [39]. Loci in this domain also present a long colocalization step at mid-cell following replication [14], [32], [39]. A specific factor involved in this colocalization step, called MatP, has been identified [40], [41]. MatP recognizes a specific sequence, matS, that is repeated 23 times in the Ter region of E. coli and interacts with the divisome. No MatP homolog is found in P. aeruginosa, and preliminary bioinformatic analysis did not identify a sequence characteristic of the Ter region of P. aeruginosa. Therefore, processes responsible for specifying the Ter region of the P. aeruginosa chromosome may rely on different molecular mechanisms. It is interesting to note that even if domains have not been characterized in C. crescentus and V. cholerae, one locus located near dif in these bacteria also remains colocalized at mid-cell after replication [42], [43].
Replication occurs at mid-cell in P. aeruginosa
This work also addressed the localization of replisomes near mid-cell in P. aeruginosa. This localization is reminiscent of that described in B. subtilis [29]. In C. crescentus, replisomes are also often colocalized, but they move from one pole to mid-cell during replication [30] following chromosomal organization. In E. coli, the two replication forks appear to follow chromosomal arms as they separate into the two halves of the cell [14], [15]. Based on our analysis of the localization of replisome proteins, it appears that replication lasts for most of the cell cycle when P. aeruginosa is grown in minimal medium supplemented with citrate (see model, Figure 5). In newborn cells, loci closest to oriC are being replicated and are found near mid-cell together with DNA polymerase. As cells grow, replisomes stay near mid-cell, where loci are successively relocated prior to their replication. This relocation may result from the replication process, which could be responsible for pulling DNA towards mid-cell.
Overall, this study allowed us to give a precise description of chromosomal organization in P. aeruginosa. This organization is original, combining large distinctive regions and a longitudinal positioning. These results pave the way for additional work that will lead to the understanding of the mechanisms responsible for such organization, which will contribute to our appreciation of the wide diversity of mechanisms used by bacteria, even when the most fundamental processes are concerned. This work might also allow us to identify drugs that would interfere with fundamental processes such as chromosomal replication and segregation in this important pathogen.
Materials and Methods
Plasmids and strains
P. aeruginosa strain PAO1 was provided by Arne Rietsch (Case Western Reserve University). This PAO1 strain does not present the inversion described for the sequenced PAO1-UW subclone resulting from homologous recombination between the rrnA and rrnB loci, which are orientated in opposite directions and separated by 2.2 Mbp [4]. This explains the discrepancies between PA numbers and positions on the chromosomal map, as gene annotation was done in the inversion containing strain. Chromosomal loci were called according to their position from oriC on each replichore. Details of plasmid and strain construction are provided in Text S1, Table S1 and Table S2.
Fluorescence microscopy analysis
Strains were grown overnight in LB, diluted 300 times in minimal medium A supplemented with either 0.25% citrate or 0.12% casamino acids and 0.5% glucose (when looking at chromosomal tags, 0.5 mM IPTG was added to growth medium) until they reach an OD600 comprised between 0.05 and 0.1. Cells were then spread on agarose pads and immediately observed using a Leica DM6000 microscope, a coolsnap HQ CCD camera (Roper) and Metamorph software. Image analysis was performed using the MATLAB-based software MicrobeTracker Suite [44].
Supporting Information
Zdroje
1. SilbyMW, WinstanleyC, GodfreySA, LevySB, JacksonRW (2011) Pseudomonas genomes: diverse and adaptable. FEMS Microbiol Rev 35 : 652–680.
2. GovanJR, DereticV (1996) Microbial pathogenesis in cystic fibrosis: mucoid Pseudomonas aeruginosa and Burkholderia cepacia. Microbiol Rev 60 : 539–574.
3. ListerPD, WolterDJ, HansonND (2009) Antibacterial-resistant Pseudomonas aeruginosa: clinical impact and complex regulation of chromosomally encoded resistance mechanisms. Clin Microbiol Rev 22 : 582–610.
4. StoverCK, PhamXQ, ErwinAL, MizoguchiSD, WarrenerP, et al. (2000) Complete genome sequence of Pseudomonas aeruginosa PAO1, an opportunistic pathogen. Nature 406 : 959–964.
5. YeeTW, SmithDW (1990) Pseudomonas chromosomal replication origins: a bacterial class distinct from Escherichia coli-type origins. Proc Natl Acad Sci U S A 87 : 1278–1282.
6. JiangY, YaoS, HelinskiD, ToukdarianA (2006) Functional analysis of two putative chromosomal replication origins from Pseudomonas aeruginosa. Plasmid 55 : 194–200.
7. de VriesR (2010) DNA condensation in bacteria: Interplay between macromolecular crowding and nucleoid proteins. Biochimie 92 : 1715–1721.
8. ThanbichlerM, ShapiroL (2006) Chromosome organization and segregation in bacteria. J Struct Biol 156 : 292–303.
9. ToroE, ShapiroL (2010) Bacterial chromosome organization and segregation. Cold Spring Harb Perspect Biol 2: a000349.
10. PossozC, JunierI, EspeliO (2012) Bacterial chromosome segregation. Front Biosci 17 : 1020–1034.
11. NikiH, YamaichiY, HiragaS (2000) Dynamic organization of chromosomal DNA in Escherichia coli. Genes Dev 14 : 212–223.
12. ValensM, PenaudS, RossignolM, CornetF, BoccardF (2004) Macrodomain organization of the Escherichia coli chromosome. EMBO J 23 : 4330–4341.
13. LemonKP, GrossmanAD (2001) The extrusion-capture model for chromosome partitioning in bacteria. Genes Dev 15 : 2031–2041.
14. BatesD, KlecknerN (2005) Chromosome and replisome dynamics in E. coli: loss of sister cohesion triggers global chromosome movement and mediates chromosome segregation. Cell 121 : 899–911.
15. Reyes-LamotheR, PossozC, DanilovaO, SherrattDJ (2008) Independent positioning and action of Escherichia coli replisomes in live cells. Cell 133 : 90–102.
16. JunS, WrightA (2010) Entropy as the driver of chromosome segregation. Nat Rev Microbiol 8 : 600–607.
17. FogelMA, WaldorMK (2006) A dynamic, mitotic-like mechanism for bacterial chromosome segregation. Genes Dev 20 : 3269–3282.
18. ToroE, HongSH, McAdamsHH, ShapiroL (2008) Caulobacter requires a dedicated mechanism to initiate chromosome segregation. Proc Natl Acad Sci U S A 105 : 15435–15440.
19. LeePS, LinDC, MoriyaS, GrossmanAD (2003) Effects of the chromosome partitioning protein Spo0J (ParB) on oriC positioning and replication initiation in Bacillus subtilis. J Bacteriol 185 : 1326–1337.
20. MinnenA, AttaiechL, ThonM, GruberS, VeeningJW (2011) SMC is recruited to oriC by ParB and promotes chromosome segregation in Streptococcus pneumoniae. Mol Microbiol 81 : 676–688.
21. SullivanNL, MarquisKA, RudnerDZ (2009) Recruitment of SMC by ParB-parS organizes the origin region and promotes efficient chromosome segregation. Cell 137 : 697–707.
22. GruberS, ErringtonJ (2009) Recruitment of condensin to replication origin regions by ParB/SpoOJ promotes chromosome segregation in B. subtilis. Cell 137 : 685–696.
23. PetrushenkoZM, SheW, RybenkovVV (2011) A new family of bacterial condensins. Mol Microbiol 81 : 881–896.
24. LasockiK, BartosikAA, MierzejewskaJ, ThomasCM, Jagura-BurdzyG (2007) Deletion of the parA (soj) homologue in Pseudomonas aeruginosa causes ParB instability and affects growth rate, chromosome segregation, and motility. J Bacteriol 189 : 5762–5772.
25. BartosikAA, MierzejewskaJ, ThomasCM, Jagura-BurdzyG (2009) ParB deficiency in Pseudomonas aeruginosa destabilizes the partner protein ParA and affects a variety of physiological parameters. Microbiology 155 : 1080–1092.
26. LauIF, FilipeSR, SoballeB, OkstadOA, BarreFX, et al. (2003) Spatial and temporal organization of replicating Escherichia coli chromosomes. Mol Microbiol 49 : 731–743.
27. NielsenHJ, OttesenJR, YoungrenB, AustinSJ, HansenFG (2006) The Escherichia coli chromosome is organized with the left and right chromosome arms in separate cell halves. Mol Microbiol 62 : 331–338.
28. ViollierPH, ThanbichlerM, McGrathPT, WestL, MeewanM, et al. (2004) Rapid and sequential movement of individual chromosomal loci to specific subcellular locations during bacterial DNA replication. Proc Natl Acad Sci U S A 101 : 9257–9262.
29. LemonKP, GrossmanAD (1998) Localization of bacterial DNA polymerase: evidence for a factory model of replication. Science 282 : 1516–1519.
30. JensenRB, WangSC, ShapiroL (2001) A moving DNA replication factory in Caulobacter crescentus. EMBO J 20 : 4952–4963.
31. LandgrafD, OkumusB, ChienP, BakerTA, PaulssonJ (2012) Segregation of molecules at cell division reveals native protein localization. Nat Methods 9 : 480–482.
32. EspeliO, MercierR, BoccardF (2008) DNA dynamics vary according to macrodomain topography in the E. coli chromosome. Mol Microbiol 68 : 1418–1427.
33. WangX, PossozC, SherrattDJ (2005) Dancing around the divisome: asymmetric chromosome segregation in Escherichia coli. Genes Dev 19 : 2367–2377.
34. BartosikAA, LasockiK, MierzejewskaJ, ThomasCM, Jagura-BurdzyG (2004) ParB of Pseudomonas aeruginosa: interactions with its partner ParA and its target parS and specific effects on bacterial growth. J Bacteriol 186 : 6983–6998.
35. Ben-YehudaS, RudnerDZ, LosickR (2003) RacA, a bacterial protein that anchors chromosomes to the cell poles. Science 299 : 532–536.
36. BowmanGR, ComolliLR, ZhuJ, EckartM, KoenigM, et al. (2008) A polymeric protein anchors the chromosomal origin/ParB complex at a bacterial cell pole. Cell 134 : 945–955.
37. EbersbachG, BriegelA, JensenGJ, Jacobs-WagnerC (2008) A self-associating protein critical for chromosome attachment, division, and polar organization in caulobacter. Cell 134 : 956–968.
38. YamaichiY, BrucknerR, RinggaardS, MollA, CameronDE, et al. (2012) A multidomain hub anchors the chromosome segregation and chemotactic machinery to the bacterial pole. Genes Dev 26 : 2348–2360.
39. LiY, YoungrenB, SergueevK, AustinS (2003) Segregation of the Escherichia coli chromosome terminus. Mol Microbiol 50 : 825–834.
40. MercierR, PetitMA, SchbathS, RobinS, El KarouiM, et al. (2008) The MatP/matS site-specific system organizes the terminus region of the E. coli chromosome into a macrodomain. Cell 135 : 475–485.
41. EspeliO, BorneR, DupaigneP, ThielA, GigantE, et al. (2012) A MatP-divisome interaction coordinates chromosome segregation with cell division in E. coli. EMBO J 31 : 3198–3211.
42. JensenRB (2006) Coordination between chromosome replication, segregation, and cell division in Caulobacter crescentus. J Bacteriol 188 : 2244–2253.
43. SrivastavaP, FeketeRA, ChattorajDK (2006) Segregation of the replication terminus of the two Vibrio cholerae chromosomes. J Bacteriol 188 : 1060–1070.
44. SliusarenkoO, HeinritzJ, EmonetT, Jacobs-WagnerC (2011) High-throughput, subpixel precision analysis of bacterial morphogenesis and intracellular spatio-temporal dynamics. Mol Microbiol 80 : 612–627.
Štítky
Genetika Reprodukční medicína
Článek Attachment Site Selection and Identity in Bxb1 Serine Integrase-Mediated Site-Specific RecombinationČlánek Bck2 Acts through the MADS Box Protein Mcm1 to Activate Cell-Cycle-Regulated Genes in Budding YeastČlánek High-Resolution Transcriptome Maps Reveal Strain-Specific Regulatory Features of Multiple IsolatesČlánek Neuropeptides Function in a Homeostatic Manner to Modulate Excitation-Inhibition Imbalance inČlánek Implicates Tyrosine-Sulfated Peptide Signaling in Susceptibility and Resistance to Root Infection
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2013 Číslo 5- Akutní intermitentní porfyrie
- Souvislost haplotypu M2 genu pro annexin A5 s opakovanými reprodukčními ztrátami
- Růst a vývoj dětí narozených pomocí IVF
- Srdeční frekvence embrya může být faktorem užitečným v předpovídání výsledku IVF
- Vztah užívání alkoholu a mužské fertility
-
Všechny články tohoto čísla
- Functional Elements Are Embedded in Structurally Constrained Sequences
- RNA–Mediated Epigenetic Heredity Requires the Cytosine Methyltransferase Dnmt2
- Loss of Expression and Promoter Methylation of SLIT2 Are Associated with Sessile Serrated Adenoma Formation
- Attachment Site Selection and Identity in Bxb1 Serine Integrase-Mediated Site-Specific Recombination
- Human Genetics in Rheumatoid Arthritis Guides a High-Throughput Drug Screen of the CD40 Signaling Pathway
- Genome-Wide Analysis in German Shepherd Dogs Reveals Association of a Locus on CFA 27 with Atopic Dermatitis
- Liver X Receptors Protect from Development of Prostatic Intra-Epithelial Neoplasia in Mice
- Chromosomal Organization and Segregation in
- A Statistical Framework for Joint eQTL Analysis in Multiple Tissues
- Cell Polarity and Patterning by PIN Trafficking through Early Endosomal Compartments in
- Bck2 Acts through the MADS Box Protein Mcm1 to Activate Cell-Cycle-Regulated Genes in Budding Yeast
- High-Resolution Transcriptome Maps Reveal Strain-Specific Regulatory Features of Multiple Isolates
- Neuropeptides Function in a Homeostatic Manner to Modulate Excitation-Inhibition Imbalance in
- A Compendium of Nucleosome and Transcript Profiles Reveals Determinants of Chromatin Architecture and Transcription
- Wnt Signaling Regulates the Lineage Differentiation Potential of Mouse Embryonic Stem Cells through Tcf3 Down-Regulation
- Filamin and Phospholipase C-ε Are Required for Calcium Signaling in the Spermatheca
- The Specificity and Flexibility of L1 Reverse Transcription Priming at Imperfect T-Tracts
- Imputation-Based Meta-Analysis of Severe Malaria in Three African Populations
- Implicates Tyrosine-Sulfated Peptide Signaling in Susceptibility and Resistance to Root Infection
- Clathrin and AP2 Are Required for Phagocytic Receptor-Mediated Apoptotic Cell Clearance in
- Encodes CDF Transporters That Excrete Zinc from Intestinal Cells of and Act in a Parallel Negative Feedback Circuit That Promotes Homeostasis
- Global Properties and Functional Complexity of Human Gene Regulatory Variation
- DNA Binding of the Cell Cycle Transcriptional Regulator GcrA Depends on N6-Adenosine Methylation in and Other
- Side Effects: Substantial Non-Neutral Evolution Flanking Regulatory Sites
- From Paramutation to Paradigm
- From Mouse to Human: Evolutionary Genomics Analysis of Human Orthologs of Essential Genes
- Distinct Translational Control in CD4 T Cell Subsets
- Female Bias in and Regulation by the Histone Demethylase KDM6A
- ATM–Dependent MiR-335 Targets CtIP and Modulates the DNA Damage Response
- HDAC7 Is a Repressor of Myeloid Genes Whose Downregulation Is Required for Transdifferentiation of Pre-B Cells into Macrophages
- The Majority of Primate-Specific Regulatory Sequences Are Derived from Transposable Elements
- Identification of Meiotic Cyclins Reveals Functional Diversification among Plant Cyclin Genes
- EGL-13/SoxD Specifies Distinct O and CO Sensory Neuron Fates in
- Congruence of Additive and Non-Additive Effects on Gene Expression Estimated from Pedigree and SNP Data
- Using Extended Genealogy to Estimate Components of Heritability for 23 Quantitative and Dichotomous Traits
- Ikbkap/Elp1 Deficiency Causes Male Infertility by Disrupting Meiotic Progression
- Analysis of the Genetic Basis of Disease in the Context of Worldwide Human Relationships and Migration
- Duplication and Retention Biases of Essential and Non-Essential Genes Revealed by Systematic Knockdown Analyses
- Strong Purifying Selection at Synonymous Sites in
- , a Susceptibility Gene for Type 1 and Type 2 Diabetes, Modulates Pancreatic Beta Cell Apoptosis via Regulation of a Splice Variant of the BH3-Only Protein
- Chromosome Movements Promoted by the Mitochondrial Protein SPD-3 Are Required for Homology Search during Meiosis
- The Secretory Pathway Calcium ATPase PMR-1/SPCA1 Has Essential Roles in Cell Migration during Embryonic Development
- The Genomic Signature of Crop-Wild Introgression in Maize
- CDK4 T172 Phosphorylation Is Central in a CDK7-Dependent Bidirectional CDK4/CDK2 Interplay Mediated by p21 Phosphorylation at the Restriction Point
- Genome-Wide Identification of Regulatory RNAs in the Human Pathogen
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Using Extended Genealogy to Estimate Components of Heritability for 23 Quantitative and Dichotomous Traits
- HDAC7 Is a Repressor of Myeloid Genes Whose Downregulation Is Required for Transdifferentiation of Pre-B Cells into Macrophages
- Female Bias in and Regulation by the Histone Demethylase KDM6A
- ATM–Dependent MiR-335 Targets CtIP and Modulates the DNA Damage Response
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



