-
Články
- Vzdělávání
- Časopisy
Top články
- Témata
- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Crystal Structure of PrgI-SipD: Insight into a Secretion Competent State of the Type Three Secretion System Needle Tip and its Interaction with Host Ligands
Many infectious Gram-negative bacteria, including Salmonella typhimurium, require a Type Three Secretion System (T3SS) to translocate virulence factors into host cells. The T3SS consists of a membrane protein complex and an extracellular needle together that form a continuous channel. Regulated secretion of virulence factors requires the presence of SipD at the T3SS needle tip in S. typhimurium. Here we report three-dimensional structures of individual SipD, SipD in fusion with the needle subunit PrgI, and of SipD:PrgI in complex with the bile salt, deoxycholate. Assembly of the complex involves major conformational changes in both SipD and PrgI. This rearrangement is mediated via a π bulge in the central SipD helix and is stabilized by conserved amino acids that may allow for specificity in the assembly and composition of the tip proteins. Five copies each of the needle subunit PrgI and SipD form the T3SS needle tip complex. Using surface plasmon resonance spectroscopy and crystal structure analysis we found that the T3SS needle tip complex binds deoxycholate with micromolar affinity via a cleft formed at the SipD:PrgI interface. In the structure-based three-dimensional model of the T3SS needle tip, the bound deoxycholate faces the host membrane. Recently, binding of SipD with bile salts present in the gut was shown to impede bacterial infection. Binding of bile salts to the SipD:PrgI interface in this particular arrangement may thus inhibit the T3SS function. The structures presented in this study provide insight into the open state of the T3SS needle tip. Our findings present the atomic details of the T3SS arrangement occurring at the pathogen-host interface.
Published in the journal: . PLoS Pathog 7(8): e32767. doi:10.1371/journal.ppat.1002163
Category: Research Article
doi: https://doi.org/10.1371/journal.ppat.1002163Summary
Many infectious Gram-negative bacteria, including Salmonella typhimurium, require a Type Three Secretion System (T3SS) to translocate virulence factors into host cells. The T3SS consists of a membrane protein complex and an extracellular needle together that form a continuous channel. Regulated secretion of virulence factors requires the presence of SipD at the T3SS needle tip in S. typhimurium. Here we report three-dimensional structures of individual SipD, SipD in fusion with the needle subunit PrgI, and of SipD:PrgI in complex with the bile salt, deoxycholate. Assembly of the complex involves major conformational changes in both SipD and PrgI. This rearrangement is mediated via a π bulge in the central SipD helix and is stabilized by conserved amino acids that may allow for specificity in the assembly and composition of the tip proteins. Five copies each of the needle subunit PrgI and SipD form the T3SS needle tip complex. Using surface plasmon resonance spectroscopy and crystal structure analysis we found that the T3SS needle tip complex binds deoxycholate with micromolar affinity via a cleft formed at the SipD:PrgI interface. In the structure-based three-dimensional model of the T3SS needle tip, the bound deoxycholate faces the host membrane. Recently, binding of SipD with bile salts present in the gut was shown to impede bacterial infection. Binding of bile salts to the SipD:PrgI interface in this particular arrangement may thus inhibit the T3SS function. The structures presented in this study provide insight into the open state of the T3SS needle tip. Our findings present the atomic details of the T3SS arrangement occurring at the pathogen-host interface.
Introduction
Bacterial infections including Salmonellosis and Shigellosis affect millions of people every year. These bacteria use a T3SS to secrete virulence factors to manipulate host cells. The T3SS is a multi-component system that forms a continuous protein transport channel through the two bacterial membranes and the periplasmatic space that extends into the surrounding medium by a needle structure [1]–[3]. Spatiotemporal control of secretion is essential for effective host invasion [4].
Tip proteins, which bind to the distal end of the T3SS needle, are thought to play an important role in this process [4]–[6]. SipD from S. typhimurium, IpaD from Shigella flexneri and BipD from Burkholderia mallei are tip proteins that are thought to interact with their corresponding needle subunits PrgI, MxiH and BsaL, respectively, to make the needle tip complex [5]. Although the mechanism is unclear, tip proteins were shown to influence secretion and invasion of bacteria [7]–[9].
Sterols like cholesterol or cholic acid derivatives found in the bile are amphipathic compounds that play important roles in cellular communication and metabolic processes. Bile salts influence the T3SS of intestinal bacteria. For instance, the presence of deoxycholate either impedes (S. typhimurium) or facilitates (S. flexneri) host invasion[10]–[13]. Noteworthy, it was recently shown that SipD and IpaD bind deoxycholate and some of its derivatives [14], [15].
To understand how T3SS are regulated it is required to analyze the structure and mechanism of proteins gating the transport channel. Here, we address the questions of how the Salmonella SipD interacts with PrgI and deoxycholate and the mechanistic consequences of the assembly of the T3SS needle tip complex.
Results
Crystal structure of the Salmonella tip protein SipD
We solved the X-ray crystal structure of SipD, the needle tip protein of S. typhimurium at 3.0 Å resolution (Figure 1A and Table 1). The crystal contained 4 copies of SipD in the asymmetric unit with structural information for 306 of 343 amino acids. In SipD crystals, as in the crystal structures of IpaD [16] and BipD [17], the N-terminal 31, 38 or 29 amino acids, respectively, were not defined. Here we report structural features of SipD chain A, which, compared with chains B to D, showed continuous electron density for most amino acids. SipD is predominantly α-helical folded and can be divided in three structurally different domains (Figure 1B). The central domain, domain 2, (green in Figure 1) adopted a coiled coil structure with two helices of 47 and 52 amino acids length, respectively. The bending of the coiled coil of domain 2 allowed us to distinguish between a concave and a convex surface. Domain 2 is joined to domain 3, an α/β-structure (yellow in Figure 1). Domain 3 is in contact with the convex surface provided by the coiled coil of the central domain (Figure 1A). Domain 2 and 3 of SipD share high sequence homology and similar three dimensional structure to the orthologs from S. flexneri (Figure S1, r.m.s. deviation 1.3 Å with IpaD) [16] or B. mallei (Figure S1, r.m.s. deviation 2.2 Å with BipD) [17]. Notably, amino acids involved in intramolecular contacts are identical or highly conserved in all three orthologs, suggesting functional relevance in T3SS tip proteins.
Fig. 1. Crystal structure of SipD. 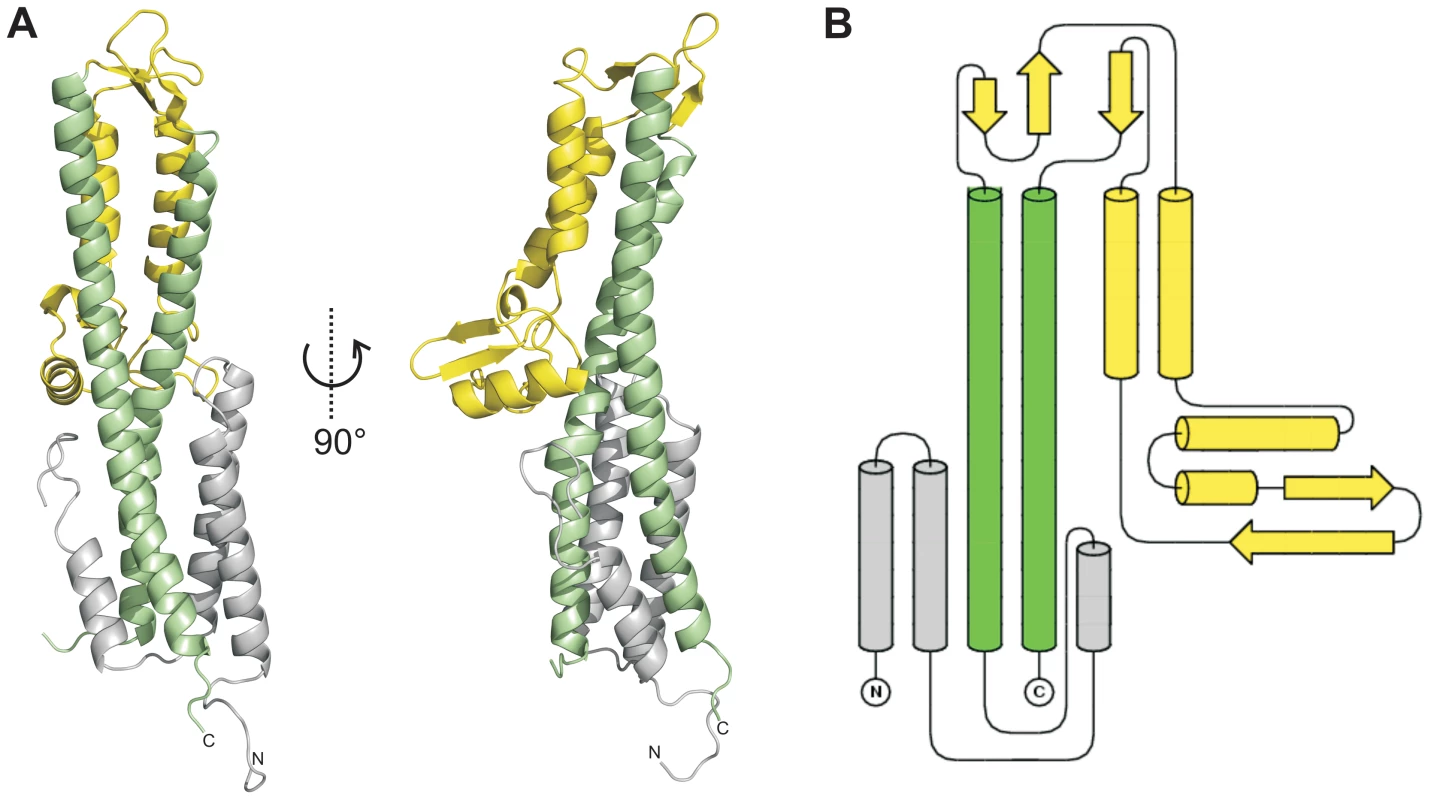
(A) X-ray crystal structure and (B) topology plot of SipD chain A from S. typhimurium: structurally distinguishable domains are coloured in grey (domain 1), green (domain 2), and yellow (domain 3), respectively. Tab. 1. Data collection and refinement statistics. 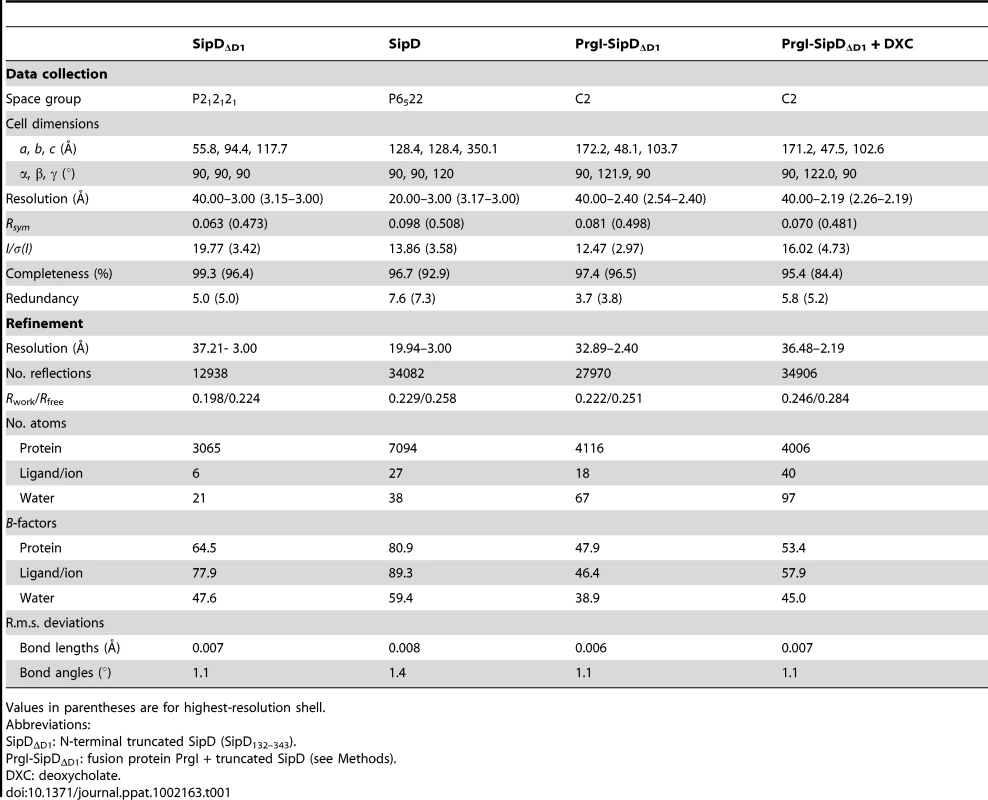
Values in parentheses are for highest-resolution shell. In contrast to domain 2 and 3, the amino acid sequence of domain 1 in SipD (grey in Figure 1) show almost no sequence conservation with IpaD or BipD. However, α-helices of domain 1 adopted a similar structure in both SipD and IpaD (Figure S2), encompassing the central coiled coil of domain 2 (Figure 1A).
PrgI displaces the N-terminal domain of SipD
In the S. flexneri cytosol, IpaD domain 1 was suggested to function as a self-chaperone avoiding either spontaneous self oligomerization or its interaction with the needle forming protein MxiH before secretion [16]. We tested the oligomerization state of purified SipD by static light scattering and found that SipD is a monomer in solution (Mw ∼37 kDa, Figure S3). Deletion of domain 1 (SipDΔD1) resulted in a mixture of SipD dimers and trimers (Figure S4). Though oligomerization of the needle tip protein changed depending on the presence of domain 1, deletion of this domain did not favour spontaneous protein polymerization as was found for PrgI and its orthologues [18]. SipD did not bind to PrgI*, a soluble and functional PrgI mutant [18], as tested by isothermic titration calorimetry (ITC, Figure 2A). In contrast, SipDΔD1 bound with a Kd of 88±3 µM to PrgI* (Figure 2B). Deletion of domain 1 did not destabilize SipD, as demonstrated by the comparative analysis of X-ray crystal structures of SipDΔD1 (Figure 2C and Table 1) and SipD (Figure 1A). Superposition of SipDΔD1 and SipD showed almost identical 3-dimensional structure reflected in an r.m.s. deviation of 0.9 Å for amino acids 149 to 328. ITC results showed that domains 2 and 3 of SipD mediate binding to PrgI*, while domain 1 impedes the interaction. In fact, prior to binding to PrgI*, the self-chaperoning domain 1 of SipD may unfold independently from the rest of the molecule as recently suggested [19], [20].
Fig. 2. Deletion of SipD domain 1 permits binding to PrgI*. 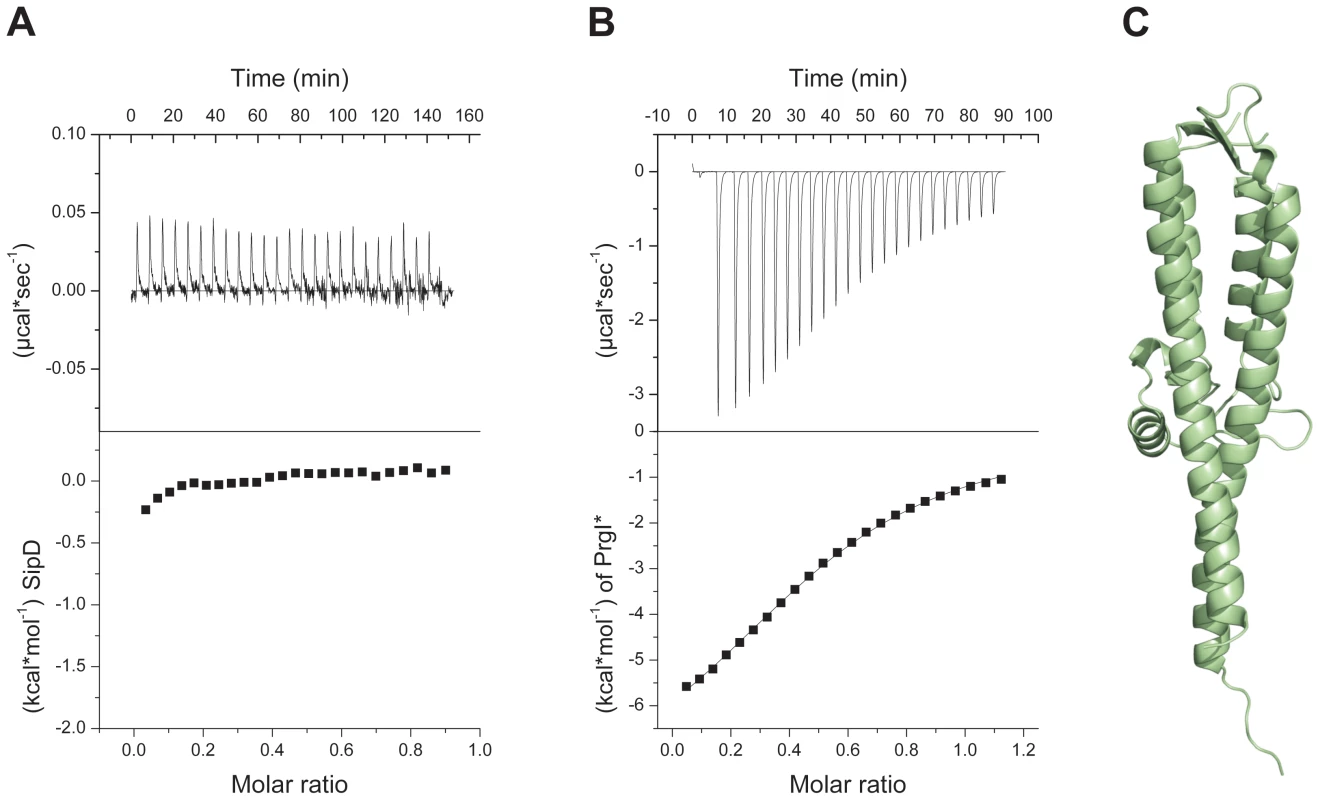
(A) Binding between S. typhimurium needle protein PrgI* and SipD was not detectable using isothermal titration calorimetry (ITC). Heat flow for each SipD injection as a function of time (upper) and integration of the thermogram (lower). (B) ITC of purified proteins PrgI* with SipDΔD1 indicated a binding affinity of 88±3 µMol. Heat flow for each PrgI* injection as a function of time (upper) and integration of the thermogram with the fitting curve (lower). (C) X-ray crystal structure of SipDΔD1. Structure of the PrgI-SipDΔD1 fusion protein
In order to decipher the 3-dimensional structure of the entire T3SS needle tip, we generated a fusion protein of N-terminal truncated SipD with PrgI (PrgI-SipDΔD1). Crystals of PrgI-SipDΔD1 contained two similar copies (chain A and chain B) in the asymmetric unit. We describe here the structure of chain A, which is more complete than chain B. The X-ray crystal structure of PrgI-SipDΔD1 solved at 2.4 Å resolution showed noteworthy features (Figure 3A and Table 1). Comparing structures of individual SipD (brown) and PrgI* molecules (grey) with the PrgI-SipDΔD1 fusion protein (SipDΔD1 in green and PrgI in blue) revealed conformational changes in both proteins (Figure 3B and Figure S5). Two of the five helices providing contact between SipD and PrgI changed conformation during complex formation. The central helix of domain 2 in SipD is kinked at Asn141 in the complex (Figure 3A–B). We observed also a kink in the C-terminal helix of PrgI at Asn63 (Figure 3A and Figure S5). Both kinked helices, together with two additional helices from SipD and PrgI, adopted a new four helix bundle in the complex.
Fig. 3. Conformational changes during interaction of SipD and PrgI. 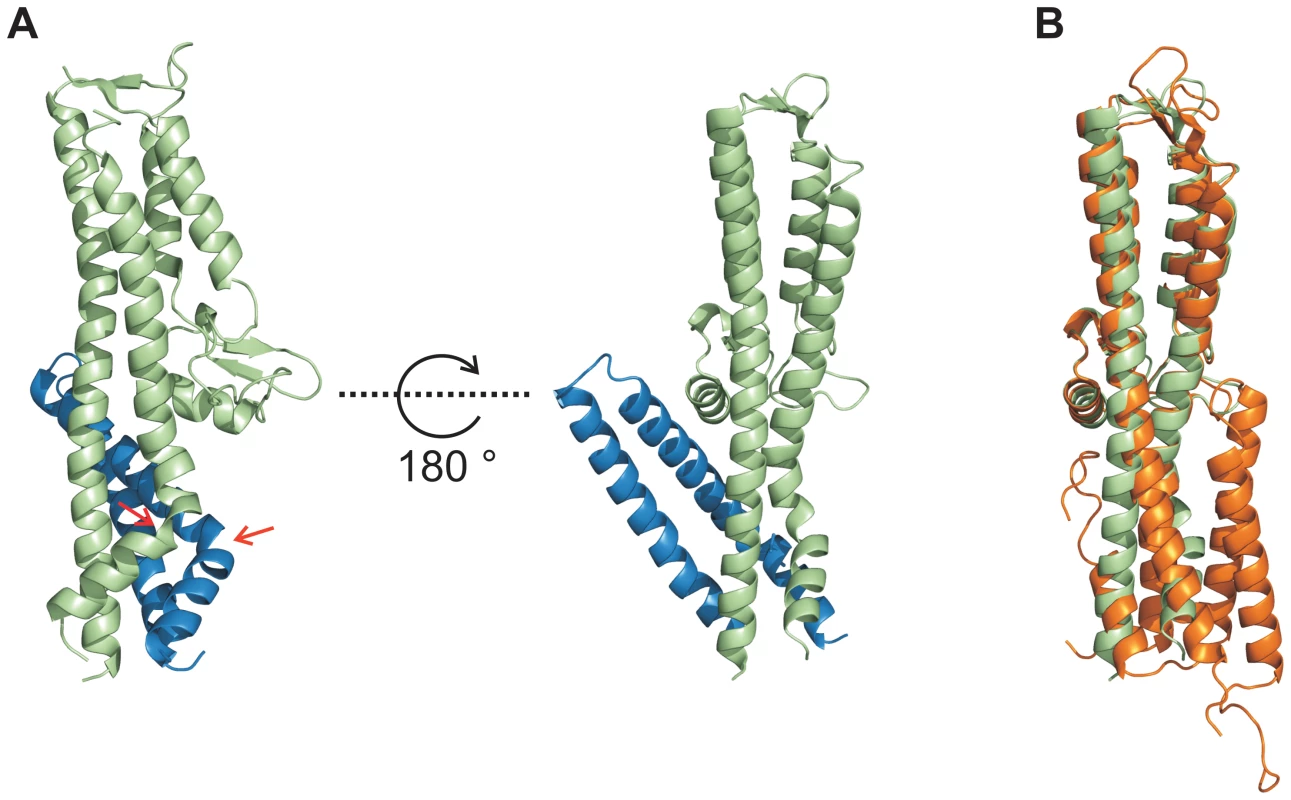
(A) Structure of the PrgI-SipDΔD1 fusion protein (SipD green and PrgI blue) from two orthogonal perspectives. The helix kink in SipD and PrgI is indicated by red arrows. (B) Superposition of complexed SipD (green) with monomeric SipD (orange) indicated conformational changes in the central coiled coil. Recent studies [9] suggest that the needle proteins could replace the two helices of domain 1 in the corresponding T3SS tip protein that are in contact with the concave side of the central coiled coil (Figure 1A). This hypothesis is in agreement with our ITC data showing that PrgI* may replace domain 1 during SipD binding (Figure 2A–B). However, the crystal structure of the complex presented here revealed PrgI binding to the convex surface of the central coiled coil in SipD (Figure 3A). Thus, in contrast to the proposed model, a single PrgI molecule may replace the helix-loop motif of SipD immediately upstream of domain 2 (Figure 1B), instead of the two helices at the N-terminus of SipD. Moreover, both proteins in the complex comprised an angle of about 45° (Figure 3A and Figure S6) due to the contact between SipD domain 3 and PrgI.
We tested the oligomerization state of the PrgI-SipDΔD1 fusion protein using static light scattering technique. The needle tip complex was monomeric in solution (Figure S7), suggesting that the supramolecular architecture of the tip complex is influenced by the PrgI assembly of the T3SS needle. This hypothesis is in agreement with our observation that deletion of domain 1 of SipD did not support polymerization of the needle tip protein (Figure S4) but rather allowed interaction with the needle protein PrgI (Figure 2).
Stabilization of the PrgI-SipDΔD1 fusion protein
The X-ray crystal structure of the PrgI-SipDΔD1 complex compared with the structure of SipD alone and with previous structural studies of needle tip proteins [18], [21]–[23] showed that both PrgI and SipD changed conformation during complex assembly. As mentioned above, the two helices which showed novel kinked conformation are part of a four helix bundle, thus providing close contact between SipD and PrgI (Figure 3A and Figure 4A). The four helix bundle stabilized by hydrophobic (Figure S8) and polar interactions encompassed a buried surface of 1113 Å2 per protein. An extended hydrogen bonding network connecting conserved amino acids of both proteins stabilizes the twisted helical arrangement found in the PrgI-SipDΔD1 fusion protein (Figure 4A). In total six hydrogen bonds and salt bridges between the C-terminal helix of PrgI and the long helices of domain 2 or the central helix of SipD domain 3 stabilized the tertiary structure of the complex (Figure 4B). A hydrogen bond between Ser333 and Asp11 of SipD and PrgI, respectively, contributed additional stabilization of the complex structure. Spin labelled amino acids Asp136, Ala144, Asp147, Leu318, Lys324, Ser328, Ser331 and Glu335 of SipD are influenced by PrgI as recently shown [24], consistently with the PrgI-SipDΔD1 crystal structure presented here.
Fig. 4. Interaction of SipD and PrgI in the T3SS needle tip complex. 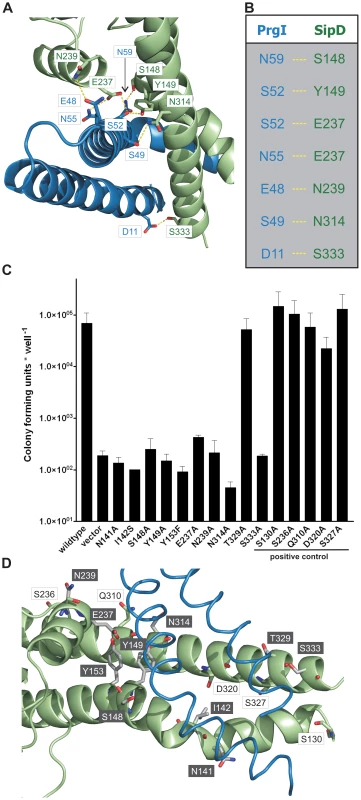
(A) Four helix bundle of the tip complex formed by two helices of SipD domain 2 (green) and the helix termini of PrgI (blue). Side chains that provided hydrogen bonding network (yellow) between PrgI-SipDΔD1 in the fusion protein are highlighted. (B) List of the amino acids that stabilized the PrgI-SipDΔD1 fusion protein by forming hydrogen bonds. (C) Host invasion assay of SipD mutant complemented S. typhimurium knockout cells. (D) Close up of the PrgI-SipDΔD1 interface: mutated SipD amino acids interacting with PrgI are highlighted in grey as stick model and corresponding labels marked with grey boxes. We tested whether the interactions between SipD and PrgI are necessary for the T3SS function in HeLa cell invasion assays. SipD knockout cells, which are not invasive, were complemented with plasmids harbouring wildtype or mutant sipD (Figure 4C). We designed sipD mutants based on the structure of the PrgI-SipDΔD1 fusion protein and on conservation of SipD amino acids (Figure S1). Including control mutants that did not hamper stabilization of the tip complex, we tested 15 SipD point mutations (Figure 4C). Except for Tyr153 and Glu237, which formed intramolecular hydrogen bonds, tested SipD mutants should not destabilize folding of the apo-protein. Most interactions between SipD and PrgI were essential for a functional T3SS as demonstrated by a dramatically reduced invasiveness (Figure 4C). In contrast, amino acids in the same region of SipD not interacting with PrgI had little or no effect (S130A, S236A, Q310A, D320A, S327A) on host cell invasion (Figure 4C–D). Circular dichroism spectroscopy of purified mutants and wildtype SipD indicated same folding, albeit the weaker spectrum of I142S indicates less α-helical content and suggests lower stability at 37°C (Figure S9). Similarly prgI mutant complemented PrgI knockout cells showed reduced invasiveness (Figure S10). Our results showed that mutations in the C-terminus of needle proteins can impede host invasion and are in agreement with previous studies in S. typhimurium and other T3SS dependent bacteria [21]–[23], [25].
π–bulge in SipD provides conformational flexibility
We showed that polar interactions at the PrgI-SipDΔD1 interface provide tight and specific contacts between the needle tip complex components. In close proximity of these contacts two helices adopted kinked conformation in the complex, but not in the individual proteins (Figure 3). By comparing the structures of PrgI-SipDΔD1 fusion protein and SipD, we can propose how assembly of the T3SS needle tip is established.
In chain A of both structures, one helix of the coiled coil (domain 2 of SipD) showed partial unwinding. This helix anomaly, usually energetically disfavoured, is stabilized in the fusion protein by hydrogen bonds formed between carbonyl oxygen of amino acid Ser148, a water molecule, and Trp234 of SipD (Figure 5A). The interaction of the Ser148 backbone carbonyl group with Trp234 shifted the typical backbone hydrogen bonding pattern stabilizing architecture of an α–helix (i→i+4) by one position (i(Ala144)→i+5(Tyr149)) (Figure 5B). Local deviation from the backbone hydrogen bonding pattern of an α–helix described as π-bulge plays an important role in many different proteins that require conformational flexibility for function [26]. In our structure of the PrgI-SipDΔD1 fusion protein the π-bulge caused local unwinding and weakening of the α–helix around Ser148. Furthermore, mutation of either of the two amino acids (Ser148 or Trp234) involved in formation of the π-bulge led to reduced or even loss of bacterial invasion in HeLa cells (Figure 4C). These results support the relevance of the π-bulge for the functionality of the T3SS needle tip complex.
Fig. 5. π–bulge in SipD domain 2 permitted the assembly of the PrgI-SipDΔD1 complex. 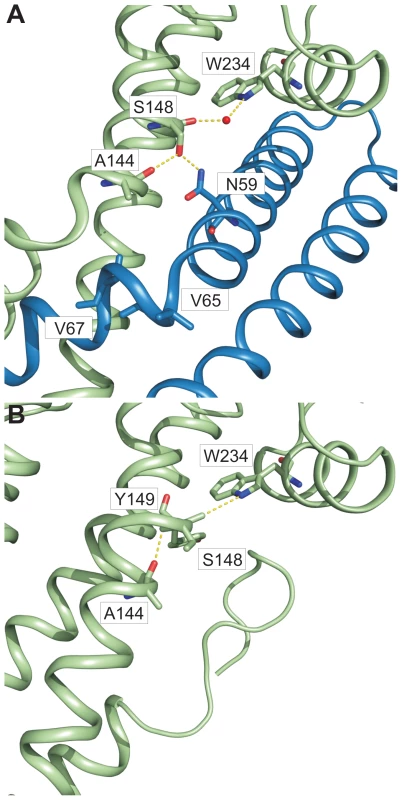
Amino acids (stick model) and interactions (yellow) that stabilize the π–bulge and helix kinks in the PrgI-SipDΔD1 fusion protein (A) and SipD (B) are highlighted. In the PrgI-SipDΔD1 fusion protein a water molecule bridges the backbone carbonyl oxygen of Ser148 and Trp234 (Figure 5A). The backbone carbonyl of Ser148 pointing towards the imino group of Trp234 suggested similar hydrogen bond stabilization in SipD as in the PrgI-SipDΔD1 fusion protein (Figure 5B). It is interesting to speculate whether the coiled coil of SipD is intrinsically destabilized even in the absence of PrgI based on the superposition of different SipD copies found in the SipD structure (Figure S11).
Structural flexibility introduced by the π-bulge may account partially for the helix kink observed in the complex. The SipD helix kink is stabilized through interactions between Ser148 side chain with Asn59 of PrgI, which is highly conserved in needle proteins (Figure 4A and Figure 5A). Notably, in the fusion protein the Ser148 side chain formed a hydrogen bond with the backbone carbonyl oxygen of Ala144, located one helix turn upstream (Figure 5A). This hydrogen bonding network stabilized the helix kink present at amino acid Ala144 of SipD in the complex. Taken together, a π–bulge in SipD may provide conformational flexibility to allow the formation of the T3SS needle tip complex. Conversely, conformational flexibility of SipD may be arrested by the folded domain 1 in cytosolic SipD.
As mentioned above, one helix of PrgI had a similar kink to SipD in the complex. Amino acids Val65 and Val67 of PrgI were located at the kink (Figure 5A). Interestingly, both amino acids were recently found to be critically involved in polymerization of the T3SS needle [18]. Briefly, substitution of valines at position 65 and 67 by alanines reduced polymerization kinetics of the needle protein, but did not abolish needle formation or affect bacterial host invasion. Assembly of the T3SS needle is coupled with α-helix-to-β-sheet change downstream of Val67. Noteworthy, both amino acids were located at the PrgI-SipDΔD1 interface, suggesting a functional importance during assembly of the T3SS needle and tip complex. The visible amino acids downstream of the helix kink in PrgI adopted a helix conformation in the complex. Conformational flexibility, depending whether PrgI molecules interact with each other to form a needle or with SipD at the T3SS needle tip, may be a prerequisite for the function of this protein.
PrgI-SipDΔD1 fusion protein binds to deoxycholate
Host invasion of enteric bacteria is often dramatically influenced by the presence of bile salts [10]–[13]. Human intestinal bile salts, including deoxycholates, taurodeoxycholates and chenodeoxycholates, can either increase or repress invasion of S. flexneri or S. typhimurium, respectively [10], [14], [15]. The bile salt effect on those bacteria is coupled to a functional T3SS. Moreover, recent studies show that bile salts bind to needle tip proteins, corroborating that a T3SS component is affected by ligands released into the human gut [27].
To measure whether the PrgI-SipDΔD1 fusion protein could also bind deoxycholate (Figure 6A), we used surface plasmon resonance assay (Biacore). The binding curve of the immobilized PrgI-SipDΔD1 fusion with increasing concentrations of deoxycholate (Figure 6B) indicated a dissociation constant of 59.0±2.7 µM, assuming a protein ligand ratio of 1 to 1. These data are in agreement with previous results indicating an affinity between the S. flexneri needle tip protein IpaD and deoxycholate in the micromolar range [14]. Next, we soaked PrgI-SipDΔD1 crystals with sodium deoxycholate and analyzed the corresponding electron density for novel features that could fit the ligand. In the co-crystal structure (Figure 6C, Figure S12 and Table 1), deoxycholate was bound through its most hydrophobic β-surface (Figure 6A) to the cleft formed by SipD and PrgI. Localization of this binding site is about 25 Å away from the previously described deoxycholate binding site in wildtype SipD [28]. The α-surface of deoxycholate, disposing two hydroxyl groups, was facing the bulk medium. Ligand binding induced only minor structural changes related to the side chain of Ser236 in SipD and Gln24 and Gln48 in PrgI. Deoxycholate is a rigid molecule that fit almost perfectly into the cleft provided by the PrgI-SipDΔD1 fusion protein. In the cocrystal, the deoxycholate carboxyl group located at the end of the binding cleft was not as tightly embedded as the rest of the ligand. Therefore bile salts with larger substituents at this position could also occupy the cleft. Indeed, taurodeoxycholate, which is deoxycholate amidated at the carboxyl group with ethansulfonic acid, also binds to SipD and IpaD [14], [15].
Fig. 6. Binding assay and cocrystal structure of PrgI-SipDΔD1 with deoxycholate. 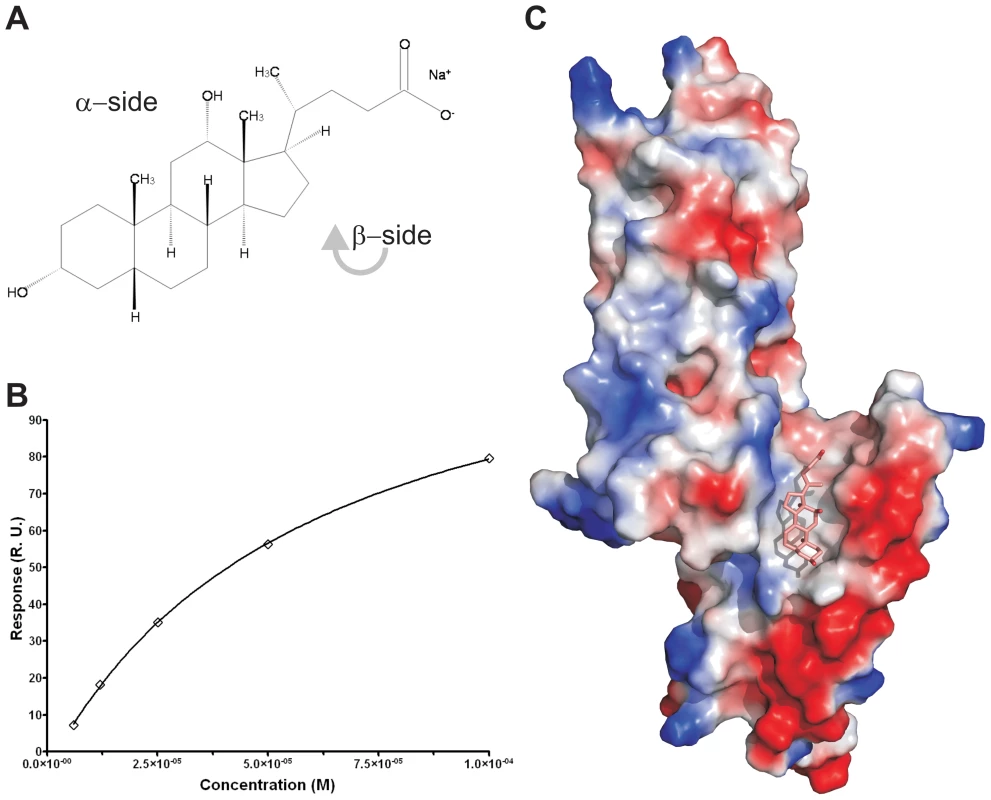
(A) Chemical formula of sodium deoxycholate, emphasizing the hydrophilic α- and the hydrophobic β-side of the molecule. (B) Binding curve of deoxycholate to PrgI-SipDΔD1: plot of the equilibrium binding response versus the concentration of deoxycholate. The solid line represents the fitting curve of the individual measurements. (C) Surface representation of the PrgI-SipDΔD1 fusion protein (chain B) coloured according the electrostatic potential (blue positive, red negative). Deoxycholate bound in the hydrophobic cleft of the PrgI-SipDΔD1 complex through its β-side is highlighted as a stick model. Results presented here are in agreement with recent NMR titration experiments showing that deoxycholate causes chemical shift changes in SipD upon binding to amino acids in the vicinity of the ligand binding site comprised by Arg232, Gln233, Ser236, Glu237 and Asn239 [15]. Moreover, mutation of amino acid Glu229 of IpaD, which is the equivalent of Glu237 in SipD, abolishes binding of deoxycholate [14].
Our data, however, indicate that bile salts bind to SipD similarly to the deoxycholate PrgI-SipDΔD1 fusion protein. The inhibitory effect of this ligand protein interaction for host invasion suggests that other proteins need to bind to the hydrophobic cleft formed by SipD and PrgI.
Discussion
We showed that the interaction of SipD with PrgI depends on the folding of domain 1 of SipD. In the PrgI-SipDΔD1 fusion protein, PrgI replaces the helix of domain 1 of SipD forming a contact with the concave side of the coiled coil. Crystal structure analysis of both SipD and of PrgI-SipDΔD1 reveal that the π-bulge in domain 2 of the tip protein contributes to the complex formation. Moreover, sterol binding to a cleft in the PrgI-SipDΔD1 complex suggests that intestinal detergents released from the gallbladder could hamper regulated secretion of virulence factors and consequently prevent invasion of S. typhimurium.
The T3SS needle tip protein from S. flexneri and Yersinia enterocolitica shows a pentameric organization [29], [30]. We found that the PrgI-SipDΔD1 fusion protein is monomeric in solution, suggesting that the architecture of the tip complex depends on the scaffold provided by the subunits of the T3SS needle.
Three dimensional model of the T3SS needle tip
The cryo-EM map of isolated needles from S. flexneri which is similar to the map obtained from isolated S. typhimurium needle [31] together with the X-ray crystal structure of a needle protomer mutant can be used to build a composite 3-dimensional model [21]. Based on this composite model of the T3SS needle we manually superimposed the similarly structured regions of the PrgI-SipDΔD1 fusion protein with the MxiH subunits of the needle. Superposition of PrgI and MxiH (PDB code 2V6L) using the program Coot [32] was feasible without structural clashes. In total, five molecules of the PrgI-SipDΔD1 fusion protein were successfully superimposed with five MxiH subunits at the distal end of the T3SS needle (Figure 7). As described above, we found that PrgI binds to the concave side of the central coiled coil in SipD. Therefore, the PrgI-SipDΔD1 fusion protein could be mounted at the distal end of the needle without inducing structural changes. Domain1 present in SipD may face the bulk medium either in an unfold state or as a folded entity. In contrast to our model, the SipD-PrgI contact predicted by a previous work [9] would require substantial structural changes at the tip of the T3SS needle.
Fig. 7. Model of the open state of the T3SS needle tip. 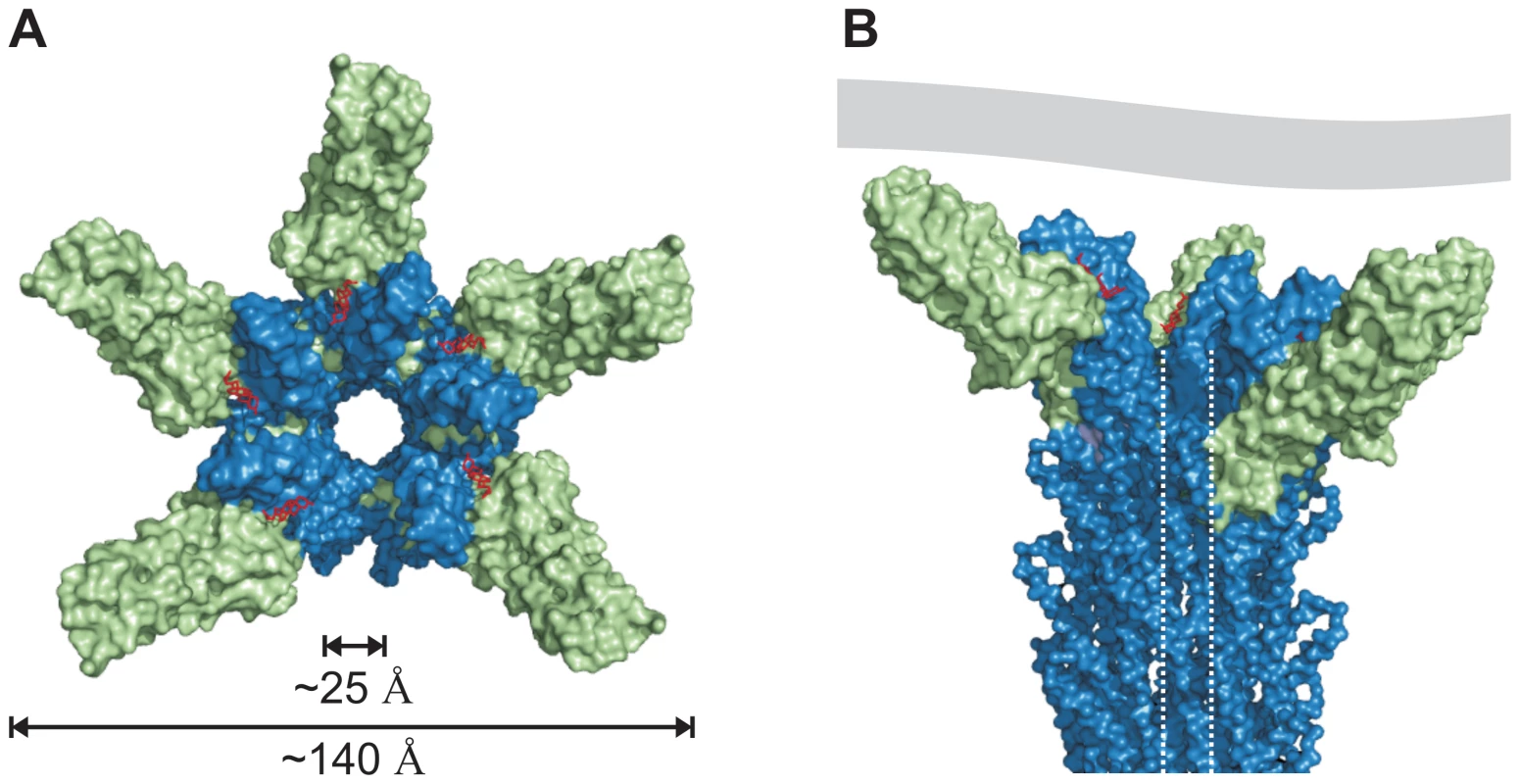
(A) View from the surface into the channel opening of the T3SS needle tip. SipD is coloured in green and PrgI in blue, respectively. Deoxycholate molecules binding to PrgI-SipDΔD1 are highlighted in red. The inner diameter of the needle tip complex (25 Å) permits passage of unfolded virulence proteins. Opening of the needle tip complex with an outer diameter of 140 Å results in a large inner cavity which could be closed through contact with the plasma membrane of host cells. (B) Side view of the T3SS needle including the tip complex. The inner channel is highlighted by dotted lines and a contacting host cell membrane is shown in grey. Open state of the T3SS needle tip
The tip complex is the distal opening of the transport channel provided by the T3SS needle. According to the proposed model, SipD bound to PrgI localizes to the outer surface of the T3SS needle without obstructing the inner channel (Figure 7). The channel is opened in the three dimensional model of the T3SS tip complex (Figure 7A), adopting a state that permits transport or release of unfolded molecules after passage through the channel inside the needle. For this reason we assign the presented structure-based model as the “open state” of the T3SS needle tip. About 25 Å for the diameter of the T3SS channel may allow the passage of a single α-helix (or even of a helix-loop-helix motif).
Deletion of the T3SS needle tip protein causes constitutive secretion of virulence factors but abolishes bacterial invasion [7]. Consequently, it was proposed that the needle tip protein blocks the secretion of virulence factors. In contrast to this hypothesis, our structure based model of the open state suggests that the T3SS tip complex is not necessarily blocking the T3SS channel. Moreover, the tip protein does not need to be released for secretion of virulence factors, as the SipD-PrgI interaction is not clogging the channel. We speculate that the presence of the tip protein enables intermittent closing of the T3SS system thus regulating the process of secretion.
The three dimensional model presented here enables the following conclusions: The open state of the SipD-PrgI needle tip must be closed to block the constitutive transport of virulence factors. The closing of the T3SS needle tip can be mediated by either a conformational change of SipD or by its interaction with other effector proteins or lipids. Notably, the structure of the needle tip complex is not in conflict with possible movement of domain 2 and 3 in SipD but further work is required to explain how SipD regulates the secretion of T3SS. Moreover, binding of other effector proteins to the T3SS needle tip was proposed for the control of the needle length [1], [33] and the translocation of virulence factors across host membranes (SipB).
Along these lines, we previously reported that the T3SS needle protein PrgI extends the needle from the distal end in the absence of tip proteins [18]. Moreover, addition of tip proteins prevented further growth of the T3SS needle [18]. It is plausible that addition of SipD avoid needle elongation.
Bile salts, including deoxycholate, can prevent S. typhimurium invasion through binding to SipD [12], [15]. We showed here that deoxycholate binds to the cleft formed by SipD and PrgI close to the constriction of the T3SS channel (Figure 6 and Figure 7a). This interaction may prevent larger conformational changes in SipD, which block closure of the T3SS channel. Likewise bound deoxycholate may impede the binding of channel blocking proteins. Future studies are needed to understand how the T3SS is regulated using deoxycholate.
The presented structural studies enable us to construct a three dimensional model of the Salmonella T3SS needle tip, which in turn suggests a secretion mechanism. In the open state of the T3SS needle tip a large cavity, maybe enclosed through contact with the host membrane, is formed. This cavity could act as a folding chamber to facilitate the folding of secreted proteins. Folding of early secreted translocator proteins at the host membrane could improve the delivery of other effector proteins into the host cytoplasm. A similar folding principle was identified in the molecular chaperones, including prefoldin which forms a cavity for the nascent protein chain at the exit channel of the ribosome [34]–[36]. Moreover, the T3SS channel could be closed by contact with host membranes. In this scenario, the host membrane could prevent waste of secreted virulence factors, which otherwise could diffuse away from the point of contact. The T3SS needle tip is crucial for bacterial invasion and searching for substances similar to deoxycholate that prevent functioning or even assembly of the complex could lead to the discovery of novel targets for the development of drugs against pathogenic enterobacteria.
Material and Methods
Cloning, gene expression and protein purification
Wildtype and mutant sipD or prgI were amplified from S. enterica serovar typhimurium strain SL1344 (S. typhimurium) by standard PCR using oligonucleotide primers with NdeI and XhoI restriction sites at either ends. Single crystallization of SipD132–343 superseded cocrystallization with PrgI in various attempts. Therefore, wildtype prgI and a sipD fragment encoding amino acids 127 to 343 were connected by fusion PCR. N-terminal PrgI was fused by the amino acids Gly-Gly-Ser-Gly-Gly to SipD127-343.
PCR products were cloned into the expression vector pET-28a(+) (Novagen) or pET-21a(+) (Novagen), both containing N-terminal His-tag, and expressed in Escherichia coli BL21(DE3) cells. Cells were induced with isopropyl-β-D-1-thiogalactopyranoside, harvested after 4 h and His-tagged protein purified using affinity chromatography (HisTrap, GE Healthcare). Bound protein was washed (40 mM imidazole) and eluted using buffer containing 500 mM imidazole. After buffer exchange (20 mM HEPES pH 7.4, 50 mM NaCl) the tag was cleaved with CleanCleave Kit (Sigma-Aldrich). The cleaved product was purified by size-exclusion chromatography (Superdex 200 or Superdex 75, GE Healthcare) and stored at 4°C until use. For functional assays, wildtype or mutant sipD or prgI were cloned into the pASK-IBA5 vector (IBA) as BsaI fragments. Point mutants were generated using QuikChange Site-Directed Mutagenesis Kit (Stratagene). All constructs were confirmed by sequencing.
Crystallization, data collection, structures determination and refinement
Crystals of SipD, SipDΔD1 and PrgI-SipDΔD1 were obtained at 18°C using hanging drop vapour diffusion technique. SipD was concentrated to ∼15 mg/ml and mixed with equal volume of reservoir solution containing 100 mM HEPES pH 7.5 and 1.5 M Li2SO4. SipDΔD1 was concentrated to 40 mg/ml and mixed with equal volume of reservoir solution 0.1 M MES pH 6.5 and 12% (w/v) polyethylene glycol 20000. PrgI-SipDΔD1 was concentrated to 30 mg/ml and mixed with equal volume of reservoir solution containing 0.49 M NaH2PO4 • H2O and 0.91 M K2HPO4, pH 6.9. To obtain cocrystals PrgI-SipDΔD1 crystals were soaked for 72 hours in mother liquor containing ∼10 mM deoxycholate. All crystals were flash frozen in liquid nitrogen in the presence of 30% glycerol (v/v). Diffraction data were collected at 100 K and wavelength 0.918 Å at BESSY II (Berlin, Germany) beamlines 14.1 or 14.2, or wavelength 1.000 Å at SLS (Villigen, Switzerland) beamline X06SA.
Diffraction data were indexed, integrated and scaled using the program package XDS [37]. The crystal structure of SipDΔD1 was solved by molecular replacement with the program Phaser [38] using the structure of truncated IpaD (pdb code: 2J0N) as template. The structures of SipD and PrgI-SipDΔD1, apo and with deoxycholate, were solved by molecular replacement using the SipDΔD1 structure as template. The initial models were refined by repeated cycles of manual building and refinement using the programs Coot [32] and CNS [39].
Crystals of SipD have 4 copies in the asymmetric unit and the following Ramachandran statistics: 82.7% of residues in most favoured regions, 16.6% in additionally allowed regions, 0.7% in generously allowed regions. Crystals of SipDΔD1 have 2 copies in the asymmetric unit and 92.7% of residues in most favoured regions, 6.5% in additionally allowed regions, and 0.8% in generously allowed regions. Crystals of PrgI-SipDΔD1 have 2 copies in the asymmetric unit and 92.9% of residues in most favoured regions, 7.1% in additionally allowed regions. The structure of PrgI-SipDΔD1 complexed deoxycolate has 93.4% of residues in most favoured regions and 6.6% in additionally allowed regions. Ramachandran statistics were calculated with PROCHECK v.3.3 [40].
Molecular graphics images, including representations of surface electrostatic potential, were produced using PyMOL version 0.99rc6 [41], except Figure S6 which was produced with UCSF Chimera package from the Resource for Biocomputing, Visualization, and Informatics at the University of California, San Francisco [42].
Generation of knockout strains
Bacterial knockouts were generated according to Datsenko and Wanner [43]. pASK-IBA5 plasmids harboring wild type or mutant sipD (psipD) were used to complement deletions of sipD in S. typhimurium strain SL1344 to generate strains SL1344ΔsipD/psipD.
HeLa cell invasion assay
HeLa cells were seeded at 1×105 cells per well and grown for 24 h at 37°C. Prior to infection, growth medium was aspirated, cells were washed twice with phosphate-buffered saline (PBS), and serum-free medium was added. To test for epithelial cell invasion and intracellular growth, HeLa cells were infected with S. typhimurium at a multiplicity of infection (MOI) of 10 : 1. Expression of sipD wild-type and sipD mutants was induced with 0.2 µg ml−1 anhydrotetracycline for 1 h. Bacterial inocula were prepared in PBS and centrifuged onto cells (2000 rpm, 10 min), and infected cultures incubated for 20 min at 37°C. Cultures were washed three times with PBS, and fresh medium containing 100 µg ml−1 gentamicin was added. After 2 h cells were washed with PBS and lysed with 0.1% Triton X-100. Numbers of viable bacteria were obtained by plating dilutions of lysates on tryptic soy agar plates and counting colonies after overnight incubation at 37°C.
Multi-angle laser light scattering (MALLS)
For mass determination a combined setup consisting of SEC and subsequent online detection by UV absorption, (three angle) static laser light scattering and differential refractive index measurement was used as described earlier [44]. SEC was performed with either a Tricorn Superdex 200 10/300 GL column or a Tricorn Superdex 75 10/300 GL (GE Healthcare) equilibrated with 20 mM HEPES (pH 7.5), 150 mM NaCl. For static light scattering and differential refractive index measurements a linear coupled miniDAWN Tristar (Wyatt Technology) system and a differential refractive index detector (RI-101, Shodex), respectively, was used. All calculations were done with the software ASTRA (Wyatt Technology). Each experiment was repeated at least in triplicate.
Isothermal Titration Calorimetry (ITC)
Titration experiments were carried out using a VP–ITC isothermal titration microcalorimeter (MicroCal, Northampton, MA, USA). Aliquots of 12 µl of SipD (1.35 mM) were injected consecutively at 20°C into the cell containing 1.4 ml of PrgI* (0.34 mM) or at 17°C by injecting consecutively 12 µl aliquots of PrgI* (1.99 mM) into the cell containing 1.4 ml of SipDΔD1 (0.37 mM). The heat of dilution of the injected protein was measured in both cases and subtracted from the heath measured at each injection. Binding stoichiometry, enthalpy, and equilibrium association constants were determined by fitting the corrected data to one set of sites model equation using the evaluation software provided by the manufacturer.
Surface plasmon resonance (Biacore)
Binding of sodium deoxycholate to the PrgI-SipDΔD1 fusion protein was measured using surface plasmon technology-based Biacore X100 biosensor (GE Healthcare) according to manufacturer's instruction. Briefly, PrgI-SipDΔD1 fusion protein was immobilized on a sensor chip CM5 (research grade) by amine coupling method. Binding experiments were performed at 25°C at continuous flow rate of HBS-N buffer (10 mM HEPES, 150 mM NaCl, pH 7.4). Deoxycholate was injected in steps with increasing concentrations in a single analysis cycle without regeneration of the surface in between injections. Affinity analysis was performed using single-cycle kinetics. Equilibrium dissociation constant (KD) was determined with Biacore evaluation Version 4.1 software. During the assays, the signal was corrected against the control surface response to eliminate refractive index changes due to buffer change.
Circular dichroism spectroscopy
CD spectra were collected with a Jasco J-500A spectropolarimeter. Samples buffered in 10 mM HEPES (pH 7.4), 25 mM NaCl were measured either at 20°C and protein concentration1.5 mg/ml between 182 and 260 nm in a quartz cuvette with optical path length of 0.1 mm or at 37°C and protein concentration 0.15 mg/ml between 198 and 260 nm in a temperature controlled quartz cuvette with optical path length of 1 mm. Wavelength scans were carried out at a scan rate of 12 nm/min, with time constant 2 sec. All the spectra were acquired in triplicates.
PDB accession codes
The atomic coordinates and structure factors of the four structures described here are available from the Protein Data Bank under the following accession codes: 2YM0 for SipDΔD1, 2YM9 for SipD, 3ZQB for PrgI - SipDΔD1, 3ZQE for PrgI - SipDΔD1 complexed with deoxycholate.
Supporting Information
Zdroje
1. CornelisGR 2006 The type III secretion injectisome. Nat Rev Microbiol 4 811 825
2. GalanJEWolf-WatzH 2006 Protein delivery into eukaryotic cells by type III secretion machines. Nature 444 567 573
3. NhieuGTSansonettiPJ 1999 Mechanism of Shigella entry into epithelial cells. Curr Opin Microbiol 2 51 55
4. DeaneJEAbrusciPJohnsonSLeaSM 2010 Timing is everything: the regulation of type III secretion. Cell Mol Life Sci 67 1065 1075
5. BlockerAJDeaneJEVeenendaalAKRoversiPHodgkinsonJL 2008 What's the point of the type III secretion system needle? Proc Natl Acad Sci U S A 105 6507 6513
6. WangYZhangLPickingWLPickingWDDe GuzmanRN 2008 Structural dissection of the extracellular moieties of the type III secretion apparatus. Mol Biosyst 4 1176 1180
7. KanigaKTrollingerDGalanJE 1995 Identification of two targets of the type III protein secretion system encoded by the inv and spa loci of Salmonella typhimurium that have homology to the Shigella IpaD and IpaA proteins. J Bacteriol 177 7078 7085
8. MuellerCABrozPCornelisGR 2008 The type III secretion system tip complex and translocon. Mol Microbiol 68 1085 1095
9. ZhangLWangYOliveAJSmithNDPickingWD 2007 Identification of the MxiH needle protein residues responsible for anchoring invasion plasmid antigen D to the type III secretion needle tip. J Biol Chem 282 32144 32151
10. OliveAJKenjaleREspinaMMooreDSPickingWL 2007 Bile salts stimulate recruitment of IpaB to the Shigella flexneri surface, where it colocalizes with IpaD at the tip of the type III secretion needle. Infect Immun 75 2626 2629
11. PopeLMReedKEPayneSM 1995 Increased protein secretion and adherence to HeLa cells by Shigella spp. following growth in the presence of bile salts. Infect Immun 63 3642 3648
12. ProutyAMGunnJS 2000 Salmonella enterica serovar typhimurium invasion is repressed in the presence of bile. Infect Immun 68 6763 6769
13. TollisonSBJohnsonMG 1985 Sensitivity to bile salts of Shigella flexneri sublethally heat stressed in buffer or broth. Appl Environ Microbiol 50 337 341
14. StensrudKFAdamPRLa MarCDOliveAJLushingtonGH 2008 Deoxycholate interacts with IpaD of Shigella flexneri in inducing the recruitment of IpaB to the type III secretion apparatus needle tip. J Biol Chem 283 18646 18654
15. WangYNordhuesBAZhongDDe GuzmanRN 2010 NMR characterization of the interaction of the Salmonella type III secretion system protein SipD and bile salts. Biochemistry 49 4220 4226
16. JohnsonSRoversiPEspinaMOliveADeaneJE 2007 Self-chaperoning of the type III secretion system needle tip proteins IpaD and BipD. J Biol Chem 282 4035 4044
17. ErskinePTKnightMJRuauxAMikolajekHWong Fat SangN 2006 High resolution structure of BipD: an invasion protein associated with the type III secretion system of Burkholderia pseudomallei. J Mol Biol 363 125 136
18. PoyrazOSchmidtHSeidelKDelissenFAderC 2010 Protein refolding is required for assembly of the type three secretion needle. Nat Struct Mol Biol 17 788 792
19. EspinaMAusarSFMiddaughCRBaxterMAPickingWD 2007 Conformational stability and differential structural analysis of LcrV, PcrV, BipD, and SipD from type III secretion systems. Protein Sci 16 704 714
20. EspinaMAusarSFMiddaughCRPickingWDPickingWL 2006 Spectroscopic and calorimetric analyses of invasion plasmid antigen D (IpaD) from Shigella flexneri reveal the presence of two structural domains. Biochemistry 45 9219 9227
21. DeaneJERoversiPCordesFSJohnsonSKenjaleR 2006 Molecular model of a type III secretion system needle: Implications for host-cell sensing. Proc Natl Acad Sci U S A 103 12529 12533
22. WangYOuelletteANEganCWRathinavelanTImW 2007 Differences in the electrostatic surfaces of the type III secretion needle proteins PrgI, BsaL, and MxiH. J Mol Biol 371 1304 1314
23. ZhangLWangYPickingWLPickingWDDe GuzmanRN 2006 Solution structure of monomeric BsaL, the type III secretion needle protein of Burkholderia pseudomallei. J Mol Biol 359 322 330
24. RathinavelanTTangCDe GuzmanRN 2011 Characterization of the interaction between the Salmonella type III secretion system tip protein SipD and the needle protein PrgI by paramagnetic relaxation enhancement. J Biol Chem 286 4922 4930
25. KenjaleRWilsonJZenkSFSauryaSPickingWL 2005 The needle component of the type III secreton of Shigella regulates the activity of the secretion apparatus. J Biol Chem 280 42929 42937
26. CartaillerJPLueckeH 2004 Structural and functional characterization of pi bulges and other short intrahelical deformations. Structure 12 133 144
27. HaywardRDCainRJMcGhieEJPhillipsNGarnerMJ 2005 Cholesterol binding by the bacterial type III translocon is essential for virulence effector delivery into mammalian cells. Mol Microbiol 56 590 603
28. ChatterjeeSZhongDNordhuesBABattaileKPLovellS 2011 The crystal structures of the Salmonella type III secretion system tip protein SipD in complex with deoxycholate and chenodeoxycholate. Protein Sci 20 75 86
29. MuellerCABrozPMullerSARinglerPErne-BrandF 2005 The V-antigen of Yersinia forms a distinct structure at the tip of injectisome needles. Science 310 674 676
30. SaniMBotteauxAParsotCSansonettiPBoekemaEJ 2007 IpaD is localized at the tip of the Shigella flexneri type III secretion apparatus. Biochim Biophys Acta 1770 307 311
31. GalkinVESchmiedWHSchraidtOMarlovitsTCEgelmanEH 2010 The structure of the Salmonella typhimurium type III secretion system needle shows divergence from the flagellar system. J Mol Biol 396 1392 1397
32. EmsleyPCowtanK 2004 Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60 2126 2132
33. MotaLJJournetLSorgIAgrainCCornelisGR 2005 Bacterial injectisomes: needle length does matter. Science 307 1278
34. BaramDPyetanESittnerAAuerbach-NevoTBashanA 2005 Structure of trigger factor binding domain in biologically homologous complex with eubacterial ribosome reveals its chaperone action. Proc Natl Acad Sci U S A 102 12017 12022
35. FerbitzLMaierTPatzeltHBukauBDeuerlingE 2004 Trigger factor in complex with the ribosome forms a molecular cradle for nascent proteins. Nature 431 590 596
36. MerzFBoehringerDSchaffitzelCPreisslerSHoffmannA 2008 Molecular mechanism and structure of Trigger Factor bound to the translating ribosome. EMBO J 27 1622 1632
37. KabschW 2010 Xds. Acta Crystallogr D Biol Crystallogr 66 125 132
38. McCoyAJGrosse-KunstleveRWAdamsPDWinnMDStoroniLC 2007 Phaser crystallographic software. J Appl Crystallogr 40 658 674
39. BrungerATAdamsPDCloreGMDeLanoWLGrosP 1998 Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr 54 905 921
40. LaskowskiRAMacarthurMWMossDSThorntonJM 1993 Procheck - a Program to Check the Stereochemical Quality of Protein Structures. J Appl Crystallogr 26 283 291
41. DeLanoWL 2006 PyMOL Incentive Product. DeLano Scientific LLC
42. PettersenEFGoddardTDHuangCCCouchGSGreenblattDM 2004 UCSF Chimera—a visualization system for exploratory research and analysis. J Comput Chem 25 1605 1612
43. DatsenkoKAWannerBL 2000 One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A 97 6640 6645
44. LunelliMLokareddyRKZychlinskyAKolbeM 2009 IpaB-IpgC interaction defines binding motif for type III secretion translocator. Proc Natl Acad Sci U S A 106 9661 9666
Štítky
Hygiena a epidemiologie Infekční lékařství Laboratoř
Článek Crystal Structure of Reovirus Attachment Protein σ1 in Complex with Sialylated OligosaccharidesČlánek A Protein Thermometer Controls Temperature-Dependent Transcription of Flagellar Motility Genes inČlánek Modulation of NKp30- and NKp46-Mediated Natural Killer Cell Responses by Poxviral Hemagglutinin
Článek vyšel v časopisePLOS Pathogens
Nejčtenější tento týden
2011 Číslo 8- Stillova choroba: vzácné a závažné systémové onemocnění
- Diagnostický algoritmus při podezření na syndrom periodické horečky
- Choroby jater v ordinaci praktického lékaře – význam jaterních testů
- Diagnostika virových hepatitid v kostce – zorientujte se (nejen) v sérologii
- Perorální antivirotika jako vysoce efektivní nástroj prevence hospitalizací kvůli COVID-19 − otázky a odpovědi pro praxi
-
Všechny články tohoto čísla
- Phenotypic Screens, Chemical Genomics, and Antimalarial Lead Discovery
- Characterisation of Regulatory T Cells in Nasal Associated Lymphoid Tissue in Children: Relationships with Pneumococcal Colonization
- Crystal Structure of Reovirus Attachment Protein σ1 in Complex with Sialylated Oligosaccharides
- Absence of Cross-Presenting Cells in the Salivary Gland and Viral Immune Evasion Confine Cytomegalovirus Immune Control to Effector CD4 T Cells
- Transcriptomic Analysis of Host Immune and Cell Death Responses Associated with the Influenza A Virus PB1-F2 Protein
- A Quorum Sensing Regulated Small Volatile Molecule Reduces Acute Virulence and Promotes Chronic Infection Phenotypes
- Autocrine Regulation of Pulmonary Inflammation by Effector T-Cell Derived IL-10 during Infection with Respiratory Syncytial Virus
- A Protein Thermometer Controls Temperature-Dependent Transcription of Flagellar Motility Genes in
- Association of Human TLR1 and TLR6 Deficiency with Altered Immune Responses to BCG Vaccination in South African Infants
- Histo-Blood Group Antigens Act as Attachment Factors of Rabbit Hemorrhagic Disease Virus Infection in a Virus Strain-Dependent Manner
- MrkH, a Novel c-di-GMP-Dependent Transcriptional Activator, Controls Biofilm Formation by Regulating Type 3 Fimbriae Expression
- Beta-HPV 5 and 8 E6 Promote p300 Degradation by Blocking AKT/p300 Association
- Modulation of NKp30- and NKp46-Mediated Natural Killer Cell Responses by Poxviral Hemagglutinin
- Transportin 3 Promotes a Nuclear Maturation Step Required for Efficient HIV-1 Integration
- Coordination of KSHV Latent and Lytic Gene Control by CTCF-Cohesin Mediated Chromosome Conformation
- A Novel Persistence Associated EBV miRNA Expression Profile Is Disrupted in Neoplasia
- The Plant Pathogen pv. Is Genetically Monomorphic and under Strong Selection to Evade Tomato Immunity
- IL-10 Blocks the Development of Resistance to Re-Infection with
- Anti-Apoptotic Machinery Protects the Necrotrophic Fungus from Host-Induced Apoptotic-Like Cell Death during Plant Infection
- Crystal Structure of PrgI-SipD: Insight into a Secretion Competent State of the Type Three Secretion System Needle Tip and its Interaction with Host Ligands
- Evades Immune Recognition of Flagellin in Both Mammals and Plants
- Tumor Cell Marker PVRL4 (Nectin 4) Is an Epithelial Cell Receptor for Measles Virus
- Provides Insights into the Evolution of the Salmonellae
- B Cell Repertoire Analysis Identifies New Antigenic Domains on Glycoprotein B of Human Cytomegalovirus which Are Target of Neutralizing Antibodies
- Thy1 Nk Cells from Vaccinia Virus-Primed Mice Confer Protection against Vaccinia Virus Challenge in the Absence of Adaptive Lymphocytes
- The Cytokine Network of Acute HIV Infection: A Promising Target for Vaccines and Therapy to Reduce Viral Set-Point?
- Dendritic Cell Status Modulates the Outcome of HIV-Related B Cell Disease Progression
- Differential Contribution of PB1-F2 to the Virulence of Highly Pathogenic H5N1 Influenza A Virus in Mammalian and Avian Species
- A Communal Bacterial Adhesin Anchors Biofilm and Bystander Cells to Surfaces
- Two Group A Streptococcal Peptide Pheromones Act through Opposing Rgg Regulators to Control Biofilm Development
- Activation of HIV Transcription by the Viral Tat Protein Requires a Demethylation Step Mediated by Lysine-specific Demethylase 1 (LSD1/KDM1)
- Unique Evolution of the UPR Pathway with a Novel bZIP Transcription Factor, Hxl1, for Controlling Pathogenicity of
- Disruption of PML Nuclear Bodies Is Mediated by ORF61 SUMO-Interacting Motifs and Required for Varicella-Zoster Virus Pathogenesis in Skin
- Flagellar Motility Is Not Directly Required to Maintain Attachment to Surfaces
- Viral Infection Induces Expression of Novel Phased MicroRNAs from Conserved Cellular MicroRNA Precursors
- Functional Cure of SIVagm Infection in Rhesus Macaques Results in Complete Recovery of CD4 T Cells and Is Reverted by CD8 Cell Depletion
- Recruitment of the Major Vault Protein by InlK: A Strategy to Avoid Autophagy
- The Steroid Catabolic Pathway of the Intracellular Pathogen Is Important for Pathogenesis and a Target for Vaccine Development
- PLOS Pathogens
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Tumor Cell Marker PVRL4 (Nectin 4) Is an Epithelial Cell Receptor for Measles Virus
- Two Group A Streptococcal Peptide Pheromones Act through Opposing Rgg Regulators to Control Biofilm Development
- Differential Contribution of PB1-F2 to the Virulence of Highly Pathogenic H5N1 Influenza A Virus in Mammalian and Avian Species
- Recruitment of the Major Vault Protein by InlK: A Strategy to Avoid Autophagy
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



