-
Články
- Vzdělávání
- Časopisy
Top články
- Témata
- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Alternative Sigma Factor σ Modulates Prophage Integration and Excision in
The prophage is one of the most important components of variable regions in bacterial genomes. Some prophages carry additional genes that may enhance the toxicity and survival ability of their host bacteria. This phenomenon is predominant in Staphylococcus aureus, a very common human pathogen. Bioinformatics analysis of several staphylococcal prophages revealed a highly conserved 40-bp untranslated region upstream of the int gene. A small transcript encoding phage integrase was identified to be initiated from the region, demonstrating that the untranslated region contained a promoter for int. No typical recognition sequence for either σA or σB was identified in the 40-bp region. Experiments both in vitro and in vivo demonstrated that σH recognized the promoter and directed transcription. Genetic deletion of sigH altered the int expression, and subsequently, the excision proportion of prophage DNAs. Phage assays further showed that sigH affected the ability of spontaneous lysis and lysogenization in S. aureus, suggesting that sigH plays a role in stabilizing the lysogenic state. These findings revealed a novel mechanism of prophage integration specifically regulated by a host-source alternative sigma factor. This mechanism suggests a co-evolution strategy of staphylococcal prophages and their host bacteria.
Published in the journal: . PLoS Pathog 6(5): e32767. doi:10.1371/journal.ppat.1000888
Category: Research Article
doi: https://doi.org/10.1371/journal.ppat.1000888Summary
The prophage is one of the most important components of variable regions in bacterial genomes. Some prophages carry additional genes that may enhance the toxicity and survival ability of their host bacteria. This phenomenon is predominant in Staphylococcus aureus, a very common human pathogen. Bioinformatics analysis of several staphylococcal prophages revealed a highly conserved 40-bp untranslated region upstream of the int gene. A small transcript encoding phage integrase was identified to be initiated from the region, demonstrating that the untranslated region contained a promoter for int. No typical recognition sequence for either σA or σB was identified in the 40-bp region. Experiments both in vitro and in vivo demonstrated that σH recognized the promoter and directed transcription. Genetic deletion of sigH altered the int expression, and subsequently, the excision proportion of prophage DNAs. Phage assays further showed that sigH affected the ability of spontaneous lysis and lysogenization in S. aureus, suggesting that sigH plays a role in stabilizing the lysogenic state. These findings revealed a novel mechanism of prophage integration specifically regulated by a host-source alternative sigma factor. This mechanism suggests a co-evolution strategy of staphylococcal prophages and their host bacteria.
Introduction
Prophages are viral cellular parasites that integrate into bacterial genomes and co-replicate with host chromosomes. A subset of bacteriophage genomes encodes additional virulence factors, and the production of these virulence factors can enhance bacterial toxicity as well as survival ability in various environments [1], [2]. Prophages are not rare in the chromosome of Staphylococcus aureus, a widely spread human pathogen. In fact, most clinical isolates harbor at least one prophage [3]. Many staphylococcal phages contain genes that encode virulence factors such as staphylokinase, enterotoxin A, chemotaxis inhibitory protein, staphylococcal complement inhibitor and leukocidin, which greatly enhance the bacterial invasiveness and help to evade host immunity in organic infection [4], [5], [6]. The transfer of toxic genes by a lysogenic bacteriophage, or phage conversion, is an important mechanism in the evolution of virulent S. aureus strains [7]. Furthermore, phages participate in the mediation of horizontal transfer of pathogenicity islands and raise intra-strain and inter-strain exchange frequency of toxic genes [8], [9].
As a member of double-strand DNA viruses, a temperate phage needs to recruit bacterial RNA polymerase with essential sigma factors to initiate its cascade. In S. aureus, only four sigma factors have been identified to date: σA, the housekeeping sigma factor, which directs the transcription of the bulk cellular RNA, and three alternative sigma factors σB [10], σH [11], and the newly defined σS [12]. The S. aureus σB protein is closely related to the σB protein of Bacillus subtilis, and is mainly involved in stress response [13], [14]. The S. aureus σH protein is a homolog of B. subtilis σH, which regulates sporulation-related genes [15].
At the turn of the 21st century, S. aureus genome nucleotide sequences were being completed at a rapid and increasing rate [16]. At least 14 S. aureus strains have been whole-genome sequenced and more are in progress. In addition, many staphylococcal phages have been identified independently and then sequenced as well [17], [18], [19], [20], [21], [22]. These sequenced genomes allowed us to comparatively analyze the staphylococcal prophage, one of the most important components of genomic variable regions and which provides numerous virulence factors to host bacteria. The majority of known staphylococcal bacteriophages belong to the order Caudovirales and the size of the phage genomes mainly ranges from 35 to 50 kb. Architectural analysis of these prophage genomes demonstrated identical gene arrangements (Figure S1). Some prophages share similar open reading frames (ORFs), especially in genes that encode products for the lytic cycle.
The integrase genes of these bacteriophages are highly conserved [23]. Some staphylococcal prophages even harbor the same integrase and insert into the same locus on the S. aureus genome. Besides integrase, the excisionase represents divergence among species. Indeed, only some of the staphylococcal prophages contain the xis gene, which encodes excisionase. The rest have a genetic structure called ORF-C, in the opposite direction of int [24]. Here we report a heretofore unrecognized manner whereby an alternative sigma factor is recruited by a staphylococcal temperate phage for the regulation of int transcription. The recognition site for σH is upstream of the ORF of int, which encodes phage integrase. Deletion of sigH resulted in a decrease of phage int mRNA level and an increase of the excised form of prophage genomes under normal growth conditions. The abilities of spontaneous lysis and lysogenization in S. aureus were also affected. These results indicate that S. aureus σH modulates the transcription of phage integrase, stabilizes the lysogeny in the host cell and may further influence the prophage life cycle and correlative bacterial virulence.
Results
Comparative analysis of S. aureus prophage genomes identified a highly conserved region upstream of the int gene
We first analyzed 43 staphylococcal prophage sequences available from NCBI GenBank by multiple sequence alignment in segments. From the results of the in silico analysis we found that a small fragment in the 5′ untranslated region (UTR) of int was extremely conserved among nearly all the prophages that were compared (Figure 1), with the only exception of Φ3A, which is int defective [3]. The region was previous mentioned as junction A when compared six S. aureus phages [25]. The conserved region was a 40-bp fragment followed by the typical Shine-Dalgarno (SD) sequence AGGAGG and was closely related to int. More interestingly, a previously reported stem-loop structure of 9-base inverted repeat with a 3-base loop (ATTTAGTACtagGTACTAAAT) found in several staphylococcal prophages [24] was also adjacent to that region. These data indicated that the region was likely to be a transcriptional regulator binding domain.
Fig. 1. Sequence analysis of 5′-UTR of int in 42 S. aureus prophages. 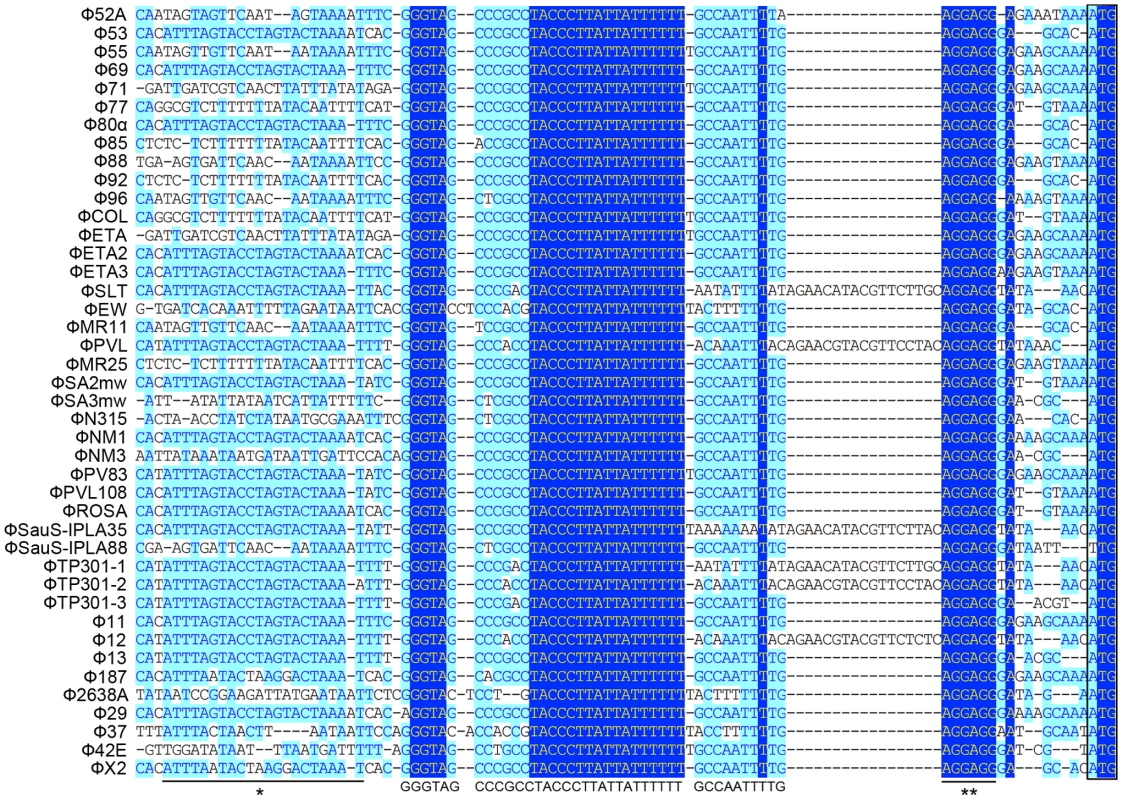
A highly conserved region (GGGTAGCCCGCCTACCCTTATTATTTTTTGCCAATTT) is identified. The previous reported stem-loop structure (*) and SD sequence (**) are located at the two sides of the newly defined region. Light blue boxes indicate the conservative nucleotide bases and dark blue boxes indicate the identical bases among all sequences compared. The start codons of int genes are boxed. The DNA sequences upstream of int gene from several other firmicutes harboring prophages and two S. aureus pathogenic islands were also compared (Figure 2A). Similar sequences were identified in Staphylococcus epidermidis prophage ФCNPH82, ФPH15, and Staphylococcus haemolyticus ФSH1, but not in other prophages. It seemed that the conserved region only existed in the staphylococci harboring prophages.
Fig. 2. Sequence analysis of int promoters in several firmicutes harboring phages and a phylogenetic tree of sigH proteins in firmicutes. 
(A) A comparison of the DNA sequences upstream of int gene from several other firmicutes harboring prophages and two S. aureus pathogenic islands showing that similar sequences were also identified in S. epidermidis prophage ФCNPH82, ФPH15, and S. haemolyticus ФSH1. SD sequences and the start codons of int genes are indicated by asterisks. (B) A phylogenetic tree of sigH orthologs in the above firmicutes showed that staphylococcal sigH proteins were separated from the others by a deep node. Protein sequences were aligned using ClustalW in Vector NTI software. Alignments were imported into Mega 4 and the tree was generated using the neighbor-joining method, ignoring positions with gaps. The scale bar represents 0.1 substitutions per nucleotide site. Analysis of the transcriptional organization of the S. aureus prophage int genes
Since the results from in silico analysis strongly suggested that the newly defined region was a transcriptional regulation domain for S. aureus prophage int gene, it would be interesting to reveal the transcriptional organization of the integrase. Northern blot assays were performed to determine the transcriptional products for integrases in Ф11, Ф12, and Ф13, respectively. Under the normal lysogenic conditions, two mRNAs encoding phage integrase were produced. The larger mRNAs (∼3-kb to ∼4-kb) were initiated from promoters in the phage immunity region according to their lengths, and the smaller transcripts (∼1-kb) were presumably started from our newly defined region (Figures 3B and S2). To identify the exact transcriptional initiation site, a primer extension assay was carried out. The signal showed that the start site was just between the conserved region and the translational start codon (Figure S3). The results confirmed our hypothesis that the conserved region harbored a promoter specific for int expression. However, no typical sequences could be predicted as recognition motifs for neither σA nor stress response sigma factor σB in the newly defined region. Therefore, another protein needs to be recruited to function as a sigma factor. The protein could be either a phage gene product or another bacterial sigma factor.
Fig. 3. Transcriptional analysis of int, cI, and xis in Φ11. 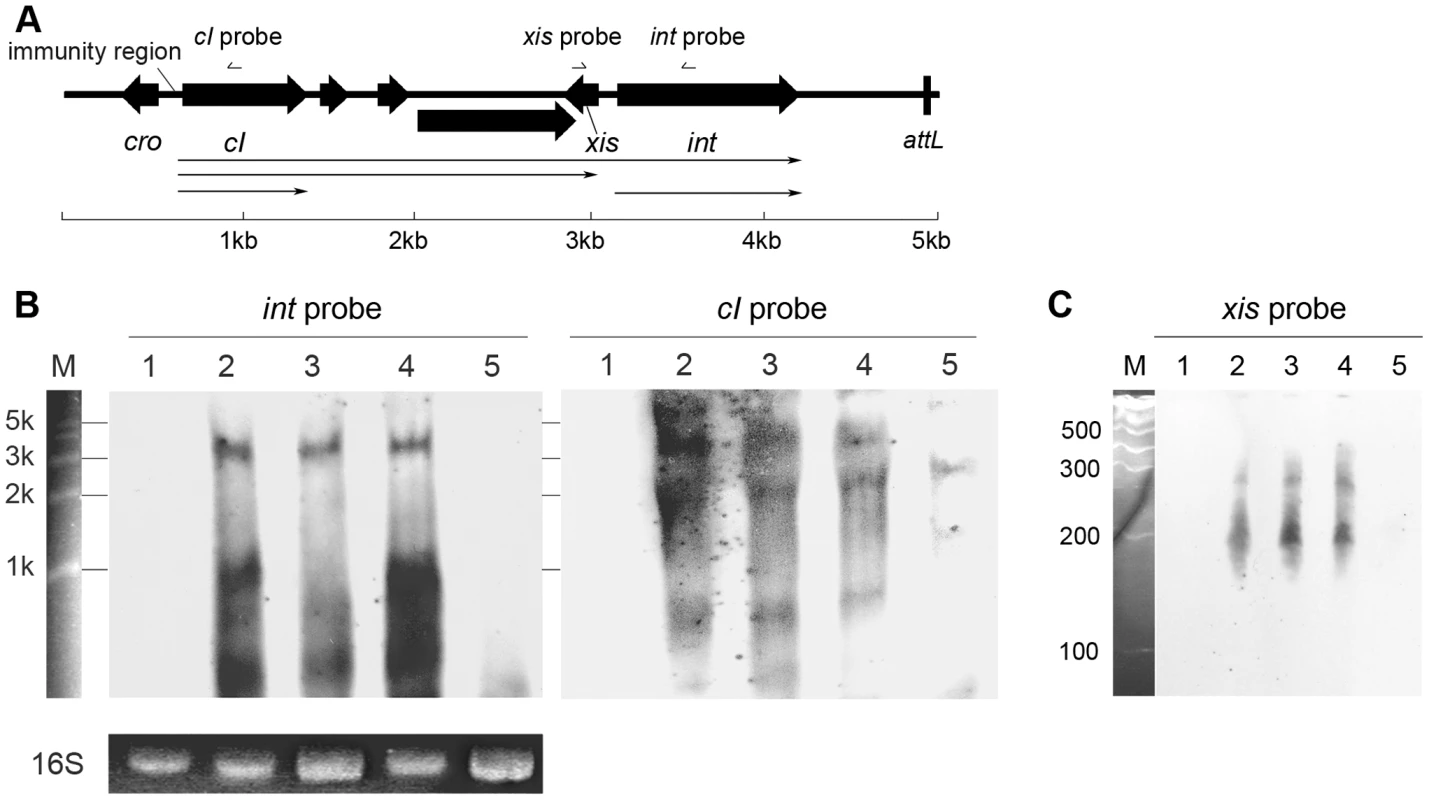
(A) Physical map of the gene organization of the left end of Ф11. The relative positions of the probes used in the Northern blot shown in panel B and C are indicated. The scale indicates the lengths of the genes. (B) Northern blots containing 5 µg of total RNA per lane were probed with Φ11 int probe (left) and reprobed with cI probe (right). Lane 1: phage cured strain RN4220; lane 2: Φ11 lysogen RN4220Φ11; lane 3: sigH mutant strain RN4220ΔsigHΦ11; lane 4: complementary strain RN4220ΔsigHcΦ11; lane 5: RN4220Φ11Δint. Ethidium bromide stained 16S rRNA patterns are shown as an indication of RNA loading. RL6000 (Takara) RNA marker (lane M) was used to estimate the molecular weight of the fragments. (C) Same RNA samples were probed with Φ11 xis probe at low molecular size. Lane 1: RN4220; lane 2: RN4220Φ11; lane 3: RN4220ΔsigHΦ11; lane 4: RN4220ΔsigHcΦ11; lane 5: RN4220Φ11Δint. RL1000 (Takara) RNA marker (lane M) was used to estimate the molecular weight of the fragments. S. aureus sigma factor σH recognized the promoter for int and regulated its transcription
Therefore, we sought to identify this sigma factor in host bacteria. The alternative sigma factor σH was conserved in firmicutes and is known for its sporulation regulation function. Interestingly, staphylococci also express sigH but do not form spores. SigH proteins from staphylococci and other firmicutes were divided into two subgroups according to a previous report [11]. The phylogenetic analysis on sigH orthologs from several firmicutes showed that staphylococcal sigH proteins were separated from the others by a deep node (Figure 2B).
Following this, the sigH gene in S. aureus MW2 [26] genome was knocked out to generate ΔsigH (SUN0802) and a decrease in int mRNA levels of both ΦSa2mw and ΦSa3mw was observed by real-time quantitative reverse transcription polymerase chain reaction (Q-RT-PCR). To determine if this phenomenon was strain-specific, another sigH deletion strain (SUN0806) was built up in S. aureus NCTC8325 [27]. Int mRNA levels of Φ11, Φ12, and Φ13 were all reduced compared with the wild type (WT) (Figure 4). The sigH gene in RN4220 was also knocked out to generate strain SUN0914 for later experiments. To obtain the complementary strains (ΔsigHc), plasmid pMADsigH was introduced into the above sigH-deficient strains and integrated into the host genomes; the backbone of the plasmid pMAD was then eliminated under the screening at 42°C. Endogenous sigH mRNA could be detected by RT-PCR in both WT and ΔsigHc but not in ΔsigH (Figure S4). As expected, int mRNA levels in ΔsigHc were recovered compared with ΔsigH (Figure 4). In the Northern blot assay, the absence of an mRNA signal of about 1-kb for Φ11 integrase was distinguished in ΔsigH compared with the WT and ΔsigHc (Figure 3B). Similarly, the hybridizing signals near 1-kb for Φ12 and Φ13 integrases were not detected in ΔsigH (Figure S2). The transcriptions of the ORF-C/xis gene on the opposite direction seemed to be unaffected without σH according to our Northern blots (Figures 3C and S2). However, when the gene locus of int was deleted, transcripts for ORF-C/xis gene could hardly be detected (Figure 3C), and the transcriptional level of cI also decreased (Figure 3B). These results demonstrated that the S. aureus alternative sigma factor σH plays a role in up-regulating the mRNA levels of prophage integrases. It was also interesting to note that the expression of sigH was growth-phase related (Figure S5).
Fig. 4. Alternative sigma factor σH up-regulates the mRNA levels of prophage integrases. 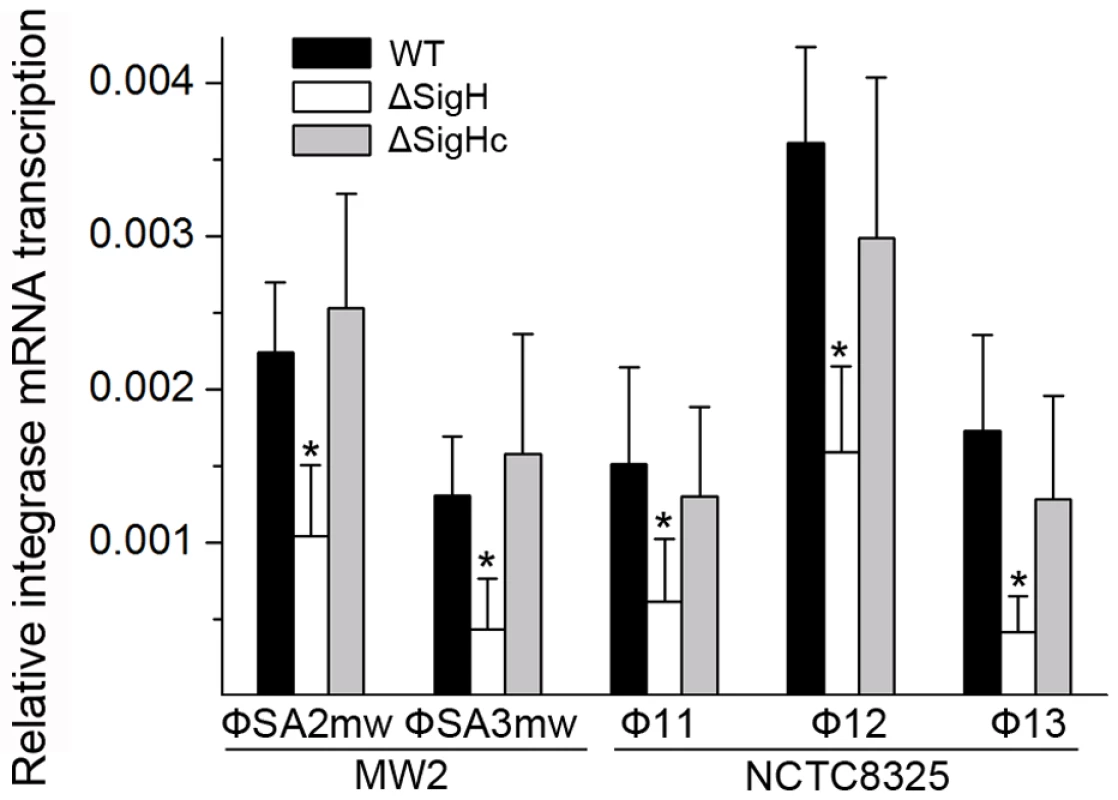
Deletion of sigH causes a reduction of prophage integrase mRNAs. The complementation of sigH gene (ΔsigHc) by transforming pMADsigH that integrated into host genome recovered the transcriptional levels of integrases. The integrases levels are indicated as the relative mRNA levels compared with the 16S RNA control (n = 3 samples/group, *p<0.05). To verify the direct interaction between σH and int promoter region, we conducted σH-directed in vitro transcription by using an amplified DNA fragment from Φ11, which contained the conserved 40-bp region and part of the int ORF as a template. The core RNA polymerase was pre-incubated with σH and then incubated with the linear DNA template. Transcription was initiated by the addition of an NTP mixture containing [α-32P]UTP at 37°C. The σA-dependent promoter of cro (pcro), an important promoter for transcription in the early period of the phage lytic cycle [28], and the S. aureus σA protein were chosen as controls. Core RNA polymerase pre-incubated with σH generated the expected signal at the presence of pint, while no corresponding signals were produced by core RNA polymerase, σA-holoenzyme, or sigma factors alone (Figure 5A). By comparison of the intensity of the signals it was also found that the transcription from this σH-dependent promoter was efficient in vitro. To investigate whether σH-holoenzyme could recognize pint and produce transcripts in vivo, a conditional replication, integration, and modular vector pAH125 [29] was used to detect the recognition of the pint with the presence of σH in an Escherichia coli model. A small DNA fragment containing pint was inserted into pAH125 to build pint-lacZ fusion. The constructed plasmid pAH125pint was then transferred into E. coli strain ZK126 [30] and integrated into the bacterial genome at the attP site of the lambda phage with the help of pINTts [31] to obtain TW0901. Strain TW0902 was later obtained by transferring the plasmid pET22btac-sigH that expresses S. aureus σH protein into TW0901 via electroporation. High activity of β-galactosidase was only exhibited in TW0902 after isopropyl β-D-1-thiogalactopyranoside (IPTG) induction (Figure 5B), confirming that S. aureus σH was specific for the int promoter recognition and gene transcription in vivo.
Fig. 5. Direct evidence of σH-dependent transcription from pint is observed both in vitro and in vivo. 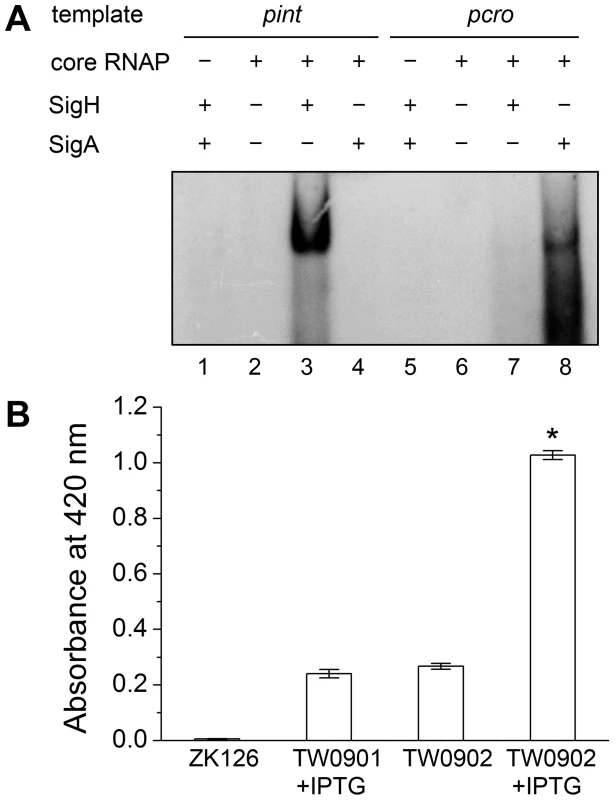
(A) In vitro transcription assay was performed using E. coli core RNA polymerase with or without σH or σA. The absence or presence of core RNAP, σH and σA are indicated. Ten µCi of [α-32P]UTP was used in each sample for radioautography. (B) A pint-lacZ fusion was deployed for the determination of σH-pint recognition in vivo. LacZ-deficient E. coli strain ZK126 was adopted as negative control. Fragment of pint-lacZ was introduced into ZK126 to generate TW0901. Strain TW0902 is derived from TW0901 by adding an IPTG-inducible plasmid pET22btac-sigH that expresses S. aureus σH. High activity of β-galactosidase was only detected in the group with pint-lacZ fusion and expressed S. aureus σH protein (n = 4 samples/group, *p<0.05 versus the controls). The deletion of sigH caused an alteration in the proportion of the excised form of the prophage in S. aureus
Two forms of prophages naturally exist in many lysogenic bacteria, mostly in the integrated form and rarely in the excised form [32], [33], [34], [35], [36]. In S. aureus, we also observed the coexistence of both integrated and excised prophage DNAs by using a set of specially designed PCR primer pairs (Figure 6). Primers check-F and check-R were used to amplify the attB sites in S. aureus genome only if the prophages were excised. Primers check-R and check-IN were used to detect the presence of integrated phage genomes. In addition, by using a real-time Q-PCR method with the above primer pairs the excision frequencies of the staphylococcal prophages were estimated.
Fig. 6. Staphylococcal prophages are mobile on host genomes by dynamic equilibrium of excision and reintegration. 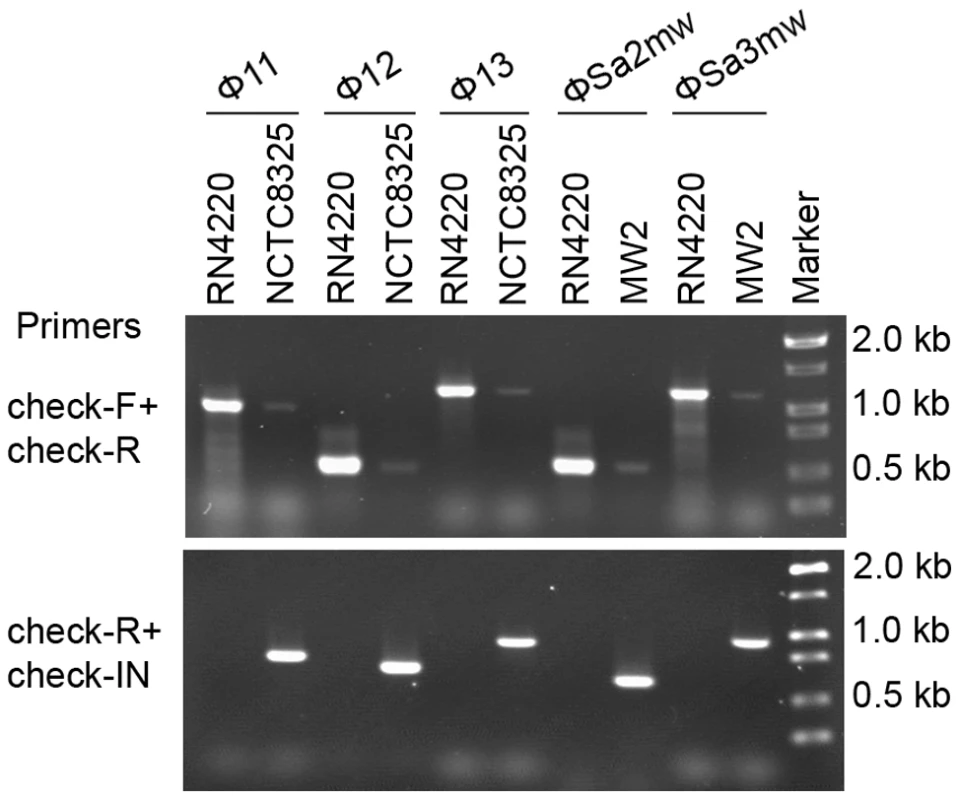
The excision of prophage from host genome is detected by PCR with primers check-Fs and check-Rs from two sides of the attB sites. Primers check-Rs outside the attB sites and check-INs inside the prophages, facing outwards, were used to detect the integrated prophage genomes. Phage-cured strain RN4220 was used as control. The int gene is required for both integration and excision of temperate phages [37] and some pathogenicity islands [38] as previously reported. We have verified the dual functions of int in the S. aureus prophages by constructing int deletion and overexpression strains. The Φ11 int mutant in S. aureus NCTC8325 (SUN0818) was generated to examine the excision of the prophage. No PCR products were observed using SUN0818 genome as templates with primer pair check11-F and check11-R (Figure 7), and the excision event could be recovered by introducing a complementary plasmid into the mutant strain (Figure S6). The Φ11 int overexpression strain was obtained by introducing PLI50Ф11int into RN4220Ф11. The Φ11 excision frequency in the overexpression strain was decreased and a lower titer was detected in the supernatant of the culture as expected (Figure S7), indicating that a higher int mRNA level had a role in stabilizing the lysogeny in the host genome.
Fig. 7. Int is essential for the process of excison. 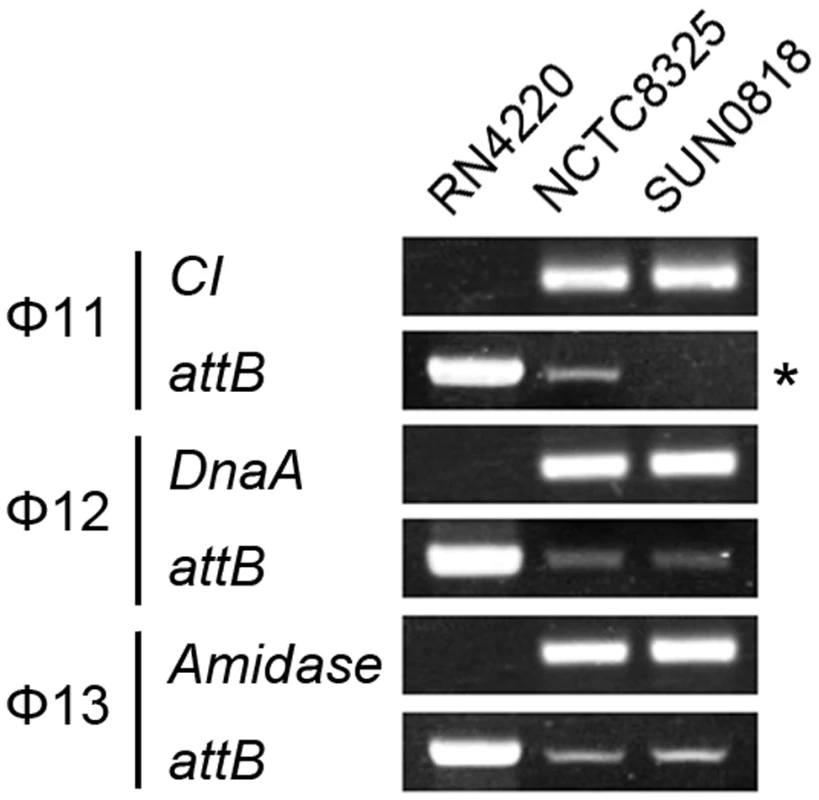
CI (Φ11), dnaA (Φ12), and amidase (Φ13) are phage genes that were used as endogenous controls. The deletion of Φ11 int makes prophage Φ11 immovable on its host genome (indicated by an asterisk) though integrases of Φ12 and Φ13 remain the same. Phage-cured strain RN4220 was used for positive control. Since sigH directly modulates the expression of phage integrase, we suspected that the defection of sigH gene would lead to a change in the number of excised circular phage DNAs. To test this, the proportion of excised prophage genomes in sigH mutant strains was compared to the WT. The proportions of the excised forms of the genomes of all five prophages tested, including ΦSa2mw, ΦSa3mw in S. aureus MW2 and Φ11, Φ12 and Φ13 in S. aureus NCTC8325, were significantly increased. In addition, the prophages excision frequencies were similar in WT and the complementation (Figure 8).
Fig. 8. SigH plays a regulatory role on prophage excision and integration via integrase. 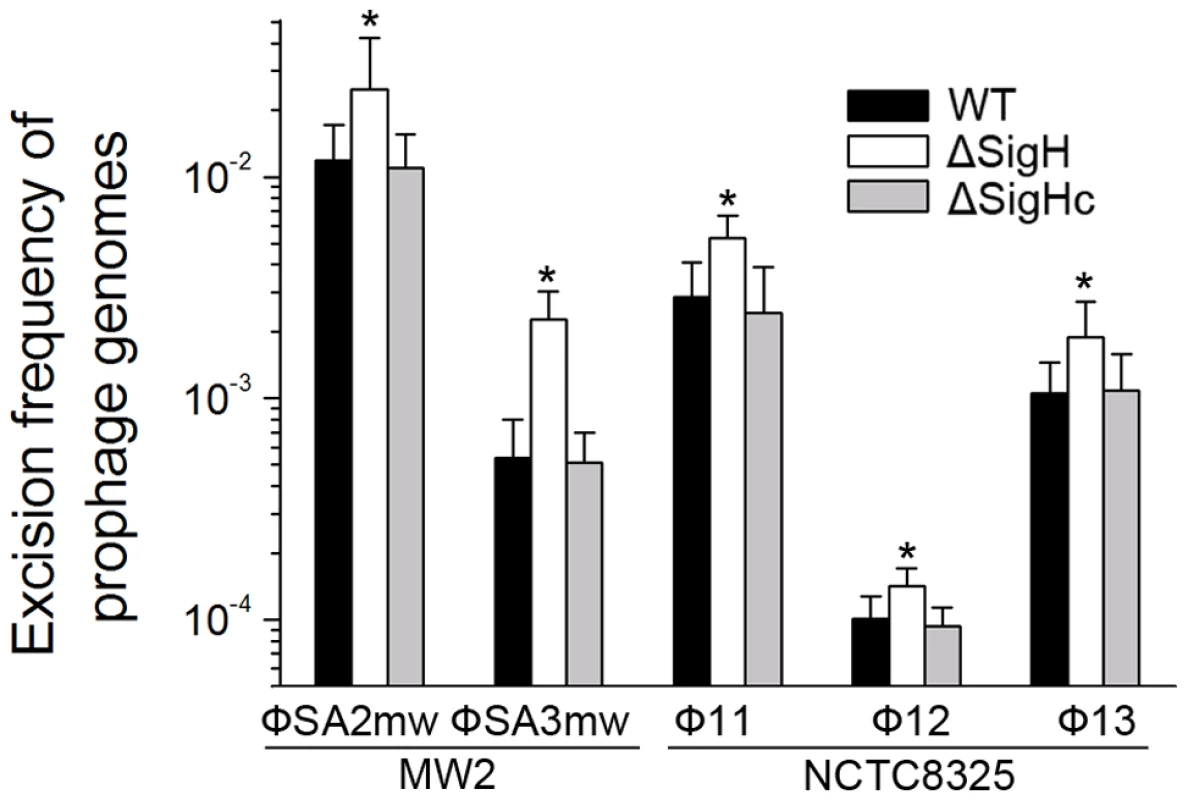
Primer pairs from two sides of attB (check-F and check-R) were used for determination of the ratio of the excised phage genome. Primer pair with one primer outside the att site and the other primer inside the prophage (check-R and check-IN) was used for determination of the ratio of the integrated prophage. SigH deletion resulted in an increase in excised phage genomes. The ratios of excised prophage genomes were similar in WT and the complementation (n = 4 samples/group, *p<0.05). Deletion of sigH altered the rate of spontaneous lysis and lysogenization in S. aureus
Integration and excision events are usually associated with the life cycle of the temperate phages [39]. We estimated the spontaneous lysis rate by determining the titer of free viral particles in S. aureus NCTC8325 culture at the exponential phase. The supernatants of the respective cultures were incubated with the indicator for adsorption and then plated onto agar plates to form plaques. A higher titer was detected in sigH mutant, suggesting that it was easier to naturally induce the prophages to a lytic cycle in the absence of sigH (Figure 9A). Besides, if the cultures were pretreated with ultraviolet (UV) radiation for phage induction, no differences of the titers could be observed between the supernatant of WT, sigH mutant and the complement cultures (Figure 9B). These results implied that sigH did not participate in the phage lytic cycle control.
Fig. 9. SigH deletion leads to an increase in spontaneous lysis. 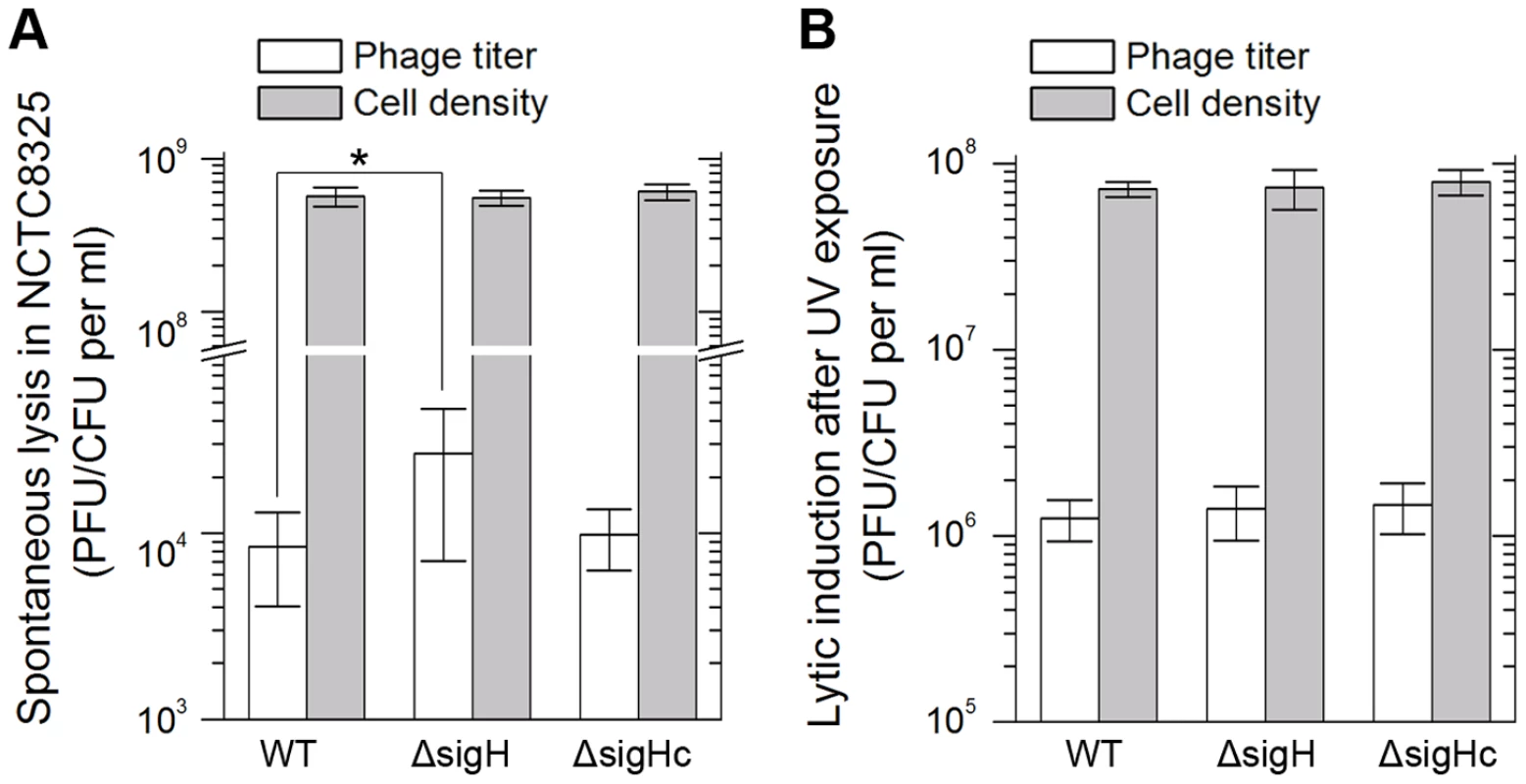
The frequencies of the spontaneous lysis of the lysogens were evaluated by determination of the phage titers of the culture supernatants. Susceptible strain RN4220 was used as the indicator. (A) The titer in the supernatant of the sigH mutant culture was much higher. (B) No difference of the titers among WT, the sigH mutant, and the complement was observed when the cells were pretreated with UV radiation for phage induction. We further investigated the effect of sigH deletion on the lysogenization ability of S. aureus. The susceptible strains RN4220, RN4220ΔsigH (SUN0914), and RN4220ΔSigHc were adsorbed by Ф11 particles with the same multiplicity and plated onto agar plates. The survival clones were checked for lysogeny by cross-streak assays. Fewer lysogens were formed in SUN0914, suggesting that the presence of sigH promoted the lysogenization in S. aureus (Table 1). These results demonstrated that sigH has an auxiliary role in maintaining the lysogeny in S. aureus by up-regulating the int mRNA level.
Tab. 1. Frequencies of lysogenization of <i>S. aureus</i> by Ф11 at the multiplicity of about 100. 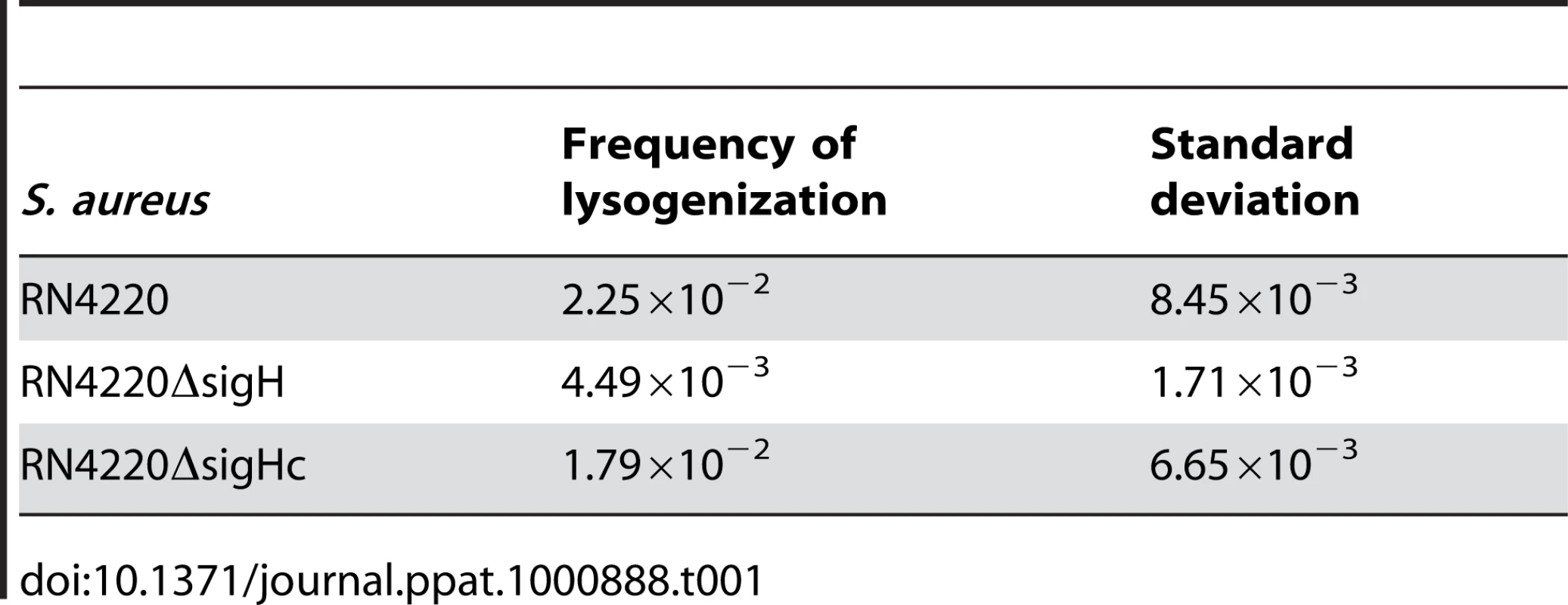
Discussion
Prophages, or lysogenic temperate viruses, provide a large group of virulence factors in bacterial pathogens. In addition, they facilitate the spread of phage genome-encoded or host genome-encoded virulence factors in different bacterial strains or even species [8]. Although the importance of prophage-related virulence in pathogenicity has gradually been realized [40], much is still unknown about how these virulence factors are modulated. The genomes of lysogenic phages do not permanently stay in its host genome. Occasionally, prophage genomes can be excised from bacterial chromosomes and integrated back later [37]. The mobility of a prophage genome usually reflects the activity of the phage although the detailed mechanism remains obscure. These reversible processes are commonly driven by the int-xis system in temperate phages, and some cofactors may participate as well [41].
The int gene is required for the process of integration, while both int and xis are required for the process of excision [37]. Structural analysis of the integrase family showed that their catalytic boxes are conserved. The major differences in the structures among the members of the integrase family mainly affect the specificity and efficiency of the reactions [42]. Although integrase has dual functions of both integration and excision, a higher int mRNA level usually promotes the reaction of integration that is required for lysogeny. The efficiency of the excision process mainly depends on the protein level of excisionase [39].
A similar regulation role was observed in S. aureus prophages. Although the xis genes are absent in some staphylococcal prophages, a gene structure called ORF-C may play a similar role instead [43]. The deletion of the gene locus of int in Ф11 resulted in a permanent lock of the prophage genome onto the host chromosome but not in a loss of the phage, while the excision and integration of Φ12 and Φ13 were not affected. No excision of the Ф11 genome could be detected though integrases of Φ12 and Φ13 still remained. The result reflected the high specificity of the integrases. The frequencies of the spontaneous excisions of prophage genomes varied among different S. aureus prophages. The ratio might partly depend on the sequence of attachment site, although some other factors are involved. For instance, the proportion of the naturally excised genome of ФSa2mw is 100 times of that of Ф12 even though they share the same attB site and integrase.
We uncovered the fact that the host alternative sigma factor σH directly modulated the transcription of S. aureus prophage integrase. The deletion of sigH caused an obvious decrease in integrase expression. This finding was confirmed by experiments both in vitro and in vivo. σH was also found to control the expression of the comG and comE operons, which may play roles in genetic competence in S. aureus [11]. Genetic competence may be defined as a physiological state for taking up high-molecular weight exogenous DNA [44]. But currently there has been no evidence that supports any connections between the phage's life cycle and the competent state of the host. While σA is commonly utilized for promoter recognition at the immunity region, the finding of the recruitment of a second sigma factor by the phage appears especially appealing. Besides, the Northern blot assay showed that integrase could be expressed by another long transcript from a σA-dependent promoter. This result demonstrated that σH presumably affected the transcriptional level of integrase in a mildly regulative manner.
The deletion of sigH would cause an increase in naturally excised phage genomes. Because the process of integration and excision is reversible, the excised forms of phage genomes may reintegrate into the host genome or occasionally enter the lytic cycle. Studies on S. aureus prophage gene arrangement showed that int genes were adjacent to the attL site and usually no ORFs were found downstream in the same direction. And Northern blot assays showed that the transcription on the opposite direction was unaffected in the sigH mutant. Therefore, we deduced that the expression of neighboring genes (i.e. xis/ORF-C) should not be modulated by the sigH gene. It can be postulated that the increase in spontaneous phage excision was more likely a result of a decrease in reintegration rather than an increase in excision due to the reduction of integrase. It was previously reported that lysis genes were more active in excised phage DNA molecules in streptococcal prophages [34]. Here we also found that the expression of cI was decreased in the int mutant probably because of the immobility of the prophage. Besides, the spontaneous lysis rate in the sigH mutant was higher. This phenomenon was probably a consequence of a higher excision frequency of free circular phage genomes. Notably, the transcriptional levels of accessory virulence genes (i.e. sak, sea) were intimately associated with the phage's life cycle as well [28].
A lower rate of lysogenization was observed in the sigH mutant, probably due to the absence of the short transcript. Although the mRNA transcribed from a σA-dependent promoter at the immunity region also expresses integrase, it is postulated that integrase expressed from the short transcript is dominant once a phage infects a bacterial cell because the σH-dependent promoter is much closer to the ORF of int. This mechanism is very similar to the one in the well-studied bacteriophage lambda. A small transcript of int is produced from pRE under the stimulation of protein CІІ, while a long transcript from pL can also express integrase in a stable lysogenic state [45], [46]. Nevertheless, the difference between them is obvious in that CII protein is encoded by a phage gene at early stage to ensure the precise regulation [47] while sigH is a host-source protein. Although the absence of sigH does not cause an intense variation in the prophage characteristics, the presence of sigH has a distinct function in promoting and stabilizing the lysogeny of prophages in S. aureus. This may also explain why the conserved promoter region has been kept in most staphylococcal prophages during evolution but has never been found in other firmicutes harboring phages.
The regulation of sigH remains largely unknown. As an alternative sigma factor, the expression level of sigH would possibly respond to certain cellular conditions and/or variations in the environment. The findings suggest that the recruitment of sigH may provide the staphylococcal temperate phages with an additional strategy to sense the host conditions and/or the change of living environments.
This study could be vital in understanding the temperate phage lysogenic strategy and phage-related virulence. Further investigations would mainly concentrate on which host physiological conditions and/or their circumstance S. aureus sigH would respond to and how that process is regulated.
Materials and Methods
Strains, plasmids, and culture conditions
The bacterial strains, plasmids, and oligonucleotides used in this study are listed in Tables S1, S2, and S3. Bacteria were routinely grown in Luria-Bertani (LB) medium (for E. coli) or Tryptone Soy Broth (TSB, Oxiod) medium (for S. aureus) with aeration (200 rpm) at 37°C. For the antibiotics supplement in cultivation, 100 µg/ml ampicillin, 50 µg/ml kanamycin or 34 µg/ml chloromycetin was used for E. coli strains; 15 µg/ml chloromycetin or 10 µg/ml erythromycin was used for S. aureus strains. All plasmids used in S. aureus were first transferred into strain RN4220 [48] and then transformed into MW2 or NCTC8325 by electroporation.
Genomic DNA extraction, RNA extraction, reverse transcription, quantitative PCR, and Northern blot assay
For genomic DNA or RNA extraction, one colony of each sample was inoculated in 5 ml of TSB medium and incubated at 37°C overnight. Each culture was started by diluting the precultures to an OD600 = 0.05 and was then incubated at 37°C (200 rpm). The cultivation was stopped at early exponential phase (OD600 = 0.2), mid-exponential phase (OD600 = 0.6), late-exponential phase (OD600 = 2.4), and stationary phase (OD600 = 4.0), respectively. S. aureus cells were pre-digested with digestion buffer containing 40 U/ml lysostaphin, 10 mg/ml lysozyme and 10% (v/v) glycerol. Genomic DNA was extracted using the EZ-10 Spin Column Genomic DNA Isolation Kit (Bio Basic Inc.). RNA extraction was performed using the SV Total RNA Isolation System (Promega). Residue DNA in extracted RNA was removed by treatment with 10 U of DNaseІ (Takara) at 37°C for 1 hour. RNA was purified by phenol-chloroform extraction and ethanol precipitation. Purified total RNA and genomic DNA were qualified and quantified by DU730 Nucleic Acid/Protein Analyzer (Beckman Coulter) for reverse transcription, Q-PCR and/or Northern blot assay. Reverse transcription was carried out following the technical manual of ImProm-ІІ Reverse Transcription System (Promega). Q-PCR was performed using StepOne Real-time System (Applied Biosystems). Northern blot assays were performed by using the BrightStar BioDetect Kit (Ambion). Probes for Northern blotting were created by using an Ambion BrightStar Psoralen-Biotin Nonisotopic Labeling Kit.
Primer extension
The promoter region of Φ11 integrase was amplified by PCR with primers pint11-F and pint11-R. The DNA ladder was created using ABI PRISM BigDye Terminators kit with primer pint11-R. 5′-FAM-labelled primer pint11-R-EX was purchased from Sangon (Shanghai, China). For primer extension assay, 50 µg of total RNA of S. aureus NCTC8325 from the exponential phase was used. The reverse transcription was performed with pint11-R-EX at 42°C for 1 hour according to the manufacture's instructions of Primer Extension System AMV Reverse Transcriptase (Promega). The product of reverse transcription and DNA ladder were then separated by capillary electrophoresis and fluorescent signals were collected by ABI3770 sequencer (Applied Biosystems).
Construction of sigH and Φ11 int mutant strains
The sigH and int genes of Φ11 in S. aureus RN4220, MW2 and/or NCTC8325 were knocked out using a temperature sensitive shuttle vector pMAD [49]. DNA sequences of about 600 bps, located at the up - and down-stream of ORFs of target genes, were amplified by PCR and inserted into pMAD consecutively to obtain pMADΔsigH and pMADΔint11. The constructed plasmids were first transformed into S. aureus strain RN4220 and later transferred into MW2 or NCTC8325 by electroporation. Mutant strains were obtained by a two-step screen method as previously described [49]. In the mutants, only the ORFs of target genes were deleted and no extra genes (i.e. antibiotics resistance genes) were introduced into the bacterial genomes.
In vitro transcription
To obtain sigH and sigA proteins for assays in vitro, sigH and sigA genes were amplified from the S. aureus NCTC8325 genome by PCR and inserted into pET22b at the site of NdeІ/XholІ to generate pET22bsigA and pET22bsigH. The plasmids were transformed into the E. coli strain Rosetta (DE3) to express his-tag fusion proteins under the induction of 1 mM IPTG at 16°C. Target proteins were purified from cell lysate by Ni-NTA resin, eluted and then dialyzed to remove imidazole.
The promoter regions of cro and int of Φ11 were amplified with primers pcro11-F, pcro11-R, pint11-F and pint11-R by PCR and purified using QIAquick PCR Purification Kit (Qiagen) to obtain the DNA templates containing pcro or pint.
One microgram of E. coli core RNA polymerase (Epicentre) was pre-incubated with or without sigA or sigH (400 ng) at 37°C for 5 min. Then, 5 µl of DNA template (1 µg), 1 µl of NTP mixture (2.5 mM each), 10 µCi of [α-32P]UTP (5000 Ci/mM) and 40 U of RNase inhibitor (Takara) were added to form a final volume of 50 µl containing 0.04 M Tris-Cl (pH 7.5), 0.15 M KCl, 10 mM MgCl2, 0.01% Triton X-100 and 0.02 M dithiothreitol. The mixture was incubated at 37°C for 20 minutes, and the reaction was stopped by 0.2 M sodium dodecyl sulfate. The reaction products were purified by phenol-chloroform extraction and then ethanol precipitation. The pellet was resuspended in formamide and denatured at 95°C for 5 minutes. Samples were then separated on an 8% polyacrylamide gel with 6 M urea for radioautography.
Detection of σH-pint recognition in vivo
To detect the σH-pint recognition in vivo, a conditional replication and integration plasmid pAH125 was deployed [29]. The promoter region of Φ11 int including SD sequence was amplified with the primers pint11-AH125-F and pint11-AH125-R by PCR. The amplicon was cloned into multiple cloning site of pAH125 at the site of PstІ/EcoRІ to create pint-lacZ fusion. Construction of pAH125pint was operated in the E. coli strain BW25142. Plasmid pAH125pint was transformed into the E. coli strain ZK126 and then integrated into the host genome with the help of pINTts to gain strain TW0901 as previously described [29].
The T7 promoter of pET22bsigH was replaced by tac promoter with lac operator from pGEX-2T at BglІІ/NdeІ site to generate pET22btac-sigH. Strain TW0902 was later obtained by introducing pET22btac-sigH into TW0902.
Overnight grown cultures of ZK126, TW0901, and TW0902 were diluted into 50 ml of LB medium, respectively, with an OD600 = 0.05 to start the incubation at 37°C (200 rpm). After OD600 reached 0.7, the cultures were transferred to 16°C, with or without 1 mM IPTG induction, for an additional 4 hours. The harvest cultures were centrifuged to remove the liquid medium and resuspended in phosphate buffer containing 60 mM Na2HPO4, 40 mM NaH2PO4 (pH 7.0), 10 mM KCl, 1 mM MgSO4 and 50 mM β-mercaptoethanol to an OD600 = 1.0 for lysis. The activity of β-galactosidase was determined as previously described [50].
Phage induction and lysogenization
Phage induction by UV treatment was performed as previously described [51]. One milliliter of culture at exponential phage (OD600 = 0.6) was diluted in 9 ml of chilled phosphate buffered saline (PBS, pH 7.4). The mixture was irradiated with a dose of 50 J/m2 on 9 cm diameter Petri dishes. Two milliliters of 5×TBS was immediately added after the irradiation. The culture was then incubated at 37°C in the dark for an additional 2 hours. Phage titers were determined by the traditional double layer method [52] using RN4220 as the indicator strain.
To obtain the phage lysate from lysogens, 2 mg/ml mitomycin-C was added into the cultures at exponential phase, followed by further incubation for 4 hours [53]. Supernatants were sterilized using 0.45 pore diameter membrane filters (Millipore). Phage particles were concentrated by precipitation with polyethylene glycol as previously described [54].
The phage lysate from NCTC8325 by treatment with mitomycin-C was spotted on the susceptible strain RN4220. Survival clones from the center of the plaques were picked and checked for lysogeny by PCR. RN4220Φ11, RN4220Φ12, and RN4220Ф13 were obtained for subsequent experiments by using this method.
To measure the lysogenization ability of the S. aureus strains, Φ11 lysate with a titer of about 5×108 was prepared. Susceptible cells were mixed with Φ11 particles with a multiplicity of 50 in PBS containing 10 mM Mg2+ at 37°C for 4 hours. The supernatants were then discarded to remove the free viral particles. Pellets were resuspended in PBS and plated onto agar plates. The survival clones were checked for lysogeny by cross-streak assays, which were performed as follows. The bacteriophage lysate was applied as a narrow band across the center of an agar plate, and bacteria were streaked across the dried lysate. After incubation at 37°C for 12 hours, a clear spot was visualized at the cross if the tested bacteria were susceptive cells but not the lysogens.
List of accession numbers
Genbank accession number of S. aureus NCTC8325: NC_007795.
Genbank accession number of S. aureus MW2: NC_003923.
Genbank available phage genomes: NC_004615 (Φ11), NC_004616 (Φ12), NC_004617 (Φ13), NC_007047 (Φ187), NC_007051 (Φ2638A), NC_007055 (Φ37), NC_007053 (Φ3A), NC_007052 (Φ42E), NC_007060 (Φ55), NC_007049 (Φ53), NC_007062 (Φ52A), NC_007061 (Φ29), NC_007048 (Φ69), NC_007059 (Φ71), NC_007050 (Φ85), NC_007063 (Φ88), NC_7064 (Φ92), NC_7057 (Φ96), NC_007065 (ΦX2), NC_005356 (Φ77), NC_009526 (Φ80α), NC_007056 (ΦEW), NC_002951 (ΦCOL), NC_003288 (ΦETA), NC_008798 (ΦETA2), NC_008799 (ΦETA3), NC_010147 (ΦMR11), NC_010808 (ΦMR25), NC_004740 (ΦN315), NC_008583(ΦNM1), NC_008617 (ΦNM3), NC_008689 (ΦPVL108), NC_011612 (ΦSauS-IPLA35), NC_011614 (ΦSauS-IPLA88), NC_002661 (ΦSTL), NC_002321 (ΦPVL), NC_002486 (ΦPV83), NC_007058 (ΦROSA), NC_007055 (Φ37), NC_008722 (ΦCNPH82), NC_008723 (ΦPH15), NC_007168 (ΦSH1), NC_007457 (ΦCherry), NC_009815 (ΦA006), NC_001884 (ΦSPBc2), NC_005822 (ΦLC3), NC_005294 (ΦEJ-1), NC_009819 (ΦP9), NC_003524 (Φ3626), NC_009231 (ΦC2).
Genbank GeneID of S. aureus NCTC8325 sigH: 3920368.
Genbank GeneID of S. aureus MW2 sigH: 1002599.
Genbank GeneID of S. epidermidis RP62A sigW: 3241810.
Genbank GeneID of S. haemolyticus JCSC1435 sigH: 3482668.
Genbank GeneID of B. anthracis Sterne sigH: 2851339.
Genbank GeneID of B. subtilis 168 sigH: 936150.
Genbank GeneID of Clostridium difficile 630 sigH: 4916669.
Genbank GeneID of Clostridium perfringens 13 sigH: 990784.
Genbank GeneID of Listeria monocytogenes Clip81459 sigH: 7703665.
Genbank GeneID of S. aureus NCTC8325 Φ11 int: 1258054.
Supporting Information
Zdroje
1. BoydEF
M.DavisB
HochhutB
2001 Bacteriophage–bacteriophage interactions in the evolution of pathogenic bacteria. Trends Microbiol 9 137 144
2. CanchayaC
FournousG
BrüssowH
2004 The impact of prophages on bacterial chromosomes. Mol Microbiol 53 9 18
3. GoerkeC
PantucekR
HoltfreterS
SchulteB
ZinkM
2009 Diversity of prophages in dominant Staphylococcus aureus clonal lineages. J Bacteriol 191 3462 3468
4. BabaT
BaeT
SchneewindO
TakeuchiF
HiramatsuK
2008 Genome Sequence of Staphylococcus aureus Strain Newman and Comparative Analysis of Staphylococcal Genomes: Polymorphism and Evolution of Two Major Pathogenicity Islands. J Bacteriol 190 300 310
5. BaeT
BabaT
HiramatsuK
SchneewindO
2006 Prophages of Staphylococcus aureus Newman and their contribution to virulence. Mol Microbiol 62 1035 1047
6. Bru″ssowH
CanchayaC
HardtW-D
2004 Phages and the Evolution of Bacterial Pathogens: from Genomic Rearrangements to Lysogenic Conversion. Microbiol Mol Biol Rev 68 560 602
7. HoldenMTG
FeilEJ
LindsayJA
PeacockSJ
DayNPJ
2004 Complete genomes of two clinical Staphylococcus aureus strains: Evidence for the rapid evolution of virulence and drug resistance. PNAS 101 9786 9791
8. ChenJ
NovickRP
2009 Phage-Mediated Intergeneric Transfer of Toxin Genes. Science 323 139 141
9. UbedaC
OlivarezNP
BarryP
WangH
KongX
2009 Specificity of staphylococcal phage and SaPI DNA packaging as revealed by integrase and terminase mutations. Mol Microbiol 72 98 108
10. WuS
HdLencastre
TomaszA
1996 Sigma-B, a putative operon encoding alternate sigma factor of Staphylococcus aureus RNA polymerase: molecular cloning and DNA sequencing. J Bacteriol 178 6036 6042
11. MorikawaK
InoseY
OkamuraH
MaruyamaA
HayashiH
2003 A new staphylococcal sigma factor in the conserved gene cassette: functional significance and implication for the evolutionary processes. Genes Cells 8 699 712
12. ShawLN
LindholmC
PrajsnarTK
MillerHK
BrownMC
2008 Identification and Characterization of sigmaS, a Novel Component of the Staphylococcus aureus Stress and Virulence Responses. PLoS ONE 3 e3844
13. SennMM
GiachinoP
HomerovaD
SteinhuberA
StrassnerJ
2005 Molecular Analysis and Organization of the sigB Operon in Staphylococcus aureus. J Bacteriol 8006 8019
14. Pane'-Farre'J
JonasB
Fo″rstnerK
EngelmannS
HeckerM
2006 The sigmaB regulon in Staphylococcus aureus and its regulation. Int J Medic Microbiol 296 237 258
15. WeirJ
PredichM
DubnauE
NairG
SmithI
1991 Regulation of spo0H, a Gene Coding for the Bacillus subtilis sigmaH Factor. J Bacteriol 173 521 529
16. HatfullGF
2008 Bacteriophage genomics. Curr Opin Microbiol 11 447 453
17. KanekoJ
KimuraT
NaritaS
TomitaT
KamioY
1998 Complete nucleotide sequence and molecular characterization of the temperate staphylococcal bacteriophage φPVL carrying Panton–Valentine leukocidin genes. Gene 215 57 67
18. KwanT
LiuJ
DuBowM
GrosP
PelletierJ
2005 The complete genomes and proteomes of 27 Staphylococcus aureus bacteriophages. PNAS 102 5174 5179
19. NaritaaS
KanekoaJ
ChibaaJ
PiémontbY
JarraudcS
2001 Phage conversion of Panton-Valentine leukocidin in Staphylococcus aureus: molecular analysis of a PVL-converting phage, φSLT. Gene 268 195 206
20. TallentSM
LangstonTB
MoranRG
ChristieGE
2007 Transducing Particles of Staphylococcus aureus Pathogenicity Island SaPI1 Are Comprised of Helper Phage-Encoded Proteins. J Bacteriol 189 7520 7524
21. YamaguchiT
HayashiT
TakamiH
NakasoneK
OhnishiM
2000 Phage conversion of exfoliative toxin A production in Staphylococcus aureus. Mol Microbiol 38 694 705
22. MaXX
ItoT
ChongtrakoolP
HiramatsuK
2006 Predominance of Clones Carrying Panton-Valentine Leukocidin Genes among Methicillin-Resistant Staphylococcus aureus Strains Isolated in Japanese Hospitals from 1979 to 1985. J Clin Microbiol 44 4515 4527
23. LucchiniS
DesiereF
BrussowH
1999 Similarly organized lysogeny modules in temperate Siphoviridae from low GC content gram-positive bacteria. Virology 263 427 435
24. IandoloJJ
WorrellV
GroicherKH
QianY
TianR
2002 Comparative analysis of the genomes of the temperate bacteriophages phi11, phi12 and phi13 of Staphylococcus aureus 8325. Gene 289 109 118
25. ZouD
KanekoJ
NaritaS
KamioY
2000 Prophage, ФPV83-pro, Carrying Panton-Valentine Leukocidin Genes, on the Staphylococcus aureus P83 Chromosome: Comparative Analysis of the Genome Structures of ФPV83-pro, ФPVL, Ф11, and Other Phages. Biosci Biotechnol Biochem 64 2631 2643
26. BabaT
TakeuchiF
Makoto KurodaHY
AokiK
OguchiA
2002 Genome and virulence determinants of high virulence community-acquired MRSA. Lancet 359 1819 1827
27. NovickR
1967 Properties of a cryptic high-frequency transducing phage in Staphylococcus aureus. Virology 33 155 166
28. SumbyP
WaldorMK
2003 Transcription of the Toxin Genes Present within the Staphylococcal Phage ΦSa3ms is Intimately Linked with the Phage's Life Cycle. J Bacteriol 185 6841 6851
29. HaldimannA
WannerBL
2001 Conditional-Replication, Integration, Excision, and Retrieval Plasmid-Host Systems for Gene Structure-Function Studies of Bacteria. J Bacteriol 183 6384 6393
30. ConnellN
HanZ
MorenoF
Kolter.R
1987 An E. coli promoter induced by the cessation of growth. Mol Microbiol 1 195
31. HasanN
KoobM
SzybalskW
1994 Escherichia coli genome targeting, I. Cre-lox-mediated in vitro generation of ori - plasmids and their in vivo chromosomal integration and retrieval. Gene 150 51 56
32. VenturaM
CanchayaC
PridmoreRD
Bru″ssowH
2004 The prophages of Lactobacillus johnsonii NCC 533: comparative genomics and transcription analysis. Virology 320 229 242
33. VenturaM
CanchayaC
BerniniV
AltermannE
BarrangouR
2006 Comparative Genomics and Transcriptional Analysis of Prophages Identified in the Genomes of Lactobacillus gasseri, Lactobacillus salivarius, and Lactobacillus casei. Appl Environ Microbiol 72 3130 3146
34. NeveH
FreudenbergW
Diestel-FeddersenF
EhlertR
HellerKJ
2003 Biology of the temperate Streptococcus thermophilus bacteriophage TP-J34 and physical characterization of the phage genome. Virology 315 184 194
35. DenouE
PridmoreRD
VenturaM
PittetA-C
ZwahlenM-C
2008 The Role of Prophage for Genome Diversification within a Clonal Lineage of Lactobacillus johnsonii: Characterization of the Defective Prophage LJ771. J Bacteriol 190 5806 5813
36. LundeM
BlatnyJM
LillehaugD
AastveitAH
NesIF
2003 Use of Real-Time Quantitative PCR for the Analysis of phiLC3 Prophage Stability in Lactococci. Appl Environ Microbiol 69 41 48
37. NashHA
1981 Integration and Excision of Bacteriophage Lambda: The Mechanism of Conservative Site Specific Recombination. Annu Rev Genet 15 143 167
38. QiuX
GurkarAU
LoryS
2006 Interstrain transfer of the large pathogenicity island (PAPI-1) of Pseudomonas aeruginosa. PNAS 103 19830 19835
39. CraigNL
1988 The Mechanism of Conservative Site-specific Recombination. Annu Rev Genet 22 77 105
40. CasjensS
2003 Prophages and bacterial genomics: what have we learned so far? Mol Microbiol 49 277 300
41. MizuuchiM
MizuuchiK
1980 Integrative recombination of bacteriophage lambda: Extent of the DNA sequence involved in attachment site function. PNAS 77 3220 3224
42. Nunes-DübySE
KwonHJ
TirumalaiRS
EllenbergerT
LandyA
1998 Similarities and differences among 105 members of the Int family of site-specific recombinases. Nucleic Acids Res 26 391 406
43. CarrollD
KehoeMA
CavanaghD
ColemanDC
1995 Novel organization of the site–specific recombination functions of the Staphylococcus aureus serotype F virulence-converting phages phi13 and phi42. Mol Microbiol 16 877 893
44. DubnauD
1991 Genetic Competence in Bacillus subtilis. Microbiol Rev 55 395 424
45. KatzirN
OppenheimA
BerfortM
OppenheimAB
1976 Activation of the Lambda int gene by the cii and ciii gene products. Virology 74 324 331
46. HoY-S
WulffDL
RosenbergM
1983 Bacteriophage λ protein cII binds promoters on the opposite face of the DNA helix from RNA polymerase. Nature 304 703 708
47. OppenheimAB
KobilerO
StavansJ
CourtDL
AdhyaS
2005 Switches in Bacteriophage Lambda Development. Annu Rev Genet 39 409 429
48. KreiswirthBN
LofdahlS
BetleyMJ
O'ReillyM
SchlievertPM
1983 The toxic shock syndrome exotoxin structural gene is not detectably transmitted by a prophage. Nature 305 709 712
49. ArnaudM
ChastanetA
De'barbouilleM
2004 New Vector for Efficient Allelic Replacement in Naturally Nontransformable, Low-GC-Content, Gram-Positive Bacteria. Appl Environ Microbiol 70 6887 6891
50. GerhardtP
KriegNR
KriegNR
MurrayRGE
WoodWA
1994 Methods for General and Molecular Bacteriology: Washington, D.C.: American Society for Microbiology
51. ThompsonJK
HartMGR
1981 Novel Patterns of Ultraviolet Mutagenesis and Weigle Reactivation in Staphylococcus aureus and Phage Ф11. J Gen Microbiol 124 147 157
52. HersheyAD
KalmansonG
BronfenbrennerJ
1943 Quantitative Method in the Study of the Phage-Antiphage Reaction. J Immunol 46 267 279
53. GoerkeC
WirtzC
FlückigerU
WolzC
2006 Extensive phage dynamics in Staphylococcus aureus contributes to adaptation to the human host during infection. Mol Microbiol 61 1673 1685
54. StewartPR
WaldronHG
LeeJS
MatthewsPR
1985 Molecular Relationships Among Serogroup B Bacteriophages of Staphylococcus aureus. J Virol 55 111 116
Štítky
Hygiena a epidemiologie Infekční lékařství Laboratoř
Článek Mouse Senile Amyloid Fibrils Deposited in Skeletal Muscle Exhibit Amyloidosis-Enhancing ActivityČlánek The Role of Intestinal Microbiota in the Development and Severity of Chemotherapy-Induced MucositisČlánek Crystal Structure of HIV-1 gp41 Including Both Fusion Peptide and Membrane Proximal External RegionsČlánek Demonstration of Cross-Protective Vaccine Immunity against an Emerging Pathogenic Ebolavirus Species
Článek vyšel v časopisePLOS Pathogens
Nejčtenější tento týden
2010 Číslo 5- Stillova choroba: vzácné a závažné systémové onemocnění
- Diagnostický algoritmus při podezření na syndrom periodické horečky
- Diagnostika virových hepatitid v kostce – zorientujte se (nejen) v sérologii
- Perorální antivirotika jako vysoce efektivní nástroj prevence hospitalizací kvůli COVID-19 − otázky a odpovědi pro praxi
- První vakcína proti klíšťové encefalitidě: vakcína FSME-IMMUN
-
Všechny články tohoto čísla
- Quorum Sensing Inhibition Selects for Virulence and Cooperation in
- The HMW1C Protein Is a Glycosyltransferase That Transfers Hexose Residues to Asparagine Sites in the HMW1 Adhesin
- Analysis of Virion Structural Components Reveals Vestiges of the Ancestral Ichnovirus Genome
- Mouse Senile Amyloid Fibrils Deposited in Skeletal Muscle Exhibit Amyloidosis-Enhancing Activity
- Global Migration Dynamics Underlie Evolution and Persistence of Human Influenza A (H3N2)
- The Type III Effectors NleE and NleB from Enteropathogenic and OspZ from Block Nuclear Translocation of NF-κB p65
- VEGF Promotes Malaria-Associated Acute Lung Injury in Mice
- Identification of a Mutant PfCRT-Mediated Chloroquine Tolerance Phenotype in
- The Early Stage of Bacterial Genome-Reductive Evolution in the Host
- Host-Detrimental Role of Esx-1-Mediated Inflammasome Activation in Mycobacterial Infection
- Elevation of Intact and Proteolytic Fragments of Acute Phase Proteins Constitutes the Earliest Systemic Antiviral Response in HIV-1 Infection
- The Pleiotropic CymR Regulator of Plays an Important Role in Virulence and Stress Response
- Alternative Sigma Factor σ Modulates Prophage Integration and Excision in
- Effect of Neuraminidase Inhibitor–Resistant Mutations on Pathogenicity of Clade 2.2 A/Turkey/15/06 (H5N1) Influenza Virus in Ferrets
- Massive APOBEC3 Editing of Hepatitis B Viral DNA in Cirrhosis
- NK Cells and γδ T Cells Mediate Resistance to Polyomavirus–Induced Tumors
- Is Genetically Diverse in Animals and Appears to Have Crossed the Host Barrier to Humans on (At Least) Two Occasions
- Adenylate Cyclase Toxin Mobilizes Its β Integrin Receptor into Lipid Rafts to Accomplish Translocation across Target Cell Membrane in Two Steps
- The Role of Intestinal Microbiota in the Development and Severity of Chemotherapy-Induced Mucositis
- HIV-1 Transmitting Couples Have Similar Viral Load Set-Points in Rakai, Uganda
- Few and Far Between: How HIV May Be Evading Antibody Avidity
- Galectin-9/TIM-3 Interaction Regulates Virus-Specific Primary and Memory CD8 T Cell Response
- Perforin Expression Directly by HIV-Specific CD8 T-Cells Is a Correlate of HIV Elite Control
- The Set3/Hos2 Histone Deacetylase Complex Attenuates cAMP/PKA Signaling to Regulate Morphogenesis and Virulence of
- Infidelity of SARS-CoV Nsp14-Exonuclease Mutant Virus Replication Is Revealed by Complete Genome Sequencing
- Combining ChIP-chip and Expression Profiling to Model the MoCRZ1 Mediated Circuit for Ca/Calcineurin Signaling in the Rice Blast Fungus
- Internalin B Activates Junctional Endocytosis to Accelerate Intestinal Invasion
- A Complex Small RNA Repertoire Is Generated by a Plant/Fungal-Like Machinery and Effected by a Metazoan-Like Argonaute in the Single-Cell Human Parasite
- Opc Invasin Binds to the Sulphated Tyrosines of Activated Vitronectin to Attach to and Invade Human Brain Endothelial Cells
- Muc2 Protects against Lethal Infectious Colitis by Disassociating Pathogenic and Commensal Bacteria from the Colonic Mucosa
- PdeH, a High-Affinity cAMP Phosphodiesterase, Is a Key Regulator of Asexual and Pathogenic Differentiation in
- Isolates with Antimony-Resistant but Not -Sensitive Phenotype Inhibit Sodium Antimony Gluconate-Induced Dendritic Cell Activation
- The Microbiota and Allergies/Asthma
- Environmental Factors Determining the Epidemiology and Population Genetic Structure of the Group in the Field
- Prolonged Antigen Presentation Is Required for Optimal CD8+ T Cell Responses against Malaria Liver Stage Parasites
- Crystal Structure of HIV-1 gp41 Including Both Fusion Peptide and Membrane Proximal External Regions
- Susceptibility to Anthrax Lethal Toxin-Induced Rat Death Is Controlled by a Single Chromosome 10 Locus That Includes
- Demonstration of Cross-Protective Vaccine Immunity against an Emerging Pathogenic Ebolavirus Species
- Effective, Broad Spectrum Control of Virulent Bacterial Infections Using Cationic DNA Liposome Complexes Combined with Bacterial Antigens
- High Multiplicity Infection by HIV-1 in Men Who Have Sex with Men
- The -Specific Human Memory B Cell Compartment Expands Gradually with Repeated Malaria Infections
- EBV Promotes Human CD8 NKT Cell Development
- Persistent Growth of a Human Plasma-Derived Hepatitis C Virus Genotype 1b Isolate in Cell Culture
- PLOS Pathogens
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Quorum Sensing Inhibition Selects for Virulence and Cooperation in
- The Role of Intestinal Microbiota in the Development and Severity of Chemotherapy-Induced Mucositis
- Crystal Structure of HIV-1 gp41 Including Both Fusion Peptide and Membrane Proximal External Regions
- Susceptibility to Anthrax Lethal Toxin-Induced Rat Death Is Controlled by a Single Chromosome 10 Locus That Includes
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



