-
Články
- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
A Polyubiquitin Chain Reaction: Parkin Recruitment to Damaged Mitochondria
article has not abstract
Published in the journal: . PLoS Genet 11(1): e32767. doi:10.1371/journal.pgen.1004952
Category: Perspective
doi: https://doi.org/10.1371/journal.pgen.1004952Summary
article has not abstract
Mutations in the E3 ubiquitin ligase Parkin or the mitochondrial kinase PINK1 cause autosomal recessive forms of Parkinson’s disease [1, 2]. Genetic and cell biological studies have implicated PINK1 and Parkin as critical elements in mitophagy, a mitochondrial quality control pathway that involves the ubiquitin-proteasome system (UPS) and the autophagy-lysosomal system [1, 2]. Under basal conditions, PINK1 is processed by mitochondrial proteases and targeted for degradation by the UPS [1, 2]. Following persistent mitochondrial damage (e.g., treatment with the mitochondrial uncoupling agent CCCP) PINK1 is stabilized and accumulates in an active form on the outer mitochondrial membrane [1, 2]. Although PINK1 activity is essential for the mitochondrial recruitment of cytoplasmic Parkin and for the subsequent ubiquitin-dependent clearance of damaged mitochondria, the mode of Parkin activation and recruitment has been elusive [1, 2].
A series of recent papers indicates that PINK1 initiates mitophagy by a two-pronged mechanism involving direct phosphorylation of ubiquitin at serine 65 [3–5] and the ubiquitin-like domain (UbL) of Parkin, also, at serine 65 [6–8] (Fig. 1A, steps 1–2). Biochemical and structural analyses of Parkin demonstrated that the unique Parkin domain (UPD):Rcat interface (previously termed RING0:RING2), the repressor element of Parkin (REP):RING1 interface, and potentially the UbL:RING1 interface mediate autoinhibition of Parkin under steady state conditions in the cell [9–14]. It is tempting to speculate that PINK1 phosphorylation of the Parkin UbL and/or the binding of phosphorylated ubiquitin releases the autoinhibitory elements to allow E2~Ub binding or facilitates conformational rearrangements to confer E2~Ub discharge, ultimately leading to exposure of an optimally aligned Parkin active site. Although the studies on PINK1 phosphorylation of ubiquitin [3–5] and Parkin [6–8] suggest a novel mechanism for PINK1 activation of Parkin, the expression of phosphomimetics of ubiquitin and Parkin was insufficient to promote the mitochondrial recruitment of Parkin [3, 8]. Thus the mechanism underlying PINK1-mediated Parkin recruitment remained a mystery.
Fig. 1. Damage-induced feedforward and positive feedback loops mediate the cellular decision to destroy mitochondria. 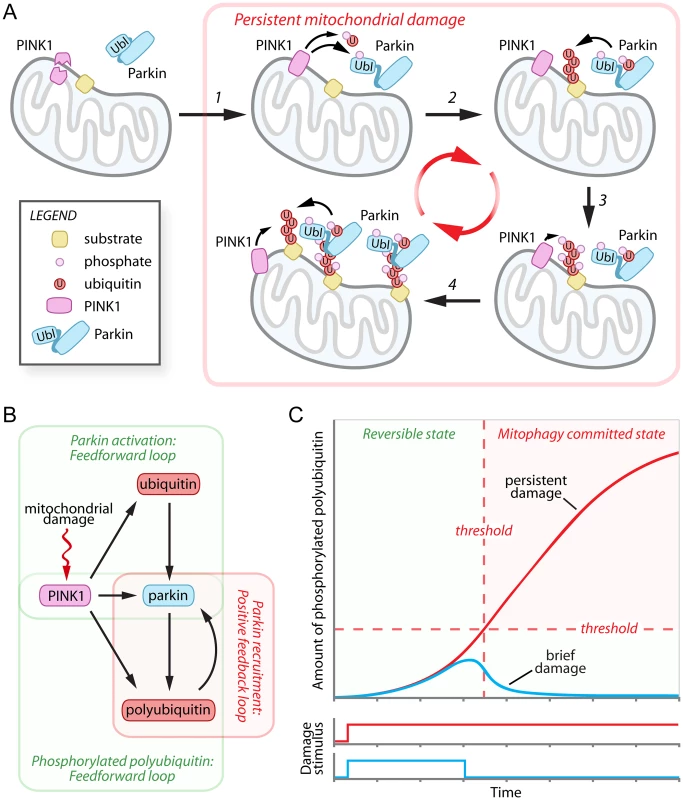
(A) In healthy cells the mitochondrial kinase PINK1 is proteolytically processed and degraded by the UPS. Following mitochondrial damage, PINK1 is stabilized and phosphorylates ubiquitin and the Parkin UbL (step 1), activating Parkin and promoting the polyubiquitination of mitochondrial outer membrane substrates (step 2). The polyubiquitin chains are then phosphorylated by PINK1 (step 3) and mediate the recruitment of additional Parkin to the mitochondria (step 4). (B) Network motifs in the PINK1-Parkin pathway. Mitochondrial damage inhibits PINK1 degradation, initiating dual coherent feedforward loops in the activation of Parkin and generation of phosphorylated polyubiquitin and a positive feedback loop in Parkin recruitment. (C) Hypothetical graph modeling the relationship between mitochondrial damage signal persistence (brief—blue line, and persistent—red line) and the accumulation of phosphorylated polyubiquitin chains during the progression towards mitophagy commitment. UPS, ubiquitin-proteasome system. In December’s issue of PLOS Genetics, Shiba-Fukushima et al. provide compelling data indicating that PINK1 directly phosphorylates polyubiquitin chains to mediate the mitochondrial recruitment and activation of Parkin (Fig. 1A, steps 3–4) [15]. In vitro, affinity purified Parkin from cells bound purified polyubiquitin chains phosphorylated by recombinant PINK1 with a preference for long K63-linked polyubiquitin over K48-linked polyubiquitin chains, although this preference was not recapitulated with Parkin purified from bacteria. To simulate mitochondrial linked polyubiquitin chains, the authors expressed four tandem copies of ubiquitin G76V fused to the mitochondrial targeting sequence of Tom70 [15]. Phosphomimetic (S65E) versions of the tandem polyubiquitin chains were bound by Parkin through its RING1-BRcat domains (previously termed RING1:IBR) and were able to promote stable mitochondrial association of Parkin even in the absence of mitochondrial damage [15]. Both cytosolic phosphomimetic ubiquitin and mitochondrially targeted phosphomimetic tandem polyubiquitin chains were sufficient to activate Parkin, as evidenced by increased Parkin C431S-ubiquitin oxyester formation, but only when the Parkin UbL phosphomimetic was used [15]. To further understand the physiological importance of their findings, Shiba-Fukushima et al. employed PINK1-/ - and Parkin -/ - Drosophila models, which are associated with severe mitochondrial swelling and matrix disorganization and age-dependent motor defects. Strikingly, the expression of mitochondrially targeted phosphomimetic tandem polyubiquitin chains significantly improve both mitochondrial morphology and motor function in PINK1-/ - flies, with little effect on mitochondrial morphology in Parkin -/ - flies [15]. These results are in excellent agreement with a recent publication that employed quantitative proteomics to study PINK1-stimulated Parkin polyubiquitination [16]. Together, these two publications [15, 16] suggest that PINK1 phosphorylation of polyubiquitin is the rate-limiting event required for the mitochondrial recruitment, and potentially also the activation, of Parkin.
The destruction of mitochondria represents an irreversible cellular decision with significant consequences for cellular physiology. The emerging data support a model in which the decision to degrade damaged mitochondria is controlled by dual coherent feedforward loops that precede a positive feedback loop (Fig. 1B). In a feedfoward loop, two input factors, one of which controls the other, jointly regulate a third target factor. In the PINK1-Parkin pathway, the first feedforward loop mediates maximal activation of Parkin by PINK1 phosphorylation of the Parkin UbL and of ubiquitin. The second feedforward loop involves the generation of mitochondrial polyubiquitin chains by PINK1-activated Parkin, and/or another mitochondrial E3 ligase, and their subsequent phosphorylation by PINK1. These phosphorylated polyubiquitin chains appear to be capable of initiating a self-propagating positive feedback loop, recruiting Parkin to the mitochondria and presumably stimulating polyubiquitination of mitochondrial substrates, which can then be phosphorylated by PINK1 and recruit additional Parkin. The organization of these network motifs predicts beneficial features with respect to the decision to degrade mitochondria, including an initial delay period and a mechanism to detect the persistence of mitochondrial damage (Fig. 1C). During the delay period (i.e., low levels of phosphorylated polyubiquitin), the decision to commit to mitophagy would be rapidly reversible, providing a useful means of filtering out brief, low levels of mitochondrial damage signals and preventing unwarranted mitochondrial destruction (Fig. 1C, blue line). Only a persistent damage stimulus that overcomes a specific threshold would be sufficient to initiate the positive feedback loop and commit mitochondria for mitophagy (Fig. 1C, red line). The actions of putative unidentified ubiquitin and Parkin UbL phosphatases, or of mitochondrial deubiquitinating enzymes USP30 [17], USP15 [18], or USP8 [19], which antagonize Parkin-mediated polyubiquitination, would be predicted to regulate the extent of the delay period and the precise commitment threshold. Interestingly, USP30 has been reported to be targeted for UPS degradation by Parkin [17], providing an elegant mechanism to gradually reduce the magnitude of UPS30’s influence during persistent mitochondrial damage.
The new study from Shiba-Fukushima et al. [15] contributes an intriguing model for the role of PINK1 in Parkin recruitment to damaged mitochondria and raises several interesting questions for future investigation. Is an unidentified Parkin UbL and/or ubiquitin phosphatase involved in mitochondrial quality control? The mitochondrial phosphatase PGAM5, which functions downstream of PINK1 [20], is a logical candidate. If polyubiquitination induces the proteasomal degradation of tagged substrates, how does the phospho-polyubiquitin mitochondrial signal persist, are they shielded by phospho-polyubiquitin binding domain-containing proteins or not efficiently recognized by the proteasome? Does Parkin self-association [21] amplify the feedforward loop? How is the binding and exchange of substrates, phospho-polyubiquitin chains and/or phospho-ubiquitin by Parkin coordinated to control the timing of substrate degradation? Finally, it will be imperative to determine the therapeutic potential of small molecule mimetics of phosphorylated ubiquitin (or Parkin UbL) and regulators of the PINK1-Parkin mitochondrial quality control pathway in the search for cures of idiopathic Parkinson’s disease.
Zdroje
1. Narendra D, Walker JE, Youle R (2012) Mitochondrial quality control mediated by PINK1 and Parkin: links to parkinsonism. Cold Spring Harb Perspect Biol 4: a011338. doi: 10.1101/cshperspect.a011338 23125018
2. Winklhofer KF (2014) Parkin and mitochondrial quality control: toward assembling the puzzle. Trends Cell Biol 24 : 332–341. doi: 10.1016/j.tcb.2014.01.001 24485851
3. Koyano F, Okatsu K, Kosako H, Tamura Y, Go E, et al. (2014) Ubiquitin is phosphorylated by PINK1 to activate parkin. Nature 510 : 162–166. 24784582
4. Kazlauskaite A, Kondapalli C, Gourlay R, Campbell DG, Ritorto MS, et al. (2014) Parkin is activated by PINK1-dependent phosphorylation of ubiquitin at Ser65. Biochem J 460 : 127–139. doi: 10.1042/BJ20140334 24660806
5. Kane LA, Lazarou M, Fogel AI, Li Y, Yamano K, et al. (2014) PINK1 phosphorylates ubiquitin to activate Parkin E3 ubiquitin ligase activity. J Cell Biol 205 : 143–153. doi: 10.1083/jcb.201402104 24751536
6. Iguchi M, Kujuro Y, Okatsu K, Koyano F, Kosako H, et al. (2013) Parkin-catalyzed ubiquitin-ester transfer is triggered by PINK1-dependent phosphorylation. J Biol Chem 288 : 22019–22032. doi: 10.1074/jbc.M113.467530 23754282
7. Kondapalli C, Kazlauskaite A, Zhang N, Woodroof HI, Campbell DG, et al. (2012) PINK1 is activated by mitochondrial membrane potential depolarization and stimulates Parkin E3 ligase activity by phosphorylating Serine 65. Open Biol 2 : 120080. doi: 10.1098/rsob.120080 22724072
8. Shiba-Fukushima K, Imai Y, Yoshida S, Ishihama Y, Kanao T, et al. (2012) PINK1-mediated phosphorylation of the Parkin ubiquitin-like domain primes mitochondrial translocation of Parkin and regulates mitophagy. Sci Rep 2 : 1002. doi: 10.1038/srep01002 23256036
9. Chaugule VK, Burchell L, Barber KR, Sidhu A, Leslie SJ, et al. (2011) Autoregulation of Parkin activity through its ubiquitin-like domain. EMBO J 30 : 2853–2867. doi: 10.1038/emboj.2011.204 21694720
10. Riley BE, Lougheed JC, Callaway K, Velasquez M, Brecht E, et al. (2013) Structure and function of Parkin E3 ubiquitin ligase reveals aspects of RING and HECT ligases. Nat Commun 4 : 1982. doi: 10.1038/ncomms2982 23770887
11. Spratt DE, Martinez-Torres RJ, Noh YJ, Mercier P, Manczyk N, et al. (2013) A molecular explanation for the recessive nature of parkin-linked Parkinson's disease. Nat Commun 4 : 1983. doi: 10.1038/ncomms2983 23770917
12. Spratt DE, Walden H, Shaw GS (2014) RBR E3 ubiquitin ligases: new structures, new insights, new questions. Biochem J 458 : 421–437. doi: 10.1042/BJ20140006 24576094
13. Trempe JF, Sauve V, Grenier K, Seirafi M, Tang MY, et al. (2013) Structure of parkin reveals mechanisms for ubiquitin ligase activation. Science 340 : 1451–1455. doi: 10.1126/science.1237908 23661642
14. Wauer T, Komander D (2013) Structure of the human Parkin ligase domain in an autoinhibited state. EMBO J 32 : 2099–2112. doi: 10.1038/emboj.2013.125 23727886
15. Shiba-Fukushima K, Arano T, Matsumoto G, Inoshita T, Yoshida S, et al. (2014) Phosphorylation of Mitochondrial Polyubiquitin by PINK1 Promotes Parkin Mitochondrial Tethering. PLoS Genet 10: e1004861. doi: 10.1371/journal.pgen.1004861 25474007
16. Ordureau A, Sarraf SA, Duda DM, Heo JM, Jedrychowski MP, et al. (2014) Quantitative Proteomics Reveal a Feedforward Mechanism for Mitochondrial PARKIN Translocation and Ubiquitin Chain Synthesis. Mol Cell 56 : 360–375. doi: 10.1016/j.molcel.2014.09.007 25284222
17. Bingol B, Tea JS, Phu L, Reichelt M, Bakalarski CE, et al. (2014) The mitochondrial deubiquitinase USP30 opposes parkin-mediated mitophagy. Nature 510 : 370–375. 24896179
18. Cornelissen T, Haddad D, Wauters F, Van Humbeeck C, Mandemakers W, et al. (2014) The deubiquitinase USP15 antagonizes Parkin-mediated mitochondrial ubiquitination and mitophagy. Hum Mol Genet 23 : 5227–5242. doi: 10.1093/hmg/ddu244 24852371
19. Durcan TM, Tang MY, Perusse JR, Dashti EA, Aguileta MA, et al. (2014) USP8 regulates mitophagy by removing K6-linked ubiquitin conjugates from parkin. EMBO J 33 : 2473–2491. doi: 10.15252/embj.201489729 25216678
20. Imai Y, Kanao T, Sawada T, Kobayashi Y, Moriwaki Y, et al. (2010) The loss of PGAM5 suppresses the mitochondrial degeneration caused by inactivation of PINK1 in Drosophila. PLoS Genet 6: e1001229. doi: 10.1371/journal.pgen.1001229 21151955
21. Lazarou M, Narendra DP, Jin SM, Tekle E, Banerjee S, et al. (2013) PINK1 drives Parkin self-association and HECT-like E3 activity upstream of mitochondrial binding. J Cell Biol 200 : 163–172. doi: 10.1083/jcb.201210111 23319602
Štítky
Genetika Reprodukční medicína
Článek Phosphorylation of Elp1 by Hrr25 Is Required for Elongator-Dependent tRNA Modification in YeastČlánek Naturally Occurring Differences in CENH3 Affect Chromosome Segregation in Zygotic Mitosis of HybridsČlánek Insight in Genome-Wide Association of Metabolite Quantitative Traits by Exome Sequence AnalysesČlánek ALIX and ESCRT-III Coordinately Control Cytokinetic Abscission during Germline Stem Cell DivisionČlánek Deciphering the Genetic Programme Triggering Timely and Spatially-Regulated Chitin Deposition
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2015 Číslo 1- Akutní intermitentní porfyrie
- Farmakogenetické testování pomáhá předcházet nežádoucím efektům léčiv
- Růst a vývoj dětí narozených pomocí IVF
- Pilotní studie: stres a úzkost v průběhu IVF cyklu
- Intrauterinní inseminace a její úspěšnost
-
Všechny články tohoto čísla
- The Combination of Random Mutagenesis and Sequencing Highlight the Role of Unexpected Genes in an Intractable Organism
- Ataxin-3, DNA Damage Repair, and SCA3 Cerebellar Degeneration: On the Path to Parsimony?
- α-Actinin-3: Why Gene Loss Is an Evolutionary Gain
- Origins of Context-Dependent Gene Repression by Capicua
- Transposable Elements Contribute to Activation of Maize Genes in Response to Abiotic Stress
- No Evidence for Association of Autism with Rare Heterozygous Point Mutations in Contactin-Associated Protein-Like 2 (), or in Other Contactin-Associated Proteins or Contactins
- Nur1 Dephosphorylation Confers Positive Feedback to Mitotic Exit Phosphatase Activation in Budding Yeast
- A Regulatory Hierarchy Controls the Dynamic Transcriptional Response to Extreme Oxidative Stress in Archaea
- Genetic Variants Modulating CRIPTO Serum Levels Identified by Genome-Wide Association Study in Cilento Isolates
- Small RNA Sequences Support a Host Genome Origin of Satellite RNA
- Phosphorylation of Elp1 by Hrr25 Is Required for Elongator-Dependent tRNA Modification in Yeast
- Genetic Mapping of MAPK-Mediated Complex Traits Across
- An AP Endonuclease Functions in Active DNA Demethylation and Gene Imprinting in
- Developmental Regulation of the Origin Recognition Complex
- End of the Beginning: Elongation and Termination Features of Alternative Modes of Chromosomal Replication Initiation in Bacteria
- Naturally Occurring Differences in CENH3 Affect Chromosome Segregation in Zygotic Mitosis of Hybrids
- Imputation of the Rare G84E Mutation and Cancer Risk in a Large Population-Based Cohort
- Polycomb Protein SCML2 Associates with USP7 and Counteracts Histone H2A Ubiquitination in the XY Chromatin during Male Meiosis
- A Genetic Strategy for Probing the Functional Diversity of Magnetosome Formation
- Interactions of Chromatin Context, Binding Site Sequence Content, and Sequence Evolution in Stress-Induced p53 Occupancy and Transactivation
- The Yeast La Related Protein Slf1p Is a Key Activator of Translation during the Oxidative Stress Response
- Integrative Analysis of DNA Methylation and Gene Expression Data Identifies as a Key Regulator of COPD
- Proteasomes, Sir2, and Hxk2 Form an Interconnected Aging Network That Impinges on the AMPK/Snf1-Regulated Transcriptional Repressor Mig1
- Functional Interplay between the 53BP1-Ortholog Rad9 and the Mre11 Complex Regulates Resection, End-Tethering and Repair of a Double-Strand Break
- Estrogenic Exposure Alters the Spermatogonial Stem Cells in the Developing Testis, Permanently Reducing Crossover Levels in the Adult
- Protein Poly(ADP-ribosyl)ation Regulates Immune Gene Expression and Defense Responses
- Sumoylation Influences DNA Break Repair Partly by Increasing the Solubility of a Conserved End Resection Protein
- A Discrete Transition Zone Organizes the Topological and Regulatory Autonomy of the Adjacent and Genes
- Elevated Mutation Rate during Meiosis in
- The Intersection of the Extrinsic Hedgehog and WNT/Wingless Signals with the Intrinsic Hox Code Underpins Branching Pattern and Tube Shape Diversity in the Airways
- MiR-24 Is Required for Hematopoietic Differentiation of Mouse Embryonic Stem Cells
- Tissue-Specific Effects of Genetic and Epigenetic Variation on Gene Regulation and Splicing
- Heterologous Aggregates Promote Prion Appearance via More than One Mechanism
- The Tumor Suppressor BCL7B Functions in the Wnt Signaling Pathway
- , A -Acting Locus that Controls Chromosome-Wide Replication Timing and Stability of Human Chromosome 15
- Regulating Maf1 Expression and Its Expanding Biological Functions
- A Polyubiquitin Chain Reaction: Parkin Recruitment to Damaged Mitochondria
- RecFOR Is Not Required for Pneumococcal Transformation but Together with XerS for Resolution of Chromosome Dimers Frequently Formed in the Process
- An Intracellular Transcriptomic Atlas of the Giant Coenocyte
- Insight in Genome-Wide Association of Metabolite Quantitative Traits by Exome Sequence Analyses
- The Role of the Mammalian DNA End-processing Enzyme Polynucleotide Kinase 3’-Phosphatase in Spinocerebellar Ataxia Type 3 Pathogenesis
- The Global Regulatory Architecture of Transcription during the Cell Cycle
- Identification and Functional Characterization of Coding Variants Influencing Glycemic Traits Define an Effector Transcript at the Locus
- Altered Ca Kinetics Associated with α-Actinin-3 Deficiency May Explain Positive Selection for Null Allele in Human Evolution
- Genetic Variation in the Nuclear and Organellar Genomes Modulates Stochastic Variation in the Metabolome, Growth, and Defense
- PRDM9 Drives Evolutionary Erosion of Hotspots in through Haplotype-Specific Initiation of Meiotic Recombination
- Transcriptional Control of an Essential Ribozyme in Reveals an Ancient Evolutionary Divide in Animals
- ALIX and ESCRT-III Coordinately Control Cytokinetic Abscission during Germline Stem Cell Division
- Century-scale Methylome Stability in a Recently Diverged Lineage
- A Re-examination of the Selection of the Sensory Organ Precursor of the Bristle Sensilla of
- Antagonistic Cross-Regulation between Sox9 and Sox10 Controls an Anti-tumorigenic Program in Melanoma
- A Dependent Pool of Phosphatidylinositol 4,5 Bisphosphate (PIP) Is Required for G-Protein Coupled Signal Transduction in Photoreceptors
- Deciphering the Genetic Programme Triggering Timely and Spatially-Regulated Chitin Deposition
- Aberrant Gene Expression in Humans
- Fascin1-Dependent Filopodia are Required for Directional Migration of a Subset of Neural Crest Cells
- The SWI2/SNF2 Chromatin Remodeler BRAHMA Regulates Polycomb Function during Vegetative Development and Directly Activates the Flowering Repressor Gene
- Evolutionary Constraint and Disease Associations of Post-Translational Modification Sites in Human Genomes
- A Truncated NLR Protein, TIR-NBS2, Is Required for Activated Defense Responses in the Mutant
- The Genetic and Mechanistic Basis for Variation in Gene Regulation
- Inactivation of PNKP by Mutant ATXN3 Triggers Apoptosis by Activating the DNA Damage-Response Pathway in SCA3
- DNA Damage Response Factors from Diverse Pathways, Including DNA Crosslink Repair, Mediate Alternative End Joining
- hnRNP K Coordinates Transcriptional Silencing by SETDB1 in Embryonic Stem Cells
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- The Global Regulatory Architecture of Transcription during the Cell Cycle
- A Truncated NLR Protein, TIR-NBS2, Is Required for Activated Defense Responses in the Mutant
- Proteasomes, Sir2, and Hxk2 Form an Interconnected Aging Network That Impinges on the AMPK/Snf1-Regulated Transcriptional Repressor Mig1
- The SWI2/SNF2 Chromatin Remodeler BRAHMA Regulates Polycomb Function during Vegetative Development and Directly Activates the Flowering Repressor Gene
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



