-
Medical journals
- Career
NMR and IR analysis of natural substances isolated from Cordyceps medicinal mushrooms
Authors: Jarmila Harvanová; Lucia Ungvarská Maľučká; Anna Uhrinová; Mária Vilková; Martin Vavra; Martin Pavlík; Martin Rajtar; Anna Furmaníková
Published in: Čes. slov. Farm., 2018; 67, 200-204
Category: Original Article
Overview
There exist about 750 species of Cordyceps at present. Ahigh price of natural Cordyceps and its lack in nature caused that the attention has been focused to its cultivation in laboratory conditions. The demand for this fungus-parasite is still quite high nowadays, as shown by the amount of commercial nutritional supplements. Phytochemical diversity has ensured that Cordyceps is used as an immunomodulatory and an antioxidant; it has anti-cancer, anti-inflammatory, anti-diabetic, antibacterial, anti-HIV effects. In the present study we focused on NMR and IR analyses of natural substances isolated from two species of Cordyceps: Cordyceps sinensis MFTCCB025/0216, MFTCCB026/0216 and Paecilomyces hepiali MFTCCB023/0216. Two types of rice substrates (Oryza sativa Indica and Oryza sativa Japonica) were used for cultivation. A total of five methanol extracts obtained by a reflux method of the ground mushroom were analysed. To determine the quality and quantity of the major chemical compounds, 1D and 2D NMR analysis has been used with 1H, 13C, COSY, NOESY, HSQC, HMBC and DEPT spectra. IR spectroscopy was chosen as a complementary analysis to determine functional groups. Linoleic acid, oleic acid and mannitol were identified as major compounds of the methanol extracts. Tyrosine, alanine, urea and the others biologically interesting substances were found as minor components.
Keywords:
Cordyceps sinensis – NMR and IR analysis – Oleic acid – D-mannitol
Introduction
Species of the genus Cordyceps (Fr.) Link are fungi that live on larvae, hoods and images of insects or on the breeds of Elaphomyces1). Western civilization has been interested in these mushrooms only for the past 30 years, and more and more scientific evidence of the possibilities of their use in medicine has been seen, although in traditional Chinese medicine it has been used for centuries2). As a part of traditional Chinese medicine, Cordyceps has been used in respiratory problems, kidney, liver and heart problems, hyposexuality, hyperlipidemia, and to combat weakening of the immune system. It has been also used as a complement to modern cancer treatments (chemotherapy, radiation and surgery)3). Modern science has confirmed the effectiveness of Cordyceps fungus treatment in most commonly used traditional medicine practices4). Modern pharmaceutical studies show that Cordyceps has a really beneficial effect on several biological systems, immune, cardiovascular, respiratory, and glandular5). A number of scientific works have described the clinical use of this parasitic fungus3, 6) with subsequent biological and pharmacological effects7). Various chemical compounds identified in these fungi, such as cordycepin, cordycepic acid, D-mannitol8–10), polysaccharides11, 12), nucleotides13), proteins and amino acids14), and unsaturated fatty acids15) are responsible for a wide range of their biological activity.
This study deals with the determination of the chemical structure of the compounds with biological activity in methanol extracts of Cordyceps and Paecilomyces hepiali, cultivated on two types of rice (Oryza sativa Japonica and Indica). Modern spectroscopic methods, such as nuclear magnetic resonance (NMR) and infrared spectroscopy (IR), were used to analyse the extracts.
Experimental part
Five specimens of Cordyceps mushrooms cultured on two types of rice substrate were analysed: Cordyceps sinensis MFTCCB026/0216 (Oryza sativa Japonica) specimen 1, C. sinensis MFTCCB026/0216 (O. sativa Indica) specimen 2, C. sinensis MFTCCB025/0216 (O. sativa Indica) specimen 3, Paeciliomyces hepiali MFTCCB023/0216 (O. sativa Japonica) specimen 4, and P. hepiali MFTCCB023/0216 (O. sativa Indica) 5. Mushroom samples were prepared in cooperation with Ing. Martin Pavlík, Ph.D. from the Technical University in Zvolen, Faculty of Forestry. Methanol (p.a., Sigma-Aldrich) was used to extract the crude, dried and milled material from samples 1–5. A rotary vacuum evaporator IKA, VWR, RV10 control was used to evaporate and thicken the extracts. NMR spectra were measured on a Varian VNMRS 600 MHz spectrometer in deuterated dimethyl sulfoxide DMSO-d6 (Merck). Infrared spectra were measured on a Thermo Scientific spectrophotometer, a NICOLET 6700 FT-IR, an ATR method on a Smart Orbit extension.
Sample preparation
The production strains came from the collections of Aloha Medicinals (USA) and Mykoforest Velčice (Slovakia). Mushroom cultures were maintained on agar and used to inoculate rice-based substrates (O. sativa Indica and O. sativa Japonica) which were sterilized in an autoclave at 121 °C at 100 kPa for 2 hours after cooking in nutrient solutions. After cooling down to room temperature, the substrates were inoculated with 3 ml of the inoculum solution and with three pieces of this 4 × 4 mm fungi from a Petri dish. Cultivation was carried out in a shaker (120 beats per minute) for 7 days. 200 mL of the nutrient solution was placed in the incubator for 30 days at 22 °C and the light/dark cycle for 12/12 hours. Afterwards, the substrate was dried in a drier at 40 °C and milled with a laboratory mill.
In total, five samples were labelled as 1‒5. The samples 1‒5 (10 g) were macerated in 50 ml of 99.9% methanol for six hours in the dark. After maceration, an additional 100 mL of methanol was added and the samples were refluxed for 4 hours, filtered and evaporated to dryness on a rotary vacuum evaporator. The yield of methanol extracts was 6.38 % for sample 1; 2.77% for sample 2; 8.10% for sample 3; 7.00% for sample 4; and 8.61% for sample 5, respectively. 30 mg of all extracts were dissolved in DMSO-d6 (0.6 mL) and were analysed spectroscopically. 1H, 13C NMR spectra were measured on a 600 MHz NMR spectrometer at room temperature. The data obtained are as follows: chemical shift in δ-scale (ppm), signal multiplicity, integrated intensity and interaction constants J in Hz. Proton and carbon assignment was performed on the basis of 1H, 13C, COSY, HSQC, HMBC and DEPT spectra. Infrared spectra were measured by the ATR method on the Smart Orbit extension. The crude, evaporated extract was transferred to the Smart Orbit measuring point. The analysed extracts were not modified before the measurement.
Results and discussion
NMR analysis
Based on the detailed analysis of 1D and 2D NMR spectra, unsaturated fatty acids (Z-oleic acid and linoleic acid) were found in all five samples. The chemical structures of Z-oleic acid, linoleic acid and D-mannitol were determined by a detailed and demanding analysis of the COSY spectra (Fig. 1), where the spin systems of all three chemical compounds were determined. The HSQC spectra analysis followed, whereby the carbon atoms signals were assigned to the respective proton of the hydrogen, based on the transfer of the magnetization from the protons to the carbon atoms via one bond. At 13C NMR spectra were assigned carbon signals that carry protons. Then HMBC spectra were analysed, in which the quaternary carbon atoms in Z-oleic and linoleic acid were assigned, based on the transfer of the magnetization from the protons to the carbon atoms via three bonds. Also, this experiment reaffirmed the correct assignment of proton and carbon signals in each of the identified substances.
1. Assignment of spin systems of Z-oleic acid, linoleic and D-mannitol based on COSY spectra sample number 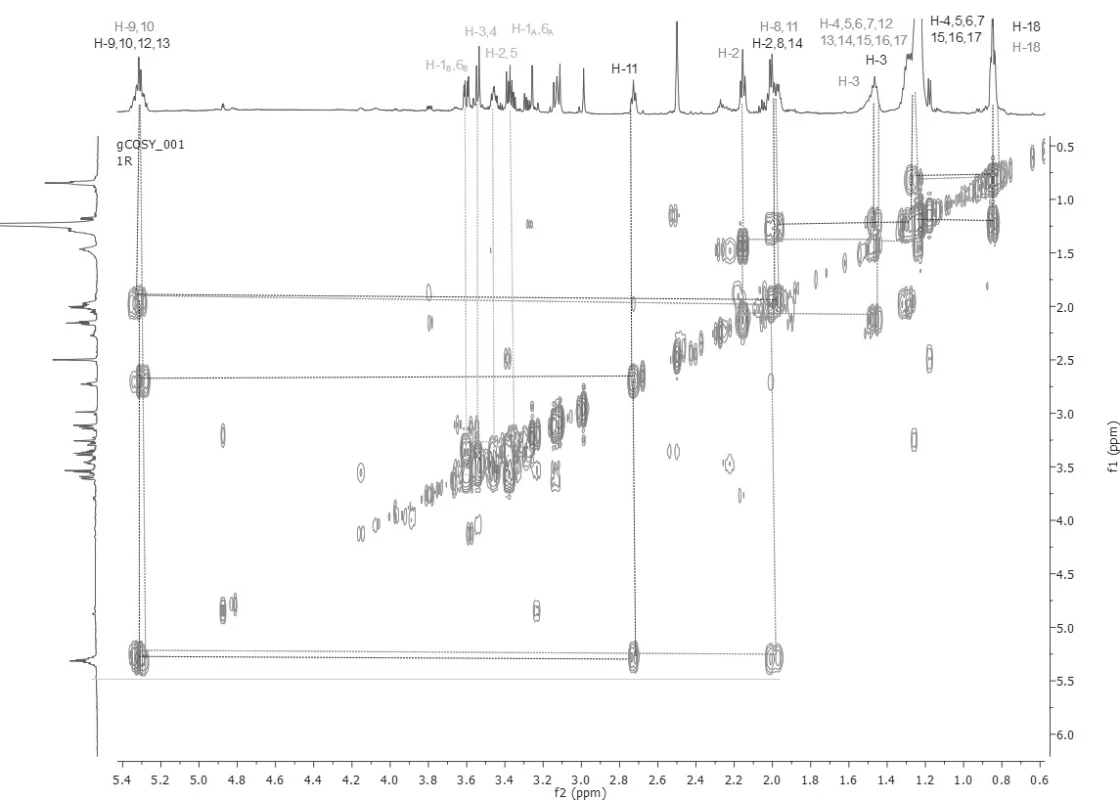
To determine the quantity (%) and the ratio of Z-oleic acid, linoleic acid and D-mannitol, proton spectra were used to compare the integrated intensity of the distinct proton signals of the individual chemical compounds (Table 1). All samples had almost double the content of Z-oleic acid compared to linoleic acid, except sample number 4, P. hepiali cultivated on O. sativa Japonica. Sample No. 1 C. sinensis cultivated on O. sativa Japonica and sample No. 2 cultivated on O. sativa Indica also contained D-mannitol in contrast to samples 3, 4 and 5.
1. Integrated intensity and percentage of Z-oleic acid, linoleic acid and D-mannitol in samples of extracts 1‒5 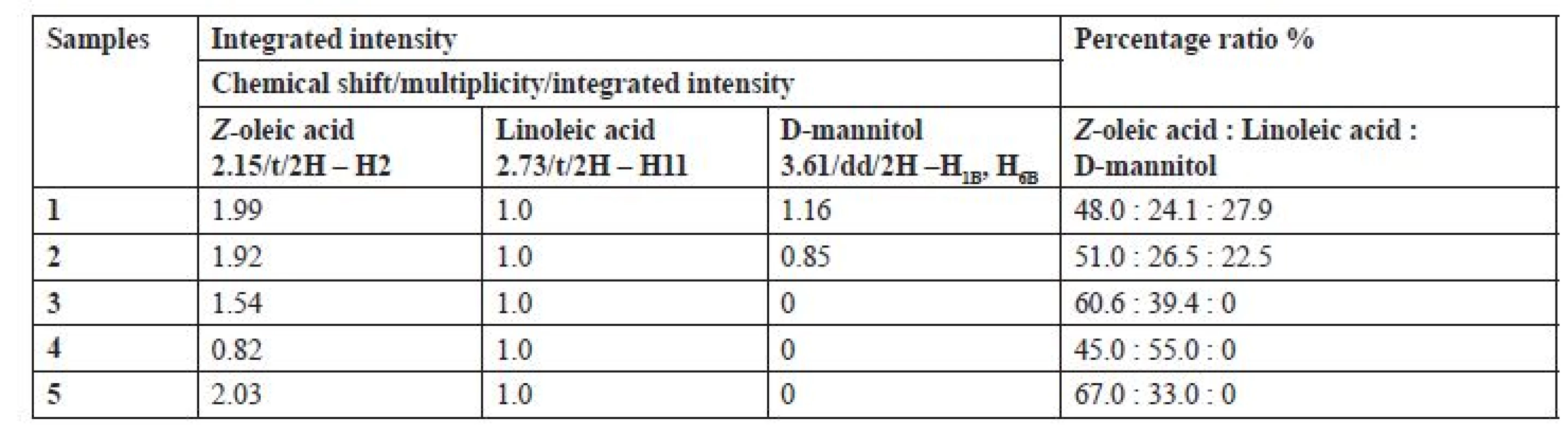
As shown in Table 1, sample extract nos. 3, 4 and 5 did not contain D-mannitol, probably due to strain variation between the individual specimens. In the case of C. sinensis 1 and 2 extracts only slight differences in the ratio of Z-oleic acid, linoleic acid and D-mannitol were observed. A more significant difference has been seen in the P. hepiali strain, where the substrate effect on the unsaturated fatty acids was more evident. Extract number 4 of P. hepiali cultivated on O. sativa Japonica contained 1.2-times more linoleic acid, whereas extract 5 cultivated on O. sativa Indica contained 2-times more Z-oleic acid. We assume that different substrates and their different lipids contents (O. sativa Indica vs. O. sativa Japonica) are responsible for different amounts of lipids which fungi can consume and metabolize to unsaturated fatty acids during their growth. As the literature shows, fatty acid composition of the rice substrate is dependent on the growing season and the ecogeographical varieties16). In addition, the difference between the individual strains (C. sinensis vs. P. hepiali) also affects the content of the chemical compounds.
These initial results indicate that not only the substrate but also the mushroom strain contains Z-oleic acid, linoleic acid and D-mannitol. We will continue to study this issue with a larger number of extracts from several strains of C. sinensis, C. militaris and P. hepiali, the results of which will give us a more comprehensive view of the problem and the relationship between the substrate, strain and chemical composition.
Table 2 shows the complete assignment of hydrogen and carbon at the mixture of substances: Z-oleic acid, linoleic acid and D-mannitol.
2. 1H a 13C NMR chemical shift in DMSO* 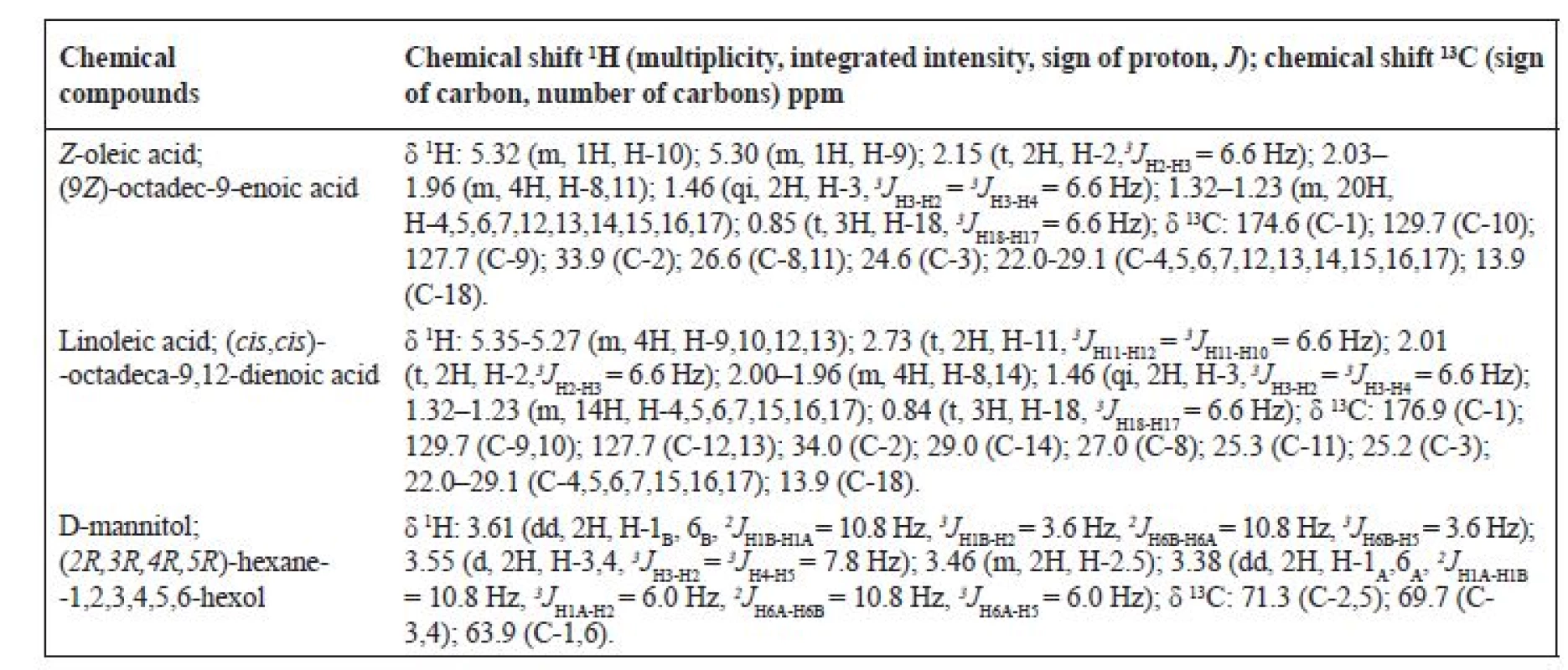
*The chemical shifts of the compounds are compared with the NMR database17)
d – doublet, t – triplet, qi – quintet, m – multipletIR analysis
In order to confirm the functional groups in the Z-oleic acid, linoleic acid and D-mannitol molecule, IR spectroscopy was used. Infrared spectra were measured by the ATR method on the Smart Orbit extension. The analysed extracts were not modified before the measurement. Table 3 shows vibrations for characteristic functional groups: hydroxyl, carbonyl, SP3 carbon methylene or SP2 carbon methyline groups. This method also confirmed the presence of the aforementioned chemical compounds identified by NMR analysis. The IR spectra of extracts 3, 4 and 5 were identical, D-mannitol was absent in these samples. In all spectra, strong bands in the 1707–1650 cm–1 region were present for the -C=O group of the carboxyl group in Z-oleic acid and linoleic acid. All samples had bands for a -OH group in the range of 3369‒3208 cm–1 in the D-mannitol or in the -COOH group.
3. Characteristic bands observed in IR spectra * of extracts 1‒5 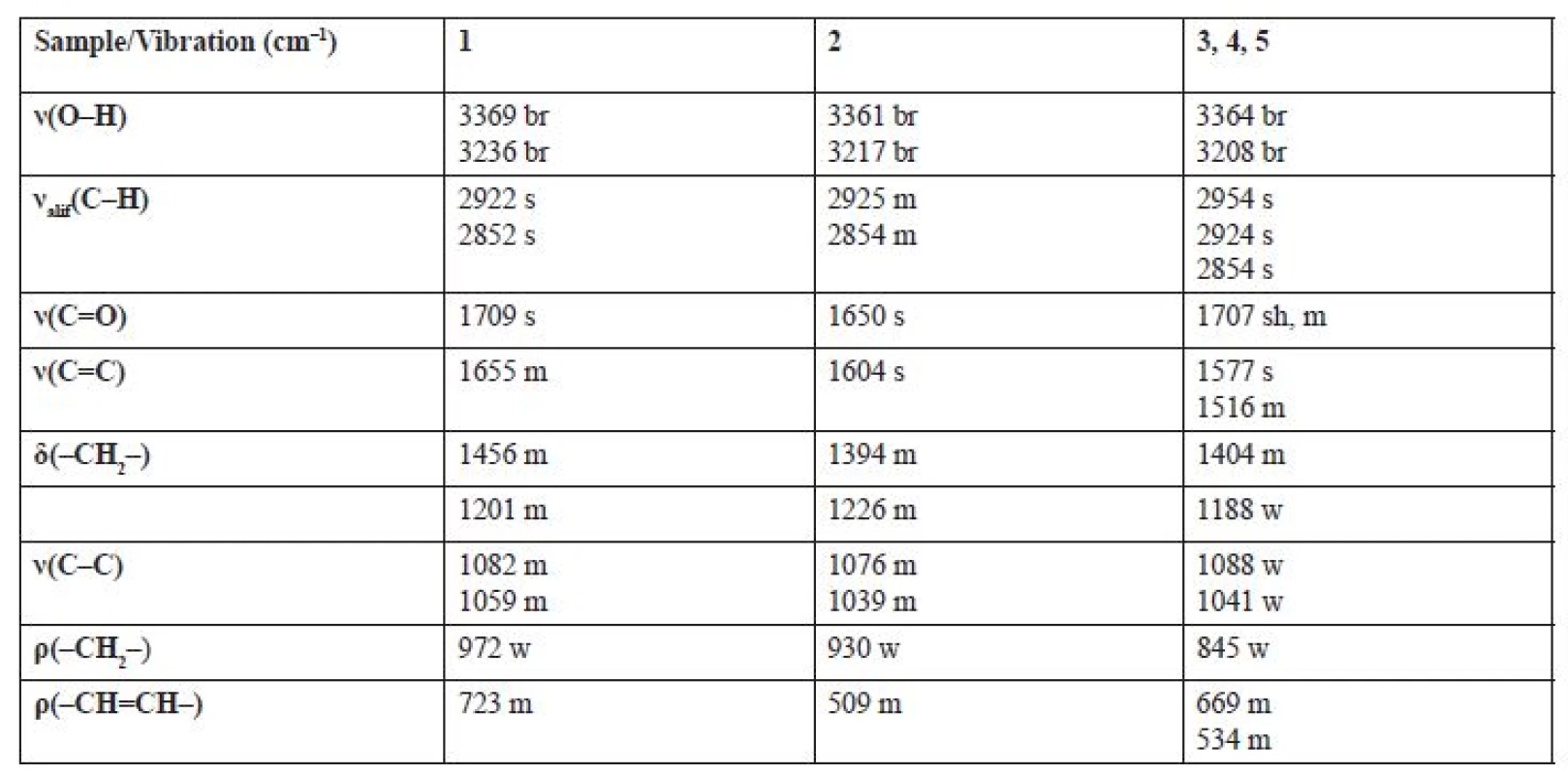
* ν – stretching vibration, δ – scissoring deformation vibration, ρ – rocking deformation vibration
br – broad, w – weak, m – middl, s – strong, sh – sharpConclusions
To determine the chemical structure of the majorities of substances in methanol extracts prepared from the fungi C. sinensis and P. hepiali, which were cultured on two rows of rice, NMR and IR spectroscopy were used. In all 1‒5 samples, unsaturated acids were present, Z-oleic acid was almost twice as high as linoleic acid. Samples 1 and 2 contained, among others, D-mannitol, which was not confirmed in other extracts. The D-mannitol content was 1.4-times higher in the sample of Cordyceps cultivated on O. sativa Japonica compared with sample No. 2, which was cultivated on O. sativa Indica.
This study was performed with the support of the project IGA-UVLF 03/2017. The authors’ thanks include Assoc. Prof. Jan Imrich, Ph.D., of Pavol Jozef Šafárik University in Košice, Faculty of Science, Nuclear Magnetic Resonance Laboratory, for the measurements of NMR spectra.
Conflict of interest: none.
Received September 24, 2018
Accepted December 7, 2018
University of Veterinary Medicine and Pharmacy
Komenského 73, 041 81 Košice, Slovak Republic
e-mail: lucia.ungvarska.malucka@uvlf.sk
M. Vilková
M. Vavra
University of Pavol Jozef Šafárik
Faculty of Science, Institute of Chemistry, Košice, Slovak Republic
M. Pavlík
Technical University, Faculty of Forestry, Department of Forest Protection and Game Management, Zvolen, Slovak Republic
M. Rajtar
Mykoforest, Velčice, Slovak Republic
Sources
1. Hobbs C. H. Medicinal mushrooms: An exploration of tradition, healing, and culture. Botanica Press 1995; 251 s.
2. Holliday J., Cleaver M., Wasser S. P. Cordyceps. In: Coates P. M., Blackman M. R., Cragg G., Levine M., Moss J., White J. (eds). Encyclopedia of dietary supplements. New York: Marcel Dekker 2005; 1–13.
3. Zhu J. S., Halpern G., Jones K. The scientific rediscovery of a precious ancient Chinese herbal regimen: Cordyceps sinensis: Part II. J. Alt. Comp. Med. 1998; 4, 429–457.
4. Holliday J., Cleaver P. Medicinal Value of the Caterpillar Fungi Species of the Genus Cordyceps (Fr.) Link (Ascomycetes). A Review. Int. J. Med. Mushrooms 2008; 10, 219–234.
5. Tuli S. H., Sandhu S. S., Sharma A. K. Pharmacological and therapeutic potential of Cordyceps with special reference to Cordycepin. Biotech. 2014; 4, 1–12.
6. Zhu J. S., Halpern G. M., Johns K. The scientific rediscovery of an ancient Chinese herbal medicine: Cordyceps sinensis: Part I. J. Alt. Comp. Med. 1998; 4, 289–303.
7. Uphof J. C. Th. Dictionary of Economic Plants. New York: Verlag von J. Cramer 1968; 152.
8. Chiang S. S., Liang Z. Ch., Wang Y. Ch., Liang Ch. H. Effect of light-emitting diodes on the production of cordycepin, and mannitol and adenosine in solid-state fermented rice by Cordyceps militaris. J. Food Compos. Anal. 2017; 60, 51–56.
9. Olatunji O. J., Tang J., Tola A., Auberon F., Oluwaniyi O., Ouyang Z. The genus Cordyceps: An extensive review of its traditional uses, phytochemistry and pharmacology. Fitoterapia 2018; Article in Press, https://doi.org/10.1016/j.fitote.2018.05.010
10. Zhang X., Liu Q., Zhou W., Li P., Alolga R. N., Qi L. W., Yin X. A comparative proteomic characterization and nutritional assessment of naturally-and artificially-cultivated Cordyceps sinensis. J. Proteomics 2018; 181, 24‒35.
11. Wang J., Nie S., Chen S., Phillips A. O., Phillips G. O., Li Y., Xie M., Cui S. W. Structural characterization of an α-1, 6-linked galactomannan from natural Cordyceps sinensis. Food Hydrocoll. 2018; 78, 77‒91.
12. Yang S., Jin L., Ren X., Lu J., Meng Q. Optimization of fermentation process of Cordyceps militaris and antitumor activities of polysaccharides in vitro. J. Food Drug Anal. 2014; 22, 468‒476.
13. Yang F. Q., Li D. Q., Feng K., Hu D. J., Li S. P. Determination of nucleotides, nucleosides and their transformation products in Cordyceps by ion-pairing reversed-phase liquid chromatography-mass spektrometry. J. Chromatogr. A 2010; 1217, 5501‒5510.
14. Zhao J., Xie J., Wang L. Y., Li S. P. Advanced development in chemical analysis of Cordyceps. J. Pharm. Biomed. Anal. 2014; 87, 271‒289.
15. Guo L. X., Xu X. M., Wu Ch. F., Lin L., Zou S. Ch., Luan T. G., Yuan J. P., Wang J. H. Fatty acid composition of lipids in wild Cordyceps sinensis from major habitats in China. Biomedicine & Preventive Nutrition 2012; 2, 42‒50.
16. OECD. www.oecd.org/science/biotrack/46815226.pdf (Organisation of Economic Co-operation and Development). 2004. Consensus document on compositional considerations for new varieties of rice (Oryza sativa): Key food and feed nutrients and anti-nutrients. OECD, Paris.
17. SDBSWeb. sdbs.db.aist.go.jp (National Institute of Advanced Industrial Science and Technology) (15. 06. 2015).
Labels
Pharmacy Clinical pharmacology
Article was published inCzech and Slovak Pharmacy

2018 Issue 5-6-
All articles in this issue
- Beer with reduced carbohydrates and alcohol content suitable for diabetics
- The beginnings of health libraries of the Czech Brothers Hospitallers in the 18th century
- Metal complexes in medicine and pharmacy – the past and the present I
- Clinical pharmacist involvement in fall management in a polymorbid geriatric patient with a history of recurrent falls
- Co-processed excipients for direct compression of tablets
- Wastage of medicines and its financial impact on the healthcare system in the Czech Republic
- NMR and IR analysis of natural substances isolated from Cordyceps medicinal mushrooms
- Czech and Slovak Pharmacy
- Journal archive
- Current issue
- Online only
- About the journal
Most read in this issue- Beer with reduced carbohydrates and alcohol content suitable for diabetics
- Metal complexes in medicine and pharmacy – the past and the present I
- Co-processed excipients for direct compression of tablets
- Clinical pharmacist involvement in fall management in a polymorbid geriatric patient with a history of recurrent falls
Login#ADS_BOTTOM_SCRIPTS#Forgotten passwordEnter the email address that you registered with. We will send you instructions on how to set a new password.
- Career

