-
Medical journals
- Career
Multilocus sequence types in Czech neonatal GBS strains from 2004 to 2008
Authors: L. Straková; M. Musílek; J. Motlová
Authors‘ workplace: National Institute of Public Health, Prague
Published in: Epidemiol. Mikrobiol. Imunol. 59, 2010, č. 1, s. 45-47
Overview
The first multilocus sequence typing results of Czech neonatal Streptococcus agalactiae isolates are presented in this paper. The aims of the study were to prove if the Czech isolates belong to potential invasive clonal complex CC17 and to further analyse serotype – sequence type relationships.
Twelve sequence types detected among 77 isolates were assigned to 5 clonal complexes. Sequence type variability was found for most of our collection serotypes. As many as 83 % of the invasive isolates were covered by as few as 3 sequence types: S17, ST23 and ST19. ST17 was the only sequence type strictly tied to serotype III. “Hyperinvasive” ST17 was identified in 85 % of the cerebrospinal fluid isolates (6 out of 7), but in 32 % of the blood isolates only.Key Words:
Streptococcus agalactiae – MLST, invasive – neonatal – serotype.Introduction
Streptococus agalactiae (Group B Streptococcus, GBS) is a well known pathogen capable of causing both mild and severe infections. Neonatal sepsis and meningitis are among the most threatening ones. In the Czech Republic, a perinatal GBS disease preventive program, based on prenatal screening and antibiotic prophylaxis, is in place [11]. Approximately one quarter of Czech pregnant women are colonised by GBS. The level of colonisation varies depending, among others, on the cultivation method and swab site (rectal / vaginal) [12]. The percentage of women fulfilling the criteria for peripartum prophylaxis is relatively high. With increasing antibiotic resistance in mind, there are attempts to specify more precisely the target groups for intervention.
One of the possibilities how to improve the prevention strategy is a better definition of „hypervirulent“ microbial clones. One of the internationally easily comparable methods capable of defining such clones is multilocus sequence typing (MLST). Isolates are assigned to sequence types (STs) based on partial sequencing of several housekeeping genes. STs are further grouped into clonal complexes (CC) based on the identity of some of the alleles.
The aims of the study were to prove if the Czech isolates belong to potential invasive clonal complex CC17 [3, 6] and to further analyse serotype – sequence type relationships.
Material and methods
In this study, 77 non-redundant GBS isolates were tested. Thirty-six isolates were obtained from neonates with mild disease and/or carriers; 12 from neonates with invasive disease likely to be caused by GBS (GBS was isolated from a primarily non-sterile site and other source of infection was not proved) and from 29 neonates with invasive disease. The invasive isolates were sent to the National Institute of Public Health, Prague, by clinical microbiology hospital laboratories throughout the Czech Republic from 2004 to 2008 and were obtained from primarily sterile sites, i.e. cerebrospinal fluid (CSF) and/or blood.
All isolates were serotyped using the Czech National Reference Laboratory for Streptococci and Enterococci in-house antisera and commercial latex agglutination set (Statensserum Institute, Denmark). MLST was carried out by the standard method [5] using a capillary sequencing machine (ABI Prism 3100). The sequences and allele combinations were compared with the MLST database (http://pubmlst. org/sagalactiae accessed on 03/08/2009). A clonal complex was defined as a maximum difference in 3 out of 7 alleles.
Results
Among 77 neonatal GBS isolates, 13 sequence types were assigned. Clonal complex CC1 involved ST1 (n=19) and ST7 (n=1). Parts of CC10 were ST10 (n=1), ST8 (n=2), ST12 (n=2) and ST358 (n=1); parts of CC19 were ST19 (n=10), ST28 (n=2), and ST182 (n=1). ST23 (n=12) and ST88 (n=1) were allocated to CC23. CC17 comprised isolates of a single only sequence type – ST17 (n=25). Seventeen (68 %) out of 25 ST17 neonatal isolates were invasive/potentially invasive. Overall distribution of CC among invasive and non-invasive isolates is shown in Table 1. Serotype – ST relationships are summarised in the same table (Table 1). ST17 was the only ST to be strictly tied to serotype III.
1. MLST clonal complexes in Czech GBS from neonates 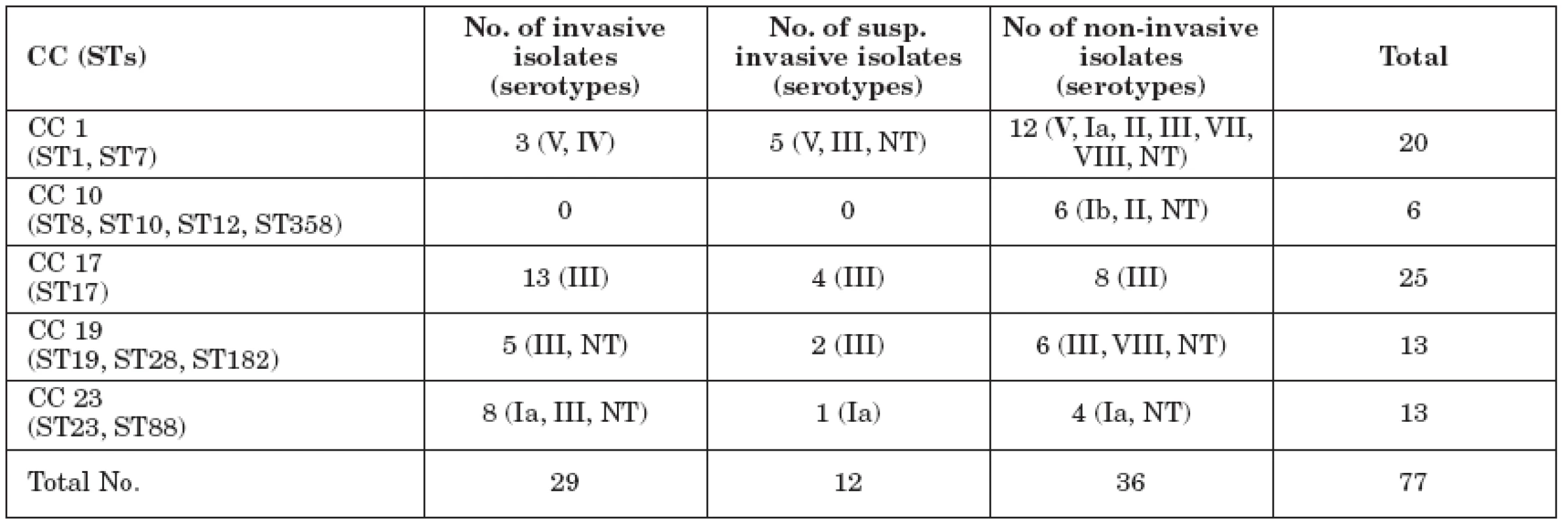
Notes: data in brackets-serotypes/sequence types detected; CC-clonal complex; NT-nontypeable; ST-sequence typ Discussion
The clonal variability of invasive GBS was relatively low in comparison with other pathogens (Streptococcus pyogenes, Streptococcus pneumoniae, Neisseria meningitidis). The Czech neonatal invasive GBS clones were similar to those studied so far in Western Europe and Northern America [4, 8, 9, 10]. Dominant representation of one serotype in one particular ST was seen (ST1-serotype V, ST19-serotype III, ST23-serotype Ia) similarly to other authors [2]. Our results confirm a „special position“ of ST17 among other STs [1]. ST17 is evolutionary distant from the other STs detected in our collection; we did not observe capsular switching (serotype change) in this particular ST. Additionally, ST17 is the most common sequence type among the Czech neonatal GBS isolates (45 %, 13 out of 29 invasive, vs. 22 %, 8 out of 36 non-invasive isolates). „Rapid detection“ methods based on PCR aimed at „hyperinvasive“ ST17 [7] would capture 85% (6 of 7) of our CSF isolates. Unfortunately, such a method would miss 68 % (15 of 22) of the Czech neonatal isolates from blood.
Conclusion
The Czech invasive neonatal GBS isolates have similar genomic characteristics as isolates from other countries studied so far. “Rapid detection” methods aimed solely at “hyperinvasive” ST17 would miss 55 % (16 of 29) of our invasive isolates tested.
Acknowledgement
Our thanks go to all microbiological and clinical staff who, from 2004 to 2008, sent GBS neonatal isolates to National Institute of Public Health from České Budějovice, Havlíčkův Brod, Hradec Králové, Kladno, Karlovy Vary, Kolín, Liberec, Písek, Plzeň, Praha, Prostějov and Šumperk. Special thanks for isolate collection are reserved to Dr. Sádlo, Dr. Srnský, Dr. Hoza, Dr. Čermáková and Dr. Jedličková from General University Hospital, Prague, Dr. Součková, Dr. Hanzl and Dr. Balejová from České Budějovice. R. Doušová and I. Mouchová from the National Institute of Public Health are to be acknowledged for precise DNA isolation and serotyping and last but not least Dr. Křížová deserves thanks for valuable advice and comments.
This study has been supported by the Czech Ministry of Health, Project grant reg. No. NR 9432-3-2007.
MUDr. Lenka Straková
Státní zdravotní ústav, Praha
Šrobárova 48
100 42 Praha 10
e-mail: lenkastrakova@yahoo.com
Sources
1. Bisharat, N., Crook, D. W., Leigh, J., Harding, R. M., et al. Hyperinvasive neonatal group B streptococcus has arisen from a bovine ancestor. J Clin Microbiol, 2004, 42, 2161-7.
2. Bohnsack, J. F., Whiting, A., Gottschalk, M., Dunn, D. M., et al. Population structure of invasive and colonizing strains of Streptococcus agalactiae from neonates of six U.S. Academic Centers from 1995 to 1999. J Clin Microbiol, 2008, 46, 1285-91.
3. Fleming, K. E., Bohnsack, J. F., Palacios, G. C., Takahashi, S., et al. Equivalence of high-virulence clonotypes of serotype III group B Streptococcus agalactiae (GBS). J Med Microbiol, 2004, 53, 505-8.
4. Gherardi, G., Imperi, M., Baldassarri, L., Pataracchia, M., et al. Molecular epidemiology and distribution of serotypes, surface proteins, and antibiotic resistance among group B streptococci in Italy. J Clin Microbiol, 2007, 45, 2909-16.
5. Jones, N., Bohnsack, J. F., Takahashi, S., Oliver, K. A., et al. Multilocus sequence typing system for group B streptococcus. J Clin Microbiol, 2003, 41, 2530-6.
6. Jones, N., Oliver, K. A., Barry, J., Harding, R. M.., et al. Enhanced invasiveness of bovine-derived neonatal sequence type 17 group B streptococcus is independent of capsular serotype. Clin Infect Dis, 2006, 42, 915-24.
7. Lamy, M. C., Dramsi, S., Billoet, A., Reglier-Poupet, H., et al. Rapid detection of the “highly virulent” group B Streptococcus ST-17 clone. Microbes Infect, 2006, 8, 1714-22.
8. Luan, S. L., Granlund, M., Sellin, M., Lagergard, T., et al. Multilocus sequence typing of Swedish invasive group B streptococcus isolates indicates a neonatally associated genetic lineage and capsule switching. J Clin Microbiol, 2005, 43, 3727-33.
9. Manning, S. D., Springman, A. C., Lehotzky, E., Lewis, M. A., et al. Multilocus sequence types associated with neonatal group B streptococcal sepsis and meningitis in Canada. J Clin Microbiol, 2009, 47, 1143-8.
10. Martins, E. R., Pessanha, M. A., Ramirez, M., and Melo-Cristino, J. Analysis of group B streptococcal isolates from infants and pregnant women in Portugal revealing two lineages with enhanced invasiveness. J Clin Microbiol, 2007, 45, 3224-9.
11. Mechurova, A., Unzeitig, V., and Vlk, R. [Guidelines for the diagnosis and treatment of group B streptococcal colonization during pregnancy and delivery. Perinatal medicine section of the CGPS CLS JEP]. Klin Mikrobiol Inf Lek, 2006, 12, 76-7. [article in Czech]
12. Motlova, J., Strakova, L., Urbaskova, P., Sak, P., et al. Vaginal & rectal carriage of Streptococcus agalactiae in the Czech Republic: incidence, serotypes distribution & susceptibility to antibiotics. Indian J Med Res, 2004, 119 Suppl, 84-7.
Labels
Hygiene and epidemiology Medical virology Clinical microbiology
Article was published inEpidemiology, Microbiology, Immunology

2010 Issue 1-
All articles in this issue
- Geography of the HIV/AIDS Pandemic: Analysis of Selected Available Papers and Studies
- Exotic Pets as a Potential Source of Salmonella
- Nocardia farcinica as the Causative Agent of a Brain Abscess in a Patient with Interstitial Lung Disease
- Changes in the Bacterial Spectrum in Severe Burn Wounds
- The Incidence of Tularemia in Slovakia in 1997-2008
- Molecular Epidemiology of Varicella Zoster Virus
- A Steady Rise in Incidence of Pertussis Since Nineties in the Czech Republic
- Multilocus sequence types in Czech neonatal GBS strains from 2004 to 2008
- Seroprevalence of Hantavirus Antibodies among Chronic Hemodialysis Patients in the Czech Republic
- Epidemiology, Microbiology, Immunology
- Journal archive
- Current issue
- Online only
- About the journal
Most read in this issue- Nocardia farcinica as the Causative Agent of a Brain Abscess in a Patient with Interstitial Lung Disease
- The Incidence of Tularemia in Slovakia in 1997-2008
- Exotic Pets as a Potential Source of Salmonella
- A Steady Rise in Incidence of Pertussis Since Nineties in the Czech Republic
Login#ADS_BOTTOM_SCRIPTS#Forgotten passwordEnter the email address that you registered with. We will send you instructions on how to set a new password.
- Career

