-
Medical journals
- Career
Correlation between integration of high-risk HPV genome into human DNA detected by molecular combing and the severity of cervical lesions: first results of the EXPL-HPV-002 study
Authors: S. Bouchilloux 1; F. Fer 1; F. Lemée 1; S. Barradeau 1; V. Dvorak 2; S. Kubickova 2; P. Ventruba 3; R. Tachezy 4; M. Trnková 5; P. Janda 6; J. Abscheidt 1; E. Annibal 1; D. El Mhali 1; F. Garcia 1; M. Kech 1; G. Pilger 1; A. Bensimon 1; F. Mahé 1
Authors‘ workplace: Genomic Vision, Bagneux, France 1; Private Gynecology Center, Brno, Czech Republic 2; Gynecological and Obstetrics Clinic, Brno, Czech Republic 3; NRL for Papillomaviruses and Polyomaviruses, IHBT, Prague, Czech Republic 4; Aeskulab Pathology, Prague, Czech Republic 5; PCS, Prague, Czech Republic 6
Published in: Ceska Gynekol 2019; 84(2): 84-92
Category: Original Article
Overview
Objectives: The aim of the EXPL-HPV-002 study is to evaluate the integration of 14 high-risk HPV as a biomarker of the severity and the progression of cervical lesions. Such a „triage biomarker“ would help to reduce the number of unnecessary colposcopies, to avoid over-treatment of lesions that spontaneously regress and to better target the lesions requiring treatment.
Design: EXPL-HPV-002 is a prospective, open-label, single arm, GCP study conducted at 2 clinical sites in the Czech Republic.
Settings: Investigations centers: Private Gynecology Center, Brno; Gynecological and Obstetrical Clinic, Brno; Genotyping central lab: NRL for Papillomaviruses and polyomaviruses, IHBT, Prague; Histology Central reading: Aeskulab Pathology, Prague; Molecular combing HPV test: Genomic Vision, Bagneux.
Methods: From June 2016 to May 2018, 688 patients aged 25–65, referred to colposcopy after an abnormal Pap-smear, were enrolled in the study. Among them 60% were found HPV high-risk. The study is divided in two phases: 1. a cross-sectional phase using data collected at first visit (colposcopy images ± histology, pap-smear for HPV genotyping and molecular combing) to study the association between HPV integration status versus colposcopy and histology grades; 2. a longitudinal phase using data collected in follow-up visits: cytology at 6, 18 and 30 months and colposcopy ± histology at 12, 24 and 36 months. A pap-smear collected at 12, 24 and 36 months allows to perform genotyping and molecular combing. HPV integration status is analyzed in comparison with the evolution of lesions, viral clearance and HPV genotype. HPV genotyping and molecular combing were performed on pap-smear samples in central laboratories. Histology data were reviewed by central reading.
Results: The transversal phase of the study is achieved and shows that the HPV integration into the human DNA, monitored by molecular combing, can significantly differentiate normal subjects from women with cervical lesions or cancer.
Conclusion: HPV integration into the host genome, monitored by Genomic Vision‘s technology, is a reliable diagnostic biomarker that will greatly help clinicians to improve their medical decision tree.
Keywords:
cervical cancer – HPV diagnostic – viral integration – biomarker – molecular combing – Genomic Morse Code
INTRODUCTION
Cervical cancer is a relatively common type of cancer in women worldwide, affecting young women in many cases. Each year, around 900 women are diagnosed with cervical cancer in the Czech Republic, and there are nearly 400 deaths from cervical cancer among Czech women.
The onset of this cancer is associated with persistent infection by one or several high-risk human papillomaviruses (HR-HPV) and has been recognized by the World Health Organization as attributable to these viruses in nearly 100% of cases. The most common genotypes associated with cervical cancer are HPV 16, 18, 31, 33 and 45, which are responsible for more than 80% of these cancers. Because of its slow progression, cervical cancer can be prevented by screening and the treatment of the precancerous lesions that precede it. The Ministry of Health of the Czech Republic initiated the national cervical cancer screening program in 2008. The aim of the program is the early detection of cervical cancer. The screening is available for all women aged over 15 years and currently based on the cytological examination known as Papanicolaou (Pap) smears performed once a year.
Recently, some countries have planned to abandon cytological screening in favor of the HPV screening test, which is considered to be more sensitive (>90%) than cytology in detecting precancerous and cancerous lesions of the uterine cervix. The slightly lower specificity of HPV testing requires the use of a triage test to avoid excessive use of colposcopy. To date, the procedure that has been best assessed is a cytological screening examination. However, other triage tests that would specifically identify patients at risk of lesion progression may be relevant in this screening strategy.
HR-HPV infection is considered to be the major cause of cervical cancer [7]. However, most infections are spontaneously cleared by the immune system, while some persist for several years and sometimes progress to cancer [3]. HR-HPV infection is therefore a necessary but not sufficient cause of cervical cancer. Integration of the HR - HPV genome into the host genome is considered to be a key event in the development of cervical cancer and one of its most important risk factors [18, 21].
During the initial infection phase, HPV is present as a nuclear episome (productive phase). However, the integration of HR-HPV DNA into the host genome is a major step in the progression of cervical neoplasia (transforming phase) [28]. The integration of the HR-HPV genome into the host cell genome gives these cells a strong selective advantage promoting the clonal expansion of this cell population. Three major mechanisms play an important role in this process. Firstly, it has been reported that the integration of the HR-HPV genome frequently and preferentially causes a deletion in the open reading frame (ORF) of the viral E2 gene [1]. The E2 protein is a negative regulator of the viral E6/E7 promoter. One of the consequences of this integration is the loss of expression control of viral oncoproteins E6 and E7, leading to their stable over-expression. E6 and E7 target the p53 and pRb tumor suppressors respectively, negatively regulating their anti-tumor function [4, 23]. Secondly, the integration of HR-HPV increases the stability of the E6 and E7 viral transcripts derived from integrated viral genome copies [15], thus increasing their level of expression and their oncogenicity [12, 13]. Finally, the integrated viral genes may activate the cellular oncogenes or inactivate tumor suppressor genes close to their integration sites. This is the case, for example, for the c-Myc oncogene, for which the protein expression increases when HR-HPV is integrated into the adjacent regions [5, 10, 20, 27]. A molecular combing study showed that integration of HR-HPV at this locus was associated with a strong genetic instability of this genomic region [8], sometimes leading to malignant progression.
From a morphological point of view, nothing distinguishes a lesion that will regress or persist from one that will progress to invasive cancer. In most cervical carcinomas, the HPV genome is integrated, whereas it is mainly in episomal form in low-grade lesions [9, 10, 14, 25, 28]. While it is recognized that integration of HR-HPV is detected in high-grade precancerous lesions and malignant tumors, an increasing number of studies support the idea that HPV integration may take place at a much earlier stage of cervical carcinogenesis in lower grade lesions [2, 6, 10, 12, 16, 17, 24, 29]. Some of these studies have even demonstrated the presence of integrated HR-HPV DNA in women with normal cytology [6, 16, 17, 24, 29].
Some data also suggest that the rapid progression of early cervical lesions to high-grade lesions is closely associated with the integration of HPV 16 and, in particular, a high integrated HPV16 viral load [19, 24]. Moreover, the HPV integration appears to be a risk factor for progression of precancerous CIN2-3 lesions to the cancer stage [9].
Detection of high-risk HPV genomes integration into the host genome may therefore provide a useful marker to identify lesions at high risk of progression. It would then be possible to adjust the patient follow-up visits, reduce the number of unnecessary colposcopies, over-treatment of lesions that spontaneously regress and better target the lesions requiring treatment.
Routine techniques for the diagnosis of HR-HPV infection (detection of HPV DNA or E6E7 mRNA) and genotyping enable us to detect a persistent infection or to assess the presence of multi-infections and estimate the specific prevalence of types. However, they give an indirect picture of the oncogenic process and none can describe the genomic integration of HR-HPV DNA in the cellular genome. Techniques for detecting the integration of HR-HPV are currently only used for research purposes and all have limitations in this application.
The molecular combing associated with a specific Genomic Morse Code (GMC) is an innovative approach that enables detection and quantification of integrated HPV sequence into the host genome with a high sensitivity as well as an unbiased and high-resolution technique. Thus, the episomal form of the virus, not involved in neoplasia progression, does not alter the test result. Molecular combing is performed according to a four-step workflow: extraction of high molecular weight DNA molecules and combing on a silane-coated glass-slide; hybridization with fluorescent probesets specific for the 14 high-risk HPV DNA sequences (HPV 16, 18, 31, 33, 35, 39, 45, 51, 52, 56, 58, 59, 66 and 68) (see Figure 1); acquisition of images using an automated epifluorescence microscope and image analysis through a dedicated software. Two recent publications [22, 26] provide evidence of the role of integration in oncogenic progression of HPV-associated cancer using molecular combing experiments.
1. GMC probe location on HPV genome – Example of HPV18. 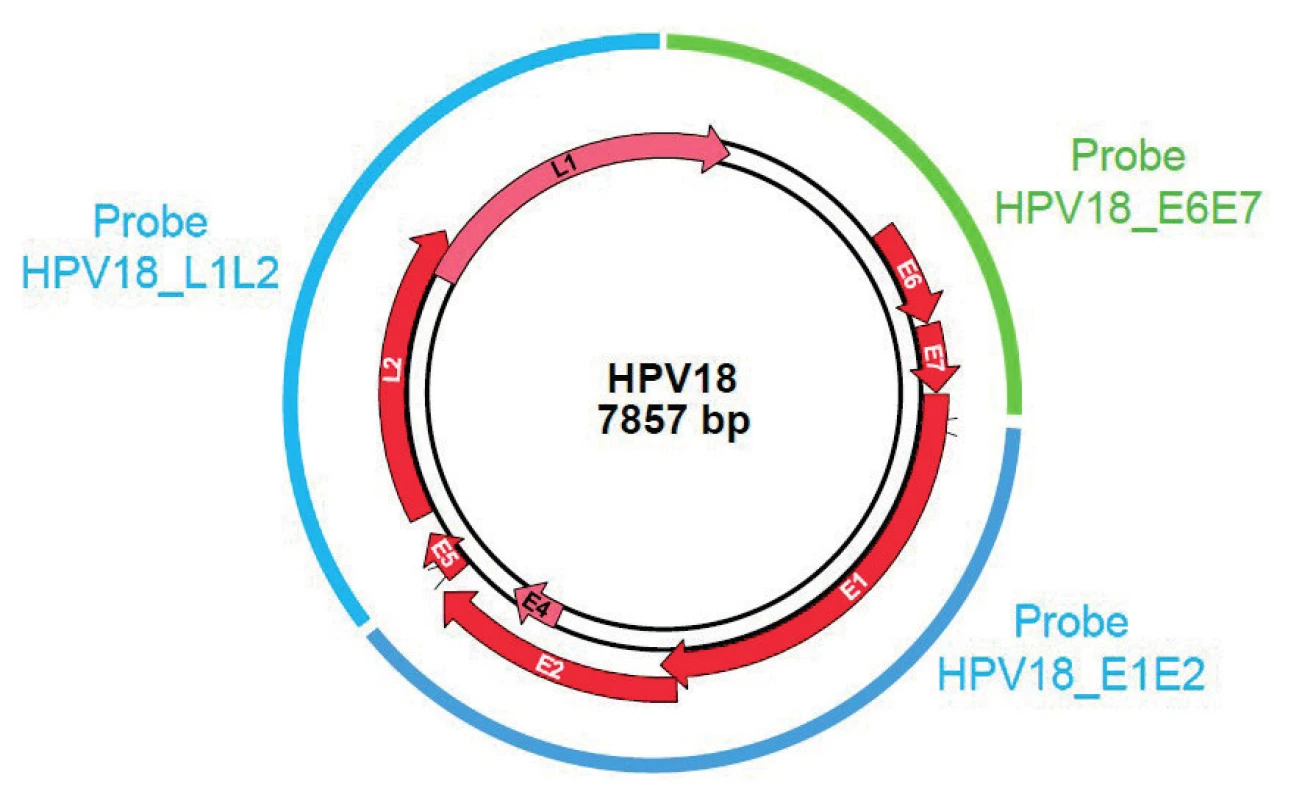
Combed DNA extracted from Pap-smears were hybridized with E6E7 specific probes revealed in cyan (blue+green), and E1E2 and L1L2 specific probes in blue. In order to evaluate the integration of 14 high-risk HPV as a biomarker of the severity of cervical lesions by molecular combing, we are conducting a prospective study called EXPL-HPV-002 at two sites in the Czech Republic (Private Gynecology Center and Gynecological and Obstetrics Clinic in Brno). It is a GCP (Good Clinical Practice), open-label and single-arm study divided into two parts: a cross-sectional part using data collected during the first visit and a longitudinal phase using data collected at the follow-up visits (up to six over 36 months). Patients included in the study are women aged 25 to 65 years, who attend the site to undergo colposcopy in the context of an abnormal cervical Pap test: Atypical Squamous Cells of Undetermined Significance (ASC-US) or high-grade (ASC-H), glandular anomalies, Low or High Grade Squamous Intraepithelial Lesions (LSIL or HSIL) performed at a maximum of six months previously. An HPV genotyping test is performed and patients are withdrawn from the study in case of negative results or low-risk HPV type. Only the high-risk HPV infected patients are fully enrolled in the study. Moreover, if the investigator decides to treat the cervical lesion, the patient is also withdrawn from the study.
We present here the results of the cross-sectional part of the study. The molecular combing analyses of HPV integration into the human genome were performed on cervical smears collected during the inclusion visit.
MATERIAL & METHODS
Pap-smear sample collection: Pap-smears collected in ThinPrep® preservative media during the inclusion visit were used as starting material. Samples were stored at -20°C until DNA extraction through agarose embedding of cells. Under these storage conditions, samples are stable for at least six months.
Preparation of agarose plugs: Smears are centrifuged at 6000 g for 10 minutes at 20°C. Supernatant is removed and 10mL of Phosphate-Buffered Saline (PBS) are added to the cell pellet. Cells are vortexed for 30s at maximum speed, then centrifuged again at 6000g for 10 minutes at 20°C. All the supernatant is removed, and the appropriate volume of PBS 1X is added to the pellet (45 µL/plug). Cell suspension is homogenized by up-and-down pipetting, then incubated for 10 seconds at 50°C. A volume of Low Melting Temperature (LMT) agarose 2% identical to the volume of PBS 1X is added to the cell suspension (45 µL/plug). Cell solution is homogenized by up-and-down pipetting, then dispensed quickly into a DNA plug mold while leaving the cell suspension at 50°C to avoid agarose solidification. DNA plug molds are set at 4°C for 30 min. Plugs are treated with FiberPrep® DNA Extraction Kit, according to the manufacturer‘s instructions. Once washed 3 times in the Buffer 4, plugs can be stored for 1 year in 1mL of Buffer 5 at 4°C.
High molecular weight genomic DNA extraction: DNA extraction is performed with FiberPrep® DNA Extraction Kit, according to the manufacturer‘s instructions. Molecular combing was performed using the Molecular Combing System (Genomic Vision S.A.) and CombiCoverslipsTM. After the combing process, the coverslips were dried for 4 h at 60°C. For direct visualization of combed DNA fibers, the coverslips were mounted with 15 µL of 1/1,000 (vol/vol) Yoyo-1 iodide (Molecular Probes) – Prolong Gold antifade reagent (Invitrogen).
Cloning of 14 HR-HPV probes: The Genomic Morse Code was designed as presented in Figure 1. 3 or 4 probes were designed on each of the 14 HR-HPV. One probe of approximately 2000 bp is covering the E6E7 region. 2 to 3 probes (≈ 3000 bp) are covering the L1L2 and E1E2 regions. HPV 16, 18, 31, 33 and 45 probes were obtained by Polymerase Chain Reaction (PCR) on genomic or plasmidic DNA. PCR fragments were purified and cloned in TOPO-TA or TOPO-XL. Sequencing was performed for verification. HPV 35, 39, 51, 52, 56, 58, 59, 66 and 68 probes were synthesized by an external manufacturer, and cloned in pUC57 vector. Sequencing was performed for verification. Probes on a reference human gene were also designed, based on Bacterial Artificial Chromosome (BAC) (CTD-2349O19), covering a part of the SOX5 region.
Hybridization and detection of HPV probes: Hybridization of labeled HPV probes was performed using the FiberProbes HPV 14HR (Genomic Vision, HPV-HYB-001-RUO) kit, according to the manufacturer‘s instructions. Labeled probes were detected using one layer of antibodies in a 1 : 25 dilution. Digoxigenin labeled probes are revealed with Mouse anti-digoxygenin Brilliant Violet 480 (Jackson Immunoresearch), biotin labeled probes with Mouse anti-biotin AF647 (Jackson Immunoresearch), and fluorescein labeled probes with Mouse anti-fluorescein-Cy3 (Jackson Immunoresearch). Coverslips are quickly washed 3×5 minutes in Saline Sodium Citrate (SSC) 2× at 60°C. Detection is performed (20 minutes incubation with the antibodies), and coverslips are washed 3×3 minutes in SSC 2×/Tween 1%. After the last washing steps, the hybridized coverslips were gradually dehydrated in 70%, 90%, and 100% ethanol solutions and air-dried.
Image acquisition and data analysis: Hybridized coverslips are mounted on FiberVision® Sample Holders, and scanned on the FiberVision® Automated Scanner. Analysis is performed with FiberStudio® Analysis Software. The HPV 16, 18, 31, 33, 45 35, 39, 51, 52, 56, 58, 59, 66 and 68 genomes are covered by L1L2 and E1E2 displayed in blue and E6E7 probes displayed in cyan (blue + green) (see Figure 2 A, B and C). The reference locus is displayed in red (See Figure 2 D).
2. Example of integrated HPV detection pattern. 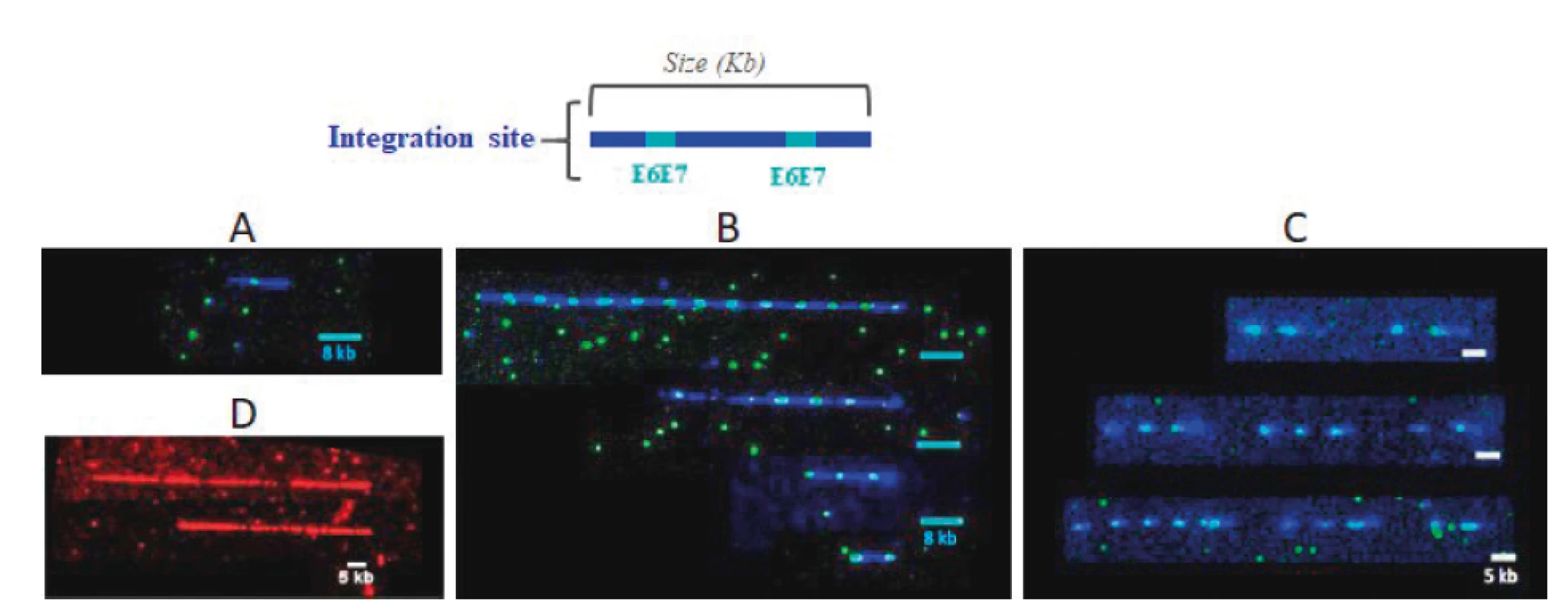
A. Single integration: Integration of a single HPV genome at the integration site. B. Tandem integration: integration of several HPV genomes at the integration site. C. Dispersed tandem integration: integration of several HPV genomes spaced by the host DNA at an integration site. D. Reference signal: Probes covering the host DNA locus of known size. Statistical analysis: For each patient, the integration profile is defined by the following variables: Estimated number of host genomes combed on the slide: It is calculated by adding the sizes (in kb) of all the reference signals displayed in red (see Figure 2 D) visible on the analyzed slide and dividing that sum by the theoretical size (in kb) of the locus, then by 2 (because there are 2 alleles per genome). Number of HPV integration for a patient: it corresponds to the number of HPV signals (blue+cyan) detected on all the coverslips hybridized for a patient. The number of HPV integration sites per patient genome corresponds to the ratio of the number of integration sites versus the number of host genome. To evaluate if two samples have the same distribution of integration per genome, the non-parametric Mann-Whitney U test was used. To compare proportion of patients with a lower integration between two groups, the Pearson‘s chi-squared two-sided test at a 5% alpha risk level was used.
RESULTS
Study population and subgroups
Patients enrolled in the study were women from 25 to 65 years old who already had a suspicious Pap smear and that were referred for a colposcopy appointment. 688 patients corresponding to those criteria were included in the study (32% in the range from 25 to 34 years old, 40% 35–44, 23% 45–54 and 5% 55–65). During the inclusion visit, a cervical cell sample is collected in liquid medium (ThinPrep®) to carry out an HPV test and the combing test for HPV integration evaluation. If the HPV test is positive for one of the 14 high-risk HPV, patients are definitively enrolled in the study. For information, 26% of the 688 women had no HPV infection, 54% were mono-infected and 20% were infected by more than one HPV. HPV16 was detected in 26% of the patients, HPV51 in 7% of the patients and each of the viruses HPV18, 33, 45, 51, 52, 56 or 58 was found in 5% of the cases. Negative results or low-risk HPV are withdrawn from the cohort. A urine pregnancy test is also performed and pregnant women are withdrawn.
The colposcopy is then performed by the clinician and photographs of the cervix are taken with a video colposcope. Biopsies are performed when it is considered necessary by the gynecologist. In this instance, the histology samples are sent to the central accredited biopsy laboratory.
The study population subgroups, according to the colposcopy and histology analysis results, are shown in Table 1. Out of the 410 HR-HPV positive patients (60% of the total enrolled population), 63 had no cervical lesion detected in colposcopy and therefore had no biopsy. 38 patients had a suspicious colposcopy result but the histology analysis didn‘t reveal any neoplasia. For 293 of the patients, a biopsy was performed and the histology revealed a cervical neoplasia. Of these, 47 were classified as low grade (CIN1) and 246 were identified as high-grade (79 CIN2 and 167 CIN3). In 16 cases, cancerous neoplasia was detected in the biopsy. We analyzed whether integration of the HPV could be detected and the number of viruses integrated per site in these different subgroups to determine if correlation with lesion severity could be highlighted.
1. Clinical study population – Subgroups in HR-HPV enrolled patients 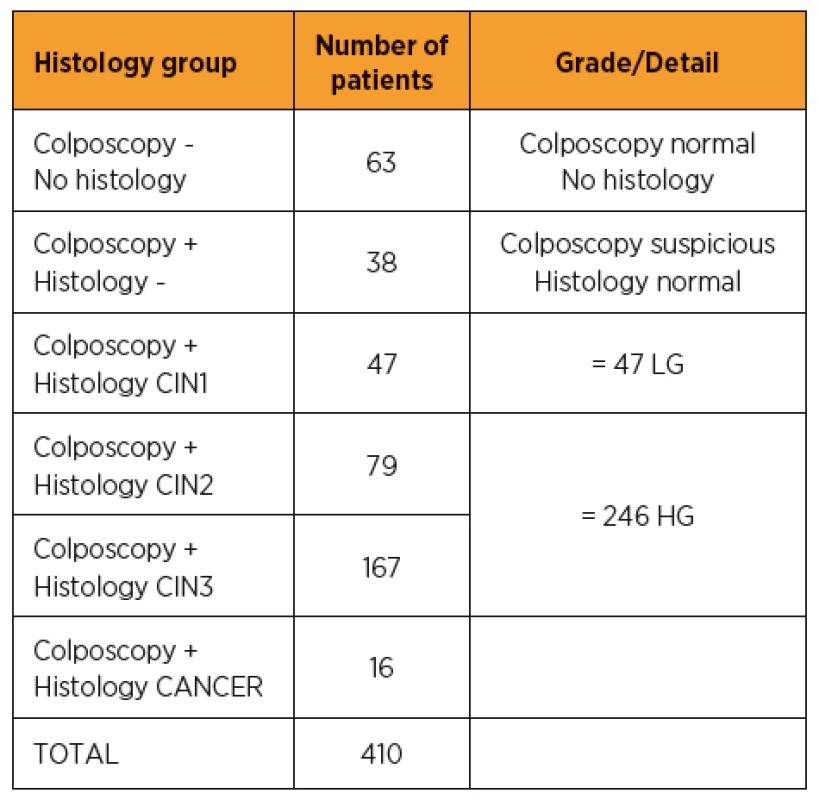
Within the 688 patients included in the study, 410 were positive for at least one of the 14 HR-HPV. The table shows the size of the various population subgroups determined by their colposcopy and histology results. [Colposcopy-] are patients whose colposcopy does not show any cervical lesion; [Colposcopy+, Histology-] are patient showing no cervical neoplasia detected in biopsy; [Colposcopy+, Histology CIN1] are patients with low-grade (LG) cervical neoplasia detected in biopsy; [Colposcopy+, Histology CIN2] and [Colposcopy+, Histology CIN3]) are patients with high-grade (HG) cervical neoplasia detected in biopsy. [Colposcopy, Histo CANCER] are patients with cancerous cervical neoplasia detected in biopsy. HPV integration in the various histology groups
It is very well documented in the literature that integration of HPV genome into human DNA is found in high grade and cancer lesions [2, 6, 10, 17]. Various publications demonstrate that virus integration could occur at earlier stages (Low Grade) or may also be detected in the DNA of patients with normal cytology [11, 16, 24, 29]. Techniques used to monitor the integration in these studies have their limitations and do not allow a direct view of what is happening inside the DNA molecule. At the beginning of the study, we estimated, according to two publications using PCR techniques [17, 25], that 16.4% of patients with normal colposcopy or low-grade histology and 29.5% of high-grade histology would have an HR-HPV integration. Those estimates were used to calculate the population size.
The results described in Figure 3 show that it is possible to visualize the HPV integration with fluorescent probes detecting the 14 HPV genomes in all the study subgroups. In fact, in the group of patients showing no lesions in colposcopy, at least one integration for 10 genomes can be detected for 49% of the patients. This percentage increases to 58% in the subgroup with suspicious colposcopy but negative biopsy. For the three groups showing a positive detection of neoplasia in histology, the percentage increases from 68% to 81% depending on the lesion severity. The percentages rise to 86% in a normal situation and 97% in CIN3 cases (data not shown) if no threshold for genome number is applied. Controls on HPV negative patients were performed to verify that no fluorescent signal was detected (data not shown). These results are significantly different from what is currently published. They highlight the high sensitivity of the molecular combing technology and reveal that HPV integration is an event that can occur early in the pre-cancerous development process. Moreover, it also confirms that a population showing progressive neoplasia is massively concerned by HPV integration into their DNA.
3. Percentage of population showing integration in histology subgroups. 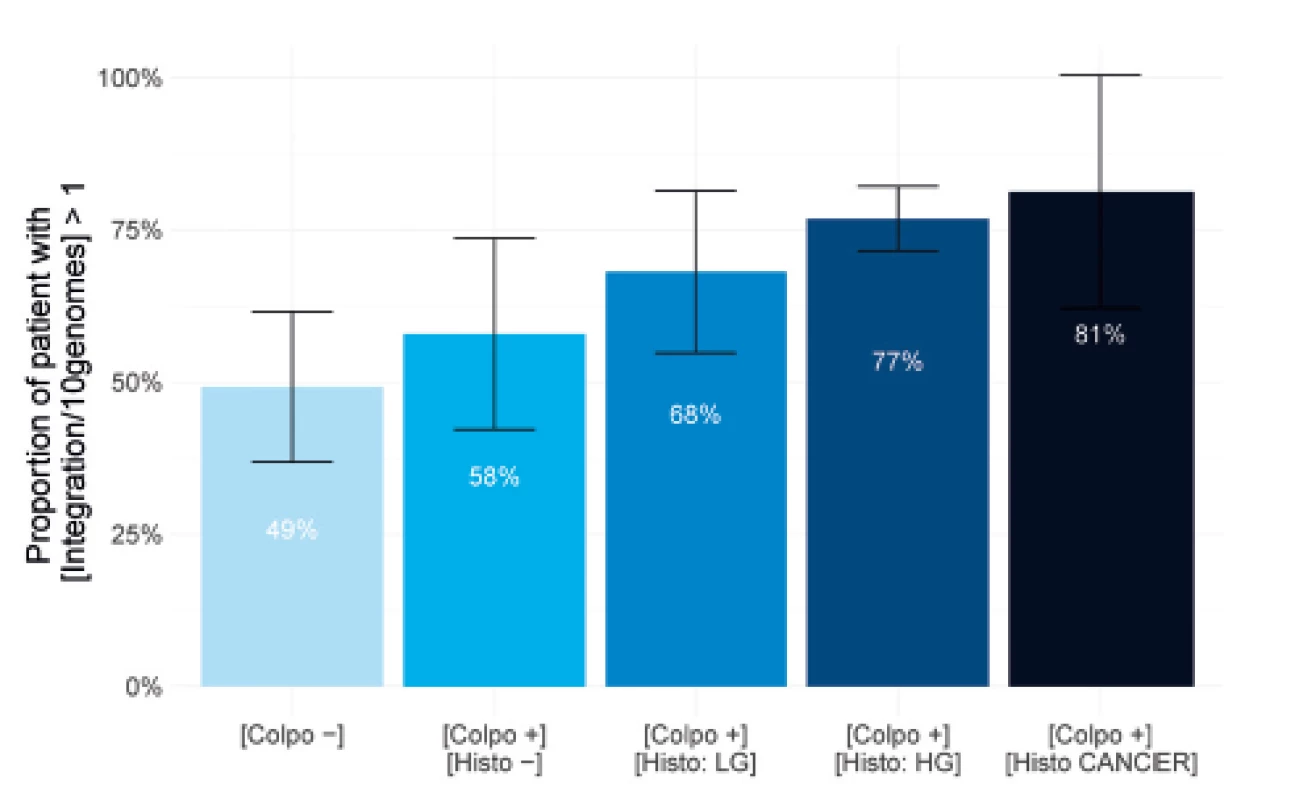
Barplots show the proportion of patients with integration level higher than 1 integration site for 10 genomes in each subgroup of the cohort. Confidence intervals on the estimate proportion are calculated at a 95% probability level. The histogram shows the following subgroups from light to dark blue: patients showing no cervical lesion in colposcopy [Colpo-], patient showing no cervical neoplasia in biopsy [Colpo+, Histo-], patients showing low-grade (LG) cervical neoplasia in biopsy [Colpo-, Histo LG], patients showing high-grade (HG) cervical neoplasia in biopsy [Colpo-, Histo HG] and patients showing cancerous neoplasia in biopsy [Colpo-, Histo CANCER]. Quantification of HPV integration level into the host genome
In order to evaluate if the HPV integration correlates with cervical lesion severity, the distribution of the number of integrations per genome was evaluated in each of the study population subgroups. The box plot in Figure 4 shows that the median value of integration per genome in normal situation is 0.09 (negative colposcopy) and 0.16 (negative histology), which equals to 1 virus integration per 10 genomes (first two plots, light blue). The median rises to 0.52 for patients with a positive cytology (CIN1, 2 or 3), equivalent to 5 integrations for 10 genomes, (third plot, marine blue). This is five times higher than for normal patients. Moreover, the median value of integration per genome in the cancer cases is 0.95 (fourth plot, dark blue), which means nearly 10 integrations per 10 genomes. This is ten times higher than in normal samples and two times higher compared to combined low and high grades samples. We conclude that the measurement of HPV integration level into the host DNA is correlated with the severity of lesions, thus measurement of HPV integration level is a powerful biomarker of the disease stage.
4. Distribution of integration per genome in various histology groups. 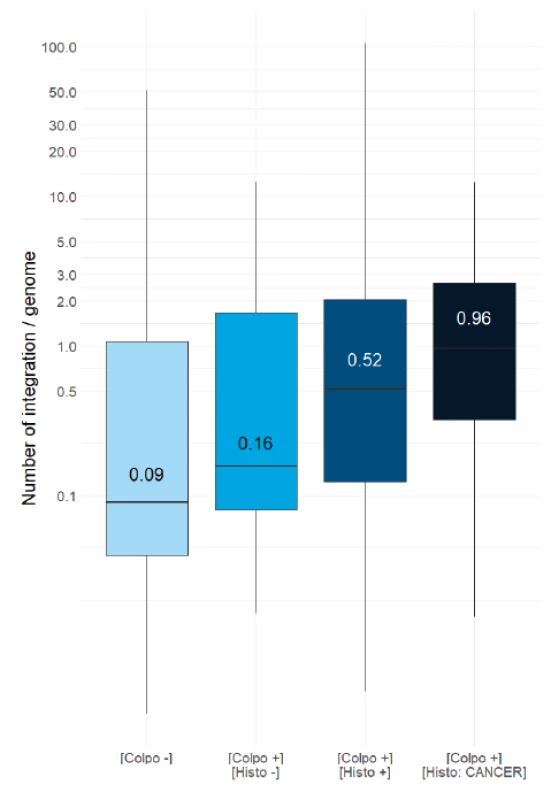
Box plot shows the distribution of HPV virus integration levels for each subgroup of the cohort. The difference of mean integration level between [colpo+, histo-] and [colpo+, histo+] is significant (Mann- Whitney alpha = 5%; p > 0.01). The histogram shows the following subgroups from light to dark blue: patients showing no cervical lesion in colposcopy [Colpo-], patient showing no cervical neoplasia in biopsy [Colpo+, Histo-], patients showing cervical neoplasia in biopsy (CIN1+CIN2+CIN3) [Colpo-, Histo+] and patients showing cancerous neoplasia in biopsy [Colpo-, Histo CANCER]. The integration median value is indicated inside each plot. Patient stratification based on integration level
Considering that HPV integration is directly correlated with cervical lesion severity, we tried to find a model that could help the clinician to make the best decision regarding patient care. The objective is to avoid unnecessary colposcopies, overtreatment of lesions that would spontaneously regress and to better target the lesions requiring treatment. We propose a three-category patient stratification based on the number of integrations per genome (see Table 2). For patients in category I, for whom no integrated HPV was detected by molecular combing, the probability of lesion is less than 30%. Category II patients, showing an HPV integration level below one per genome, have a probability of the lesion increasing to 60%, thus with a relative risk factor of two compared to category I. For patients in category III, who show more than one integration per genome, the risk factor is close to 2.5 compared to patients with no integration. Such a classification tool could be of very significant help to the clinician to adapt his or her prescription and/or follow-up of the patient.
2. Patient stratification based on HPV integration level 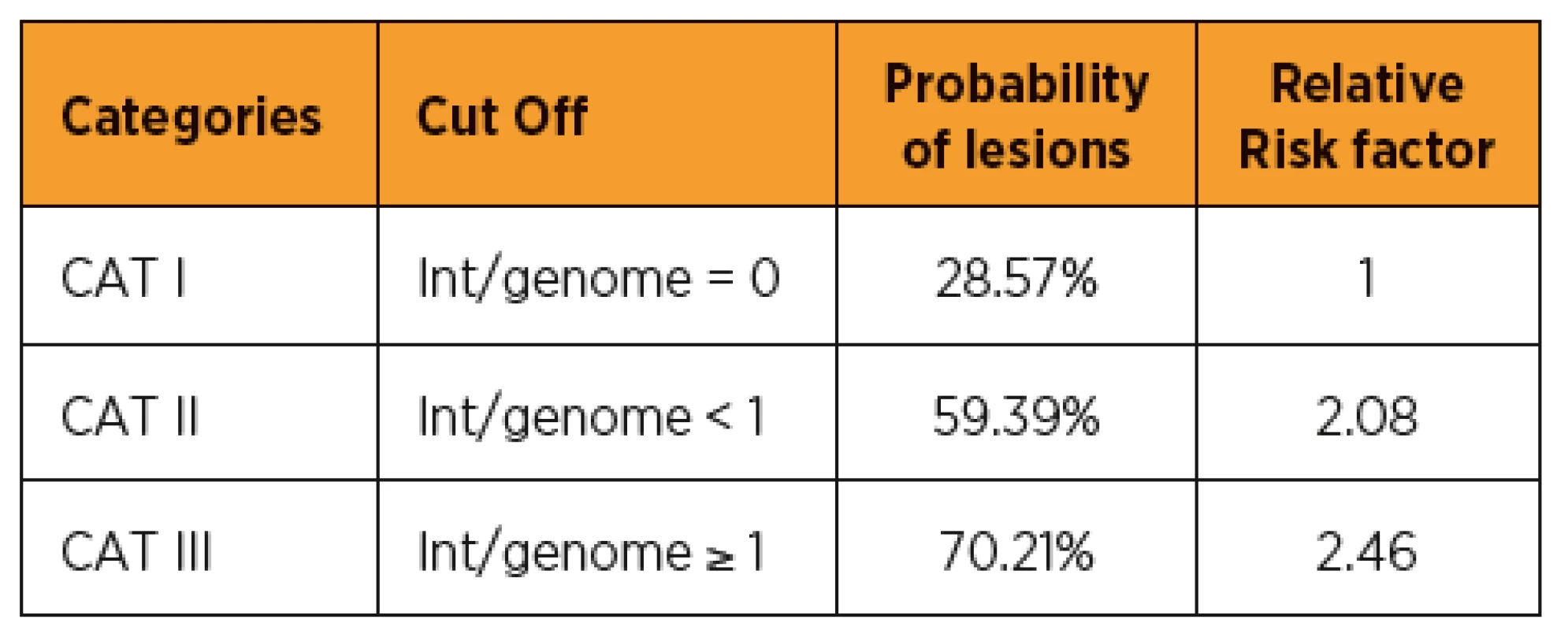
Three categories of patients (CAT I, CAT II and CAT III) were defined depending on the number of virus found integrated per host genome. CAT I corresponds to patients with no HPV integration detected by molecular combing and GMC. CAT II corresponds to patients showing less than one integration per host genome. CAT III corresponds to patients showing one or more integration per host genome. The probability of lesions is the number of high-grade out of the total for each category. The relative risk factor is the ratio of the probability of lesions in CAT II or III versus the probability of lesions in CAT I. DISCUSSION
Current cervical cancer diagnosis is a combination of Pap-smear, HPV testing, colposcopy and histology, each of these techniques has their limitations. Pap-smear remains the primary screening test for cervical cancer but this technique shows a limited sensitivity, is highly operator-dependent and the results are often ambiguous. The consequence is a patient management that could be improved to avoid unnecessary colposcopies, overtreatment, economical and psychological impacts. Our opinion is that there is a critical medical need here.
The various commercially available HPV tests are based on analysis of the viral load, genotyping, E6/E7 mRNA expression or oncoproteins detection but none of them focuses on HPV virus integration into the human genome, although this is the critical starting point of neoplasia progression.
Genomic Vision has developed a technology that allows direct visualization of DNA fibers. With specific fluorescent probes covering the 14 high-risk HPV, we are able to visualize and quantify the HPV integration events into the host genome. The whole HPV genome can be monitored and the various patterns can be detected and quantified. Unlike many commercial quantitative PCR tests, this test is not biased by episomal form of the virus, responsible for the productive phase of the infection but not the transforming phase.
Thus, we have decided to evaluate the diagnostic and prognostic value of HPV integration by molecular combing in a correlation study. Our prospective clinical study EXPL-HPV-002 was initiated in June 2016, the cross-sectional phase is achieved and the longitudinal phase (follow-up) is ongoing. So far, we have started to analyze the diagnostic phase data and we conclude that: 1) HPV integrations are events that can be visualized in all stages of the pre-cancerous process; 2) the number of patients that show integration events is more significant in the high - grade group; 3) the median number of integrations per genome is four times higher in patients showing abnormal histology versus normal and ten times higher in cancerous cervical cells; 4) HPV integration threshold can be set to classify patients and help the clinician to improve his or her medical decision tree.
We conclude that HPV integration into the human DNA is an efficient biomarker for cervical cancer diagnosis and we propose the molecular combing test as a potential new and powerful diagnostic test.
The longitudinal phase of the study is in progress. We are collecting cytological, histological, colposcopy and molecular combing data in up to six follow-up visits over 36 months. In-depth analysis of those data will allow us to identify whether HPV integration measurement is also a good prognostic marker of cervical lesions outcome and will help to refine the biomarker parameters (integration pattern, HPV genotype impact, viral clearance etc.). If the results are positive, the molecular combing HPV test could become a reference for the early triage of patients. This would greatly facilitate the clinician‘s decision-making and would also greatly benefit women by improving the quality, safety and comfort of cervical cancer screening and follow-up throughout their lives.
Florence Mahe
Director Medical Research
Green Square – Bât.E
80-84 rue des Meuniers
92220 Bagneux
France
e-mail: f.mahe@genomicvision.com
Sources
1. Choo, KB., Pan, CC., Liu, MS., et al. Presence of episomal and integrated human papillomavirus DNA sequences in cervical carcinoma. J Med Virol, 1987, 21(2), p. 101–107.
2. Cricca, M., Morselli-Labate, AM., Venturoli, S., et al. Viral DNA load, physical status and E2/E6 ratio as markers to grade HPV16 positive women for high-grade cervical lesions. Gynecol Oncol, 2007, 106(3), p. 549–557.
3. Crosbie, EJ., Einstein, MH., Franceschi, S., Kitchener, HC. Human papillomavirus and cervical cancer. Lancet, 2013, 382(9895), p. 889–899.
4. Dyson, N., Howley, PM., Munger, K., Harlow, E. The human papilloma virus-16 E7 oncoprotein is able to bind to the retinoblastoma gene product. Science, 1989, 243(4893), p. 934–937.
5. Ferber, MJ., Thorland, EC., Brink, AA., et al. Preferential integration of human papillomavirus type 18 near the c-myc locus in cervical carcinoma. Oncogene, 2003, 22(46), p. 7233–7242.
6. Gradissimo Oliveira, A., Delgado, C., Verdasca, N., Pista, A. Prognostic value of human papillomavirus types 16 and 18 DNA physical status in cervical intraepithelial neoplasia. Clin Microbiol Infect, 2013, 19(10), p. E447–450.
7. Hausen, zur H. Papillomaviruses and cancer: from basic studies to clinical application. Nat Rev Cancer, 2002, 2, 342–350.
8. Herrick, J., Conti, C., Teissier, S., et al. Genomic organization of amplified MYC genes suggests distinct mechanisms of amplification in tumorigenesis. Cancer Res, 2005, 65(4), p. 1174–1179.
9. Hopman, AH., Smedts, F., Dignef, W., et al. Transition of high-grade cervical intraepithelial neoplasia to micro-invasive carcinoma is characterized by integration of HPV 16/18 and numerical chromosome abnormalities. J Pathol, 2004, 202(1), p. 23–33.
10. Hu, Z., Zhu, D., Wang, W., et al. Genome-wide profiling of HPV integration in cervical cancer identifies clustered genomic hot spots and a potential microhomology-mediated integration mechanism. Nat Genet, 2015, 47(2), p. 158–163.
11. Huang, LW., Chao, SL., Lee, BH. Integration of human papillomavirus type-16 and type-18 is a very early event in cervical carcinogenesis. J Clin Pathol, 2008, 61(5), p. 627–631.
12. Jeon, S., Allen-Hoffmann, BL., Lambert, PF. Integration of human papillomavirus type 16 into the human genome correlates with a selective growth advantage of cells. J Virol 1995, 69(5), p. 2989–2997.
13. Jeon, S., Lambert, PF. Integration of human papillomavirus type 16 DNA into the human genome leads to increased stability of E6 and E7 mRNAs: implications for cervical carcinogenesis. Proc Natl Acad Sci U S A, 1995, 92(5), p. 1654–1658.
14. Klaes, R., Woerner, SM., Ridder, R., et al. Detection of high-risk cervical intraepithelial neoplasia and cervical cancer by amplification of transcripts derived from integrated papillomavirus oncogenes. Cancer Res, 1999, 59(24), p. 6132–6136.
15. Knebel von Doeberitz, M., Bauknecht, T., Bartsch, D., zur Hausen, H. Influence of chromosomal integration on glucocorticoid-regulated transcription of growth-stimulating papillomavirus genes E6 and E7 in cervical carcinoma cells. Proc Natl Acad Sci U S A, 1991, 88(4), p. 1411–1415.
16. Kulmala, SM., Syrjanen, SM., Gyllensten, UB., et al. Early integration of high copy HPV16 detectable in women with normal and low-grade cervical cytology and histology. J Clin Pathol, 2006, 59(5), p. 513–517.
17. Marongiu, L., Godi, A., Parry, JV., Beddows, S. Human papillomavirus type 16 long control region and E6 variants stratified by cervical disease stage. Infect Genet Evol, 2014, 26, p. 8–13.
18. McBride, AA., Warburton, A. The role of integration in oncogenic progression of HPV-associated cancers. PLoS Pathog, 2017, 13(4), e1006211.
19. Peitsaro, P., Johansson, B., Syrjanen, S. Integrated human papillomavirus type 16 is frequently found in cervical cancer precursors as demonstrated by a novel quantitative real-time PCR technique. J Clin Microbiol, 2002, 40(3), p. 886–891.
20. Peter, M., Rosty, C., Couturier, J., et al. MYC activation associated with the integration of HPV DNA at the MYC locus in genital tumors. Oncogene, 2006, 25(44), p. 5985–5993.
21. Pett, M., Coleman, N. Integration of high-risk human papillomavirus: a key event in cervical carcinogenesis? J Pathol, 2007, 212(4), p. 356–367.
22. Redmond, CJ., Fu, H., Aladjem, MI., McBride, AA. Human Papillomavirus Integration: Analysis by Molecular Combing and Fiber-FISH. Curr Protoc Microbiol, 2018, 51(1), p. e61.
23. Scheffner, M., Werness, BA., Huibregtse, JM., et al. The E6 oncoprotein encoded by human papillomavirus types 16 and 18 promotes the degradation of p53. Cell, 1990, 63(6), p. 1129–1136.
24. Vega-Pena, A., Illades-Aguiar, B., Flores-Alfaro, E., et al. Risk of progression of early cervical lesions is associated with integration and persistence of HPV-16 and expression of E6, Ki-67, and telomerase. J Cytol, 2013, 30(4), p. 226–232.
25. Vinokurova, S., Wentzensen, N., Kraus, I., et al. Type-dependent integration frequency of human papillomavirus genomes in cervical lesions. Cancer Res, 2008, 68(1), p. 307–313.
26. Warburton, A., Redmond, CJ., Dooley, KE., et al. HPV integration hijacks and multimerizes a cellular enhancer to generate a viral-cellular super-enhancer that drives high viral oncogene expression. PLoS Genet, 2018, 14(1), p. e1007179.
27. Wentzensen, N., Ridder, R., Klaes, R., et al. Characterization of viral-cellular fusion transcripts in a large series of HPV16 and 18 positive anogenital lesions. Oncogene, 2002, 21(3), p. 419–426.
28. Wentzensen, N., Vinokurova, S., von Knebel Doeberitz, M. Systematic review of genomic integration sites of human papillomavirus genomes in epithelial dysplasia and invasive cancer of the female lower genital tract. Cancer Res, 2004, 64(11), p. 3878–3884.
29. Zubillaga-Guerrero, MI., Illades-Aguiar, B., Leyva-Vazquez, MA., et al. The integration of HR-HPV increases the expression of cyclins A and E in cytologies with and without low-grade lesions. J Cytol, 2013, 30(1), p. 1–7.
Labels
Paediatric gynaecology Gynaecology and obstetrics Reproduction medicine
Article was published inCzech Gynaecology

2019 Issue 2-
All articles in this issue
- Operative vaginal deliveries and their impact on maternal and neonatal outcomes – prospective analysis
- Střednědobé výsledky chirurgické léčby recidivující cystokély po hysterektomii s využitím transvaginálního implantátu
- Sakrospinous fixation sec. Miyazaki – complications and long-term results
- Pilot study comparing tolerance of transperineal and endoanal ultrasound examination of anal sphincter
- Is it possible to estimate urethral mobility based on maximal urethral closure pressure measurements?
- Uterine rupture during pregnancy and delivery: risk factors, symptoms and maternal and neonatal outcomes – restrospective cohort
- Maternal morbidity and mortality in Slovak Republic in the years 2007–2015
- Sacrococcygeal teratoma
- Embolic event in the puerperium with tragic end
- Gynecological and urological aspects of pelvic vasculitis
- Latest findings on the placenta from the point of view of immunology, tolerance and mesenchymal stem cells
- Bisphenols in the pathology of reproduction
- Correlation between integration of high-risk HPV genome into human DNA detected by molecular combing and the severity of cervical lesions: first results of the EXPL-HPV-002 study
- Czech Gynaecology
- Journal archive
- Current issue
- Online only
- About the journal
Most read in this issue- Uterine rupture during pregnancy and delivery: risk factors, symptoms and maternal and neonatal outcomes – restrospective cohort
- Sacrococcygeal teratoma
- Operative vaginal deliveries and their impact on maternal and neonatal outcomes – prospective analysis
- Latest findings on the placenta from the point of view of immunology, tolerance and mesenchymal stem cells
Login#ADS_BOTTOM_SCRIPTS#Forgotten passwordEnter the email address that you registered with. We will send you instructions on how to set a new password.
- Career

