-
Medical journals
- Career
EPIGENETIC CHANGES IN MALIGNANT UVEAL MELANOMA AND POSSIBILITIES OF THEIR THERAPEUTIC TARGETING
Authors: B. Smolková; L. Demková
Authors‘ workplace: Ústav experimentálnej onkológie, Biomedicínske centrum Slovenskej akadémie vied, Bratislava
Published in: Čes. a slov. Oftal., 76, 2020, No. 2, p. 55-60
Category: Review Article
doi: https://doi.org/10.31348/2020/12Overview
Uveal melanoma (UM) is a deadly cancer that leads to metastatic disease in more than 50 % of the patients. Despite the improvement in the treatment of primary disease, there is still no effective therapy to prevent the development of metastases. Therefore, the disease requires intensive research to identify new treatment strategies.
In preclinical UM models, epigenetic drugs have been shown to increase the sensitivity of resistant tumour cells to treatment. The successful use of histone deacetylase inhibitors, which induced cell cycle arrest, reprogramming consistent with melanocyte differentiation and inhibition of tumour growth in preclinical models, demonstrates the role of epigenetic regulation in UM metastasis. Identification of epigenetic changes associated with UM development an progression could contribute to the discovery of more effective drugs that, in combination with traditional approaches, may yield better therapeutic results for high-risk patients.
Keywords:
malignant uveal melanoma – epigenetic changes – DNA methylation – histone modifications – non-coding RNAs
Declaration
The authors of the article declare that no conflict of interest exists in the compilation, theme and subsequent publication of this professional communication, and that it is not supported by any pharmaceuticals company.
The study has not been submitted to any other journal or printed elsewhere.
INTRODUCTION
Malignant uveal melanoma (UM) is the most commonly occurring intraocular tumour in adults. Its incidence in Slovakia is 5.1 per million of the population, which means 32 to 54 newly diagnosed patients per year [1]. Tumours most often originated from melanocytes localised in the uveal layer of the eye, primarily in the choroidea (85 %), but may also originate from the corpus ciliare (5–8 %) or iris (3–5 %) [2].
At present there is a turn away from radical procedures in the treatment of primary UM. Treatment by ionising radiation and combined approaches predominates, and treatment by proton and photon therapy is possible. In our conditions brachytherapy using episcleral radiators is used, e.g. Ru106, treatment by gamma knife or stereotactic radiosurgery on a linear accelerator [3–5]. The growth of a choroidal melanoma into the eye socket leads to a radical procedure, not only enucleation of the eyeball but also exenteration of the orbit [6]. No significant differences have been determined upon observation of patient survival following enucleation and following treatment by ionising radiation [7,8]. Despite the low incidence of local recurrences, at present no treatment is available that would increase the overall survival of metastatic patients. There is also no explanation relating to primary resistance of UM with regard to any systemic therapy. Tyrosine kinase inhibitors or Met (hepatocyte growth factor receptor protein), histone deacetylase inhibitors (HDAC) or immune therapy (NCT02068586, NCT02223819, NCT02068586, NCT01585194) are in the stage of clinical trials [9].
UM metastasises in the liver in as many as 90 % of cases. The mechanism which is the basis of liver tropism is not yet known. Metastatic disease, which occurs in almost 50 % of patients, is associated with a poor prognosis. A recently published meta-analysis conducted on a cohort of 912 patients with metastatic UM states the median of average survival without progression at 3.3 months and overall survival at 10.2 months [10]. The risk of occurrence of metastases is associated with tumour size, extraocular growth, high mitotic activity and epitheloid type of tumour cells (Table 1). The most significant molecular markers of metastatic risk include monosomy of chromosome 3 and specific expression profile of mRNA in the tumour tissue. An expression profile of a panel of 15 genes, which is commercially available under the name DecisionDX-UM, enables division of patients into 3 groups on the basis of metastatic risk: Class 1A – very low risk with 2 % probability of metastatic dissemination: Class 1B – low risk with 21 % probability of occurrence of metastases: Class 2 – high risk with 72 % probability of metastasis during the course of the following five years [11]. A key role in metastasis is played by somatic mutations of the gene BAP1 (BRCA1-Associated Protein 1), leading to a reduction or complete loss of expression of the Bap1 protein [12]. BAP1, localised in the region of 3p21, is a tumour-suppressor gene, which contributes to the epigenetic regulation of genes that are significant during development and differentiation. Its inactivity leads to the formation of cells which have the properties of tumour stem cells. This phenotype is associated with aggressive behaviour of tumours and a poor prognosis [13]. Abnormalities of chromosomes 1, 6, 8 and 9 are linked with metastatic risk, together with specific mutations in other genes such as SF3B1 (Splicing Factor 3B Subunit 1A) or EIF1AX (eukaryotic translation initiation factor 1A, X-linked) [14]. Mutations of BRAF (B-Raf Proto-Oncogene, Serine/Threonine Kinase) and NRAS (NRAS Proto-Oncogene, GTPase), which are typical of skin melanomas (40–50 % and 15–20 %) do not occur in UM [15, 16].
1. Clinical, histological and molecular markers for the prediction of risk of metastasis 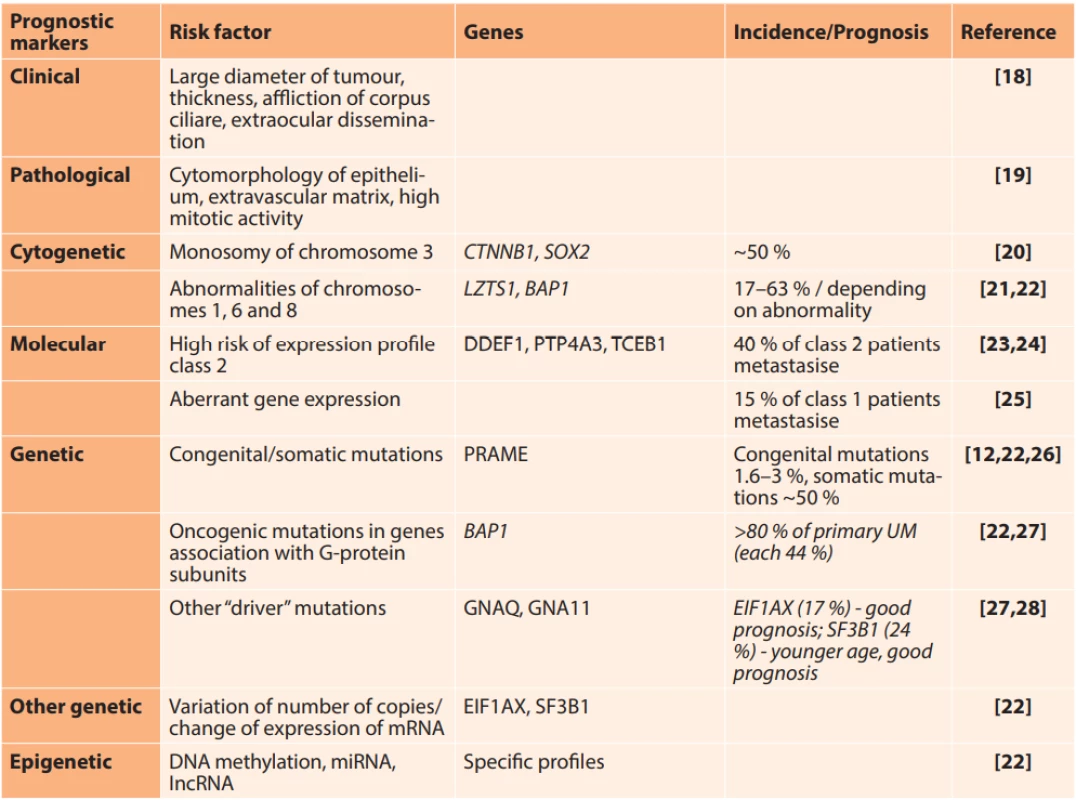
Modified according to [17] Epigenetic mechanisms
Epigenetic mechanisms such as DNA methylation, histone modifications and non-coding RNAs are essential for the normal development and homeostasis of the organism. They mutually interreact and have a key role in the maintenance of tissue-specific gene expression and protection against genetic instability. Their impairment may lead to changes in the function of genes, malignant transformation, and may have an effect on the individual signal pathways subject to metastasis. The role of epigenetic deregulation in the pathogenesis of UM has been confirmed by the results of several studies. One of the most significant of these is an integrated multi-platform analysis of 80 UMs, which demonstrated that in addition to known genetic changes, changes in DNA methylation and the expression of multiple microRNAs (miRNA) and long non-coding RNAs (IncRNA) are also associated with a poor prognosis for patients [22]. In tumours with a high risk of metastasis, the expression of genes coding the actual epigenetic regulation of enzymes is deregulated [29]. Epigenetic inactivity of expression may have a particularly significant role primarily in genes localised in chromosomes with typical abnormalities in the number of copies, such as the chromosomes 1, 3, 6 or 8. Chromosome 3, monosomy of which is present in approximately 50 % of patients with UM, contains a number of tumour-suppressor genes, as well as genes which have a key role in haematogenous dissemination. These include for example the genes RASSF1A (RAS association domain family 1), FHIT (Fragile Histidine Triad), BAP1 (BRCA1 - Associated Protein 1), CTNNB1 (Catenin Beta 1) or SOX2 (Sex-determining region Y (SRY)-Box2).
DNA methylation
DNA methylation ranks among the best studied epigenetic mechanisms. It concerns covalent bonding of the methyl group (-CH3) to DNA bases, primarily cytosine residue in the dinucleotide sequence CpG. It is catalysed by DNA methytransferase enzymes (DNMTs) with the presence of S-adenosylmethionine (SAM), which is applied as a donor of the methyl group. Methylation / demethylation is an important mechanism for maintaining the integrity of the genome and safeguarding tissue-specific gene expression (Fig. 1). In comparison with normal cells, tumour cells have an impaired DNA methylation formula either through a reduction (hypomethylation) or increase (hypermethylation) of the number of methyl groups. Significant above all in the initiation of oncological diseases are hypermethylation of promoters of tumor suppressor genes, hypermethylation of proto-oncogenes and global hypomethylation (influencing extragenetic and intragenetic regions), which leads to an increase of chromosome instability. In patients with UM, DNA hypermethylation has been identified as a cause of inactivation of a number of genes, of which the majority contribute to the regulation of the cellular cycle. These include the genes APC, RASSF1A, RARB, LZTS1, CDH1, RB1, CDKN2A, PRAME and others [17,30].
1. Schematic illustration of differences in DNA methylation in gene promoters, in extragenetic and intragenetic regions in a normal and tumour cell. Methylation in extragenetic regions protects the cell against chromosome instability, the tumour cell has a reduced number of methyl groups in this region. Active genes do not have methylated promoters, unprescribed genes have them hypermethylated. Methylated CpG inucleotides are indicated by a black dot, non-methylated CpG dinucleotides by a white dot. 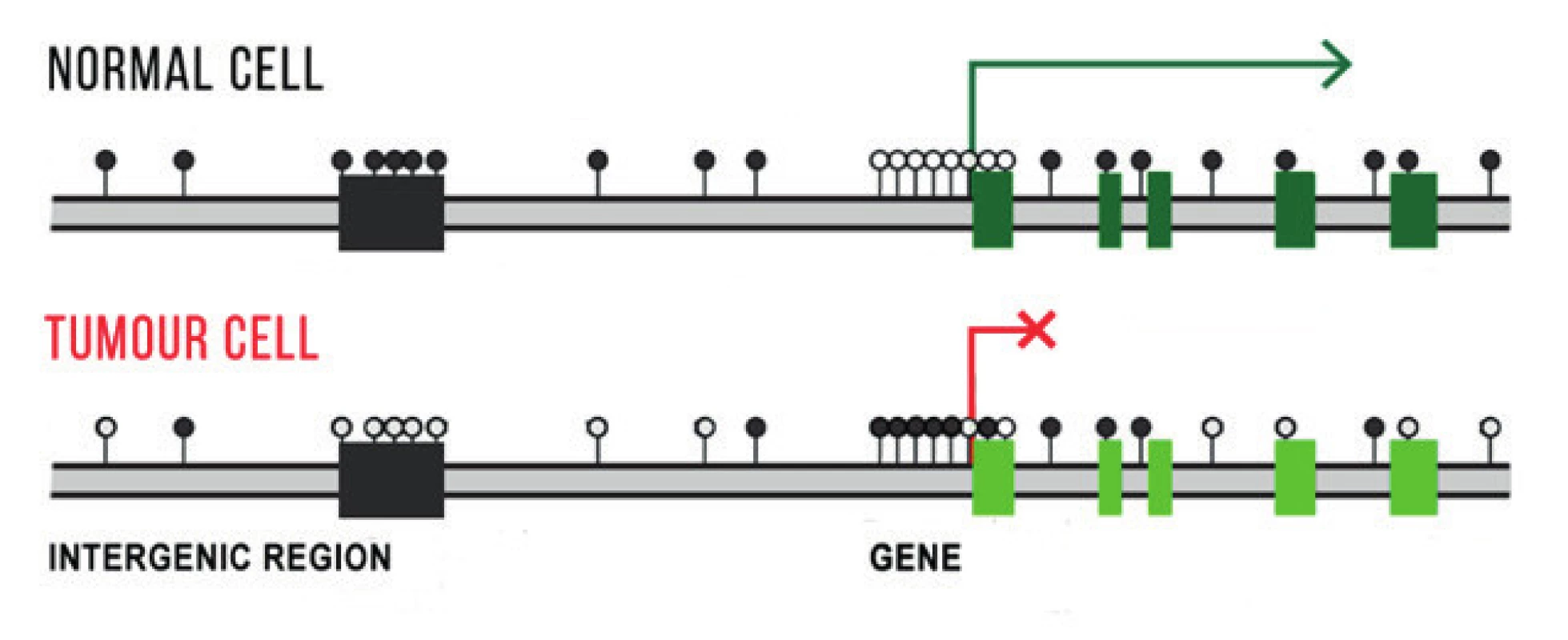
Histone modifications
Chemical modifications of histone proteins, which together with tightly coiled DNA fibre form a complex referred to as a nucleosome, also have a large influence on gene activity. This is the basic unit of chromatin, a fibrous structure which, depending on the degree of condensation, plays a key role in controlling gene expression (Fig. 2). The formation of an inactive, condensed heterochromatin is linked with a low degree of acetylation and a high degree of methylation of histone molecules. Less condensed euchromatin is transcriptionally active [31]. In order to understand the role of histone modifications in the progression of UM, a groundbreaking discovery was that of the role of the Bap1 protein in the regulation of differentiation of embryonic cells. Loss of expression of Bap1 prevents acetylation of histone H3K27 in the promoters of key genes, regulating the differentiation of the ectoderm, mesoderm and neural crest, which leads to a reduction of their expression [13]. The balance between acetylation and deacetylation of a gene is determined by the relative activities of histones of acetyltransferase and HDAC. HDAC inhibitors enable re-expression of epigenetically inactivated genes.
2. Schematic illustration of the influence of histone modifications and DNA methylation on the condensation of chromatin. The loose structure of chromatin, referred to as euchromatin, with acetylation and methylation of histone molecules – without DNA methylation – is transcriptionally active. Condensed heterochromatin with DNA methylation is transcriptionally inactive. Methylated CpG dinucleotides are indicated by a black dot, non-methylated CpG dinucleotides by a white dot. Ac – acetylation, Met -histone methylation. Adjusted according to [19]. ![Schematic illustration of the influence of histone modifications and DNA methylation on the condensation of chromatin. The loose structure of chromatin, referred
to as euchromatin, with acetylation and methylation of histone molecules – without
DNA methylation – is transcriptionally active. Condensed heterochromatin with DNA
methylation is transcriptionally inactive. Methylated CpG dinucleotides are indicated
by a black dot, non-methylated CpG dinucleotides by a white dot. Ac – acetylation,
Met -histone methylation. Adjusted according to [19].](https://www.prolekare.cz/media/cache/resolve/media_object_image_small/media/image_pdf/9a98eeb32a2c2205379263855d638bb1.jpeg)
Non-coding RNAs
The best studied non-coding RNAs include small, 19–25 nucleotide RNA molecules, known as microRNA (miRNA). The primary mechanism of their functioning is binding to complementary mRNA, by which they inhibit its translation or degrade mRNA (Fig. 3) [32]. In cell lines, in tumour tissues and peripheral blood of patients with UM, changes of the expression of several miRNAs have been described, including increased expression of let-7b, miR-20a, miR-125b, miR-143, miR-146a, miR-155, miR-181, miR-193b, miR-199a, miR-223, miR-367, miR-454, miR-652, or reduced expression of miR-9, miR-34b/c, miR-124a, miR-137, miR4-144, miR-145, miR-182, miR-204 and others (see summary article [17]). Aberrantly exprimated miRNAs play an important role in the deregulation of oncological pathways in UM, and may support metastatic dissemination [33]. In addition to the fact that they may provide an interesting diagnostic and prognostic biomarker, they also offer us a promising therapeutic target. It has been demonstrated that it is possible to inhibit the function of specific miRNAs by means of complementary, chemically modified oligonucleotides, which are referred to as anti-miRs or antagomiRs. These have shown promising results in pre-clinical development [34], and could compensate for increased expression of genes of oncogenic pathways and thereby assist in the management and treatment of UM. The expression of miRNAs can be regulated by means of methylation of their promoters. For example, Chen et al. stated that the hypomethylating agent 5-aza-2’-deoxycytidine (decitabine) enabled increased expression of miR-137 [35]. Similarly, the expression of miR-124a in UM cells was restored after the use of decitabine and the histone deacetylase inhibitor trichostatin A [36]. These findings provide evidence that individual epigenetic mechanisms, in addition to fulfilling their individual role, mutually act upon one another on several levels and mutually interreact.
3. Mechanism of effect of miRNA. By binding to a complementary sequence in 3 areas of the target mRNA it prevents their transcription. 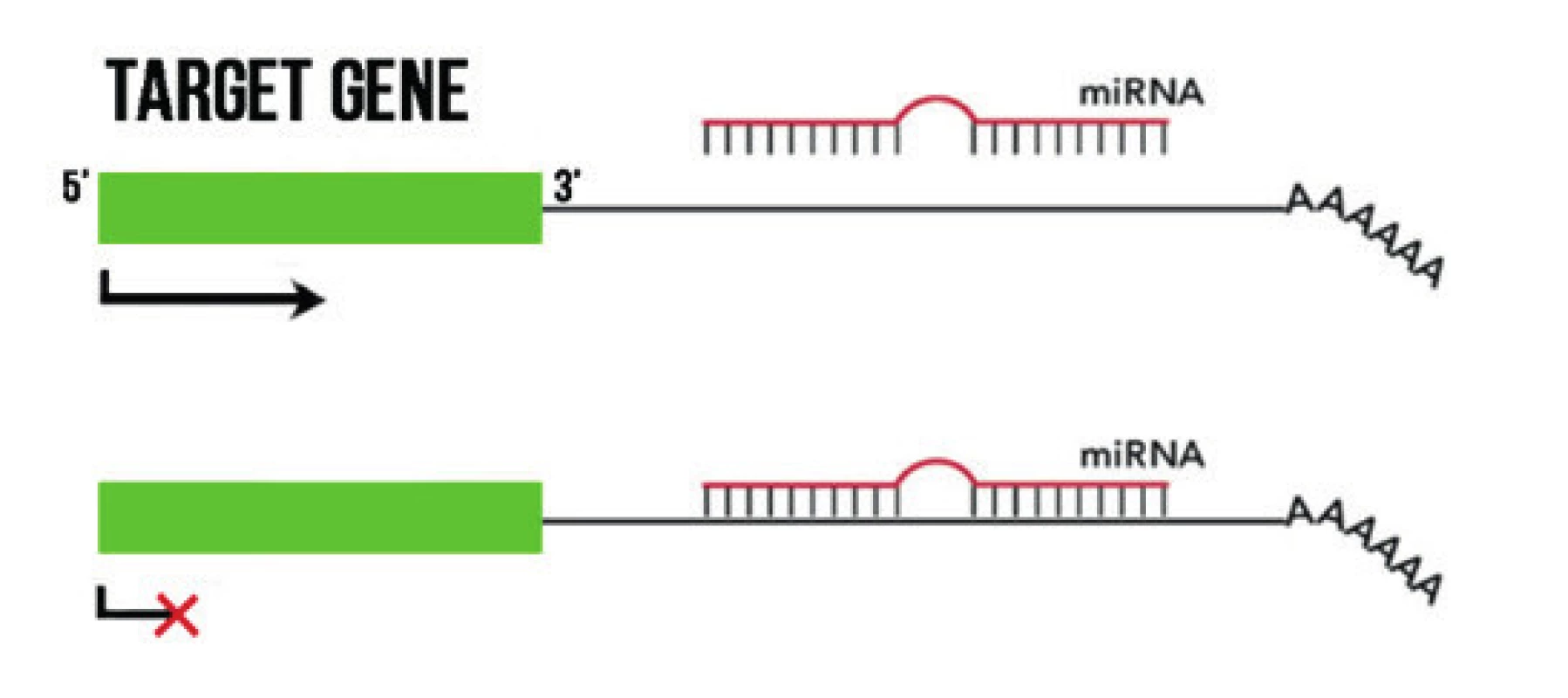
Long non-coding RNAs are defined as transcripts of RNA with a length of more than 200 nucleotides, without the capacity to code proteins. They intervene in tumorigenesis, contribute in processes of angiogenesis, cell proliferation, migration and apotosis. In the case of UM, reduced expression of PAUPAR and increased expression of multiple IncRNAs has been described, for example ROR, HOXA11-AS, FTH1P3, PVT1, CYTOR, BANCR, PVT1/NEAT1 and MALAT1 (see summary article [17]).
Potential for epigenetic therapy
The loss of BAP1 is linked with a loss of melanocyte differentiation and an increase of metastatic potential. HDAC inhibitors have succeeded in reversing the phenotype effects of inactivation of BAP1 through the induction of morphological differentiation and change of high-risk to low-risk profile of gene expression in UM cells [37]. Valproic acid, trichostatin A, tenovin-6, depsipeptide, panobinostat (LBH-589), vorinostat (suberoylanilide hydroxamic acid), entinostat (MS-275), quisinostat, NaB, JSL-1, MC1568 and MC1575 have demonstrated promising anti-tumour effects also in other pre-clinical trials in the case of UM [38]. They may block the proliferation of tumour cells, induce halting of their growth, terminal differentiation, cell death and inhibition of angiogenesis. It was thereby demonstrated that HDAC and DNMT inhibitors could represent an alternative adjuvant therapy for prolonging tumour dormancy. At present clinical trials are under way with valproic acid and vorinostat (NCT02068586, NCT01587352), entinostat (PEMDAC study with pembrolizumab, entinostat, NCT02697630) and bromodomain inhibitor BRD4 PLX2853 (NCT03297424) [39]. Inhibition of bromodomain and extraterminal (BET) proteins offers a new therapeutic approach for UM. This concerns third generation epigenetic regulators, which influence DNA replication, remodelling and transcription of chromatin. Despite regulatory approval for the treatment of certain haematological malignancies, the problem of epigenetic therapy in solid tumours remains controversial. According to certain authors, global hypomethylation of DNA as a consequence of the use of the inhibitor DNMT1 5-aza-2’-deoxycytidine in mice leads to chromosomal instability and an increased incidence of secondary malignancies [40,41]. Although assertions concerning the risk in connection with treatment by hypomethylating substances have been criticised by other authors [42], the effectiveness of first generation epigenetic medications in patients with solid tumours has been disappointing [43]. Thanks to the development of new compounds and better understanding of the molecular basis of tumorous pathologies, however, it appears that epigenetic medicaments could play an important role in synergy with classic therapeutic approaches [43]. The use of nanotechnologies could contribute to increasing the effectiveness and reducing the tissue toxicity of such combined treatment.
CONCLUSION
Epigenetic changes play a significant role in the pathogenesis of oncological diseases. They are of a reversible nature and as a result represent a good therapeutic target. In several pre-clinical trials it has been demonstrated that epigenetic drugs enable the restoration of expression of aberrantly inactivated tumour-suppressor genes, and increase the sensitivity of resistant tumour cells to treatment. In order to discover more effective medicaments for adjuvant therapy of UM and the treatment of metastatic diseases, it is essential to accept the significance of epigenetic changes and understand their role in the pathogenesis and progression of this disease. In combination with traditional therapeutic approaches such as immune, chemo or radiotherapy, epigenetic drugs could bring better results for hitherto untreatable, advanced stages of UM.
Sources
1. Furdova, A., Olah, Z., Svetlosakova, Z., et al.: The current state of the evidence of malignant tumors of the eye and its adnexa (dg. C69) in the Slovak Republic and in the Czech Republic. Cesk Slov Oftalmol, 68(5);2012;195–201.
2. Vivet-Noguer, R., Tarin, M., Roman-Roman, S., et al.: Emerging Therapeutic Opportunities Based on Current Knowledge of Uveal Melanoma Biology. Cancers, 11(7);2019 : 1019 s.
3. Furdova, A., Sramka, M., Chorvath, M., et al.: Clinical experience of stereotactic radiosurgery at a linear accelerator for intraocular melanoma. Melanoma Res, 27(5);2017 : 463–8.
4. Furdova, A., Sramka, M., Chorvath, M., et al.: Stereotactic radiosurgery in intraocular malignant melanoma--retrospective study. Neuro Endocrinol Lett, 35(1);2014 : 28–36.
5. Furdova, A., Sramka, M., Waczulikova, I., et al.: Stereotactic Rediosurgery for Uveal Melanoma; Postradiation Complications. Cesk Slov Oftalmol, 71(5);2015 : 134–42.
6. Furdová, A., Ferková, A., Krásnik, V., et al.: Exenterácia očnice pre malígny melanóm choroidey v štádiu T4; možnosti epitetického riešenia. Čes a slov Oftal,71(3);2015.
7. Furdova, A., Babal, P., Kobzova, D., et al.: Uveal melanoma survival rates after single dose stereotactic radiosurgery. Neoplasma, 65(6);2018 : 965–71.
8. Furdova, A., Slezak, P., Chorvath, M., et al.: No differences in outcome between radical surgical treatment (enucleation) and stereotactic radiosurgery in patients with posterior uveal melanoma. Neoplasma, 57(4);2010 : 377–81.
9. Yang, J., Manson, D. K., Marr, B. P., et al.: Treatment of uveal melanoma: where are we now? Therapeutic advances in medical oncology, 10;2018 : 1–17.
10. Khoja, L., Atenafu, E. G., Suciu, S., et al.: Meta-Analysis in Metastatic Uveal Melanoma to Determine Progression-Free and Overall Survival Benchmarks: an International Rare Cancers Initiative (IRCI) Ocular Melanoma study. Ann Oncol, 30;2019 : 1370–1380.
11. Onken, M. D., Worley, L. A., Tuscan, M. D., et al.: An accurate, clinically feasible multi-gene expression assay for predicting metastasis in uveal melanoma. J Mol Diagn, 12(4);2010 : 461–8.
12. Harbour, J. W., Onken, M. D., Roberson, E. D., et al.: Frequent mutation of BAP1 in metastasizing uveal melanomas. Science, 330; 2010 : 1410–3.
13. Kuznetsov, J. N., Aguero, T. H., Owens, D. A., et al.: BAP1) regulates epigenetic switch from pluripotency to differentiation in developmental lineages giving rise to BAP1-mutant cancers. Science Advances, 5(9);2019: eaax1738.
14. Dogrusöz, M., Jager, M. J., Damato, B. Uveal melanoma treatment and prognostication. Asia Pac J Ophthalmol (Phila), 6(2); 2017 : 186–96.
15. Horkovicova, K., Markus, J., Krcova, I., et al.: Mutácia BRAF a možnosti identifikácie prognostických markerov metastázovania uveálneho melanómu. Cesk Slov Oftalmol, 72(4); 2016 : 149-56.
16. Cruz, F., 3rd, Rubin, B. P., Wilson, D., et al.: Absence of BRAF and NRAS mutations in uveal melanoma. Cancer Res, 63(18);2003 : 5761–6.
17. Smolkova, B., Horvathova Kajabova, V., Zmetakova, I., et al.: Role of epigenetic deregulation in hematogenous dissemination of malignant uveal melanoma. Neoplasma, 65(6);2018 : 840–54.
18. Shields, C. L., Kaliki, S., Furuta, M., et al.: American Joint Committee on Cancer classification of posterior uveal melanoma (tumor size category) predicts prognosis in 7731 patients. Ophthalmology, 120(10);2013 : 2066–71.
19. Kaliki, S., Shields, C. L., Shields, J. A. Uveal melanoma: Estimating prognosis. Indian Journal of Ophthalmology, 63(2);2015 : 93–102.
20. Damato, B., Coupland, S. E. Translating uveal melanoma cytogenetics into clinical care. Arch Ophthalmol, 127(4);2009 : 423–9.
21. Coupland, S. E., Lake, S. L., Zeschnigk, M., et al.: Molecular pathology of uveal melanoma. Eye (Lond), 27(2);2013 : 230–42.
22. Robertson, A. G., Shih, J., Yau, C., et al.: Integrative Analysis Identifies Four Molecular and Clinical Subsets in Uveal Melanoma. Cancer Cell, 32(2);2017 : 204–20 e15.
23. Tschentscher, F., Husing, J., Holter, T., et al.: Tumor classification based on gene expression profiling shows that uveal melanomas with and without monosomy 3 represent two distinct entities. Cancer Res, 63(10);2003 : 2578–84.
24. Onken, M. D., Worley, L. A., Ehlers, J. P., et al.: Gene expression profiling in uveal melanoma reveals two molecular classes and predicts metastatic death. Cancer Res, 64(20);2004 : 7205–9.
25. Field, M. G., Decatur, C. L., Kurtenbach, S., et al.: PRAME as an Independent Biomarker for Metastasis in Uveal Melanoma. Clinical Cancer Research, 22(5);2016 : 1234–42.
26. Gupta, M. P., Lane, A. M., DeAngelis, M. M., et al.: Clinical Characteristics of Uveal Melanoma in Patients With Germline BAP1 Mutations. JAMA Ophthalmol, 133(8);2015 : 881–7.
27. Decatur, C. L., Ong, E., Garg, N., et al.: Driver mutations in uveal melanoma: associations with gene expression profile and patient outcomes. JAMA ophthalmology, 134(7);2016 : 728–33.
28. Helgadottir, H., Höiom, V. The genetics of uveal melanoma: current insights. The Application of Clinical Genetics, 9;2016 : 147–55.
29. Herlihy, N., Dogrusoz, M., van Essen, T. H., et al.: Skewed expression of the genes encoding epigenetic modifiers in high-risk uveal melanoma. Invest Ophthalmol Vis Sci, 56(3);2015 : 1447–58.
30. Li, Y., Jia, R., Ge, S. Role of Epigenetics in Uveal Melanoma. International journal of biological sciences, 13(4);2017 : 426–33.
31. Svoreňová, M. L., Smolková, M. B. Úloha epigenetickej regulácie v procese epiteliálno-mezenchýmového prechodu, 12;2017 : 410–413.
32. Catalanotto, C., Cogoni, C., Zardo, G. MicroRNA in Control of Gene Expression: An Overview of Nuclear Functions. International journal of molecular sciences, 17(10); 2016 : 1712.
33. Smit, K. N., Chang, J., Derks, K., et al.: Aberrant MicroRNA Expression and Its Implications for Uveal Melanoma Metastasis. Cancers, 11(6);2019 : 815.
34. Chakraborty, C., Sharma, A. R., Sharma, G., et al.: Therapeutic miRNA and siRNA: Moving from Bench to Clinic as Next Generation Medicine. Mol Ther Nucleic Acids, 8;2017 : 132–43.
35. Chen, X., Wang, J., Shen, H., et al.: Epigenetics, microRNAs, and carcinogenesis: functional role of microRNA-137 in uveal melanoma. Invest Ophthalmol Vis Sci, 52(3);2011 : 1193–9.
36. Chen, X., He, D., Dong, X. D., et al.: MicroRNA-124a is epigenetically regulated and acts as a tumor suppressor by controlling multiple targets in uveal melanoma. Invest Ophthalmol Vis Sci, 54(3); 2013 : 2248–56.
37. Landreville, S., Agapova, O. A., Matatall, K. A., et al.: Histone deacetylase inhibitors induce growth arrest and differentiation in uveal melanoma. Clin Cancer Res, 18(2);2012 : 408–16.
38. Moschos, M. M., Dettoraki, M., Androudi, S., et al.: The role of histone deacetylase inhibitors in uveal melanoma: current evidence. Anticancer research, 38(7);2018 : 3817–24.
39. Piperno-Neumann, S., Piulats, J. M., Goebeler, M., et al.: Uveal Melanoma: A European Network to Face the Many Challenges of a Rare Cancer. Cancers (Basel), 11(6);2019 : 817.
40. Gaudet, F., Hodgson, J. G., Eden, A., et al.: Induction of Tumors in Mice by Genomic Hypomethylation. Science, 300(5618);2003 : 489–92.
41. Eden, A., Gaudet, F., Waghmare, A., et al.: Chromosomal Instability and Tumors Promoted by DNA Hypomethylation. Science, 300(5618);2003 : 455.
42. Yang, A. S., Estecio, M. R., Garcia-Manero, G., et al.: Comment on „Chromosomal instability and tumors promoted by DNA hypomethylation“ and „Induction of tumors in nice by genomic hypomethylation“. Science, 302(5648);2003 : 1153.
43. Morel, D., Jeffery, D., Aspeslagh, S., et al.: Combining epigenetic drugs with other therapies for solid tumours - past lessons and future promise. Nat Rev Clin Oncol;2019.
Labels
Ophthalmology
Article was published inCzech and Slovak Ophthalmology

2020 Issue 2-
All articles in this issue
- Zemřela prof. MUDr. Jarmila Boguszaková DrSc.
- Vzpomínka na doc. MUDr. Cigánka, CSc.
- EPIGENETIC CHANGES IN MALIGNANT UVEAL MELANOMA AND POSSIBILITIES OF THEIR THERAPEUTIC TARGETING
- Primary Intrabulbar Neurofibroma
- Does adjuvant intracameral triamcinolone acetonide increase the effectiveness of phacotrabeculectomy? A Case-Control Study
- Non-arteritic anterior ischaemic optic neuropathy: treatment and risk factors
- Aflibercept for Vascularised Serous Pigment Epithelial Detachment: One-Year Anatomical and Functional Results
- Betaxolol, Brimonidin and Carteolol in the Therapy of Normal-Tension Glaucoma
- Czech and Slovak Ophthalmology
- Journal archive
- Current issue
- Online only
- About the journal
Most read in this issue- Non-arteritic anterior ischaemic optic neuropathy: treatment and risk factors
- Vzpomínka na doc. MUDr. Cigánka, CSc.
- Betaxolol, Brimonidin and Carteolol in the Therapy of Normal-Tension Glaucoma
- Aflibercept for Vascularised Serous Pigment Epithelial Detachment: One-Year Anatomical and Functional Results
Login#ADS_BOTTOM_SCRIPTS#Forgotten passwordEnter the email address that you registered with. We will send you instructions on how to set a new password.
- Career

