-
Medical journals
- Career
Colistin resistance and heteroresistance in Klebsiella pneumoniae & Escherichia coli clinical isolates from intensive care units
Authors: M. A. Meheissen 1; S. M. Hendawy 1; F. S. Shabaan 2; A. M. Elmenshawy 3; R. A. Harfoush 1
Authors‘ workplace: Medical Microbiology and Immunology Department, Faculty of Medicine, Alexandria University, Egypt 1; Community Department, Faculty of Medicine, Alexandria University, Egypt 2; Critical care Department, Faculty of Medicine, Alexandria University, Egypt 3
Published in: Epidemiol. Mikrobiol. Imunol. 71, 2022, č. 2, s. 86-92
Category: Original Papers
Overview
Aim: Colistin resistance poses a serious clinical problem. This study aims to investigate the presence of plasmid-mediated resistance and heteroresistance among Escherichia coli (E. coli) and Klebsiella pneumoniae (K. pneumoniae) clinical isolates at intensive care units of Main Alexandria University Hospital.
Material and Methods: Seventy colistin-resistant and 30 colistin-susceptible K. pneumoniae and E. coli isolates constituted the material of the study. The rapid polymyxin NP (Nordmann Poirel) test was performed. Heteroresistance in colistin-susceptible isolates was investigated by population analysis profile. mcr-1 and mcr-2 genes were amplified by polymerase chain reaction (PCR).
Results: The rapid polymyxin NP test showed 100% concordance with the results of BMD (broth microdilution method). Heteroresistance to colistin was detected in 23.3% of colistin-susceptible K. pneumoniae isolates. All colistin-resistant isolates were negative for mcr-1 and mcr-2 genes.
Conclusions: The rapid polymyxin NP test is a reliable screening tool for colistin resistance, but not for heteroresistance. Other colistin resistance mechanisms should be investigated.
Keywords:
colistin – plasmid resistance – heteroresistance – rapid polymyxin NP
INTRODUCTION
The emergence of extensive drug resistant gram-negative bacilli, especially the carbapenem resistant ones, had led to increase use of colistin in the last decade [1].
Colistin resistance has been reported worldwide either via chromosomal genes or plasmids. The plasmid-encoded resistance was first described by Liu and colleagues [2], in Escherichia coli (E. coli) and Klebsiella pneumoniae (K. pneumoniae) animal and human isolates recovered from China. It is mediated by mcr-1 (mobile colistin resistance) gene that encodes phosphoethanolamine transferase enzyme, conferring resistance to colistin. This type of resistance is responsible for horizontal transfer and dissemination of stable colistin resistance among gram-negative bacilli posing a major public health concern [2].
To date, ten mcr variants (mcr-1 to mcr-10) showing nucleotide sequence similarity to each other have been described in Enterobacterales and other genera [3].
Heteroresistance is the presence of an antibiotic-resistant subpopulation of microbes within a larger population that is susceptible. Detection of heteroresistance is problematic in-vitro and most probably promotes resistance to antibiotics in-vivo. Because of lack of uniform standards to test colistin heteroresistance, the rates from different countries varied greatly [4].
Colistin resistance is reported sporadically among extensive drug resistant gram-negative isolates at our university hospital and no data is available about the mechanism of resistance in those isolates. Moreover, as colistin is considered the last line treatment in those cases, therefore, the present study was conducted to investigate the presence of plasmid-mediated resistance (mcr-1 and mcr-2 genes) and heteroresistance among K. pneumoniae and E. coli clinical isolates at intensive care units (ICUs) of Main Alexandria University Hospital (MAUH), Egypt. The study also aimed to evaluate the performance of the rapid polymyxin Nordmann Poirel (NP) test for phenotypic detection of colistin resistance in K. pneumoniae and E. coli clinical isolates.
MATERIAL AND METHODS
Seventy colistin-resistant and 30 colistin-susceptible K. pneumoniae and E. coli clinical isolates, randomly selected, constituted the material of this study. A sample size of 70 isolates, calculated using GPower version 3.1.9.2, was enough to detect the prevalence of plasmid mediated colistin resistance in K. pneumoniae and E. coli, to reach 80% power of the study and at a significance level of 0.05. The isolates were isolated from different ICUs of MAUH during the study period starting from the first of July 2018 till the end of June 2020. All included isolates were extensive drug resistant (XDR). XDR was defined as non-susceptibility to at least one agent in all but two or fewer antimicrobial categories (i.e. bacterial isolates remain susceptible to only one or two categories) [5]. Out of the 30 colistin-susceptible isolates, 20 were K. pneumoniae and ten were E. coli. While out of 70 colistin-resistant isolates, 65 were K. pneumoniae and 5 were E. coli.
The study was approved by Medical Research Ethics Committee of Alexandria Faculty of Medicine.
Selection of isolates
During the study period, a total of 3369 Enterobacterales isolates (1826 Klebsiella spp., 1149 E. coli, 307 Proteaee, 60 Citrobacter spp., and 27 Enterobacter spp.) were collected from ICU patients. Out of Klebsiella spp. and E. coli isolates, only 70 isolates (70/2975; 2.3%) were colistin resistant and were included in the study. Thus, it took almost two years to collect those 70 isolates. The authors randomly selected only 30 colistin susceptible isolates for testing heteroresistance as the test is very laborious.
Clinical data collection
All relevant clinical data corresponding to the study isolates were recorded including: age, sex, previous antibiotic use data, type of specimen, type of ICU.
Microbiological processing
1. Identification to the species level
Identification of the study isolates was performed using VITEK 2 compact system according to the manufacturer’s instructions.
2. Antimicrobial susceptibility testing (AST)
AST was performed by Kirby Bauer disc diffusion method according to Clinical Laboratory Standards Institute (CLSI) 2020 recommendations [6]. The interpretive zone breakpoints that are recommended for cefoperazone discs (75 μg) were used for interpretation of cefoperazone/sulbactam (75/10 μg) susceptibility. Tigecycline (15 μg) zone diameters were interpreted according to European Committee on Antimicrobial Susceptibility Testing (EUCAST), 2020 [7].
3. Detection of colistin resistance by phenotypic methods
3.1. Broth microdilution (BMD) reference method was done according to CLSI recommendations [8]. Minimum inhibitory concentration (MIC) was interpreted according to the recommended MIC breakpoints for Enterobacterales of EUCAST [7]: > 2 μg/mL (Resistant), ≤ 2 μg/mL (Susceptible).
3.2. Rapid polymyxin NP (Nordmann Poirel) test: It was performed as previously described [9]. Briefly, a rapid polymyxin reagent containing a mixture of 2.5% of cation adjusted Muller Hinton powder, 0.005% of phenol red indicator and 1% of d (+)-glucose was prepared. Bacterial suspension equivalent to 109 CFU/mL was inoculated in parallel into 2 wells of a microtiter plate, with and without colistin (5 μg/mL) and mixed to the reactive medium. A colistin-susceptible (E. coli ATCC 25922) isolate and a colistin-resistant (intrinsically polymyxin-resistant Proteus mirabilis ATCC 14153) were included as controls. The inoculated microtiter plate was incubated up to 4 hours at 37 °C in ambient air, not sealed and without agitation. A positive test was indicated by a color change (from orange to yellow) when bacteria metabolize glucose in the medium in the presence of colistin.
4. Detection of heteroresistance to colistin using population analysis profile (PAP)
It was performed for the 30 colistin-susceptible isolates using the method described by Meletis et al. [10] by spreading aliquot of 50 μL from tenfold serially diluted bacterial suspension, equivalent to 108 CFU/mL, on Mueller Hinton agar plates containing colistin sulfate concentrations of 0, 0.5, 1, 2, 3, 4, 5, 6, 8 and 10 μg/ mL. The plates were incubated at 37 °C for 48 hours then colonies were counted. The frequency of resistant subpopulations at the highest drug concentration was calculated by dividing the number of colonies grown on an antibiotic - containing plate by the colony counts from the same bacterial inoculum plated onto antibiotic - free plates. The MIC for heteroresistant colonies was reassessed by serial daily subcultures on antibiotic-free medium for one week in order to evaluate whether this resistance was stable. K. pneumoniae ATCC 700603 was included as a negative control (colistin-susceptible) strain in the test.
5. Detection of plasmid mediated colistin resistance by multiplex PCR
The 70 colistin-resistant isolates and heteroresistant isolates were screened for the presence of mcr-1 [2] and mcr-2 [11] genes by multiplex PCR as previously described. Despite there are 10 mcr variants, the current work tested for presence of only the most common two of them for screening purposes. The primers used were as follows: mcr-1 forward: CGGTCAGTCCGTTTGTTC, mcr-1 reverse: CTTGGTCGGTCTGTAGGG, mcr-2 forward: TGTTGCTTGTGCCGATTGGA, mcr-2 reverse: AGATGGTATTGTTGGTTGCTG, using a cycling condition of initial denaturation at 94 °C for 15 min, 25 cycles of 94°C for 30 sec, 58°C for 90 sec and 72°C for 60 sec, and a final elongation at 72°C for 10 min. An internal control gene (16SrRNA) was included in each PCR run.
Statistical analysis
After completing data collection, data entry was performed using Statistical Package for Social Sciences (SPSS ver. 22). Parametric and non-parametric tests were used as appropriate. The level of significance of the results was 0.05.
RESULTS
Clinical data of the isolates are demonstrated in table 1. None of the patients had history of travel within the preceding 12 months. All patients received antibiotics in the previous one month or were already on antibiotic treatment while taking the sample. Around 50% of colistin-resistant and -susceptible groups received previous combination of a glycopeptide and a carbapenem. Four patients from the resistant group received previous colistin treatment (Figure 1).
1. The characteristics of the study isolates 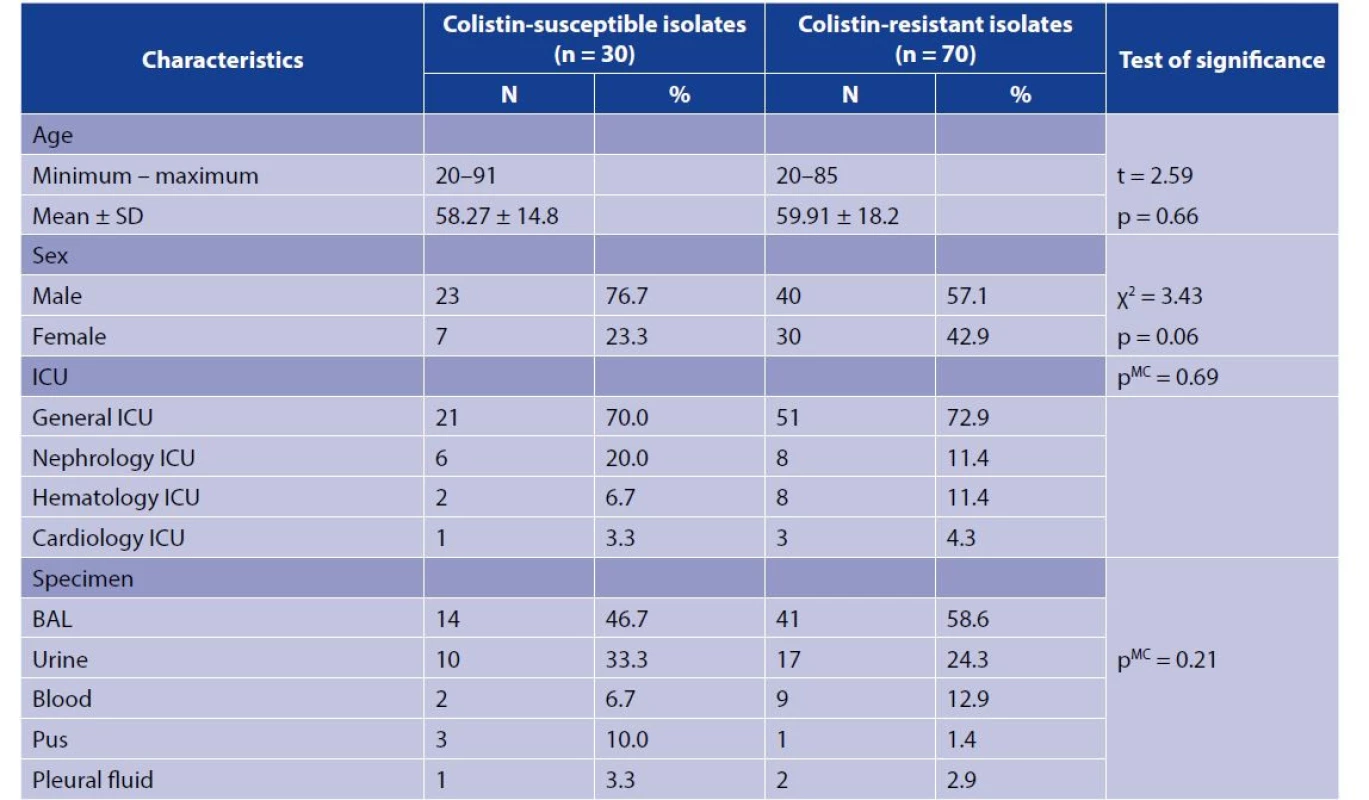
*Significant (p < 0.05)
t = Independent sample t test
pMC = p value of Monte Carlo test
χ2 = Chi-square testFigure 1. Types of previously used antibiotics in the two study groups 
The isolates of the current study were selected to be XDR. Regarding colistin-resistant isolates, they were 100% resistant to all antibiotics except meropenem (69/70; 98.6%), amikacin (68/70; 97.1%), trimethoprim/ sulfamethoxazole (64/70; 91.4%), tetracycline (63/70; 90%), gentamicin (62/70; 86.7%), doxycycline (59/70; 84.3%), and tigecycline (43/70; 61.4%). Similarly, all colistin-susceptible isolates were 100% resistant to most antibiotics except gentamicin (26/30; 86.7%), amikacin (27/30; 90%), tetracycline (25/30; 83.3%), doxycycline (24/30; 80%), tigecycline (19/30; 63.3%), and sulfamethoxazole-trimethoprim (29/30; 96.7%). Out of 17 urine isolates in colistin-resistant group, 15 (88.2%) were resistant to nitrofurantoin, while all (100%) urine isolates from colistin-susceptible group were resistant to nitrofurantoin. As recommended by CLSI 2020, fosfomycin was not tested for K. pneumoniae. All E. coli isolates (colistin-susceptible and colistin-resistant) were susceptible to fosfomycin.
Regarding the performance of the rapid polymyxin NP test, all K. pneumoniae and E. coli colistin-resistant isolates gave positive NP test results. Similarly, the 30 colistin-susceptible isolates (K. pneumoniae and E. coli) gave negative rapid NP results, including the seven colistin heteroresistant K. pneumoniae isolates. Thus, the test failed to detect all heteroresistants. Therefore, excluding the heteroresistant isolates, the rapid NP test showed an excellent correlation with BMD test, providing sensitivity, specificity, positive, negative predictive values and accuracy of 100%. The color change of the NP test wells was inspected and recorded every hour, all positive isolates gave final results from 2–3 hours incubation at 37 °C under aerobic conditions. All negative isolates remained negative ≥ 4 hours (Figure 2).
Figure 2. Results of rapid polymyxin NP test in the present study 
The upper row: colistin-free solution. The lower row: colistin- containing solution. 1A, 1B: non-inoculated wells, saline controls (first column). 2A, 2B: colistin-susceptible reference strain (second column) showing a positive reaction in upper row and negative result in the lower row. 3A, 3B: Colistin resistant reference strain (third column) showing positive results in the upper and the lower rows. 4A, 4B: A tested (resistant) isolate showing a positive test result. 5A, 5B: A tested (susceptible) isolate showing a negative test result. Population analysis profile revealed colistin heteroresistance in seven clinical K. pneumoniae isolates initially identified as colistin-susceptible based on MIC value of BMD test (≤ 2 μg/mL). Resistant subpopulations in colistin heteroresistant isolates grew in colistin concentrations ranging from 3 to 10 μg/mL. The mean frequency of resistant subpopulations was 7.6 x 107 (0.0000076). Resistant subpopulations had MICs ranging from 4 to 32 μg/mL. The resistant subpopulations of the seven isolates demonstrated unstable resistance after repeated subculture for one week on colistin free medium (Table 2, Figure 3). The colistin-susceptible ATCC 700603 isolate survived up to 0.5 μg/mL colistin sulfate, with no heteroresistant subpopulations observed. None of the patients from whom those isolates were recovered has received previous colistin treatment. All heteroresistant K. pneumoniae isolates were negative for rapid polymyxin NP test and all were negative for mcr-1 and mcr-2 genes.
2. Characteristics of the seven heteroresistant K. pneumoniae clinical isolates studied by population analysis profile (PAP) 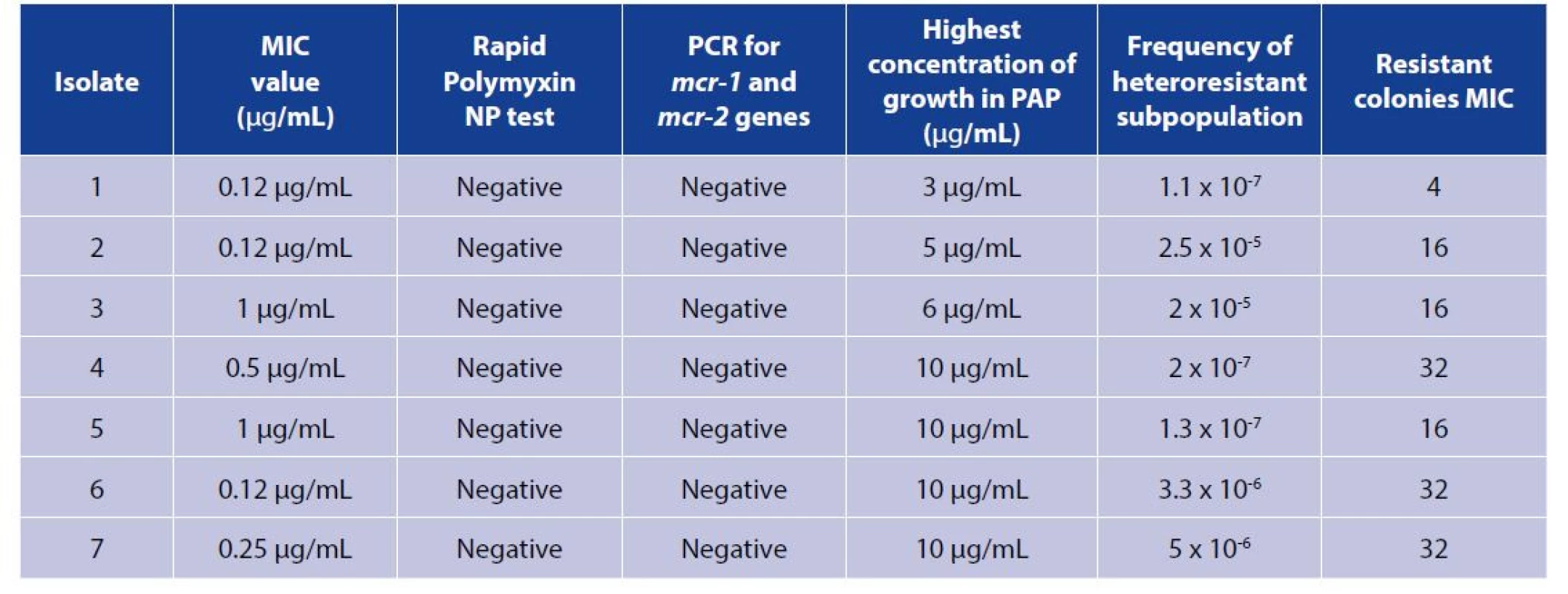
Figure 3. Population analysis profiles (PAP) of seven K. pneumoniae heteroresistant clinical isolates and one colistin susceptible K. pneumoniae ATCC 700603 
PCR results showed that mcr-1 and mcr-2 genes were not detected in any of the 70 colistin-resistant isolates.
DISCUSSION
Concerning the clinico-microbiological profile of the 70 colistin-resistant isolates in this study, respiratory samples (58.5%), were the most prevalent, followed by urine (24.3%), and blood (13%). This agreed with the results of Emara et al. [12] where sputum (50%), and urine (30%) were the predominant samples in the 10 patients with colistin resistant isolates.
While comparing the performance of the rapid polymyxin NP test and the BMD reference method in the current study, the colistin-resistant group gave positive NP test results from 2–3 hours after incubation, showing an excellent agreement with BMD, with no VME; very major error (false susceptibility) detected. Similarly, Bakthavatchalam et al. [13], studied 232 non-duplicate Enterobacteriaceae bloodstream isolates in India, they reported a high degree of concordance between BMD and the rapid polymyxin NP test in detection of colistin-resistant isolates in less than 3 hours.
On the other hand, all our 30 colistin-susceptible isolates gave negative rapid polymyxin NP results, missing the seven colistin heteroresistant K. pneumoniae isolates that were further detected by PAP. So, by excluding the heteroresistant isolates, the rapid polymyxin NP test provided 100% sensitivity, specificity, positive, negative predictive values and accuracy. In the study of Jayol et al. [14], out of the 40 colistin-susceptible enterobacterial isolates, only a single susceptible K. pneumoniae isolate with a colistin MIC value of 2 μg/mL was identified as colistin-resistant with the Rapid Polymyxin NP test, at a rate of 2.5%. The study group did not perform PAP test to detect heteroresistance, thus they interpreted this result as major error (false resistance). Interestingly, in an Indian study, the authors reported one heteroresistant K. pneumoniae isolate that was found to be positive by rapid polymyxin NP test [13].
Heteroresistance to colistin, or the presence of resistant subpopulations within phenotypically susceptible populations is known to occur in K. pneumoniae. The real prevalence of colistin heteroresistance is unknown; due to difficulty in its detection by routine susceptibility methods [10]. Although based on relatively few isolates, two studies showed remarkably high prevalence of colistin heteroresistance; 75% (12 out of 16) and 94% (15 out of 16) respectively [10, 15]. In contrary to our results, that revealed a low prevalence 23.3% (7 out of 30) with MIC values ranging from 4 to 32 μg/mL. The absence of colistin pressure as evidenced by the absence of previous colistin intake among patients from whom our isolates were recovered might explain this low heteroresistance rate.
The rate of the resistant subpopulations varies in different studies; Poudyal et al. [15] reported the proportion of resistant subpopulations among K. pneumoniae heteroresistant isolates as ranging from 6.03x10-9 – 1.29x10-5 from the main populations. Meletis et al. [10] similarly, reported the range of the resistant subpopulations as ranging from 10-7 to 10-5 in isolates exposed or not exposed to colistin. This was in accordance with the present work, where the mean frequency of resistant subpopulations in colistin heteroresistant K. pneumoniae isolates was 7.6 x 10-7. The genetic background and the dynamics of heteroresistant isolates in the present study should be included in a further study. Time kill assay should be performed to confirm or negate the stability of the resistant subpopulation. Furthermore, failure of detection of resistance in the seven heteroresistant isolates by BMD may be due to the fact that the antibiotic resistant subpopulation was present at a very low frequency (1 in 7.6 x 107 cells) to be detected by routine tests.
We could not relate the heteroresistance in K. pneumoniae isolates of the present study to a specific resistance mechanism, as all heteroresistant isolates were negative for mcr-1 and mcr-2 genes, and other colistin resistance mechanisms (including mutations) were not investigated. Regardless of the mechanism, it is likely that resistant subpopulations may become dominant during colistin treatment, by selective pressure, eventually leading to treatment failure. This was previously confirmed in an in-vivo murine model, where carbapenem resistant K. pneumoniae having resistant colistin subpopulations exhibited treatment failure [16]. Thus, heteroresistance phenomenon merits further investigations as it points to a serious clinical problem.
The use of PCR in the present study failed to detect mcr-1 and mcr-2 genes in any isolate of the colistin-resistant group. The resistance in these isolates could be due to other resistance mechanisms (chromosome encoded resistant gene, alteration in the two-component system, other plasmid mediated genes). This agreed with the results of a previous study, which could not detect mcr-1 gene in 11 colistin-resistant K. pneumoniae isolates out of 219 gram-negative isolates [17]. Previous studies reported a very low prevalence of mcr-1 gene in Enterobacteriaceae (0.1–1%) that was mainly detected in E. coli [18, 19]. The prevalence of the mcr-1 gene was notably lower in isolates from human sources than from animal source [2]. The absence of mcr-1 and mcr-2 genes in the present study may be due to the inclusion criteria of the included isolates; all were XDR, or the very low number of resistant E. coli tested (5 isolates). The other mechanisms of colistin resistance were not investigated as this was not the objective of the current study. The authors‘ aim was to investigate the presence of the mobile transmissible type of resistance (plasmid-mediated) which poses more risk in healthcare settings.
CONCLUSIONS
The rapid polymyxin NP test was found to be a reliable, cheap, rapid screening tool for detection of colistin resistance, regardless of the molecular mechanism of colistin resistance (plasmid, chromosomal), and it can be used as an alternative of BMD when MIC value determination is not mandatory. However, rapid polymyxin NP is not reliable for detection of colistin heteroresistance in K. pneumoniae and E. coli clinical isolates in our settings. The absence of mcr-1 and mcr-2 genes from the tested isolates does not exclude their presence in our hospital setting. Conducting further studies, with large number of clinical isolates, to determine the responsible possible molecular mechanisms (chromosomal and plasmid mediated) of colistin resistance is crucial.
Acknowledgment
None.
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Conflicts of interest
None.
Do redakce došlo dne 27. 6. 2021.
Correspondence address:
Dr. Marwa Ahmed Meheissen
Medical Microbiology & Immunology Department
Faculty of Medicine, Alexandria University
Kartoum Square
Azarita, 21512, Egypt
E-mail: marwa.meheissen@alexmed.edu.eg
Sources
1. Poirel L, Jayol A, Nordmann P. Polymyxins: antibacterial activity, susceptibility testing, and resistance mechanisms encoded by plasmids or chromosomes. Clin Microbiol Rev, 2017; 30(2):557 – 596. doi: 10.1128/CMR.00064-16.
2. Liu Y-Y, Wang Y, Walsh TR, et al. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect Dis, 2016;16 : 161–168. doi:10.1016/S1473 - 3099(15)00424-7.
3. Wang C, Feng Y, Liu L, et al. Identification of novel mobile colistin resistance gene mcr-10. Emerg Microbes Infect, 2020;9(1):508 – 516. doi:10.1080/22221751.2020.1732231.
4. El-Halfawy OM, Valvano MA. Antimicrobial heteroresistance: an emerging field in need of clarity. Clin Microbiol Rev, 2015;28(1):191–207. doi: 10.1128/CMR.00058-14.
5. Magiorakos AP, Srinivasan A, Carey RB, et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect, 2012;18(3):268 – 281. doi: 10.1111/j.1469-0691.2011.03570.x.
6. Clinical and Laboratory Standards Institute (CLSI). Performance Standards for Antimicrobial Susceptibility Testing. 30th ed. CLSI supplement M100. Wayne, Pennsylvania, USA: Clinical and Laboratory Standards Institute; 2020.
7. The European Committee on Antimicrobial Susceptibility Testing. Breakpoint tables for interpretation of MICs and zone diameters. Version 10.0; 2020. Dostupné na www: http://www. eucast.org.
8. Clinical and Laboratory Standards Institute (CLSI). Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically; Approved Standard – Ninth edition. CLSI document M07-A9. Wayne, PA: Clinical and Laboratory Standards Institute; 2012.
9. Nordmann P, Jayol A, Poirel L. Rapid detection of polymyxin resistance in Enterobacteriaceae. Emerg Infect Dis, 2016;22(6):1038 – 1043. doi: 10.3201/eid2206.151840.
10. Meletis G, Tzampaz E, Sianou E, et al. Colistin heteroresistance in carbapenemase-producing Klebsiella pneumoniae. J Antimicrob Chemother, 2011;66(4):946–947. doi: 10.1093/jac/dkr007.
11. Xavier BB, Lammens C, Ruhal R, et al. Identification of a novel plasmid-mediated colistin - resistance gene, mcr-2, in Escherichia coli, Belgium. Euro Surveill, 2016;21(27):30280. doi: 10.2807/1560-7917.ES.2016.21.27.30280.
12. Emara MM, Abd-Elmonsef ME, Abo Elnasr LM, et al. Study of mcr-1 gene-mediated colistin-resistance in Gram-negative isolates in Egypt. Egyp J Med Microbiol, 2019;28(3):9–16.
13. Bakthavatchalam YD, Veeraraghavan B, Mathur P, et al. Polymyxin Nordmann/Poirel test for rapid detection of polymyxin resistance in Enterobacteriaceae: Indian experience. Indian J Med Microbiol, 2016;34(4):564–565. doi: 10.4103/0255-0857.195382.
14. Jayol A, Nordmann P, Lehours P, Poirel L, Dubois V. Comparison of methods for detection of plasmid-mediated and chromosomally encoded colistin resistance in Enterobacteriaceae. Clin Microbiol Infect, 2018;24(2):175–179. doi: 10.1016/j.cmi.2017.06.002.
15. Poudyal A, Howden BP, Bell JM, et al. In vitro pharmacodynamics of colistin against multidrug-resistant Klebsiella pneumoniae. J Antimicrob Chemother, 2008;62 : 1311–1318. doi:10.1093/jac/ dkn425.
16. Band V, Satola SW, Burd EM, et al. Carbapenem-resistant Klebsiella pneumoniae exhibiting clinically undetected colistin heteroresistance leads to treatment failure in a murine model of infection. MBio, 2018;9(2). pii: e02448–17. doi: 10.1128/ mBio.02448-17.
17. Kalaiselvi T. Study of colistin sensitivity among clinical isolates of extensively drug resistant gram negative bacilli. PSG institute of medical sciences and research, peelamedu, coimbatore, tamilnadu, india; 2018. Dostupé na www: www.semanticscholar.org.
18. Prim N, Turbau M, Rivera A, et al. Prevalence of colistin resistance in clinical isolates of Enterobacteriaceae: a four-year crosssectional study. J Infect, 2017;75(6):493–498. doi: 10.1016/j. jinf.2017.09.008.
19. Castanheira M, Griffin MA, Deshpande LM, et al. Detection of mcr-1 among Escherichia coli clinical isolates collected worldwide as part of the SENTRY Antimicrobial Surveillance Program in 2014 and 2015. Antimicrob Agents Chemother, 2016;60 : 5623 – 5624. doi: 10.1128/AAC.01267-16.
Labels
Hygiene and epidemiology Medical virology Clinical microbiology
Article was published inEpidemiology, Microbiology, Immunology

2022 Issue 2-
All articles in this issue
- Monitoring changes in invasive disease caused by Haemophilus influenzae in the Czech Republic between 1999 and 2020
- Norovirus infections in the Czech Republic in 2008–2020
- Incidence of tuberculosis among HIV-positive persons in the Czech Republic between 2000 and 2020
- Genetic diversity of human strains of Listeria monocytogenes in the Czech Republic in 2016–2020
- Candida glabrata – basic characteristics, virulence, treatment, and resistance
- First evaluation of completeness and sensitivity of the measles surveillance system in the Czech Republic, January 1, 2018 until June 30, 2019
- Colistin resistance and heteroresistance in Klebsiella pneumoniae & Escherichia coli clinical isolates from intensive care units
- Epidemiology, Microbiology, Immunology
- Journal archive
- Current issue
- Online only
- About the journal
Most read in this issue- Candida glabrata – basic characteristics, virulence, treatment, and resistance
- Incidence of tuberculosis among HIV-positive persons in the Czech Republic between 2000 and 2020
- Norovirus infections in the Czech Republic in 2008–2020
- Monitoring changes in invasive disease caused by Haemophilus influenzae in the Czech Republic between 1999 and 2020
Login#ADS_BOTTOM_SCRIPTS#Forgotten passwordEnter the email address that you registered with. We will send you instructions on how to set a new password.
- Career

