-
Medical journals
- Career
Clinical values of two estrogen receptor signaling targeted lncRNAs in invasive ductal breast carcinoma
Authors: S. Ilbeigi 1; Y. Naeimzadeh 2; M. Davoodabadi Farahani 1; M. Ee Monjezi Rafi 3; H. Dastsooz 4; A. Daraei 5; F. Farahani 1; A. Dastgheib 1; Y. Mansoori 6; M. Tabei S. Bagher 1
Authors‘ workplace: Department of Medical Genetics, School of Medicine Shiraz University of Medical Science, Shiraz, Iran 1; Department of Biology, Faculty of Sciences, Yazd University, Yazd, Iran 2; Department of Medical Immunology, Shiraz University of Medical Science, Shiraz, Iran 3; Department of life science and system biology, University of Turin, Turin, Italy 4; Department of Genetics, Faculty of Medicine, Babol University of Medical Sciences, Babol, Iran 5; Department of Medical Genetics, Fasa University of Medical Science, Fasa, Iran 6
Published in: Klin Onkol 2021; 34(5): 382-391
Category: Original Articles
doi: https://doi.org/10.48095/ccko2021382Overview
Background: Invasive ductal carcinoma (IDC) is the most frequent type of breast cancer (BC) in women, with a high clinical burden due to its high invasive properties. Despite of quickly emerging new data regarding the molecular heterogeneity of invasive cancers, far less is known about the molecular patterns among cases of IDC. An expanding body of evidence has demonstrated that dysregulation of long noncoding RNAs (lncRNAs) is involved in the heterogeneity feature of BC. Methods: In this study, we analyzed the expression levels of two novel lncRNAs LOC100288637 and RP11-48B3 in 51 IDC tissues in comparison with adjacent non-cancerous tissues. And finally, bioinformatic evaluation has been done. Results: The results of quantitative polymerase chain reaction showed that LOC100288637 and RP11-48B3 were significantly overexpressed in tumor tissues compared to normal samples (P = 0.0085 and P = 0.0002, respectively). Also, the two lncRNAs were overexpressed in both MDA-MB-231 and MCF-7 BC cell lines, nevertheless, with a higher expression pattern in MDA-MB-231 than MCF7 cell line. Furthermore, LOC100288637 had an elevated expression level in HER-2 positive tumors compared to HER-2 negative tumors (P = 0.031). Interestingly, the lncRNA RP11-48B3.4 was upregulated in IDC subjects with the age at menarche < 14 years compared to patients with the age at menarche ≥ 14 (P = 0.041). It was observed in another result that lncRNA RP11-48B3.4 is significantly upregulated in tumors with a lower histological grade compared to tumor samples with higher grades (P = 0.047). And finally, using bioinformatic evaluation, we found a predicted interaction between RP11-48B3.4 and mRNA zinc finger and BTB domain containing 10 (ZBTB10). Conclusion: Altogether, our findings suggest that these lncRNAs with potential oncogenic roles are involved in the pathogenesis of IDC with clinical significance and they may therefore serve as novel markers for the diagnosis and treatment of IDC.
Keywords:
Prognosis – invasive ductal breast carcinoma – long noncoding RNAs – LOC100288637 – RP11-48B3
Introduction
Breast cancer (BC) is the second cause of death among women worldwide [1]. BC is a highly heterogeneous disease which is characterized by different phenotypic features, diverse response to existing therapy and unpredictable clinical outcome [2]. Invasive ductal carcinoma (IDC) and invasive lobular carcinoma (ILC) are the most common variants of the BC [3]. IDC, also known as infiltrating carcinoma, is accounted for about 80% of all invasive BC [4]. The relevant molecular mechanism of IDC is not elucidated and thus the optimal management of IDC has become increasingly complex. Further investigation is urgent to identify effective approaches for the diagnosis and treatment of IDC [5].
Long noncoding RNA (lncRNA) are a class of RNA molecules longer than 200 nucleotides [6]. They regulate gene expression at the transcriptional, post-transcriptional, and epigenetic levels and also play an important role in pathological processes such as neurological disorders, diabetes, tumors [7,8] and more. Recent studies have clarified vital roles of several lncRNAs in BC pathobiology. Accordingly, lncRNAs could be a potential biomarker for BC prognosis, diagnosis, and therapeutic management [9]. Although cell - specific expression of lncRNAs and their role in regulating the expression of oncogenic and tumor suppressor genes have been shown in recent investigations, their underlying molecular mechanisms in IDC remains undetermined [10–12]. In recent years, a number of lncRNAs including LOC100288637 and RP11-48B3 were found to be associated with abnormal estrogen receptor (ER) signaling pathway in BC, suggesting that lncRNAs can be applied as prognosis biomarker in patients with breast cancer [13,14]. AS mentioned previously, the information on their roles in IDC remains unclear. Therefore, in the presented study, we assessed the expression levels of LOC100288637 and RP11-48B3 in patients with IDC as well as BC cell lines and analyzed their correlation with clinicopathological parameters.
Materials and methods
Study population
A total of 51 invasive ductal carcinoma breast cancer patients who had received no chemotherapy or radiotherapy were included in this study. All subjects were selected from individuals referred to Faghihie Hospital affiliated to Shiraz University of Medical Sciences. Written informed consent was obtained from all patients, and the study was approved by the Ethics Committee of Shiraz University of Medical Sciences, Shiraz, Iran. The demographic, reproductive and clinical information as well as the pathological data of tumor samples were obtained from each participant in the current study. Tab. 1 shows the characteristics of the study participants. The participants were 27–68 years old, 15 (30%) participants were aged < 40 years, and 36 (70%) were ≥ 40 years old. Among the 51 participants, 36 (70%) were parous and 15 (30%) were nulliparous. Regarding the menopausal status, 4 (8%) participants were premenopausal and 47 (92%) were in postmenopausal status. Moreover, the participants were divided into 2 subgroups based on the age at menarche of < 14 or ≥ 14 years. The age at the first full term pregnancy and breast-feeding duration were also recorded. Parous participants were divided into 2 subgroups according to the age at the first full-term pregnancy of < 25 or ≥ 25 years.
1. Demographic and reproductive characteristics of the participant subjects. 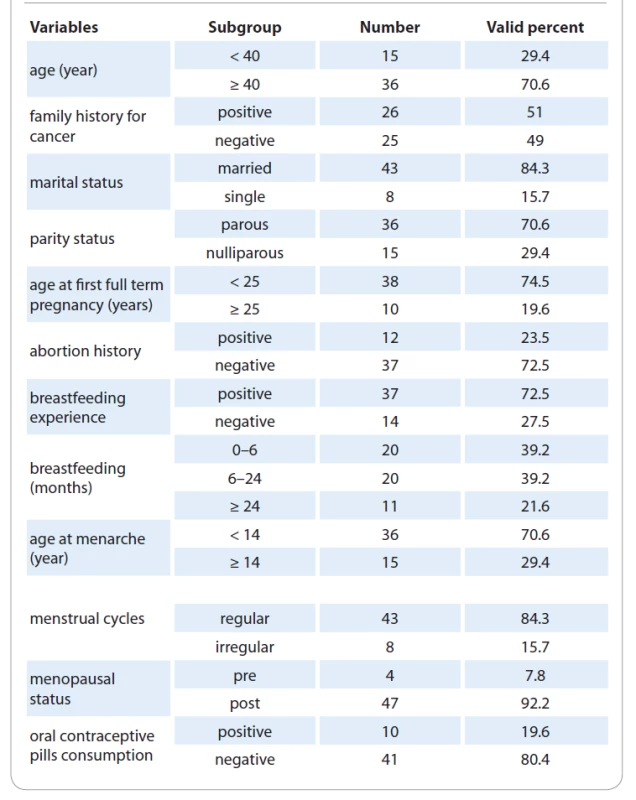
Breast tissue sampling
Fresh breast cancer tissues and their adjacent noncancerous tissues were snap-freeze in liquid nitrogen after resection and stored at −80 °C until RNA extraction.
Estrogen receptor, progesterone receptor and HER2 determination
The estrogen receptor (ER), progesterone receptor (PR), and HER2 status were determined according to the patient’s histopathological data, following immunohistochemistry (IHC) staining. If ≥ 1% of tumor cells show positive ER/PR staining, the ER/PR interpretation is positive. The IHC HER2 score 3+ was considered positive as well.
Cell culture
For the purpose of this study, we chose MCF7 and MDA-MB-231 (triple negative) human breast cell lines. All cells were obtained from the cell bank of Pasteur Institute of Iran, and cultured in RPMI 1640 medium (Sigma 42 Aldrich, St. Louis, MO, USA), supplemented with 10% fetal bovine serum (Gibco, Carlsbad, CA, USA), 100 U/ml penicillin, and 100 μg/ml streptomycin at 37 °C in 5% CO2 at 95% humidity.
RNA isolation and real time polymerase chain reaction
Total RNA was extracted from all tissue samples and cell lines using the TRizol as was recommended by the manufacturer (Life Technologies, Carlsbad, CA) and the final concentration was quantified using a NanoDrop at 260 and 280 nm. In our experiment, 500 ng RNA was used for each sample with a purity range 1.8–2 for both 260/280 and 260/230 nm.
The RNA integrity was confirmed by gel electrophoresis and to remove the probable DNA contamination, the total RNA was treated with DNase (Takara Bio Inc, Otsu, Japan) according to the manufacturer’s instruction. The cDNA synthesis was carried out using approximately 500 ng of total RNA with the Prime Script-RT kit (Takara, Japan) according to the manufacturer’s protocol. In the next step, real time polymerase chain reaction (PCR) was carried out in a QuantStudio™ 3 system (Applied Biosystems, USA by Thermo Fisher Scientific) with SYBR Premix Ex Taq II kit (Takara, Japan) according to the manufacturer’s protocol. Real-time specific primer pairs used were as follows: RP11-48B3, forward 5’-CAAGCCCTGATCAACTAGGAATA-3’, reverse 5’-GGAAAGTTGGTTGCTGTGTAAG-3’; LOC100288637, forward 5’-CTAAGCCCTGCTTCTGGTATG-3’, reverse 5’-GGAGGCAGATCCAGTTCATTAG-3’; B2M, forward 5’-AGATGAGTATGCCTGCCGTG-3’, reverse 5’-GCGGCATCTTCAAACCTCCA-3’. For each reaction, 10 µl SYBR® Green Master Mix with 0.8 µL (40 nM) Primer 1 and 0.8 µL (40 nM) Primer 2 with 2 µL was used and the final volume was adjusted to a total of 18 µL using distilled H2O. All the reactions were carried out in triplicates. The real time PCR was performed in the following conditions: at 95 °C for 30 s followed by 40 repetitive cycles at 95 °C for 30 s and then at 60 °C for 30 s. No template controls were included in each run. Relative mRNA expression levels of LOC100288637 and RP11-48B3 were normalized to b2-microglobulin expression level as a housekeeping gene. The expression level (i.e. fold change) for each gene was calculated using the 2-DDCT method.
Bioinformatic analysis
In the current study, we also conducted different bioinformatics analyses, mainly by using the data of the Cancer Genome Atlas (TCGA) [15], to get more information about RP11-48B3.4 and LOC100288637. In this regard, we investigated the expression correlation between these two lncRNAs and mRNAs in the TCGA-BRCA dataset through using TANRIC web server [16]. Subsequently, we used the possible correlated mRNAs for any possible interactions between these mRNAs and lncRNA using the lncRRI search web [17].
Statistical analysis
The data are expressed as the mean ± ± SD and analyzed with SPSS version 20.0 software. P < 0.05 are considered statistically significant. The comparisons between groups were made using unpaired Student’s t-test or Mann--Whitney test. One-way ANOVA followed by Bonferroni or Kruskal-Wallis tests were used for multiple comparisons. The correlations were analyzed using Spearman’s rank correlation coefficient.
Results
Demographics, reproductive and clinical data of the patients
The average patient age was 45.7 with the range 27–68 years. Fifty-one percent of the subjects had a family history of BC and other cancers while 49 % were negative for this variable. Among the 51 enrolled women, 36 participants were parous and 15 cases were nulliparous. The 37 women had experienced breastfeeding in their life and the rest were negative for the breastfeeding experience. Besides, 47 patients were in postmenopausal status and 4 patients were in premenopausal condition. Other demographics and reproductive characteristics of the patients are shown in Tab. 1. Clinically, all 51 tumor samples from BC patients had invasive ductal breast carcinoma. Most tumors were positive for ER (+45 vs. –6) and PR (+34 vs. –17) receptors, but negative for HER-2/neu marker (–30 vs. +21). A total of 33.3% of the tumor samples had a size < 2 cm, 64.7% had 2–5 cm, and 2% had > 5 cm. The tumor size was measured as the largest dimension of the microscopic invasive component in pathologic sections. The data on other tumor features including histological grade and lymph node metastasis (TNM) are indicated in Tab. 2. The tumor grade and TNM stage of the cells were determined by the WHO standard [18].
2. Pathological data of the evaluated tumor samples from breast cancer patients. 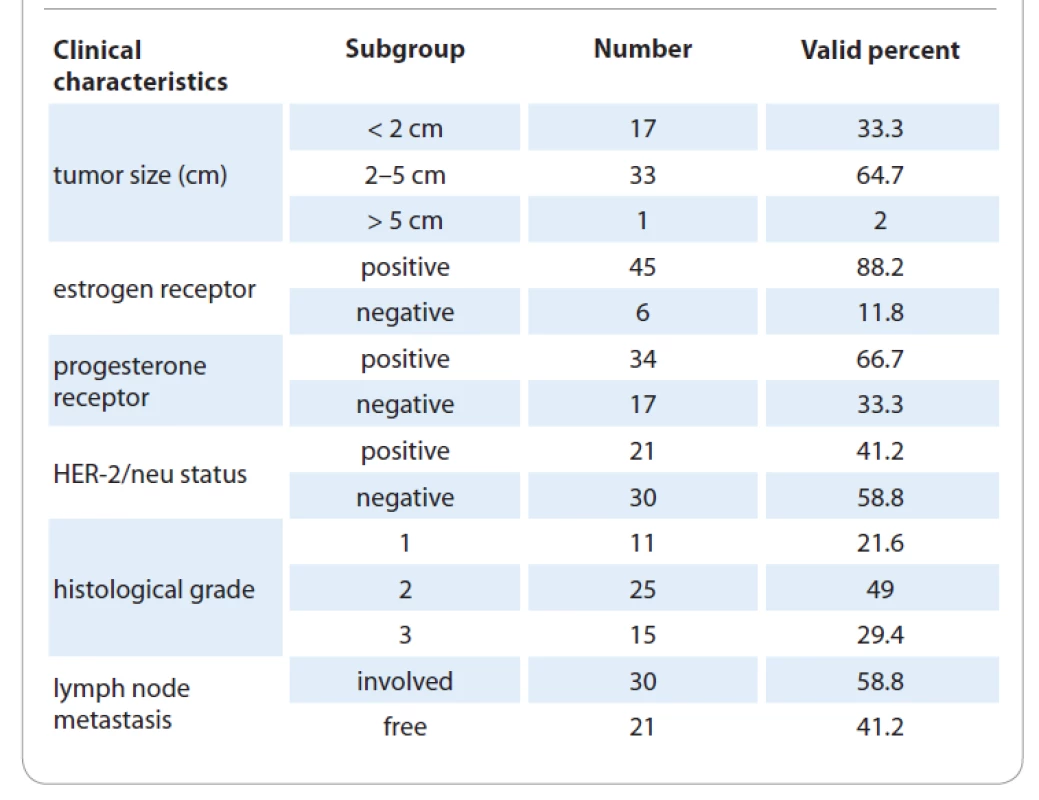
Expression levels of LOC100288637 and RP11-48B3 in BC tissues and different subgroups of the samples regarding the clinicopathological, demographic, and reproductive characteristics
In the next step, we investigated the expression profile of these two lncRNAs in 51 BC tissues and their adjacent normal tissues. The quantitative PCR (qPCR) results showed that LOC100288637 (P = 0.0085) was significantly upregulated in tumor tissues compared to normal samples. Furthermore, RP11-48B3 had a similar significant overexpression pattern in tumor tissues versus normal tissues (P = 0.0002). Fig. 1 represents the results on relative expression levels of both LOC100288637 and RP11-48B3 in tumor tissues compared to normal samples. In the next step, we compared the expression levels of the two lncRNAs in different subgroups of the patients with the various demographic, reproductive, and clinicopathological features of the subjects (Tab. 3 and 4). The findings showed that the lncRNA RP11-48B3.4 is upregulated in patients with age at menarche < 14 years compared to patients with age at menarche ≥ 14 (P = 0.041). The expression of this lncRNA was not significantly different among the different subgroups of the demographic and reproductive variables of the participants. Also, it was not observed any significant data on the difference in expression of the lncRNA LOC100288637 among various levels of the demographic and reproductive characteristics. From the clinicopathological point of view, our analyses indicated that lncRNA RP11-48B3.4 was significantly upregulated in tumors with a lower histological grade (grade 1) compared to tumor samples with higher grades, including grades 2 and 3 (P = 0.047). Regarding LOC100288637, its expression showed a higher level in HER-2 positive tumors than HER-2 negative tumors (P = 0.031). For other different subgroups of the clinical characteristics, any noteworthy results were not found among them in terms of differences in expression of the two studied lncRNAs (Tab. 4). In another of our statistic evaluations, we determined the correlation of LOC100288637 and RP11-48B3 with clinicopathological features of the BC patients. The Spearman‘s correlation analysis disclosed that the expression level of RP11-48B3 was negatively correlated with histological grade (r = −282, P = 0.045). The Spearman‘s analysis did not find any other significant correlation between the expression level of these lncRNAs and the other studied variables (Tab. 5).
Fig. 1. Expression levels of lncRNAs LOC100288637 and RP11-48B3 in invasive ductal carcinomas and adjacent normal tissues. T and N denote tumor tissue and normal breast samples, respectively. 
3. Quantitative expression data of target lncRNAs in tumor tissues in relation to demographic and reproductive variables. 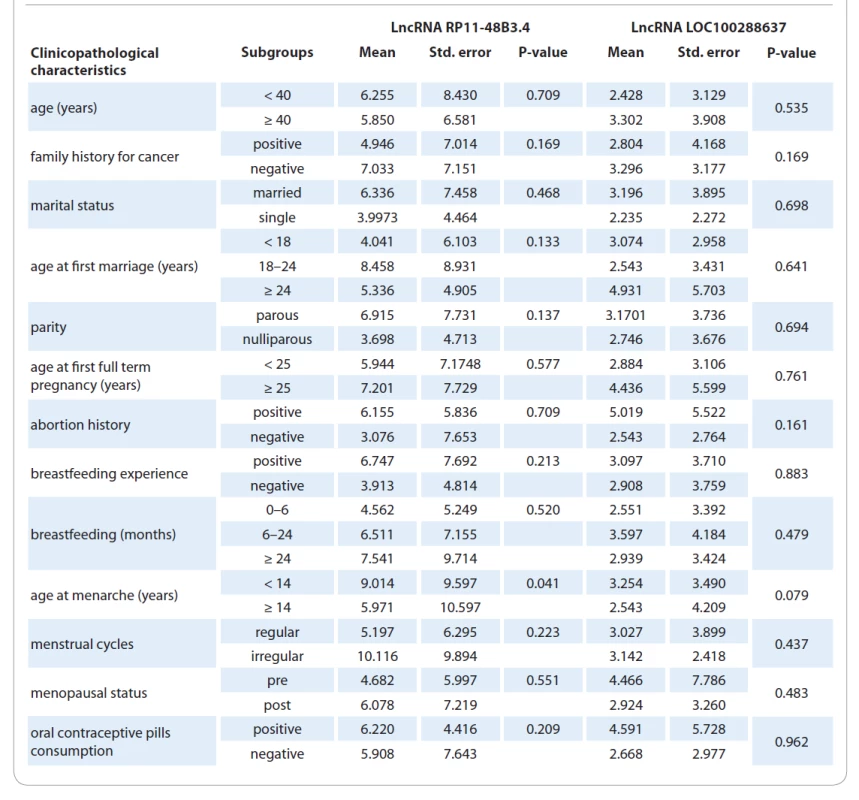
4. Expression levels of the studied lncRNAs in the tumor samples of the patients regarding to the clinicopathological characteristics. 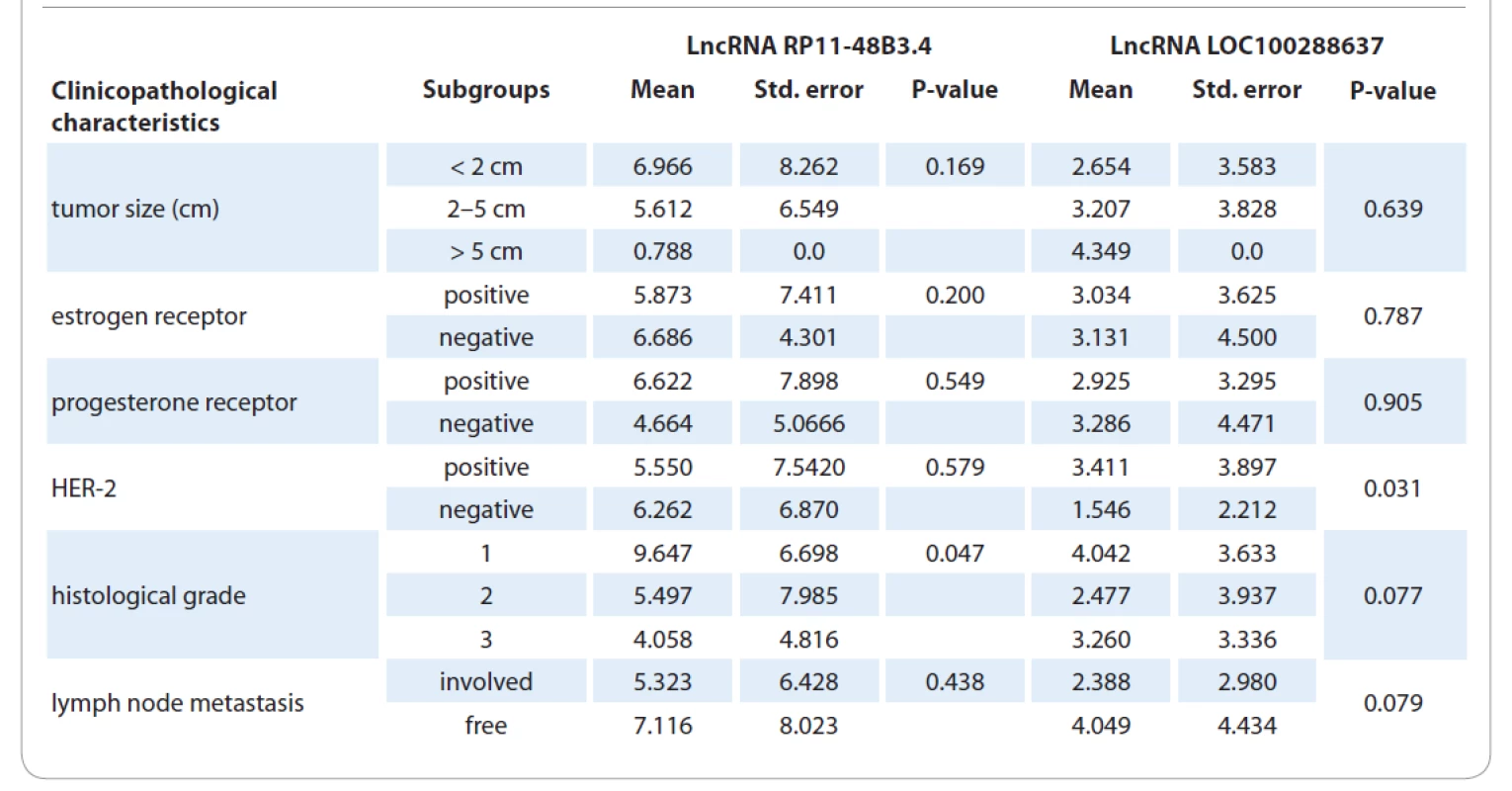
5. Correlation of lncRNAs LOC100288637 and RP11-48B3.4 expressions with pathological variables. 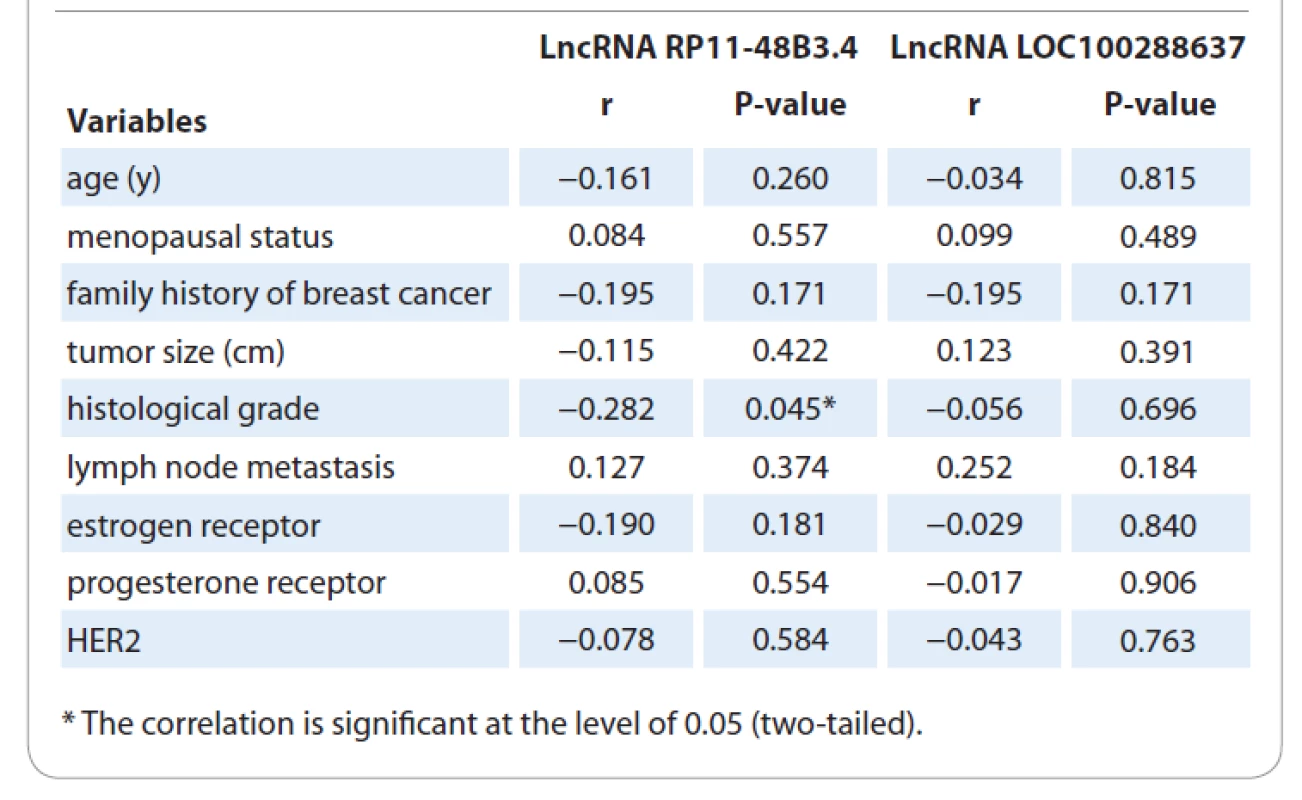
Expression levels of LOC100288637 and RP11-48B3 in MDA-MB-231 and MCF-7 cell lines
For getting more information about involving lncRNAs LOC100288637 and RP11-48B3 in the pathogenesis of IDC, especially metastasis, their expression levels were compared in human BC cell lines MDA-MB-231 and MCF-7 BC. The quantitative PCR (qPCR) data showed that the two lncRNAs were overexpressed in both MDA-MB-231 and MCF-7 BC cell lines; nevertheless, both lncRNAs showed a higher expression pattern in MDA-MB-231 than MCF7 cell line (P = 0.0013 and P = 0.0003, respectively) (Fig. 2).
Fig. 2. Relative expression of target lncRNAs in MDA-MB-231 and MCF-7 cell lines compared to normal cell lines. 
Bioinformatic evaluations
Through expression correlation analysis, we found some correlations between RP11-48B3.4 and mRNA expression in TCGA-BRCA genes, with negative correl - ation for mRNA MR1 (r = −0.409, P = 0) and positive correlations for mRNAs MRPS28 (r = 0.426, P = 0), NSMCE2 (r = 0.444, P = 0), POLR2K (r = 0.436, P = 0), TCEB1 (r = 0.476, P < 10–47), UQCRB (r = 0.418, P = 0), YWHAZ (r = 0.464, P = 0), ZBTB10 (r = 0.567, P = 0), and ZNF706 (r = 0.471, P = 0) (Fig. 3). Regarding the interaction between the correlated mRNAs and RP11-48B3.4, we found that only zinc finger and BTB domain containing 10 (ZBTB10) were predicted to have interaction with this lncRNA (Fig. 4). It was not any observable interaction between lncRNA LOC100288637 and evaluated mRNAs.
Fig. 3. Correlation analysis between RP11-48B3.4 and mRNAs expression in TCGA-BRCA. 
Fig. 4. Prediction of possible interaction between RP11-48B3.3 and ZBTB10. 
Discussion
Invasive ductal carcinoma (IDC), as the most frequent form of BC in women, has a high clinical burden due to its high invasive properties. Although recent investigations have made significant advances in revealing some key molecular mechanisms regarding its pathogenesis, it shows a very heterogeneous and complex etiology with many unknown aspects [19,20]. Thus, it is extremely expedient to clarify the underlying molecular mechanisms through which IDC develops. lncRNA transcripts are emerging as key players in cancer initiation and pathobiology of BC, with both oncogenic and tumor-suppressive roles. In this regard, new experimental studies have revealed some novel molecular mechanisms by which lncRNAs involved in BC malignancy, providing a new avenue of investigation for characterizing the different hallmarks of BC [21]. Moreover, these small noncoding RNAs not only play an important role in BC development, but also have some links with BC risk factors in the breast tissue of healthy women [22,23]. And interestingly, lncRNAs have been shown to exhibit diagnostic and prognostic biomarker properties as well as therapeutic targets for BC [24].
In the current study, we determined the expression levels of two novel ER signaling pathway – targeted lncRNAs LOC100288637 and RP11-48B3 in clinical samples of BC tumors type IDC. To the best of our knowledge, this is the first study to explore the links of expression signatures of these two lncRNAs to IDC type of BC as well as its clinicopathological characteristics. Our results showed a significant overexpression of these two ER-related lncRNAs in IDC tumor tissues compared to normal breast samples. Emerging studies have revealed that dysregulation of ER expression and its signaling pathway is intensely linked to the development and pathophysiology of the BC. Furthermore, provided evidence from different investigations is available for involving some lncRNAs in regulating the ER signaling and their aberrant expressions play key roles in the development of BC malignancy, especially the progression and endocrine-resistance of ER-positive subtype. The findings of the present study are in agreement with the results from a previous array-based study that revealed dysregulation of LOC100288637 and RP11-48B3.4 through regulating signaling pathways in ER+ BC patients. They indicated that the expression pattern of these two lncRNAs were significantly correlated with endocrine resistance-free survival and distant metastasis-free survival as well as disease-free survival of ER+ BC patients. Of note, the most of samples included in the current study were ER+ that is consistent with this observation that the majority of the BC tumors are molecularly fallen into ER+ subtype. Therefore, these observations highlight the clinical significance of these lncRNAs in the ER+ BC subtype via regulating the ER signaling pathway.
Furthermore, our study demonstrated a significant overexpression of the lncRNAs LOC100288637 and RP11-48B3.4 in human BC cell lines, including MDA-MB-231 (TNBC for ER and PR expression, as well as HER2 amplification) and MCF-7 (positive for ER, PR expression). However, the results represented a higher expression of both lncRNAs in MDA-MB-231 cells than MCF7 cells. Thus, it could be thought that they might have oncogenic roles in BC tumorigenesis through dysregulating the ER signaling pathway and also give invasiveness and metastatic properties to BC tumor cells.
Notably, our analyses further showed a significantly increased level of LOC100 288637 in HER-2 positive tumors compared to HER-2 negative samples. This finding is consistent with the results of previous work by Fan and colleagues that indicated the elevated expression level of LOC100288637 was strongly correlated with Her2/neu positive status in BC through next-generation sequencing and bioinformatics. Accordingly, we supposed that this lncRNA may play crucial roles in the pathogenesis of IDC via oncogenic functions. However, its exact mechanism needs more investigation by future studies. Furthermore, we observed a higher expression of the RP11-48B3 in lower grade tumors in comparison with the higher-grade tumors which also correlatively indicated by correlation analysis. Although this may be inconsistent with the abovementioned observation that this lncRNA had a higher expression in MDA-MB-231 cell line with high aggressive, invasive, and poorly differentiated properties, such a finding may occur due to our relatively small sample size or unknown complex nature of RP11-48B3 function during BC tumorigenesis which requires more investigation to disclose.
Interestingly, another result of the current study was that the lncRNA RP11-48B3.4 had an elevated expression pattern in BC patients with the age at menarche < 14 years in comparison to patients with the age at menarche ≥ 14 years. It has been reported that lower age at menarche increases the risk of BC through estrogen-related mechanisms [25,26]; however, little is known about its molecular mechanisms behind the risk of BC. Therefore, it can be suggested that a lower age at menarche may increase the risk of IDC partly through affecting the expression level of estrogen-linked lncRNA RP11-48B3.4. Although, confirmation of such assumptions requires conducting more functional studies.
Lastly, the current study also bioinformatically demonstrated some correlations between lncRNA RP11-48B3.4 and expression of several mRNAs in TCGA-BRCA, including negative expression correlation for mRNA MR1 and positive correlations for mRNAs MRPS28, NSMCE2, POLR2K, TCEB1, UQCRB, YWHAZ, ZBTB10, and ZNF706. However, only the mRNA ZBTB10 were predicted to have interaction with the lncRNA RP11-48B3.4. Regarding LOC100288637, the results were not detected any evident interaction with given mRNAs. This highlights that the novel lncRNA RP11-48B3.4 may be involved in the pathogenesis of the IDC via interacting with some encoding genes by different mechanisms. There is evidence that lncRNAs play key roles in the development of BC through influencing the expression of other coding and non-coding genes [27]. In this way, one suggested mechanism is competing endogenous RNA (ceRNA) function through which lncRNAs regulate expression of mRNAs via sponging miRNAs in regulatory molecular networks whose roles in cancer development, especially in BC, are emerging with clinical significance [28,29].
Conclusion
The presented study showed the elevated expression levels of LOC100288637 and RP11-48B3 lncRNA in IDC breast tumors as well as BC cell lines have some important significance on their clinical outcome. This suggested them as putative oncogenic markers at the molecular level of IDC. However, it is important to analyze the correlation between their expression as well as progression-free survival time to conclude these lncRNAs as prognostic biomarkers. In our study, it was not possible to do such evaluation, since we used samples from newly diagnosed cancers. These findings can also be useful to candidate these two lncRNAs as targets for BC treatment. Future studies are warranted to analyze the expression of these two lncRNAs in various type of cancers to pro - pose them as candidates of tumor biomarkers in combination with biomarker panels.
Acknowledgment
This study has been supported by the funding from the Shiraz University of Medical Sciences (No.16897). The authors would like to thank the staff members of Faghihi Hospital for their contributions to breast tissue sampling and data collection, and Autoimmune Disease Research Center of Shiraz University of Medical Science, for their help and support and also the participants who took part in this study.
Data availability
The data that support the findings of this study are available from the corresponding authors upon reasonable request.
Author contributions
A.D, Y.M, and S.I contributed to the conception and design of the research. F.F, S.I, M.D.F, and M.R.M performed the experiments. F.F., S.I, H.D., M.D. F, and Y. M interpreted the results of the experiments, analyzed data, and prepared the figures. S.I, M.R.M, and A.D drafted the manuscript. A.D, H.D, S.M.B.T, and S.I. edited and revised the manuscript. M.B.T and Y.M, made study supervision and technical or material support. All authors read and approved the final manuscript.
The authors declare they have no potential conflicts of interest concerning drugs, products, or services used in the study.
Autoři deklarují, že v souvislosti s předmětem studie nemají žádné komerční zájmy.
The Editorial Board declares that the manuscript met the ICMJE recommendation for biomedical papers.
Redakční rada potvrzuje, že rukopis práce splnil ICMJE kritéria pro publikace zasílané do biomedicínských časopisů.
Dr Seyed Mohammad Bagher Tabei
Department of Medical Genetics
Faculty of Medicine
Shiraz University of Medical Sciences
Zand St, Shiraz
Iran, post code: 7134845794
and
Dr Yaser Mansoori
Fasa University of Medical Sciences
Ave Sina Square
Fasa, Fars
Iran, post code:7461686688
Submitted/Obdrženo: 31. 12. 2020
Accepted/Přijato: 25. 4. 2021
Sources
1. Bray F, Ferlay J, Soerjomataram I et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018; 68 (6): 394–424. doi: 10.3322/caac.21 492.
2. Cancer Genom Atlas Network. Comprehensive molecular portraits of human breast tumors. Nature 2012; 490 (7418): 61–70. doi: 10.1038/nature11412.
3. Tulinius H, Bjarnason O, Sigvaldason H et al. Tumours in Iceland 10. Malignant tumors of the female breast: a histological classification, laterality, survival, and epidemiological considerations. APMIS 1988; 96 (3): 229–238. doi: 10.1111/j.1699-0463.1988.tb05296.x.
4. Tan PH and Ellis IO. Myoepithelial and epithelial-myoepithelial, mesenchymal, and fibroepithelial breast lesions: updates from the WHO classification of tumours of the breast 2012. J Clin Pathol 2013; 66 (6): 465–470. doi: 10.1136/jclinpath-2012-201078.
5. DeSantis CE, Lin CC, Mariotto AB et al. Cancer treatment and survivorship statistics, 2014. CA Cancer J Clin 2014; 64 (4): 252–271. doi: 10.3322/caac.21235.
6. Carninci P, Kasukawa T, Katayama S et al. The transcriptional landscape of the mammalian genome. Science 2005; 309 (5740): 1559–1563. doi: 10.1126/science. 1112014.
7. Du T, Zhang B, Zhang S et al. Decreased expression of long non-coding RNA WT1-AS promotes cell proliferation and invasion in gastric cancer. Biochim Biophys Acta 2016. 1862 (1): 12–19. doi: 10.1016/j.bbadis.2015.10. 001.
8. Kurokawa R. Long noncoding RNAs: structures and functions. Springer 2015.
9. Iranpour M, Soudyab M, Geranpayeh L et al. Expression analysis of four long noncoding RNAs in breast cancer. Tumour Biol 2016; 37 (3): 2933–2940. doi: 10.1007/s13277-015-4135-2.
10. Nie Z-L, Wang Y-S, Mei Y-P et al. Prognostic significance of long noncoding RNA Z38 as a candidate biomarker in breast cancer. J Clin Lab Anal 2018; 32 (1): e22193. doi: 10.1002/jcla.22193.
11. Calin GA, Liu C-G, Ferracin M et al. Ultraconserved regions encoding ncRNAs are altered in human leukemias and carcinomas. Cancer Cell 2007; 12 (3): 215–229. doi: 10.1016/j.ccr.2007.07.027.
12. Gupta RA, Shah N, Wang KC et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature 2010; 464 (7291): 1071–1076. doi: 10.1038/nature08975.
13. Wu L, Xu Q, Zhang H et al. A new avenue for obtaining insight into the functional characteristics of long noncoding RNAs associated with estrogen receptor signaling. Sci Rep 2016; 6 : 31716. doi: 10.1038/srep31716.
14. Yang F, Lyu S, Dong S et al. Expression profile analysis of long noncoding RNA in HER-2-enriched subtype breast cancer by next-generation sequencing and bioinformatics. Onco Targets Ther 2016; 9 : 761–772. doi: 10.2147/OTT.S97664.
15. The Cancer Genome Atlas. [online]. Available from: https: //www.cancer.gov/aboutnci/organization/ccg/research/structuralgenomics/tcga.
16. The atlas of non-coding RNA in cancer. [online]. Available from: https: //bioinformatics.mdanderson.org/publicsoftware/tanric/.
17. lncRNASNP2. [online]. Available from: http: //bioinfo.life.hust.edu.cn/lncRNASNP#!/lncrna_info?lncrna=NON HSAT127417.2.
18. Reis-Filho J, Ellis I. WHO classification of tumors of the breast. IARC, Lyon 2012 : 48–52.
19. Kominsky SL, Argani P, Korz D et al. Loss of the tight junction protein claudin-7 correlates with histological grade in both ductal carcinoma in situ and invasive ductal carcinoma of the breast. Oncogene 2003; 22 (13): 2021–2033. doi: 10.1038/sj.onc.1206199.
20. Harris M, Howell A, Chrissohou M et al. A comparison of the metastatic pattern of infiltrating lobular carcinoma and infiltrating duct carcinoma of the breast. Br J Cancer 1984; 50 (1): 23–30. doi: 10.1038/bjc.1984.135.
21. Wang J, Ye C, Xiong H et al. Dysregulation of long non-coding RNA in breast cancer: an overview of mechanism and clinical implication. Oncotarget 2017; 8 (3): 5508–5522. doi: 10.18632/oncotarget.12537.
22. Mansoori Y, Tabei MB, Askari A et al. Expression levels of breast cancer-related GAS 5 and LSINCT 5 lnc RNA s in cancer-free breast tissue: molecular associations with age at menarche and obesity. Breast J 2018; 24 (6): 876–882. doi: 10.1111/tbj.13067.
23. Mansoori Y, Zendehbad Z, Askari A et al. Breast cancer-linked lncRNA u-Eleanor is upregulated in breast of healthy women with a lack or short duration of breastfeeding. J Cell Biochem 2019; 120 (6): 9869–9876. doi: 10.1002/jcb.28269.
24. Tian T, Wang M, Lin S et al. The impact of lncRNA dysregulation on clinicopathology and survival of breast cancer: a systematic review and meta-analysis. Mol Ther Nucleic Acids 2018; 12 : 359–369. doi: 10.1016/j.omtn.2018.05.018.
25. Burgess S, Thompson DJ, Rees JMB et al. Dissecting causal pathways using Mendelian randomization with summarized genetic data: application to age at menarche and risk of breast cancer. Genetics 2017; 207 (2): 481–487. doi: 10.1534/genetics.117.300191.
26. Day FR, Thompson DJ, Helgason H et al. Genomic analyses identify hundreds of variants associated with age at menarche and support a role for puberty timing in cancer risk. Nat Genet 2017. 49 (6): 834–841. doi: 10.1038/ng.3841.
27. Arshi A, Sharifi FS, Ghahfarokhi MK et al. Expression analysis of MALAT1, GAS5, SRA, and NEAT1 lncRNAs in breast cancer tissues from young women and women over 45 years of age. Mol Ther Nucleic Acids 2018; 12 : 751–757. doi: 10.1016/j.omtn.2018.07.014.
28. Abdollahzadeh R, Daraei A, Mansoori Y et al. Competing endogenous RNA (ceRNA) cross talk and language in ceRNA regulatory networks: a new look at hallmarks of breast cancer. J Cell Physiol 2019; 234 (7): 10080–10100. doi: 10.1002/jcp.27941.
29. Zhong Z, Huang M, Lv M et al. Circular RNA MYLK as a competing endogenous RNA promotes bladder cancer progression through modulating VEGFA/VEGFR2 signaling pathway. Cancer Lett 2017; 403 : 305–317. doi: 10.1016/j.canlet.2017.06.027.
Labels
Paediatric clinical oncology Surgery Clinical oncology
Article was published inClinical Oncology

2021 Issue 5-
All articles in this issue
- Myelodysplastický syndrom, refrakterní anemie a O2 sensing
- Promising treatment modalities in the therapy of myelodysplastic syndromes
- Informace z České onkologické společnosti
- eHealth in medicine and oncology – new horizons of clinical practice
- Personalities and their contribution to the development of radiation oncology
- Anotace knihy
- Expert opinion on the care of patients with implanted pacemakers and cardioverters-defi brillators with an indication for radiotherapy – a summary for professions in the field of radiation oncology
- Aktuality z odborného tisku
- Skupina léků anti-HER2 pro pacientky s karcinomu prsu se rozrostla o preparát Phesgo
- Spomienka na prof. MUDr. Ivana Kozu, DrSc.
- The place and importance of hyaluronic acid in radiotherapy side effects
- Association of polymorphisms in nucleotide excision repair pathway genes with susceptibility to cutaneous melanoma
- Clinical values of two estrogen receptor signaling targeted lncRNAs in invasive ductal breast carcinoma
- Cabozantinib in the treatment of metastatic renal cell carcinoma – final data analysis from four oncology centers in the Czech Republic
- Metal artifact-free MRI-guided re-irradiation for recurrent spinal metastases from thyroid cancer
- Clinical Oncology
- Journal archive
- Current issue
- Online only
- About the journal
Most read in this issue- Personalities and their contribution to the development of radiation oncology
- Skupina léků anti-HER2 pro pacientky s karcinomu prsu se rozrostla o preparát Phesgo
- Promising treatment modalities in the therapy of myelodysplastic syndromes
- The place and importance of hyaluronic acid in radiotherapy side effects
Login#ADS_BOTTOM_SCRIPTS#Forgotten passwordEnter the email address that you registered with. We will send you instructions on how to set a new password.
- Career

