-
Medical journals
- Career
Genetic and epigenetic bases of prostate tumor cell radioresistance
Authors: Denis Kutilin
Authors‘ workplace: National Medical Research Oncology Center, Rostov-on-Don, Russian Federation
Published in: Klin Onkol 2021; 34(3): 220-234
Category: Original Article
doi: https://doi.org/10.48095/ccko2021220Overview
Background: Radiation therapy plays a leading role in the treatment of prostate cancer, but the emergence of radioresistant forms of this disease dictates the need for a personalized approach based on the data from genetic and epigenetic markers. Such markers include the copy number variation as well as gene and microRNA expression. Purpose: The aim of the study was to validate the list of potential predictors of radioresistance of prostate tumor cells in a model experiment based on the determination of gene copy number variation, gene transcriptional activity and microRNA expression. Material and methods: The study used a PC-3 prostate cancer cell culture. The determination of the relative copy number variation and expression of 32 genes (BRCA1, BRCA2, PTEN, CASP3, CASP8, BAX, BCL2, CASP9, P53, MDM2, AKT1, ATM, BRIP1, CDK1, CDKN1B, CCND1, CCND3, FGFR2, KU70, RAD50, RAP80, Rif1, RNF168, TopBP1, HIST, H2AX, EXO1, XRCC4, RBBP8, EP300, LIG4, C-FLIP), as well as 15 microRNAs (let-7, miR15a/ 16, miR-17, miR-18a, miR-21, miR-24, miR-26b, miR-99a, miR-100, miR-101, miR-106a, miR-663a, miR-143, miR-145) was performed using the real-time quantitative polymerase chain reaction method. It was found that daily irradiation of PC-3 cells on a Novalis TX linear accelerator at doses of 6 and 7 Gy for 5 days leads to a significant decrease in the total number of cells and the number of viable cells. Nevertheless, after 5 days of irradiation, about 15% of the initial number of prostate tumor cells retained their viability, which is due to their special genetic and epigenetic characteristics: increased copy number and expression of genes BRCA2, CDK1, CDKN1B, H2AX, RAD50, XRCC4, RBBP8 and EP300 and reduced copy number and expression of CCND3, TP53, and BCL2 genes, as well as differential expression of six microRNAs (hsa-miR-18a-5p, hsa-miR-24-5p, hsa-miR-99a-5p, hsa-miR-100-5p, hsa-miR-145-5p, hsa-let-7a-3p). Conclusion: This study enabled to identify genetic and epigenetic markers of prostate tumor cells resistance to radiation therapy.
Keywords:
Radiation therapy – prostate cancer – cell culture – copy number variation of genes – transcriptional activity of genes – micro-RNA – apoptosis – DNA repair
Background
Prostate cancer ranks second in the structure of cancer incidence in men of all age groups. The increase in the incidence over the past 10 years is 143%, which is the highest rate among all oncological pathologies [1].
Radical prostatectomy and radiation therapy are the main therapeutic approaches for prostate cancer [2]. RT in the independent embodiment, and in combination with other methods, plays a leading role in the treatment of this pathology [3], but the appearance of radioresistant forms necessitates personalized approach based on the data of molecular genetic markers [1].
The reasons for radioresistance are being actively studied. One of them may be hyperactivation of DNA repair mechanisms in tumor cells [4]. It is known that the balance between the mechanisms of DNA damage and DNA repair determines the cells viability after exposure to ionizing radiation [5]. An equally important factor affecting the sensitivity to RT is hypoxia, taking into account the important role of oxygen in the reaction of reactive oxygen species formation induced by RT [6].
Despite the evolution of RT methods, and despite the fact that the main mechanisms of radioresistance formation are currently known, the oncologist is unable to determine predictors of RT sensitivity with a high level of reliability. Therefore, the problem of predicting radioresistance remains relevant. To solve it, model experiments on appropriate cell cultures are needed, the first stage of which is to assess the effect of RT on the tumor cells survival.
The second stage of such experiments involves molecular markers of radioresistance identification. Such markers include the copy number variation (CNV), a type of genetic polymorphism that can result in a decrease or increase in the copy number of a particular gene, and, as a consequence, decreased or increased expression of a gene product – a protein or non-coding RNA (including micro-RNA) [7]. Accordingly, the expression indices of these genes and microRNAs also have a certain potential as markers.
A meta-analysis carried out on the basis of data published in the years 2000–2020, summarizing the experience of radiation therapy for patients with localized prostate cancer and data on gene copy numbers, gene expression and micro-RNA, allowed us to form a list of potential molecular markers, consisting of 32 genetic loci – components of signaling cascades involved in DNA repair, cell cycle regulation and apoptosis – as well as a list of 15 microRNAs [8].
Therefore, the study aim was to validate the list of potential predictors of prostate tumor cells radioresistance in a model experiment based on the determination of genes copy number, genes transcriptional activity and microRNA expression.
Materials and methods
The study used a human prostate cancer cells culture – PC-3. Cells genotyping was carried out using the COrDIS Sprint kit (Gordiz, Russia), the compliance with the standard genotype and absence of contamination of the given cell line was checked (comparison with the ATCC database) [9]. Cultivation was carried out in sterile flat-bottomed flasks in RPMI-1640 medium with 10% fetal bovine serum and 50 μg/ mL gentamicin concentration under conditions of 5% CO2 and 95% humidity at 37 ºC [10].
For the model experiment, doses of 6 and 7 Gy were used (irradiation was performed 5 times every 24 hours on a Novalis TX linear accelerator). The cells were counted in a Goryaev chamber (0.4% trypan blue solution). On the 5th day of irradiation, PC-3 cells were removed from the substrate with trypsin/ versene solution. The estimation of the number of cells in different stages of apoptosis was carried out on a FacsCantoII flow cytometer (BD, USA) using the Annexin V-FITC Apoptosis Detection Kit (BD Pharminogen, USA) [10].
Nucleic acids isolation
The cell mass (PC-3 cell culture) was washed from the medium with Dulbecco’s phosphate buffered saline (Biolot, Russia), divided into two tubes, and precipitated by centrifugation. A total of 900 μL of QIAzol reagent (QIAGEN) was added to the pellets of tube 1. Further isolation and purification of total RNA fraction was performed using the RNeasy Plus Universal Kits (QIAGEN) according to the manufacturer’s protocol. If necessary, prepared RNA samples were treated with DNase I to eliminate genomic DNA traces according to enzyme manufacturer’s protocol. RNazine was added to prepared total RNA to solution final concentration of 1 U/ μL. For long-term storage, the RNA samples were precipitated with 80% ethanol, transferred into Eppendorf tubes, placed in cryoboxes and stored in a low-temperature vertical freezer MDF-U500VX (temperature mode −80 °C).
The obtained RNA quality was checked by electrophoretic method in agarose gel according to the ratio of the intensity of 28S and 18S rRNA bands luminescence (Fig. 1)
Assessment of gene copy number
DNA extraction was carried out from the sediment in tube 2 using the phenol-chloroform method according to the protocol described by Kit O.I. and coauthors [11]. To determine the relative copy number of genes using the NCBI GenBank database, we developed sequences of 32 pairs of synthetic oligonucleotides (AKT, ATM, BRIP, BRCA1, BRCA2, CDK1, CDKN1B, CCND1, CCND3, EXO1, FGFR2, HIST1, H2AX, KU70, PTEN, RAD50, RAP80, RIF1, RNF168, TOPB1, TP53, XRCC4, BAX, CASP8, CASP3, CASP9, MDM2, BCL2, RBBP8, EP300, LIG4, C-FLIP) and 3 pairs for reference loci (ACTB, B2M, GAPDH) (Tab. 1). Determination of the relative copy number of genetic loci was performed using the real-time quantitative polymerase chain reaction (RT-qPCR) method.
RT-qPCR amplification was performed using the CFX96 thermal cycler (Bio-Rad, USA). A reaction mix containing 1× PCR buffer, 0.2 mM dNTP mix, 1.5–2.0 mM MgCl2, 400 nM primers, 0.1 U/ μL Taq polymerase (Synthol) and 10 ng DNA was used for amplification. EvaGreen® Dye (Biotium, USA) was used as an intercalating dye. The amplification of each sample was carried out in triplicate. The averaged data for each genetic locus were normalized according to the averaged index of reference genes to obtain the ΔCt-value.
The relative copy number (rC) was calculated by the formula [12]:
rC = rCir/ rCc = = E-ΔCt(irradiated cells)/ E-ΔCt(intact cells (control))
where E is the amplification efficiency calculated by the formula E = 10-1/ k where k is the coefficient from the equation of the straight line C(T) = k × logP0+b, obtained by linear approximation of the experimental data (Eav = 1.914), and ΔCt = geometric mean of Ct (test gene) − geometric mean of Ct (reference gene) [12].
Fig.1. Electropherogram of RNA isolated from PC-3 cell mass. The system of gel documentation Gel Doc XR PLUS (BioRad, USA) was used for visualization. 
A reaction mixture containing 5 μM random primers, 1× RT buffer, 0.5 mM dNTP mix, 0.5 U/ μL RNase Inhibitor (Thermo scientific), 5 U/ μL ReverseTranscriptase ММLV (Syntol) was prepared for cDNA production. The resulting mix was gently mixed, incubated in a thermostat at 44 °C for 1 hour, then reverse transcriptase was inactivated for 10 minutes at 92 °C. The resulting preparations were subjected to concentration measurement and stored at −80 °C until RT-qPCR.
RT-qPCR was used to determine the relative expression values of 32 genetic loci (BRCA1, BRCA2, PTEN, CASP3, CASP8, BAX, BCL2, CASP9, P53, MDM2, AKT1, ATM, BRIP1, CDK1, CDKN1B, CCND1 CCND3, FGFR2, KU70, RAD50, RAP80, Rif1, RNF168, TopBP1, HIST, H2AX, EXO1, XRCC4, RBBP8, EP300, LIG4, C-FLIP). After a series of preliminary experiments, GAPDH, ACTB and B2M were used as reference genes. Expression stability for reference genes selection was assessed using the geNorm program [13]. The design of specific oligonucleotide primers was carried out using the reference sequences of the NCBI GenBank and the Primer-BLAST program. The primer sequence is shown in Tab. 2.
Each locus was positioned in three technical repetitions (as well as for CNV determination). The reaction mix containing 1× PCR buffer, 0.2 mM dNTP mix, 2.0 mM MgCl2, 500 nM primers, 0.05 U/ μL Taq polymerase (Synthol) and 10 ng DNA was used for amplification. The resulting mixture was incubated at RT-thermocycler CFX 96 (Bio-Rad, USA) in accordance with temperature conditions: initial denaturation t = 95 °C for 240 s, 40 cycles: t = 95 °C for 10 s, t = 58 °C for 30 s, t = 72 °C for 30 s.
Relative expression (RE) was calculated using the formula RE = E-ΔΔCt. The results were normalized for three reference genes GAPDH, ACTB and B2M, and the level of corresponding target genes expression in intact cell samples, sequentially according to the scheme below:
1. normalization by the geometric mean of the reference genes: ΔC (t) = gC (t) target – gC (t) reference;
2. calculation of the median ΔC (t) for each gene for the control (intact cells) and experimental (irradiated) groups;
3. normalization for the control group and the final result (fold difference): E-ΔΔC (t) (ΔΔC (t) = DC (t) median of the experimental group – ΔC (t) median of the control group) [14]. E is the amplification efficiency calculated by the formula E = 10-1/ k, where k is the coefficient from the equation of the straight line C(T) = k × logP0 + b, obtained by linear approximation of the experimental data (Eav = 1.94).
1. Panel of primers for determining the copy number of genes. 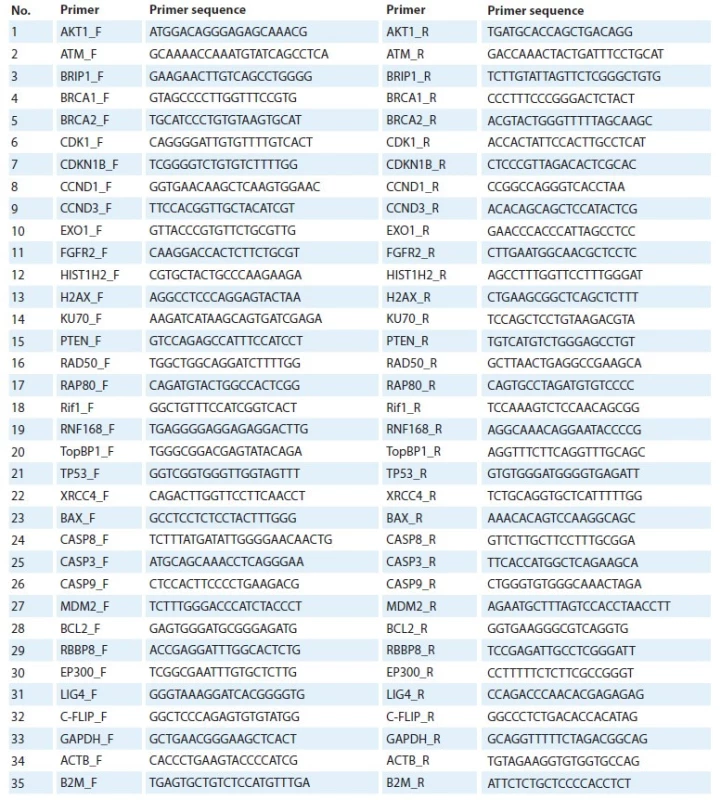
F – forward primer, R – reverse primer 2. Primer panel for determining relative gene expression. 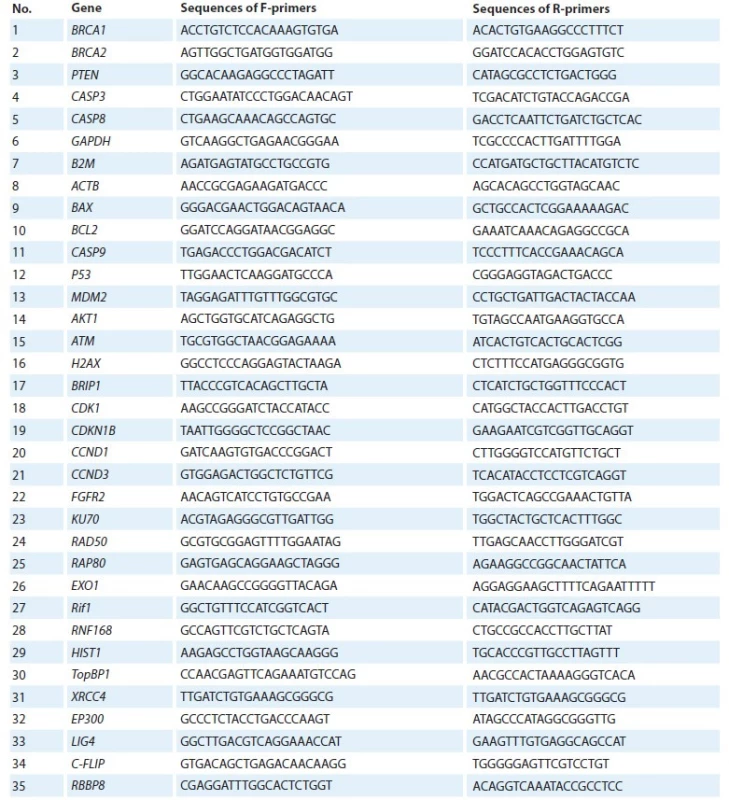
F – forward primer, R – reverse primer Using the MirTarBase, miRDB, and TargetScan databases, 15 microRNAs associated with the sensitivity of tumor cells to radiation therapy were selected: let-7, miR15a / 16, miR-17, miR-18a, miR-21, miR-24, miR -26b, miR-99a, miR-100, miR-101, miR-106a, miR-663a, miR-143 and miR-145. MicroRNAs hsa-miR-126-5p and hsa-miR-7-5p were used as reference. Expression stability for the selection of reference microRNAs was also assessed using the geNorm program [13]. The design of specific oligonucleotide primers was carried out using the Balcells I. algorithm (Tab. 3). The isolated total RNA was used in a reverse transcription reaction, which was carried out simultaneously with RNA polyadenylation, using specific RT primers [15].
3. Sequences of micro-RNA primers. 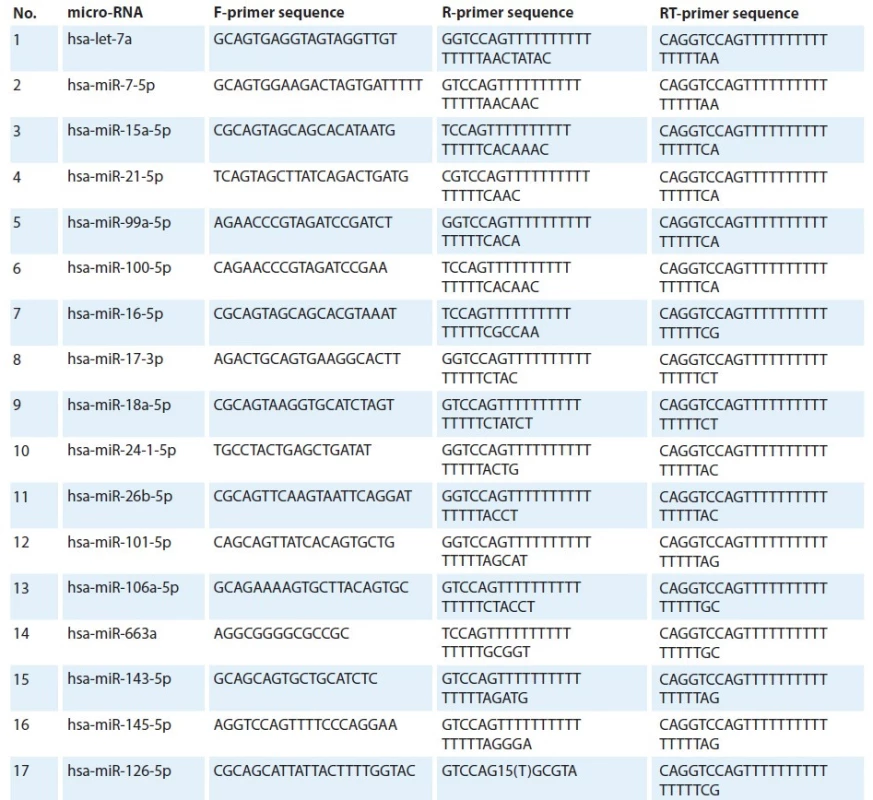
F – forward primer, R – reverse primer, RT – primer for reverse transcription reaction The microRNA relative expression changing was evaluated by RT-qPCR. Amplification was carried out in 20 μL PCR mixture containing 1× PCR buffer, 0.25 mM dNTPs, 2.5 mM MgCl2, 1 U/ μL Taq-DNA polymerase, 0.6 μM forward and reverse primers. RT-qPCR of each sample was performed in triplicate. The resulting mixtures were incubated in CFX 96 RT-amplifier according to the following program: 5 minutes at 95 °C, 40 cycles: 15, 30 and 30 s at 95, 64 and 72 °C, respectively. The calculation of the relative expression of microRNA was carried out in the manner of the calculation for mRNA described above.
Statistical analysis
Statistical analysis was performed using the one-way analysis of variance (one-way ANOVA). To carry out cluster analysis (hierarchical clustering, euclidean distance) and build heat maps, we used our own scripts in the R language (R-Studio 8.10.173.987). To construct the Edwards-Venn diagrams, we used an algorithm for comparing a larger number of sets implemented in JavaScript. The clustering of genes by function was performed using the functional module detection (FMD) algorithm. The Q value of each term associated with functional modules is calculated using the one-tailed Fisher’s exact test and the Benjamini-Hochberg correction to correct for multiple comparisons [16]. The online service GIANT (genome-scale integrated analysis of gene networks in tissues) [17,18] was used to build a network of tissue-specific functional interactions of genes by combining the data from full-genome experiments.
Fig. 2. Cell monolayer micrograph after 5-day irradiation (from left to right: control and doses of 6 and 7 Gy, respectively). 
The target gene search was performed using the TarPmiR algorithm with TargetScan, mirDB and miRTarBase databases. To predict a target microRNA site, the TarPmiR uses a “random forest” approach – a machine learning an algorithm that uses a set of decision trees (combines the random subspace and Breiman bagging methods). The result of the “random forest” model is predicted probability that the candidate target site is a true target site [19].
The analysis of the overrepresentation of miRNAs in signaling pathways (over-representation analysis – ORA) was also performed. This method measures the percentage of microRNAs in the signaling pathway that are differentially expressed. The goal of ORA is to obtain a list of the most important signaling pathways, ordered by P-value. The statistical significance of the overlap between the genes from the signaling pathway and the list of differentially expressed genes is calculated using the Fisher’s exact test [20].
Results
In the course of a 5-day experiment with irradiation of prostate gland tumor cells, there were obtained data characterizing the potential of external beam radiation therapy and stereotactic radiation therapy.
Regardless the dose, no statistically significant differences were found in the number of dead cells in the nutrient medium on the first day of irradiation. Relative to the control, the number of dead cells increased 7.0 times at a dose of 7 Gy on the second day of irradiation, 12.0 and 7.3 times at doses of 6 and 7 Gy on the third day, respectively, and 4.0 and 7.0 times at doses of 6 and 7 Gy on the fourth day, respectively. It should be noted that the number of dead cells in control flasks with culture medium did not change during the experiment. At the same time, a statistically significant (P < 0.05) decrease (1.4 times) in the number of dead cells was recorded at a dose of 6 Gy on the second day of irradiation; an increase (2.4 times) in the number of dead cells was recorded at doses of 6 and 7 Gy on the third day, respectively; and an increase (2.3 times) was recorded at a dose of 7 Gy on the fourth day relative to the first day of the experiment. This significant effect of irradiation on PC-3 cells appears at doses of 7 and 6 Gy on the second and third day of irradiation, respectively.
The study results of the influence of selected irradiation modes on PC-3 cell culture after 5-day irradiation are shown in Fig. 2 and Graph 1.
Graph 1. Total number of cells. 
* statistically significant differences (P < 0.05) relative to control Daily irradiation of PC-3 cell culture on a Novalis TX linear accelerator at doses of 6 and 7 Gy for 5 days led to a significant decrease in the total number of cells by 71.6 and 71.4%, respectively (P < 0.05) (Fig 2).
Tab. 4 and Fig. 3 show the data, obtained on a flow cytometer, on the distribution of PC-3 cells at different stages of apoptosis.
In Fig. 3 (from left to right), the first graph shows cells according to their size and granularity, unstained. In the second graph, the tumor cells are highlighted, excluding debris (non-living cells). The third graph shows the location of tumor cells stained with propidium and annexin.
Q1 – cells stained with propidium only, dead cells.
Q2 – cells stained with propidium and annexin, this is late apoptosis / necrosis.
Q3 – living cells, neither annexin nor propidium sits on them.
Q4 – cells stained with annexin only, early apoptosis.
4. Assessment of the level of apoptosis in cell culture PC-3 after different modes of irradiation for 5 days (percentage of cells of different functional states). 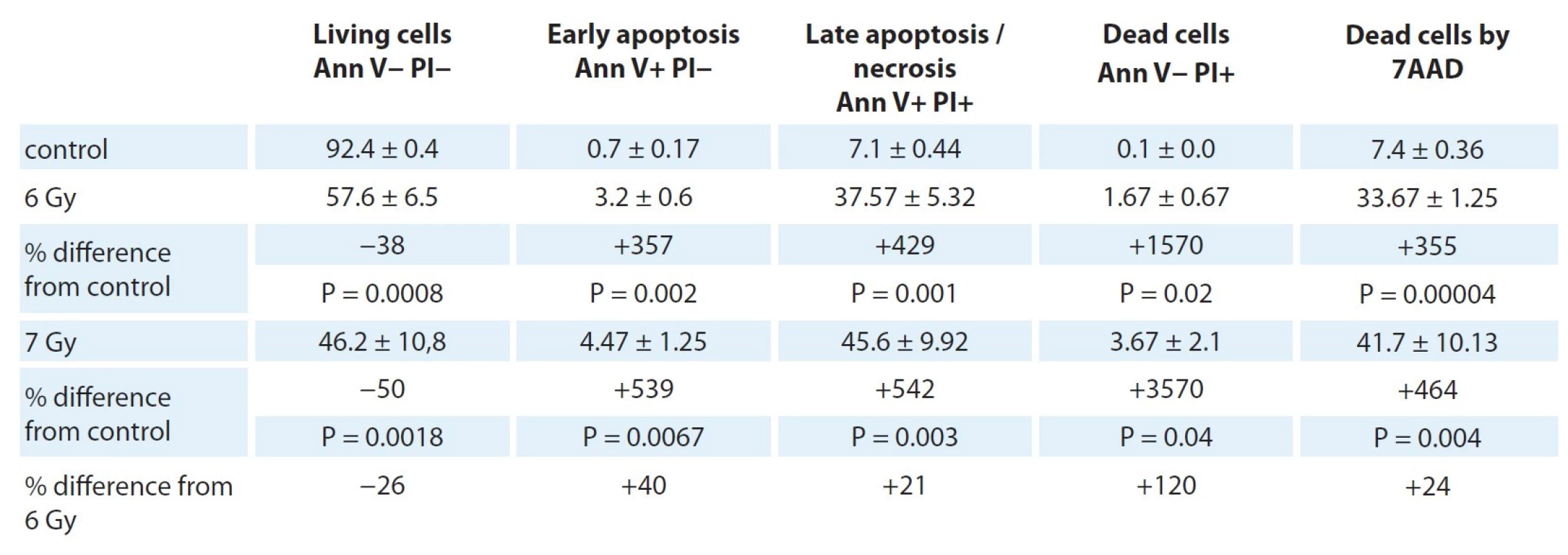
− negative, + positive, 7AAD – 7-aminoactinomycin D, Ann V – annexin V, PI – propidium Fig. 4. Heatmap and cluster analysis of the differential in the copy number variation of 32 genes in intact (control) and irradiated PC-3 cells. 
C – control, 6Y– dose of 6 Gy; 7Y – dose of 7 Gy From the results presented in Tab. 4, it can be seen that the proportion of living cells in the control was −92.4 ± 0.4%, at the same time, there were also cells at different stages of death in these samples – early apoptosis (0.7 ± 0.17%), late apoptosis / necrosis (7.1 ± 0.44%), and dead cells (0.1 ± 0.0%).
After РС-3 culture irradiation at a dose of 6 Gy, changes in the ratios of cells at different stages of apoptosis were observed. Naturally, a decrease in the percentage of living cells to 57.6 ± 6.5% was observed, which is 1.6 times less than in the control (P = 0.0008). At the same time, the proportion of cells in early apoptosis increased to 3.2 ± 0.6%, which is 4.5 times higher than the control values (P = 0.002), see Tab. 4. The proportion of cells in the late apoptosis stages (necrosis) and dead cells after irradiation reached values of 37.57 ± 5.32% and 1.67 ± 0.67%, which is 5.3 times (P = 0.001) and 16.7 times (P = 0.02) higher than the control, respectively.
An increase in the radiation dose to 7 Gy led to even greater changes in the ratio of cells at different stages of apoptosis. Compared with the control, the number of living cells decreased 2 times (P < 0.05) and reached 46.2 ± 10.8%, which is also 1.2 times lower (P < 0.05) than the values after irradiation at a dose of 6 Gy. The percentage of cells in the stage of early apoptosis exceeded the control values 6.4 times (P < 0.05) and reached values of 4.47 ± 1.25%, which is also 1.4 times higher in comparison with an irradiation level of 6 Gy (P < 0.05). The proportion of cells in the late stages of apoptosis (necrosis) and dead cells after irradiation of the cells of this culture reached the values 45.6 ± 9.92% (6.4 times more (P < 0.05) than the control values and 1,2 times more (P < 0.05) than the proportion after exposure to 6 Gy) and 3.67 ± 2.1% (36.7 times more (P < 0.05) than the control values and 2.2 times more (P < 0.05) than the proportion after exposure to 6 Gy), respectively (Tab. 4).
The study enabled to establish that daily irradiation of PC-3 cells on a Novalis TX linear accelerator at doses 6 and 7 Gy for 5 days led to a significant decrease in both the total number of cells and the number of viable cells. In this case, the irradiation effect at a dose of 7 Gy manifested itself earlier and, ultimately, after 5 days of irradiation was more pronounced than from irradiation at a dose of 6 Gy. Nevertheless, after 5 days of irradiation, about 15% of the initial number of prostate tumor cells retained their viability, which may be associated with the development or initial presence of radioresistance in some cells.
After a 5-day experiment on PC-3 cells irradiation, there were obtained data on the differential copy number of 32 genes in intact (control) and irradiated cells (Fig. 4).
According to the features of copy numbers in intact and irradiated samples, four main gene clusters were identified:
1. BRCA1, BRCA2, BRIP, RNF168, C-FLIP, EXO1, LIG4;
2. ATM, CDKN1B, HIST1, CASP8, TP53, RIF1, BAX;
3. AKT, CCND3, H2AX, CCND1, RAD50, FGFR2, RBBP8, EP300, RAP80, BCL2;
4. PTEN, KU70, TOPB1, MDM2, XRCC4, CASP3.
At the same time, the intact and irradiated samples themselves are combined into clusters according to the similarity of gene CNV (Fig. 4). The similarity in the copy number of some genes in intact and irradiated samples at different clustering levels (Fig. 4, 5 and Tab. 5) can be explained by initial presence in PC-3 line cell subpopulation with a certain copy number of genes.
5. Similarities and differences in the copy number of genes between clusters. 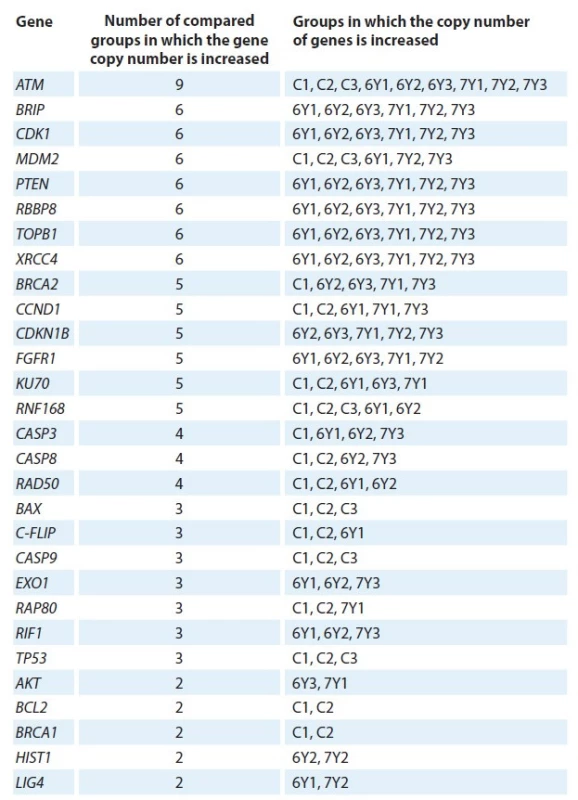
C – control, 6Y – dose of 6 Gy, 7Y – dose of 7 Gy Obviously, decreased or increased copy numbers of genes (mentioned above) under the conditions of RT can ensure the survival of these cells subpopulation.
After copy number normalization in the irradiated cells relative to intact ones, there were obtained data shown in Graph 2. Relative to control group cells, in PC-3 cells exposed to 6 Gy irradiation, the copy numbers of CDK1, CDKN1B, H2AX, PTEN, XRCC4 and RBBP8 genes were significantly increased 1.9, 2.5, 1.9, 1.7, 1.5 and 2.0 times (P < 0.05), and the copy numbers of genes BCL2, CCND3, TP53 and BAX were reduced 2.6, 2.4, 1.9 and 1.8 times (P < 0.05), respectively.
Graph 2. Relative copy number of genes in PC-3 cells with increased radioresistance (after 5-day irradiation at a dose of 6 Gy). 
* statistically significant differences relative to intact (control) cells (P < 0.05) Graph 3. Relative copy number of genes in PC-3 cells with increased radioresistance (after 5-day irradiation at doses of 7 Gy). 
* statistically significant differences relative to intact (control) cells (P < 0.05) Relative to control group cells, in PC-3 cells exposed to 7 Gy irradiation, the copy numbers of genes CDK1, CDKN1B, PTEN, XRCC4, EP300 and RBBP8 were statistically significantly increased 1.8, 2.6, 1.7, 1.7, 2.0 and 1.7 times (P < 0.05), while the copy numbers of CCND3, TP53 and BCL2 genes were reduced 2.6, 1.8 and 2.4 times (P < 0.05), respectively (Graph 3).
Similar data were obtained for the expression of 32 genes in intact and irradiated samples (Graph 4). There was a statistically significant (P < 0.05–0.005) overexpression of BRCA2, CDK1, CDKN1B, H2AX, RAD50, XRCC4, RBBP8 and EP300 genes, and hypoexpression of CCND3, TP53 and BCL2 genes. A strong positive correlation was observed between the copy number and expression of these genes: r = 0.906, 0.889 and 0.803 for control group cells, cells irradiated at a dose of 6 Gy and cells irradiated at a dose of 7 Gy, respectively.
However, significant differences between expression and copy number variation of BRCA2 and RAD50 genes were found, indicating that the change in the expression of these genes after irradiation at doses of 6 and 7 Gy is not associated with their copy number variation, but is probably regulated by other mechanisms.
Using Gene Set Enrichment Analysis, the participation of genes BRCA2, CDK1, CDKN1B, H2AX, RAD50, XRCC4, RBBP8, EP300, CCND3, TP53 and BCL2 in 10 sig-naling pathways was shown according to the Kyoto Encyclopedia of Genes and Genomes database (Tab. 6).
The analysis of the signaling pathways that involve genes AKT, ATM, BRIP, BRCA1, BRCA2, CDK1, CDKN1B, CCND1, CCND3, EXO1, FGFR2, HIST1, H2AX, KU70, PTEN, RAD50, RAP80, RIF1, RNF168, TOPB1, TP53, XRCC4, BAX, CASP8, CASP3, CASP9, MDM2, BCL2, RBBP8, EP300, LIG4 and C-FLIP using the FMD algorithm enabled to identify six functional modules, which are visually presented in Fig. 6.
We also analyzed the interaction network and the relationship index of genes changing their expression and CNV – BRCA2, CDK1, CDKN1B, H2AX, RAD50, XRCC4, RBBP8, EP300, CCND3, TP53, BCL2 (Fig. 7).
Graph 4. Relative expression of genes in PC-3 cells with increased radioresistance (after 5-day irradiation at doses of 6 and 7 Gy). 
* statistically significant differences relative to intact (control) cells (P < 0.05) 6. Data from Gene Set Enrichment Analysis. 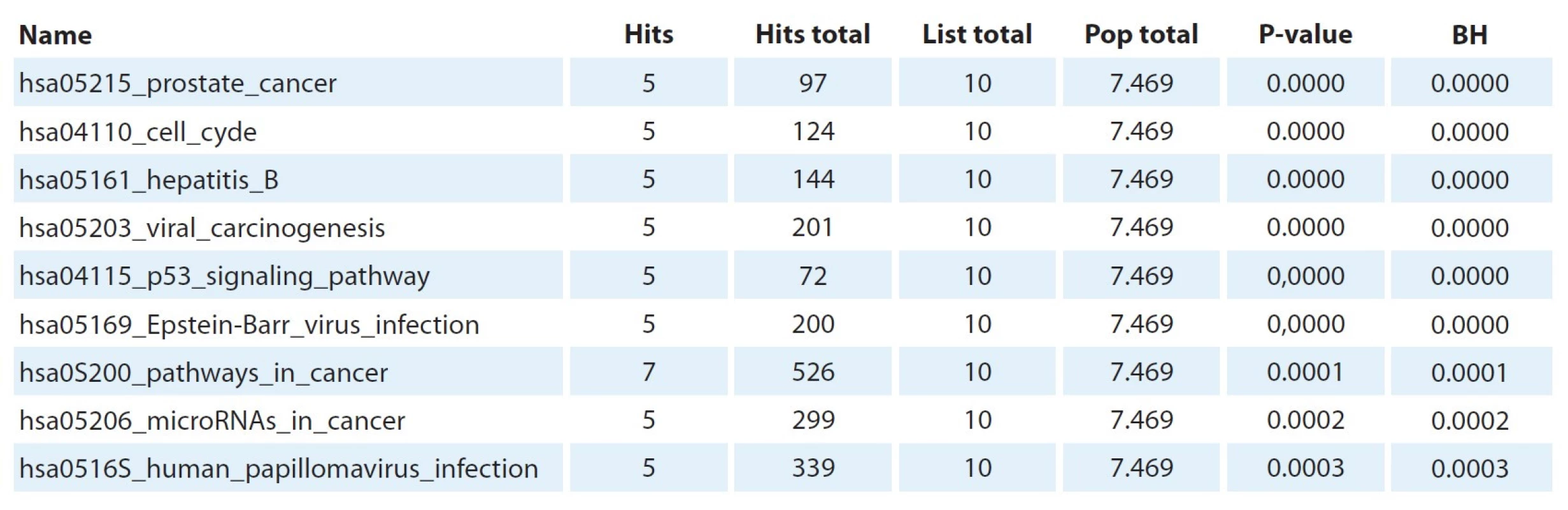
BH – Benjamin-Hochberg amendment 7. Micro-RNAs that altered expression in irradiated cells and their target genes. 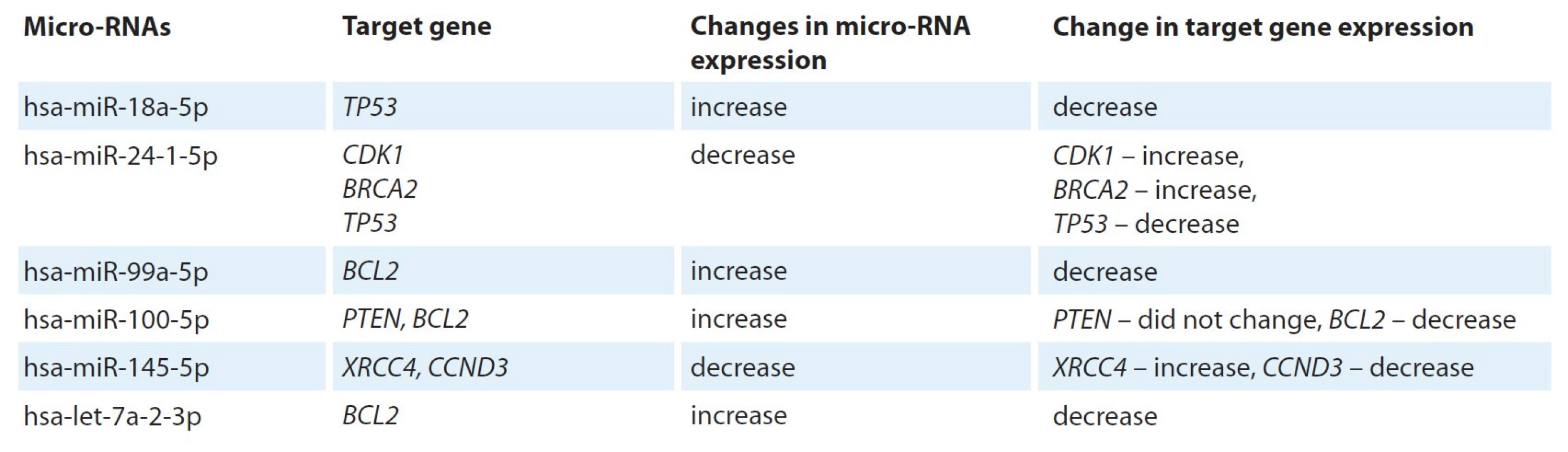
Graph 5. Changes in micro-RNA expression upon irradiation of PC-3 cells at doses of 6 and 7 Gy. 
* statistically significant differences relative to intact (control) cells (P < 0.05) The Multi-association network integration algorithm [21] was used for established interactions between BRCA2, CDK1, CDKN1B, H2AX, RAD50, XRCC4, RBBP8, EP300, CCND3, TP53, BCL2 and RNF168, RIF1, EXO1, LIG4, XRCC6, UIMC1, BAX, PTEN, CASP9, ATM, MDM2, CDKN1A, XRCC5, APAF1, XIAP, CCND2 genes (Fig. 7). The algorithm predicts the gene function in a complex network of many genes, using a kind of machine learning algorithms, assigns an estimate (discriminant value) to each node-point of the constructed network. This score reflects the calculated bond strength (Fig. 8).
Thus, BRCA2, CDK1, CDKN1B, H2AX, RAD50, XRCC4, RBBP8, EP300, CCND3, TP53 and BCL2 genes are components of various signaling cascades of normal and tumor cells, and their expression changes mediated by copy number changes lead to the changes in the expression of a number of other genes (Fig. 7, 8) [22].
The analysis of 15 microRNAs expression (let-7, miR15a/16, miR-17, miR-18a, miR-21, miR-24, miR-26b, miR-99a, miR-100, miR-101, miR-106a , miR-663a, miR-143, miR-145) in intact and irradiated PC-3 cells showed statistically significant (P < 0.05) changes in the expression of six microRNA: relative to intact cells, the expression of hsa-miR-18a-5p increased 64 times under irradiation doses of 6 and 7 Gy, the expression of hsa-miR-24-1-5p decreased 16 times under irradiation doses of 6 and 7 Gy, the expression of hsa-miR-99a-5p increased 16 times under irradiation doses of 6 and 7 Gy, the expression of hsa -miR-100-5p increased 32 and 64 times under irradiation doses of 6 and 7 Gy, the expression of hsa-miR-145-5p decreased 32 and 64 times under irradiation doses of 6 and 7 Gy, and the expression of hsa-let-7a-2 -3p increased 4 times at irradiation doses of 6 and 7 Gy, respectively (Graph 5).
For differentially expressed micro-RNAs, a search for target genes was carried out using the TarPmiR algorithm. For six microRNAs that alter the expression of PC-3 cells upon irradiation, 6,129 target genes were found, including 7 genetic loci, expression of which was observed in this study: TP53, CDK1, BRCA2, BCL2, PTEN, XRCC4 and CCND3 (Tab. 7)
Discussion
In the course of the study, it was demonstrated that the effect of various doses of ionizing radiation on in vitro prostate tumor cells leads to their significant elimination. The fundamental mechanism by which ionizing radiation has a therapeutic effect is DNA damage initiation in tumor cells, especially double strand breaks (DSBs) [23]. It is known from the literature that single irradiation doses 4–8 Gy cause a short-term delay in the cell cycle in G2/ M phase with a maximum effect approx. 16 hours after irradiation [24]. However, tumor cells are able to use DSB repair systems to overcome radiation-induced cytotoxicity [25], which is counterproductive for therapy. As a result, radioresistance may develop.
Other signaling pathways that are not part of the cell’s repair systems can also influence the radiation response of prostate tumor cells [26]. In our experiment, about 50% of the remaining tumor cells (15% of the initial number) still remained viable after 5 days of irradiation. This is caused by the development of radioresistance due to certain molecular genetic and epigenetic features of these cells.
Scheme 1. Scheme of genetic and epigenetic changes in tumor cells under irradiation at doses of 6 and 7 Gy. 
Thus, the changes in the transcriptional activity and copy number of BRCA2, CDK1, CDKN1B, H2AX, RAD50, XRCC4, RBBP8, EP300, CCND3, TP53 and BCL2 genes are likely to provide an increased survival of prostate tumor cells under radiation therapy, due to the changes in the work of key signaling pathways providing DNA repair, apoptosis and cell cycle regulation. For DNA repair after irradiation, chromatin must be decondensed [27]. The H2AX gene encodes a histone protein from the H2A family. In response to DSBs in DNA caused by ionizing radiation, H2AX becomes phosphorylated (gH2AX). Due to this modification, DNA becomes less condensed, freeing up space for the attachment of protein complexes necessary during the repair. Then gH2AX binds to MDC1, and ubiquitin ligases RNF8 and RNF168 are attached to the gH2AX/ MDC1 complex, ubiquitinating other chromatin components. This allows BRCA1 bind to chromatin modified by gH2AX/ MDC1 [28]. At the site of gH2AX-modified chromatin, the MRN complex (a protein complex consisting of Mre11, Rad50, and Nbs1) is also assembled [29].
The BRCA2 gene product is required for the repair of damaged DNA; it binds single-stranded DNA and directly interacts with the RAD51 recombinase to stimulate an important step in homologous recombination [30]. The movement of RAD51 to a double-stranded DNA break requires the BRCA1-PALB2-BRCA2 complex formation [31]. BRCA2 also plays an important role in protection against Mre11-dependent nucleolytic degradation of reversed replication forks, which are formed when the DNA replication fork is stopped by mutation, intercalating substance, or radiation [32]. RBBP8 encodes a protein that regulates cell proliferation. This protein forms complexes with the transcriptional co-repressor of C-terminal binding protein and modulates the functions of BRCA1 in the regulation of DNA transcription and repair [33].
Two more genes, the expression and copy number of which are increased in radioresistant prostate cells are XRCC4 and EP300. Together with DNA ligase IV and DNA-dependent protein kinase, the protein encoded by XRCC4 gene participates in the repair of DNA double-strand breaks (non-homologous recombination) [34]. Histone acetyltransferase p300 is an enzyme that is encoded by EP300 gene in humans. This enzyme regulates gene transcription through chromatin remodeling and plays an important role in the regulation of cell growth and division, causing cells to take on specialized functions (to differentiate). EP300 is also a co-activator of hypoxia-inducible factor 1-alpha and plays a role in the stimulation of hypoxia-induced genes such as VEGF. Mutations in EP300 gene have been identified in some types of cancer. Somatic mutations in EP300 gene have been found in a small number of solid tumors, including cancer of the colon and rectum, stomach, breast, and prostate. In tumor cells, mutations in EP300 gene do not allow functional protein production, without which cells cannot effectively inhibit growth and division [35]. The likely increased copy number of XRCC4, as well as RBBP8, provides effective DNA repair after irradiation. Today, the role of EP300 gene in radioresistance is difficult to determine, perhaps its copy number simply reflects the molecular genetic characteristics of the surviving clones of PC-3 cells and is not associated with CNV sensitivity to radiation.
Respectively, increased CNV and expression of BRCA2, XRCC4, RAD50, H2AX, and RBBP8 genes in individual clones of PC-3 cell line gives them the advantage of a more efficient DNA repair system and ensures survival when exposed to radiation therapy.
The BCL2 gene product is involved in apoptosis regulation. BCL2 suppresses apoptosis in many cell systems by controlling the permeability of mitochondrial membrane, inhibits caspases by preventing the release of cytochrome C from mitochondria and by binding the apoptosis-activating factor APAF1 [36]. It can be suggested that the surviving clones of the cell line could initially have a more powerful DNA repair system, as well as a more efficient apoptosis regulation system.
Interestingly, an increase in CNV and expression of two genes, CDK1 and CDKN1B, in PC-3 cells was found. Cyclin-dependent kinase 1 (CDK1) is a highly conserved protein that plays a key role in cell cycle regulation [37]. When it binds to cyclin, the cell cycle progresses. Binding to cyclin alters access to the active center of CDK1, ensuring its activity; in addition, cyclins can target CDK1 to certain subcellular locations [37]. The product of CDKN1B gene, p27 protein, is an inhibitor of cyclin-dependent kinase 1, regulates the cell cycle, being responsible for its arrest in G1 phase. Protein p27 inhibits the activity of cyclin (A, E, or D)/ cyclin-dependent kinase complexes, while the positive or negative regulation of activity depends on the phosphorylation status of the corresponding complex [38]. At the same time, the copy number of the gene that starts the cell cycle and that of the gene that stops it increases. The biological effect of this phenomenon and its role in the formation of the radioresistance of tumor cells have yet to be clarified. The protein encoded by CCND3 gene belongs to a highly conserved cyclin family, whose members also function as regulators of cyclin-dependent kinases. Different cyclins show different patterns of expression and degradation that facilitate the temporal coordination of each mitotic event [39]. Apparently, the special patterns of the expression of these genes ensure the survival of cells under the conditions of radiation therapy.
Another interesting molecular genetic change concerns the increased copy number of PTEN genetic locus encoding the enzyme phosphatase, which can be substituted by proteins and phosphatidylinositol-3-phosphates. The phosphate group cleavage from phosphatidylinositol-3-phosphates, catalyzed by phosphatase and tensin homologue deleted on chromosome 10 (PTEN), deprives functions of secondary messengers these compounds in cellular signaling cascades. Moreover, PTEN is an anti-oncoprotein that negatively regulates the PI3K/ AKT/ mTOR signaling pathway [40]. At the same time, the transcriptional activity of this gene is not statistically significantly increased. Based on the obtained data, it means that the prostate tumor cells that survived after irradiation could have increased the expression of PTEN gene and anti-oncoprotein due to the increased number of corresponding gene copies. However, other mechanisms obviously affect PTEN gene expression. Interaction with microRNA may be one of these mechanisms.
MicroRNAs are short non-coding RNAs that regulate gene expression either by catalyzing the destruction of mRNA or by inhibiting the translation of mRNA into protein [41]. Mature miRNA is a single-stranded RNA of about 22 nucleotides in length, sequentially derived from the primary transcript (pri-miRNA) [42]. miRNAs are epigenetic regulators. Many microRNAs are tissue specific and modulate gene expression by interacting with complementary nucleotide sequences of target mRNAs [43].
The obtained data on CNV, gene and micro-RNA expression enabled to form a scheme of genetic and epigenetic changes in tumor cells that survived after irradiation at doses of 6 and 7 Gy (Scheme 1).
According to this scheme and Tab. 7, BRCA2, CDK1, CDKN1B, H2AX, RAD50, XRCC4, RBBP8 and EP300 gene overexpression is associated with an increase in the copy number of CDK1, CDKN1B, H2AX, XRCC4, RBBP8 and EP300 genes, as well as with decrease in hsa-miR-24-1-5p and hsa-miR-145-5p expression. At the same time, hypoexpression of CCND3, TP53, BCL2 and the absence of changes in the expression of PTEN gene (with an increased copy number of this) are associated with a decrease in the copy number of the corresponding genes (CCND3, TP53, BCL2), as well as with an increase in the expression of hsa-miR-18a-5p (for TP53), hsa-miR-99a-5p (for BCL2), hsa-miR-100-5p (for BCL2 and PTEN) and hsa-let-7a-2-3p (for BCL2).
Conclusion
Thereby, the study enabled to establish that daily irradiation of PC-3 cells on a Novalis TX linear accelerator at doses of 6 and 7 Gy for 5 days led to a significant decrease in both the total number of cells and the number of viable cells. In this case, the effect of irradiation at a dose of 7 Gy was manifested earlier and, ultimately, was more pronounced after 5 days of irradiation than after irradiation at a dose of 6 Gy. Nevertheless, after 5 days of irradiation, about 15% of the initial number of prostate tumor cells of PC-3 line retained their viability, which is associated with the initial presence of radioresistance in some cells. It was found that PC-3 cells that retained their viability after 5 days of radiation therapy have special genetic and epigenetic characteristics ensuring their survival: increased copy number and expression of BRCA2, CDK1, CDKN1B, H2AX, RAD50, XRCC4, RBBP8 and EP300 genes and reduced copy number and expression of CCND3, TP53 and BCL2 genes, as well as differential expression of six microRNAs (hsa-miR-18a-5p, hsa-miR-24-1-5p, hsa-miR-99a-5p, hsa -miR-100-5p, hsa-miR-145-5p3, hsa-let-7a-2-3p).
Acknowledgments
We express our gratitude to Potemkin DS and Vasilchenko NG (staff of National Medical Research Oncology Center, laboratory of molecular oncology) for their help with the cell culture, and to Sagakyants AB and Bondarenko ES (staff of National Medical Research Oncology Center, laboratory of immunophenotyping of tumors) for their help in conducting studies on a flow cytometer.
Submitted/Obdrženo: 27. 8. 2020
Accepted/Přijato: 22.10.2020Denis Kutilin, PhD
National Medical Research
Oncology Center
14 line str., 63
Rostov-on-Don
Rostov region
344037, Russian Federation
e-mail: k.denees@yandex.ru
Sources
- Zinkovich MS, Maksimov AY, Rozenko LY et al. Radioresistance as a factor of evolution of radiation therapy for prostate cancer. [online]. Available from: http://science-education.ru/ru/article/view?id=28627.
- Chaiswing L, Weiss HL, Jayswal RD. Profiles of radioresistance mechanisms in prostate cancer. Crit Rev Oncog 2018; 23(1–2): 39–67. doi: 10.1615/CritRevOncog.2018025946.
- Mottet N, Bellmunt J, Bolla M. EAU-ESTRO-SIOG Guidelines on prostate cancer. Part 1: Screening, diagnosis, and local treatment with curative intent. Eur Urol 2017; 71(4): 618–629. doi: 10.1016/j.eururo.2016.08.003.
- Negre-Salvayre A, Coatrieux C, Ingueneau C et al. Advanced lipid peroxidation end products in oxidative damage to proteins. Potential role in diseases and therapeutic prospects for the inhibitors. Br J Pharmacol 2008; 153(1): 6–20. doi: 10.1038/sj.bjp.0707395.
- Li Z, Pearlman AH, Hsieh P. DNA mismatch repair and the DNA damage response. DNA Repair (Amst) 2016; 38 : 94–101. doi: 10.1016/j.dnarep.2015.11.019.
- Sonveaux P. ROS and radiotherapy: More we care. Oncotarget 2017; 8(22): 35482–35483. doi: 10.18632/oncotarget.16613.
- Kutilin DS, Airapetova TG, Anistratov PA et al. Copy number variation in tumor cells and extracellular dna in patients with lung adenocarcinoma. Bull Exp Biol Med 2019; 167(6): 771–778. doi: 10.1007/s10517-019-04620-y.
- Kutilin DS, Zinkovich MS, Gusareva MA et al. Gene copy number variation as a factor of radiation resistance of prostate tumor cells. Modern problems of science and education 2020. [online]. Available from: http://science-education.ru/ru/article/view?id=29866.
- ATCC, Standards Development Organization is accredited by the American National Standards Institute (ANSI). [online]. Available from: http://www.lgcstandards-atcc.org.
- Kutilin DS, Sagakyants AB, Zinkovich MS et al. Effects of various radiotherapy doses on survival of prostate tumor cells line PC-3. [online]. Available from: http://www.science-education.ru/article/view?id=28740.
- Kit OI, Vodolazhsky DI, Kutilin DS et al. Changes in the number of copies of genetic loci in gastric cancer. Mol Biol (Mosk) 2015; 49(4): 658–666. doi: 10.7868/S0026898415040096.
- Kutilin DS. Regulation of gene expression of cancer/testis antigens in colorectal cancer patients. Mol Biol (Mosk) 2020; 54(4): 580–595. doi: 10.31857/S0026898420040096.
- Vandesompele J, De Preter K, Pattyn F et al. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. [online]. Available from: https://rdcu.be/b6uOX. doi: 10.1186/gb-2002-3-7-research0034.
- Kutilin DS, Mogushkova KA. Effect of anthracycline antitumor antibiotics upon transcription activity of cancer-testis antigens in model experiments with HeLa cells. Medical Immunology (Russia) 2019; 21(3): 539–546. [online]. https://doi.org/10.15789/1563-0625-2019-3-539-546.
- Balcells I, Cirera S, Busk PK. Specific and sensitive quantitative RT-PCR of miRNAs with DNA primers. BMC Biotechnol 2011; 11(1): 70. doi: 10.1186/1472-6750-11-70.
- Krishnan A, Zhang R, Yao V et al. Genome-wide prediction and functional characterization of the genetic basis of autism spectrum disorder. Nature Neuroscience 2016; 19(11):1454–1462. doi: 10.1038/nn.4353.
- Genome-scale integrated analysis. [online]. Available from: http://giant.princeton.edu/.
- Greene CS, Krishnan A, Wong AK et al. Understanding multicellular function and disease with human tissue-specific networks. Nature Genet 2015; 47(6): 569–576. doi: 10.1038/ng.3259w.
- Ding J, Li X, Hu H. TarPmiR: a new approach for microRNA target site prediction. Bioinformatics 2016; 32(18): 2768–2775. doi:10.1093/bioinformatics/btw318.
- Backes C, Khaleeq QT, Meese E et al. miEAA: microRNA enrichment analysis and annotation. Nucleic Acids Res 2016; 44(W1):W110–W116. doi: 10.1093/nar/gkw345.
- Warde-Farley D, Donaldson SL, Comes O et al. The GeneMANIA prediction server: biological network integration for gene prioritization and predicting gene function. Nucleic Acids Res 2010; 38(2): W214–W220. doi: 10.1093/nar/gkq537.
- Wu G, Feng X, Stein L. A human functional protein interaction network and its application to cancer data analysis. Genome Biol 2010; 11(5): R53. doi: 10.1186/gb-2010-11-5-r53.
- Sandler HM, Mirhadi AJ. Radical radiotherapy for prostate cancer is the only way to go. Oncology (Williston Park) 2009; 23(10): 840–843.
- Geldof AA, Plaizier M, Duivenvoorden I et al. Cell cycle perturbations and radiosensitization effects in a human prostate cancer cell line. J. Cancer Res Clin Oncol 2003; 129(3): 175–182. doi: 10.1007/s00432-002-0412-8.
- Lomax M, Folkes L, O’Neill P. Biological consequences of radiation-induced DNA damage: relevance to radiotherapy. Clin Oncol 2013; 25(10): 578–585. doi: 10.1016/j.clon.2013.06.007.
- Yao M, Rogers L, Suchowerska N et al. Sensitization of prostate cancer to radiation therapy: Molecules and pathways to target. Radiother Oncol 2018;128(2): 283–300. doi: 10.1016/j.radonc.2018.05.021.
- Mailand N, Bekker-Jensen S, Faustrup H et al. RNF8 ubiquitylates histones at DNA double-strand breaks and promotes assembly of repair proteins. Cell 2007; 131(5): 887–900. doi: 10.1016/j.cell.2007.09.040.
- Scully R, Xie A. Double strand break repair functions of histone H2AX. Mutat Res 2013; 750(1–2): 5–14. doi:10.1016/j.mrfmmm.2013.07.007.
- Bekker-Jensen S, Lukas C, Kitagawa R et al. Spatial organization of the mammalian genome surveillance machinery in response to DNA strand breaks. J. Cell Biol 2006; 173(2): 195–206. doi: 10.1083/jcb.200510130.
- Wang CX, Jimenez-Sainz J, Jensen RB et al. The post-synaptic function of Brca2. Scientific Reports 2019; 9(1): 4554. doi:10.1038/s41598-019-41054-y.
- Holloman WK. Unraveling the mechanism of BRCA2 in homologous recombination. Nat Struct Mol Biol 2011; 18(7): 748–754. doi: 10.1038/nsmb.2096.
- Mijic S, Zellweger R, Chappidi N et al. Replication fork reversal triggers fork degradation in BRCA2-defective cells. Nat Commun 2017; 8(1): 859. doi: 10.1038/s41467-017-01164-5.
- Sartori AA, Lukas C, Coates J et al. Human CtIP promotes DNA end resection. Nature 2007; 450(7169): 509–514. doi: 10.1038/nature06337.
- Watson J. Molecular biology of the gene. New York: Cold Spring Harbor Laboratory Press 2008 : 265–278.
- Ntranos A, Casaccia P. Bromodomains: translating the words of lysine acetylation into myelin injury and repair. Neurosci Lett 2016; 625 : 4–10. doi: 10.1016/j.neulet.2015.10.015.
- Chao DT, Korsmeyer SJ. BCL-2 family: regulators of cell death. Annu Rev Immunol 1998; 16 : 395–419. doi: 10.1146/annurev.immunol.16.1.395.
- Morgan DL. The cell cycle: principles of control. London: New Science Press 2007 : 30–31.
- Mitrea DM, Yoon MK, Ou L et al. Disorder-function relationships for the cell cycle regulatory proteins p21 and p27. Biol Chem 2012; 393(4): 259–274. doi: 10.1515/hsz-2011-0254.
- Inaba T, Matsushime H, Valentine M et al. Genomic organization, chromosomal localization, and independent expression of human cyclin D genes. Genomics 1992; 13(3): 565–574. doi: 10.1016/0888-7543(92)90 126-d.
- McCubrey JA, Steelman LS, Chappell WH et al. Mutations and deregulation of Ras/Raf/MEK/ERK and PI3K/PTEN/Akt/mTOR cascades which alter therapy response. Oncotarget 2012; 3(9): 954–987. doi: 10.18632/oncotarget.652.
- Dimitriadi TA, Burtsev DV, Dzhenkova EA et al. Differential expression of microRNAs and their target genes in cervical intraepithelial neoplasias of varying severity. Advances in Molecular Oncology 2020; 7(2): 30–44. doi: 10.17650/2313-805X-2020-7-2-47-61.
- Palanichamy JK, Rao DS. miRNA dysregulation in cancer: towards a mechanistic understanding. Front Genet 2014; 5 : 54. doi: 10.3389/fgene.2014.00054.
- Dimitriadi TA, Burtsev DV, Dzhenkova EA et al. Micro-RNA as markers of pre-cancer diseases progression in cervical cancer. [online]. Available from: http://science-education.ru/ru/article/view?id=29529. doi 10.17513/spno.29529
Labels
Paediatric clinical oncology Surgery Clinical oncology Paediatric gynaecology Gynaecology and obstetrics Reproduction medicine
Article was published inClinical Oncology

2021 Issue 3-
All articles in this issue
- Léčba nemetastatického kastračně rezistentního karcinomu prostaty
- Social factors influence the course and prognosis of malignat diseases
- Current treatment of non-metastatic castration-refractory prostate cancer
- Indications for venous access in oncology – recommendations of national professional societies and current state in the Czech Republic
- Mechanobiology of cancerogenesis
- Epithelioid hemangioendothelioma on PET/CT scan
- The use of PET/ CT with a hypoxia marker (18F-MISO) in radiotherapy treatment planning
- Lipozomální irinotekan – nová možnost systémové léčby karcinomu pankreatu
- COVID-19 and oncological disease
- Aktuality z odborného tisku
- Životné jubileum doc. MU Dr. Juraja Kaušitza, CSc.
- Jubileum prvej dámy radiačnej onkológie na Slovensku – MUDr. Eva Siracká, DrSc.
- Genetic and epigenetic bases of prostate tumor cell radioresistance
- Clinical Oncology
- Journal archive
- Current issue
- Online only
- About the journal
Most read in this issue- Indications for venous access in oncology – recommendations of national professional societies and current state in the Czech Republic
- Epithelioid hemangioendothelioma on PET/CT scan
- COVID-19 and oncological disease
- Mechanobiology of cancerogenesis
Login#ADS_BOTTOM_SCRIPTS#Forgotten passwordEnter the email address that you registered with. We will send you instructions on how to set a new password.
- Career

