-
Medical journals
- Career
Association of XPG rs17655G>C and XPF rs1799801T>C Polymorphisms with Susceptibility to Cutaneous Malignant Melanoma: Evidence from a Case-Control Study, Systematic Review and Meta-Analysis
Authors: Niktabar Mohammadreza Seyed 1; Dastgheib Alireza Seyed 2; Heiranizadeh Naeimeh 1; Kargar Saeed 1; Raee-Ezzabadi Ali 3; Jarahzadeh Hossein Mohammad 4; Miresmaeili Mohsen Seyed 5; Zare-Shehneh Masoud 6; Neamatzadeh Hossein 6,7
Authors‘ workplace: Department of General Surgery, Shahid Sadoughi University of Medical Sciences, Yazd, Iran 1; Department of Medical Genetics, School of Medicine, Shiraz University of Medical Sciences, Shiraz, Iran 2; Department of Emergency Medicine, Shahid Sadoughi University of Medical Sciences, Yazd, Iran 3; Department of Anesthesiology and Critical Care, Shahid Sadoughi University of Medical Sciences, Yazd, Iran 4; Department of Biology, Science and Arts University, Yazd, Iran 5; Department of Medical Genetics, Shahid Sadoughi University of Medical Sciences, Yazd, Iran 6; Mother and Newborn Health Research Center, Shahid Sadoughi University of Medical Sciences, Yazd, Iran 7
Published in: Klin Onkol 2020; 33(3): 184-194
Category:
doi: https://doi.org/10.14735/amko2020184Overview
Background: Previous studies have evaluated associations of XPG rs17655G>C and XPF rs1799801T>C polymorphisms with a risk of cutaneous malignant melanoma (CMM). However, their results thus remained inconsistent or even contradictory. Thus, the aim of this meta-analysis was to evaluate association of XPG rs17655G>C and XPF rs1799801T>C polymorphism with a risk of CMM.
Methods: A comprehensive literature search was performed on PubMed, Web of Science, Scopus, SciELO and CNKI databases up to October 15, 2019 to identify relevant studies. Moreover, a case-control study was conducted to evaluate association of XPF rs1799801T>C with CMM risk in the Iranian population. The odds ratio (OR) and 95% confidence interval (CI) values were used to estimate the strength of the associations.
Results: Total of 12 studies including 9 studies with 5,362 cases and 7,195 controls on XPG rs17655G>C and 3 studies with 803 CMM cases and 737 controls on XPF rs1799801T>C were selected. Pooled data revealed that XPF rs1799801T>C polymorphism was significantly associated with an increased risk of CMM under the heterozygote model (CT vs. TT: OR = 1.313; 95% CI 1.062–1.624; P = 0.012). However, XPG rs17655G>C polymorphism was not significantly associated with the risk of CMM in the overall population and by ethnicity. The subgroup analysis showed a significant association between XPG rs17655G>C polymorphism and CMM in polymerase chain reaction-based restriction fragments length polymorphism (PCR-RFLP) group of studies.
Conclusion: This meta-analysis result revealed that XPF rs1799801T>C polymorphism may be a risk factor for developing of CMM. However, our pooled data inconsistence with the previous meta-analyses revealed that XPG rs17655G>C polymorphism was not associated with the risk of CMM.
Keywords:
polymorphism – meta-analysis – cutaneous malignant melanoma – XPG – XPF – association
Introduction
Cutaneous malignant melanoma (CMM) is a heterogeneous malignant neoplastic disease with high morbidity rates [1,2]. CMM is characterized by a high mutational load than other tumor types due to extensive UV damage and has a vastly variable prognosis depending on disease stage [3]. CMM arise upon malignant transformation of melanocytes, the pigment-producing cells within the skin that generally protect skin cells from UV-radiation induced DNA-damage [4]. Although CMM only accounts for less than 3% of all skin cancers, it is responsible for the vast majority of skin cancer related deaths highlighting the aggressiveness of this cancer type and the need for the improvement of the therapies [5]. The worldwide incidence of CMM has risen rapidly over the course of the last 50 years despite public efforts to promote sun protection behaviors among populations at risk [6]. It is reported as the 19th most common cancer worldwide, with estimated age-standardized incidence rates of 2.8–3.1 cases per 100,000 people [7]. Moreover, CMM incidence and also mortality rates vary by gender, ethnicity and age, which is also associated with differences in melanoma anatomic sites [8]. The annual costs of melanoma treatment are substantial; in USA, they have raised by 288 % in less than a decade [5,9]. Moreover, they comprise 3.3 billion USD of the total 8.1 billion USD in all direct skin cancer annual costs [5,8].
CMM is a heterogeneous malignancy with a complex biology. Genetic aberrations in the mitogen-activating protein kinase pathway is a central event in the etiology of CMM, of which mutations in BRAF and NRAS genes are the most common [10,11]. NRAS was the first gene in this pathway to be found mutated in CMM in a considerable rate (approximately 20%) [12]. Additionally, recurrent mutations in the promoter of telomerase reverse transcriptase, the catalytic subunit of telomerase, have been associated with a poor prognosis in primary melanoma [13,14]. Moreover, approximately 10% of all CMM occur in cases with a family history of an autosomal dominant genodermatosis named familial atypical multiple mole and melanoma syndrome [15]. Over the past decade, studies have been revealed that failure in the nucleotide excision repair (NER) pathway may result in many human diseases such as CMM [16]. NER is responsible for repairing bulky DNA damage, such as DNA adducts caused by UV radiation, mutagenic chemicals or chemotherapeutic drugs [16,17]. In NER pathway at least eight genes including ERCC1, XPA, XPB/ERCC3, XPC, XPD/ERCC2, XPE/DDB1, XPF/ERCC4 and XPG/ERCC5 have a vital role in DNA repair and capable of preserving genetic integrity to prevent cells from malignant transformation [18,19].
The xeroderma pigmentosum group G (XPG), also known as the ERCC5 gene, functions to cut DNA lesions during DNA repair. XPG also participates in other cell processes such as transcription‐coupled DNA repair and RNA polymerase II transcription [20]. ERCC5/XPG is located on chromosome 13q22-33, consisting of 15 exons and 14 introns [21]. Moreover, the xeroderma pigmentosum complementation group F (XPF), also called excision repair cross-complimentary group 4 (ERCC4), plays a central role in NER of DNA, by nicks on the 5΄ side of the damaged DNA strand, by cleaving an open “bubble” intermediate [22,23]. ERCC4/XPF gene is located on chromosome 16p13.12, comprises 10 exons and spanning about 15 kb [22].
Human XPG and XPF genes are highly polymorphic and among known single nucleotide polymorphisms within these genes; XPG rs17655G>C and XPF rs1799801T>C polymorphisms are most frequently studied in association with risk of different type of cancers. To date, a number of molecular epidemiological studies have been performed to evaluate the association of XPG rs17655G>C and XPF rs1799801T>C polymorphisms with a risk of CMM in diverse populations. However, results thus far have remained inconsistent or even contradictory, partially because of the possible small effect of the polymorphism on CMM risk and the relatively small sample size in each of published study. Thus, we conducted a case-control study and comprehensive meta-analysis by including the most recent and relevant published articles to identify statistical evidence of the association of XPG rs17655G>C and XPF rs1799801T>C polymorphisms with the risk of CMM.
Materials and methods
Case-control study
A total of 300 participants including 150 cases with CMM and 150 healthy controls were recruited between June 2015 and September 2018. The age and sex of matched controls were randomly selected from the same hospital among patients who refereed for health check‐-up. The study was approved by the Medical Research Ethics Committee and a written informed consent was obtained from the study participants. Genomic DNA was extracted from blood samples using a QIAamp DNA Blood Mini Kit (Qiagen, Hilden, Germany) according to the instructions of the manufacturer. Genotype analyses of XPG rs17655G>C and XPF rs1799801T>C polymorphisms were performed by a polymerase chain reaction-based restriction fragments length polymorphism (PCR-RFLP) method as described previously [24–26].
Meta-analysis
Search strategy and study selection
Online electronic databases including PubMed, EMBASE, Scopus, Cochrane Library database, Springer Link, SciELO, Chinese Biomedical Database (CBD), China National Knowledge Infrastructure (CNKI), WanFang, OVID, SID, EBSCO and VIP were searched to collect all the eligible studies evaluating association of XPG rs17655G>C and XPF rs1799801T>C polymorphisms with the risk of CMM up to October 15, 2019. The following terms, keywords and their combinations were used: (“Cutaneous Malignant Melanoma” OR ”Melanoma”) AND (“Xeroderma Pigmentosum Group G” OR “XPG” OR “ERCC5” OR “rs17655G>C” OR “c.3310G>C” OR “p.Asp1104His”) AND (“Xeroderma Pigmentosum Group F” OR “XPF” OR “ERCC4 “ OR “rs1799801T>C” OR “30028T>C” OR “c.2505T>C” OR “p.Ser835=”) AND (“Gen” OR “Single Nucleotide Polymorphisms” OR “SNPs” OR “Polymorphism” OR “Mutation” OR “Variation” OR “Allele”). The search was limited to English, Chinese and Farsi language papers. The search was limited to human studies. Additional studies were identified by hand searching references in original articles and review articles. Of the studies with the same or overlapping data published by the same investigators, we selected the most recent ones with the largest number of subjects
Selection criteria
The studies eligible for the inclusion in our meta-analysis had to fulfill the criteria:
- studies with case-control design;
- studies evaluating association of any or both polymorphisms of XPG rs17655G>C and XPF rs1799801T>C polymorphisms with the risk of CMM;
- sufficient data for estimating the odds ratio (OR) with 95% confidence interval (CI).
The studies were excluded for the following reasons:
- No control population (case only studies);
- animal studies;
- data unavailable for calculating the genotype or allele frequencies;
- linkage studies, twin, sibling and other family-based studies;
- abstracts, reviews, case reports, posters, editorials, conference articles;
- overlapping data or duplicate of previous publication.
Studies overlapping with other studies should be eliminated and the largest study should be included in the final analysis.
Data extraction
The eligible studies were screened by 2 authors and all data were extracted carefully according to the inclusion criteria. Any disagreement was resolved by consultation. The following information was extracted from the eligible studies: first author’s surname, year of publication, ethnicity (categorized as Asian, Caucasian or mixed populations), source of controls (hospital based or population based), genotyping method, sample size, genotype distributions for both XPG rs17655G>C and XPF rs1799801T>C polymorphisms in cases and controls, minor allele frequency and Hardy-Weinberg equilibrium (HWE) in controls.
Statistical analysis
Crude OR values together with their corresponding 95% CI values were used to assess the strength of association between XPG rs17655G>C and XPF rs1799801T>C polymorphisms and the risk of CMM. The pooled OR values were performed under all 5 genetic models, i.e. allele (B vs. C), homozygote (BB vs. AA), heterozygote (BA vs. AA), dominant (BB + BA vs. AA) and the recessive (BB vs. BA + AA). The Cochran’s Q statistic test was employed to test between-study heterogeneity, and the heterogeneity was considered significant when P < 0.05 for Q-statistic. Moreover, I2 statistic was used to quantify inconsistency, which describes the proportion of variation in the log that is attributed to genuine differences across studies rather than to random error (range 0–100 %: I2 = 0–25 %, no heterogeneity; I2 = 25–50 %, moderate heterogeneity; I2 = 50–75 %, large heterogeneity; I2 = 75–100 %, extreme heterogeneity). The fixed-effects model (Mantel-Haenszel method) was used to calculate the pooled OR values when no significant heterogeneity was detected; otherwise, the random-effects model (DerSimonian-Laird method) was applied. The goodness‐of‐fit chi‐squared test was applied to test the Hardy‐Weinberg equilibrium (HWE) for XPG genotype distributions in controls, P > 0.05 were considered to have reliable and representative controls. Furthermore, meta-regression analysis was performed to investigate potential sources of heterogeneity including ethnicity, source of controls, genotyping methods and HWE status. The sensitivity analysis was mainly performed by sequential omission of individual studies to reflect the influence of the individual data set to the pooled OR values. An estimate of potential publication bias was carried out by the funnel plot. Funnel plot asymmetry was also assessed by the method of Egger’s linear regression test. For the interpretation of the Begg’s test, the statistical significance was defined as P < 0.1. All of the statistical calculations were performed using Comprehensive Meta-Analysis (CMA) software, version 2.0 (Biostat, USA). Two-sided P-values < 0.05 were considered statistically significant.
Results
Characteristics of included studies
Fig. 1 shows the flowchart of literature search and selection process. Through comprehensive literature search and study selection procedures, a total of 113 studies were retrieved based on the search criteria for cancer susceptibility related to XPG rs17655G>C and XPF rs1799801T>C polymorphisms. After reading the titles and abstracts, 78 full-text articles were preliminarily identified for further detailed evaluation. Then, 66 articles were excluded because they clearly did not meet the inclusion criteria or included overlapping references. The selected studies characteristics are summarized in Tab. 1. A total of 12 studies including 9 studies with 5,362 CMM cases and 7,195 controls on XPG rs17655G>C [24,27–32] and 3 studies with 803 CMM cases and 737 controls on XPF rs1799801T>C [25,26] were selected. All included studies were conducted between 2005 and 2019. The studies have been carried out in Germany, USA, UK, Spain, Brazil, Poland and Iran. Two PCR-RFLP and TaqMan genotyping methods were used in the included studies. For the XPG rs17655G>C polymorphism, there were seven studies of Caucasian population, one of Asian population and one of mixed population. For the XPF rs1799801T>C polymorphism, there was one of Caucasian population, one of Asian population, and one of mixed population. The genotype and minor allele frequency distributions in the studies considered in the present meta-analysis are shown in Tab. 1. Moreover, the distribution of genotypes in controls was in agreement with the Hardy-Weinberg equilibrium (HWE) for all selected studies, except for two studies for XPG rs17655G>C polymorphism and one study for XPF rs1799801T>C polymorphism (Tab. 1).
1. Flowchart of literature search and selection process. SNPs – single nucleotid polymorphisms 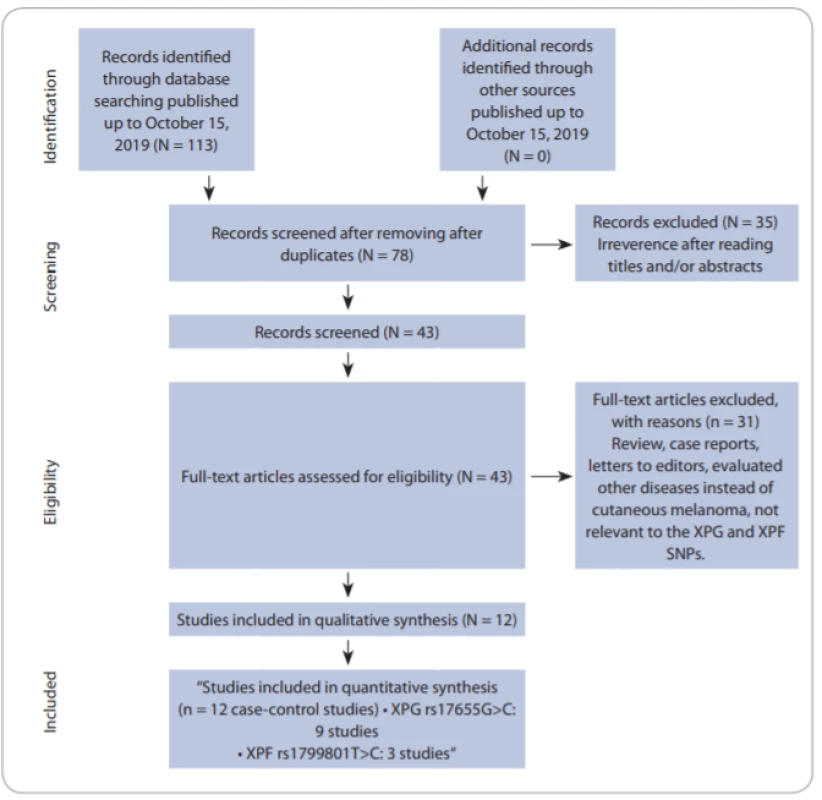
1. Characteristics of studies included in meta-analysis. 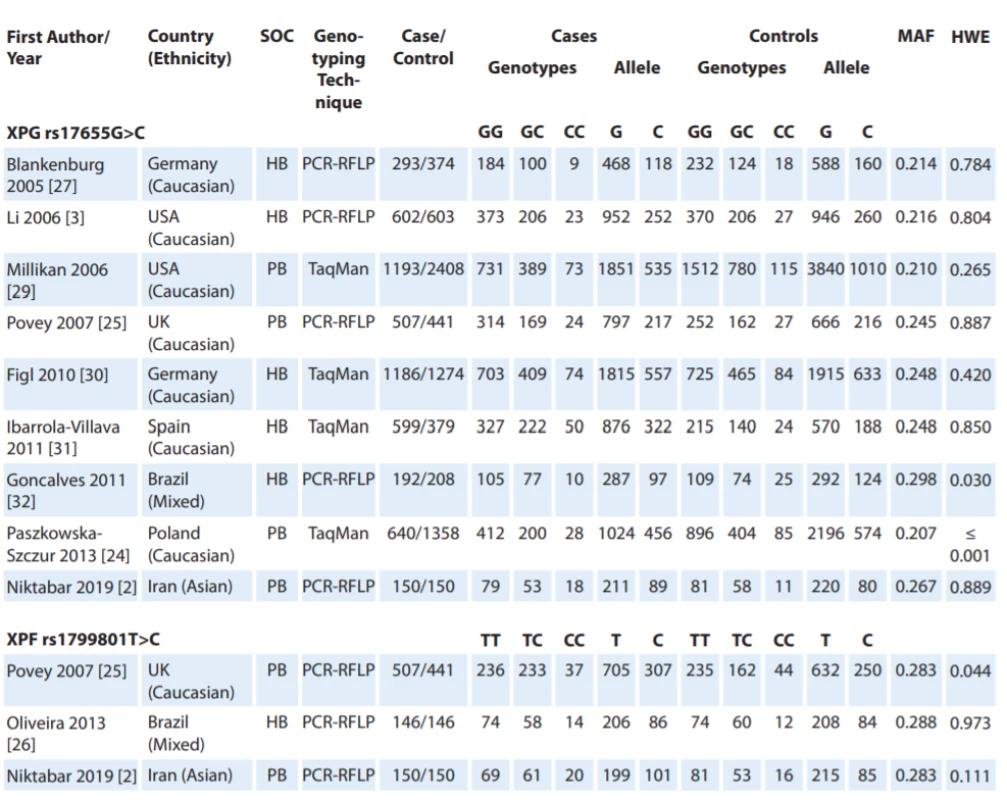
HB – hospital-based, HWE – Hardy-Weinberg equilibrium, MAF – minor allele frequency, NA – not available, PB – population-based, PCR – polymerase chain reaction, RFLP – restriction fragment length polymorphism, SOC – source of control Quantitative data synthesis
XPG rs17655G>C polymorphism
In Tab. 2, there are listed the main results of the meta-analysis of XPG rs17655G>C polymorphism and risk of CMM. When all the eligible studies were pooled into the meta-analysis of XPG rs17655G>C polymorphism, there was no evidence of significant association between XPG rs17655G>C polymorphism and CMM risk under all 5 genetic models, i.e. allele (C vs. G: OR = 1.397; 95% CI 0.619–3.152; P = 0.420) (Fig. 2A), homozygote (CC vs. GG: OR = 0.883; 95% CI 0.689–1.131; P = 0.324), heterozygote (CG vs. GG: OR = 0.988; 95% CI 0.915–1.068; P = 0.765) (Fig. 2B), dominant (CC + CG vs. GG: OR = 1.015; 95% CI 0.890–1.157; P = 0.901) and recessive (CC vs. CG + GG: OR = 0.880; 95% CI 0.686–1.129; P = 0.315). Moreover, we have performed the subgroup analysis by ethnicity (Caucasians), source of controls (HB and PB) and genotyping methods (PCR-FRLP and TaqMan). In the stratified analysis, there was not still significant association by ethnicity (Fig. 2C) and a source of controls. However, the subgroup analysis by genotyping methods revealed a significant association between XPG rs17655G>C polymorphism and CMM in PCR-RFLP subgroup studies under 2 genetic models, i.e. homozygote (CC vs. GG: OR = 0.674; 95% CI 0.485–0.938; P = 0.019) and recessive (CC vs. CG + GG: OR = 0.681; 95% CI 0.492–0.942; P = 0.020), but not in TaqMan group of studies.
2. Forest plot for association of XPG rs17655G>C polymorphism with a risk of CMM: A) in the overall population (allele model: C vs. G); B) in the overall population (heterozygote model: CG vs. GG); C) in the Caucasian populations (dominant model: CC + CG vs. GG). CMM – cutaneous malignant melanoma, CI – confi dence interval 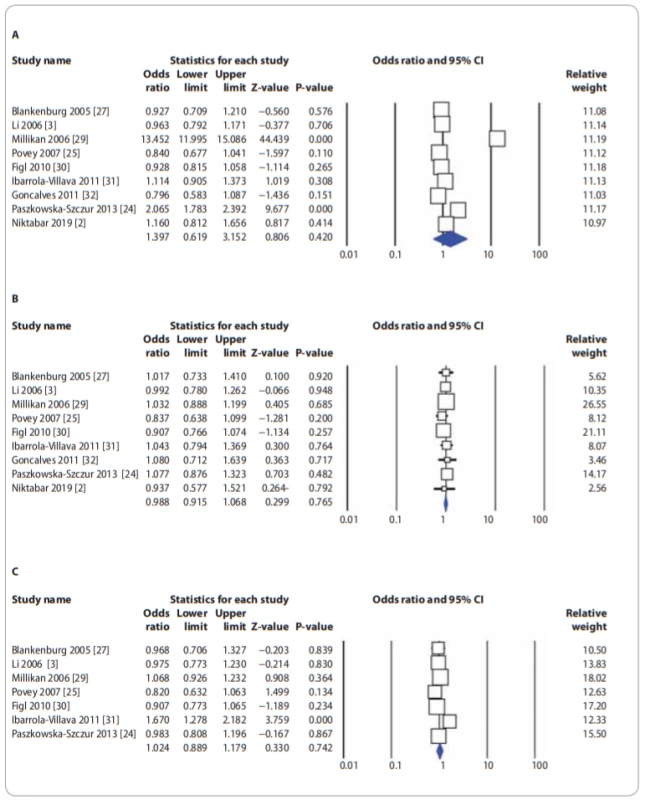
2. Summary of meta-analysis for the association of XPG rs17655G>C and XPF rs1799801T>C polymorphisms with a risk of CMM. 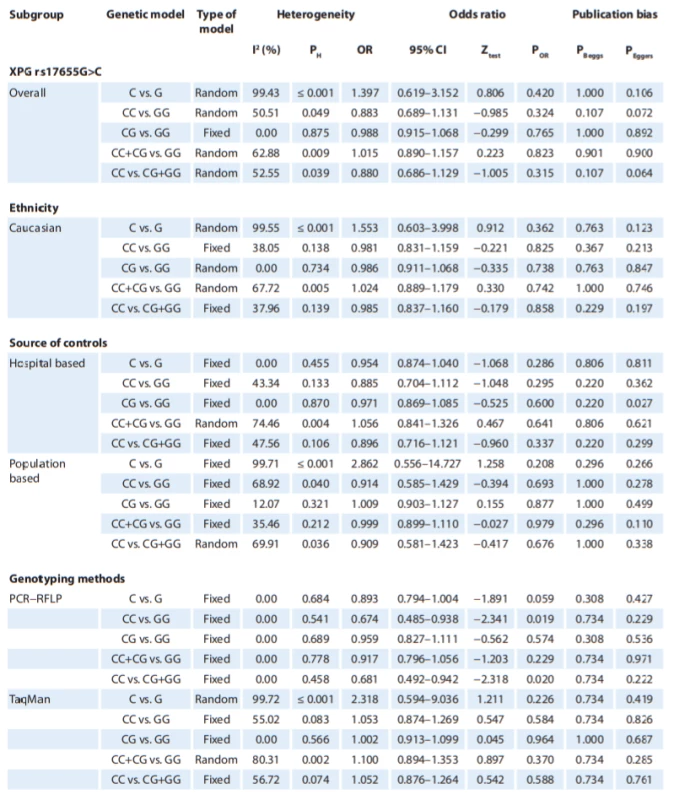
*By excluding HWE−violating studies
CI – confi dence interval, HWE – Hardy-Weinberg equilibrium, PCR – polymerase chain reaction, RFLP – restriction fragment length polymorphismTab. 2 – continuing. Summary of meta-analysis for the association of XPG rs17655G>C and XPF rs1799801T>C polymorphisms with a risk of CMM. 
*By excluding HWE−violating studies
HWE – Hardy-Weinberg equilibrium, PCR – polymerase chain reaction, RFLP – restriction fragment length polymorphism, CI – confi dence intervalXPF rs1799801T>C polymorphism
In Tab. 2, there are also listed the main results of the meta-analysis of XPF rs1799801T>C polymorphism and CMM risk. When all the eligible studies were pooled into the meta-analysis of XPF rs1799801T>C polymorphism, there was a significant association between XPF rs1799801T>C polymorphism and CMM risk under the heterozygote model (CT vs. TT: OR = 1.313; 95% CI 1.062–1.624; P = 0.012) (Fig. 3).
3. Forest plot for association of XPF rs1799801T>C polymorphism with a risk of CMM in the overall population under the heterozygote model (CT vs. TT). CMM – cutaneous malignant melanoma, CI – confi dence interval 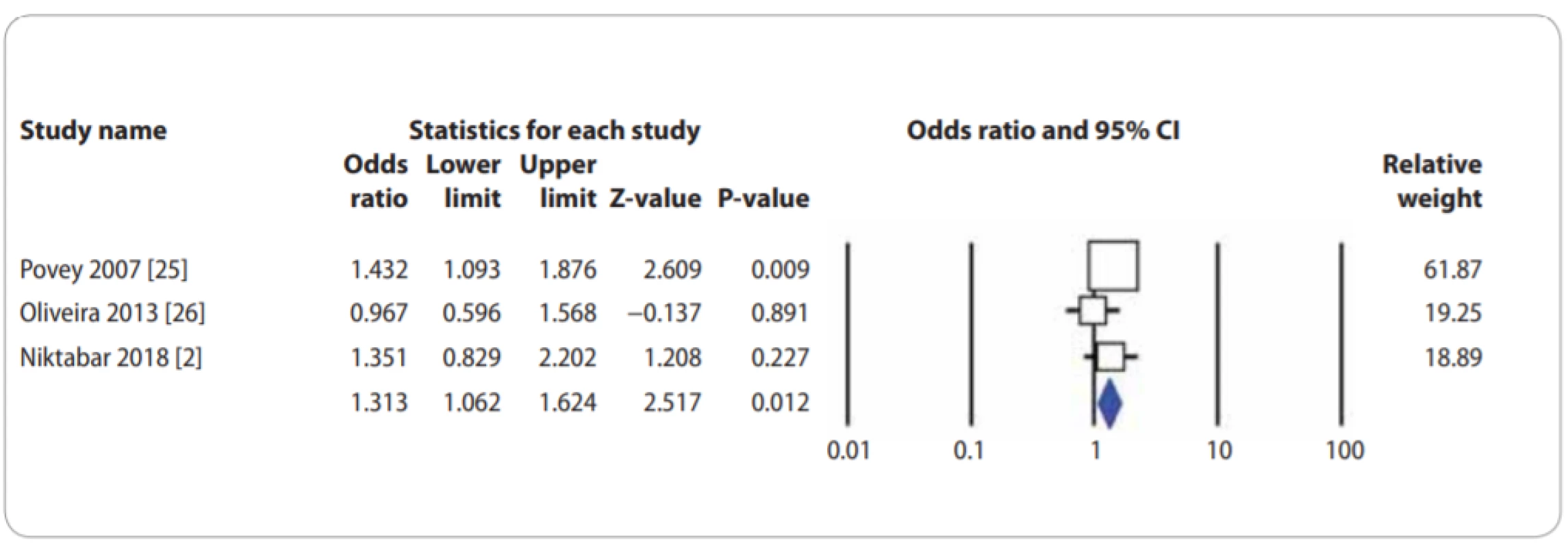
Between-study heterogeneity test
There was a moderate to high variation under most genetic models when we performed overall analysis for XPG rs17655G>C polymorphism. Thus, we conducted subgroup analyses by ethnicity, source of controls, genotyping methods and HWE status to explore the potential source of heterogeneity. After the meta-regressions in the subgroup of population based, hospital based, PCR-RFLP, and TaqMan, there was almost no variation, indicating that the source of controls and genotyping methods might be the major source of heterogeneity in this meta-analysis.
Sensitivity analysis and publication bias
We performed a sensitivity analysis through sequentially excluded individual studies. The results showed that no individual study affected the pooled OR values and statistically and similar results were obtained, suggesting the stability of this meta-analysis. Subsequently, we conducted sensitivity analysis by excluding those studies departure from HWE. Similarly, the results suggested that the current meta-analysis was relatively consistent even by excluding HWE-violating studies. In the current meta-analysis, Begg’s funnel plot and Egger’s test were applied to evaluate the publication bias of the literature on XPG rs17655G>C and XPF rs1799801T>C polymorphisms. As showed in Tab. 2 and Fig. 4, the results did not reveal any publication bias with either the Begg’s funnel plot or the Egger’s tests under all five genetic models in the overall populations. However, the results of Begg’s funnel plots and Egger’s regression test suggested evidence of publication bias in stratified analysis among hospital based group of studies under heterozygote model (PBeggs = 0.220; PEggers =0.027). Therefore, to adjust the literature bias among the HB group of studies, the trim-and-fill method developed by Duval and Tweedie was applied. However, after trimming we have yield similar results, indicating that the results were statistically reliable.
4. Funnel plot for publication bias in the meta-analysis of XPG rs17655G>C polymorphism with CMM risk: A) allele model (C vs. G); B) heterozygote model (CG vs. GG). CMM – cutaneous malignant melanoma 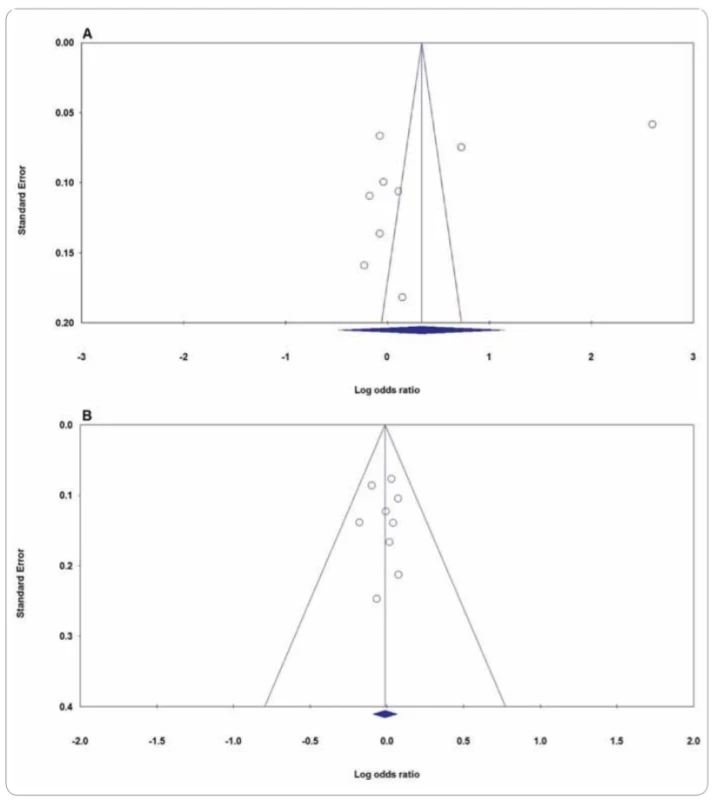
Discussion
A single study cannot be sufficient enough to confirm the association of XPG rs17655G>C and XPF rs1799801T>C polymorphisms with CMM risk convincingly, especially for small-sample size studies. This analysis is the most updated one to provide an evaluation of XPG rs17655G>C and XPF rs1799801T>C polymorphisms with CMM risk. To the best of our knowledge, this is the first meta-analysis to assess the association between XPF rs1799801T>C polymorphism with CMM risk. When 3 eligible case-control studies with 803 CMM cases and 737 controls were pooled into the meta-analysis of XPF rs1799801T>C polymorphism, there was a significant association between XPF rs1799801T>C polymorphism and CMM risk under the heterozygote model (CT vs. TT: OR = 1.313; 95% CI 1.062–1.624; P = 0.012). However, our pooled data failed to demonstrate any significant association between XPG rs17655G>C polymorphism and an increased risk of CMM. Moreover, the subgroup analysis by ethnicity also showed that there was no significant association among Caucasians. However, the stratified analysis by genotyping methods revealed a significant association between XPG rs17655G>C and CMM risk in PCR-RFLP group of studies under 2 genetic models, i.e. homozygote (CC vs. GG: OR = 0.674; 95% CI 0.485–0.938; P = 0.019) and recessive (CC vs. CG + GG: OR = 0.681; 95% CI 0.492–0.942; P = 0.020).
To date, the meta-analysis has evaluated evaluated the association between XPG rs17655G>C polymorphism and risk of CMM. However, our pooled data was inconsistent with the previous meta--analysis. In 2015, Xu et al. evaluated the association between XPG Asp1104His (rs17655G>C) polymorphism and melanoma susceptibility in a meta-analysis of eight published case-control studies containing 5,212 cases and 7,045 controls. Their results showed that XPG Asp1104His polymorphism was significantly associated with an increased risk of melanoma under a dominant model (CC + GC vs. GG: OR = 2.42; 95% CI; P = 2.26–2.60). Moreover, their subgroup analysis showed a significant association by source of controls among population-based (CC + GC vs. GG: OR 2.51; 95% CI 2.28–2.77) and hospital-based (CC + GC vs. GG: OR 2.34; 95% CI 2.12–2.58) groups of studies. Thus, they have indicated that the XPG Asp1104His polymorphism was a risk factor for melanoma susceptibility [33]. However, their meta-analysis has some limitation, as the meta-analysis study did not carry out further subgroup analysis by ethnicity. When stratified by ethnicity, the results showed the same association between XPG rs17655G>C polymorphism and CMM susceptibility in the overall population. Moreover, compared with their meta-analysis, we have performed subgroup analysis by genotyping methods and by excluding those HWE-violating studies. By excluding those studies, the results still showed that there was not association between XPG rs17655G>C polymorphism and CMM risk. Furthermore, they did not use all 5 genetic models (allele model, dominant model, recessive model, homozygous model, and heterozygous model) to assess the strength of association between XPG rs17655G>C polymorphism and CMM risk. These limitations revealed that our pooled OR values have good reliability than the previous meta-analysis results.
The between-study heterogeneity significantly affects a genetic association of meta-analysis result [34,35]. Several factors such as ethnicity, sample size, source of controls, genotyping method and Hardy-Weinberg equilibrium (WHE) might contribute to potential sources of heterogeneity [34–37]. In the current meta-analysis, there was a moderate to high heterogeneity in most genetic models for XPG rs17655G>C polymorphism. The subgroup analyses revealed that the source of controls and genotyping methods might be the major source of heterogeneity in this meta-analysis.
Several limitations exist in the current meta-analysis, which have to be acknowledged:
- We have included only published studies in the meta-analysis, and there may still be some unpublished studies with negative results that may have been missed. Therefore, publication bias may exist; even no statistical evidence suggested publication bias in the meta-analysis.
- We have mostly focused on studies published in English, Chinese and Farsi, which might have biased the results and causing a language bias.
- Only 3 studies on XPF rs1799801T>C polymorphism were included to this meta-analysis, which might reduce the statistical power for identifying the potential association of XPF rs1799801T>C polymorphism with susceptibility to CMM, and the conclusions may be biased.
- For XPG rs17655G>C and XPF rs1799801T>C polymorphisms, almost all of the included studies were conducted among Caucasians. Therefore, we could not assess the association stratified by ethnicity among other population, so these results should be interpreted with caution. Thus, data from large-scale and different ethnicities (especially from Asians and Africans) epidemiological studies are still needed to confirm the association of XPG rs17655G>C and XPF rs1799801T>C polymorphisms with CMM risk.
- Due to lack of individual original data, we could not assess CMM risk stratified by other covariates including age, gender, environment, lifestyle and other risk factors. Finally, like most malignancies, CMM is mainly caused by gene-gene and gene-environment interactions. However, no appropriate data was available to further analysis and sorting data. Therefore, further large-scale studies in different populations with different environmental background are required to validate gene-gene and gene-environment interactions on association of XPG rs17655G>C and XPF rs1799801T>C polymorphisms with susceptibility to CMM.
Conclusion
Our results suggested that XPF rs1799801T>C polymorphism might be a risk factor to development of CMM. However, XPG rs17655G>C polymorphism was not significantly associated with an increased risk of CMM. Nevertheless, further large-scale, multicenter, epidemiological studies are required to confirm this finding and the molecular mechanism for the associations need to be elucidated in future studies.
The authors declare they have no potential conflicts of interest concerning drugs, products, or services used in the study.
Autoři deklarují, že v souvislosti s předmětem studie nemají žádné komerční zájmy.
The Editorial Board declares that the manuscript met the ICMJE recommendation for biomedical papers.
Redakční rada potvrzuje, že rukopis práce splnil ICMJE kritéria pro publikace zasílané do bi omedicínských časopisů.
Dr. Naeimeh Heiranizadeh
Department of General Surgery Shahid Sadoughi Hospital Ave-Sina Blvd, Shahid Ghandi Blvd Yazd, Iran
e-mail: naiemehheiranizadeh@gmail.com
Submitted/Obdrženo: 21. 10. 2019
Accepted/Přijato: 31. 12. 2019
Sources
1. Cummins DL, Cummins JM, Pantle H et al. Cutaneous malignant melanoma. Mayo Clinic proceedings 2006; 81 (4): 500–507. doi: 10.4065/81.4.500.
2. Niktabar SM, Latifi SM, Moghimi M et al. Association of vitamin D receptor gene polymorphisms with risk of cutaneous melanoma. A meta-analysis based on 40 case-control studies. Dermatology Review/Przegląd Dermatologiczny 2019; 106 (3): 268–279. doi: 10.5114/dr.2019.86909.
3. Mar VJ, Wong SQ, Li J et al. BRAF/NRAS wild-type melanomas have a high mutation load correlating with histologic and molecular signatures of UV damage. Clin Cancer Res 2013; 19 (17): 4589–4598. doi: 10.1158/1078-0432.CCR-13-0398.
4. von Thaler AK, Kamenisch Y, Berneburg M. The role of ultraviolet radiation in melanomagenesis. Exper Dermatol 2010; 19 (2): 81–88. doi: 10.1111/j.1600 0625.2009.01025.x.
5. Matthews NH, Li W-Q, Qureshi AA et al. Cutaneous Melanoma: Etiology and therapy. Brisbane: Codon Publications 2017 : 3–22.
6. Yang K, Fung TT, Nan H. An epidemiological review of diet and cutaneous malignant melanoma. Cancer Epidemiol Biomarkers Prev 2018; 27 (10): 1115–1122. doi: 10.1158/1055-9965.EPI-18-0243.
7. Ali Z, Yousaf N, Larkin J. Melanoma epidemiology, biology and prognosis. EJC Suppl 2013; 11 (2): 81–91. doi: 10.1016/j.ejcsup.2013.07.012.
8. Pollack LA, Li J, Berkowitz Z et al. Melanoma survival in the United States, 1992 to 2005. J Am Acad Dermatol 2011; 65 (Suppl 1): 78–86. doi: 10.1016/j.jaad.2011.05.030.
9. Guy GP, Machlin SR, Ekwueme DU et al. Prevalence and costs of skin cancer treatment in the U.S., 2002–2006 and 2007–2011. Am J Prev Med 2015; 48 (2): 183–187. doi: 10.1016/j.amepre.2014.08.036.
10. Kunz M, Vera J. Modelling of protein kinase signaling pathways in melanoma and other cancers. Cancers (Basel) 2019; 11 (4): E465. doi: 10.3390/cancers11040465.
11. Zhang XY, Zhang PY. Genetics and epigenetics of melanoma. Oncol Lett 2016; 12 (5): 3041–3044. doi: 10.3892/ol.2016.5093.
12. Muñoz-Couselo E, Adelantado EZ, Ortiz C et al. NRAS-mutant melanoma: Current challenges and future prospect. OncoTargets Ther 2017; 10 : 3941–3947. doi: 10.2147/OTT.S117121.
13. Griewank KG, Murali R, Puig-Butille JA et al. TERT promoter mutation status as an independent prognostic factor in cutaneous melanoma. J Natl Cancer Inst 2014; 106 (9): 246. doi: 10.1093/jnci/dju246.
14. Leão R, Apolónio JD, Lee D et al. Mechanisms of human telomerase reverse transcriptase (hTERT) regulation: Clinical impacts in cancer. J Biomed Sci 2018; 25 (1): 22. doi: 10.1186/s12929-018-0422-8.
15. Eckerle Mize D, Bishop M, Resse E et al. Familial atypical multiple mole melanoma syndrome in cancer syndromes. In: Riegert-Johnson DL, Boardman LA, Hefferon T (eds) Cancer syndromes online. Available from: https: //www.ncbi.nlm.nih.gov/pubmed/21249754.
16. Murray HC, Maltby VE, Smith DW et al. Nucleotide excision repair deficiency in melanoma in response to UVA. Exp Hemat Oncol 2015; 5 : 6. doi: 10.1186/s40164-016-0035-4.
17. Mehdinejad M, Sobhan MR, Mazaheri M et al. Genetic association between ERCC2, NBN, RAD51 gene variants and osteosarcoma risk: a systematic review and meta--analysis. Asian Pac J Cancer Prev 2017; 18 (5): 1315–1321. doi: 10.22034/APJCP.2017.18.5.1315.
18. Zhao J, Chen S, Zhou H et al. XPG rs17655 G>C polymorphism associated with cancer risk: Evidence from 60 studies. Aging 2018; 10 (5): 1073–1088. doi: 10.18632/aging.101448.
19. Zhu ML, Wang M, Cao ZG et al. Association between the ERCC5 Asp1104His polymorphism and cancer risk: A meta-analysis. PLoS ONE 2012; 7 (7): e36293. doi: 10.1371/journal.pone.0036293.
20. Zhang Z, Yin J, Xu Q et al. Association between the XPG gene rs2094258 polymorphism and risk of gastric cancer. J Clin Lab Anal 2018, 32 (8): e22564. doi: 10.22034/APJCP.2017.18.10.2611.
21. Namazi A, Forat-Yazdi M, Jafari MA et al. Association between polymorphisms of ERCC5 gene and susceptibility to gastric cancer: a systematic review and meta-analysis. Asian Pacific J Cancer Prev 2017; 18 (10): 2611–2617. doi: 10.22034/APJCP.2017.18.10.2611.
22. Manandhar M, Boulware KS, Wood RD. The ERCC1 and ERCC4 (XPF) genes and gene products. Gene 2015; 569 (2): 153–161. doi: 10.1016/j.gene.2015.06.026.
23. Liu J, Zheng B, Li Y et al. Genetic polymorphisms of DNA repair pathways in sporadic colorectal carcinogenesis. J Cancer 2019; 10 (6): 1417–1433. doi: 10.7150/jca.28406.
24. Paszkowska-Szczur K, Scott RJ, Serrano-Fernandez P et al. Xeroderma pigmentosum genes and melanoma risk. Int J Cancer 2013; 133 (5): 1094–1100. doi: 10.1002/ijc.28123.
25. Povey JE, Darakhshan F, Robertson K et al. DNA repair gene polymorphisms and genetic predisposition to cutaneous melanoma. Carcinogenesis 2007; 28 (5): 1087–1093. doi: 10.1093/carcin/bgl257.
26. Oliveira C, Rinck-Junior JA, Lourenço GJ et al. Assessment of the XPC (A2920C), XPF (T30028C), TP53 (Arg72Pro) and GSTP1 (Ile105Val) polymorphisms in the risk of cutaneous melanoma. J Cancer Res Clin Oncol 2013; 139 (7): 1199–1206. doi: 10.1007/s00432-013-1430-4.
27. Blankenburg S, König IR, Moessner R et al. Assessment of 3 xeroderma pigmentosum group C gene polymorphisms and risk of cutaneous melanoma: a case–control study. Carcinogenesis 2005; 26 (6): 1085–1090. doi: 10.1093/carcin/bgi055.
28. Li C, Hu Z, Liu Z et al. Polymorphisms in the DNA Repair Genes XPC, XPD, and XPG and risk of cutaneous melanoma: a case-control analysis. Cancer Epidemiol Biomarkers Prev 2006; 15 (12): 2526–2532. doi: 10.1158/1055-9965.EPI-06-0672.
29. Millikan RC, Hummer A, Begg C et al. Polymorphisms in nucleotide excision repair genes and risk of multiple primary melanoma: the genes environment and melanoma study. Carcinogenesis 2006; 27 (3): 610–618. doi: 10.1093/carcin/bgi252.
30. Figl A, Scherer D, Nagore E et al. Single-nucleotide polymorphisms in DNA-repair genes and cutaneous melanoma. Mutat Res 2010; 702 (1): 8–16. doi: 10.1016/j.mrgentox.2010.06.011.
31. Ibarrola-Villava M, Peña-Chilet M, Fernandez LP et al. Genetic polymorphisms in DNA repair and oxidative stress pathways associated with malignant melanoma susceptibility. Eur J Cancer 2011; 47 (17): 2618–2625. doi: 10.1016/j.ejca.2011.05.011.
32. GPaPonçalves FT, Francisco G, de Souza SP et al. European ancestry and polymorphisms in DNA repair genes modify the risk of melanoma: A case–control study in a high UV index region in Brazil. J Dermatol Sci 2011; 64 (1): 59–66. doi: 10.1016/j.jdermsci.2011.06.003.
33. Xu Y, Jiao G, Wei L et al. Current evidences on the XPG Asp1104His polymorphism and melanoma susceptibility: a meta-analysis based on case–control studies. Mol Genet Genomics 2014; 290 (1): 273–279. doi: 10.1007/s00438-014-0917-2.
34. Moghimi M, Ahrar H, Karimi-Zarchi M et al. Association of IL-10 rs1800871 and rs1800872 polymorphisms with breast cancer risk: A systematic review and meta-analysis. Asian Pac J Cancer Prev 2018; 19 (12): 3353–3359. doi: 10.31557/APJCP.2018.19.12.3353.
35. Moghimi M, Kargar S, Jafari MA et al. Angiotensin converting enzyme insertion/deletion polymorphism is associated with breast cancer risk: a meta-analysis. Asian Pac J Cancer Prev 2018; 19 (11): 3225–3231. doi: 10.31557/APJCP.2018.19.11.3225.
36. Moghimi M, Sobhan MR, Jarahzadeh MH et al. Association of GSTM1, GSTT1, GSTM3, and GSTP1 genes polymorphisms with susceptibility to osteosarcoma: a case-control study and meta-analysis. Asian Pac J Cancer Prev 2019; 20 (3): 675–682. doi: 10.31557/APJCP.2019.20.3.675.
37. Aflatoonian M, Moghimi M, Akbarian-Bafghi MJ et al.Association of TNF-a-308G>A polymorphism with susceptibility to celiac disease: a systematic review and meta-analysis. Arquivos de gastroenterologia 2019; 56 (1): 88–94. doi: 10.1590/S0004-2803.201900000-20.
Labels
Paediatric clinical oncology Surgery Clinical oncology
Article was published inClinical Oncology

2020 Issue 3-
All articles in this issue
- Importance of Aberrantly Activated Hedgehog/Gli Pathway in Tumour Progression
- Assessment of Quality of Life in Patients with Head and Neck Cancer
- Current Possibilities of Early Detection of Cardiotoxicity of Cytostatic Treatment
- Prognostic Survival Factors of Hepatocellular Carcinoma Treated with Transarterial Chemoembolization
- Complete Response to Chemotherapy in Metastatic Pancreatic Carcinoma Associated with Double Heterozygous Germline Mutation in BRCA2 and CHEK2 Genes – a Case Report
- Long-Term Effect of Erlotinib Therapy in the Third Line of Anticancer Treatment in a Patient with Non-Small-Cell Lung Cancer – a Case Report
- Therapy with Trifluridine/Tipiracil and Regorafenib in Patientswith Pre-treated Metastatic Colorectal Cancer – Experience from the Czech Republic
- Editorial
- Aktuality z odborného tisku
- Predstavujeme nové knihy
- Oncology in pictures
- Levels of NT-proBNP and Troponin T in Cancer Patients – Mini-Review
- Association of XPG rs17655G>C and XPF rs1799801T>C Polymorphisms with Susceptibility to Cutaneous Malignant Melanoma: Evidence from a Case-Control Study, Systematic Review and Meta-Analysis
- The Relationship of FOXR2 Gene Expression Profile with Epithelial-Mesenchymal Transition Related Markers in Epithelial Ovarian Cancer
- Clinical Oncology
- Journal archive
- Current issue
- Online only
- About the journal
Most read in this issue- Levels of NT-proBNP and Troponin T in Cancer Patients – Mini-Review
- Complete Response to Chemotherapy in Metastatic Pancreatic Carcinoma Associated with Double Heterozygous Germline Mutation in BRCA2 and CHEK2 Genes – a Case Report
- Assessment of Quality of Life in Patients with Head and Neck Cancer
- Importance of Aberrantly Activated Hedgehog/Gli Pathway in Tumour Progression
Login#ADS_BOTTOM_SCRIPTS#Forgotten passwordEnter the email address that you registered with. We will send you instructions on how to set a new password.
- Career

