-
Medical journals
- Career
Association of IL-8 -251T>A and IL-18 -607C>A polymorphisms with susceptibility to breast cancer – a meta-analysis
Authors: M. Farbod 1; S. A. Dastgheib 2; F. Asadian 3; M. Karimi-Zarchi 4,5; S. Sayad 6; M. Barahman 7; S. Kargar 8; M.- Mazaheri 9 11; H. Neamatzadeh 9,10
Authors‘ workplace: Cancer Institute, Imam Khomeini Hospital, Tehran University of Medical Sciences, Tehran, Iran 1; Department of Medical Genetics, School of Medicine, Shiraz University of Medical Sciences, Shiraz, Iran 2; Department of Medical Laboratory Sciences, School of Paramedical Science, Shiraz University of Medical Sciences, Shiraz, Iran 3; Department of Obstetrics and Gynecology, Iran University of Medical Sciences, Tehran, Iran 4; Endometriosis Research Center, Firoozgar Hospital, Iran University of Medical Sciences, Tehran, Iran 5; Department of Surgery, Tehran University of Medical Sciences, Tehran, Iran 6; Firoozgar Clinical Research Development Center (FCRDC), Firoozgar Hospital, Iran University of Medical Sciences, Tehran, Iran 7; Department of Surgery, Shahid Sadoughi University of Medical Sciences, Yazd, Iran 8; Department of Medical Genetics, Shahid Sadoughi University of Medical Sciences, Yazd, Iran 9; Mother and Newborn Health Research Center, Shahid Sadoughi University of Medical Sciences, Yazd, Iran 10; Stem Cell Biology Research Center, Yazd Reproductive Sciences Institute, Shahid Sadoughi University of Medical Sciences, Yazd, Iran 11
Published in: Klin Onkol 2022; 35(3): 181-189
Category: Review
doi: https://doi.org/10.48095/ccko2022181Overview
Background: Previous studies have evaluated the association of IL-8 -251T>A and IL-18 -607C>A polymorphisms with a risk of breast cancer in different populations, but the results remain inconsistent and inconclusive. Thus, we performed this meta-analysis to explore the associations. Methods: A comprehensive literature search in PubMed, EMBASE, Web of Science, Scopus, SciELO, SID, and CNKI for all eligible studies published up to October 1, 2020. The pooled odds ratios (ORs) with 95% confidence intervals (CIs) were used to evaluate the intensity of associations. Results: A total of 12 case-control studies including seven studies with 2,370 cases and 2,314 controls on IL-8 -251T>A, and five studies with 900 cases and 882 controls on IL-18 -607C>A polymorphism were selected. Pooled data showed that IL-8 -251T>A (AT vs. TT: OR= 1.187; 95% CI 1.038–1.356; P = 0.012) and IL-18 -607C>A polymorphisms (A vs. T: OR = 1.205; 95% CI 1.055–1.377; P = 0.006; AA vs. TT: OR = 1.379; 95% CI 1.056–1.802; P = 018; and AA vs. AT+TT: OR = 1.329; 95% CI 1.053–1.678; P = 0.017) were associated with increased risk of breast cancer in overall. Moreover, when the studies were stratified by ethnicity, the IL-8 -251T>A was significantly associated with breast cancer risk in Africans. Publication bias tests provide no evidence of presence of publication bias in a meta-analysis. Conclusion: This meta-analysis results revealed that the IL-8 -251T>A and IL-18 -607C>A polymorphisms are associated with susceptibility to breast cancer. However, further multicenter studies with larger sample sizes in different ethnicities are required to make a better assessment of these associations.
Keywords:
meta-analysis – breast cancer – association – interleukin-8 – interleukin-18
Introduction
Breast cancer is the most common form of cancer and the leading cause of death in women of all ethnic groups [1,2], especially in case of low - and middle-income developing countries due to screening barriers [3]. In 2018, an estimated 266,000 new cases of breast cancer will be diagnosed with 41,000 cases of deaths, were estimated to occur in women in the United States [4]. In the literature, obesity, hormone replacement therapy, radiation, age, and family history has been described as a pivotal risk factor for breast cancer development [5–8]. Approximately 4% of cases diagnosed with breast cancer in the United States are younger than 40 years and a report by National Cancer Institute showed that the breast cancer incidence rate in women aged 21–54 years was 33.6% of all breast cancer cases [9,10].
Despite its high prevalence and rapid progress in molecular biology of cancer achieved in the last few decades, the exact mechanism of breast cancer is still poorly understood [11,12]. It is evident that genetic and environmental factors play a role in the development of breast cancer [13,14]. Moreover, there is also a relationship between social environment and breast cancer development and woman cope with a breast cancer diagnosis. Epidemiological studies have demonstrated that single nucleotide polymorphisms (SNPs) in different interleukins might be involved in development of breast cancer [15,16].
Recently, the roles of interleukin 8 (IL-8) and IL-18 have been extensively studied in the development of breast cancer. Human IL-8 enhances the immune system against cancer cells and also modifies the tumor microenvironment facilities [17]. Increasing evidence demonstrates that human IL-8 is considerably expressed in ER-, PR - and HER-2/neu+ breast cancer cells, but highly correlated with invasiveness and metastatic of both ER - and ER+ cells by twofold, indicating the invasion-promoting role of IL-8 [17–20]. IL-8 is associated with growth receptors expressed on the surface of breast cancer cells. However, the mechanisms by which IL-8 contributes to breast cancer progression have remained poorly understood [21]. Moreover, IL‐18 may also have role in development of breast cancer. Previous studies revealed that serum levels of IL-18 were higher in advanced than in early stages and higher in metastatic than in non-metastatic breast cancer cases [22]. In the recent decade, several studies have investigated the influence of IL-8 -251T>A and IL-18 -607C>A polymorphisms in breast cancer, but their results remained controversial. Therefore, we performed meta-analyses to evaluate and summarize the contribution of the IL-8 -251T>A and IL-18 -607C>A polymorphisms to breast cancer susceptibility.
Materials and methods
Identification and eligibility of relevant studies
This study was performed according to the Preferred reporting items for systematic reviews and meta-analyses (PRISMA) statement on reporting meta - -analysis. We conducted a comprehensive literature on PubMed, EMBASE, Web of Science, Science Citation Index (SCI), Springer Link, CNKI, Wanfang, OVID, SID, EBSCO and Science Direct databases to identify potential studies evaluated the association of IL-8 -251T>A and IL-18 -607C>A polymorphisms with a risk of breast cancer. The search keywords used were as follows: (‘’Breast Cancer’’ OR ‘‘Tumor’’ OR ‘’Cancer’’ OR ‘’Neoplasm’’) AND (‘’Interlukine-8 ‘’ OR ‘’IL-8’’ OR ‘’-251A>T’’ OR ‘’rs4073’’) AND (‘’Interlukine-18 ‘’ OR ‘’IL-18’’ OR ‘’-607C>A’’ OR ‘’rs1946518’’) AND (‘’Gene’’ OR ‘’Polymorphism’’ OR ‘’SNPs’’ OR ‘’Mutation’’ OR ‘’Variation’’ OR ‘’Allele’’). Then, the references of all retrieved publications and reviews to identify other potential relevant studies were checked manually. The final literature search was updated on October 1, 2020, with no restrictions on publication year or methodological filter. Data for the largest sample set or most recent published articles were included when data from the same set of cases were used more than once within a publication.
Selection criteria
The eligible studies included in the current meta-analysis, must meet all the following criteria:
1) genetic association studies on association of IL-8 -251T>A and IL-18 -607C>A polymorphisms with breast cancer;
2) in human beings;
3) studies with case-control or cohort design;
4) allele and genotype distribution for both IL-8 -251 T>A and IL-18 -607 C>A polymorphisms provided to estimate the odds ratios (ORs) and 95% confidence intervals (CIs);
5) articles published in English, Chinese, Portuguese and Farsi (with an English abstract).
The studies were excluded if one of the following criteria was fulfilled:
1) studies not about IL-8 -251T>A and IL-18 -607C>A polymorphisms and breast cancer;
2) studies on other polymorphisms at IL-8 and IL-18 genes;
3) animal models, cell lines and in vitro studies;
4) no control population;
5) linkage and family-based studies;
6) data unavailable or insufficient for calculating allele and genotypes frequencies;
7) abstracts, posters, case reports, case series, editorials, letters, editorial articles, conference presentations, comments, reviews, meta-analyses;
8) overlapping data or duplicate of previous publication.
Data extraction
The necessary data were carefully extracted by two observers from eligible studies according to the mentioned criteria. Any disagreement between two authors was discussed with the third author until a consensus was achieved. If the data were not presented, the corresponding authors were contacted to request extra information. The following characteristics were collected from each study: last name of first author, year of publication, country of origin, ethnicity of study participants, source of controls (hospital based or population based), genotyping methods, genotypic/allelic distributions for both IL-8 -251T>A and IL-18 -607C>A polymorphisms in cases and controls, the number of cases and control genotypes, minor allele frequency (MAFs) and Hardy-Weinberg equilibrium (HWE) in controls.
Statistical Analysis
The strength of associations of IL-8 -251T>A and IL-18 -607C>A polymorphisms with susceptibility to breast cancer was estimated by ORs with 95% confidence intervals (95% CIs). The significance of the pooled effect size was examined by the Z-test. The associations were evaluated under all five genetic models, i.e. allele (B vs. A), homozygote (BB vs. AA), heterozygote (BA vs. AA), dominant (BB+BA vs. AA), and recessive (BB vs. BA+AA). To take into account the possibility of between-study heterogeneity, the Cochran’s Q test was used, in which P < 0.10 indicated a significant heterogeneity. Moreover, I2 statistic (range of 0–100%) was also employed to qualify the heterogeneity, where a lower value represents non-significant heterogeneity and a higher value represents a high degree of between study heterogeneity. A fixed-effect method (the Mantel-Haenszel method) was applied to calculate the pooled ORs and the corresponding 95% CIs for all genetic models which did not show significant heterogeneity; otherwise a random-effect method (the DerSimonian and Laird method) was adopted. Subgroup analyses were conducted by stratification of ethnicity to identifying potential source of heterogeneity. HWE was tested in control groups of each study by c2 test to assess the latent bias resulting from the deviation of the genotype distribution; P > 0.05 were considered to have reliable and representative controls. The sensitivity analysis was carried out by omitting one study at a time to determine the underlying effects of each single study on the pooled data. Furthermore, we conducted the sensitivity analyses again to delete those studies deviating from HWE and calculate the pooled ORs for the remainder of the studies. Publication bias was performed by the construction of Begg’s funnel plot and Egger’s regression analysis. All of the statistical calculations were performed using Comprehensive Meta-Analysis (CMA) software, version 2.0 (Biostat, USA). For all analyses, statistical significance was assumed at P < 0.05, unless otherwise stated.
Results
Selected studies characteristics
The selection process of eligible studies is presented in Scheme 1. Initially, 108 studies were obtained through publication search in electronic databases, and one study was identified from other sources. Irrelevant articles were excluded by evaluating the titles and abstracts. Therefore, 76 publications were deleted for obvious irrelevance. Finally, 12 case-control studies including seven studies [23–29] with 2,370 cases and 2,314 controls on IL-8 -251T>A polymorphism, and 5 studies [30–34] with 900 cases and 882 controls on IL-18 -607C>A polymorphisms were selected to the meta-analysis. Tab. 1 describes principal characteristics of selected studies. All the studies were published between December 2004 and March 2018. The studies have been carried out in UK, Tunisia, Iran, China, and Brazil. Among these studies, eight studies were conducted among Asians, two studies among Caucasians and two studies among Africans. Seven different genotyping methods were used: ARMS-PCR, AS-PCR, TaqMan, ASO-PCR, PCR-RFLP, MALDI-TOF and direct sequencing. The genotype, allele and MAF in each study for both IL-8 -251T>A and IL-18 -607C>A polymorphisms are shown in Tab. 1. More - over, the distribution of genotypes in the controls was in agreement with HWE for all selected studies, except for one study on IL-8 -251T>A polymorphism (Tab. 1).
Scheme 1. Flowchart of literature search and selection process. 
1. Main characteristics of studies included in this meta-analysis. 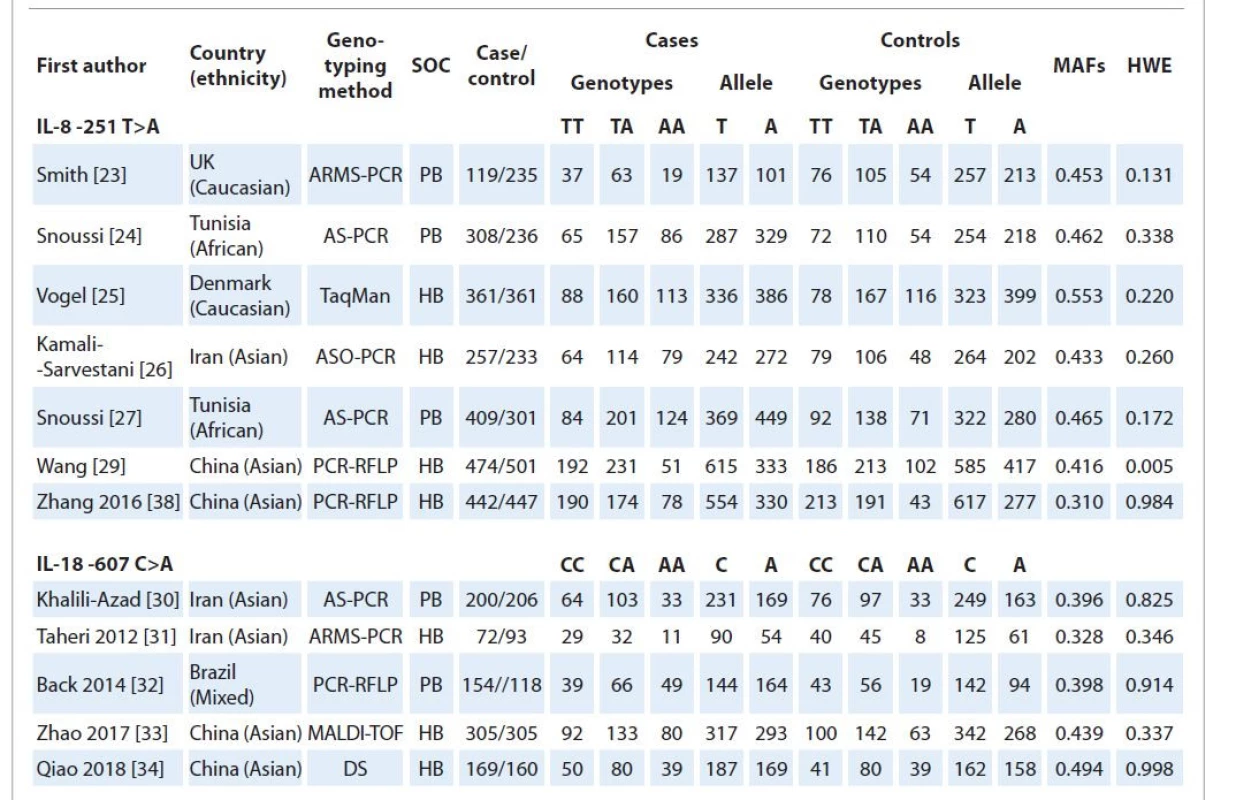
ARMS – amplification refractory mutation system, AS-PCR – allele-specific polymerase chain reaction, ASO-PCR – allele-specific oligonucleotide polymerase chain reaction, RFLP – restriction fragment length polymorphism, DS – direct sequencing, SOC – source of controls, PB – population-based, HB – hospital-based, MAF – minor allele frequency, HWE – Hardy-Weinberg equilibrium Quantitative synthesis
IL-8 -251T>A polymorphism
The association between IL-8 -251T>A polymorphism and breast cancer risk is shown in Tab. 2. Pooled data showed that there was a significant association between IL-8 -251T>A polymorphism and an increased risk of breast cancer under heterozygote model (AT vs. TT: OR = 1.187; 95% CI 1.038–1.356, P = 0.012, Fig. 2A). When stratified analysis by ethnicity, a significant association was found between IL-8 -251T>A polymorphism and breast cancer risk in Africans under all five genetic models, i.e. allele (A vs. T: OR = 1.398; 95% CI 1.174–1.663; P ≤ 0.001), homozygote (AA vs. TT: OR = 2.083; 95% CI 1.529–2.839; P ≤ 0.001), heterozygote (AT vs. TT: OR = 1.411; 95% CI 1.066–1.867; P = 0.016), dominant (AA+AT vs. TT: OR = 1.676, 95% CI 1.295–2.168; P ≤ 0.001), and recessive (AA vs. AT+TT: OR = 1.364; 95% CI 1.055–1.764; P = 0.018), but not in Asians and Caucasians (Tab. 2).
Fig. Forest plot of IL-8 -251T>A and IL-18 -607C>A polymorphisms and risk of breast cancer. A) IL-8 -251T>A (heterozygote model: AT vs. TT); B) IL-18 -607C>A (allele model: A vs. C). 
2. Summary of meta-analysis for the association of IL-8 -251T>A and IL-18 -607C>A polymorphisms with breast cancer risk. 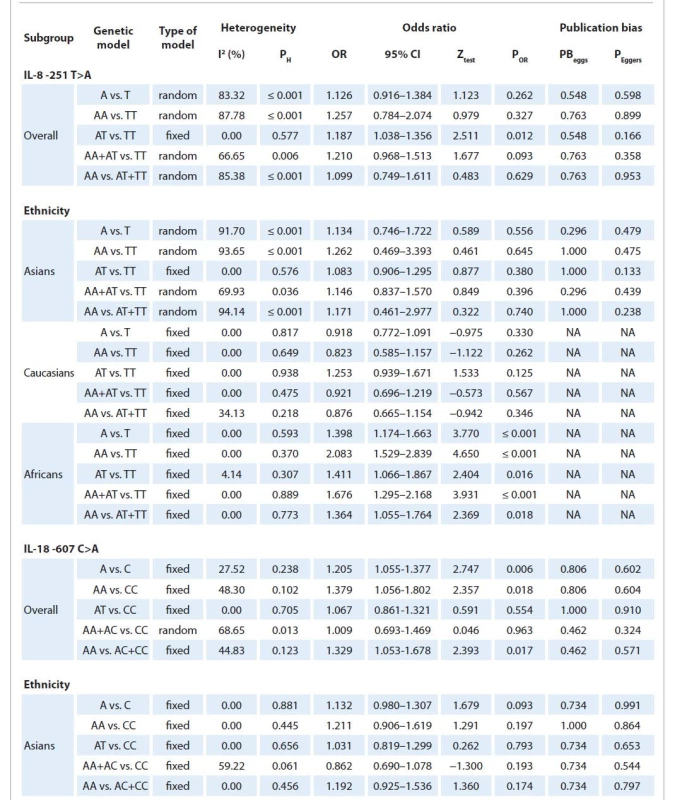
NA – not applicable IL-18 -607C>A polymorphism
Results of pooled analysis for IL-18 -607C>A polymorphism and breast cancer risk are summarized in detail in Tab. 2. Pooled data showed that the IL-18 -607C>A polymorphism was significantly associated with an increased risk of breast cancer under three genetic models, i.e. allele (A vs. T: OR = 1.205; 95% CI 1.055–1.377; P = 0.006, Fig. 2B), homozygote (AA vs. TT: OR = 1.379; 95% CI 1.056–1.802, P = 018), and recessive (AA vs. AT+TT: OR = 1.329; 95% CI 1.053–1.678; P = 0.017). However, stratified analysis by ethnicity failed to show a significant association between the IL-18 -607C>A polymorphism and breast cancer in Asians (Tab. 2).
Heterogeneity test
In this meta-analysis there was statistical significance between-study heterogeneity for IL-8 -251T>A polymorphism under four genetic models, i.e. allele (A vs. T: I2 = 83.32; PH ≤ 0.001), homozygote (AA vs. TT: I2 = 87.78; PH ≤ 0.001), dominant (AA+AT vs. TT: I2 = 66.65; PH = ?) and recessive (AA vs. AT+TT: I2 = 85.38; PH ≤ 0.001), models and IL-18 -607 C>A polymorphism under dominant model (AA+AC vs. CC: I2 = 68.65; PH = 0.013). Therefore, we conducted subgroup analyses by ethnicity to explain the potential source of heterogeneity. Results showed that the heterogeneity disappeared in the subgroup analysis among Asians, Caucasians and Africans, indicating that ethnicity might be the major source of heterogeneity for IL-8 -251T>A polymorphism (Tab. 2).
Sensitivity analysis
While omitting each individual study any time, sensitivity analysis was applied to detect the influence of each study on the pooled OR by repeating the meta-analysis. Sensitivity analyses results showed that the significance of the pooled data for both for IL-8 -251 T>A and IL-18 -607 C>A polymorphisms was not affected by any single study in the overall population. In addition, when we excluded the studies out of HWE, the statistical significance of the results did not change.
Publication bias
Begg’s funnel plot and Egger’s test were used to evaluate the publication bias of the literature for IL-8 -251T>A and IL-18 -607C>A polymorphisms. No publication bias was detected with either the Begg’s funnel plot or the Egger’s tests under all five genetic models in overall and subgroup analysis by ethnicity (Tab. 2). Funnel plots for IL-8 -251T>A polymorphism in allele model and IL-18 -607C>A polymorphism in recessive model were showed in Graphs 3A, B.
Graph Begg’s funnel plot of the Egger test for publication bias of the IL-8 -251T>A and IL-18 -607C>A polymorphisms and risk of breast cancer. A) IL-8 -251T>A (allele model: A vs. T); B) IL-18 -607C>A (recessive model: AA vs. AC+CC). 
Discussion
Human IL-8, also known as neutrophil chemotactic factor, has significant potential as a prognostic and predictive biomarker in various inflammatory conditions and malignancies [35]. It plays a key role in the recruitment of neutrophils and other immune cells to the site of infection [36]. IL-8 is mapped on chromosome 4q13-q21, contains four exons, a proximal promoter region and has a length of 5191 bp [37,38]. Moreover, human IL-18, a member of the IL-1 family, was initially identified as a protein that induces interferon g (IFNg) production [39,40]. It is a pro-inflammatory chemokine, plays a key role in the initiation, modulation, and maintenance of the gastrointestinal inflammatory response. Human IL-18 gene is mapped on 11q22.2–22.3, contains six exons and several genetic polymorphisms, especially in the promoter region [41]. A functional polymorphism at position -251 of the IL-8 promoter region has been identified in 2000, which has effect on IL-8 gene expression or secretion [42].
In this study, we evaluated the association of IL-8 -251T>A and IL-18 -607C>A polymorphisms with breast cancer risk based on all available studies. By pooling all eligible studies, we found the IL-8 -251T>A and IL-18 -607C>A polymorphisms are significantly associated with an increased risk of breast cancer in the global population. Previous epidemiological studies have reported that these polymorphisms were associated with an increased risk of breast cancer in different populations such as Chinese, Iranian and Danish. However, subgroup analyses by ethnicity showed lack of association between IL-8 -251T>A and IL-18 -607C>A polymorphisms and a risk of breast cancer in Asians and Caucasians. This difference is a common finding in a meta-analysis and could be explained by two reasons. First, because of the complex nature of breast cancer, it is unlikely that a single nucleotide polymorphism in IL-8 and IL-18 genes would be associated with an increased risk of breast cancer, without an interaction from other polymorphic susceptibility genes. Second, other factors, such as age, hormone therapy, family history, life style and different environmental exposure can also influence the development of breast cancer. In 2014, Wang et al evaluated the association between IL-8 -251T>A polymorphism and breast cancer risk in a meta-analysis of five case-control studies [15]. Their results showed that IL-8 -251T>A polymorphism was significantly associated with an increased risk of breast cancer under four genetic models, i.e. allele (A vs. T, OR = 1.21; 95% CI 1.01–1.45), heterozygote (AT vs. TT, OR = 1.28; 95% CI 1.07–1.53), dominant (AA+TT vs. TT: OR = 1.34, 95% CI 1.03–1.74) and recessive (AA vs. AT+TT: OR = 1.25; 95% CI 1.05–1.49). In 2015, Li et al have examined the association of IL-18 -607C>A polymorphism with risk of breast cancer in a meta-analysis of three case-control studies [43]. They have found that IL-18 -607C>A polymorphism was significantly associated with an increased risk of breast cancer under three genetic models, i.e. allele (A vs. C: OR =1.33; 95% CI 1.00–1.75; PH = 0.155), homozygote (AA vs. CC: OR = 1.80; 95% CI 1.02–3.21, PH = 0.162) and dominant (AA+CA vs. CC: OR = 1.33; 95% CI 1.00–1.78; PH = 0.546). Considering that the samples of breast cancer cases and controls in the previous meta-analyses were quite small, their results might not have enough statistical power. Moreover, their estimates were not adjusted OR values such as ethnicity, which might be caused to inaccurate results. Therefore, our meta-analysis gave strong evidence of an association of IL-8 -251T>A and IL-18 -607C>A polymorphisms with a risk of breast cancer in global population and by ethnicity.
Between-study heterogeneity significantly affects a meta-analysis result. Several factors such as diversity in study populations, study design, sample size, source of controls, genotyping method, and WHE might contribute to potential sources of heterogeneity [44–46]. There was a significant heterogeneity in this meta-analysis mostly for IL-8 -251T>A polymorphism. Thus, due to the significant heterogeneity, we applied the random-effects model to calculate the pooled ORs, which could provide stable results. Subgroup analyses by ethnicity showed that the heterogeneity significantly decreased in Asians, Caucasians and Africans, indicating that ethnicity might be the major source of heterogeneity for IL-8 -251T>A polymorphism.
We did not observe any publication bias in both IL-8 -251T>A and IL-18 -607C>A polymorphisms, demonstrating that the results of this meta-analysis are stable. However, it is important to note the limitations of our meta-analysis. First, although an increased risk of breast cancer was observed to be associated with IL-8 -251T>A and IL-18 -607C>A polymorphisms; the sample size for both polymorphisms was not large enough to provide enough statistical power. Second, in the current meta-analysis studies from Caucasians, Asians, Africans and mixed populations were involved, although the number of studies was relatively small and results might not have enough statistical power to obtain the association of IL-8 -251T>A and IL-18 -607C>A polymorphisms with breast cancer. Third, only published studies in English were included; it is possible that some relevant published or unpublished studies with null results were missed, which might have biased the results and causing a language bias. Fourth, several important confounding factors, such as age, smoking, drinking, family history of breast cancer, environmental exposures and lifestyle, were not considered for stratification analysis because relevant data was insufficient in the primary reports. Finally, the lack of original data in the eligible studies limited the evaluation of the effects of gene-gene and gene-environment interactions on IL-8 -251T>A and IL-18 -607C>A polymorphisms and breast cancer risk.
Conclusion
Our results showed that IL-8 -251T>A and IL-18 -607C>A polymorphisms are significantly associated with increased susceptibility to breast cancer. Our findings also indicate that the IL-8 -251T>A polymorphism also plays an important role in the development of breast cancer in Africans. Future studies with large sample sizes and more ethnic groups are needed to confirm our findings. Moreover, IL-8 -251T>A, IL-18 -607C>A polymorphisms, other interleukin polymorphisms and gene-gene interactions should also be considered in future studies.
Ethical approval and consent to participate
The ethical approval was not required for this study, as it is a systematic review and meta-analysis.
Availability of data and material
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Funding
No funding was obtained for this study.
Authors Contribution
Conceived and designed the study and experiments: MF, FA and SAD
Performed the experiments: HN and MKZ
Analyzed the data: SHS, SK and HN
Contributed reagents/materials/analysis tools: MZS, SAD and FA
Wrote the paper: MF, SAD and HN
All authors have read and approved the manuscript.
Acknowledgements
I would like to express my sincere gratitude to Professor Seyed Mehdi Kalantar for his motivation, knowledge and support during the course of this research.
The authors declare they have no potential conflicts of interest concerning drugs, products,or services used in the study.
Autoři deklarují, že v souvislosti s předmětem studie nemají žádné komerční zájmy.
The Editorial Board declares that the manuscript met the ICMJE recommendation for biomedical papers.
Redakční rada potvrzuje, že rukopis práce splnil ICMJE kritéria pro publikace zasílané do biomedicínských časopisů.Submitted/Obdrženo: 16. 8. 2020
Accepted/Přijato: 24. 1. 2021Prof. Seyed Alireza Dastgheib
Department of Medical Genetics,
School of Medicine,
Shiraz University of Medical
Sciences, Shiraz, Iran
e-mail: dastgheibsa@gmail.com
Sources
1. Elobaid YE, Aw TC, Grivna M et al. Breast cancer screening awareness, knowledge, and practice among Arab women in the United Arab Emirates: a cross-sectional survey. PLoS ONE 2014; 9 (9): e105783. doi: 10.1371/journal.pone.0105783.
2. Momenimovahed Z, Salehiniya H. Epidemiological characteristics of and risk factors for breast cancer in the world. Breast Cancer (Dove Med Press) 2019; 11 : 151–164. doi: 10.2147/BCTT.S176070.
3. O’Donovan J, Newcomb A, Macrae MC et al. Community health workers and early detection of breast cancer in low-income and middle-income countries: a systematic scoping review of the literature. BMJ Global Health 2020; 5 (5): e002466. doi: 10.1136/bmjgh-2020-002466.
4. Yan C, Sun C, Ding X et al. Association of CAV1 polymorphisms with the risks of breast cancer: a systematic review and meta-analysis. Pathol Res Pract 2019; 215 (9): 152518. doi: 10.1016/j.prp.2019.152518.
5. Jin T-F, Zhang W-T, Zhou Z-F. The 6q25.1 rs2046210 polymorphism is associated with an elevated susceptibility to breast cancer: a meta-analysis of 261,703 subjects. Mol Gen Genomic Med 2019; 7 (3): e553. doi: 10.1002/mgg3.553.
6. Yoo KY, Tajima K, Park SK et al. Postmenopausal obesity as a breast cancer risk factor according to estrogen and progesterone receptor status (Japan). Cancer Lett 2001; 167 (1): 57–63. doi: 10.1016/s0304-3835 (01) 00463-3.
7. Weiderpass E, Meo M, Vainio H. Risk factors for breast cancer, including occupational exposures. Saf Health Work 2011; 2 (1): 1–8. doi: 10.5491/SHAW.2011.2.1.1.
8. Feng Y, Spezia M, Huang S et al. Breast cancer development and progression: risk factors, cancer stem cells, signaling pathways, genomics, and molecular pathogenesis. Genes Dis 2018; 5 (2): 77–106. doi: 10.1016/j.gendis.2018.05.001.
9. DeSantis C, Ma J, Bryan L et al. Breast cancer statistics, 2013. CA Cancer J Clin 2014; 64 (1): 52–62. doi: 10.3322/caac.21203.
10. Anders CK, Johnson R, Litton J et al. Breast cancer before age 40 years. Semin Oncol 2009; 36 : 237–249. doi: 10.1053/j.seminoncol.2009.03.001.
11. Golemis EA, Scheet P, Beck TN et al. Molecular mechanisms of the preventable causes of cancer in the United States. Genes Dev 2018; 32 (13–14): 868–902. doi: 10.1101/gad.314849.118.
12. Fares J, Fares MY, Khachfe HH et al. Molecular principles of metastasis: a hallmark of cancer revisited. Signal Transduct Target Ther 2020; 5 (1): 28. doi: 10.1038/s41392-020-0134-x.
13. Ge J, Liu H, Qian D et al. Genetic variants of genes in the NER pathway associated with risk of breast cancer: a large-scale analysis of 14 published GWAS datasets in the DRIVE study. Int J Cancer 2019; 145 (5): 1270–1279. doi: 10.1002/ijc.32371.
14. Han H, Guo W, Shi W et al. Hypertension and breast cancer risk: a systematic review and meta-analysis. Sci Rep 2017; 7 : 44877. doi: 10.1038/srep44877.
15. Wang Z, Liu Y, Yang L et al. The polymorphism interleukin-8 -251A/T is associated with a significantly increased risk of cancers from a meta-analysis. Tumour Biol 2014; 35 (7): 7115–7123. doi: 10.1007/s13277-014-1881-5.
16. Huang Q, Wang C, Qiu LJ et al. IL-8-251A>T polymorphism is associated with breast cancer risk: a meta-analysis. J Cancer Res Clin Oncol 2011; 137 (7): 1147–1150. doi: 10.1007/s00432-011-0981-5.
17. Liu Q, Li A, Tian Y et al. The CXCL8-CXCR1/2 pathways in cancer. Cytokine Growth Factor Rev 2016; 31 : 61–71. doi: 10.1016/j.cytogfr.2016.08.002.
18. Sheikhpour R. The role of interleukin-8 and its mechanism in patients with breast cancer: its relation with oxidative stress and estrogen receptor. Int J Cancer Man 2017; 10. doi: 10.5812/ijcm.8791.
19. Todorović-Raković N, Milovanović J. Interleukin-8 in breast cancer progression. J Interferon Cytokine Res 2013; 33 (10): 563–570. doi: 10.1089/jir.2013.0023.
20. Freund A, Chauveau C, Brouillet JP et al. IL-8 expression and its possible relationship with estrogen-receptor-negative status of breast cancer cells. Oncogene 2003; 22 (2): 256–265. doi: 10.1038/sj.onc.1206113.
21. Singh JK, Simões BM, Clarke RB et al. Targeting IL-8 signalling to inhibit breast cancer stem cell activity. Expert Opin Ther Targets 2013; 17 (11): 1235–1241. doi: 10.1517/14728222.2013.835398.
22. Nicolini A, Carpi A, Rossi G. Cytokines in breast cancer. Cytokine Growth Factor Rev 2006; 17 (5): 325–337. doi: 10.1016/j.cytogfr.2006.07.002.
23. Smith KC, Bateman AC, Fussell HM et al. Cytokine gene polymorphisms and breast cancer susceptibility and prognosis. Eur J Immunogenet 2004; 31 (4): 167–173. doi: 10.1111/j.1365-2370.2004.00462.x.
24. Snoussi K, Mahfoudh W, Bouaouina N et al. Genetic variation in IL-8 associated with increased risk and poor prognosis of breast carcinoma. Hum Immunol 2006; 67 (1–2): 13–21. doi: 10.1016/j.humimm.2006.03. 018.
25. Vogel U, Christensen J, Dybdahl M et al. Prospective study of interaction between alcohol, NSAID use and polymorphisms in genes involved in the inflammatory response in relation to risk of colorectal cancer. Mutat Res 2007; 624 (1–2): 88–100. doi: 10.1016/j.mrfmmm.2007.04.006.
26. Kamali-Sarvestani E, Aliparasti MR, Atefi S. Association of interleukin-8 (IL-8 or CXCL8) -251T/A and CXCR2 +1208C/T gene polymorphisms with breast cancer. Neoplasma 2007; 54 (6): 484–489.
27. Snoussi K, Mahfoudh W, Bouaouina N et al. Combined effects of IL-8 and CXCR2 gene polymorphisms on breast cancer susceptibility and aggressiveness. BMC Cancer 2010; 10 : 283. doi: 10.1186/1471-2407-10-283.
28. Liu S, Cai H, Cheng W et al. Association of VDR polymorphisms (Taq I and Bsm I) with prostate cancer: a new meta-analysis. J Int Med Res 2017; 45 (1): 3–10. doi: 10.1177/0300060516668939.
29. Wang Z, Liu Q-L, Sun W et al. Genetic polymorphisms in inflammatory response genes and their associations with breast cancer risk. Croat Med J 2014; 55 (6): 638–646. doi: 10.3325/cmj.2014.55.638.
30. Khalili-Azad T, Razmkhah M, Ghiam AF et al. Association of interleukin-18 gene promoter polymorphisms with breast cancer. Neoplasma 2009; 56 (1): 22–25. doi: 10.4149/neo_2009_01_22.
31. Taheri M, Hashemi M, Eskandari-Nasab E et al. Association of -607 C/A polymorphism of IL-18 gene (rs1946518) with breast cancer risk in Zahedan, Southeast Iran. Prague Med Rep 2012; 113 (3): 217–222. doi: 10.14712/23362936.2015.19.
32. Back LK d C, Farias TDJ, da Cunha PA et al. Functional polymorphisms of interleukin-18 gene and risk of breast cancer in a Brazilian population. Tissue Antigens 2014; 84 (2): 229–233. doi: 10.1111/tan.12367.
33. Zhao Y, Wang S, Zhang Z et al. Association of IL-18 genetic polymorphisms and haplotypes with breast cancer risk in a Chinese population. Biomed Res 2017; 28 : 8433–8437. doi: 10.1371/journal.pone.0073671.
34. Qiao X, Xu D, Sun D et al. Association analysis of interleukin-18 gene promoter region polymorphisms and susceptibility to sporadic breast cancer in Chinese Han women. J Clin Lab Anal 2018; 32 (9): e22591. doi: 10.1002/jcla.22591.
35. Todorović-Raković N, Milovanović J. Interleukin-8 in breast cancer progression. J Interferon Cytokine Res 2013; 33 : 563–570. doi: 10.1089/jir.2013.0023.
36. El Ayadi A, Herndon DN, Finnerty CC. Biomarkers in burn patient care. Total burn care (fifth ed.). Elsevier Inc. 2018; 232–235.e2.
37. Charrad R, Kaabachi W, Rafrafi A et al. IL-8 gene variants and expression in childhood asthma. Lung 2017; 195 (6): 749–757. doi: 10.1007/s00408-017-0058-6.
38. Zhang M, Fang T, Wang K et al. Association of polymorphisms in interleukin-8 gene with cancer risk: a meta-analysis of 22 case–control studies. Onco Targets Ther 2016; 9 : 3727–3737. doi: 10.2147/OTT.S103159.
39. Salimi E, Karimi-Zarchi M, Dastgheib SA et al. Association of promoter region polymorphisms of IL-6 and IL-18 genes with risk of recurrent pregnancy loss: a systematic review and meta-analysis. Fetal Pediatr Pathol 2019; 39 (4): 346–359. doi: 10.1080/15513815.2019.1652379.
40. Rex DAB, Agarwal N, Prasad TSK et al. A comprehensive pathway map of IL-18-mediated signalling. J Cell Commun Signal 2020; 14 (2): 257–266. doi: 10.1007/s12079-019-00544-4.
41. Zhang M-J, Zhou Y, Wang X et al. Interleukin-18 gene promoter 607A polymorphism, but not 137C polymorphism, is a protective factor for ischemic stroke in the Chinese population: a meta-analysis. Meta Gene 2016; 9 : 165–172. doi: 10.1016/j.mgene.2016.06.006.
42. Yin YW, Hu AM, Sun QQ et al. Association between interleukin-8 gene -251 T/A polymorphism and the risk of peptic ulcer disease: a meta-analysis. Hum Immunol 2013; 74 (1): 125–130. doi: 10.1016/j.humimm.2012.09.006.
43. Li X, Ren D, Li Y et al. Increased cancer risk associated with the -607C/A polymorphism in interleukin-18 gene promoter: an updated meta-analysis including 12,502 subjects. J BUON 2015; 20 (3): 902–917.
44. Bahrami R, Dastgheib SA, Niktabar SM et al. Association of BMP4 rs17563 polymorphism with nonsyndromic cleft lip with or without cleft palate risk: literature review and comprehensive meta-analysis. Fetal Pediatr Pathol 2020; 40 (4): 305–319. doi: 10.1080/15513815.2019.1707916.
45. Veisian M, Tabatabaei RS, Javaheri A et al. Association of interleukin-10 -1082G > a polymorphism with susceptibility to preeclampsia: a systematic review and meta--analysis based on 21 studies. Fetal Pediatr Pathol 2020; 39 (6): 518–532. doi: 10.1080/15513815.2019.1683919.
46. Abbasi H, Dastgheib SA, Hadadan A et al. Association of endothelial nitric oxide synthase 894G > T polymorphism with preeclampsia risk: a systematic review and meta--analysis based on 35 studies. Fetal Pediatr Pathol 2021; 40 (5): 455–470. doi: 10.1080/15513815.2019.1710880.
Labels
Paediatric clinical oncology Surgery Clinical oncology
Article was published inClinical Oncology

2022 Issue 3-
All articles in this issue
- Oslepení
- Changes of serum protein N-glycosylation in cancer
- New approaches in palliative systemic therapy of anal squamous cell carcinoma
- Metabolic plasticity of cancer cells
- Neurobiology of cancer – the role of cancer tissue innervation
- Direct and indirect impacts of the COVID-19 pandemic on patients with pulmonary and pleural malignancies – a retrospective analysis of patient outcomes treated at Department of Respiratory Diseases, University Hospital Brno, during the 2nd and 3rd coronavirus waves
- Meigs’ syndrome
- Informace z České onkologické společnosti
- Bone remineralization after palliative radiotherapy
- Aktuality z odborného tisku
- Prof. MUDr. Luboš Petruželka, CSc.
- Association of IL-8 -251T>A and IL-18 -607C>A polymorphisms with susceptibility to breast cancer – a meta-analysis
- Analysis of the results of radiotherapy and chemoradiotherapy on the background of immunotherapy of patients with cancer of the oral cavity and oropharynx
- Diffuse large B-cell lymphoma associated ileocecal intussusception in adulthood
- Clinical Oncology
- Journal archive
- Current issue
- Online only
- About the journal
Most read in this issue- Meigs’ syndrome
- Analysis of the results of radiotherapy and chemoradiotherapy on the background of immunotherapy of patients with cancer of the oral cavity and oropharynx
- Changes of serum protein N-glycosylation in cancer
- Metabolic plasticity of cancer cells
Login#ADS_BOTTOM_SCRIPTS#Forgotten passwordEnter the email address that you registered with. We will send you instructions on how to set a new password.
- Career

