-
Medical journals
- Career
Median method for determining cortical brain activity in a near infrared spectroscopy image
Authors: Ondřej Klempíř 1; Radim Krupička 1; Robert Jech 2
Authors‘ workplace: Department of Biomedical Informatics, Faculty of Biomedical Engineering, Czech Technical University in Prague, Kladno, Czech Republic 1; Department of Neurology, First Medical Faculty and General University Hospital, Charles University, Prague, Czech Republic 2
Published in: Lékař a technika - Clinician and Technology No. 1, 2018, 48, 11-16
Category: Original research
Overview
Near-InfraRed-Spectroscopy (NIRS) is a neuroimaging method of brain cortical activity using low-energy optical radiation to detect local changes in (de)oxyhaemoglobin concentration. A methodology consisting of a raw signal pre-processing phase, followed by statistical analysis based on a general linear model, is currently being used to determine signal activity. The aim of this research is to define the median modification of the standard method usually used for the estimation of cortical activity from the NIRS signal and to verify its applicability in measuring motor tasks for patients with Parkinson's disease. Individual examinations were conducted in 10 cycles, during which finger tapping, and rest phases were alternating. Changes in oxyhaemoglobin concentration were calculated from the native NIRS signal using the modified Lambert-Beer equation. The signals were filtered in the 0.015–0.3 Hz band and fitted by the physiological response function of the brain tissue for each finger tapping cycle separately. The median value from the 10 cycles was then computed. Activity values obtained in individual subjects have been used in Brain Mapping visualizations. These describe motor task patterns during the ON and OFF deep brain stimulation of the subthalamic nucleus in Parkinson's disease, which demonstrates activation in accordance with the current state of knowledge in functional imaging.
Keywords:
near infrared spectroscopy, neuroimaging, Parkinson's disease, neurophotonics, brain mapping, neuromodulationIntroduction
Near-InfraRed-Spectroscopy (NIRS) is a neuroimaging method using low-energy optical radiation (800–2500 nm) to measure extinction changes in subsurface tissue caused by changes of haemoglobin concentration which correlates with cortical activity. It detects changes in concentrations of oxy - (HbO) and deoxyhaemoglobin (HbR), which are the correlates of functional brain neural activity BOLD (Blood Oxygenation Level Dependent) [1, 2]. To achieve a spatial mapping of brain activity, pairs of source-detectors are used and placed in a head cap similar to a multichannel EEG. The optimal distance between the nearest pairs of sources and detectors is 30–35 mm. The penetration depth for an adult is in between 15–25 mm [2]. The NIRS system is safe, versatile, portable and can measure most of the cortex.
The hemodynamic response function (HRF) is similar to a functional magnetic resonance (fMRI) [3]. The HR starts with a latency of 1–2 seconds after the beginning of the event and reaches a maximum after 7 seconds. The frequency band associated with cognitive or motor tasks corresponds in a range from 0.01 Hz to 0.1 Hz in NIRS imaging [4]. In comparison to a fMRI, NIRS as a medical device provides lower spatial resolution, but it has better time resolution.
In addition to measuring the physiological activity [5], sport activity [6] and pathophysiological states [7], NIRS has also been used in causal connectivity research [8].
To determine signal activity, the most commonly used methodology consists of two main phases. The first part is raw signal preprocessing, followed by the statistical analysis of the preprocessed signal based on the defined model. This leads to determining the quantitative value of activity. Generally, the NIRS signal can be considered as stochastic within one measuring state (resting state). However, the response to a stimulus appears to be deterministic in the time series. This fact is used to define the appropriate model, usually a general linear model (GLM), which is fitted to the observed signal. A detailed description, including technical details and ways to (pre)process NIRS can be found in the extensive review article [9]. Most of the above defined procedures for the complete processing pipeline including GLM are implemented in the Matlab Open Source package for Statistical Parametric Mapping (SPM) [10]. Previous phases are often subsequently visualized in the form of brain mapping. This is a topographic mapping of brain activity, where spatial (surface) properties are analyzed [11].
The aim of this research is to define the median modification of the standard method of determining cortical activity from the NIRS signal and to verify its imaging capabilities in the measurement of motor task in patients with Parkinson's disease (PD), who underwent implantation of deep brain stimulation for treatment of motor symptoms. Knowledge of cortical activity plays a key role in understanding the pathophysiology.
Methods
Clinical dataset and signals acquisition
This study includes data from the Department of Neurology, 1st Faculty of Medicine and General University Hospital in Prague. The study was conducted in compliance with the Declaration of Helsinki and was approved by the Ethics Committee of the General University Hospital in Prague. We included 9 patients with advanced PD (1F, 8M, mean age: 64.2 ± (SD) 3.4 years) chronically treated with doublesided deep brain stimulation of the subthalamic nucleus (STN-DBS 130 Hz). DBS electrodes were bilaterally implanted using standard stereotactic methods and incorporating intraoperative microelectrode recording. Every participant signed an informed consent before enrolment in the study.
Brain activity was scanned by 22 partially dependent channels of the multichannel-system NIRSport (by NIRX Medizintechnik, Berlin, Germany, Fs = 8.93 Hz), which consists of 7 sources and 8 detectors of infrared light. The device emits NIR radiation at 2 wavelengths, 760 and 850 nm. Photodetectors were arranged on the scalp using the international 10-20 EEG system for positioning: F1, FC1, CP1, FC3, F2, FC2, CP2, FC4 (Fig. 1). An accurate 3D neuroanatomic position is available through the coordinate atlas of the MNI standard (Montreal Neurological Institute). The NIRS cap was placed that the source Cz was located halfway between the nasioninion (Fig. 1, Fig. 2).
1. Scheme of the distribution of detectors, sources and channels above the cortical regions of the brain with respect to the EEG 10-20 system. 22 channels cover from the frontal cortex to the parietal cortex. 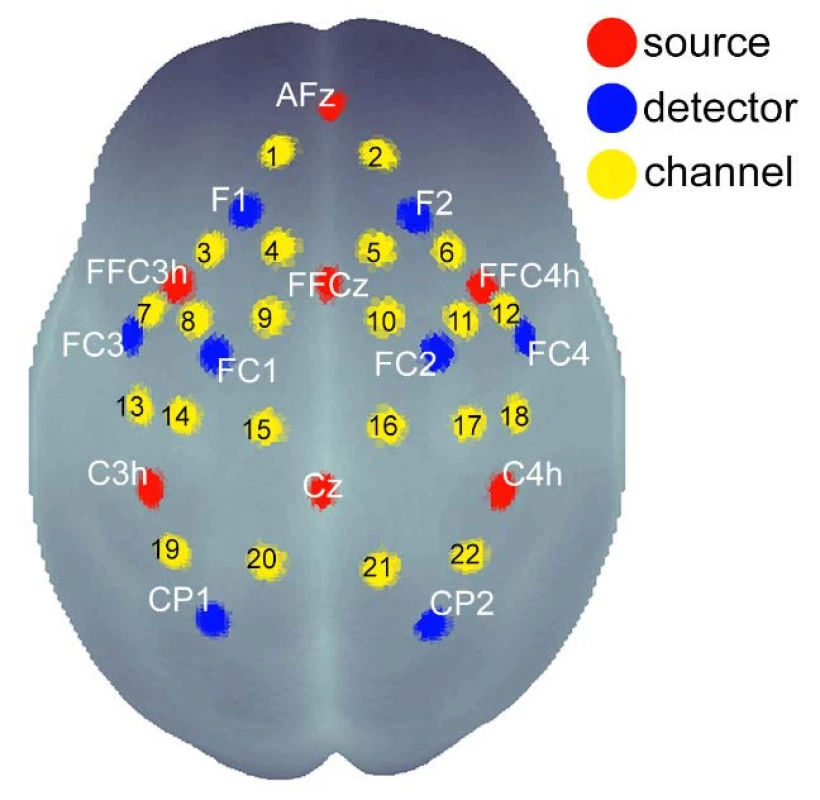
2. Illustration photo with a NIRS cap placed. 
The signal was recorded in a block experiment for both STN-DBS ON and STN-DBS OFF states. Each task was repeated in 10 continuous cycles. Each cycle consisted of 15 seconds of an alternating finger motion (finger tapping) rotated by a 15-second rest phase. The start and end of the events were marked with markers in the signals.
NIRS signal preprocessing
The optical signals are strongly attenuated when passing through biological tissue. The intensity decreases by several orders of magnitude at a distance of several centimeters. A physical model derived from the modified Lambert-Beer law was used for the purposes of determining the changes in oxyhaemoglobin concentrations from raw centred signals. The following equation can be defined based on knowledge of recorded time series for 2 wavelengths and extinction coefficient values for 2 chromophores (HbO and HbR) [12]:
CHbO denotes the HbO concentration, ei represents the extinction coefficient of the given chromophore for a particular wavelength (Table 1), Ai corresponds to the raw signal and is proportional to the negative logarithm of the ratio of detected and emitted light intensity, Li represents the mean free path length describing the most likely path of the photon towards the detector (set to 3 cm, see Introduction). The signals were filtered in the range from 0.015 to 0.3 Hz to attenuate slow drifts and extracerebellar artefacts generated by respiration and cardiac variation (3rd order Butterworth bandpass filter). Records with other visible movement artefacts or subjects with incomplete measurements were not included in the study.
1. Values of extinction coefficients (M-1cm-1) used to calculate time series of HbO and HbR concentrations for the NIRS device (taken from the device documenttation). 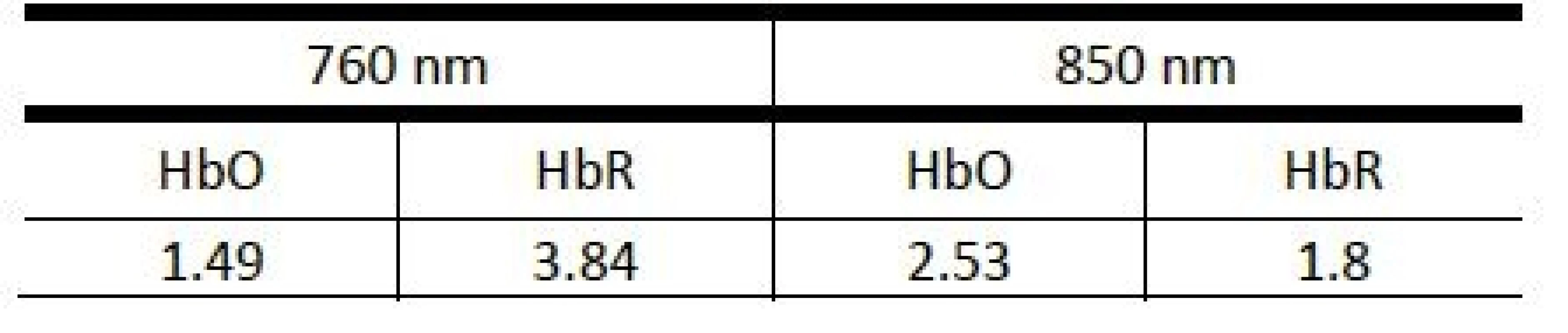
Method for determining NIRS signal activity
The most commonly used method for determining signal activity and for examining the relationships between variables in neuroscience has become a general linear model (2), in a better interpretable case (3).
The simplest GLM design matrix consists of 2 explanatory variables: a variable containing the only ones for the computation of the absolute member and a variable for estimating the size of the experiment factor (i.e. motor task). It is determined by the value 1 in the case of a stimulus and the value 0 in its absence. The modification of the boxcar course closer to the known physiological BOLD nature of the response can be reached by convolution with Boynton HRF [13]. Estimation of parameter values β is calculated using the least squares (LS) method:
Two parameters β are normally considered for describing the signal activity in the case of the time series obtained by the NIRS technique, characterizing the entire time series for 10 cycles of the entire finger tapping examination. The innovative proposed method consists of approaching the LS fitting for 10 cycles of the motor task individually and then calculating medians from 10 pairs of estimated parameters β.
Results
Statistical properties of the proposed method
Offline scripts with the proposed median method for NIRS signal activity determination have been implemented in Matlab R2015a (Mathworks, USA). The method was applied to 396 signals (22 channels, 9 patients, 2 DBS-STN states) in the motor task of PD patients. The accuracy of the automatic procedure for the entire dataset was inspected visually during the computations. A specific example of median activity determination for the real signal is shown in the Fig. 3.
3. An example of determining the NIRS signal activity based on median methodconsists of LS fitting for 10 HRF models into individual cycles and calculating the median. 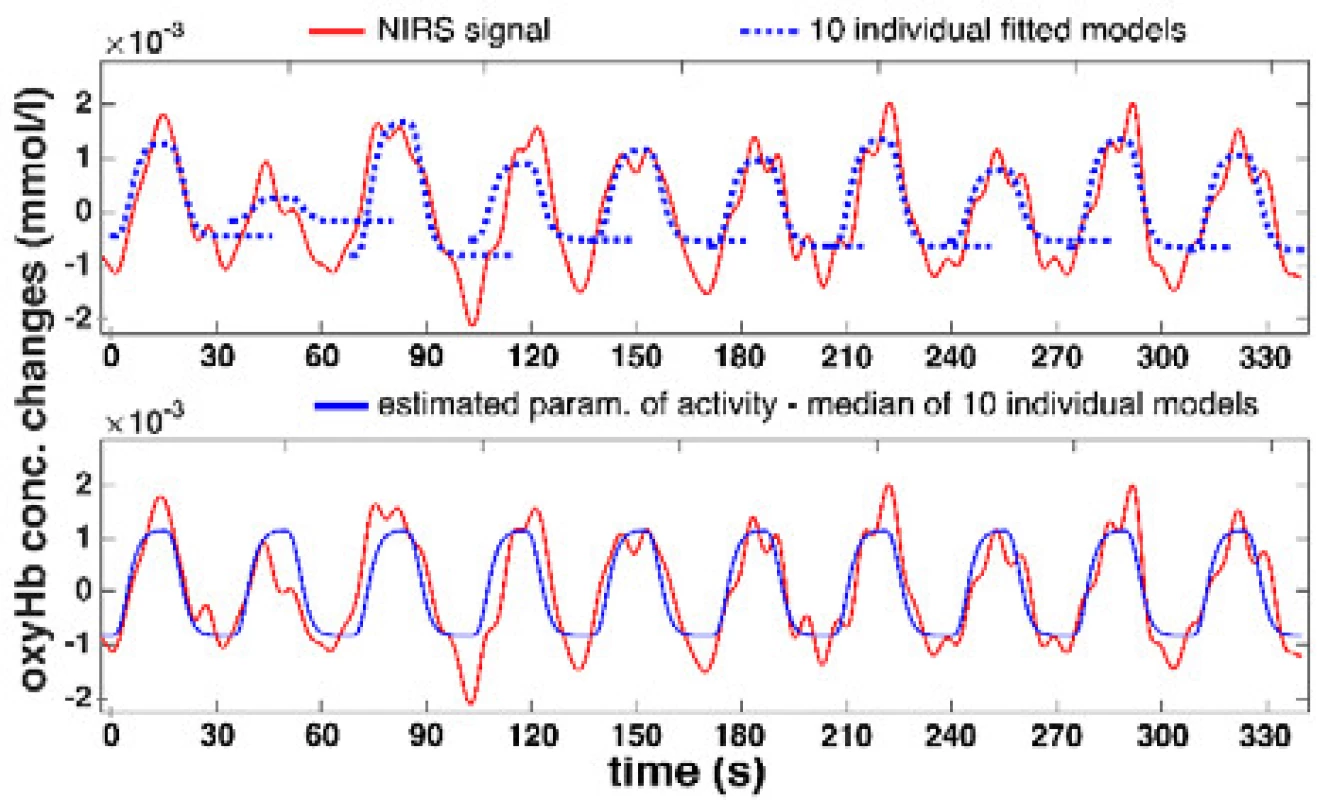
From the statistical error properties point of view, the results show that the mean value estimates are slightly deviated from zero at the expense of the median, but the method errors are symmetrical, and the normality is not too disturbed (Fig. 4). The activity values of both methods, standard GLM and proposed median GLM, do not differ for regular NIRS signals.
4. An example of a histogram of error distribution obtained by the median method in the selected signal. 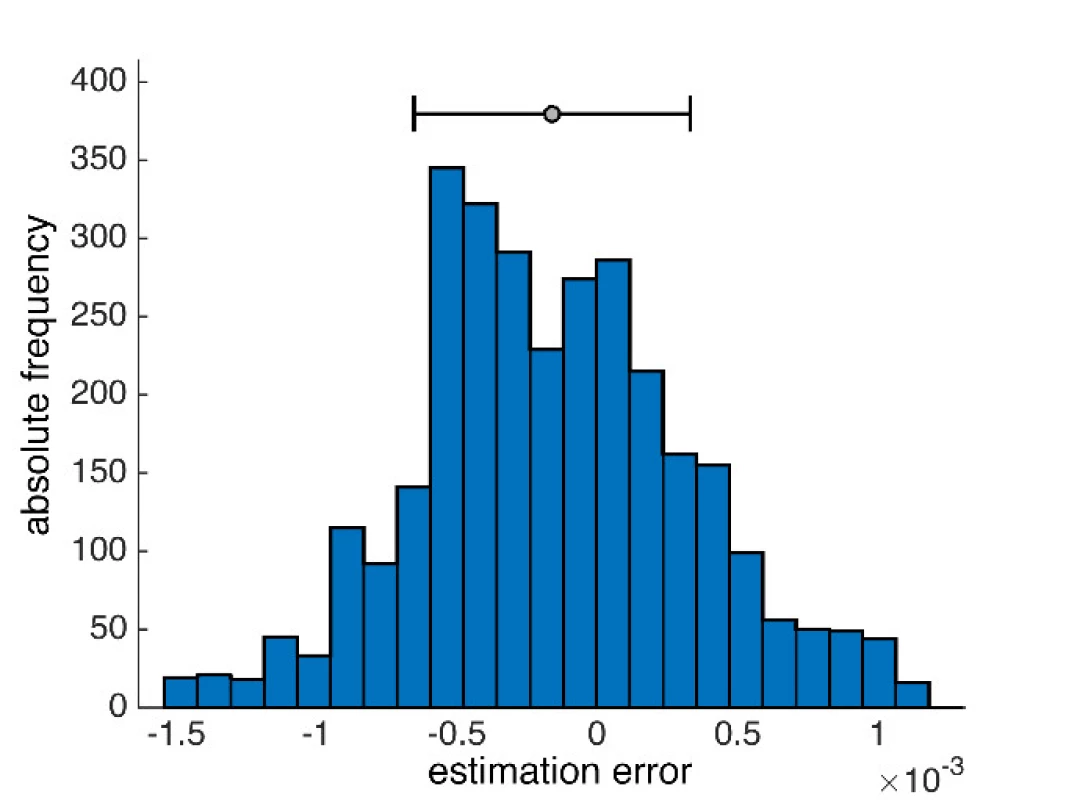
Clinical application of motor activity mapping
The activity values obtained within individual subjects were visualized in the form of NIRS brain mapping under ON vs. OFF DBS-STN conditions. The NIRS Brain Mapping tool was implemented on top of Matlab based on the knowledge of accurate neuroanatomical standardized MNI coordinates for individual channels. Tool NIRS Brain Mapping is based on triangulation-cubic interpolation of activity values in channels according to MNI coordinates (Fig. 5). The visualization (Fig. 6) shows that in 8 out of 9 patients, ON DBS-STN led to an increase in HbO activity in the contralateral regions of the brain during right-hand finger tapping associated with planning and performing the motion. Anatomical mapping of NIRS channels to EEG 10-20 positions was evaluated by an expert and can be performed according to Table 2.
5. Comparison of linear (A) and cubic (B) interpolation. (C) 3D view of the interpolation curve that passes through all MNI coordinates with median activity for an average of 9 patients DBS-STN ON–OFF differential map. Input grid in the form of a matrix has 375 rows and 329 columns. Blackpoints represent the position of individual channels. Linear and cubic give a comparable result of interpolation. Points define a network between which activity is interpolated. The griddata() Matlab function has been used, which interpolates with a triple grid resolution. The color scale represents activity intensity. The 2D map is created by displaying the scalp into 2D space, which thus causes a distortion of the position of some optodes. A) linear interpolation leads to a sharper map compared to cubic. B) contains the representation of statistically significant areas according to the Wilcoxon paired test 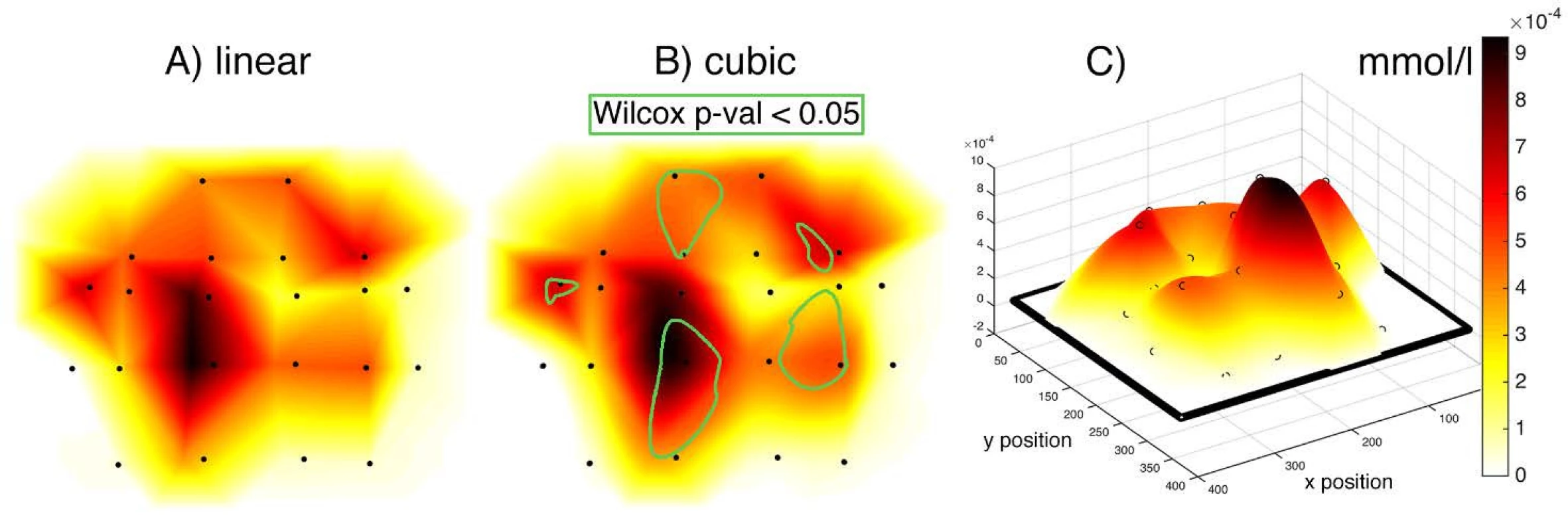
6. NIRS Brain Mapping visualization. Cubic interpolation patterns of HbO activity in the cortical regions of the brain in the motor task (finger tapping) ON vs. OFF DBS-STN. 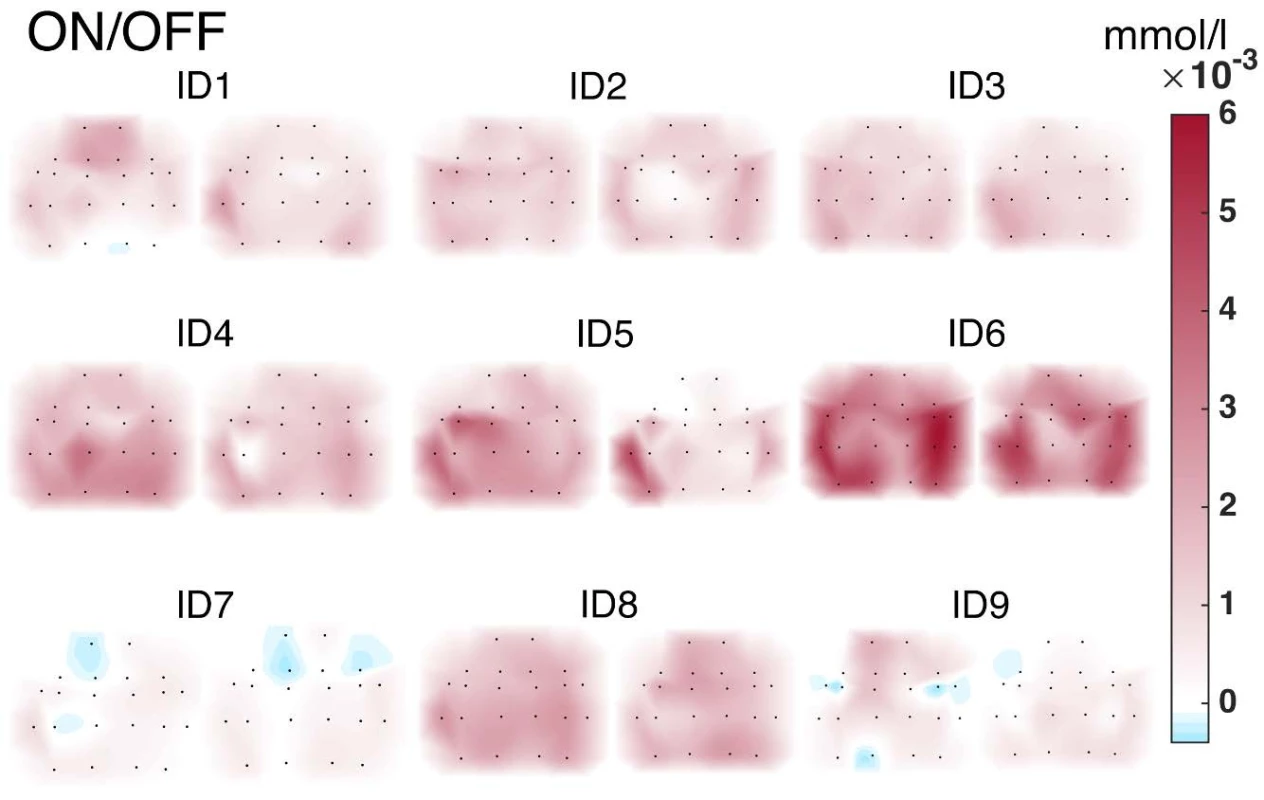
2. Result of an expertise mapping of NIRS channels to anatomical brain areas [14]. ![Result of an expertise mapping of NIRS channels to anatomical brain areas [14].](https://www.prolekare.cz/media/cache/resolve/media_object_image_small/media/image_pdf/497ebb0754014229282fcd2a6a16ef9b.jpeg)
Discussion
Study shows the design, properties and applicability of a new median method for NIRS functional changes detection in the motor cortex in PD and indicated its future use for applications that can not be done using PET or fMRI. The proposed method is based on a simple model of 2 independent regressors. For this reason, it does not have issues with the multicollinearity problem. However, we have not yet dealt with autoregression of median method residues. It offers to perform group statistics on the 2nd level and provides detailed information on the potential significance of differences between DBS-STN conditions in addition to brain mapping visualizations for ON vs. OFF, for example including the use of clinical data in the form of GLM covariates.
Currently, there are many relevant available packages, tools and libraries that can easily be integrated into the work of biomedical software programmers. They are usually downloadable via GitHub. The following list of tools for NIRS time series analysis and fMRI brain mapping is worth mentioning: NFRI toolbox [15], xTopo [16], AFNI [17], Nitime [18], NIfTI [19] and Nilearn [20]. The advantage of the proposed method over the available implementations is easy to understand; a clear algorithm of the method. Also, easy script scalability. The use of available tools often leads users to access a black box, while selfrealization helps deeper understanding of the analysis procedures. The proposed median method leads to robust estimates for irregular signals, which are often obtained from patients with advanced progressive pathophysiology. In such cases, the estimate behaves empirically better and considers the time change of the parameter β values during a block experiment.
The unique 22-channel NIRS system has shown an increase in the activity of cortical contralateral areas associated with motion, which according to literature and the opinion of experts from the Department of Neurology, First Medical Faculty and General University Hospital in Prague is in accordance with the current state of knowledge in functional imaging [21–23].
In this study, we designed a new method for determining NIRS signal activity and scripts for its implementation were developed, including NIRS Brain Mapping visualization. In addition, for work needs, a mapping of NIRS channels into space EEG 10-20 was performed. This work contributes to the field of neuroimaging, especially for patient records, when the signal values during a block experiment have a greater variance over healthy controls. The study supports the hypothesis for NIRS applicability in various movement tasks, which cannot be examined via fMRI or PET scans and patients can only imagine it (e.g. gait). NIRS Brain Mapping source codes are released under the MIT license and are available on request.
Acknowledgement
This study is a result of activities performed within the AZV Grant no. 16-28119a project "Analysis of movement disorders for the study of extrapyramidal diseases mechanism using motion capture camera systems".
Ing. Ondřej Klempíř
Department of Biomedical Informatics
Faculty of Biomedical Engineering
Czech Technical University in Prague
Nám. Sítná 3105, CZ-272 01 Kladno
E-mail: klempond@gmail.com
Phone: +420 605 536 419
Sources
- Glover, H.: Overview of Functinal Magnetic Resonance Imaging. Clinical Neurosurgery, 2011, vol. 22, no. 2, p. 133–139.
- Agbangla, N. F., Audiffren, M., Albinet, C. T.: Use of near-infrared spectroscopy in the investigation of brain activation during cognitive aging: A systematic review of an emerging area of research. Aging research reviews, 2017, vol. 38, p. 52–66.
- Cui, X., Bray, S., Bryant, D. M., Glover, G. H., Reiss, A. L.: Quantitative comparison of NIRS and fMRI accros multiple tasks. Neuroimage, 2011, vol. 54, no. 4, p. 2808–2821.
- Tohka, J., Foerde, K., Aron, A. R., Tom, S. M., Toga, A. W., Poldrack, R. A.: Automatic independent component labeling for artifact removal in fMRI. Neuroimage, 2008, vol. 39, no. 3, p. 1227–45.
- McKendrick, R., Mehta, R., Ayaz, H., Scheldrup, M., Parasuraman, R.: Prefrontal Hemodynamics of Physical Activity and Environmental Complexity During Cognitive Work. Human factors, 2017, vol. 59, no. 1, p. 147–162.
- Oussaidene, K., Prieur, F., Tagougui, S., Abaidia, A., Matran, R., Mucci, P.: Aerobic fitness influences cerebral oxygenation response to maximal exercise in healthy subjects. Respiratory physiology & neurobiology, vol. 205, no. 1, p. 53–60.
- Bick, S., Folley, B., Mayer, J., Park, S., Charles, P. D., Camalier, C. R., Pallavaram, S., Konrad, P. E., Neimat, J. S.: Subthalamic Nucleus Deep Brain Stimulation Alters Prefrontal Correlates of Emotion Induction. Neuromodulation, 2016, vol. 20, no. 3, p. 233–237.
- Arizono, N., Ohmura, Y., Yano, S., Kondo, T.: Functional Con-nectivity Analysis of NIRS Data under Rubber Hand Illusion to Find a Biomarker of Sense of Ownership. Neural Plasticity, 2016, vol. 2016, p. 1–9.
- Tak, S., Ye, J. C.: Statistical analysis of fNIRS data: A compre-hensive review. Neuroimage, 2014, vol. 85, no. 1, p. 72–91.
- Statistical Parametric Mapping. www.fil.ion.ucl.ac.uk/spm/ [cit. 2. 12. 2017].
- Krajca, V., Petranek, S.: Wave-Finder: A new system for an automatic processing of long-term EEG recordings. Quantitative EEG analysis clinical utility and new methods, 1993, p. 103–106.
- Cope, D., Delpy, D. T.: System for long-term measurement of cerebral blood oxygenation. Medical Computing, 1988, vol. 26, no. 3, pp. 5.
- Boynton, G. M., Engel, A. S., Glover, G. H., Heeger, D. J.: Linear systems analysis of functional magnetic resonance imaging in human. Journal of Neuroscience, 1996, vol. 16, no. 13, p. 4207–4221.
- Electrode 10-20 positions, mapping, anatomical brain areas. http://www.brainm.com/software/pubs/dg/BA_10-20_ROI_
Talairach/nearesteeg.htm [cit. 3. 12. 2017]. - NFRI toolbox. http://www.jichi.ac.jp/brainlab/tools.html [cit. 3. 12. 2017].
- xTopo. http://www.alivelearn.net/?p=785 [cit. 3. 12. 2017].
- AFNI. https://afni.nimh.nih.gov [cit. 3. 12. 2017].
- Nitime. http://nipy.org/nitime/ [cit. 3. 12. 2017].
- NIfTI. https://nifti.nimh.nih.gov [cit. 3. 12. 2017].
- Nilearn. http://nilearn.github.io [cit. 3. 12. 2017].
- Payoux, P., Brefel-Courbon, Ch., Julian, A.: Motor activity in parkinsonism and levodopa effect: A PET study. Journal of nuclear medicine, 2007, vol. 48, no. supplement 2 : 8P.
- Naoki, I., Takefumi, M., Akira, S., Eiji, K., Fumiko, I., Koji, T., Yasuki, K., Takayuki, T., Toshio, H.: Monitoring Local Regional Hemodynamic Signal Changes during Motor Execution and Motor Imagery Using Near-Infrared Spectroscopy. Frontiers in physiology, 2015, vol. 6.
- Hou, L. B., Bhatia, S., Carpentera, J. S.: Quantitative compari-sons on hand motor functional areas determined by resting state and task BOLD fMRI and anatomical MRI for pre-surgical planning of patients with brain tumors. Neuroimage: clinical, 2016, vol. 11, p. 378–387.
Labels
Biomedicine
Article was published inThe Clinician and Technology Journal

2018 Issue 1-
All articles in this issue
- Median method for determining cortical brain activity in a near infrared spectroscopy image
- Patient´s respiratory curve synchronization by visual feedback application
- A comparsion of the quality of dental crowns from TI-6AL-4V and cocr alloys made with SLM technology
- Dimensionality reduction methods for biomedical data
- Agcu bimetallic nanoparticles modified by polyvinyl alcohol - the cells viability study in vitro
- The Clinician and Technology Journal
- Journal archive
- Current issue
- Online only
- About the journal
Most read in this issue- Median method for determining cortical brain activity in a near infrared spectroscopy image
- A comparsion of the quality of dental crowns from TI-6AL-4V and cocr alloys made with SLM technology
- Dimensionality reduction methods for biomedical data
- Patient´s respiratory curve synchronization by visual feedback application
Login#ADS_BOTTOM_SCRIPTS#Forgotten passwordEnter the email address that you registered with. We will send you instructions on how to set a new password.
- Career




