-
Medical journals
- Career
Association of polymorphisms in nucleotide excision repair pathway genes with susceptibility to cutaneous melanoma
Authors: A. Hashemzehi 1; M. Ghadyani 2; F. Asadian 3; S. A. Dastgheib 4; S. Kargar 5; H. Neamatzadeh 6,7; E. Akbarian 8; A. Emarati 8
Authors‘ workplace: Department of Pharmacology, Faculty of Pharmacology, Tehran University of Medical Sciences, Tehran, Iran 1; Department of Advanced Medical Sciences and Technologies, Islamic Azad University, Science and Research Branch, Tehran, Iran 2; Department of Medical Laboratory Sciences, School of Paramedical Science, Shiraz University of Medical Sciences, Shiraz, Iran 3; Department of Medical Genetics, School of Medicine, Shiraz University of Medical Sciences, Shiraz, Iran 4; Department of General Surgery, Shahid Sadoughi University of Medical Sciences, Yazd, Iran 5; Department of Medical Genetics, Shahid Sadoughi University of Medical Sciences, Yazd, Iran 6; Mother and Newborn Health Research Center, Shahid Sadoughi University of Medical Sciences, Yazd, Iran 7; Children Growth Disorder Research Center, Shahid Sadoughi University of Medical Sciences, Yazd, Iran 8
Published in: Klin Onkol 2021; 34(5): 350-355
Category: Reviews
doi: https://doi.org/10.48095/ccko2021350Overview
Background: The effects of single nucleotide polymorphisms (SNPs) at nucleotide excision repair (NER) pathway on susceptibility to cutaneous melanoma (CM) are of great interest. To date, several epidemiological studies have evaluated whether the XPC, XPD, XPG and XPF polymorphisms are associated with CM. However, those studies results are controversial or inconclusive. Therefore, we conducted a study to evaluate the association of seven frequently investigated NER pathway polymorphisms with CM risk. Methods: A total of 150 patients diagnosed with CM and 150 healthy controls were enrolled in the study. Seven SNPs in the NER pathway including XPC (Lys939Gln and Ala499Val), XPD (Lys157Gln, Asp272Asn, and Arg751Arg), XPG (Asp1104His) and XPF (Arg415Gln) were analyzed by polymerase chain reaction restriction fragment length polymorphism assay. Results: There was no a significant association between XPC Lys939Gln, Ala499Val; XPD Asp272Asn, Arg751Arg, Arg751Arg; XPF Arg415Gln; and XPG Asp1104His polymorphisms and an increased risk of CM. Conclusions: This study results revealed that the XPC, XPD, XPG and XPF polymorphisms were not risk factor for susceptibility to CM. However, more well-designed with larger sample size studies in different populations are necessary to further evaluate and validate our results. Future studies which take into account gene-gene and gene-environment interactions are warranted for more precise evidence and further elucidation of the underlying mechanism of CM.
Keywords:
association – cutaneous melanoma – nucleotide excision repair – single nucleotide polymorphism
Introduction
Each year, tens of millions of cases are diagnosed with a malignancy worldwide, and more than half of them eventually die from it [1–5]. Cutaneous melanoma (CM) is an aggressive tumor of melanocytes in the skin [6]. CM causes more deaths than any other skin tumor, and the annual incidence of this disease has increased dramatically, which makes it one of the fastest growing cancers worldwide [7]. Estimates of the worldwide incidence revealed 232,000 new cases and 55,000 deaths for CM in 2012, with a 15th place among most common cancers worldwide [8]. The incidence and mortality rates of cutaneous melanoma differ widely by ethnic background [9]. In the United States, CM is among the top ten of leading cancer sites with a 5th place for men and a 6th place for women [10]. In less than 10 years, melanoma treatment has been revolutionized. However, the outcome for melanoma is highly dependent on the stage of the disease [6,11]. The etiology of melanoma is multifactorial and includes both environmental and genetic factors [6]. The most important risk factor for developing melanoma is exposure to skin color and ultraviolet radiation intensity [12]. Approximately 10% of CM cases have affected relatives, thus positive family history of CM may be associated with an increased risk of CM [13]. Recent advances in molecular genetic technology, notably next-generation sequencing (NGS), have led to the identification of multiple loci involved in the development and progression of CM such as genetic variations at nucleotide excision repair (NER) pathway [14,15].
The human genome is frequently subjected to damage from several deleterious environmental agents, e. g. ultraviolet (UV) light, chemotherapeutic agents or radiation, and endogenous weak mutagens, e. g. reactive oxygen species and metabolites like alkylating agents [16,17]. NER is the major mechanism in the process of DNA repair, which is mainly implicated in the replacement of bulky and helix distorting adducts with newly synthesized DNA segments. The major proteins involved in NER include XPA, XPC, XPD, XPE, XPF, XPG and ERCC [18]. Previously published epidemiological studies have indicated the association between single nucleotide polymorphisms (SNPs) in NER pathway genes and development of different cancers [19,20]. Hence, genetic alteration of NER-related genes may be closely related to the occurrence and development of CM [21–23]. In this study, we have evaluated association of seven potential functional polymorphisms including XPC (Lys939Gln and Ala499Val), XPD (Lys157Gln, Asp272Asn, and Arg751Arg), XPG (Asp1104His) and XPF (Arg415Gln) with a risk of CM.
Materials and methods
Subjects
A total of 150 patients diagnosed with CM were enrolled between February 2015 and September 2018. The cases were eligible if they had newly diagnosed and histologically confirmed CM without undergoing radiotherapy and chemotherapy treatment. During the same period, 150 age - and gender-matched healthy subjects were randomly selected as controls from the same centers. All the research subjects were unrelated ethnic Iranian population from six different cities. The study was reviewed and approved by the ethical review boards and a written informed consent was received from the guardian of each participant.
SNPs selection and genotyping
Seven potentially functional polymorphisms widely evaluated in various types of cancer including XPC (Lys939Gln and XPC Ala499Val), XPD (Asp272Asn, Arg751Arg and Arg751Arg), XPF (Arg415Gln) and XPG (Asp1104His) genes were selected. The selected SNPs had a high mutation frequency and were located in the noncoding or coding regions of the NER pathway. Genomic DNA was extracted from 2 mL of peripheral blood of cases and controls using the YTA Blood DNA Kit (Yekta-Tajhiz Azma, Tehran, Iran) according to the manufacturer’s instructions and the samples were frozen at −80 °C. Then, the selected SNPs were genotyped using polymerase chain reaction-restriction fragment length polymorphism assay as described previously [24–29]. The volume of PCR reaction was performed in a final volume of 25 µL containing 50 ng of genomic DNA, 0.24 µM of each primer, 1X of Taq DNA polymerase buffer, 2.5 mM of MgCl2, 0.20 mM of dNTP, and 1U of Taq DNA polymerase (Promega, USA). The PCR fragments of the selected SNPs were subsequently digested with their specific restriction enzyme. Then, the digested products were separated by electrophoresis on EtBr stained agarose gel and visualized under UV light.
Statistical analysis
We used the c2 test to evaluate the differences in the frequency distributions of the genotypes between CM cases and healthy subjects. The genotype distributions in the study population were tested to ensure conformity with Hardy-Weinberg equilibrium by means of Pearson’s c2 test. Odds ratios (ORs) and 95% confidence intervals (CIs) were used to assess the correlations between the three polymorphisms and neuroblastoma susceptibility with the unconditional multivariate logistic regression analysis. All statistical analyses were performed with SPSS 21.0 package (SPSS Inc., Chicago, IL, USA). The P-value was considered statistically significant at < 0.05.
Results
The clinical and epidemiological parameters of CM cases and controls were presented in Tab. 1. The mean ages of patients with CM and healthy subjects were 52.11±16.4 and 51.18±17.0 years, respectively. No significant differences were observed in terms of age (P = 0.684) and gender (P = 0.816) between the case and the control groups. The genotype frequencies of the three polymorphisms are shown in Tab. 2. Genotype frequency distributions of the two SNPs in controls were in agreement with the Hardy-Weinberg equilibrium (HWE) (P = 0.492 for XPC Lys939Gln; P = 0.698 for XPC Ala499Val; P = 0.351 for XPD Asp272Asn; P = 0.371 for XPD Arg751Arg; P = 0.763 for XPD Arg751Arg; P = 0.889 for XPF Arg415Gln; and P = 0.111 for XPG Asp1104His). Our results showed that none of the XPC (Lys939Gln and Ala499Val), XPD (Lys157Gln, Asp272Asn, and Arg751Arg), XPG Asp1104His, and XPF Arg415Gln polymorphisms was significantly associated with an increased risk of CM.
1. Clinical and demographic characteristics of cases with cutaneous melanoma and healthy subjects. 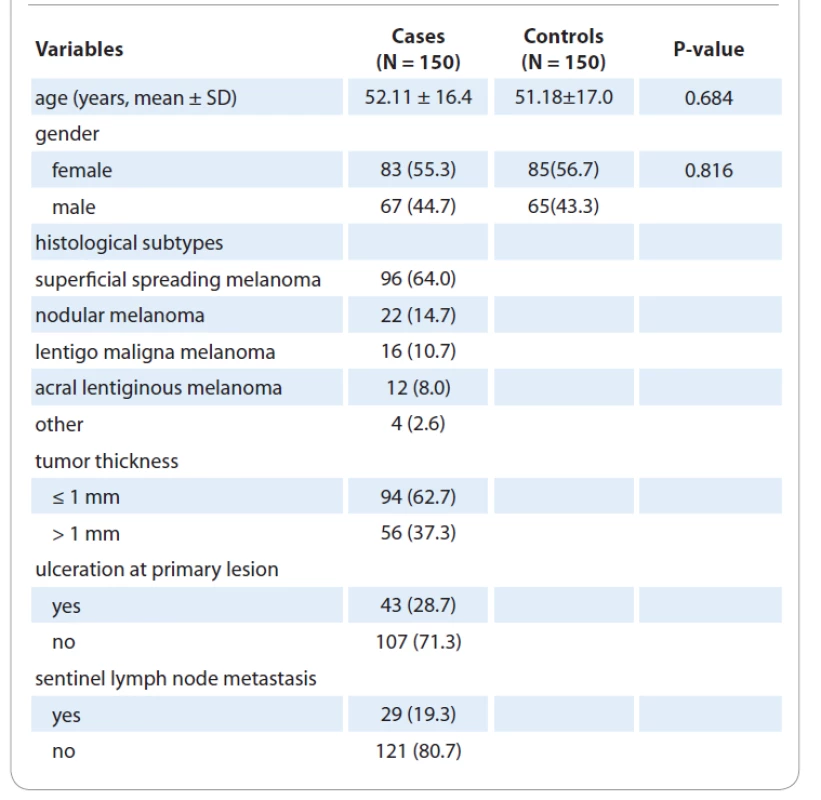
2. Genotype and allele frequencies of the polymorphisms at nucleotide excision repair pathway and cutaneous melanoma risk. 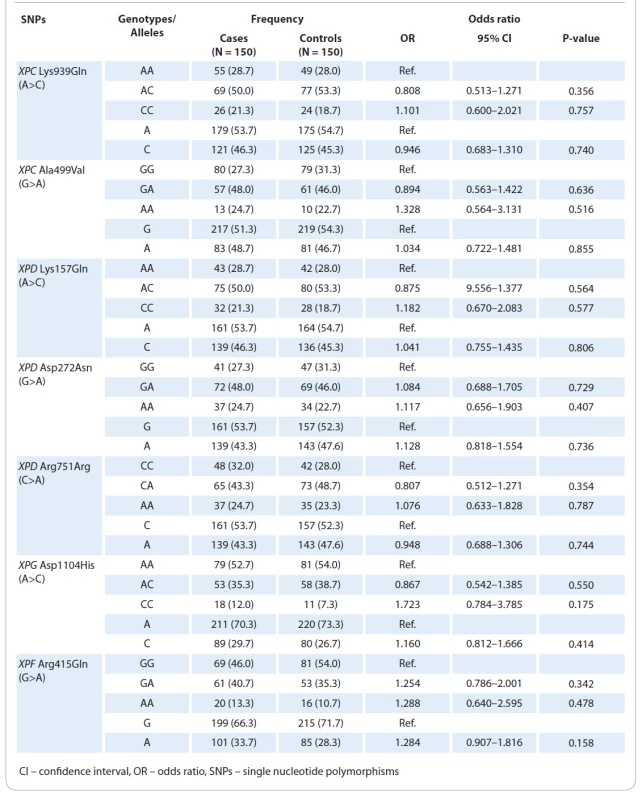
Discussion
To the best knowledge, this is the first study to evaluate the association of XPC (Lys939Gln and Ala499Val), XPD (Lys157Gln, Asp272Asn, and Arg751Arg), XPG Asp1104His and XPF Arg415Gln polymorphisms with susceptibility to CM in Iranian population. The current case-control study results revealed that none of the XPC Lys939Gln, XPC Ala499Val; XPD Asp272Asn, Arg751Arg and Arg751Arg; XPF Arg415Gln and XPG Asp1104His polymorphisms were associated with an increased risk of CM. Povey et al evaluated the association of DNA repair gene polymorphisms and genetic predisposition to CM in a Scottish population. Their results showed that polymorphisms at ERCC1 and XPF genes that act together in a complex to make an incision to the 5‘ side of a DNA lesion were associated with CM risk [30].
The XPC gene plays an important role in the early step of Global Genome Repair pathway (GGR), especially in damage recognition, open complex formation, and reparation via a complex composed by HR23B-XPC-CEN during GGR pathway [31,32]. XPC gene is localized on chromosome 3p25 and consists of 16 exons and 15 introns. To date, more than 2,582 polymorphic variants in the XPC gene have been identified [33]. Two common polymorphic loci of the XPC gene, rs2228000 (G>A, Ala499Val) and rs2228001 (G>T, Lys939Gln) have been investigated in association with the risk of CM. Our results showed that XPC Ala499Val and Lys939Gln polymorphisms were not associated with CM risk in our population, confirming the results reported for the same polymorphism by Povey et al in the Scottish population. Previous meta-analyses also reported inconsistent results about the association between these polymorphisms and risk of CM [30]. In a meta-analysis of eight studies with 4,631 CM cases and 5,111 controls, Jiang et al evaluated the association of XPC Lys939Gln polymorphism with CM risk. Their pooled data suggested that XPC Lys939Gln polymorphism was not significantly associated with CM risk [32]. In a meta-analysis, Zhou et al found an association between XPC Lys939Gln polymorphism and CM risk [34].
The human XPD gene is located at chromosome 19q13.3, possesses both single strand DNA-dependent ATPase and 5’-3’ DNA helicase activities and participates in DNA unwinding during NER. It is reported that genetic variants at XPD gene reduced the helicase activity, causing repair activity of NER pathway in a lower DNA and increasing cancer susceptibility [35]. Our study results failed to show an association between three Asp272Asn, Arg751Arg, Asp272Asn polymorphisms at XPD gene and susceptibility to CM. The Scottish study involving 596 CM patients and 441 population-based controls also reported no association between the XPD Lys751Gln polymorphism and CM risk [30]. Other two studies in Poland and UK did not report an association between single XPD polymorphisms and CM [36,37]. However, Debniak et al found that XPD haplotypes were associated with susceptibility to CM in a Polish population [37]. In a meta-analysis of eleven studies with 5,961 cases and 8,669 controls, Sun et al evaluated an association between XPD Lys751Gln polymorphism and CM among Caucasians. Their pooled data suggest that there was no evidence for a major role of XPD/ERCC2 Lys751Gln polymorphism in the pathogenesis of CM among Caucasians [38]. However, in other meta-analysis including 3,492 cases and 5,381 controls, Liu et al revealed that the XPD Lys751Gln polymorphism has contributed to CM susceptibility, and XPD is a possible drug target [39]. Zhu et al assessed an association of XPD Lys751Gln and Asp312Asn polymorphisms with skin cancer risk in a meta-analysis. They have reported that XPD Lys751Gln polymorphism was not associated with a risk of skin cancer under all genetic models. Their stratified analysis by tumor type revealed that XPD Lys751Gln polymorphism was not associated with an increased risk of non-melanoma skin cancer, but was significantly associated with an increased risk of CM. However, they have showed that XPD Asp312Asn polymorphism is not significantly associated with a risk of non-melanoma and melanoma skin cancers [40].
The human XPG gene encodes a structure-specific endonuclease that cuts the damaged DNA strand 3‘ to the lesion near the junction between the unpaired damaged strand and downstream undamaged duplex DNA [24,41] and XPG has a noncatalytic role in NER which is necessary in the incision complex to permit the XPF/ERCC1 heterodimer to make the 5‘-cut [42]. The human XPG gene is located on chromosome 13q32-33, encodes a protein with a predicted molecular mass of 133 kDa [43]. Previous studies indicated that XPG Asp1104His polymorphism can influence the DNA repair ability for tobacco and alcohol-induced DNA damage, thereby increasing the susceptibility to cancer. In this study, we have not found a significant association between XPG rs17655 polymorphism and development of CM. In a meta-analysis of eight published case-control studies with 5,212 cases and 7,045 controls, Xu et al evaluated the association of XPG Asp1104His polymorphism with CM susceptibility. Their results revealed that the XPG Asp1104His polymorphism was a risk factor for CM susceptibility [41]. However, Li et al did not found evidence of an association between XPD genotypes and development of CM [44].
The human XPF gene is involved in the 5‘ incision made during nucleotide excision repair and maintaining chromosome stability [45,46]. It is located at chromosome 16p13.12, consists of 11 exons and spanning about 28.2 kb [42]. Further, XPF/ERCC1 heterodimer participates in the repair of „crosslink“ damage that harmfully links the two DNA strands [47]. Our results failed to show a significant association between XPF Arg415Gln polymorphism and CM risk. However, Oliveira et al revealed in a case-control study that polymorphisms at XPC, XPF, TP53 and GSTP1 pathways of the DNA repair genes are important determinants of CM in individuals from south-eastern Brazil [48]. Moreover, Gomez et al found that inherited abnormalities in DNA repair pathway related to XPF Arg415Gln polymorphism might be a prognostic factor for overall survival of Brazilian CM patients [49]. Povey et al also supported that XPF Arg415Gln polymorphism is associated with a risk of CM Scottish patients [30].
Conclusions
This study results showed that all the seven polymorphisms at XPC (Lys939Gln and Ala499Val), XPD (Lys157Gln, Asp272Asn, and Arg751Arg), XPG (Asp1104His) and XPF (Arg415Gln) genes may not associate with an increased risk of CM. However, more well-designed studies with larger sample size in different ethnicities are necessary to further evaluate and verify our results. Future studies which take into account gene-gene and gene-environment interactions are warranted for more precise evidence and further elucidation of the underlying mechanism of CM.
Acknowledgments: We thank the anonymous reviewers for reviewing this manuscript.
Funding: This work was supported by Islamic Azad University, Science and Research Branch, Tehran, Iran
Availability of data and material: The datasets generated and/or analyzed during this study are available from the corresponding author on reasonable request.
Authors‘ contributions: FA and MAG conceptualized the study. SK and HN designed the study and the interview guide. Data analysis was done by SAD and HN. Manuscript was written and critically reviewed by GRD, HN, EA and AE. All authors read and approved the final manuscript.
The authors declare they have no potential conflicts of interest concerning drugs, products, or services used in the study.
Autoři deklarují, že v souvislosti s předmětem studie nemají žádné komerční zájmy.
The Editorial Board declares that the manuscript met the ICMJE recommendation for biomedical papers.
Redakční rada potvrzuje, že rukopis práce splnil ICMJE kritéria pro publikace zasílané do biomedicínských časopisů.
Fatemeh Asadian, MD
Department of Medical Laboratory
Sciences
School of Paramedical Science
Shiraz University of Medical
Sciences
Shiraz
Iran
e-mail: asadian@sums.ac.ir
Submitted/Obdrženo: 22. 7. 2020
Accepted/Přijato: 24. 1. 2021
Sources
1. Ghaemmaghami F, Zarchi MK, Gilani MM et al. Uterine sarcoma: clinicopathological characteristics, treatment and outcome in Iran. Asian Pac J Cancer Prev 2008; 9 (3): 421–426.
2. Behtash N, Karimi Zarchi M, Deldar M. Preoperative prognostic factors and effects of adjuvant therapy on outcomes of early stage cervical cancer in Iran. Asian Pac J Cancer Prev 2009; 10 (4): 613–618.
3. Karimi Zarchi M, Akhavan A, Gholami H et al. Evaluation of cervical cancer risk-factors in women referred to Yazd-Iran hospitals from 2002 to 2009. Asian Pac J Cancer Prev 2010; 11 (2): 537–538.
4. Ghaemmaghami F, Zarchi MK, Mousavi A. Surgical management of primary vulvar lymphangioma circumscriptum and postradiation: case series and review of literature. J Minim Invasive Gynecol 2008; 15 (2): 205–208. doi: 10.1016/j.jmig.2007.09.005.
5. Binesh F, Zarchi MK, Vahedian H et al. Primary malignant lymphoma of the uterine cervix. BMJ Case Rep 2012; 2012: bcr2012006675. doi: 10.1136/bcr-2012-006675.
6. Niktabar SM, Latifi SM, Moghimi M et al. Association of vitamin D receptor gene polymorphisms with risk of cutaneous melanoma. A meta-analysis based on 40 case-control studies. Dermatol Rev/Przegl Dermatol 2019; 106 : 268–279. doi: 10.5114/dr.2019.86909.
7. Iglesias-Pena N, Paradela S, Tejera-Vaquerizo A et al. Cutaneous melanoma in the elderly: review of a growing problem. Actas Dermosifiliogr 2019; 110 (6): 434–447. doi: 10.1016/j.ad.2018.11.009.
8. Ferlay J, Soerjomataram I, Dikshit R et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer 2015; 136 (5): E359–386. doi: 10.1002/ijc.29210.
9. Cormier JN, Xing Y, Ding M et al. Ethnic differences among patients with cutaneous melanoma. Arch Intern Med 2006; 166 (17): 1907–1914. doi: 10.1001/archinte.166.17.1907.
10. Melanoma stats, facts, and figures n.d. [online]. Available from: http: //www.optisigma.pt/editor/wp-content/uploads/2016/11/Melanoma-Stats-Facts-and-Figures-AIM-at-Melanoma.pdf.
11. Leonardi GC, Falzone L, Salemi R et al. Cutaneous melanoma: from pathogenesis to therapy (review). Int J Oncol 2018; 52 (4): 1071–1080. doi: 10.3892/ijo.2018.4287.
12. Yu J, Luo X, Huang H et al. Clinical characteristics of malignant melanoma in southwest China: a single-center series of 82 consecutive cases and a meta-analysis of 958 reported cases. PLoS One 2016; 11 (11): e0165591. doi: 10.1371/journal.pone.0165591.
13. Read J, Wadt KAW, Hayward NK. Melanoma genetics. J Med Genet 2016; 53 (1): 1–14. doi: 10.1136/jmedgenet - 2015-103150.
14. Leibeling D, Laspe P, Emmert S. Nucleotide excision repair and cancer. J Mol Histol 2006; 37 (5–7): 225–238. doi: 10.1007/s10735-006-9041-x.
15. Paszkowska-Szczur K, Scott RJ, Serrano-Fernandez P et al. Xeroderma pigmentosum genes and melanoma risk. Int J Cancer 2013; 133 (5): 1094–1100. doi: 10.1002/ijc.28123.
16. Jackson SP, Bartek J. The DNA-damage response in human biology and disease. Nature 2009; 461 (7267): 1071–1078. doi: 10.1038/nature08467.
17. Chatterjee N, Walker GC. Mechanisms of DNA damage, repair, and mutagenesis. Environ Mol Mutag 2017; 58 (5): 235–263. doi: 10.1002/em.22087.
18. Petruseva IO, Evdokimov AN, Lavrik OI. Molecular mechanism of global genome nucleotide excision repair. Acta Naturae 2014; 6 (1): 23–34. doi: 10.32607/20758251-2014-6-1-23-34.
19. Han C, Huang X, Hua R et al. The association between XPG polymorphisms and cancer susceptibility: evidence from observational studies. Medicine 2017; 96 (32): e7467. doi: 10.1097/MD.0000000000007467.
20. Ge J, Liu H, Qian D et al. Genetic variants of genes in the NER pathway associated with risk of breast cancer: a large-scale analysis of 14 published GWAS datasets in the DRIVE study. Int J Cancer 2019; 145 (5): 1270–1279. doi: 10.1002/ijc.32371.
21. Blankenburg S, König IR, Moessner R et al. Assessment of 3 xeroderma pigmentosum group C gene polymorphisms and risk of cutaneous melanoma: a case–control study. Carcinogenesis 2005; 26 (6): 1085–1090. doi: 10.1093/carcin/bgi055.
22. Wu K-G, He X-F, Li Y-H et al. Association between the XPD/ERCC2 Lys751Gln polymorphism and risk of cancer: evidence from 224 case-control studies. Tumour Biol 2014; 35 (11): 11243–11259. doi: 10.1007/s13277-014-2379-x.
23. Li C, Hu Z, Liu Z et al. Polymorphisms in the DNA repair genes XPC, XPD, and XPG and risk of cutaneous melanoma: a case-control analysis. Cancer Epidemiol Biomarkers Prev 2006; 15 (12): 2526–2532. doi. 10.1158/1055-9965.EPI-06-0672.
24. ElMahgoub IR, Gouda HM, Samra MA et al. Polymorphisms of xeroderma pigmentosum genes (XPC, XPD, and XPG) and susceptibility to acute leukemia among a sample of Egyptian patients. J Hematopathol 2017; 10 (6): 3–7. doi: 10.1007/s12308-017-0290-2.
25. López-Cima MF, González-Arriaga P, García-Castro L et al. Polymorphisms in XPC, XPD, XRCC1, and XRCC3 DNA repair genes and lung cancer risk in a population of Northern Spain. BMC Cancer 2007; 7 : 162. doi: 10.1186/1471-2407-7-162.
26. Yang ZH, Liang WB, Jia J et al. The xeroderma pigmentosum group C gene polymorphisms and genetic susceptibility of nasopharyngeal carcinoma. Acta Oncol 2008; 47 (3): 379–384. doi: 10.1080/02841860701558815.
27. Yousaf S, Khan MI, Micheal S et al. XRCC1 and XPD DNA repair gene polymorphisms: a potential risk factor for glaucoma in the Pakistani population. Mol Vis 2011; 17 : 1153–1163.
28. Zhou C, Xie L-P, Lin Y-W et al. Susceptibility of XPD and hOGG1 genetic variants to prostate cancer. Biomed Rep 2013; 1 (4): 679–683. doi: 10.3892/br.2013.123.
29. Zhu S, Wang A, Xia Z. Polymorphisms of DNA repair gene XPD and DNA damage of workers exposed to vinylchloride monomer. Int J Hyg Environ Health 2005; 208 (5): 383–390. doi: 10.1016/j.ijheh.2005.05.002.
30. Povey JE, Darakhshan F, Robertson K et al. DNA repair gene polymorphisms and genetic predisposition to cutaneous melanoma. Carcinogenesis 2007; 28 (5): 1087–1093. doi: 10.1093/carcin/bgl257.
31. Torres SM, Luo L, Lilyquist J et al. DNA repair variants, indoor tanning, and risk of melanoma. Pigment Cell Melanoma Res 2013; 26 (5): 677–684. doi: 10.1111/pcmr.12 117.
32. Jiang W, Zhang H, Chen QW et al. A meta-analysis of XPC Lys939Gln polymorphism and melanoma susceptibility. J Eur Acad Dermatol Venereol 2016; 30 (8): 1327–1331. doi: 10.1111/jdv.13477.
33. Hua RX, Zhu ZJ, Shen GP et al. Polymorphisms in the XPC gene and gastric cancer susceptibility in a Southern Chinese population. OncoTargets Ther 2016; 9 : 5513–5519. doi: 10.2147/OTT.S113055.
34. Zhou L, Lu Y, Yang G et al. Quantitative assessment of the association between XPC Lys939Gln polymorphism and cutaneous melanoma risk. Tumor Biol 2014; 35 (2): 1427–1432. doi: 10.1007/s13277-013-1196-y.
35. Zhao F, Shang Y, Zeng C et al. Association of single nucleotide polymorphisms of DNA repair genes in NER pathway and susceptibility to pancreatic cancer. Int J Clin Exp Pathol 2015; 8 (9): 11579–11586.
36. Winsey SL, Haldar NA, Marsh HP et al. A variant within the DNA repair gene XRCC3 is associated with the development of melanoma skin cancer. Cancer Res 2000; 60 (20): 5612–5616.
37. Dębniak T, Scott RJ, Huzarski T et al. XPD common variants and their association with melanoma and breast cancer risk. Breast Cancer Res Treat 2006; 98 (2): 209–215. doi: 10.1007/s10549-005-9151-2.
38. Sun Y, Zhang H, Ying H et al. A meta-analysis of XPD/ERCC2 Lys751Gln polymorphism and melanoma susceptibility. Int J Clin Exp Med 2015; 8 (8): 13874–13878.
39. Liu J, Song J, Wang M-Y et al. Association of EGF rs4444903 and XPD rs13181 polymorphisms with cutaneous melanoma in Caucasians. Med Chem 2015; 11 (6): 551–559. doi: 10.2174/1573406410666141224115516.
40. Zhu H-L, Bao J-M, Lin P-X et al. XPD Lys751Gln and Asp312Asn polymorphisms and susceptibility to skin cancer: a meta-analysis of 17 case-control studies. Asian Pac J Cancer Prev 2014; 15 (16): 6619–6625. doi: 10.7314/apjcp.2014.15.16.6619.
41. Xu Y, Jiao G, Wei L et al. Current evidences on the XPG Asp1104His polymorphism and melanoma susceptibility: a meta-analysis based on case-control studies. Mol Genet Genomics 2015; 290 (1): 273–279. doi: 10.1007/s00438-014-0917-2.
42. Manandhar M, Boulware KS, Wood RD. The ERCC1 and ERCC4 (XPF) genes and gene products. Gene 2015; 569 (2): 153–161. doi: 10.1016/j.gene.2015.06.026.
43. Emmert S. The human XPG gene: gene architecture, alternative splicing and single nucleotide polymorphisms. Nucleic Acids Res 2001; 29 (7): 1443–1452. doi: 10.1093/nar/29.7.1443.
44. Li C, Hu Z, Liu Z et al. Polymorphisms in the DNA repair genes XPC, XPD, and XPG and risk of cutaneous melanoma: a case-control analysis. Cancer Epidemiol Biomarkers Prev 2006; 15 (12): 2526–2532. doi: 10.1158/1055-9965.EPI-06-0672.
45. He X-F, Liu L-R, Wei W et al. Association between the XPG Asp1104His and XPF Arg415Gln polymorphisms and risk of cancer: a meta-analysis. PLoS One 2014; 9 (5): e88490. doi: 10.1371/journal.pone.0088490.
46. Wang M, Li Q, Gu C et al. Polymorphisms in nucleotide excision repair genes and risk of primary prostate cancer in Chinese Han populations. Oncotarget 2017; 8 (15): 24362–24371. doi: 10.18632/oncotarget.13848.
47. Schärer OD. ERCC1/XPF endonuclease-positioned to cut. EMBO J 2017; 36 (14): 1993–1995. doi: 10.15252/embj.201797489.
48. Oliveira C, Rinck-Junior JA, Lourenço GJ et al. Assessment of the XPC (A2920C), XPF (T30028C), TP53 (Arg72Pro) and GSTP1 (Ile105Val) polymorphisms in the risk of cutaneous melanoma. J Cancer Res Clin Oncol 2013; 139 (7): 1199–1206. doi: 10.1007/s00432-013-1430-4.
49. Gomez GVB, de Oliveira C, Rinck-Junior JA et al. XPC (A2920C), XPF (T30028C), TP53 (Arg72Pro), and GSTP1 (Ile105Val) polymorphisms in prognosis of cutaneous melanoma. Tumour Biol 2016; 37 (3): 3163–3171. doi: 10.1007/s13277-015-4123-6.
Labels
Paediatric clinical oncology Surgery Clinical oncology
Article was published inClinical Oncology

2021 Issue 5-
All articles in this issue
- Myelodysplastický syndrom, refrakterní anemie a O2 sensing
- Promising treatment modalities in the therapy of myelodysplastic syndromes
- Informace z České onkologické společnosti
- eHealth in medicine and oncology – new horizons of clinical practice
- Personalities and their contribution to the development of radiation oncology
- Anotace knihy
- Expert opinion on the care of patients with implanted pacemakers and cardioverters-defi brillators with an indication for radiotherapy – a summary for professions in the field of radiation oncology
- Aktuality z odborného tisku
- Skupina léků anti-HER2 pro pacientky s karcinomu prsu se rozrostla o preparát Phesgo
- Spomienka na prof. MUDr. Ivana Kozu, DrSc.
- The place and importance of hyaluronic acid in radiotherapy side effects
- Association of polymorphisms in nucleotide excision repair pathway genes with susceptibility to cutaneous melanoma
- Clinical values of two estrogen receptor signaling targeted lncRNAs in invasive ductal breast carcinoma
- Cabozantinib in the treatment of metastatic renal cell carcinoma – final data analysis from four oncology centers in the Czech Republic
- Metal artifact-free MRI-guided re-irradiation for recurrent spinal metastases from thyroid cancer
- Clinical Oncology
- Journal archive
- Current issue
- Online only
- About the journal
Most read in this issue- Personalities and their contribution to the development of radiation oncology
- Skupina léků anti-HER2 pro pacientky s karcinomu prsu se rozrostla o preparát Phesgo
- Promising treatment modalities in the therapy of myelodysplastic syndromes
- The place and importance of hyaluronic acid in radiotherapy side effects
Login#ADS_BOTTOM_SCRIPTS#Forgotten passwordEnter the email address that you registered with. We will send you instructions on how to set a new password.
- Career

